Abstract
Background
This study investigated the effect of xeroderma pigmentosum group D (XPD) silencing on the growth of hepatoma cells and assessed the mechanisms.
Matreial/Methods
XPD gene was silenced by siRNA in hepatoma cells. The experiments were randomly divided into a control group, a liposome control group, a negative control (NC) group, an XPD siRNA group, and an XPD siRNA + P53 inhibitor group. 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) was used to detect cell viability 24 h after gene silencing and treatments. Terminal deoxynucleotidyl transferases (TdT)-mediated dUTP nick-end labeling (TUNEL) and flow cytometry were used to detect apoptosis. Invasive ability was detected by Transwell assay. Additionally, the expression of mouse double-minute 2 homolog (Mdm2), mouse double-minute 4 homolog (Mdm4), CyclinD1, P21, Bax, P53, C-sis, and Bcl-2 was detected by real-time polymerase chain reaction and Western blotting.
Results
Compared with the NC group, XPD siRNA significantly reduced XPD expression at both mRNA and protein levels. XPD siRNA significantly promoted cell proliferation, reduced apoptosis, and promoted cell invasive ability. Expression of CyclinD1, Bcl-2, and C-sis increased significantly after XPD silencing, while the expression of P21, Mdm2, Mdm4, Bax, and P53 significantly decreased (vs. NC, P<0.05). Importantly, P53 inhibitor (1 μM bpV) further enhanced the effect of XPD silencing (vs. XPD silencing, P<0.05).
Conclusions
Our data revealed that XPD silencing promoted growth of hepatoma cells by reducing P53 expression.
MeSH Keywords: Carcinoma, Hepatocellular; Tumor Suppressor Protein p53; Xeroderma Pigmentosum Group D Protein
Background
Liver cancer is a malignant tumor that seriously affects human health and has a high mortality rate. Hepatocellular carcinoma (HCC) accounts for the vast majority of liver cancer cases [1,2]. With the development of diagnostic imaging technology, early diagnosis and monitoring of liver cancer have extensively improved [3]. Multidisciplinary treatment combined with radiotherapy and chemotherapy surgery also increases the survival rate of patients with liver cancer. However, the recurrence rate of liver cancer is still high [4].
Xeroderma pigmentosum group D (XPD) is a component of transcription factor II H (TFIIH) that plays important roles in the initiation of transcription and nucleotide excision repair (NER) in eukaryotes [5]. DNA repair capacity is a major obstacle related to tumor occurrence and development [6]. XPD is involved in the process of DNA repair after nucleic acid damage, and indirectly regulates the expression of oncogenes and tumor suppressor genes [7]. XPD can also activate the checkpoint of DNA damage, promote the repair of nucleic acids, and increase gene stability, as well as preventing gene mutation and formation and growth and metastasis of breast tumor cells [8]. However, the regulation of XPD in liver cancer and the underlying mechanisms are still unclear.
P53 gene is a well-known tumor suppressor gene that plays important regulatory roles in response to DNA damage [9]. When DNA was damaged for various reasons, P53 first induces the cells to enter the G1 phase and inhibits the proliferation of cells until DNA repair. When the DNA is not repaired, P53 activates the gene transcription that elicits apoptosis [10]. Therefore, P53 is critical in maintaining normal cell growth and inhibiting malignant proliferation. Its deletion and mutation are closely related to the occurrence and development of most tumors [11]. XPD can combine with P53 and acts on P53, thus participating in apoptosis [12]. It is unclear whether XPD is an oncogene of hepatoma cells and the downstream mechanisms require investigation. In this study, XPD was downregulated by XPD siRNA and the biological activities of hepatoma cells were evaluated thereafter. This study may provide an experimental basis for gene therapy of liver cancer.
Material and Methods
Cell culture
Hepatoma cell line (HepG2) was purchased from Shanghai Cell Bank of the Chinese Academy of Science (China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) in 5% CO2 at 37°C.
The experiments were divided into a control group, a liposome control group, a negative control (NC) group, an XPD siRNA group, and an XPD siRNA+P53 inhibitor [1 μM, bpV (phen)] (EMD Chemicals, Inc., Gibbstown, NJ, USA) group. The liposome was used in accordance with the instruction of Lipofectamine 3000 reagent. siRNA-liposome complexes were prepared at low temperature environment at the dark at room temperature after 1-h incubation of siRNA with liposome complexes.
The sequence of XPD siRNA was designed by Shanghai SANGON according to the gene sequence of XPD: Sense, GGAAGACAGUAUCCCUGUUTT (5′-3′); Antisense, AACAGGG AUACUGUCUUCCTT (5′-3′). At 24 h after silencing, XPD expression at mRNA and protein levels was confirmed by real-time PCR and Western blotting.
MTT assay
Hepatoma cells were seeded in 96-well plates and treated with XPD siRNA for 24 h. Thereafter, 10 μL 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was added into each well. After an additional 4-h incubation in CO2 incubator at 37°C, the absorbance was detected by a microplate reader (Thermo Fisher Scientific, Inc.) at 570 nm. Cell viability was calculated according to the optical density (OD) values.
Flow cytometry
Hepatoma cells were seeded in 6-well plates and treated with XPD siRNA for 24 h. The cells were collected after digestion by trypsin (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cells were incubated with Annexin V-FITC and PI (C1062, Beyotime, Ningbo, China) for 30 min in the dark, after which apoptosis was detected by flow cytometry (NovoCyte 2060R, ACEABIO, China) and data were analyzed by FlowJo 10.
Transwell assay
Hepatoma cells were seeded in 6-well plates and treated with XPD siRNA for 24 h. The cells were digested after 24-h treatment and collected. After that, the cells were seeded into the upper Transwell chamber with 1-day starvation of serum. The lower chamber contained DMEM medium with 10% FBS. The cells were cultured in a CO2 incubator for 24 h and the cells in the lower chamber were taken out and fixed with polyformaldehyde for 20 min and stained with crystal violet (0.1%) for 5 min. Light microscopy was used to count cells in at least 5 fields and the cell numbers represented the capacity of cell invasion.
Quantitative real-time polymerase chain reaction (QRT-PCR)
Hepatoma cells were seeded in 6-well plates and treated with XPD siRNA for 24 h. mRNA in different groups was extracted using an Ultrapure RNA Extraction Kit (CW0581M, CWBIO, China). Subsequently, mRNA was transcribed into cDNA according to the instruction of the HiFiScript cDNA Synthetic Kit (CW2569M, CWBIO, China). UltraSYBR Mixture (CW0957M, CWBIO, China) was used to detect the expression level of the targeted genes using cDNA as template. The expression of XPD, P21, CyclinD1, Mdm2, Mdm4, Bax, P53, C-sis, and Bcl-2 was normalized to GAPDH. The primers were:
XPD-F: CCCGCTGGCATCTACAA
XPD-R: TCCTGAGCACCGTCTTCTG
P21-F: AGCGACCTTCCTCATCCACC
P21-R: AAGACAACTACTCCCAGCCCCATA
CyclinD1-F CCCTCGGTGTCCTACTTCAAATGT
Cyclin-D1-R GGAAGCGGTCCAGGTAGTTCAT
Mdm2-F AATCAGCAGGAATCATCGG
Mdm2-R GTGGCGTTTTCTTTGTCGT
Mdm4-F CATTTCGGCTCCTGTCGTT
Mdm4-R CCGTCTCGTGGTCTTTTCTC
Bax-F AGACACTCGCTCAGCTTCTTG
Bax-R CTTTTGCTTCAGGGTTTCATC
P53-F AGTGCTCGCTTAGTGCTCCCT
P53-R GTGCATGTTTGTGCCTGTCCT
C-sis-F ATGGAGTTTGCTGTTGAGGTGG
C-sis-R GCAGGGTGGAGGTAGAGAGATG
Bcl-2-F GTGCCTGCTTTTAGGAGACCGA
Bcl-2-R GAGACCACACTGCCCTGTTGATC
GAPDH-F GAAGGTCGGAGTCAACGGAT
GAPDH-R CCTGGAAGATGGTGATGGG
Western blotting
Hepatoma cells were seeded in 6-well plates and treated with XPD siRNA for 24 h. Protein was extracted from the treated cells. The mouse monoclonal antibody against GAPDH (1: 10000, TA-08, Zsbio, China), XPD (1: 5000, ab167418, Abcam), rabbit polyclonal antibody against Mdm2 (1: 1000, abs130145, Absin Bioscience), Mdm4 (1: 1000, abs137018, Absin Bioscience), P53 (1: 1000, #2524, Cell Signaling), rabbit monoclonal antibody against P21 (1: 1000, ab109199, Abcam), C-sis (1: 1000, ab178409, Abcam), Cyclin D1 (1: 1000, ab134175, Abcam), Bcl-2 (1: 1000, A11025, ABclonal), and Bax (1: 1000, ab32503, Abcam) were incubated overnight at 4°C. After washing, the membranes were incubated with goat anti-mouse IgG (ZB-2305, Zsbio, China) or goat anti-rabbit IgG (ZB-2301, Zsbio, China) for 2 h at room temperature. The ECL kit (cat. no. RPN2133; GE Healthcare Life Sciences) was used and the membrane visualized by Chemi DocTM XRS+ (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Data were analyzed using SPSS 17.0 and results are presented as means and standard deviations (SD). Statistical significance was calculated by one-way analysis of variance (ANOVA) with Newman-Keuls as the post hoc test. P values less than 0.05 were considered significant.
Results
XPD siRNA decreased XPD expression in hepatoma cells
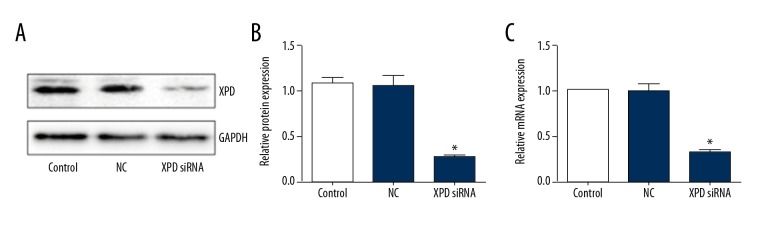
XPD expression was detected by real-time PCR and Western blotting. As shown in Figure 1, XPD siRNA significantly reduced XPD expression in hepatoma cells compared with the control group (P<0.05), but NC did not influence XPD expression.
Figure 1.
XPD siRNA reduced XPD expression in hepatoma cells. (A) Representative blots; (B) Quantification data the protein expression; (C) Relative mRNA expression of XPD after siRNA treatment. Compared with control, * P<0.05.
XPD siRNA promoted cell viability of hepatoma cells
After XPD silencing, cell viability was detected. As shown in Figure 2, XPD siRNA significantly promoted cell viability compared with NC (P<0.05). Moreover, p53 inhibitor further increased the cell viability (vs. XPD siRNA, P<0.05).
Figure 2.
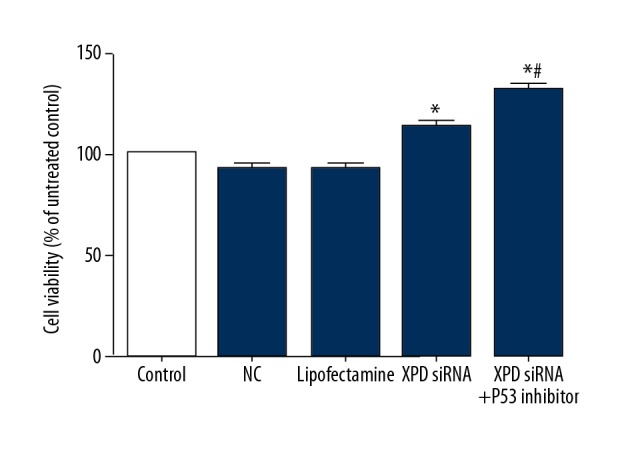
XPD siRNA promoted cell viability of hepatoma cells. Compared with NC, * P<0.05; Compared with XPD siRNA, # P<0.05.
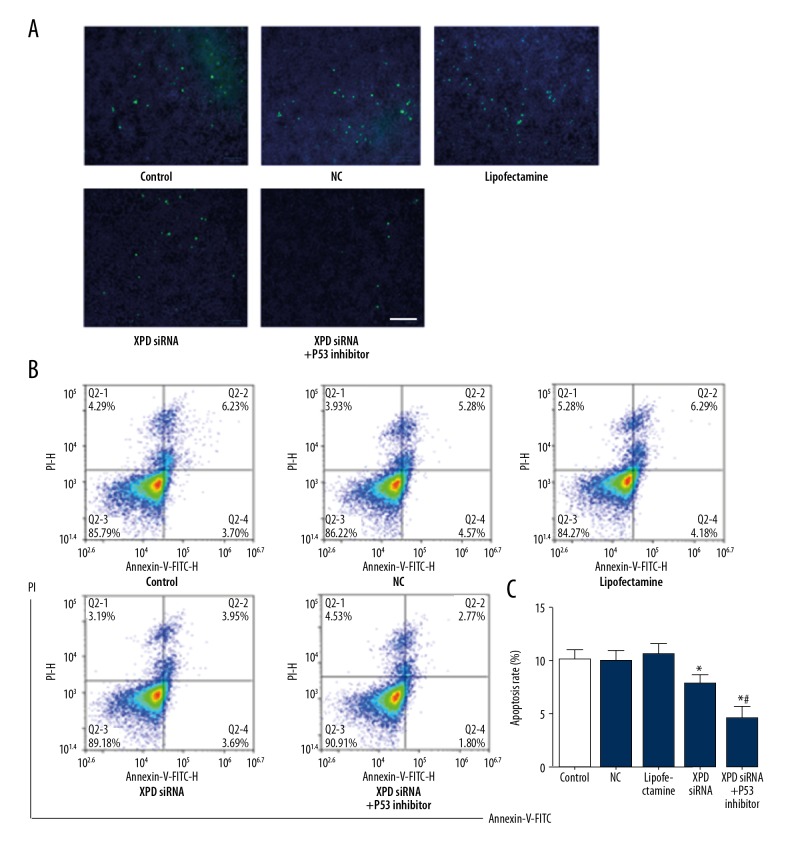
XPD siRNA reduced apoptosis of hepatoma cells
After XPD silencing, apoptosis was detected by flow cytometry and TUNEL assay. As shown in Figure 3, XPD siRNA remarkably reduced apoptosis compared with NC (P<0.05). Moreover, P53 inhibitor further reduced apoptosis with the treatment of XPD siRNA (vs. XPD siRNA, P<0.05).
Figure 3.
XPD siRNA reduced apoptosis of hepatoma cells. (A) Representative images of TUNEL assay. Scale bar: 100 μm; (B) Representative images of flow cytometry; (C) Quantification data of apoptosis. Compared with NC, * P<0.05; Compared with XPD siRNA, # P<0.05.
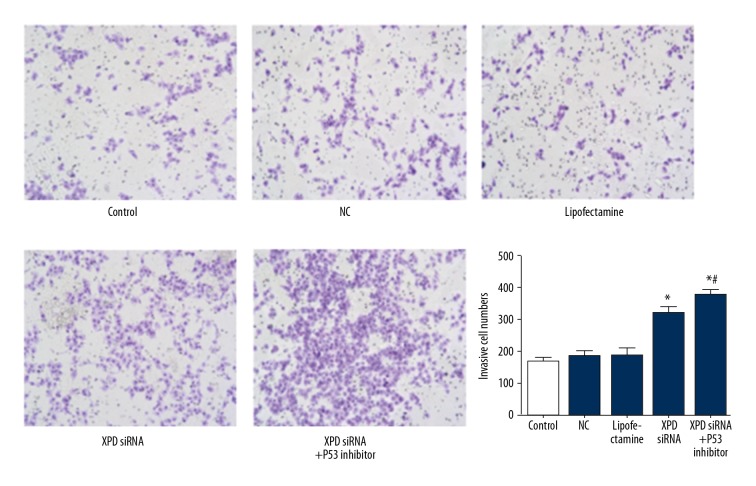
XPD siRNA promoted cell invasion of hepatoma cells
After XPD silencing, the cell invasive ability was detected. As shown in Figure 4, XPD siRNA significantly promoted cell invasion compared with NC. Moreover, p53 inhibitor further increased cell invasion with the treatment of XPD siRNA.
Figure 4.
XPD siRNA promotes cell invasion of hepatoma cells. Magnification: ×200. Compared with NC, * P<0.05; Compared with XPD siRNA, # P<0.05.
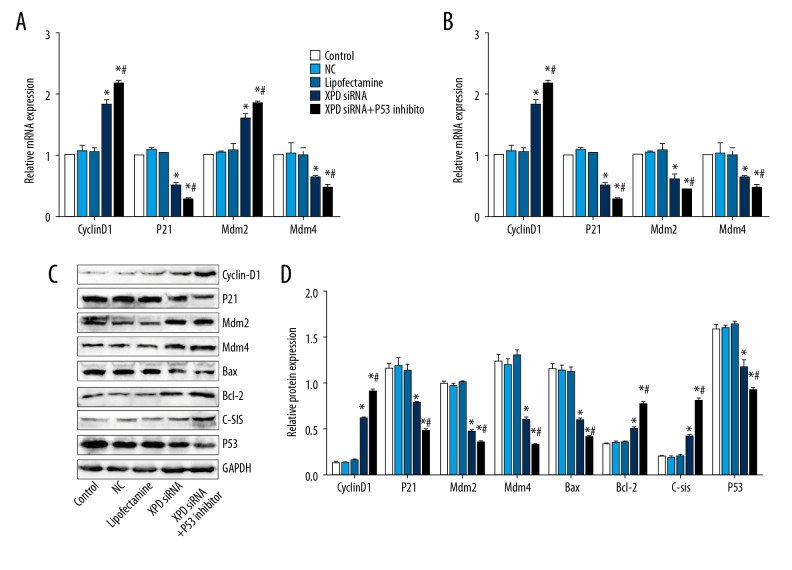
XPD siRNA promotes CyclinD1, Bcl-2, C-sis, reduces P21, Mdm2, Mdm4, Bax, and P53 expression
After XPD silencing, the expression of cell proliferation and apoptosis-related genes were detected. As shown in Figure 5, CyclinD1, Bcl-2, and C-sis were promoted after XPD siRNA treatment, while P21, Mdm2, Mdm4, Bax, and P53 expression was reduced. Interestingly, P53 inhibitor further promoted CyclinD1, Bcl-2, and C-sis expression, and reduced P21, Mdm2, Mdm4, Bax, and P53 expression.
Figure 5.
XPD siRNA promotes Cyclin D1, Bcl-2, C-sis and reduces P21, Mdm2, Mdm4, Bax, and P53 expression. (A) Relative expression of CyclinD1, P21, Mdm2, and Mdm4 at mRNA level; (B) Relative expression of Bax, Bcl-2, C-sis, and P53; (C) Representative blots of the related proteins; (D) Quantification data of protein expression. Compared with NC, * P<0.05; Compared with XPD siRNA, # P<0.05.
Discussion
Liver carcinoma is a serious malignant tumor. HCCs are the most common type of liver cancer, and the morbidity and mortality have been gradually increasing [13]. With the development of chemotherapy, surgical resection, and liver transplantation, application of molecular-targeted drug sorafenib in the treatment of liver cancer, the survival rate of HCCs has improved [14]. However, recurrence and metastasis after surgery still severely affect patient quality of life [13]. Gene therapy is a new approach in the treatment of liver cancer [15,16]. XPD plays an important role in repairing damaged DNA. In a previous study, we constructed an XPD overexpression vector to promote XPD expression in hepatoma cells, and the results showed that overexpression of XPD promoted apoptosis and inhibited the growth of hepatoma cells [17]. In the present study we further clarified the effect of XPD in hepatoma cells by silencing XPD.
In this study, real-time PCR and Western blotting were used to verify the effect of XPD silencing, and the expression of XPD was obviously reduced after transfection of XPD siRNA. XPD siRNA achieved effective silencing effects and was applied in the following experiments. P53 can induce apoptosis by directly combining with XPD [12]. Our previous study showed that overexpression of XPD combined with P53 inhibitor inhibited cell apoptosis, increased cell viability, and enhanced cell invasion ability [17]. The study further revealed that apoptosis was significantly decreased after silencing XPD and/or P53 blockage, while proliferation and invasive ability were promoted. These results revealed a synergistic action of XPD silence and P53 blockage in hepatoma cells.
In the present study, the expression of P53 was downregulated after the silencing of XPD, which was consistent with the previous results [17]. The main function of CyclinD1 is to promote cell proliferation through activating cyclin-dependent kinase 4 (CDK4) [18]. The cyclinD1-CDK4 complex facilitates passage through the G1 phase by inhibiting retinoblastoma protein (pRb) expression [19]. CyclinD1-CDK4 inhibits pRb through phosphorylation, allowing E2F transcription factors to transcribe genes required for entry into the S phase. The results of our study also demonstrated that CyclinD1 expression increased after silencing XPD and blocking P53. Therefore, XPD silencing and P53 inhibition can promote cell proliferation by increasing the expression of CyclinD1.
P21, a cell cycle regulator, is one of the most important downstream regulators of the P53 [20]. The reduction of P21 can lead to loss of cell proliferation and increase tumorigenesis [21]. Previous studies have revealed that XPD can increase the expression of P21; concurrently, the growth of hepatoma cells is inhibited and apoptosis is promoted [17]. The present study demonstrated that down-regulation of XPD also reduced the expression of P21; growth of hepatoma cells was promoted and apoptosis was inhibited.
Bax and Bcl-2 belong to the Bcl-2 gene family. The former is a tumor suppressor gene and the latter is an oncogene [22]. The ratio of Bax to Bcl-2 regulates the balance of cell proliferation and apoptosis [23]. P53 can directly downregulate the expression of Bcl-2 and activate the expression of Bax [24]. The present study found that silencing XPD downregulated Bax expression and upregulated Bcl-2 expression, which also proves that XPD has antitumor effects.
Mdm2 is widely expressed in mammals, indicating that it plays a role in basic cellular physiological processes. According to the analysis of Mdm2 oncogene products, it also suggests that Mdm2 plays a role in controlling cell growth [25]. Mdm4 is a key regulator of P53, which plays the role in apoptosis by regulating the transcriptional function of P53 or the mitochondrial pathway [26]. Mdm4 promotes apoptosis depending on the mitochondrial pathway of P53 [27]. We demonstrated that the expression of Mdm2 and Mdm4 was downregulated by the expression of XPD and the blocking of P53. C-sis is an oncogene [28], and the expression of C-sis was upregulated after XPD silencing. It is further demonstrated that XPD has a tumor suppression effect.
Conclusions
In conclusion, our data reveal that XPD silencing promotes growth of hepatoma cells through P53-mediated expression of oncogenes and downregulating the tumor suppression genes.
Footnotes
Source of support: This project was funded by the National Natural Science Foundation of China (No. 81300348)
References
- 1.Wang J, Shen JL. Spectral CT in evaluating the therapeutic effect of transarterial chemoembolization for hepatocellular carcinoma: A retrospective study. Medicine. 2017;96:e9236. doi: 10.1097/MD.0000000000009236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han H, Deng H, Han T, et al. Association between hepatocellular carcinoma and type 2 diabetes mellitus in Chinese hepatitis B virus cirrhosis patients: A case-control study. Med Sci Monit. 2017;23:3324–34. doi: 10.12659/MSM.902440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He X, Wu J, Holtorf AP, et al. Health economic assessment of Gd-EOB-DTPA MRI versus ECCM-MRI and multi-detector CT for diagnosis of hepatocellular carcinoma in China. PLoS One. 2018;13:e0191095. doi: 10.1371/journal.pone.0191095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Y, He J, Janssen HLA, et al. Peroxiredoxin 1, restraining cell migration and invasion, is involved in hepatocellular carcinoma recurrence. J Dig Dis. 2018;19:155–69. doi: 10.1111/1751-2980.12580. [DOI] [PubMed] [Google Scholar]
- 5.Vashisht AA, Yu CC, Sharma T, et al. The association of the Xeroderma Pigmentosum Group D DNA Helicase (XPD) with transcription factor IIH is regulated by the cytosolic iron-sulfur cluster assembly pathway. J Biol Chem. 2015;290:14218–25. doi: 10.1074/jbc.M115.650762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torgovnick A, Schumacher B. DNA repair mechanisms in cancer development and therapy. Front Genet. 2015;6:157. doi: 10.3389/fgene.2015.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann AR. XPD structure reveals its secrets. DNA Repair. 2008;7:1912–15. doi: 10.1016/j.dnarep.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Zhang Z, Yan W. Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta. 2005;359:150–55. doi: 10.1016/j.cccn.2005.03.047. [DOI] [PubMed] [Google Scholar]
- 9.Graur F, Furcea L, Mois E, et al. Analysis of p53 protein expression in hepatocellular carcinoma. J Gastrointestin Liver Dis. 2016;25(3):345–49. doi: 10.15403/jgld.2014.1121.253.p53. [DOI] [PubMed] [Google Scholar]
- 10.Gu MJ, Choi JH. Correlation between p53 and Rb protein expression and clinicopathologic features in hepatocellular carcinoma. Cancer Res Treat. 2003;35:514–20. doi: 10.4143/crt.2003.35.6.514. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein I, Marcel V, Olivier M, et al. Understanding wild-type and mutant p53 activities in human cancer: New landmarks on the way to targeted therapies. Cancer Gene Ther. 2011;18:2–11. doi: 10.1038/cgt.2010.63. [DOI] [PubMed] [Google Scholar]
- 12.Wang HY, Xiong GF, Zhang JX, et al. The role of XPD in cell apoptosis and viability and its relationship with p53 and cdk2 in hepatoma cells. Med Oncol. 2012;29:161–67. doi: 10.1007/s12032-011-9818-y. [DOI] [PubMed] [Google Scholar]
- 13.Kimhofer T, Fye H, Taylor-Robinson S, et al. Proteomic and metabonomic biomarkers for hepatocellular carcinoma: A comprehensive review. Br J Cancer. 2015;112:1141–56. doi: 10.1038/bjc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng Z, Chen S, Wei M, et al. Advanced recurrent hepatocellular carcinoma: Treatment with sorafenib alone or in combination with transarterial chemoembolization and radiofrequency ablation. Radiology. 2018;287:705–14. doi: 10.1148/radiol.2018171541. [DOI] [PubMed] [Google Scholar]
- 15.Wang ZY, Yang J, Liu CK, Shen SQ. High expression of Retinoblastoma-Binding Protein 2 (RBP2) in patients with hepatocellular carcinoma and its prognostic significance. Med Sci Monit. 2017;23:2736–44. doi: 10.12659/MSM.905262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Zhang M, Peng Y, He J. Ubiquitin Associated Protein 2-Like (UBAP2L) overexpression in patients with hepatocellular carcinoma and its clinical significance. Med Sci Monit. 2017;23:4779–88. doi: 10.12659/MSM.907071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding H, Xu JJ, Huang Y, et al. XPD could suppress growth of HepG2.2.15 and down-regulate the expression of hepatitis B virus x protein through P53 pathway. Biochem Biophys Res Commun. 2012;419:761–67. doi: 10.1016/j.bbrc.2012.02.097. [DOI] [PubMed] [Google Scholar]
- 18.Gillett CE, Lee AH, Millis RR, Barnes DM. Cyclin D1 and associated proteins in mammary ductal carcinoma in situ and atypical ductal hyperplasia. J Pathol. 1998;184:396–400. doi: 10.1002/(SICI)1096-9896(199804)184:4<396::AID-PATH1259>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 19.Wilson CS, Butch AW, Lai R, et al. Cyclin D1 and E2F-1 immunoreactivity in bone marrow biopsy specimens of multiple myeloma: relationship to proliferative activity, cytogenetic abnormalities and DNA ploidy. Br J Haematol. 2001;112:776–82. doi: 10.1046/j.1365-2141.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, Zhang L. The transcriptional targets of p53 in apoptosis control. Biochem Biophys Res Commun. 2005;331:851–58. doi: 10.1016/j.bbrc.2005.03.189. [DOI] [PubMed] [Google Scholar]
- 21.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 22.Zhu L, Han MB, Gao Y, et al. Curcumin triggers apoptosis via upregulation of Bax/Bcl-2 ratio and caspase activation in SW872 human adipocytes. Mol Med Rep. 2015;12:1151–56. doi: 10.3892/mmr.2015.3450. [DOI] [PubMed] [Google Scholar]
- 23.Zhao B, He T. Chidamide, a histone deacetylase inhibitor, functions as a tumor inhibitor by modulating the ratio of Bax/Bcl-2 and P21 in pancreatic cancer. Oncol Rep. 2015;33:304–10. doi: 10.3892/or.2014.3595. [DOI] [PubMed] [Google Scholar]
- 24.Aboutaleb N, Shamsaei N, Khaksari M, et al. Pre-ischemic exercise reduces apoptosis in hippocampal CA3 cells after cerebral ischemia by modulation of the Bax/Bcl-2 proteins ratio and prevention of caspase-3 activation. J Physiol Sci. 2015;65:435–43. doi: 10.1007/s12576-015-0382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffill CR, Lee AP, Siau JW, et al. The p53-Mdm2 interaction and the E3 ligase activity of Mdm2/Mdm4 are conserved from lampreys to humans. Genes Dev. 2016;30:281–92. doi: 10.1101/gad.274118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi DL, Cobrinik D. MDM2 but not MDM4 promotes retinoblastoma cell proliferation through p53-independent regulation of MYCN translation. Oncogene. 2017;36:1760–69. doi: 10.1038/onc.2016.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancini F, Pieroni L, Monteleone V, et al. MDM4/HIPK2/p53 cytoplasmic assembly uncovers coordinated repression of molecules with anti-apoptotic activity during early DNA damage response. Oncogene. 2016;35:228–40. doi: 10.1038/onc.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maass T, Thieringer FR, Mann A, et al. Liver specific overexpression of platelet-derived growth factor-B accelerates liver cancer development in chemically induced liver carcinogenesis. Int J Cancer. 2011;128:1259–68. doi: 10.1002/ijc.25469. [DOI] [PubMed] [Google Scholar]