In luminal B tumors LATS2 depletion results in metabolic rewiring whereas LATS1 depletion promotes the expression of basal-like features.
Abstract
Deregulated activity of LArge Tumor Suppressor (LATS) tumor suppressors has broad implications on cellular and tissue homeostasis. We examined the consequences of down-regulation of either LATS1 or LATS2 in breast cancer. Consistent with their proposed tumor suppressive roles, expression of both paralogs was significantly down-regulated in human breast cancer, and loss of either paralog accelerated mammary tumorigenesis in mice. However, each paralog had a distinct impact on breast cancer. Thus, LATS2 depletion in luminal B tumors resulted in metabolic rewiring, with increased glycolysis and reduced peroxisome proliferator-activated receptor γ (PPARγ) signaling. Furthermore, pharmacological activation of PPARγ elicited LATS2-dependent death in luminal B-derived cells. In contrast, LATS1 depletion augmented cancer cell plasticity, skewing luminal B tumors towards increased expression of basal-like features, in association with increased resistance to hormone therapy. Hence, these two closely related paralogs play distinct roles in protection against breast cancer; tumors with reduced expression of either LATS1 or LATS2 may rewire signaling networks differently and thus respond differently to anticancer treatments.
Introduction
Breast cancer is a heterogeneous disease, with a wide spectrum of clinical, pathological, and prognostic subtypes. Although for many decades breast cancer classification was solely based on histology, nowadays studies integrating gene expression and histology are in the forefront of research (Perou et al, 2000; Sørlie et al, 2001; Curtis et al, 2012; The Cancer Genome Atlas Network, 2012), aiming to better classify and predict the clinical outcome of different tumors. Nevertheless, there remains a strong need to expand our knowledge about molecular pathways that contribute to breast cancer progression and response to therapy.
Breast cancer is broadly categorized into subtypes, with luminal and basal-like being the most common. Luminal tumors (consisting of luminal A and luminal B) express hormone receptors, underpinning their response to hormone therapies such as tamoxifen (Osborne, 1998). Luminal B (lumB) tumors tend to be of higher grade and convey a significantly worse prognosis than luminal A (lumA) tumors (Sørlie et al, 2001; Tran & Bedard, 2011; Haque et al, 2012). In contrast, basal-like tumors are mostly estrogen receptor (ER), progesterone receptor (PR), and HER2 negative (triple negative breast cancer [TNBC]) (Perou et al, 2000; Brenton et al, 2005). Importantly, decreased ER expression in lumB breast cancer has been associated with tumor recurrence and TNBC-like resistance to hormone therapy (Viale et al, 2007; Li et al, 2016). Moreover, transition of luminal tumors to invasive behavior, critical for metastasis, involves the phenotypic conversion of a luminal subpopulation to basal-like cells (Cheung et al, 2013), advocating against a rigorous division between luminal and basal-like tumors. Because of this plasticity, resistance to hormone therapy, associated with disease progression, remains a substantial therapeutic challenge.
In recent years, the LATS1 and LATS2 (LArge Tumor Suppressor [LATS]) Hippo pathway kinases have become the focus of intense research (Furth & Aylon, 2017). Classically, LATS1 and LATS2 are viewed as redundant paralogs that phosphorylate and inactivate the transcriptional cofactors YAP and TAZ (Moroishi et al, 2015). Both LATS are down-regulated in human breast cancer (Furth et al, 2015), and both have recently been implicated in modulating ER protein stability (Britschgi et al, 2017). Yet, evidence of distinct functions and differential impacts of the two paralogs is accumulating (Furth & Aylon, 2017). For instance, Lats1 knockout mice are highly sensitive to carcinogens and display pituitary dysfunction (St John et al, 1999), whereas conditional Lats2 knockout results in metabolic defects, such as fatty liver disease (Aylon et al, 2016).
Metabolic control is key to tumor suppression, reflecting the need of tumor cells to adapt their metabolism to support rapid growth. ER+ tumors often have increased fatty acid transport and elevated levels of short- and medium-chain fatty acids (Tang et al, 2014), which may affect their metabolic state, in part by regulating the activity of the nuclear peroxisome proliferator-activated receptor γ (PPARγ [Liberato et al, 2012]). This suggests a key role for PPARγ in luminal breast cancer (Zhou et al, 2009). Activation of PPARγ alters the expression of a large set of target genes, affecting adipogenesis, lipid metabolism, inflammation, and metabolic homeostasis (El Akoum, 2014). Furthermore, PPARγ activation can exert antiproliferative effects in a variety of cancer types, including breast cancer (Kersten et al, 2000; Fenner & Elstner, 2005).
Here, we show that a LATS2-associated gene expression pattern is specifically down-regulated in lumB breast cancer. Deletion of Lats2 in the mouse mammary gland results in increased lumB tumorigenesis and metabolic rewiring of the tumor cells. Conversely, LATS2 stimulates PPARγ signaling and promotes death of lumB-derived cells. In contrast, deletion of Lats1 reprograms lumB tumors towards basal-like characteristics. Concordantly, low LATS1 correlates with increased resistance to hormone therapy (tamoxifen). Thus, each LATS paralog exerts distinct tumor suppressive effects in the context of breast cancer, in a subtype-specific manner.
Results
To gain insight into the impact of LATS1 and LATS2 deregulation on breast cancer, we examined the correlation between the expression levels of LATS1 and LATS2 in human breast cancer samples (TCGA- BRCA dataset). Although there was an overall positive correlation between the two paralogs, a subset of tumors displayed selective down-regulation of LATS2 mRNA while retaining relatively high LATS1 mRNA (LATS2L quadrant, Fig 1A). By comparing gene expression patterns between LATS2L and LATSH tumors, we generated a signature of genes most down-regulated selectively in LATS2L tumors (LATS2L signature). Comparative analysis of breast cancer subtypes revealed strong down-regulation of the LATS2L signature particularly in lumB tumors (Fig 1B). Correspondingly, LATS2 mRNA itself was significantly lower in lumB tumors, compared with other subtypes (Figs 1C and S1A). Importantly, decreased expression of the LATS2L signature was correlated with reduced survival of luminal breast cancer patients (Fig 1D); furthermore, low LATS2 mRNA was associated with decreased probability of relapse-free survival among lumB patients (Fig S1B). Together, these observations suggest that LATS2 is a tumor suppressor in lumB breast cancer.
Figure 1. LATS2-associated gene expression pattern is down-regulated specifically in lumB breast tumors.
(A) Scatter plot of LATS1 and LATS2 expression levels in breast cancer tumors (TCGA-BRCA dataset). Pearson’s correlation coefficient 0.44. A cutoff of the 20% of tumors expressing the lowest levels of each LATS gene was used to divide the tumors into three groups: LATS1L, LATS2L, and LATSL, compared with LATSH. (B) Heatmap depicting the expression levels of a 20-gene LATS2L signature (the 20 most down-regulated genes exclusively in LATS2L tumors, compared with LATSH tumors; see Table S4 and the Materials and Methods section) in breast tumors (TCGA-BRCA) sorted according to PAM50 subtype classification. (C) Distribution of LATS2 mRNA expression levels in different breast cancer subtypes (PAM50, TCGA-BRCA); ***P-value < 0.001, t test comparing lumB tumors with all other subtypes. Number of tumors of each subtype is indicated at the bottom. (D) Kaplan-Meier analysis of survival probability of luminal breast cancer patients (METABRIC dataset, n = 1139; Cox proportional hazards model) divided according to expression levels of the LATS2L signature (as in B).
Figure S1.
(A) Distribution of LATS2 mRNA expression levels in different breast cancer subtypes (PAM50, METABTIC dataset); ***P-value < 0.001, t test comparing lumB tumors with all other subtypes. Number of tumors of each subtype is indicated at the bottom. (B) Kaplan–Meier plot of relapse-free survival (RFS) probability of lumB breast cancer patients separated according to LATS2 expression levels (n = 407, KM-plotter [Györffy et al, 2010]).
Mice harboring mammary gland-specific expression of the polyomavirus middle T antigen (MMTV-PyMT) develop breast tumors that recapitulate the progression of human ER+ cancer and resemble lumB tumors (Maglione et al, 2001; Herschkowitz et al, 2007; Cai et al, 2017). Hence, to explore more directly the role of LATS2 in lumB cancer, we generated MMTV-PyMT mice with mammary-specific deletion of Lats2 (Lats2-CKO PyMT; Fig S2A–C). Compared with nondeleted littermate controls (WT-PyMT), deletion of Lats2 significantly augmented mammary tumor burden (Fig 2A), formally validating the tumor suppressive function of LATS2 in mammary tumors. Importantly, by 3 mo of age, WT-PyMT mice displayed mainly adenoma/mammary intraepithelial neoplasia (MIN, [Lin et al, 2003]) and benign hyperplasia, or even no detectable pathology at all. In contrast, most of the Lats2-CKO PyMT mice displayed full-blown carcinomas (Figs 2B and C, and S2D). Interestingly, in line with its tumor suppressive role, Lats2 expression declined progressively as WT-PyMT tumors became more aggressive (Fig 2D).
Figure S2.
(A) Schematic representation of the conditional Lats2 locus. Upon mammary-specific CRE expression, exon 5 (colored blue) is deleted. (B) Genotyping of the Lats2 and the Cre alleles. Asterisks designate nonspecific bands. (C) Expression levels of Lats2 mRNA in WT-PyMT and Lats2-CKO PyMT tumors, quantified using RT-qPCR. Numbers of mice are indicated at the bottom; mean ± SEM; **P-value < 0.01. (D) Representative H&E-stained sections of tumors from three month old Lats2-CKO PyMT and WT-PyMT littermate controls. Inset: immunohistochemical (IHC) staining for smooth muscle actin (SMA), a marker for myoepithelial cells surrounding the adenoma/MIN. Scale bar = 200 μm. (E) Kaplan–Meier plot of survival probability of luminal breast cancer patients (METABRIC, n = 1138) divided according to relative expression of the genes commonly down-regulated in LATS2L human lumB and Lats2-CKO PyMT tumors (n = 20, see Table S4).
Figure 2. LATS2 is a tumor suppressor in a mouse lumB breast cancer model.
(A) Relative total tumor weight, as percentage of total body weight of 3-mo-old Lats2-CKO PyMT and WT-PyMT littermate controls (n = number of mice); mean ± SEM; * P-value < 0.05. (B) Three mammary glands (see the Materials and Methods section) from Lats2-CKO PyMT and WT-PyMT littermate control mice were histologically scored. The most advanced pathological lesion from each mouse was tallied. (C) Representative H&E-stained sections from WT-PyMT (adenoma/MIN) and Lats2-CKO PyMT (carcinoma); scale bar = 500 μm. (D) Expression levels of Lats2 mRNA in WT-PyMT tumors of different histological stages, analyzed by RT-qPCR; mean ± SEM. (E) Left panel: Heatmap representing hierarchical clustering of global expression patterns of tumors from Lats2-CKO PyMT (n = 4) and WT-PyMT littermate controls (n = 3). Each tumor was taken from a different mouse. Standardized rld values are shown for differentially expressed genes (P-value < 0.05, n = 1131); ad = adenoma/MIN, car = carcinoma. Right panel: PCA of the most differentially expressed genes between Lats2-CKO PyMT and WT-PyMT tumors (adjP-value < 0.05), deduced from RNA-seq analysis. (F) GSEA of 1,131 genes ranked by fold change (red to blue gradient) between WT-PyMT and Lats2-CKO PyMT tumors (P-value < 0.05) and compared with genes down-regulated in LATS2L (versus LATSH) human lumB tumors (vertical black lines).
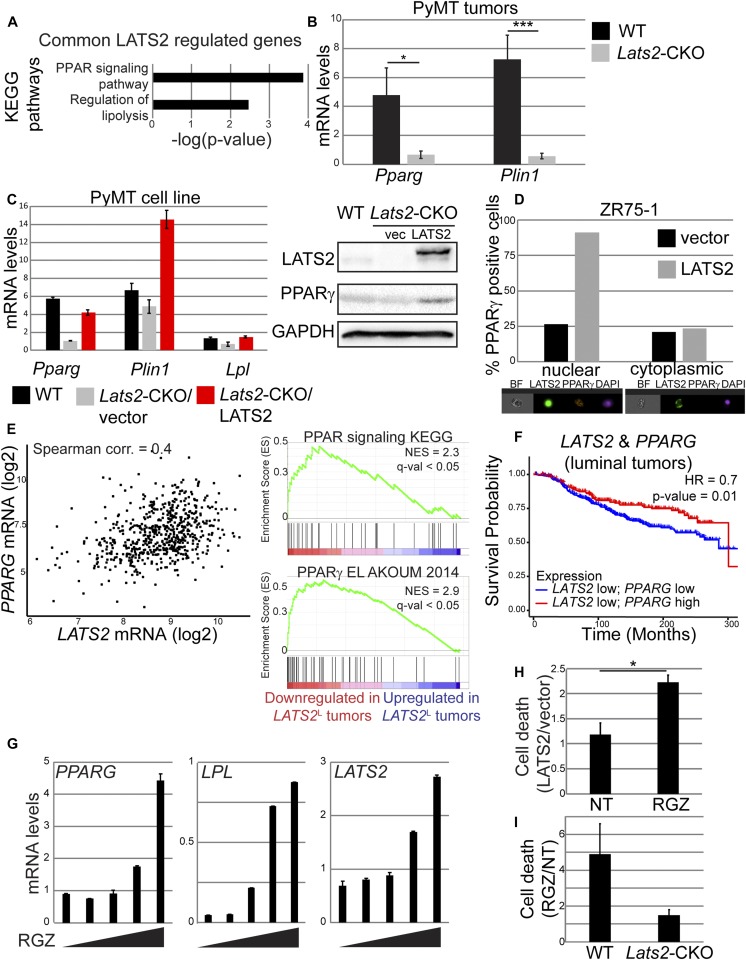
To further elucidate the means by which LATS2 might exert its breast tumor suppressive activity, we performed RNA sequencing (RNA-seq) analysis of WT- and Lats2-CKO PyMT tumors. WT-PyMT adenoma/MIN displayed a distinct transcriptional profile compared with WT-PyMT carcinoma (Fig 2E, left panel: lanes 1, 2, 3 and right panel: PCA). Interestingly, the transcription pattern of the Lats2-CKO PyMT adenoma/MIN displayed remarkable resemblance to that of the WT-PyMT carcinoma, suggesting that deletion of Lats2 facilitates a carcinoma-like gene expression pattern even at early stages of tumorigenesis. Importantly, gene set enrichment analysis (GSEA) indicated that gene expression changes in Lats2-CKO PyMT tumors correlated with the transcription profile of human LATS2L lumB tumors (Fig 2F), confirming the similarity of this mouse model to human lumB cancer. Moreover, genes commonly down-regulated in LATS2L human lumB and Lats2-CKO PyMT tumors were also found significantly associated with worse outcome in luminal breast cancer (Fig S2E). Overall, these findings demonstrate that deletion of Lats2 facilitates PyMT-driven tumorigenesis, further supporting the role of LATS2 as a tumor suppressor in human lumB breast cancer.
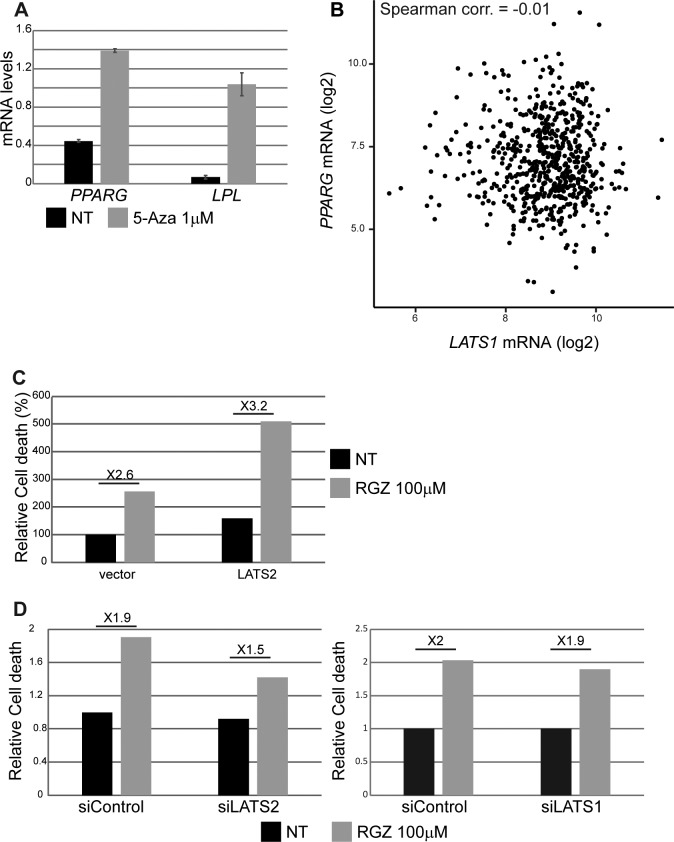
To further explore the impact of LATS2 down-regulation on human lumB cancer, we used our LATS2L signature to probe breast cancer cell lines in the Cancer Cell Line Encyclopedia (Barretina et al, 2012). Consistent with the human patient data, the LATS2L signature was down-regulated in cell lines derived from luminal cancers, including MCF7 and ZR75-1 (Fig S3A, Holliday & Speirs, 2011). Decreased LATS2 expression in breast cancer has been associated with promoter hypermethylation (Takahashi et al, 2005). Specifically, the CpG island surrounding the LATS2 transcription start site displayed increased methylation in LATS2L lumB tumors relative to LATS2H tumors (Fig S3B). Indeed, inhibition of DNA methylation by 5-aza-2′deoxycytidine (5-Aza) treatment increased the levels of LATS2 mRNA and protein in lumB-derived ZR75-1 cells (Fig 3A), suggesting that lumB breast cancers might undergo selective pressure to silence LATS2 expression.
Figure S3.
(A) Relative expression levels of the top most down-regulated genes in LATS2L breast tumors (TCGA-BRCA dataset, see Fig 1) in the panel of breast cancer cell lines included in the Cancer Cell Line Encyclopedia. Representative luminal cell lines are marked. (B) Upper panel: schematic representation of the LATS2 genomic locus. Positions of CpG island and methylation probes with differential methylation (P-value < 0.05) between LATS2L and LATS2H lumB tumors are shown. Lower panel: mean methylation of each probe in LATS2L versus LATS2H tumors. Line thickness corresponds to the P-value of the difference in methylation levels of the indicated probe between the two groups. For each probe, Pearson's correlation coefficient between expression and methylation in all lumB tumors is listed. (C) Left panel: ZR75-1 cells were transiently transfected with the indicated expression plasmids and harvested 72 h later. Cell extracts were subjected to Western blot analysis of LATS1, LATS2, and GAPDH as loading control. Right panel: ZR75-1 cells were transduced with MYC-LATS2 expression plasmid or vector control and selected with blasticidin to obtain stable expression. Cell extracts were subjected to Western blot analysis with the indicated antibodies. (D) MDA-MB-468 cells were transfected with the indicated expression plasmids and subjected 48 h later to PI exclusion analysis followed by flow cytometry analysis. “Relative cell death” represents the ratio between the percentages of dead (PI+) cells in the GFP-positive population versus the GFP-negative population. Values within each experiment were normalized to GFP transfected sample; mean ± SEM of two independent experiments.
Figure 3. LATS2 promotes death of lumB cells.
(A) ZR75-1 cells were treated with 1 μM 5-aza-2′-deoxycytidine (5-Aza) for 4 d. Upper panel: RT-qPCR analysis of LATS2 mRNA; mean ± SD of two technical replicates. Lower panel: Western blot analysis of LATS2 protein. (B) Functional enrichment of cell viability-related terms for differentially expressed genes in luminal cancer cell lines with transient silencing (siLATS2) or stable overexpression (OE) of LATS2, compared with controls. Ingenuity Pathway Analysis (IPA); Diseases or Functions annotations. (C) ZR75-1 cells were transiently transfected with GFP-LATS1 or GFP-LATS2, stained with anti-cleaved caspase 3 (CC3) antibody after 48 h, and subjected to imaging flow cytometry (ImageStreamX). Only cells with intact nucleus and positive GFP signal were analyzed; mean ± SEM of staining intensity; ***P-value < 0.001. (D) ZR75-1 cells were transfected with GFP-LATS1 or GFP-LATS2 or GFP only (vector). 72 h later, cell viability was assessed by PI exclusion followed by flow cytometry analysis. “Cell death” represents the ratio between the percentages of dead cells (PI+) in the GFP-positive population versus the GFP-negative population; mean ± SEM of three independent experiments; ***P-value < 0.001. Representative FACS results are shown on the right; in each case, upper and lower panels represent the GFP-positive and GFP-negative subpopulation, respectively, of the same transfected culture. (E) ZR75-1 cells stably transduced with MYC-LATS2 plasmid or control vector were subjected to PI exclusion analysis followed by flow cytometry. Left panel: percentage of dead (PI+) cells. Right panel: cell count of live (PI−) cells; mean ± SEM of three independent experiments; *P-value < 0.05, **P-value < 0.01. (F) Histological samples from carcinomas of Lats2-CKO PyMT and WT-PyMT littermate controls were immunostained for γH2AX and cleaved caspase 3 (CC3, left panel, scale bar = 100 μm). Right panel: mean ± SEM of percentage of positive cells, based on at least nine sections of each genotype; ***P-value < 0.001.
Next, we assessed the transcriptional impact of LATS2 modulation in cell lines derived from human luminal cancers. To that end, we performed RNA-seq analysis on ZR75-1 and MCF7 cells subjected to siRNA-mediated silencing of LATS2, as well as ZR75-1 cells overexpressing exogenous LATS2. Modulation of LATS2 levels in these cells resulted in expression changes relating to different signaling pathways (Table S1A). Unexpectedly, YAP/TAZ gene signatures were not among the most significantly enriched, rather, in all cases, LATS2 modulation elicited expression changes in genes related to cell death (Fig 3B and Table S1B). Concordantly, transient or stable overexpression of LATS2 (Fig S3C) in ZR75-1 cells elicited increased apoptosis, assessed by cleaved caspase 3 (CC3), and reduced cell viability, assessed by propidium iodide (PI) exclusion (Fig 3C–E); this was not seen with LATS1. In contrast, LATS2 overexpression did not reduce the viability of MDA-MB-468 cells, derived from basal-like breast cancer (Fig S3D), suggesting a luminal cancer-specific proapoptotic role of LATS2. In line with the increased death upon LATS2 overexpression in human luminal cancer cells, mouse Lats2-CKO PyMT tumors displayed reduced numbers of CC3-positive cells, relative to WT-PyMT tumors (Fig 3F). Interestingly, this was concomitant with augmented γH2AX staining, indicative of sustained DNA damage (Fig 3F). Notably, inability to clear DNA-damaged cells has been associated with aggressive clinicopathological features and poor patient outcome (Yang et al, 2017). Together, this implies that LATS2 might suppress tumorigenesis in part by promoting the elimination of damaged cells.
To further elucidate the underpinnings of LATS2-mediated tumor suppression, we analyzed the functional differences between the transcriptional profiles of Lats2-CKO PyMT and WT-PyMT tumors (see Fig 2E). Enrichment analysis (Fig 4A) revealed pronounced deregulation of numerous metabolic terms, including “regulation of lipolysis,” “TCA cycle,” and “glycolysis/gluconeogenesis,” with “PPAR signaling” being the most significantly altered. Likewise, the top predicted upstream regulator of LATS2-dependent genes in this comparison was PPARγ (ingenuity activation z-score 3.7; P-value of overlap 7.8 × 10−17). Similar metabolism-associated changes in gene expression were seen also when LATS2 was transiently overexpressed in ZR75-1 cells or silenced by siRNA in either ZR75-1 or MCF7 cells (Table S2). In line with LATS2-dependent metabolic homeostasis, stable overexpression of LATS2 in ZR75-1 cells (ZR75-1/LATS2) augmented cell death under regular culture conditions but dampened the further increase in cell death upon glucose deprivation when compared with control ZR75-1 cells (ZR75-1/vector, Fig 4B). These observations suggested that the inherently low levels of LATS2 in ZR75-1 cells might render them more dependent on extracellular glucose to maintain high aerobic glycolysis (“Warburg effect”). Furthermore, high LATS2 may favor oxidative phosphorylation, conferring less dependence on supplemented glucose. Indeed, Seahorse metabolic analysis confirmed that the glycolytic rate was diminished in ZR75-1/LATS2 relative to control ZR75-1/vector cells, whereas respiratory capacity was augmented (Fig 4C). In agreement, cell lines derived from Lats2-CKO PyMT tumors (Fig S4A and B) mirrored these observations, performing less respiration than WT-PyMT cells (Fig 4D). Overall, these results suggest that down-regulation of LATS2 may facilitate the survival of cells that have undergone metabolic rewiring towards a Warburg-like profile, favoring glycolysis over oxidative respiration.
Figure 4. LATS2 augments oxidative phosphorylation.
(A) KEGG pathways enrichment analysis for genes differentially regulated in Lats2-CKO PyMT compared with WT-PyMT tumors (adjP-value < 0.05, n = 114). (B) ZR75-1 cells stably transfected with an MYC-LATS2 plasmid (LATS2) or with control vector were cultured with or without glucose for 3 d. Cell death was measured by PI exclusion followed by FACS analysis. Values from each experiment were normalized to % PI+ of control cells grown in glucose-containing medium; mean ± SEM of three independent experiments. (C) Extracellular acidification rate (ECAR, indicative of glycolysis) and oxygen consumption rate (OCR, indicative of respiration) of ZR75-1 cells stably expressing MYC-LATS2, relative to vector control cells, determined by Seahorse; mean ± SEM of five independent experiments. Representative tracks of Seahorse measurements are shown on the right. (D) Respiration measured by OCR in WT-PyMT and Lats2-CKO PyMT tumor-derived cell lines; mean ± SEM in log 10 scale of three independent experiments; P-value < 0.05. Bottom: representative Seahorse track.
Figure S4.
(A) Genotyping of the Lats2 and Cre alleles in the PyMT-derived cell lines (see primers location in Fig S2A). (B) Relative expression of Lats2 mRNA in cell lines derived from WT- or Lats2-CKO-PyMT tumors, analyzed by RTqPCR; mean ± SEM of 2 and three independently isolated cell lines, respectively, originating from different tumors. ***P-value < 0.001.
Dampening of PPARγ activity might reflect an energetic necessity of highly glycolytic LATS2-depleted cancer cells. Indeed, genes commonly down-regulated in both Lats2-CKO PyMT tumors and LATS2L lumB human breast cancers were significantly enriched for “PPAR signaling” (Fig 5A), confirming the link between LATS2 and PPARγ activity in both mouse and human tumors. Decreased expression of Pparg (subject to positive autoregulation [Wakabayashi et al, 2009]) and its transcriptional target Plin1 was confirmed by RT-qPCR in an expanded number of Lats2-CKO PyMT tumors (Fig 5B). Furthermore, Pparg and its target genes Plin1 and Lpl were down-regulated also in cultured Lats2-CKO PyMT cells (Fig 5C), implicating a cell-autonomous regulatory interaction between LATS2 and PPARγ. Of note, stable expression of human LATS2 in the Lats2-CKO PyMT cells (Lats2-CKO/LATS2) resulted in increased expression of Pparg and its target genes (Fig 5C). Interestingly, in LATS2-overexpressing ZR75-1 cells, augmented PPARγ was selectively associated with nuclear localization of LATS2 (Fig 5D), suggesting that a nuclear function of LATS2 is responsible for promoting PPARγ expression.
Figure 5. PPARγ signaling correlates with LATS2 in human and mouse tumors and promotes cell death in a LATS2-dependent manner.
(A) KEGG pathways significantly enriched within the list of genes commonly down-regulated in Lats2-CKO PyMT (compared with WT-PyMT) tumors and LATS2L lumB human tumors (TCGA). (B) RT-qPCR quantification of Pparg and Plin1 expression in tumors derived from 3 mo old Lats2-CKO PyMT (n = 9) and WT-PyMT littermate controls (n = 6); mean ± SEM; *P-value < 0.05, ***P-value < 0.001. (C) Expression levels of the indicated genes (left) and proteins (right) in cultured WT PyMT and Lats2-CKO PyMT tumor-derived cells stably transduced with LATS2 or vector control; mean ± SD of 2 technical repeats. (D) ZR75-1 cells were transfected with GFP-LATS2 or control GFP plasmid, stained 48 h later with anti-PPARγ and cleaved caspase 3 (CC3) antibodies, and subjected to imaging flow cytometry (ImageStreamX). Only GFP-positive cells with intact nuclei were analyzed. Cells with nuclear localization of the transfected protein were identified by similarity between GFP and DAPI staining. Percentage of cells positively stained for PPARγ in each subpopulation is presented. Representative images are shown at the bottom. BF = bright field. (E) Left panel: scatter plot depicting PPARG and LATS2 expression in luminal tumors in the TCGA-BRCA dataset (n = 613). Right panel: GSEA of genes ranked according to expression fold change between LATS2L tumors and LATSH tumors in the TCGA lumB dataset. (F) Kaplan-Meier analysis of survival probability of luminal breast cancer patients (METABRIC dataset, n = 1139; Cox proportional hazards model) defined according to expression levels of LATS2 and PPARG. (G) RT-qPCR quantification of PPARG and LPL (left) and LATS2 (right) mRNA in ZR75-1 cells treated for 48 h with increasing concentrations of RGZ (0, 10, 25, 50, and 100 μM, respectively); mean ± SD of two technical replicates. (H) ZR75-1/vector (vector) and ZR75-1/LATS2 (LATS2) cells were treated with 100 μM RGZ for 48 h, followed by PI exclusion analysis; mean ± SEM of the ratio between % PI+ cells in each cell type, from three independent experiments. (I) Cell lines derived from WT-PyMT and Lats2-CKO PyMT tumors were treated with 100 μM RGZ for 48 h, followed by PI exclusion analysis; mean ± SEM of the ratio between % PI+ cells in treated versus untreated cells, from two independent experiments.
Analysis of human luminal tumors revealed that LATS2 and PPARG mRNA levels are positively correlated (Fig 5E, left); moreover, LATS2L lumB tumors displayed reduced PPARγ signaling (Fig 5E, right). Of note, similar to LATS2 overexpression in ZR75-1 cells (see Fig 3A), 5-Aza treatment increased the expression of both PPARG and LPL (Fig S5A), suggesting that both LATS2 expression and PPARγ activity are epigenetically down-regulated during lumB tumorigenesis. In contrast, PPARG expression in luminal tumors was not significantly correlated with LATS1 expression (Fig S5B). Interestingly, luminal breast cancer patients with low expression of both LATS2 and PPARG displayed impaired overall survival, compared with patients with low LATS2 but high PPARG expression (Fig 5F). Altogether, our observations support the notion that LATS2 down-regulation, which leads to reduced PPARγ signaling, may facilitate metabolic rewiring towards a more favorable energetic state, associated with more lethal tumors.
Figure S5.
(A) ZR75-1 cells were treated with 1 μM 5-aza-2′-deoxycytidine (5-Aza) for 4 d. PPARG and LPL mRNA levels were analyzed by RT-qPCR; mean ± SD of two technical replicates. (B) Scatter plot depicting PPARG and LATS1 mRNA expression levels in luminal tumors (TCGA BRCA dataset, n = 613). (C) ZR75-1 cells were transfected with either control vector (GFP only) or GFP-LATS2 expression plasmid and treated with 100 μM RGZ for 4 d. Cell death was measured as in Fig 3D. Values were normalized relative to nontreated cells transfected with vector only. (D) ZR75-1 cells were transfected with the indicated siRNA oligonucleotides and treated with 100 μM RGZ for 4 d. The percentage of dead cells was measured by PI exclusion followed by flow cytometry analysis, and values were normalized to nontreated siControl cells.
PPARγ activation has been shown to inhibit proliferation and induce cell cycle arrest in breast cancer cells (Mueller et al, 1998; Suh et al, 1999; Mehta et al, 2000). Rosiglitazone (RGZ) is an FDA-approved antidiabetic drug (Soccio et al, 2014) that selectively activates PPARγ (Willson et al, 1996). As expected, treatment of ZR75-1 cells with RGZ resulted in increased PPARγ activity, reflected by augmented expression of the PPARγ target gene LPL as well as of the PPARG mRNA itself (Fig 5G, left panel). Interestingly, activation of PPARγ also resulted in up-regulation of LATS2 mRNA (Fig 5G, right panel), implying a positive feedback between the two. Re-expression of LATS2 conferred hypersensitivity to RGZ (Figs 5H and S5C), suggesting that high LATS2 may further sensitize these cells to excessive PPARγ activation. In agreement, partial silencing of LATS2, but not LATS1, reduced sensitivity to RGZ (Fig S5D). Moreover, deletion of Lats2 nearly abolished the death of cultured PyMT mouse tumor cells upon RGZ treatment (Fig 5I), further confirming that sensitivity of lumB breast cancer cells to PPARγ activation is LATS2 dependent.
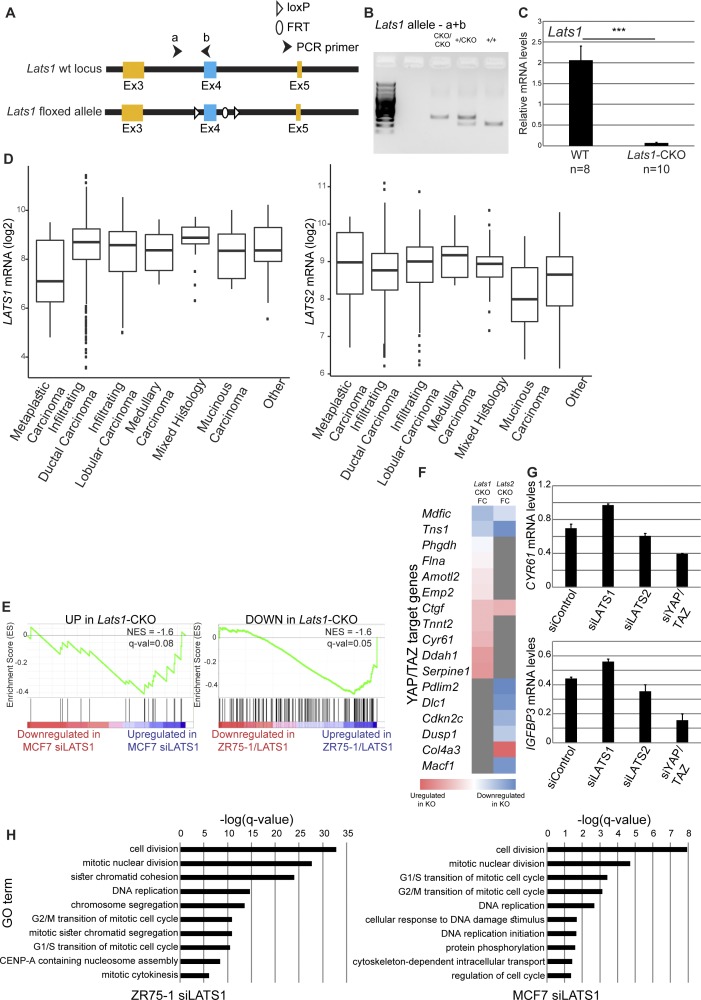
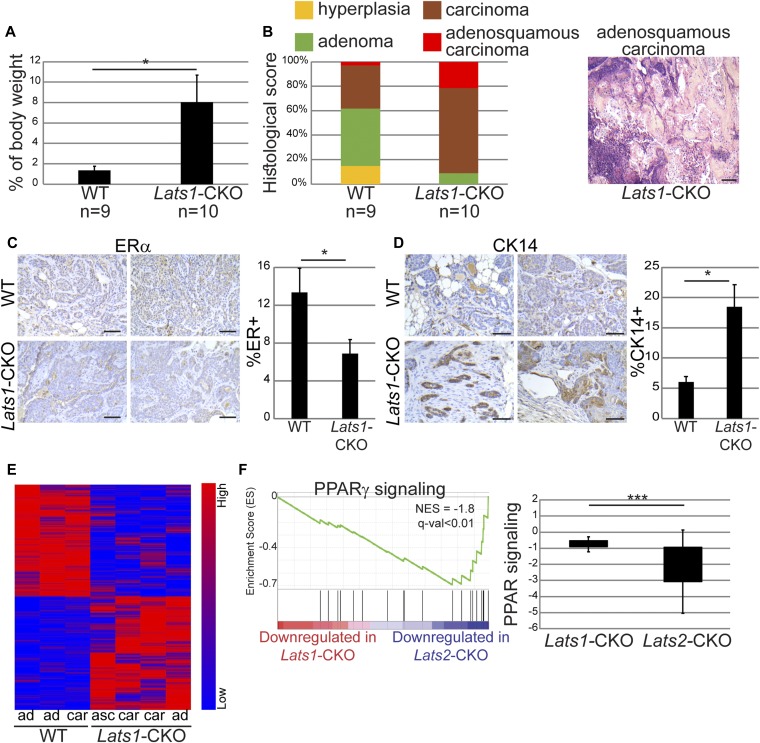
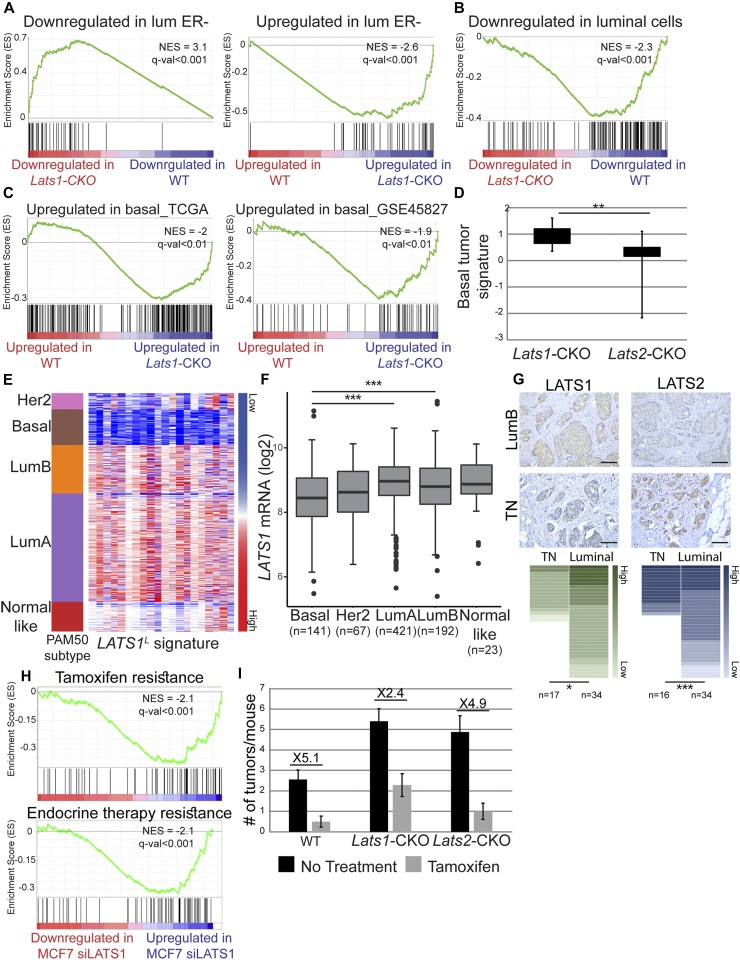
Next, we wished to examine the impact of Lats1 on breast cancer development in PyMT mice. To that end, we generated MMTV-Cre PyMT mice with mammary glandspecific knockout of Lats1 (Lats1-CKO PyMT) (Fig S6A–C). Similar to Lats2 deletion, conditional knockout of Lats1 caused a significant increase in tumor burden relative to WT-PyMT littermate controls (Fig 6A). Surprisingly, a notable portion of Lats1-CKO PyMT mice developed adenosquamous carcinoma (Fig 6B), a tumor type not observed in Lats2-CKO PyMT mice (see Fig 2B). This was particularly striking because the MMTV-PyMT model typically represents ER+ lumB cancer (Maglione et al, 2001; Herschkowitz et al, 2007; Cai et al, 2017), whereas human adenosquamous carcinoma is invariably triple negative (Geyer et al, 2017). In line with this, Lats1-CKO PyMT tumors displayed significantly reduced ER positivity (Fig 6C) and increased abundance of the basal cell marker CK14 (Fig 6D). This suggests that differently from LATS2, LATS1 might be important for maintaining the expression of luminal cell markers and restricting cell plasticity. Notably, LATS1, but not LATS2, was down-regulated in human breast metaplastic carcinoma (Fig S6D), which is thought to occur via transdifferentiation of a subpopulation of cancer cells (van Deurzen et al, 2011). Metaplastic carcinomas are typically negative of hormone receptors (ER/PR) and HER-2/neu, and are histologically characterized by mixed epithelial and transdifferentiated components (Aydiner et al, 2015), thus resembling the adenosquamous carcinomas detected in the Lats1-CKO PyMT tumors. Altogether, this suggests that down-regulation of LATS1, but not LATS2, favors partial loss of luminal identity of tumor cells.
Figure S6.
(A) Schematic representation of the conditional Lats1 locus. Upon mammary-specific Cre expression, exon 4 (colored blue) is excised. (B) Genotyping of the Lats1 allele. (C) Expression of Lats1 mRNA in WT- and Lats1-CKO PyMT tumors, analyzed by RT-qPCR. Mean ± SEM of tumors from different mice. Mouse numbers indicated at the bottom; ***P-value < 0.001. (D) LATS1 and LATS2 mRNA expression levels in human breast cancer tumors (TCGA-BRCA dataset). Tumors were stratified according to histological type (n = 1095). (E) Genes were ranked according to fold change between LATS1 siRNA (siLATS1) and control siRNA in transiently transfected MCF7 cells (left) or between LATS1 overexpression and vector control in ZR75-1 cells (ZR75-1/LATS1, right) and compared with differentially expressed genes in Lats1-CKO PyMT tumors (adjP-value < 0.05, FC > 1.5). (F) YAP/TAZ target genes (Dupont gene set, Dupont et al, 2011), differentially expressed in either Lats1-or Lats2-CKO PyMT tumors relative to their WT-PyMT littermate controls (P-value < 0.05). The heat map depicts the log ratio between Lats-CKO and WT-PyMT littermate controls; grey indicates genes not significantly differentially expressed between the two conditions. (G) ZR75-1 cells were transfected with the indicated siRNA oligonucleotides. CYR61 and IGFBP3 mRNA levels were analyzed by RT-qPCR; mean ± SD of two technical replicates. (H) Functional enrichment analysis (GO biological processes) for genes differentially regulated upon LATS1 silencing in either ZR75-1 (FC > 1.5, adjP-value < 0.05, n = 733, left panel) or MCF7 (FC > 1.5, adjP-value < 0.05, n = 285, right panel). The 10 most enriched terms for each cell line are shown.
Figure 6. Deletion of Lats1 is phenotypically distinct from Lats2 deletion in PyMT tumors.
(A) Relative tumor weight, as percentage of total body weight, in three month old Lats1-CKO PyMT and WT-PyMT littermate controls (n = number of mice); mean ± SEM; *P-value < 0.05. (B) Three mammary glands from Lats1-CKO PyMT and WT-PyMT littermate controls were histologically scored and tallied. Right panel: representative H&E-stained adenosquamous carcinoma sample from a Lats1-CKO PyMT tumor (scale bar = 200 μm). (C) Immunohistochemistry analysis of ERα protein expression in tumors from 3-mo-old Lats1-CKO PyMT and WT-PyMT littermate controls. Left panel: two representative tumor sections from each genotype (scale bar = 100 μm). Right panel: mean ± SEM of % ER+ cells in tumors from eight Lats1-CKO PyMT and 6 WT-PyMT mice; *P-value < 0.05. (D) Immunohistochemistry analysis of CK14, performed as in (C). Right panel depicts mean ± SEM of percentage of slide area with intense membranous CK14 staining in the invasive front of the tumor; *P-value < 0.05. (E) Heatmap representing hierarchical clustering of global expression patterns of tumors from 3-mo-old Lats1-CKO PyMT and WT-PyMT littermate controls. Standardized rld values are shown for differentially expressed genes (P-value < 0.05, n = 2029). ad = adenoma/MIN, car = carcinoma, asc = adenosquamous carcinoma; (F) Left panel: GSEA assessing PPARγ transcriptional activity in Lats1-CKO PyMT versus Lats2-CKO PyMT tumors. Genes differentially expressed (adjP-value < 0.05) in either Lats1-CKO PyMT (compared with littermate controls) or Lats2-CKO PyMT (compared with littermate controls) were ranked according to fold change differences (log ratio) and compared with a gene set comprising PPARγ target genes (El Akoum, 2014). Right panel: box plot representing mean fold change (log 2) of genes comprising the PPAR pathway (KEGG database) in Lats1-CKO PyMT and Lats2-CKO PyMT, each compared with their WT-PyMT littermate controls. Only genes with P-value of comparison < 0.05 were included; ***P-value < 0.001.
We next compared the global gene expression patterns of Lats1-CKO PyMT tumors and their WT-PyMT littermate control tumors (Fig 6E). Overall, the expression pattern changes in the mouse Lats1-CKO PyMT tumors agreed well with those seen in human luminal cancer cell lines (MCF7 and ZR75-1) subjected to either up- or down-modulation of LATS1 (Fig S6E), supporting the human relevance of the Lats1-CKO PyMT model. Of note, the altered signaling pathways pattern in Lats1-CKO PyMT tumors was distinct from that observed upon depletion of Lats2. For example, unlike in Lats2-CKO PyMT tumors, PPARγ signaling was only mildly deregulated in Lats1-CKO PyMT tumors (Fig 6F). Also, up-regulation of YAP/TAZ target genes was more apparent in Lats1-CKO PyMT tumors and following LATS1 silencing in ZR75-1 cells, as compared with Lats2-CKO PyMT tumors (Fig S6F) or LATS2 silencing, respectively (Fig S6G). This is consistent with canonical YAP/TAZ inhibition by LATS1 in the human breast cancer setting (Zhang et al, 2008; Cordenonsi et al, 2011; Britschgi et al, 2017). Furthermore, in agreement with the known ability of hyperactive YAP/TAZ to augment cell proliferation, LATS1 silencing in human luminal breast cancer-derived cells yielded a transcriptional signature strongly enriched for cell cycle and mitosis-related pathways (Fig S6H).
Congruous with the changes in ER and CK14 levels in Lats1-CKO PyMT tumors (see Fig 6C and D), the expression pattern of Lats1-CKO PyMT tumors was similar to other ER-negative mouse mammary tumors (Fig 7A) and dissimilar to mature luminal cells (Fig 7B). Furthermore, it was significantly similar to that of human basal-like tumors, compared with luminal tumors (Fig 7C). In addition, we derived a basal-like expression signature from human breast cancer samples (TCGA dataset, see the Materials and Methods section) and found it to be up-regulated selectively in the Lats1-CKO PyMT, but not Lats2-CKO tumors (each compared with WT-PyMT littermate control tumors, Fig 7D and Table S3). Overall, this implies that LATS1 is important for maintaining luminal breast cancer-associated expression patterns; without it, tumors may become more basal-like, even within an otherwise lumB-predisposed in vivo setting. To further examine this possibility, we generated a LATS1L 20-gene signature, comprising genes that were selectively down-regulated in LATS1-low human breast tumors (see Fig 1A) and evaluated its expression in different breast cancer subtypes. Notably, this signature (Fig 7E), as well as LATS1 mRNA itself (Fig 7F), was strongly down-regulated particularly in basal-like tumors. Furthermore, LATS1 protein levels were lower in TNBC tumors relative to luminal tumors, whereas the opposite trend was seen for LATS2 (Fig 7G).
Figure 7. Down-regulation of LATS1 promotes the formation of tumors enriched in basal-like features.
(A) Genes were ranked according to fold change between Lats1-CKO PyMT and WT-PyMT tumors (P-value < 0.05), and compared with genes differentially expressed in ER-negative PyMT tumors (GSE64453), using GSEA. (B) Genes were ranked as in (A) and compared with genes down-regulated in mature luminal cells relative to mammary stem cells (GSE19446), using GSEA. (C) Genes were ranked as in (A) and compared with genes up-regulated in human basal-like tumors relative to luminal tumors (TCGA, see the Materials and Methods section) (left panel) or genes up-regulated in human basal tumors relative to lumB (GSE45827), using GSEA. (D) Box plot representing mean fold change (log2) of genes comprising the basal tumor signature (see the Materials and Methods section and Table S3), in Lats1-CKO PyMT and Lats2-CKO PyMT tumors compared with their WT-PyMT littermate controls. (E) Heatmap depicting the expression levels of the 20 most down-regulated genes in LATS1L tumors relative to LATSH tumors (see Table S4 and the Materials and Methods section). Tumors were sorted according to the PAM50 classification. (F) Relative expression levels of LATS1 in different breast cancer subtypes (PAM50 and TCGA-BRCA). Numbers of samples are indicated at the bottom. ANOVA coupled with Dunnett's test; ***P-value < 0.01. (G) Immunohistochemistry (IHC) analysis of LATS1 and LATS2 in luminal and triple negative (TN) human tumors. Representative tumor sections of human lumB and TN tumors are shown in the upper panel (scale bar = 100 μm). Lower panel: human tumor samples (n = number of samples in each subtype) were scored by IHC (see the Materials and Methods section) for LATS1 and LATS2; *P-value < 0.05, ***P-value < 0.001. (H) Genes were ranked according to fold change between control siRNA-transfected and LATS1 siRNA-transfected (siLATS1) MCF7 cells and compared with the indicated gene sets (Creighton et al, 2008; Massarweh et al, 2008), using GSEA. (I) WT-PyMT, Lats1-CKO PyMT, and Lats2-CKO PyMT mice were injected IP with 4-hydroxytamoxifen (Tamoxifen) or left untreated (no treatment); n = 12, 18, 7, 10, 9, and 9, respectively.
Silencing of LATS1 in MCF7 cells resulted in increased expression of genes associated with resistance to hormone therapy, particularly tamoxifen (Fig 7H), supporting a role for LATS1 in maintaining estrogen dependence of luminal breast cancer cells. To assess more directly whether deletion of Lats1 might facilitate hormone therapy resistance, we monitored the response of Lats1-CKO PyMT tumors to tamoxifen treatment. Tamoxifen treatment retarded tumor growth in all genotypes (Fig 7I); however, Lats1-CKO PyMT tumors were relatively less inhibited than WT-PyMT or Lats2-CKO PyMT tumors. Overall, our observations suggest that the partial loss of luminal identity upon LATS1 down-regulation may promote resistance to hormone therapy, paralleling the clinical manifestations of tamoxifen-resistant human breast tumors (Kuukasjärvi et al, 1996).
Discussion
Previous studies have demonstrated that reduced expression of LATS kinases is associated with breast cancer characteristics in vitro (Zhang et al, 2008; Furth et al, 2015; Li & Gumbiner, 2016). We provide evidence that both LATS1 and LATS2 are bona fide tumor suppressors in an in vivo breast cancer setting. In a lumB breast cancer mouse model, conditional deletion of either paralog increases tumorigenesis, both in magnitude and severity. Importantly, in both human and mouse breast tumors, down-regulation of each paralog is associated with a distinct breast cancer subtype and deregulation of different signaling pathways; decreased LATS1 compromises the strict maintenance of luminal cell fate and favors a drift towards a more basal-like state, whereas decreased LATS2 rewires metabolism towards reduced PPARγ activity and increased glycolysis.
Differential impact of LATS1 and LATS2 on distinct molecular signaling pathways is in line with a growing body of evidence that each paralog can operate within a functional spectrum that includes also unique, nonredundant activities (Furth & Aylon, 2017). Within this spectrum, LATS may sometimes even contribute to cancer progression, for example, by suppressing anticancer immunity (Moroishi et al, 2016). Yet, most of our current knowledge is more consistent with a tumor suppressive role of these proteins. Notably, both LATS proteins inhibit the tumor-promoting activities of YAP and TAZ (Moroishi et al, 2015). Indeed, loss of YAP has been shown to suppress the growth of PyMT-driven mammary tumors (Chen et al, 2014). Interestingly, we observed that deletion of Lats1, but not Lats2, results in a moderate increase of YAP/TAZ-associated transcriptional activity in such tumors. Nevertheless, in other contexts, LATS2 does contribute to the inhibition of YAP/TAZ-mediated phenotypes (Meng et al, 2015). Thus, the contribution of each LATS kinase to the regulation of YAP/TAZ activity is markedly context dependent.
We found that LATS2 augments PPARγ activity in vitro and in vivo. Of note, PPARγ was reported to suppress glycolysis and induce apoptosis in breast cancer cells by repressing the expression of several glycolytic enzymes (Shashni et al, 2013). Hence, the attenuated PPARγ signaling in LATS2-depleted breast cancer cells may account, at least in part, for the observed increase in glycolysis and the increased dependence on glucose for survival. Moreover, we observed that LATS2 is required for induction of luminal breast cancer cell death by the PPARγ agonist RGZ. We propose that inefficient clearance of highly glycolytic, genomically altered cells with low levels of LATS2 might represent a devious evolutionary adaptation of lumB tumors. Conversely, simultaneous activation of LATS2 and PPARγ may instigate a metabolic catastrophe, particularly in cancer cells with high energy demands, possibly by increasing mitochondrial burden (Zolezzi et al, 2013). PPARγ agonists have shown efficacy against some solid tumors in clinical trials (Demetri et al, 1999); our data suggest that LATS2 status might be a determinant of the success of such agents. More broadly, the ability of LATS2 to modulate master regulators of lipid metabolism such as PPARγ and SREBP (Aylon et al, 2016) expands the functional consequences of the deregulation of Hippo pathway components in cancer and may indicate a metabolic Achilles' heel of particular tumors.
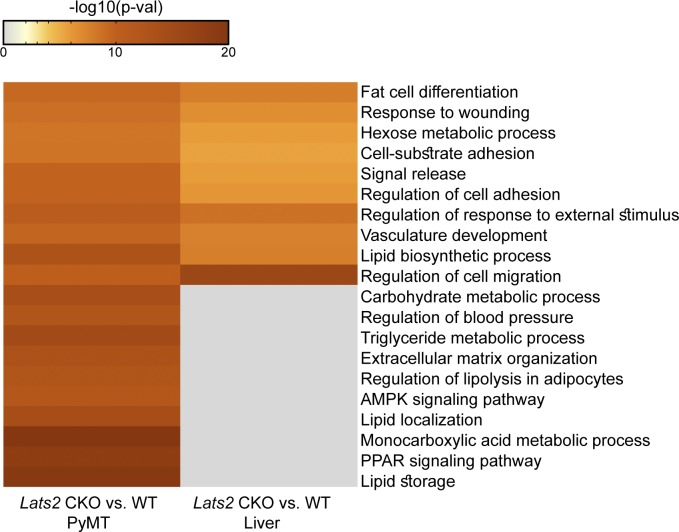
Although PPARγ itself was not functionally enriched by depletion of hepatic Lats2 (Aylon et al, 2016), several other metabolic pathways were commonly enriched by deletion of Lats2 in both breast and liver (Fig S7). One such pathway is SREBF1 (also known as SREBP1), which is hyperactivated upon Lats2 depletion in both mammary tumors (ingenuity activation z-score 0.64; P-value of overlap 3.6 × 10−8) and mouse livers (Aylon et al, 2016). This resonates with our previous finding that in the liver, LATS2 (but not LATS1) interacts with SREBP1 and its paralog SREBP2 and inhibits their transcriptional activity (Aylon et al, 2016). By similar reasoning, it is plausible that LATS2 and LATS1 may also affect PPARγ activity and cell identity, respectively, in other tissues beyond mammary glands. These effects of LATS2 and LATS1 may be mediated, at least in part, by YAP/TAZ. However, as is often the case for important signaling pathways, multiple components may act in parallel to converge on a common biological outcome. Thus, LATS proteins might execute their functions via a combination of YAP/TAZ-dependent and YAP/TAZ-independent molecular processes. For example, LATS2-mediated phosphorylation of TAZ can indirectly promote PPARγ activity in adipocytes by relieving the inhibitory activity of TAZ on PPARγ (Hong et al, 2005; An et al, 2013), although also within the context of adipogenesis, the upstream Hippo components MST1/2 can promote SAV1–PPARγ complex formation and PPARγ activation (Park et al, 2012). Together with our observations, this implies that multiple Hippo pathway components may modulate PPARγ activity in different biological settings.
Figure S7.
Metascape Tripathi et al 2015pathways enrichment analysis (Tripathi et al 2015, http://metascape.org) of differentially expressed genes (counts > 5, FC > 1.5, adjP-value < 0.05) in Lats2-CKO mammary tumors (PyMT) or Lats2-CKO livers, relative to their WT littermate controls. Heatmap shows the enrichment score (−log10(P-value)) for each pathway in each of the differentially expressed gene lists.
Likewise, LATS1 activity may affect cellular identity in contexts beyond mammary tumorigenesis. For instance, liver-specific combined Lats1 and Lats2 knockout forces hepatoblasts to commit to biliary epithelial cell lineage (Lee et al, 2016). This occurs via YAP/TAZ repression of Hnf4a expression. Furthermore, in preimplantation embryos, LATS-mediated YAP phosphorylation alters cell fate to inhibit the development of the trophectoderm lineage (Nishioka et al, 2009). A similar role in cell fate specification was observed in Drosophila for the LATS ortholog warts during photoreceptor differentiation (Mikeladze-Dvali et al, 2005). Although our transcriptomic analysis revealed activation of YAP/TAZ transcriptional program preferentially upon Lats1 deletion, the exact contribution of this activation to the observed phenotypes remains to be elucidated.
Although the robust categorization of breast tumors into subtypes has both molecular and clinical implications, there may exist substantial plasticity in many tumors, confounding unequivocal assignment to a defined subtype (Wahl & Spike, 2017). We observed that deletion of Lats1 in a lumB-prone breast cancer model results in tumors that exhibit histology and gene expression patterns partially resembling ER-negative, basal-like tumors. Likewise, most of the human BRCA1-associated tumors, typically classified as basal-like, have been shown to originate from luminal progenitors rather than from basal stem cells (Lim et al, 2009; Molyneux et al, 2010). More broadly, the properties of breast cancer propagating cells and the nature of the tumors that they spawn can be shaped extensively by epigenetic and microenvironmental influences (Wahl & Spike, 2017). Of note, reduction in LATS kinase levels in primary human breast epithelial cells increases the number of bipotent and luminal progenitor cells (Britschgi et al, 2017). Interestingly, although we find that depletion of LATS1 in a luminal cancer model enables the increased expression of genes characteristic of a basal-like transcriptional program, Britschgi et al (Britschgi et al, 2017) observed that loss of LATS in basal cells induced the expression of luminal markers. Thus, tumor suppressors such as LATS1 may constrain the inherent plasticity of luminal progenitor cells, and their loss may augment plasticity in the context of tumorigenesis. Alternatively, it remains possible that in some contexts, mature luminal cells might undergo transdifferentiation, acquiring basal-like characteristics and gene expression patterns (Doherty et al, 2016). Future lineage tracing studies hopefully would resolve this important issue. Either way, we propose that compromised LATS1 expression may be conducive to enhanced tumor cell plasticity, facilitating escape from hormone therapy. Importantly, patients with lumB tumors harboring low levels of LATS1 might be less responsive to tamoxifen treatment alone and might benefit from more aggressive therapeutic options.
Selective pressure to decrease a particular LATS paralog in breast cancer may depend on distinct subtype-specific signaling events. In particular, LATS proteins were shown to modulate estrogen signaling (Lit et al, 2013; Britschgi et al, 2017) and sustain the canonical functions of the p53 tumor suppressor (Aylon et al, 2006, 2010; Furth et al, 2015). Interestingly, ERα can bind p53 and inhibit p53-dependent transcription (Liu et al, 2006; Konduri et al, 2010). In this scenario, diminished expression of LATS proteins in ER+ breast cancers may have a two-pronged effect in compromising p53 tumor suppressive functionality. In contrast, ER- tumors frequently harbor TP53 mutations, often resulting in accumulation of mutant p53 proteins (Curtis et al, 2012). Such mutant p53 proteins may acquire oncogenic gain-of-function features (Muller & Vousden, 2014; Shetzer et al, 2016), which can be further augmented by binding to YAP (Di Agostino et al, 2016), whose activity is typically suppressed by the LATS kinases. Altogether, our findings imply that deregulation of LATS1 and LATS2 can exert both common and distinct effects on breast cancer progression, by interaction with a variety of regulatory pathways. Deciphering the nuances of those interactions will be critical for advancing our ability to apply Hippo pathway knowledge towards improving therapeutic options for breast cancer patients.
Materials and Methods
Human cancer gene expression and methylation data
The TCGA breast invasive carcinoma gene expression (IlluminaHiSeq log (normalized counts + 1)), methylation (β values from Infinium Methylation 450 k), and clinical data were downloaded from the Xena cancer genome browser (http://xena.ucsc.edu).
A cutoff of the 20% of tumors expressing the lowest levels of each LATS gene was used to divide the tumors into groups (i.e., high versus low expression of each gene). Differences between the four groups of tumors were examined by ANOVA.
LATS1- and LATS2-associated gene signatures were generated based on the above described comparison. The most down-regulated genes in LATS2L (compared with LATSH) tumors were extracted (foldchange > 2, adjP-value < 0.05). The 20 genes most differentially down-regulated exclusively in the LATS2L group of tumors (i.e., not significantly down-regulated in the LATS1L or LATSL groups) were used. The same process was used to generate the LATS1-associated gene signature (Table S4).
Table S4 Gene signatures used for human tumor expression and survival analysis. (9.9KB, xlsx)
For LATS2 promoter methylation analysis, lumB tumors were divided according to LATS2 expression (median cutoff).
Xena cancer genome browser was used to visualize expression data from the TCGA Breast cancer (BRCA) dataset. Tumors were clustered according to PAM50 subtype (PAM50Call_RNAseq). Only tumors with classification were included.
Survival plots were generated either by KM plotter (LATS2 probe 227013_at, Györffy et al, 2010) or by the R packages “survival” and “survminer,” based on the data available in the METABRIC dataset (Curtis et al, 2012). Tumors were divided according to expression (median cutoff). When comparing groups of genes as a signature, z-scores of the expression were calculated per each gene, and the average of these z-scores per sample was used as the signature.
Correlation between methylation and expression was calculated by Pearson's correlation coefficient.
Animals
All mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the Weizmann Institute (approval 14521114-1). Genotyping was used to classify littermates into the different experimental groups, and all comparisons of the different genotypes were done between littermates. After sacrificing, the mice were weighed, and tumors were extracted, weighed, and measured. Three tumors consisting of the largest, smallest, and average “representative” tumors were processed further for sample. For analysis of tamoxifen sensitivity, 6-8-wk-old females were injected IP with 20 mg/kg tamoxifen free base (#T5648; Sigma-Aldrich) every other day for 14 d, until the end of the experiment.
Human tumor samples
Collection of the clinical samples and their experimental use were approved by the Bioethics Committee of the Medical School of Athens, in accordance with the Declaration of Helsinki and local laws and regulations, following also written patient consent. None of the patients received any cancer therapy before surgical resection of the lesions.
Histopathological evaluation
Histological assessment was performed by three experienced pathologists (ISP, VV, and VGG) based on previously published criteria (Cardiff et al, 2000; Lin et al, 2003; Rudmann et al, 2012).
Immunohistochemistry
Immunohistochemistry analysis was performed on paraffin-embedded tissues using the primary antibodies listed in Table S5. Heat-mediated antigen retrieval was performed in 10 mM citric acid (pH 6.0). The UltraVision Quanto Detection System was used (#TL-060-QHD; Thermo Fisher Scientific, Bioanalytica) according to the manufacturer's instructions. Hematoxylin was used as counterstain. Evaluation of γΗ2ΑΧ was performed as previously described (Tarcic et al, 2016). For cleaved caspase 3 immunostaining, the % of positive cells was calculated. ERα was evaluated by calculating the % of cancer cells exhibiting intense nuclear staining. For cytokeratin 14, we counted the % of cancer cells exhibiting intense staining at areas with invasive carcinoma, to avoid counting myoepithelial cells. Evaluation of LATS1 and LATS2 in human breast sporadic carcinomas was based on the method described by Xu et al (2015), using a mixed score (intensity score > 0: negative; 1: weak; 2: moderate; 3: strong and proportion score > 0: negative; 1: < 10%; 2: 11–50%, 3: 51–80%; 4: >80%). In all cases, three independent observers carried out slide examination, with minimal interobserver variability. When analyzed for specific markers, tumors from the same histological subtype were tallied and statistically analyzed, to prevent confounding data due to different tumor spectra in different subtypes.
Table S5 Reagents used throughout this study. (12.3KB, xlsx)
Isolation of total RNA, reverse transcription, and RTqPCR
RNA was isolated using the NucleoSpin kit (Macherey Nagel), RNeasy Mini kit (QIAGEN), or RNeasy Microkit (QIAGEN). 1-2 μg of each RNA sample was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Promega) and random hexamer primers (Applied Biosystems). Real-time qPCR was performed using SYBR Green PCR Supermix (Invitrogen) with a StepOne real-time PCR instrument (Applied Biosystems). For each gene, values for the standard curve were measured and the relative quantity was normalized to HPRT or GAPDH mRNA.
Library preparation, RNA-seq, and analysis
PyMT tumors
RNA was isolated from Lats1-CKO and Lats2-CKO PyMT tumors. For each genotype, the corresponding WT littermate controls were used. For RNA-seq analysis, 500 ng of total RNA was processed using the TruSeq RNA Sample Preparation Kit v2 protocol (Illumina) (Part #15026495). Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of seven samples per lane (three WT tumors and four Lats-CKO tumors). Samples were sequenced on an Illumina HiSeq 2500 V4 instrument. 29–38 million and 35–35 million single-end 60-bp reads were sequenced for the Lats1-CKO and the Lats2-CKO experiments, respectively.
Reads were mapped using STAR (Dobin and Gingeras, 2015) to mm10 assembly and quantified using mm10 RefSeq annotation. Differential expression analysis was performed on genes with a sum value of at least five counts in all samples using DESeq2 (Love et al, 2014). Raw P-values were adjusted for multiple testing using the procedure of Benjamini and Hochberg.
Expression values of differentially expressed genes (rld-log normalized counts) were used to generate hierarchical clustering heatmaps (Pearson’s correlation and complete linkage using Partek Genomics Suite). PCA plots of RNA-seq data were generated using rld values with R ggplot.
Human cell lines
RNA from ZR75-1 and MCF7 cells transfected with siControl, siLATS1, or siLATS2 oligonucleotides was isolated and subjected to RNA-seq as above. Three and two independent biological replicates were used for ZR75-1 and MCF7 cells, respectively. ZR75-1 cells were transfected with either GFP only, GFP-LATS1, or GFP-LATS2. GFP-positive cells were sorted by FACS 24 h following transfection (∼80,000 cells per sample). Two independent replicates were used. For RNA-seq analysis, 500 ng of total RNA was fragmented, followed by reverse transcription and second strand cDNA synthesis. The double-stranded cDNA was subjected to end repair, A base addition, adapter ligation, and PCR amplification to create libraries. Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of multiple samples over three lanes. ∼15 million single-end 60-bp reads were sequenced per sample on Illumina HiSeq 2500 V4 instrument.
Reads were trimmed using cutadapt (Martin, 2011) and mapped to assembly hg38. Counting proceeded over genes annotated in RefSeq release hg38 using STAR (Dobin and Gingeras, 2015). Differential expression analysis was performed using DESeq2 (Love et al, 2014) with the cooksCutoff = FALSE, independentFiltering = FALSE set to False. Raw P-values were adjusted for multiple testing using the procedure of Benjamini and Hochberg.
The data have been deposited in NCBI's Gene Expression Omnibus (GEO) and are available through GEO series accession number GSE116818.
Cell lines, transfections, and treatments
All cell lines were maintained at 37°C with 5% CO2. ZR75-1 cells were cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin + streptomycin (P/S). MDA-MB-468 and MCF7 cells were cultured in DMEM supplemented with 10% FBS and 1% P/S. PyMT-derived cell lines were generated from freshly minced tissue after digestion with 10 mg collagenase A (Roche 10103586001) and 1.5 mg hyluronidase (H4272-30MG; Sigma-Aldrich). After dissociation, the cells were filtered through 70-μm strainers; washed with DMEM, Hanks' Balanced Salt Solution [+Ca +Mg] supplemented with 2% FBS and 2% Hepes, and Hanks' solution with 8.29 g NH4Cl/l Tris (pH 7.2); and finally resuspended in DMEM. Epithelial cell pellets were obtained by twice centrifuging at 800 g for 5 s. The following day, adherent cells were washed aggressively to detach fibroblasts. Initially, the cells were cultured in DMEM supplemented with 15% FBS, 2 mM glutamine, 1× nonessential amino acids, and 1% P/S. After the cultures stabilized, they were acclimated to and propagated in DMEM supplemented with 10% FBS and 1% P/S.
For siRNA-mediated knockdown, the indicated SMARTpools (Dharmacon, see Table S5) were used with Dharmafect #1 transfection reagent according to the manufacturer's instructions. Final siRNA concentration was 25 nM in all cases. Plasmid transfections were performed using jetPRIME DNA transfection reagent (Polyplus Transfection) according to the manufacturer's instructions. A list of plasmids used can be found in Table S5. Retroviral packaging was performed by jetPEI-mediated transfection of HEK293T or HEK293T Pheonix cells with the appropriate plasmids together with pMD2.G DNA encoding VSV-G envelopes proteins (when infecting human cell lines). Virus-containing supernatants were collected 48 h following transfection, filtered, and supplemented with 8 μg/ml polybrene. Infected ZR75-1 and PyMT cells were selected with 1.5 or 1 μg/ml blasticidin, respectively. The cells were treated with 1 μM or 5 μM 5-aza-2′deoxycytidine each day for four consecutive days. Glucose starvation and treatments with RGZ were conducted in media containing 1% serum.
Western blots
Cell pellets were resuspended in protein sample buffer and boiled. The samples were resolved by SDSPAGE. Imaging was performed using a ChemiDoc MP imaging system (Bio-Rad) with the Image Lab 4.1 program (Bio-Rad).
Imaging flow cytometry (ImageStream)
Cells were collected with trypsin, washed with PBS, and fixed in 3.5% PFA followed by permeabilization with 0.1% Triton. Washes were done in PBS supplemented with 1% FCS and 2 mM EDTA. Cells were incubated with the indicated primary antibody for 1 h at room temperature, followed by washes and 45 min of incubation with fluorescent-conjugated secondary antibody (GaR Alexa 647, #A21244; LifeTech or GaM Alexa 595, #A11032; LifeTech) and DAPI (#D1306; LifeTech). The cells were imaged by ImageStreamX mark II (Amnis, part of EMD Millipore) using bright field 488 nm, 561 nm, and 642 nm lasers. At least 30,000 cells were collected from each sample and data were analyzed using image analysis software (IDEAS 6.2; Amnis Corporation). Images were compensated for fluorescent dye overlap by using single-stain controls. Gating was done for single cells, using the area and aspect ratio features, and for focused cells using the gradient RMS feature, as previously described (George et al, 2006). Data were analyzed with the IDEAS 6.1 software (Amnis, part of EMD Millipore). Only cells with intact nucleus (according to DAPI staining) were analyzed. Nuclear localization was determined by the similarity feature on the nuclear mask of the DAPI staining and the GFP signal (the log transformed Pearson's correlation coefficient in the two input images). PPARγ-positive cells were gated on the basis of comparison with nonstained sample.
PI exclusion assay
Cells, including floaters, were collected using trypsin, washed once with PBS, and resuspended in PBS containing 12.5 μg/ml PI. Percentage of dead cells (positive for PI staining) was measured for each sample using Guava EasyCyte Flow Cytometer (Merck Millipore).
Measurement of oxygen consumption rate (OCR) and extracellular acidification rates (ECAR) (Seahorse analysis)
OCR and ECAR assays were performed according to the manufacturer’s instructions using the XFe96 or XFp Extracellular Flux analyzers (Agilent Technologies).
ZR75-1 and PyMT cells were plated at 20,000 and 5,000 cells in 80 μl per well, respectively, in Agilent Seahorse cell culture microplates (Cat No. 101085-004). The cells were incubated for 48 h in a humidified 37°C incubator with 5% CO2 in RPMI (ZR75-1 cells) or DMEM (PyMT cells) media supplemented with 1% FCS. One hour before performing the assay, growth medium in the wells of an XF cell plate was exchanged with the appropriate assay medium (pH 7.4 ± 0.1 at 37°C) by three consecutive washes leaving 180 μl of medium per well in the end. For ECAR measurements, XF base medium 4 (Agilent Technologies) supplemented with 2 mM L-glutamine and for OCR measurements, 2 mM L-glutamine, 1 mM sodium pyruvate, and 10 mM D+ glucose. The cells were incubated at 37°C (no CO2) for 60 min to allow temperature and pH equilibration. Meanwhile, hydrated XFe96/XFp sensor cartridge ports were loaded with respective assay compounds diluted in the assay medium. For ECAR, four injection cycles consisted of: 10 mM glucose (#G7528; Sigma-Aldrich) 1 μM oligomycin (#O4876; Sigma-Aldrich) and then 50 mM 2-DG (#D6134; Sigma-Aldrich). For OCR measurements, injection cycles were as follows: 1 μM oligomycin, 1 μM FCCP (#C2920; Sigma-Aldrich), and 0.5 μM rotenone/antimycin A (#R8875/#A8674; Sigma-Aldrich) were injected consecutively. Each cycle consisted of 3-min mixing and 3-min measurement. The average rates of each cycle were used for computations.
Nuclear stain with CyQuant (#C35011; Thermo Fisher Scientific) was used for normalization. Basal respiration was calculated by subtracting the nonmitochondrial respiration rate (minimum rate measurement after rotenone/antimycin A addition) from the last measurement rate before oligomycin addition. Basal glycolysis was calculated by subtracting the last rate measurement before glucose addition from the maximum rate measurement after glucose addition.
Functional analysis of gene expression data
For GSEA analysis (Subramanian et al, 2005), genes were ranked according to fold change between the two described conditions, with only significant differences considered (PyMT CKO experiments P-value < 0.05; siRNA in human cell lines adjP-value < 0.05; overexpression in human cell lines average FC > 1.5). Comparison with different gene sets (Table S6) was done using GSEA preranked tool. Similar ranking was used to analyze gene expression patterns by Ingenuity Pathway Analysis software (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). KEGG pathways and GO-BP enrichment analysis was performed using DAVID (Huang et al, 2008). Pathway enrichment analysis was also performed using GeneAnalytics (Ben-Ari Fuchs et al, 2016).
Table S6 List of gene sets used for GSEA analysis. (210KB, xlsx)
To derive a basal-associated gene signature, tumors in the BRCA dataset of TCGA were divided according to tumor type, and the genes most differentially expressed between luminal and basal tumors were extracted (adjP-value < 0.05, FC > 1.5) and analyzed for enriched biological pathways (Metascape). Both differentially expressed genes and genes associated with the most differential pathways were compared with a list of differentially expressed genes in either Lats1- or Lats2-CKO PyMT tumors (each compared with its littermate WT-PyMT controls). All overlapping genes were used to define the signature.
Statistics
Unless noted otherwise, P-values were determined using two-tailed t tests (paired or two-sample, depending on experimental setting).
Supplementary Material
Acknowledgements
We thank Giuseppe Mallel for fruitful discussions and valuable inputs; Sima Benjamin, Dana Robbins, Orit Zion, and Shlomit Gilad (Israel National Center for Personalized Medicine, Weizmann Institute of Science [WIS]) for their help with RNA-seq; Gil Stelzer (Life Sciences Core Facilities, WIS) for assisting with pipeline analysis of human cell line RNA-seq results; Ori Brenner (Veterinary Resources, WIS) for help with histological analysis; and Ziv Porath (Life Sciences Core Facilities, Faculty of Biochemistry, WIS) for ImageStream data acquisition and analysis. We thank James Martin (Baylor College of Medicine, Houston, TX, USA) for the generous sharing of conditional knockout mice. This work was supported by grants from the Rising Tide Foundation, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, a Center of Excellence grant from the Israel Science Foundation, the Comisaroff Family Trust, the Estate of John Hunter, and the Moross Integrated Cancer Center. M Oren is incumbent of the Andre Lwoff chair in molecular biology.
Author Contributions
N Furth: conceptualization, formal analysis, funding acquisition, validation, investigation, visualization, methodology, and writing—original draft, review, and editing.
IS Pateras: formal analysis and investigation.
R Rotkopf: formal analysis, investigation, and methodology.
V Vlachou: formal analysis and investigation.
I Rivkin: investigation.
I Schmitt: investigation.
D Bakaev: investigation.
A Gershoni: investigation.
E Ainbinder: investigation.
D Leshkowitz: data curation and formal analysis.
RL Johnson: resources.
VG Gorgoulis: supervision, funding acquisition, and investigation.
M Oren: conceptualization, supervision, funding acquisition, methodology, and writing—review and editing.
Y Aylon: conceptualization, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, and writing—original draft, review, and editing.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- An Y, Kang Q, Zhao Y, Hu X, Li N (2013) Lats2 modulates adipocyte proliferation and differentiation via Hippo signaling. PLoS One 8: e72042 10.1371/journal.pone.0072042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydiner A, Sen F, Tambas M, Ciftci R, Eralp Y, Saip P, Karanlik H, Fayda M, Kucucuk S, Onder S, et al. (2015) Metaplastic breast carcinoma versus triple-negative breast cancer: Survival and response to treatment. Medicine 94: e2341 10.1097/md.0000000000002341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Gershoni A, Rotkopf R, Biton IE, Porat Z, Koh AP, Sun X, Lee Y, Fiel M-I, Hoshida Y, et al. (2016) The LATS2 tumor suppressor inhibits SREBP and suppresses hepatic cholesterol accumulation. Genes Dev 30: 786–797. 10.1101/gad.274167.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M (2006) A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev 20: 2687–2700. 10.1101/gad.1447006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Ofir-Rosenfeld Y, Yabuta N, Lapi E, Nojima H, Lu X, Oren M (2010) The Lats2 tumor suppressor augments p53-mediated apoptosis by promoting the nuclear proapoptotic function of ASPP1. Genes Dev 24: 2420–2429. 10.1101/gad.1954410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. (2012) The cancer cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Fuchs S, Lieder I, Stelzer G, Mazor Y, Buzhor E, Kaplan S, Bogoch Y, Plaschkes I, Shitrit A, Rappaport N, et al. (2016) GeneAnalytics: An integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS 20: 139–151. 10.1089/omi.2015.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenton JD, Carey LA, Ahmed AA, Caldas C (2005) Molecular classification and molecular forecasting of breast cancer: Ready for clinical application? J Clin Oncol 23: 7350–7360. 10.1200/jco.2005.03.3845 [DOI] [PubMed] [Google Scholar]
- Britschgi A, Duss S, Kim S, Couto JP, Brinkhaus H, Koren S, De Silva D, Mertz KD, Kaup D, Varga Z, et al. (2017) The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERα. Nature 541: 541 10.1038/nature20829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Nogales-Cadenas R, Zhang Q, Lin J-R, Zhang W, O'Brien K, Montagna C, Zhang ZD (2017) Transcriptomic dynamics of breast cancer progression in the MMTV-PyMT mouse model. BMC Genomics 18: 185 10.1186/s12864-017-3563-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, Rehm S, Russo J, Tavassoli FA, Wakefield LM, et al. (2000) The mammary pathology of genetically engineered mice: The consensus report and recommendations from the Annapolis meeting. Oncogene 19: 968 10.1038/sj.onc.1203277 [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhang N, Gray RS, Li H, Ewald AJ, Zahnow CA, Pan D (2014) A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev 28: 432–437. 10.1101/gad.233676.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, Gabrielson E, Werb Z, Ewald AJ (2013) Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155: 1639–1651. 10.1016/j.cell.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti Anna R, Poletti A, et al. (2011) The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147: 759–772. 10.1016/j.cell.2011.09.048 [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Massarweh S, Huang S, Tsimelzon A, Shou J, Malorni L, Hilsenbeck SG, Osborne CK, Schiff R (2008) Development of resistance to targeted therapies transforms the clinically-associated molecular profile subtype of breast tumor xenografts. Cancer Res 68: 7493–7501. 10.1158/0008-5472.can-08-1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetri GD, Fletcher CDM, Mueller E, Sarraf P, Naujoks R, Campbell N, Spiegelman BM, Singer S (1999) Induction of solid tumor differentiation by the peroxisome proliferator-activated receptor-γ ligand troglitazone in patients with liposarcoma. Proc Natl Acad Sci U S A 96: 3951–3956. 10.1073/pnas.96.7.3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S, Sorrentino G, Ingallina E, Valenti F, Ferraiuolo M, Bicciato S, Piazza S, Strano S, Del Sal G, Blandino G (2016) YAP enhances the pro‐proliferative transcriptional activity of mutant p53 proteins. EMBO Rep 17: 188–201. 10.15252/embr.201540488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR (2015) Mapping RNA-seq reads with STAR. Curr Protoc Bioinform 51: 11.14.1–11.14.19. 10.1002/0471250953.bi1114s51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty MR, Smigiel JM, Junk DJ, Jackson MW (2016) Cancer stem cell plasticity drives therapeutic resistance. Cancers 8: 8 10.3390/cancers8010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474: 179 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- El Akoum S. (2014) PPAR gamma at the crossroads of health and disease: A masterchef in metabolic homeostasis. Endocrinol Metab Syndr 03: 1–12. 10.4172/2161-1017.1000126 [DOI] [Google Scholar]
- Fenner MH, Elstner E (2005) Peroxisome proliferator-activated receptor-γ ligands for the treatment of breast cancer. Expert Opin Investig Drugs 14: 557–568. 10.1517/13543784.14.6.557 [DOI] [PubMed] [Google Scholar]
- Furth N, Aylon Y (2017) The LATS1 and LATS2 tumor suppressors: Beyond the Hippo pathway. Cell Death Differ 24: 1488 10.1038/cdd.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N, Bossel Ben-Moshe N, Pozniak Y, Porat Z, Geiger T, Domany E, Aylon Y, Oren M (2015) Down-regulation of LATS kinases alters p53 to promote cell migration. Genes Dev 29: 2325–2330. 10.1101/gad.268185.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George TC, Fanning SL, Fitzgeral-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, et al. (2006) Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods 311: 117–129. 10.1016/j.jim.2006.01.018 [DOI] [PubMed] [Google Scholar]
- Geyer FC, Pareja F, Weigelt B, Rakha E, Ellis IO, Schnitt SJ, Reis-Filho JS (2017) The spectrum of triple-negative breast disease. Am J Pathol 187: 2139–2151. 10.1016/j.ajpath.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123: 725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Haque R, Ahmed SA, Inzhakova G, Shi J, Avila C, Polikoff J, Bernstein L, Enger SM, Press MF (2012) Impact of breast cancer subtypes and treatment on survival: An analysis spanning two decades. Cancer Epidemiol Biomarkers Prev 21: 1848–1855. 10.1158/1055-9965.epi-12-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. (2007) Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8: R76–R76. 10.1186/gb-2007-8-5-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday DL, Speirs V (2011) Choosing the right cell line for breast cancer research. Breast Cancer Res 13: 215–215. 10.1186/bcr2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-H, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, et al. (2005) TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309: 1074–1078. 10.1126/science.1110955 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kersten S, Desvergne B, Wahli W (2000) Roles of PPARs in health and disease. Nature 405: 421 10.1038/35013000 [DOI] [PubMed] [Google Scholar]
- Konduri SD, Medisetty R, Liu W, Kaipparettu BA, Srivastava P, Brauch H, Fritz P, Swetzig WM, Gardner AE, Khan SA, et al. (2010) Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc Natl Acad Sci U S A 107: 15081–15086. 10.1073/pnas.1009575107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuukasjärvi T, Kononen J, Helin H, Holli K, Isola J (1996) Loss of estrogen receptor in recurrent breast cancer is associated with poor response to endocrine therapy. J Clin Oncol 14: 2584–2589. 10.1200/jco.1996.14.9.2584 [DOI] [PubMed] [Google Scholar]
- Lee D-H, Park JO, Kim T-S, Kim S-K, Kim T-h, Kim M-c, Park GS, Kim J-H, Kuninaka S, Olson EN, et al. (2016) LATS-YAP/TAZ controls lineage specification by regulating TGFβ signaling and Hnf4α expression during liver development. Nat Commun 7: 11961 10.1038/ncomms11961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gumbiner BM (2016) Deregulation of the Hippo pathway in mouse mammary stem cells promotes mammary tumorigenesis. Mamm Genome 27: 556–564. 10.1007/s00335-016-9662-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZH, Hu PH, Tu JH, Yu NS (2016) Luminal B breast cancer: Patterns of recurrence and clinical outcome. Oncotarget 7: 65024–65033. 10.18632/oncotarget.11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberato MV, Nascimento AS, Ayers SD, Lin JZ, Cvoro A, Silveira RL, Martínez L, Souza PCT, Saidemberg D, Deng T, et al. (2012) Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) γ activators and pan-PPAR partial agonists. PLoS One 7: e36297 10.1371/journal.pone.0036297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat M-L, Gyorki DE, Ward T, Partanen A, et al. (2009) Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15: 907 10.1038/nm.2000 [DOI] [PubMed] [Google Scholar]
- Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW (2003) Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 163: 2113–2126. 10.1016/s0002-9440(10)63568-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lit LC, Scott S, Zhang H, Stebbing J, Photiou A, Giamas G (2013) LATS2 is a modulator of estrogen receptor alpha. Anticancer Res 33: 53–63. [PubMed] [Google Scholar]
- Liu W, Konduri SD, Bansal S, Nayak BK, Rajasekaran SA, Karuppayil SM, Rajasekaran AK, Das GM (2006) Estrogen receptor-α binds p53 tumor suppressor protein directly and represses its function. J Biol Chem 281: 9837–9840. 10.1074/jbc.c600001200 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglione JE, Moghanaki D, Young LJT, Manner CK, Ellies LG, Joseph SO, Nicholson B, Cardiff RD, MacLeod CL (2001) Transgenic polyoma middle-T mice model premalignant mammary disease. Cancer Res 61: 8298–8305. [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, Weiss H, Rimawi M, Schiff R (2008) Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res 68: 826–833. 10.1158/0008-5472.can-07-2707 [DOI] [PubMed] [Google Scholar]
- Mehta RG, Williamson E, Patel MK, Koeffler HP (2000) A ligand of peroxisome proliferator-activated receptor γ, retinoids, and prevention of preneoplastic mammary lesions. J Natl Cancer Inst 92: 418–423. 10.1093/jnci/92.5.418 [DOI] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, Park HW, Mo J-S, Lu W, Lu S, et al. (2015) MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun 6: 8357 10.1038/ncomms9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen Y-W, Cohen S, Desplan C (2005) The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122: 775–787. 10.1016/j.cell.2005.07.026 [DOI] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay F-A, McCarthy A, Kendrick H, Natrajan R, MacKay A, Grigoriadis A, Tutt A, Ashworth A, et al. (2010) BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7: 403–417. 10.1016/j.stem.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Moroishi T, Hansen CG, Guan K-L (2015) The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer 15: 73 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Hayashi T, Pan W-W, Fujita Y, Holt MV, Qin J, Carson DA, Guan K-L (2016) The Hippo pathway kinases LATS1/2 suppress cancer immunity. Cell 167: 1525–1539. e1517 10.1016/j.cell.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller E, Sarraf P, Tontonoz P, Evans RM, Martin KJ, Zhang M, Fletcher C, Singer S, Spiegelman BM (1998) Terminal differentiation of human breast cancer through PPARγ. Mol Cell 1: 465–470. 10.1016/s1097-2765(00)80047-7 [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH (2014) Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25: 304–317. 10.1016/j.ccr.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Inoue K-i, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. (2009) The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 16: 398–410. 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Osborne CK. (1998) Steroid hormone receptors in breast cancer management. Breast Cancer Res Treat 51: 227–238. 10.1023/a:1006132427948 [DOI] [PubMed] [Google Scholar]
- Park BH, Kim DS, Won GW, Jeon HJ, Oh B-C, Lee Y, Kim E-G, Lee YH (2012) Mammalian Ste20-like kinase and SAV1 promote 3T3-L1 adipocyte differentiation by activation of PPARγ. PLoS One 7: e30983 10.1371/journal.pone.0030983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- Rudmann D, Cardiff R, Chouinard L, Goodman D, Küttler K, Marxfeld H, Molinolo A, Treumann S, Yoshizawa K (2012) Proliferative and nonproliferative lesions of the rat and mouse mammary, zymbal's, preputial, and clitoral glands. Toxicol Pathol 40: 7S–39S. 10.1177/0192623312454242 [DOI] [PubMed] [Google Scholar]
- Shashni B, Sakharkar KR, Nagasaki Y, Sakharkar MK (2013) Glycolytic enzymes PGK1 and PKM2 as novel transcriptional targets of PPARγ in breast cancer pathophysiology. J Drug Target 21: 161–174. 10.3109/1061186x.2012.736998 [DOI] [PubMed] [Google Scholar]
- Shetzer Y, Molchadsky A, Rotter V (2016) Oncogenic mutant p53 gain of function nourishes the vicious cycle of tumor development and cancer stem-cell formation. Cold Spring Harb Perspect Med 6: a026203 10.1101/cshperspect.a026203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio RE, Chen ER, Lazar MA (2014) Thazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cel Metab 20: 573–591. 10.1016/j.cmet.2014.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98: 10869–10874. 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John MAR, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, McGrath J, Xu T (1999) Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet 21: 182 10.1038/5965 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Wang Y, Williams CR, Risingsong R, Gilmer T, Willson TM, Sporn MB (1999) A new ligand for the peroxisome proliferator-activated receptor-γ (PPAR-γ), GW7845, inhibits rat mammary carcinogenesis. Cancer Res 59: 5671–5673. [PubMed] [Google Scholar]
- Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, Noguchi S (2005) Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res 11: 1380–1385. 10.1158/1078-0432.ccr-04-1773 [DOI] [PubMed] [Google Scholar]
- Tang X, Lin C-C, Spasojevic I, Iversen ES, Chi J-T, Marks JR (2014) A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Res 16: 415 10.1186/s13058-014-0415-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarcic O, Pateras Ioannis S, Cooks T, Shema E, Kanterman J, Ashkenazi H, Boocholez H, Hubert A, Rotkopf R, Baniyash M, et al. (2016) RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep 14: 1462–1476. 10.1016/j.celrep.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran B, Bedard PL (2011) Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res 13: 221–221. 10.1186/bcr2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LCF, et al. (2015) Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18: 723–735. 10.1016/j.chom.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deurzen CH, Lee AH, Gill MS, Menke‐Pluijmers MB, Jager A, Ellis IO, Rakha EA (2011) Metaplastic breast carcinoma: Tumour histogenesis or dedifferentiation? J Pathol 224: 434–437. 10.1002/path.2872 [DOI] [PubMed] [Google Scholar]
- Viale G, Regan MM, Maiorano E, Mastropasqua MG, Dell'Orto P, Rasmussen BB, Raffoul J, Neven P, Orosz Z, Braye S, et al. (2007) Prognostic and predictive value of centrally reviewed expression of estrogen and progesterone receptors in a randomized trial comparing letrozole and tamoxifen adjuvant therapy for postmenopausal early breast cancer: BIG 1-98. J Clin Oncol 25: 3846–3852. 10.1200/jco.2007.11.9453 [DOI] [PubMed] [Google Scholar]
- Wahl GM, Spike BT (2017) Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. NPJ Breast Cancer 3: 14 10.1038/s41523-017-0012-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, Kitakami J, Ihara S, Hashimoto Y, Hamakubo T, et al. (2009) The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-set7/setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 29: 3544–3555. 10.1128/mcb.01856-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson TM, Cobb JE, Cowan DJ, Wiethe RW, Correa ID, Prakash SR, Beck KD, Moore LB, Kliewer SA, Lehmann JM (1996) The Structure−Activity relationship between peroxisome proliferator-activated receptor γ agonism and the antihyperglycemic activity of thiazolidinediones. J Med Chem 39: 665–668. 10.1021/jm950395a [DOI] [PubMed] [Google Scholar]
- Xu B, Sun D, Wang Z, Weng H, Wu D, Zhang X, Zhou Y, Hu W (2015) Expression of LATS family proteins in ovarian tumors and its significance. Hum Pathol 46: 858–867. 10.1016/j.humpath.2015.02.012 [DOI] [PubMed] [Google Scholar]
- Yang SX, Polley EC, Nguyen D (2017) Association of γH2AX at diagnosis with chemotherapy outcome in patients with breast cancer. Theranostics 7: 945–951. 10.7150/thno.19102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Smolen GA, Haber DA (2008) Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res 68: 2789–2794. 10.1158/0008-5472.can-07-6205 [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhang W, Liang B, Casimiro MC, Whitaker-Menezes D, Wang M, Lisanti MP, Lanza-Jacoby S, Pestell RG, Wang C (2009) PPARγ activation induces autophagy in breast cancer cells. Int J Biochem Cell Biol 41: 2334–2342. 10.1016/j.biocel.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolezzi JM, Silva-Alvarez C, Ordenes D, Godoy JA, Carvajal FJ, Santos MJ, Inestrosa NC (2013) Peroxisome proliferator-activated receptor (PPAR) γ and PPARα agonists modulate mitochondrial fusion-fission dynamics: Relevance to reactive oxygen species (ROS)-related neurodegenerative disorders? PLoS One 8: e64019 10.1371/journal.pone.0064019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4 Gene signatures used for human tumor expression and survival analysis. (9.9KB, xlsx)
Table S5 Reagents used throughout this study. (12.3KB, xlsx)
Table S6 List of gene sets used for GSEA analysis. (210KB, xlsx)