Abstract
Single-nucleotide polymorphisms (SNPs) in CYP2B6 have been shown to predict variation in plasma efavirenz concentrations, but associations between these SNPs and efavirenz-mediated depression and viral suppression are less well described. We evaluated three SNPs in CYP2B6 (rs3745274, rs28399499, and rs4803419) in Ugandan persons living with HIV. To define exposure, we used previously published pharmacokinetic modeling data to categorize participants as normal, intermediate, and poor efavirenz metabolizers. Our outcomes were probable depression in the first 2 years after antiretroviral therapy (ART) initiation (mean score of >1.75 on the Hopkins Symptom Depression Checklist) and viral suppression 6 months after ART initiation. We fit generalized estimating equation and modified Poisson regression models adjusted for demographic, clinical, and psychosocial characteristics with or without individuals with depression at the time of ART initiation. Among 242 participants, there were no differences in the pre-ART depression or viral load by efavirenz metabolism strata (p > .05). Participants were classified as normal (32%), intermediate (50%), and poor (18%) metabolizers. Seven percent (56/242) of follow-up visits met criteria for depression. Eighty-five percent (167/202) of participants who completed a 6-month visit achieved viral suppression. CYP2B6 metabolizer strata did not have a statistically significant association with either depression [adjusted risk ratio (aRR) comparing intermediate or poor vs. normal, 1.46; 95% confidence interval (CI), 0.72–2.95] or 6-month viral suppression (aRR, 1.01; 95% CI, 0.88–1.15). However, in analyses restricted to participants without pre-ART depression, poorer CYP2B6 metabolism was associated with increased odds of depression (adjusted odds ratio, 4.11; 95% CI, 1.04–16.20). Efavirenz-metabolizing allele patterns are strongly associated with risk of incident depression. Future work should elucidate further region-specific gene–environment interactions and whether alternate polymorphisms may be associated with efavirenz metabolism.
Keywords: : HIV, efavirenz, CYP2B6, single-nucleotide polymorphisms, depression, viral suppression
Introduction
Efavirenz (EFV) is widely used as a first-line antiretroviral therapy (ART) agent in sub-Saharan Africa, where 70% of persons living with HIV (PLHIV) reside.1 There is wide between-patient variability in the metabolism of efavirenz, spanning almost two orders of magnitude of concentration in controlled pharmacokinetic studies conducted largely in high-income countries.2–4
Approximately 90% of the active form of efavirenz are inactivated by the cytochrome P450 enzyme CYP2B6.5 Three single-nucleotide polymorphisms (SNPs) in the gene have been associated with efavirenz metabolism in populations composed of participants of European and African descent in resource-rich countries: 516G>T (rs3745274),6,7 983T>C (rs28399499),8,9 and 15582C>T (rs4803419).9 These SNPs are observed at minor allele frequencies of 23.6%, 0.0%, and 32.0%, respectively, in populations of European descent, and 37.4%, 8.2%, and 8.2%, respectively, in participants of African descent, as shown in the 1000 Genomes project.10 In one multiethnic study including participants from Caucasian (50%), African (33%), and Hispanic (18%) descent, the combination of these three genotypes predicted 33% of the variation in trough efavirenz level variation.9 The association of rs3745274,11–13 rs28399499,13–15 and rs480341916 with lower efavirenz clearance has also been confirmed in populations in sub-Saharan Africa, with one study in South Africa estimating that the three genotypes predicted 34% of the variation in mid-dose efavirenz concentrations.16
The variability in efavirenz levels due to CYP2B6 polymorphisms may have important clinical relevance for virologic outcomes and the wide spectrum of neuropsychiatric effects implicated in efavirenz use, including dizziness, insomnia, anxiety, headache, impaired concentration, suicidal ideation, and depressive symptoms.17–19 However, there are few data on relationships between these previously identified allelic patterns and efavirenz-related clinical outcomes or neuropsychiatric events in sub-Saharan Africa. One study in Botswana showed an unexpected association between poorer CYP2B6 metabolism alleles and lower central nervous system (CNS) toxicity scores.20 In this analysis, we aimed to estimate the association between CYP2B6 allele genotype combinations and two treatment outcomes in participants of African ancestry in Uganda: depression in the first 2 years and viral suppression after 6 months of ART suppression with an efavirenz-containing regimen in ART-naive individuals. We hypothesized that genotypic combinations that have been historically correlated with lower trough EFV levels (i.e., indicative of faster metabolism of EFV) would decrease the risk of viral suppression. Conversely, we hypothesized that genotypic combinations that historically correlated with higher trough EFV levels (i.e., indicative of poorer metabolism of EFV) would be correlated with increased reporting of neuropsychiatric side effects.8,20–22
Methods
Study population, design, and data collection
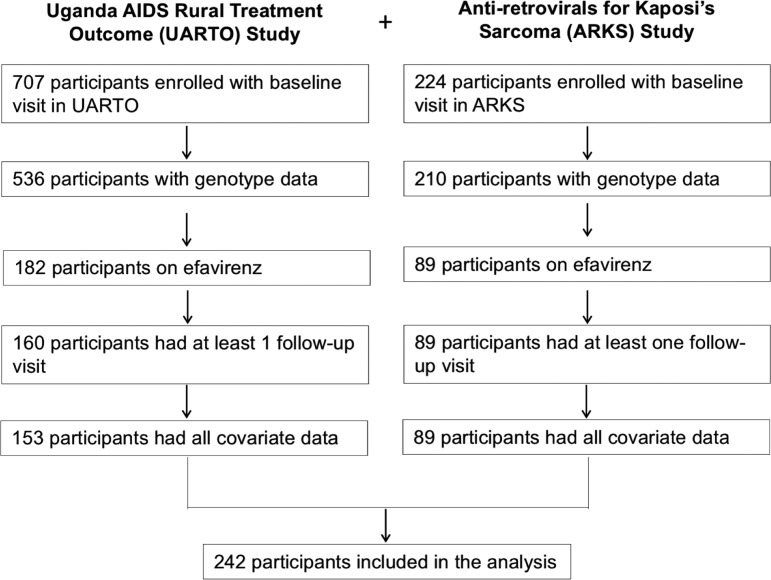
Data for this study were pooled from two prospective cohorts: the Uganda AIDS Rural Treatment Outcome Study (UARTO, NCT01596322) and the Antiretrovirals in Kaposi Sarcoma Study (ARKS, NCT00444379). The UARTO study was an observational cohort of adult PLHIV recruited from the Mbarara Regional Referral Hospital HIV Clinic at the time of ART initiation.23,24 The ARKS study recruited adult PLHIV with mild-to-moderate Kaposi sarcoma from a specialty treatment center in Kampala (NCT00444379). Participants were randomized to initiate ART with either efavirenz-based or boosted protease inhibitor–based regimen. Participants underwent a depression screen three to four times each year in the UARTO cohort and three times each year in the ARKS cohort. For the purposes of this analysis, only those visits in which the participant was recorded as receiving EFV-based ART were included in the longitudinal depression analysis, and only those participants enrolled in the efavirenz arm were included in the 6-month viral suppression analysis.
Outcomes
Our outcomes of interest were probable depression in the first 2 years and viral suppression at 6 months after ART initiation. Viral suppression was defined as an undetectable viral load, with limit of detection varying by type of assay. As viral detection technology improved over the course of the UARTO and ARKS studies, the viral detection limit decreased from 400 [Amplicor assay (Roche)] to 20 copies/mL [Cobas TaqMan assay (Roche)]. Because study visits were not conducted exactly at 3 month intervals, for the 6-month follow-up, we selected the study visit closest to 6 months after ART initiation, provided it was within the range of 3–9 months of follow-up.
Probable depression was defined with an adapted version of the 15-item Hopkins Symptom Checklist Depression subscale (HSCL-D),25 which we have previously validated among adult PLHIV in Uganda.26–28 This adapted version adds a 16th item (“feeling like I don't care about my health”) to the instrument.29 A participant was considered to have probable depression if the mean score on the items was >1.75.30
Predictors
Our primary predictor of interest was the combination of three CYP2B6 SNPs: rs3745274, rs28399499, and rs4803419. Individual CYP2B6 SNP genotype combinations were grouped into three categories corresponding to normal, intermediate, and poor EFV metabolizer phenotypes. These classifications (Appendix Table 1) are based on previously published pharmacokinetic modeling data9 and have been used in previous studies.31,32 SNPs were genotyped as part of genome-wide genotyping analyses performed at the RIKEN Center for Genomic Medicine using the Human Omni Express Bead Chip (Illumina, San Diego, CA) including over 700,000 SNPs.23 Two SNPs (rs3745274 and rs28399499) were directly genotyped, whereas rs4803419 was imputed using the 1000 Genomes database. We performed quality control steps with checks of identity-by-descent and sex. As previously described, only imputed SNPs that passed quality control analyses with an info score of >0.8 were included in the final analysis.23
Statistical models
We first characterized the distributions of variables at baseline and compared them between participants of the three EFV-metabolizing strata. Differences in continuous and categorical variables at baseline were tested by using the Wilcoxon rank-sum and Pearson's chi-square tests, respectively.
We next estimated the minor allelic frequency of each SNP in our cohort. We fit a Poisson regression model with robust standard errors to estimate the association between EFV-metabolizing strata and achievement of viral suppression at 6 months, specifying normal metabolizers as the referent exposure category. We adjusted the model for demographic variables, including age, sex, marital status, educational attainment, household asset wealth, and year of study enrollment. The index of household asset wealth was derived by applying the method of principal component analysis to 25 household asset and housing characteristic variables as suggested by Filmer and Pritchett.33 The first component was extracted and used to define the index, which we then categorized into quintiles of relative household asset wealth. Year of enrollment was included to adjust for secular trends,34 defined as a categorical variable with 3-year increments from 2005 until 2013, when enrollment concluded. We additionally adjusted for pre-ART clinical variables, including probable depression at enrollment, CD4+ T lymphocyte cell count, viral suppression, ART duration, tuberculosis (TB) coinfection, health status, and heavy alcohol use. ART duration was measured as cumulative weeks since ART initiation. TB coinfection was determined by a combination of self-report and clinical record abstraction. Health status was measured by the Physical Health Summary (PHS) score from the Medical Outcome Survey-HIV (MOS-HIV) questionnaire and was categorized into quartiles.35 Heavy alcohol use was determined by the three-item consumption subset of the Alcohol Use Disorders Identification Test.36
We also fit generalized estimating equation (GEE) Poisson regression models with an exchangeable correlation matrix and cluster-correlated robust standard errors to estimate the association between EFV-metabolizing strata and probable depression. For this analysis, we used time-updated covariates and restricted estimation to visits within 2 years of ART initiation in the UARTO cohort and to the 1-year follow-up in the ARKS cohort, as these were the minimum time period of scheduled follow-up for all participants. In all the models, we included a binary covariate for study (ARKS vs. UARTO) in the model if its Wald p-value was <.25 in fully adjusted models. We fit similar models for both viral suppression and probable depression outcomes comparing slow and intermediate metabolizers combined versus normal metabolizers. We also fit models for probable depression excluding participants with pre-ART baseline probable depression.
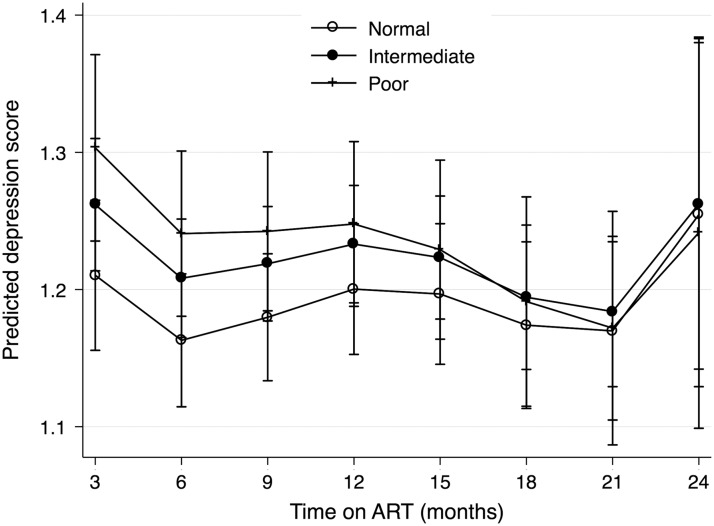
To graphically depict relationships between EFV-metabolizing strata and depression symptom severity, we modeled the association between metabolizer strata and depressive symptom score using a mixed-effects linear regression model, adjusting for all aforementioned covariates (but substituting pre-ART depressive symptom score for pre-ART probable depression). The regression model also included product terms for the interaction between EFV-metabolizing strata and time on ART and a random intercept by participant. We then used postestimation margins to plot the predicted depression scores by visit for each metabolizer strata.
In post hoc exploratory analyses, we fit adjusted GEE logistic regression models estimating the association between minor allele copy number and probable depression in the first 2 years for each SNP. Due to problems with model convergence, we used a logistic instead of a Poisson regression model, collapsed participants heterozygous or homozygous for the minor allele into the same category (due to small counts in each strata), and fit models with and without a exposure by time interaction terms and plotted relationships with postestimation margins. We also fit adjusted mixed-effects linear regression models and plotted predicted depression scores by visit for each SNP. We further fit adjusted Poisson regression models with robust standard errors estimating the association between each of the SNP combinations and viral suppression and plotted relationships with postestimation margins.9
Results
Of the 746 participants with genotype data, the minor allelic frequencies for rs3745274, rs28399499, and rs4803419 were 34.7%, 7.1%, and 7.0%, respectively. A total of 242 participants taking efavirenz-based regimens met inclusion criteria and were included in the final analysis of depression (Appendix Fig. 1); of these, 202 participants had 6-month data for the viral suppression analysis.
Of the 242 total participants in the depression analytic sample, 78 (32.2%), 120 (49.6%), and 44 (18.2%) had CYP2B6 genotypic combinations that corresponded to normal, intermediate, and poor efavirenz-metabolizing strata, respectively. The median age of the sample was 35 years (interquartile range [IQR], 29–42), and the median CD4+ T lymphocyte count at enrollment was 160 cells/mm3 (IQR, 59–284). The median log10 viral load at enrollment was 5.1 (IQR, 4.7–5.6). Sixty-two participants (25.6%) screened positive for probable depression at enrollment, and the median Hopkins Symptom Checklist score was 1.38 (IQR, 1.12–1.82).
There were no pre-ART differences between the three strata in any of the factors measured, including CD4+ T lymphocyte count, viral load, probable depression, and depressive symptom score (all p > .05; Table 1). There were a larger proportion of total visits missed in the normal metabolizer strata than the intermediate or poor strata (8.5%, 5.0%, and 5.1% of visits, respectively; p = .040). There was no statistically significant difference in the proportions of participants lost to follow-up, which was defined as having the last study visit more than 6 months before the end of 2-year right censoring in the UARTO study or less than 3 months before the end of the 1-year ARKS study (p = .11).
Table 1.
Summary Characteristics for Study Cohort at Enrollment
| Metabolizer level | ||||
|---|---|---|---|---|
| Variable | Normal (n = 78) | Intermediate (n = 120) | Poor (n = 44) | p |
| Demographic characteristics | ||||
| Age (years), median (IQR) | 35 (29, 41) | 35 (28, 43) | 35 (30, 44) | .87 |
| Female, n (%) | 40 (51.3%) | 65 (54.2%) | 21 (47.7%) | .75 |
| Married, n (%) | 50 (64.1%) | 71 (59.2%) | 27 (61.4%) | .78 |
| Secondary education, n (%) | 27 (34.6%) | 46 (38.3%) | 15 (34.1%) | .82 |
| Asset index, n (%) | ||||
| 1st quintile (most poor) | 13 (16.7%) | 17 (14.2%) | 9 (20.5%) | .97 |
| 2nd quintile | 14 (17.9%) | 26 (21.7%) | 8 (18.2%) | |
| 3rd quintile | 17 (21.8%) | 22 (18.3%) | 9 (20.5%) | |
| 4th quintile | 14 (17.9%) | 27 (22.5%) | 8 (18.2%) | |
| 5th quintile (least poor) | 20 (25.6%) | 28 (23.3%) | 10 (22.7%) | |
| Year of enrollment | ||||
| 2005–2007 | 24 (30.8%) | 28 (23.3%) | 13 (29.5%) | .81 |
| 2008–2010 | 32 (41.0%) | 53 (44.2%) | 18 (40.9%) | |
| 2011–2013 | 22 (28.2%) | 39 (32.5%) | 13 (29.5%) | |
| Clinical characteristics | ||||
| CD4 count (cells/mm3), median (IQR) | 159 (56, 278) | 173 (68, 289) | 132 (60, 256) | .70 |
| Viral load (log10 copies/mL), median (IQR) | 5.1 (4.6, 5.4) | 5.2 (4.7, 5.6) | 5.3 (4.9, 5.8) | .068 |
| Depressed, n (%) | 19 (24.4%) | 32 (26.7%) | 11 (25.0%) | .93 |
| Depressive symptom score, median (IQR) | 1.41 (1.19, 1.75) | 1.38 (1.12, 1.88) | 1.38 (1.12, 1.78) | .89 |
| Physical health summary score, n (%) | .36 | |||
| 1st quartile (least healthy) | 38 (49.4%) | 63 (52.5%) | 28 (65.1%) | |
| 2nd quartile | 25 (32.5%) | 28 (23.3%) | 8 (18.6%) | |
| 3rd quartile | 8 (10.4%) | 19 (15.8%) | 6 (14.0%) | |
| 4th quartile (most healthy) | 6 (7.8%) | 10 (8.3%) | 1 (2.3%) | |
| Heavy drinking, n (%) | 14 (28%) | 12 (16%) | 3 (12%) | .15 |
| Follow-up characteristics | ||||
| Enrolled in the ARKS study | 27 (34.6%) | 44 (36.7%) | 18 (40.9%) | .79 |
| No. of visits per participant, median (IQR) | 6 (4,7) | 7 (4,7) | 7 (4,7) | .62 |
| Total No. of visits missed, n (%) | 38 (8.5%) | 36 (5.0%) | 13 (5.1%) | .040 |
| Participants lost to follow-up, n (%) | 1 (1.3%) | 2 (1.7%) | 3 (6.8%) | .11 |
Primary analyses
The 242 participants contributed a total of 1,021 follow-up study visits. At 53 visits (6.7%), there was a positive screen for probable depression. The adjusted risk ratio (aRR) of probable depression for participants in the intermediate metabolism strata compared with the normal strata was 1.53 (95% confidence interval [CI], 0.75–3.12; Table 2). The aRR for those in the poor strata compared with the normal strata was 1.19 (95% CI, 0.42–3.33). There was a nonsignificant stepwise association between poorer metabolizer strata and higher predicted mean depressive at symptom scores at 3 months that dissipated over time (Fig. 1). In analyses restricted to participants without pre-ART probable depression (180/242 participants), the adjusted odds ratio (aOR) of probable depression for participants in the intermediate metabolism strata compared with the normal strata was 4.15 (95% CI, 1.05–16.51) and 3.86 (95% CI, 0.65–22.99) for the poor strata compared with the normal strata (Table 2).
Table 2.
Generalized Estimated Equations Regression Estimates for Probable Depression by Metabolizer Strata
| All participants (n = 242) | Excluding participants with pre-ART probable depression (n = 180) | |||||||
|---|---|---|---|---|---|---|---|---|
| Strata | Unadjusted RR (95% CI) | p | Adjusted RRa(95% CI) | p | Unadjusted OR (95% CI) | p | Adjusted ORb(95% CI) | p |
| Normal | REF | — | REF | — | REF | — | REF | — |
| Intermediate | 1.80 (0.78–4.15) | .170 | 1.53 (0.75–3.12) | .24 | 3.97 (0.83–19.05) | .084 | 4.15 (1.05–16.51) | .043 |
| Poor | 1.63 (0.61–4.30) | .33 | 1.19 (0.42–3.33) | .75 | 5.17 (0.99–26.96) | .051 | 3.86 (0.65–22.99) | .137 |
| Normal | REF | — | REF | — | REF | — | REF | — |
| Intermediate or poor | 1.75 (0.79–3.90) | .171 | 1.46 (0.72–2.95) | .30 | 4.29 (0.95–19.32) | .058 | 4.11 (1.04–16.20) | .043 |
Adjusted for baseline depression, baseline suicidal ideation, sex, year of enrollment, time-updated age, marital status, education, asset index, CD4+ T lymphocyte count, viral suppression, health status, and heavy drinking.
Adjusted for sex, year of enrollment, time-updated age, marital status, education, asset index, CD4+ T lymphocyte count, viral suppression, and health status. Not adjusted for baseline suicidal ideation or heavy drinking, as these variables were too collinear with the outcome. Third and fourth quartiles of health status score were also collapsed due to zero counts in the fourth quartile. Logistic regression was used due to problems with convergence with Poisson regression.
FIG. 1.
Model-adjusted predicted depressive symptom scores over visits by metabolizer strata.
Approximately 82% (167/202) of participants achieved viral suppression at 6 months. In the adjusted model, the aRR of 6-month viral suppression for participants in the intermediate metabolism strata compared with the normal strata was 1.01 (95% CI, 0.99–1.17; Table 3) and 1.04 (95% CI 0.85–1.26) in the poor versus normal strata. Results were similar when comparing combined slow and intermediate metabolizers versus normal metabolizers for both probable depression and viral suppression outcomes (Tables 2 and 3).
Table 3.
Poisson Regression Model Estimates for 6-Month Viral Suppression by Metabolizer Strata
| Strata | Unadjusted RR (95% CI) | p | Adjusted RRa(95% CI) | p |
|---|---|---|---|---|
| Normal | REF | — | REF | — |
| Intermediate | 1.03 (0.89–1.19) | .70 | 1.01 (0.88–1.15) | .94 |
| Poor | 1.01 (0.84–1.22) | .92 | 1.00 (0.83–1.22) | .96 |
| Normal | REF | — | REF | — |
| Intermediate or poor | 1.02 (0.89–1.18) | .74 | 1.01 (0.88–1.15) | .94 |
Adjusted for the following baseline covariates: age, sex, marital status, education, asset index, year of enrollment, CD4+ T lymphocyte count, log10 viral load, probable depression, health status, heavy drinking, and study data source.
Exploratory analyses
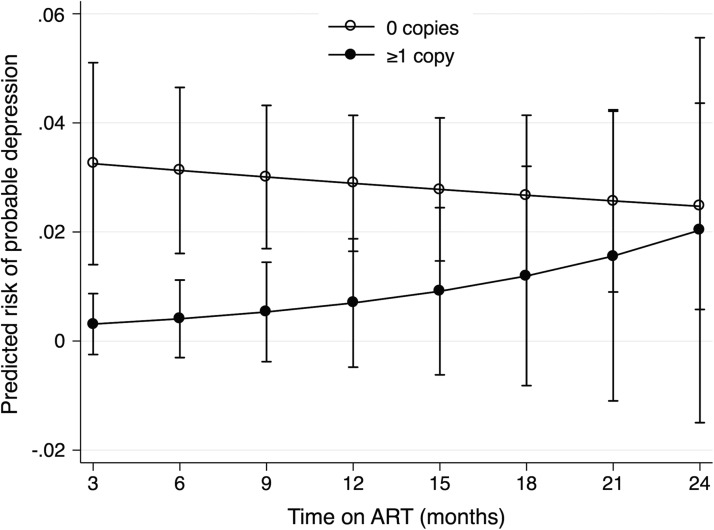
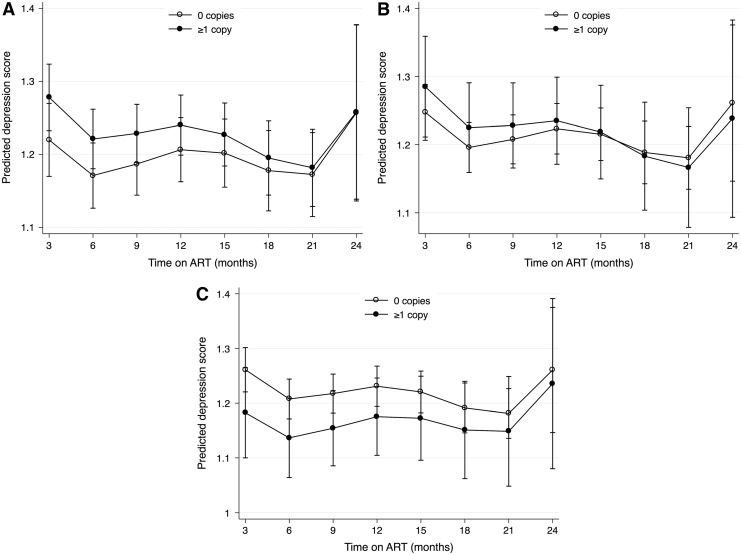
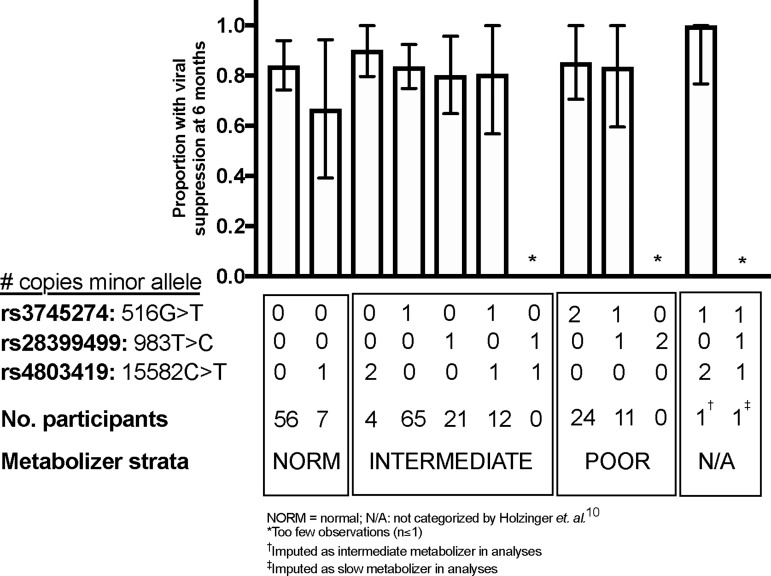
In post hoc GEE regression analyses, ≥1 copy of the allele copy number for the rs4803419 allele was associated with a nonsignificant decreased risk of probable depression (aOR, 0.27; 95% CI, 0.48–1.47). After graphical exploration and the inclusion of a allele-by-time interaction term (Wald p-value for interaction = .004), ≥1 copy of the rs4803419 allele was significantly associated with a decreased risk of probable depression before 1 year of follow-up but converged to have similar risk compared with those with wild-type alleles at later follow-up times (Appendix Tables 2 and 3, Appendix Fig. 2). In the adjusted mixed-effects regression models, we estimated no statistically significant difference between depressive symptom score over time (Appendix Fig. 3). We also estimated no statistically significant association between minor allele copy number and viral suppression for each of the SNP (all p > .20; Appendix Table 4; Appendix Fig. 4).
Discussion
In this pooled analysis of data on African adult PLHIV on EFV-containing ART participating in prospective cohort and experimental studies conducted in Uganda, we found that CYP2B6 polymorphisms associated with poorer EFV metabolism was significantly associated with an approximately fourfold increase in the odds of probable depression in those without pre-ART baseline probable depression but not in the cohort as a whole, which may have important therapeutic implications for patients starting ART in the region.
Prior studies about the relationship between EFV-metabolizing genotypes and CNS adverse effects have been conflicting; some have reported an association between poor efavirenz-metabolizing genotypes and increased CNS toxicity in participants of both Caucasian and African ancestry,6,37 whereas others have found no significant association between CYP2B6 genotype and increased risk of various symptoms of CNS toxicity among participants of African origin.8,20,22 In our study, we hypothesize that we were only able to observe a statistically significant relationship in those without depression at baseline because there may be other much stronger predictors of depression in those with pre-ART baseline depression that might modify and mask this association.
We did also observe a nonsignificant association between poorer metabolism strata and higher predicted depressive symptom scores at 3 months that dissipated with longer follow-up; this could be consistent with studies showing attenuation of neuropsychiatric effects with prolonged EFV use,38 although other studies suggest that EFV could have longer term neuropsychiatric effects.18,39
In post hoc analyses in which the SNPs were evaluated individually, we found no evidence of an association between poor-metabolizing SNPs and increased risk of probable depression in all participants. In contrast, we found some evidence in a post hoc analysis that ≥1 copy of the rs4803419 allele, a SNP that has been associated with poor metabolism in both Caucasian and African populations,9,16 may be associated with a lower odds of depression in the first year of follow-up after starting efavirenz-containing ART compared with those with no copies of the minor allele. This unexpected, inverse association is similar to that found in a cohort study in Botswana, in which Gross et al. found an association between a composite exposure of two SNPs examined in this study (rs3745274 and rs28399499) and both lower efavirenz clearance and a lower CNS adverse experience score.20 However, we note that the association between rs4803419 and CNS adverse effects is not well established, has unclear biological or pharmacologic plausibility, and that additional data are needed to better elucidate these relationships.
The evidence is also conflicting for the association of CYP2B6 alleles and virologic outcomes. Ribaudo et al. found an inverse association between the presence of the rs3745274 allele and virologic failure in a subgroup analysis of African Americans.8 However, other studies, including ours, have not corroborated this finding.20,31
Limitations of this study include lack of pharmacokinetic measures of efavirenz metabolism to support mechanistic relationships between allelic patterns and our clinical outcomes. In addition, we only assessed three SNPs within the CYP2B6 gene; other polymorphisms including rare or novel variants identified by genetic sequencing, rather than genotyping, might be able to identify a more detailed association between this gene and viral suppression and probable depression. Nonetheless, these three SNPs have been reported to explain approximately one third of the variability in efavirenz concentrations in both studies in the United States and South Africa,9,16 and although we did not have genetic sequencing data, imputation was performed to estimate one of the three SNPs that was not directly genotyped. We may also have limited power for assessing our outcomes, given our sample size and outcome distribution. There is also potential for measurement bias given that depression symptoms severity was assessed by participant self-report rather than observer rating. However, previous studies have found this measure to be valid and reliable when administered to study participants in rural Uganda.26–28 There is also potential for observational bias in our study, given the differential proportion of missed visits among the metabolism strata in our studies. However, the proportion of missed visits was relatively small (<10% of all visits missed in all strata).
In summary, we found a strong association between efavirenz metabolism–determining CYP2B6 genotypic combinations and probable depression in the first 2 years among participants without depression at baseline and no association with viral suppression at 6 months. Future work should reassess these relationships with pharmacologic data and explore other potential gene–environment interactions that may be associated with efavirenz metabolism in this population.
Appendix
Appendix Table 1.
Definitions of Metabolizer Categories and Number of Participants with Each SNP Combination
| Allele copy numbers defining each SNP combination and metabolizer category | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Fast | Intermediate | Slow | Not categorizeda | ||||||||
| Ordinal level of EFV metabolismb (1 = fastest, 10 = slowest) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | — | — |
| rs3745274 (516G>T) | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 1 | 0 | 1 | 1 |
| rs28399499 (983T>C) | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 2 | 0 | 1 |
| rs4803419 (15582C>T) | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 1 |
| No. of participants in whole study by SNP combination | 206 | 45 | 6 | 249 | 59 | 43 | 2 | 91 | 42 | 1 | 1 | 1 |
| No. of participants in final cohort by SNP combination | 69 | 9 | 2 | 77 | 22 | 18 | 0 | 30 | 14 | 0 | 1c | 0d |
Two SNP combinations were unable to be categorized as fast, intermediate, or slow based on the previously published pharmacokinetic framework,10 and metabolizer strata were imputed for these SNP combinations.
Based on previously published pharmacokinetic modeling study by Holzinger et al.9
Imputed as intermediate metabolize.
Imputed as slow metabolizer.
SNP, single-nucleotide polymorphism.
Appendix Table 2.
GEE Logistic Regression Models for Depression by the Presence of rs3745274, rs28399499, and rs4803419 Minor Alleles
| No. of copies minor allele | Unadjusted OR (95% CI) | p | Adjusted ORa(95% CI) | p | Wald p-value for added allele × time interaction term |
|---|---|---|---|---|---|
| By the presence of rs3745274 allele | |||||
| 0 | REF | — | REF | — | .79 |
| 1 or 2 | 1.28 (0.63–2.63) | .49 | 1.03 (0.49–2.19) | .93 | |
| By the presence of rs28399499 allele | |||||
| 0 | REF | — | REF | — | .36 |
| 1 or 2 | 1.42 (0.66–3.09) | .37 | 1.34 (0.59–3.08) | .48 | |
| By the presence of rs4803419 allele | |||||
| 0 | REF | — | REF | — | .004 |
| 1 or 2 | 0.43 (0.06–3.22) | .41 | 0.27 (0.48–1.47) | .129 | |
Estimated OR based on main effects model adjusted for the following baseline covariates: age, sex, marital status, education, asset index, year of enrollment, CD4+ T lymphocyte count, log10 viral load, probable depression, health status, heavy drinking, and study data source.
OR, odds ratio.
Appendix Table 3.
Crude Risk of Depression by the Presence of rs4803419 Minor Allele Over Time
| No. of depressed/no. of participants (%) stratified by time on antiretroviral therapy | ||||||||
|---|---|---|---|---|---|---|---|---|
| No. of copies minor allele | Baseline | 3 months | 6 months | 9 months | 12 months | 15 months | 18 months | 21 months |
| 0 | 33/110 (30%) | 15/176 (8.5%) | 7/150 (4.46%) | 6/73 (7.6%) | 10/163 (5.8%) | 13/112 (10.4%) | 3/117 (2.5%) | 7/100 (7.0%) |
| 1 or 2 | 3/14 (21.4%) | 0/23 (0.0%) | 0/19 (0.0%) | 0/14 (0.0%) | 1/23 (4.3%) | 2/13 (15.4%) | 0/17 (0.0%) | 1/13 (7.7%) |
Appendix Table 4.
Modified Poisson Regression Models for Viral Suppression by rs3745274, rs28399499, and rs4803419 Minor Allele Copy Number
| No. of copies | Unadjusted RR (95% CI) | p | AdjustedaRR (95% CI) | p |
|---|---|---|---|---|
| By rs3745274 allele copy number | ||||
| 0 | REF | — | REF | — |
| 1 | 1.01 (0.88–1.15) | .92 | 1.00 (0.88–1.14) | .96 |
| 2 | 1.03 (0.85–1.25) | .75 | 1.04 (0.85–1.28) | .69 |
| By rs28399499 allele copy number | ||||
| 0 | REF | — | REF | — |
| 1 | 0.95 (0.79–1.15) | 0.95 (0.79–1.14) | .56 | |
| 2 | —b | — | — | — |
| By rs4803419 allele copy number | ||||
| 0 | REF | — | REF | — |
| 1 | 0.84 (0.62–1.13) | .24 | 0.85 (0.65–1.11) | .24 |
| 2 | 1.20 (1.12–1.28) | <.001 | 1.09 (0.95–1.24) | .239 |
Adjusted for the following baseline covariates: age, sex, marital status, education, asset index, year of enrollment, CD4+ T lymphocyte count, log10 viral load, probable depression, health status, heavy drinking, and study data source.
No participants with two copies of rs28399499 minor allele.
RR, risk ratio.
APPENDIX FIG. 1.
Flow chart for included participants.
APPENDIX FIG. 2.
Model-adjusted predicted probability of probable depression by copy number of rs4803419 minor alleles with exposure by time interaction term.
APPENDIX FIG. 3.
Predicted depression scores over visits by copy number of (A) rs3745274, (B) rs28399499, and (C) rs4803419 minor alleles.
APPENDIX FIG. 4.
Adjusted risk of viral suppression by allele combinations. N/A, not categorized by Holzinger et al9; *Too few observations (n ≤ 1).
Acknowledgments
This study was supported by the National Institutes of Health (R01 MH054907, U01 CA066529, K23 MH099916), University of California, San Francisco—Gladstone Center for AIDS Research (P30AI027763), Harvard Center for AIDS Research (P30AI060354), and the Doris Duke Charitable Foundation. P.W.H. discloses consulting fees from Merck, Gilead, and Viiv.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Fettig J, Swaminathan M, Murrill CS, Kaplan JE: Global epidemiology of HIV. Infect Dis Clin North Am 2014;28:323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starr SE, Fletcher CV, Spector SA, et al. : Combination therapy with efavirenz, nelfinavir, and nucleoside reverse-transcriptase inhibitors in children infected with human immunodeficiency virus type 1. Pediatric AIDS Clinical Trials Group 382 Team. N Engl J Med 1999;341:1874–1881 [DOI] [PubMed] [Google Scholar]
- 3.Staszewski S, Morales-Ramirez J, Tashima KT, et al. : Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. Study 006 Team. N Engl J Med 1999;341:1865–1873 [DOI] [PubMed] [Google Scholar]
- 4.van Luin M, Bannister WP, Mocroft A, et al. : Absence of a relation between efavirenz plasma concentrations and toxicity-driven evafirenz discontinuations in the EuroSIDA study. Antivir Ther 2009;14:75–83 [PubMed] [Google Scholar]
- 5.Ward BA, Gorski CJ, Jones DR, Hall SD, Flockhart DA, Zeruesenay D: The Cytochrome P450 2B6 (CYP2B6) Is the Main Catalyst of Efavirenz Primary and Secondary Metabolism: Implication for HIV/AIDS Therapy and Utility of Efavirenz as a Substrate Marker of CYP2B6 Catalytic Activity. J Pharmacol Exp Ther 2003;306:287–300 [DOI] [PubMed] [Google Scholar]
- 6.Haas DW, Ribaudo HJ, Kim RB, et al. : Pharmacogenetics of efavirenz and central nervous system side effects: An Adult AIDS Clinical Trials Group study. AIDS 2004;18:2391–2400 [PubMed] [Google Scholar]
- 7.Wyen C, Hendra H, Vogel M, et al. : Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother 2008;61:914–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribaudo HJ, Liu H, Schwab M, et al. : Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: An AIDS Clinical Trials Group study. J Infect Dis 2010;202:717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holzinger ER, Grady B, Ritchie MD, et al. : Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genomics 2012;22:858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auton A, Abecasis GR, Altshuler DM, et al. : A global reference for human genetic variation. Nature 2015;526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwara A, Lartey M, Sagoe KW, Rzek NL, Court MH: CYP2B6 (c.516G???T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol 2009;67:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyakutira C, Röshammar D, Chigutsa E, et al. : High prevalence of the CYP2B6 516G→T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur J Clin Pharmacol 2008;64:357–365 [DOI] [PubMed] [Google Scholar]
- 13.Maimbo M, Kiyotani K, Mushiroda T, Masimirembwa C, Nakamura Y: CYP2B6 genotype is a strong predictor of systemic exposure to efavirenz in HIV-infected Zimbabweans. Eur J Clin Pharmacol 2012;68:267–271 [DOI] [PubMed] [Google Scholar]
- 14.Jamshidi Y, Moreton M, McKeown DA, et al. : Tribal ethnicity and CYP2B6 genetics in Ugandan and Zimbabwean populations in the UK: Implications for efavirenz dosing in HIV infection. J Antimicrob Chemother 2010;65:2614–2619 [DOI] [PubMed] [Google Scholar]
- 15.Mutwa PR, Fillekes Q, Malgaz M, et al. : Mid-dosing interval efavirenz plasma concentrations in HIV-1-infected children in Rwanda: Treatment efficacy, tolerability, adherence, and the influence of CYP2B6 polymorphisms. J Acquir Immune Defic Syndr 2012;60:400–404 [DOI] [PubMed] [Google Scholar]
- 16.Sinxadi PZ, Leger PD, McIlleron HM, et al. : Pharmacogenetics of plasma efavirenz exposure in HIV-infected adults and children in South Africa. Br J Clin Pharmacol 2015;80:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abers MS, Shandera WX, Kass JS: Neurological and psychiatric adverse effects of antiretroviral drugs. CNS Drugs 2014;28:131–145 [DOI] [PubMed] [Google Scholar]
- 18.Fumaz CR, Muñoz-Moreno JA, Moltó J, et al. : Long-term neuropsychiatric disorders on efavirenz-based approaches: Quality of life, psychologic issues, and adherence. J Acquir Immune Defic Syndr 2005;38:560–565 [DOI] [PubMed] [Google Scholar]
- 19.Sütterlin S, Vögele C, Gauggel S: Neuropsychiatric complications of efavirenz therapy: Suggestions for a new research paradigm. J Neuropsychiatr Clin Neurosci 2010;22:361–369 [DOI] [PubMed] [Google Scholar]
- 20.Gross R, Bellamy SL, Ratshaa B, et al. : CYP2B6 genotypes and early efavirenz-based hiv treatment outcomes in botswana. AIDS 2017;31:2107–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas DW, Smeaton LM, Shafer RW, et al. : Pharmacogenetics of long-term responses to antiretroviral regimens containing Efavirenz and/or Nelfinavir: An Adult Aids Clinical Trials Group Study. J Infect Dis 2005;192:1931–1942 [DOI] [PubMed] [Google Scholar]
- 22.Mukonzo JK, Okwera A, Nakasujja N, et al. : Influence of efavirenz pharmacokinetics and pharmacogenetics on neuropsychological disorders in Ugandan HIV-positive patients with or without tuberculosis: A prospective cohort study. BMC Infect Dis 2013;13:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SA, Mefford JA, Huang Y, et al. : Host Genetic Predictors of the Kynurenine Pathway of Tryptophan Catabolism Among Treated HIV-Infected Ugandans. AIDS 2016;30:1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez P, Tsai AC, Muzoora C, et al. : Reversal of the kynurenine pathway of tryptophan catabolism may improve depression in ART-treated HIV-infected ugandans. JAIDS J Acquir Immune Defic Syndr 2014;65:456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derogatis LR, Lipman RS, Rickels K, Uhlenhuth EH, Covi L: The hopkins symptom checklist (HSCL): A self‐report symptom inventory. Behav Sci 1974;19:1–15 [DOI] [PubMed] [Google Scholar]
- 26.Ashaba S, Kakuhikire B, Vořechovská D, et al. : Reliability, Validity, and Factor Structure of the Hopkins Symptom Checklist-25: Population-Based Study of Persons Living with HIV in Rural, Uganda. AIDS Behav 2018;22:1467–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai AC, Bangsberg DR, Frongillo EA, et al. : Food insecurity, depression and the modifying role of social support among people living with HIV/AIDS in rural Uganda. Soc Sci Med 2012;74:2012–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai AC, Weiser SD, Steward WT, et al. : Evidence for the reliability and validity of the internalized aids-related stigma scale in rural Uganda. AIDS Behav 2013;17:427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolton P, Ndogoni L: Cross-Cultural Assessment Of Trauma-Related Mental Illness. Johns Hopkins Univ, Baltimore, MD, 2000 [Google Scholar]
- 30.Winokur A, Winokur DF, Rickels K, Cox DS: Symptoms of emotional distress in a family planning service: Stability over a four-week period. Br J Psychiatry 1984;144:395–399 [DOI] [PubMed] [Google Scholar]
- 31.Haas DW, Severe P, Juste MAJ, Pape JW, Fitzgerald DW: Functional CYP2B6 variants and virologic response to an efavirenz-containing regimen in Port-au-Prince, Haiti. J Antimicrob Chemother 2014;69:2187–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollan KR, Tierney C, Hellwege JN, et al. : Race/Ethnicity and the pharmacogenetics of reported suicidality with efavirenz among clinical trials participants. J Infect Dis 2017;216:554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filmer D, Pritchett LH: Estimating wealth effects without expenditure data—or tears: An application to educational enrollments in states of India. Demography 2001;38:115–132 [DOI] [PubMed] [Google Scholar]
- 34.Chan BT, Weiserd SD, Boum Y, et al. : Declining prevalence of probable depression among patients presenting for antiretroviral therapy in rural Uganda: The role of early treatment initiation. AIDS Behav 2016;19:19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu AW, Rubin HR, Mathews WC, et al. : A Health Status Questionnaire Using 30 Items From The Medical Outcomes Study: Preliminary Validation in Persons With Early HIV Infection. Med Care 1991;29:786–798 [DOI] [PubMed] [Google Scholar]
- 36.Bush K, Kivlahan DR, McDonell MB, Fihn SD, Bradley KA: The AUDIT alcohol consumption questions (AUDIT-C): An effective brief screening test for problem drinking. Arch Intern Med 1998;158:1789–1795 [DOI] [PubMed] [Google Scholar]
- 37.Rotger M, Colombo S, Furrer H, et al. : Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics 2005;15:1–5 [DOI] [PubMed] [Google Scholar]
- 38.Kenedi CA, Goforth HW: A systematic review of the psychiatric side-effects of efavirenz. AIDS Behav 2011;15:1803–1818 [DOI] [PubMed] [Google Scholar]
- 39.Gutiérrez F, Navarro A, Padilla S, et al. : Prediction of neuropsychiatric adverse events associated with long-term efavirenz therapy, using plasma drug level monitoring. Clin Infect Dis 2005;41:1648–1653 [DOI] [PubMed] [Google Scholar]