Abstract
Backgrounds
Recent studies have shown that some members of the tripartite motif-containing protein (TRIM) family function as important regulators in several tumors. However, the clinical significance of TRIM15 in gastric adenocarcinoma has not been elucidated. In the present study, we aimed to examine the expression pattern of TRIM15 and explore whether the TRIM15 expression is correlated with clinicopathological characteristics of patients with gastric adenocarcinoma.
Material/Methods
The expression pattern of TRIM15 was examined in gastric adenocarcinoma tissues and adjacent normal stomach tissues by using immunohistochemistry staining. The prognostic role of TRIM15 in gastric cancer patients was evaluated by univariate and multivariate analyses. Clinical outcomes were assessed by the Kaplan-Meier analysis and log-rank test. The effects of TRIM15 on cancer cell proliferation and invasion were tested through cellular experiments.
Results
TRIM15 was highly expressed in normal stomach tissues compared to tumor tissues. TCGA database showed that higher TRIM15 RNA transcription indicates poorer overall survival of gastric cancer patients. Besides, low expression of TRIM15 was significantly associated with advanced tumor invasion depth and advanced TNM stage. Moreover, gastric cancer patients with lower KDM5B expression had poorer overall survival, and TRIM15 was identified as an independent prognosis factor according to multivariate analysis. Using the gastric cancer cell lines, we found that overexpression of TRIM15 can inhibits tumor cell invasion.
Conclusions
Our study demonstrated that low expression of TRIM15 in gastric adenocarcinoma tissues was significantly associated with poorer prognosis of patients, indicating the potential of TRIM15 as a novel clinical biomarker and therapeutic target.
MeSH Keywords: Neoplasm Invasiveness, Prognosis, Stomach Neoplasms
Background
Gastric adenocarcinoma is one of the most frequent malignancy that occurs in stomach and contributes to most of cancer-associated mortality [1,2]. Current treatment therapies toward gastric adenocarcinoma include surgical resection, chemotherapy, radiotherapy, and immune therapy [3,4]. Despite previous improvements in therapies, patients with unresectable gastric cancer have very unfavorable prognosis, particularly those with advanced disease [5,6]. Therefore, it is in urgent need to explore the progression mechanisms of gastric adenocarcinoma and identify novel diagnostic biomarkers for more effective treatment strategies to improve prognosis.
The tripartite motif containing (TRIM) protein family contains 3 similar conserved domains in the N-terminal region: a RING-finger domain, zinc-binding motifs, and an associated coiled-coil region [7]. There are now more than 60 known TRIM proteins in humans and they are involved in a broad range of biological processes, and their alterations are correlated with several pathological disease such as developmental disorders, viral infections, and cancer [8,9]. Most of the TRIM family members play critical roles in innate immunity by regulating immune signaling pathways, and several TRIM proteins are involved in diverse oncogenic processes, such as transcriptional regulation, cell differentiation, and apoptosis [10]. For example, TRIM59 was found to promote the proliferation and metastasis of colorectal carcinoma through the PI3K/AKT pathway [11]. Knockdown of TRIM65 can inhibit lung cancer cell migration and invasion [12]. In contrast, TRIM19 was found to function as an anti-tumor protein in acute promyelocytic leukemia [13]. TRIM24 can also function as a potent tumor suppressor protein in liver cancer [14].
TRIM15, also called as ZNFB7 or RNF93, was found to be involved in numerous cellular processes [8]. TRIM15 has been recently identified as a tumor suppress gene of colon cancer, because expression level of TRIM15 was downregulated in tumor tissues of colon cancer patients and TRIM15 protein level was closely related to the migration ability of colon cancer cell [15]. In addition, TRIM15 was reported to function as a restriction factor especially in the human immunodeficiency virus (HIV) 1 through inhibiting the process of viral releasee [16]. However, little is known about the clinical roles and potential functions of TRIM15 in gastric adenocarcinoma.
In the current study, we first examined the expression levels of TRIM15 in gastric adenocarcinoma tissues together with adjacent normal stomach tissues. Furthermore, we found the positive correlation between low TRIM15 expression and poorer clinical outcomes by statistical analyses, thus we identified TRIM15 as an independent prognostic factor for overall survival time of patients with gastric adenocarcinoma. Finally, we conducted cellular experiments to explore the potential function of TRIM15 in gastric cancer cell lines, which revealed that overexpression of TRIM15 can suppress the invasion capacity of tumor cells.
Material and Methods
Patients and samples
This study was approved by the Ethic Committee of Zhangzhou Affiliated Hospital of Fujian Medical University and written informed consents were obtained from all participants. There were 134 formalin-fixed paraffin-embedded (FFPE) gastric adenocarcinoma tissues and matched adjacent normal stomach tissues randomly selected from patients who underwent surgical resection during June 2010 to September 2015 in Zhangzhou Affiliated Hospital of Fujian Medical University. Only patients who survived more than 12 months after surgery were enrolled and the median age was 56 years old. The overall survival time for all patients was 53 months and the 5-year survival rate were 58.0%. All enrolled patients were followed up until death or the end of our study. Until the end of follow-up, a total 52 patients had succumbed to gastric adenocarcinoma. All the specimens used in the present study were confirmed based on pathology examination. The invasion depth (T stage) was defined according to the American Joint Committee on Cancer TNM tumor staging system. Briefly, T1 is defined as tumor invades lamina propria, muscularis mucosae, or submucosa; T2 means tumor invades muscularis propria; T3 means tumor penetrates subserosal connective tissue without invasion of visceral peritoneum or adjacent structures; T4 means tumor invades serosa (visceral peritoneum) or adjacent structures.
Immunohistochemistry (IHC) staining
Immunohistochemistry (IHC) staining for TRIM15 was carried out by using the standard protocol as described by others. Briefly, 6-μm serial sections were dried at 70°C and then deparaffinized with xylene and rehydrated in alcohols. The microwave antigen retrieval was carried out in pH 6.0 citrate buffer. Subsequently, specimens were incubated with the rabbit anti-human TRIM antibody (1: 200 dilution; Cat. No. SAB1408159; Sigma-Aldrich, St. Louis, MO, USA) overnight. On the next day, the slide sections were washed, and their immunoreactivities were visualized by using poly HRP IgG and 3,3′-diaminobenzidine substrates [17]. Primary antibody was replaced with phosphate-buffered saline as the negative control.
IHC scoring
The stained slides were examined and scored by 2 independent investigators at 400× magnification, and 10 fields of each section were randomly selected. Staining intensity was divided into 4 grades as follows: 1 (negative); 2 (weak); 3 (moderate); 4 (strong). The staining percentage was scored as follows: 1 (<25%); 2 (25% to 50%); 3 (51% to 75%); 4 (>75%). The IHC score was finally determined by multiplying the intensity score by the staining percentage score. In this study, the expression of TRIM15 was defined as low expression when the score was <8, and high expression when the score was ≥8.
Cell culture and transfection
The gastric cancer cell lines AGS and MKN-1 were purchased from the American Type Culture Collection (ATCC, Rockville, IN, USA). The single silence TRIM15 siRNA (siRNA-TRIM15: 5′-CCCAAUCCUCGGGCAAGAU-3′) was obtained from ABM (Richmond, BC, Canada). The overexpression plasmid of TRIM15 was synthesized by Qiagen (Valencia, CA, USA) and verified by DNA sequencing (Genewiz, Suzhou, China). AGS and MKN-1 cells were transfected with TRIM15 plasmid or siRNA by using FuGENE HD transfection reagent according to the manufacturer’s instructions [18].
MTT assay
Cell proliferation was examined by using the MTT assay. Briefly, 6×103 cells were added to 96-well plates and cultured for different time points. The MTT solution was added to each well and incubated for 3 hours at 37°C followed by measuring OD570 nm absorbance using an automated plate reader [19]. All the experiments were repeated for 3 times.
Cell invasion assay
The invasion was measured by the Matrigel-coated Transwell assay. Briefly, 5 × 104 transfected cells were added to the upper chamber and cultured for 48 hours. Invaded cells were fixed and stained. Cell counting was carried out in 6 random fields of view. All the experiments were repeated for 3 times [20].
Statistical analysis
Statistical analyses were conducted by using the SPSS version 20.0 (IBM, New York, NY, USA). The association between TRIM15 expression and clinicopathologic features were analyzed by chi-square test. The overall survival curves of gastric adenocarcinoma patients were plotted using the Kaplan-Meier method. Statistical validation of independent prognostic factors was tested with multivariate analysis. P<0.05 was considered statistically significant [21].
Results
Higher TRIM15 expression in normal stomach tissues than in tumor tissues
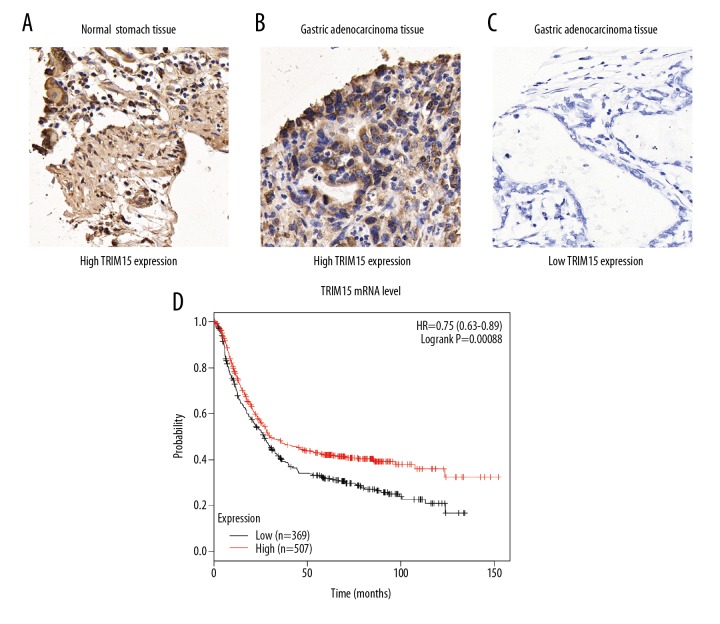
To investigate the role of TRIM15 in gastric adenocarcinoma, we first examined its expression patterns in tumor tissues together with adjacent normal stomach tissues by IHC staining and we identified the cytoplasm localization of TRIM15 protein (Figure 1A–1C). Therefore, IHC data revealed that HOXD4 was lowly expressed in normal gastric tissues, but significantly elevated in gastric adenocarcinoma tissues.
Figure 1.
Analyses of TRIM15 expression pattern in patients with gastric adenocarcinoma. (A–C) Immunohistochemistry staining of TRIM15 in adjacent normal stomach tissues (A) and gastric adenocarcinoma tissues (B, C). (D) TCGA database showed that lower TRIM15 RNA transcription indicates poor overall survival of gastric cancer patients. Magnification, 400×. *, P=0.00088 by log-rank t-test.
Furthermore, TCGA database showed higher TRIM15 RNA transcription indicates poor overall survival of gastric cancer patients (Figure 1D, P=0.00088). Taken together, these results suggested that the TRIM15 expression level was downregulated in tumor tissues and may be involved in gastric adenocarcinoma development.
Lower TRIM15 protein level indicates more aggressive phenotypes of gastric adenocarcinoma patients
To better investigate the predictive role of TRIM15 in clinical application, 71 patients were grouped into low TRIM15 expression group (IHC score <8), and the other 63 patients were classified into high TRIM15 expression (IHC score ≥8) group. Subsequently, we tested the correlations of TRIM15 expression with clinicopathological features in gastric adenocarcinoma patients (Table 1). We found that the lower level of TRIM15 expression was significantly correlated with advanced invasion depth (P=0.002) and advanced TNM stage (P<0.001). Besides, no significant correlation was identified between TRIM15 expression and patients’ age, gender, tumor localization, and tumor size (all P>0.05).
Table 1.
Correlations between TRIM15 expression with patients’ features.
| Variables | Cases | TRIM15 level | P value | |
|---|---|---|---|---|
| (n=134) | Low (n=71) | High (n=63) | ||
| Age | 0.844 | |||
| ≤50 yrs | 52 | 27 | 25 | |
| >50 yrs | 82 | 44 | 38 | |
| Gender | 0.112 | |||
| Female | 44 | 19 | 25 | |
| Male | 90 | 52 | 38 | |
| Localization | ||||
| Upper | 24 | 16 | 8 | 0.282 |
| Middle | 61 | 29 | 32 | |
| Lower | 49 | 26 | 23 | |
| Tumor size | 0.136 | |||
| ≤5.0 cm | 76 | 26 | 40 | |
| >5.0 cm | 58 | 35 | 23 | |
| Invasion depth | 0.002* | |||
| T1/T2 | 44 | 15 | 29 | |
| T3/T4 | 90 | 56 | 34 | |
| TNM stage | <0.001* | |||
| I/II | 51 | 14 | 37 | |
| III/IV | 83 | 57 | 26 | |
Low expression level of TRIM15 indicates poor prognosis of gastric adenocarcinoma patients
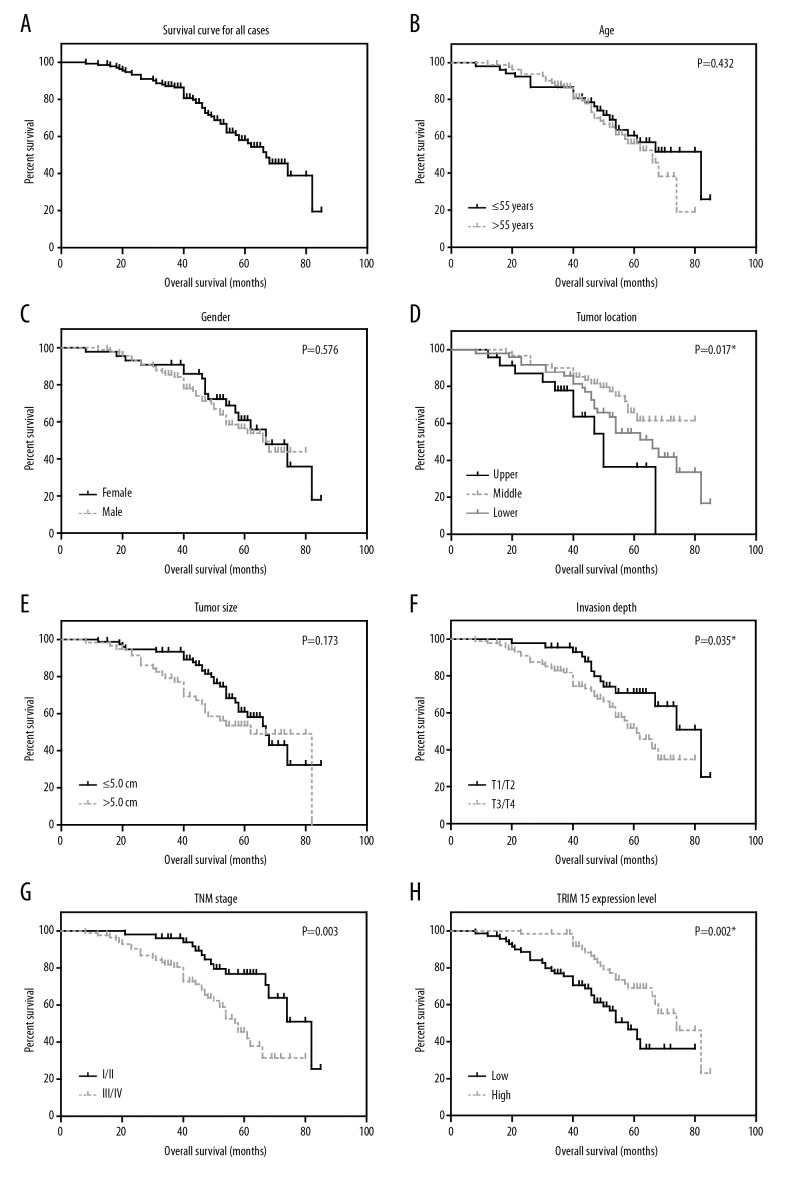
The role of tissue TRIM15 expression in overall survival of gastric adenocarcinoma patients was tested by Kaplan-Meier analysis and compared by the log-rank test. Gastric adenocarcinoma patients that expressed low protein levels of TRIM15 had a poorer mean overall survival time (55.5±3.1 months) compared to those with high expression levels of HOXD4 (69.0±2.5 months; P=0.002, Table 2). Well-known conventional prognostic factors were also found to be associated with the patients’ overall survival. For example, tumor localization (P=0.017), advanced tumor invasion depth (P=0.035) and advanced TNM stage (P=0.003, Figure 2, Table 2) can both indicate poorer clinical outcomes.
Table 2.
Kaplan-Meier overall survival analysis.
| Variables | Cases (n=134) | OS months (mean ±S.D.) | 5-year OS (%) | P value |
|---|---|---|---|---|
| Age | 0.432 | |||
| ≤50 yrs | 52 | 64.3±3.5 | 60.5% | |
| >50 yrs | 82 | 59.5±2.5 | 56.2% | |
| Gender | 0.576 | |||
| Female | 44 | 63.8±3.6 | 60.9% | |
| Male | 90 | 60.8±2.4 | 56.5% | |
| Localization | 0.017* | |||
| Upper | 24 | 48.5±4.3 | 36.4% | |
| Middle | 61 | 66.6±2.7 | 66.0% | |
| Lower | 49 | 60.9±3.3 | 54.8% | |
| Tumor size | 0.173 | |||
| ≤5.0 cm | 76 | 64.9±2.7 | 61.0% | |
| >5.0 cm | 58 | 59.6±3.4 | 53.6% | |
| Invasion depth | 0.035* | |||
| T1/T2 | 44 | 69.4±3.2 | 70.8% | |
| T3/T4 | 90 | 58.0±2.5 | 51.8% | |
| TNM stage | 0.003* | |||
| I/II | 51 | 70.2±2.9 | 75.2% | |
| III/IV | 83 | 56.3±2.6 | 46.1% | 0.002* |
| TRIM15 level | ||||
| Low | 71 | 55.5±3.1 | 46.6% | |
| High | 63 | 69.0±2.5 | 69.0% | |
Figure 2.
Analyses of the overall survival of gastric adenocarcinoma patients. The overall survival curves of gastric adenocarcinoma patients were plotted by Kaplan-Meier analysis and assessed by log-rank test, based on entire cohort (A), patients’ age (B), gender (C), tumor location (D), tumor size (E), invasion depth (F), TNM stage (G) and TRIM15 expression level (H), respectively. * P<0.05 by log-rank test.
In addition, we conducted multivariate analysis using a Cox hazard regression model to test their independent effects on patients’ overall survival (Table 3). All significant prognostic factors (P<0.05) identified by univariate analyses were subjected to the model, and we found that TRIM15 expression was an independent prognostic factor (hazard ratio=0.560, 95% confident interval=0.307–0.922, P=0.039). In addition, tumor location and advanced TNM stage were also independent factors of decreased overall survival.
Table 3.
Multivariate analysis.
| Variables | HR | 95% CI | P value |
|---|---|---|---|
| Location (middle/lower vs. upper) | 0.389 | 0.195–0.776 | 0.007* |
| Invasion depth (T3/T4 vs. T1/T2) | 1.125 | 0.477–2.651 | 0.788 |
| TNM stage (III/IV vs. I/II) | 2.952 | 1.409–4.707 | 0.026* |
| TRIM15 (high vs. low) | 0.560 | 0.307–0.922 | 0.039* |
Overexpression of TRIM15 inhibits the invasion ability of gastric cancer cell lines
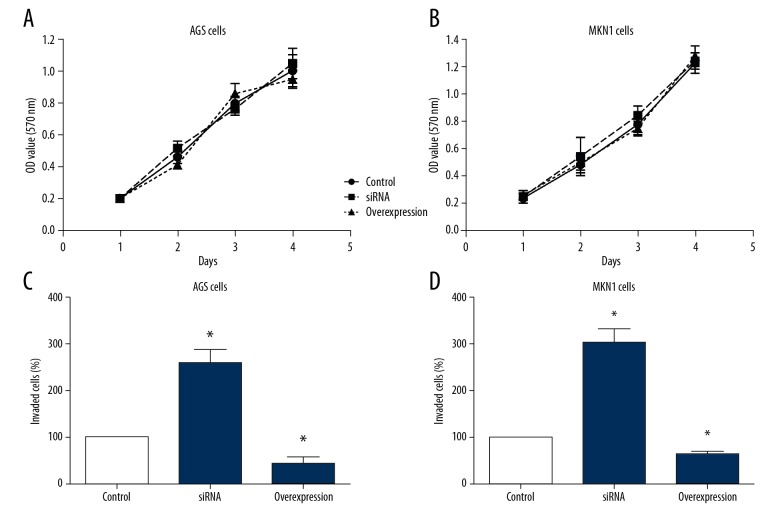
We conducted cellular experiments to test the effects of TRIM15 in gastric cancer. We overexpressed or knocked down the TRIM15 respectively in gastric cancer cell lines AGS and MKN1, then we tested the characteristics of different transfected cells to investigate the role of TRIM15 in tumor progression. The results demonstrated that TRIM15 overexpression or knockdown had no effect on the proliferation capacity in gastric cancer cells (Figure 3A, 3B). However, overexpression of TRIM15 inhibited cancer cell invasion compared to knockdown cells (Figure 3C, 3D). These data suggested that downregulated TRIM15 expression might contribute to the progression of human gastric adenocarcinoma by enhancing tumor cell invasion.
Figure 3.
TRIM15 inhibits the invasion capacity of gastric cancer cell lines. (A, B) Overexpression or knockdown of TRIM15 has on effect on the proliferation capacity of AGS and MKN-1 cells. (C, D) Overexpression of TRIM15 inhibited the invasion capacity of AGS and MKN-1 cells, whereas knockdown of TRIM15 promoted cancer cell invasion.
Discussion
Ubiquitylation belongs to one of the most important post-translational modification, which can regulate a broad range of cellular physiology processes in the organism [22]. The ubiquitin-dependent proteolytic pathway plays critical roles especially in the degradation of numerous functional proteins, such as those correlated with DNA repairing, transcriptional regulation, and cellular signaling [23]. Approximately all oncoproteins and anti-tumor genes are mediated by different post-translational modifications, including the ubiquitin-dependent proteolytic pathway [24]. TRIM family proteins have been defined as a subfamily of the RING type E3 ubiquitin ligase family and include more than 60 members in humans [25]. Those different TRIM family proteins play multiple roles in a broad range of physiological processes, including cell differentiation, organ development, and tumorigenesis [26]. It has been acknowledged that several TRIM family proteins can positively or negatively regulate cancer initiation and progression through affecting some important pathways such as DNA repairing, cell proliferation, and apoptosis [27,28]. In gastric carcinoma, it has been reported that some TRIM members such as TRIM28 and TRIM31 have alterations in the proteins which were found to be correlated with poor clinical outcomes and unfavorable prognosis of patients [29,30]. As a member of the TRIM protein family, TRIM15 has also been implicant in several disease including cancer [31]. However, the clinical role of TRIM15 in the progression of gastric adenocarcinoma has not been elucidated up to now.
Here in the present study, we found that TRIM15 expression was significantly downregulated in gastric adenocarcinoma tissues compared to that in adjacent normal stomach tissues by IHC staining. Furthermore, we dissect the associations between TRIM15 expression level and clinical characteristics of enrolled patients. The lower expression of TRIM15 was closely related to advanced tumor invasion depth and advanced TNM stage in gastric adenocarcinoma patients. We identified TRIM15 expression as an independent prognosis factor for gastric adenocarcinoma patients by using multivariate analysis. Finally, we conducted cellular experiments to investigate the potential effect of TRIM15 on gastric cancer cells. Overexpression of KDM5B in AGS and MKN-1 cells suppressed the invasion ability of tumor cells, whereas knockdown of TRIM15 promoted tumor cell invasion. Taken together, these data indicated that downregulated TRIM15 expression was related to unfavorable clinicopathological outcomes of gastric adenocarcinoma patients, possibly by promoting cancer cell invasion.
Consistent with our findings, it was previously reported that TRIM15 can function as a tumor suppress gene in colon cancer [15]. Some other TRIM family members have shown different roles in gastric cancer. For example, TRIM29 was identified as an oncogene in gastric cancer and was regulated by miR-185 [32]. High expression of TRIM44 was found to contribute to malignant outcomes in gastric cancer patients [33]. Therefore, it seems that different TRIM proteins might function through different signaling pathways in gastric cancer, and more intensive studies focused on exploring the underlying mechanisms of TRIM15 in progression of gastric adenocarcinoma was needed in the future.
Conclusions
Our study demonstrated that the expression level of TRIM15 was downregulated in gastric adenocarcinoma tissues and closely related to the aggressive phenotypes of gastric adenocarcinoma. Furthermore, TRIM15 was identified as a potential independent prognostic factor by univariate and multivariate analyses. Our data thus provided initial evidence for TRIM15 serving as a novel prognostic biomarker in gastric adenocarcinoma and could be helpful for clinical prediction and personal treatment improvement.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Clemente-Gutierrez U, Sanchez-Morales G, Santes O, Medina-Franco H. Clinical usefulness of extending the proximal margin in total gastrectomies for gastric adenocarcinoma. Rev Gastroenterol Mex. :2018. doi: 10.1016/j.rgmx.2018.03.006. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Wang R, Chen J, et al. miR-340 inhibits proliferation and induces apoptosis in gastric cancer cell line SGC-7901, possibly via the AKT pathway. Med Sci Monit. 2017;23:71–77. doi: 10.12659/MSM.898449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De B, Rhome R, Jairam V, et al. Gastric adenocarcinoma in young adult patients: Patterns of care and survival in the United States. Gastric Cancer. :2018. doi: 10.1007/s10120-018-0826-x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Ma H, Tian Y, Yu X. Targeting smoothened sensitizes gastric cancer to chemotherapy in experimental models. Med Sci Monit. 2017;23:1493–500. doi: 10.12659/MSM.903012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saldana M, Montes de Oca G, Tirado-Sanchez A, et al. Acquired ichthyosis associated with gastric adenocarcinoma. Int J Dermatol. 2018;57(6):713–14. doi: 10.1111/ijd.13988. [DOI] [PubMed] [Google Scholar]
- 6.Zheng R, Deng Q, Liu Y, Zhao P. Curcumin inhibits gastric carcinoma cell growth and induces apoptosis by suppressing the Wnt/β-catenin signaling pathway. Med Sci Monit. 2017;23:163–71. doi: 10.12659/MSM.902711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Cao S, Sun Y, Li C. Gene expression profiling of the TRIM protein family reveals potential biomarkers for indicating tuberculosis status. Microb Pathog. 2018;114:385–92. doi: 10.1016/j.micpath.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Crawford LJ, Johnston CK, Irvine AE. TRIM proteins in blood cancers. J Cell Commun Signal. 2018;12(1):21–29. doi: 10.1007/s12079-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatakeyama S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Patel LR, Barton MC. TRIM-ing ligand dependence in castration-resistant prostate cancer. Cancer Cell. 2016;29(6):776–78. doi: 10.1016/j.ccell.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Zhan W, Han T, Zhang C, et al. TRIM59 promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS One. 2015;10(11):e0142596. doi: 10.1371/journal.pone.0142596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang XL, Shi WP, Shi HC, et al. Knockdown of TRIM65 inhibits lung cancer cell proliferation, migration and invasion: A therapeutic target in human lung cancer. Oncotarget. 2016;7(49):81527–40. doi: 10.18632/oncotarget.13131. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Masroori N, Merindol N, Berthoux L. The interferon-induced antiviral protein PML (TRIM19) promotes the restriction and transcriptional silencing of lentiviruses in a context-specific, isoform-specific fashion. Retrovirology. 2016;13:19. doi: 10.1186/s12977-016-0253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Zhao L, Shi K, et al. TRIM24 promotes hepatocellular carcinoma progression via AMPK signaling. Exp Cell Res. 2018;367(2):274–81. doi: 10.1016/j.yexcr.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Lee OH, Lee J, Lee KH, et al. Role of the focal adhesion protein TRIM15 in colon cancer development. Biochim Biophys Acta. 2015;1853(2):409–21. doi: 10.1016/j.bbamcr.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Uchil PD, Pawliczek T, Reynolds TD, et al. TRIM15 is a focal adhesion protein that regulates focal adhesion disassembly. J Cell Sci. 2014;127(Pt 18):3928–42. doi: 10.1242/jcs.143537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Xu Y, Zhang Q, et al. Correlations between TBL1XR1 and recurrence of colorectal cancer. Sci Rep. 2017;7:44275. doi: 10.1038/srep44275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Fan H, Zou Q, et al. TEAD 4 exerts pro-metastatic effects and is negatively regulated by miR6839-3p in lung adenocarcinoma progression. J Cell Mol Med. 2018;22(7):3560–71. doi: 10.1111/jcmm.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou S, Du P, Wang P, et al. Significance of MNK1 in prognostic prediction and chemotherapy development of epithelial ovarian cancer. Clin Transl Oncol. 2017;19(9):1107–16. doi: 10.1007/s12094-017-1646-x. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Liu Z, Li K, et al. TBL1XR1 predicts isolated tumor cells and micrometastasis in patients with TNM stage I/II colorectal cancer. J Gastroenterol Hepatol. 2017;32(9):1570–80. doi: 10.1111/jgh.13749. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Yang X, Li Z, et al. Sprouty2 correlates with favorable prognosis of gastric adenocarcinoma via suppressing FGFR2-induced ERK phosphorylation and cancer progression. Oncotarget. 2017;8(3):4888–900. doi: 10.18632/oncotarget.13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baranes-Bachar K, Levy-Barda A, Oehler J, et al. The Ubiquitin E3/E4 Ligase UBE4A adjusts protein ubiquitylation and accumulation at sites of DNA damage, facilitating double-strand break repair. Mol Cell. 2018;69(5):866–78e7. doi: 10.1016/j.molcel.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jbara M, Sun H, Kamnesky G, Brik A. Chemical chromatin ubiquitylation. Curr Opin Chem Biol. 2018;45:18–26. doi: 10.1016/j.cbpa.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol. 2018;19(1):59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 25.Kamanova J, Sun H, Lara-Tejero M, Galan JE. The salmonella effector protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS Pathog. 2016;12(4):e1005552. doi: 10.1371/journal.ppat.1005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cambiaghi V, Giuliani V, Lombardi S, et al. TRIM proteins in cancer. Adv Exp Med Biol. 2012;770:77–91. doi: 10.1007/978-1-4614-5398-7_6. [DOI] [PubMed] [Google Scholar]
- 27.Cammas F, Khetchoumian K, Chambon P, Losson R. TRIM involvement in transcriptional regulation. Adv Exp Med Biol. 2012;770:59–76. doi: 10.1007/978-1-4614-5398-7_5. [DOI] [PubMed] [Google Scholar]
- 28.Petrera F, Meroni G. TRIM proteins in development. Adv Exp Med Biol. 2012;770:131–41. doi: 10.1007/978-1-4614-5398-7_10. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Zhang Y, Zhang Y, et al. TRIM31 is downregulated in non-small cell lung cancer and serves as a potential tumor suppressor. Tumour Biol. 2014;35(6):5747–52. doi: 10.1007/s13277-014-1763-x. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Zhao E, Li C, et al. TRIM28, a new molecular marker predicting metastasis and survival in early-stage non-small cell lung cancer. Cancer Epidemiol. 2013;37(1):71–78. doi: 10.1016/j.canep.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Meroni G. Preface. TRIM/RBCC proteins. Adv Exp Med Biol. 2012;770:vii–viii. [PubMed] [Google Scholar]
- 32.Liu C, Huang X, Hou S, et al. Silencing of tripartite motif (TRIM) 29 inhibits proliferation and invasion and increases chemosensitivity to cisplatin in human lung squamous cancer NCI-H520 cells. Thorac Cancer. 2015;6(1):31–37. doi: 10.1111/1759-7714.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu X, Wu Y, Miao X, et al. High expression of TRIM44 is associated with enhanced cell proliferation, migration, invasion, and resistance to doxorubicin in hepatocellular carcinoma. Tumour Biol. 2016;37(11):14615–28. doi: 10.1007/s13277-016-5316-3. [DOI] [PubMed] [Google Scholar]