The authors apply telomeric chromatin analysis to identify factors that accumulate at telomeres during cellular transformation, promoting telomere replication and repair and counteracting oncogene-borne telomere replication stress.
Abstract
Telomeres play crucial roles during tumorigenesis, inducing cellular senescence upon telomere shortening and extensive chromosome instability during telomere crisis. However, it has not been investigated if and how cellular transformation and oncogenic stress alter telomeric chromatin composition and function. Here, we transform human fibroblasts by consecutive transduction with vectors expressing hTERT, the SV40 early region, and activated H-RasV12. Pairwise comparisons of the telomeric proteome during different stages of transformation reveal up-regulation of proteins involved in chromatin remodeling, DNA repair, and replication at chromosome ends. Depletion of several of these proteins induces telomere fragility, indicating their roles in replication of telomeric DNA. Depletion of SAMHD1, which has reported roles in DNA resection and homology-directed repair, leads to telomere breakage events in cells deprived of the shelterin component TRF1. Thus, our analysis identifies factors, which accumulate at telomeres during cellular transformation to promote telomere replication and repair, resisting oncogene-borne telomere replication stress.
Introduction
Telomeres play critical roles in the progression of human cancer (Maciejowski & de Lange, 2017). Most somatic cells in the human body do not express telomerase (Kim et al, 1994). Therefore, telomeres shorten with every round of DNA replication because of the end replication problem and the nucleolytic processing of chromosome ends (Soudet et al, 2014) by approximately 50–100 bp. Upon reaching a critically short length, telomeres elicit a DNA damage response (DDR) involving the DNA checkpoint protein kinases ATM and ATR (d’Adda di Fagagna et al, 2003; Denchi & de Lange, 2007). The telomeric DDR induces permanent cell cycle arrest referred to as cellular senescence with a G1 DNA content. This block to proliferation of precancerous cells can be prevented through inactivation of the p53 and RB tumor suppressors (Shay & Wright, 2005). Cells that bypass cellular senescence will hit telomere crisis during which telomeres lose their protective roles from end-to-end chromosome fusions by classical and alternative nonhomologous end joining (Jones et al, 2014). Thus, telomere crisis leads to chromosome fusions, mitotic missegregation, and chromosome breakage events that give rise to extensive chromosome instability. In cancer, telomere crisis is mostly overcome through up-regulation of the telomerase catalytic subunit hTERT, which frequently involves mutations in the hTERT promoter (Horn et al, 2013; Huang et al, 2013). Thus, telomerase becomes active, stabilizing telomere length of partially rearranged chromosomes.
In addition to gradual telomere shortening induced by the lack of telomerase, telomeres can be damaged and lost because of stochastic replication defects occurring during semiconservative replication of telomeric DNA (Miller et al, 2006; Chang et al, 2007; Sfeir et al, 2009). Telomere replication defects can give rise to a fragile phenotype, which is characterized by discontinuities in the telomeric signal detected on metaphase chromosome spreads (Sfeir et al, 2009). Telomeres are difficult to replicate and fragile for at least four reasons. First, the single-stranded TTAGGG repeat (G-rich)–containing strand may adopt highly stable G-quadruplex structures that need to be unwound to serve as a template during replication (Sfeir et al, 2009; Paeschke et al, 2011; Vannier et al, 2012). Second, telomeres can fold into t-loop structures in which the telomeric 3′ overhang is tucked into the double-stranded part of the telomere which need to be unwound during replication (Vannier et al, 2012; Doksani et al, 2013). Third, telomeres are transcribed into the long noncoding RNA TERRA that can form DNA/RNA hybrid structures and as such can interfere with replication (Balk et al, 2013; Pfeiffer et al, 2013; Sagie et al, 2017). Fourth, telomere replication is driven from origins of replication that are present in the subtelomeric DNA. Origin firing occurs only rarely from within telomeric repeat sequences (Drosopoulos et al, 2015). Therefore, telomere replication is unidirectional and stalled forks may not be rescued from converging forks coming from the end of the chromosome. Telomere fragility may become particularly pronounced during oncogenic transformation. Oncogene-induced hyperproliferation leads to replication stress, causing DNA damage at telomeres and elsewhere in the genome leading to cellular senescence (Bartkova et al, 2006; Di Micco et al, 2006; Suram et al, 2012). Remarkably, oncogene-induced damage at telomeres appears to persist, enforcing oncogene-induced cellular senescence (Fumagalli et al, 2012).
Despite the extensive knowledge on the critical roles of telomere length for cellular senescence and telomere crisis during carcinogenesis, very little is known about changes in telomeric protein components that occur during transformation. The telomerase catalytic subunit hTERT, which associates with telomeres during extension, is a notable exception having well-described key roles to allow cell immortality of cancer cells (Bodnar et al, 1998). In addition, mutations affecting the single-strand telomeric DNA-binding protein POT1 have been reported in a range of malignancies (Quesada et al, 2013; Ramsay et al, 2013; Robles-Espinoza et al, 2014; Calvete et al, 2015). However, a systematic analysis of the telomeric proteome during transformation is missing.
Here, we exploit the established in vitro transformation protocol of Hahn and Weinberg (Hahn et al, 1999, 2002) for oncogenic transformation of human lung fibroblasts (HLFs) by introducing defined genetic elements. We apply the previously developed quantitative telomeric chromatin isolation protocol (QTIP) (Grolimund et al, 2013; Majerska et al, 2017) to compare the telomeric proteomes during different stages of transformation. The telomeric proteome changes most notably upon transduction with the SV40 early region expressing the large T and small t antigens inhibiting p53 and the protein phosphatase 2A, respectively. Up-regulated telomeric proteins include factors that safeguard telomeres from fragility and, therefore, may suppress oncogene-induced replication stress and senescence. We also discover a crucial function for SAM domain and HD domain-containing protein 1 (SAMHD1), which counteracts telomere breakage events in cells depleted of telomere repeat-binding factor 1 (TRF1).
Results
Experimental system to study the telomeric proteome during transformation
We transformed HLFs (Ducrest et al, 2001) using the following consecutive steps. First, HLFs were transduced with a retroviral vector containing the hTERT cDNA. Thus, HLFs expressed active telomerase, stabilized their telomere length around 13 kb (Fig S1), and became immortal. These cells are referred to as HLF-T. Second, we transduced the HLF-T with retrovirus expressing the SV40 early region, encoding large T and small t antigens. These cells, referred to as HLF-TS suppressed p53 and RB function via large T and phosphatase 2A via small t antigens (Hahn et al, 2002). In a final step, HLF-TS cells were transduced with retrovirus containing the H-RasV12 allele, which is a constitutively active version of the H-Ras GTPase, stimulating uncontrolled cell growth in the absence of external signals. These cells were referred to as HLF-TSR. Upon large-scale cell expansion, HLF-T, HLF-TS, and HLF-TSR were indistinguishable with regard to their telomere length, whereas the telomeres in HLFs with inactive telomerase were markedly shorter (Fig S1), because of progressive telomere attrition (Cristofari & Lingner, 2006).
Figure S1. Telomere length analysis of HLF-derived cell lines.
In-gel hybridization analysis of telomeric ssDNA and total DNA in cells used for QTIP experiments. SILAC labeling with light (L) and heavy (H) amino acids for the label-swap replicates is indicated.
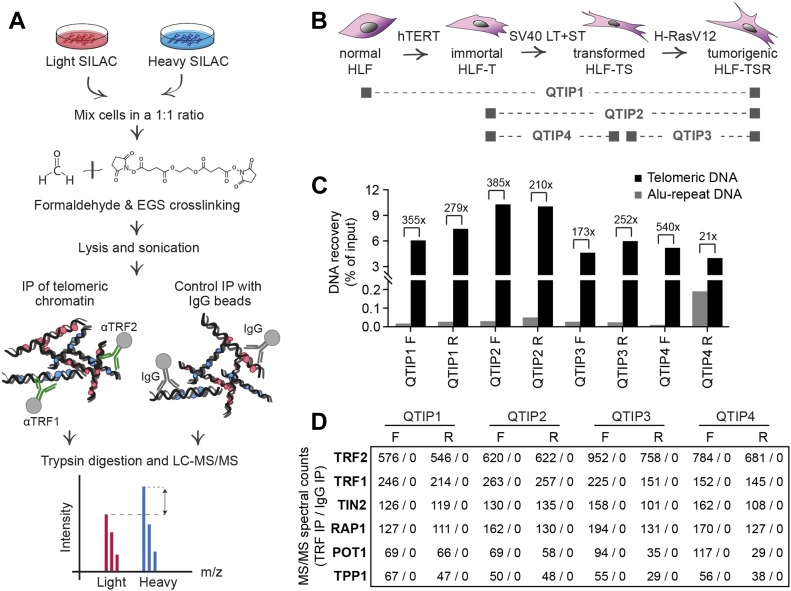
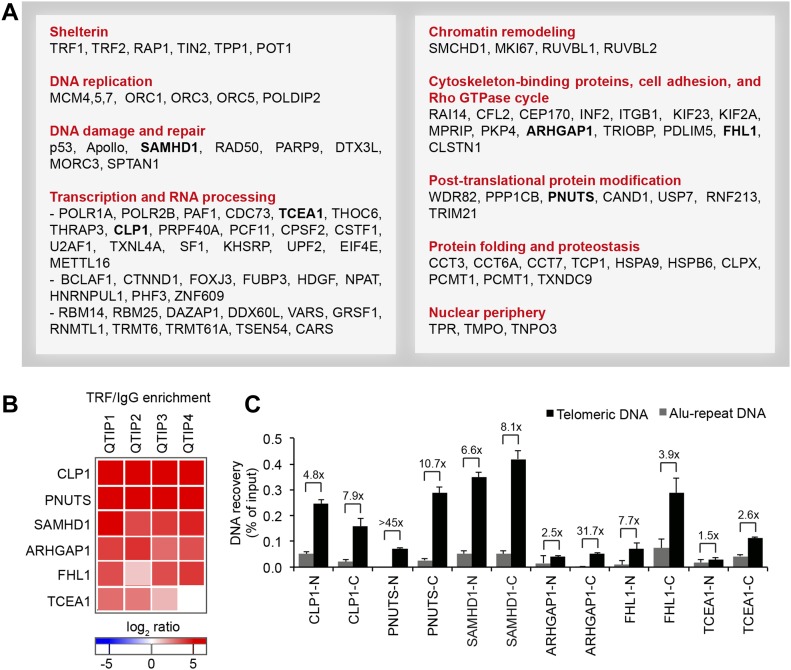
We compared the telomeric protein composition of the four HLF cell lines by QTIP (Grolimund et al, 2013; Majerska et al, 2017) in a pairwise fashion (Fig 1A). Briefly, cells to be compared were grown in different SILAC (stable isotope labeling by amino acids in cell culture; Ong et al, 2002) media to distinguish identical proteins from the pair of cells by their mass and be able to identify quantitative differences in telomere protein abundance. Cell populations to be compared were mixed (Fig 1B) and cross-linked with formaldehyde and ethylene glycol bis(succinimidyl succinate). Chromatin was sonicated and telomeric chromatin isolated using antibodies against the abundant telomeric proteins TRF1 and TRF2. Protein amounts were compared by LC-MS/MS. Replicates were done in which the heavy and light SILAC media were swapped between the two cell populations (Fig S2). The recovery of telomeric DNA was approximately 5–10% and the enrichment of telomeric DNA over Alu repeat DNA around 300-fold (Figs 1C and S3). The spectral counts of shelterin proteins obtained with TRF1/TRF2 antibodies varied from roughly 50 for POT1 and TPP1 to more than 500 for TRF2 (Fig 1D). Shelterin proteins were completely absent from control IgG pulldowns, confirming highest specificity of the purification. Proteins were considered as putatively telomeric when their spectral counts obtained with TRF1/2 antibodies over those obtained with IgG were significantly higher in at least two of the four QTIPs using a Significance A left-sided test (P < 0.05). 134 proteins passed the filtration criteria (Table S1). The identified proteins overlapped partially with proteins that were identified in previous telomeric proteomic screens (Dejardin & Kingston, 2009; Grolimund et al, 2013; Bartocci et al, 2014). These include most prominently the six shelterin proteins but also proteins involved in DNA replication (MCM4, MCM5, and MCM7), repair and end processing (RAD50; Apollo), chromatin remodeling (SMCHD1), and RNA processing (TCEA1) (Fig 2A). In addition, several new telomeric protein candidates were discovered in HLFs. To test their association with telomeric DNA, we expressed selected candidates (Fig 2B) as 3xHA-tagged proteins upon transient transfection in HEK293T cells and tested association with telomeric DNA and Alu repeat DNA by chromatin immunoprecipitation (ChIP) via the HA tags (Figs 2C and S4). HA-tagged CLP1, PNUTS, SAMHD1, ARHGAP1, and TCEA1 all immunoprecipitated with the anti-HA antibody significantly more telomeric DNA than the empty vector control, indicating that they are genuine telomere-associated proteins. The considerably lower but detectable association with Alu repeat DNA suggested association also with other regions of the genome.
Figure 1. QTIP method was used to characterize changes in telomere protein composition during cellular transformation.
(A) Workflow of QTIP. (B) Schematic of the cell line model and overview of the four pairwise QTIP experiments. HLFs were serially transduced with retroviral vectors expressing hTERT, the SV40 large T (LT) and small t (ST) antigens, and H-RasV12. (C) Quantification of precipitated DNA in QTIPs, based on a dot blot hybridized with a specific telomeric probe and a control Alu repeat probe (Fig S3). To determine IP efficiency, the amounts of telomeric DNA in QTIP eluates were quantified and compared with the telomeric DNA in inputs. Fold enrichment of precipitated telomeric DNA compared with precipitated Alu repeat DNA is used as an indicator of IP specificity. Plotted are values from the forward (F) and reverse (R) TRF IP replicates. (D) Enrichment of shelterin subunits in QTIPs. The mean of spectral counts of two replicates is indicated.
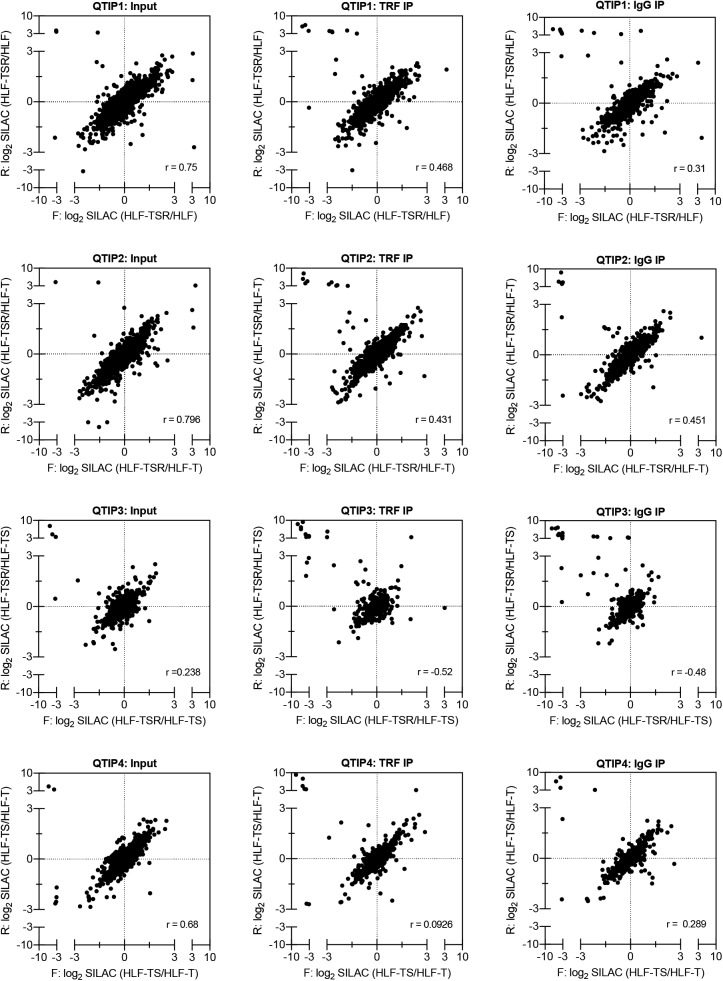
Figure S2. Correlation analysis of label-swap QTIP replicates.
Scatter plots of all identified proteins showing correlation of log2 SILAC ratios from the reciprocal, forward (F) and reverse (R), labeling experiments. One of the two technical replicates of the input sample is shown. External contaminants appear in the upper left quadrant of the scatter plot. r = Pearson’s correlation coefficient.
Figure S3. Dot blots used for quantification of DNA precipitated in QTIP experiments shown in Fig 1C.
Dashed lines indicate omitted or rearranged samples.
Figure 2. Telomeric proteome in human fibroblasts.
(A) Overview of proteins with significant TRF/IgG enrichment in at least two of four QTIPs (Significance A left-sided test, P < 0.05). The full protein list is in Table S1, list B. (B, C) Validation of telomeric localization of selected QTIP candidates by anti-HA ChIP against N- and C-terminally tagged proteins. The precipitated DNA was analyzed as described for Fig 1C. The background of empty vector control was subtracted. Plotted is mean + SD from two to three technical replicates. The images of dot blots are provided in Fig S4.
Figure S4. Dot blots used for quantification of DNA precipitated in anti-HA ChIP experiments shown in Fig 2C.
Dashed lines indicate omitted or rearranged samples.
Identification of telomeric proteins that change during transformation
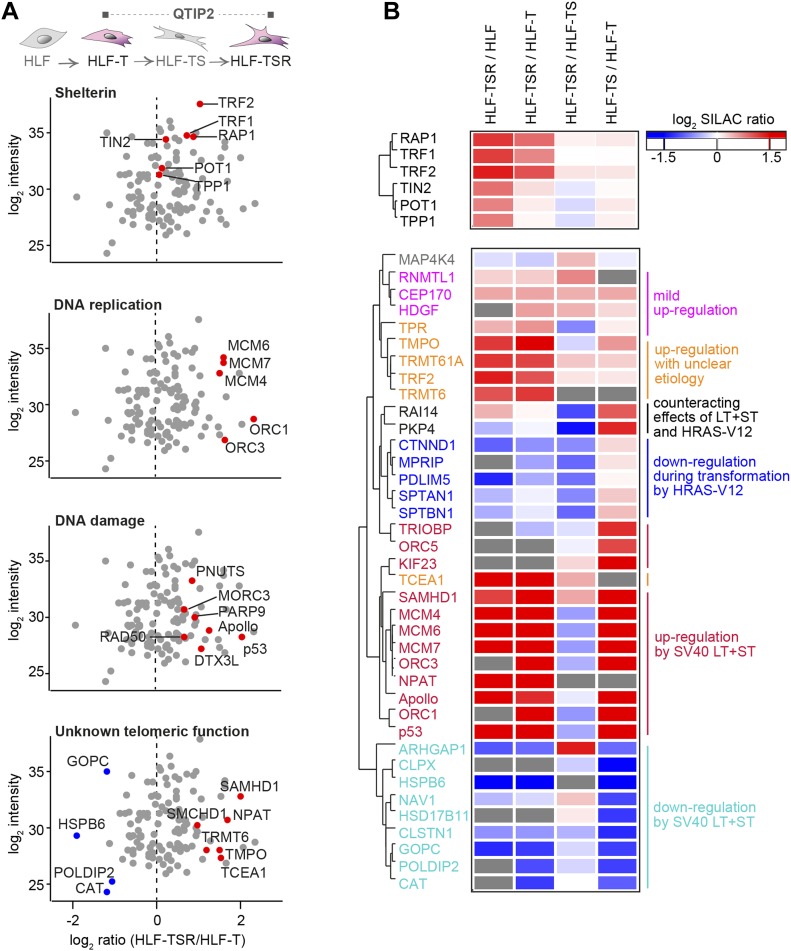
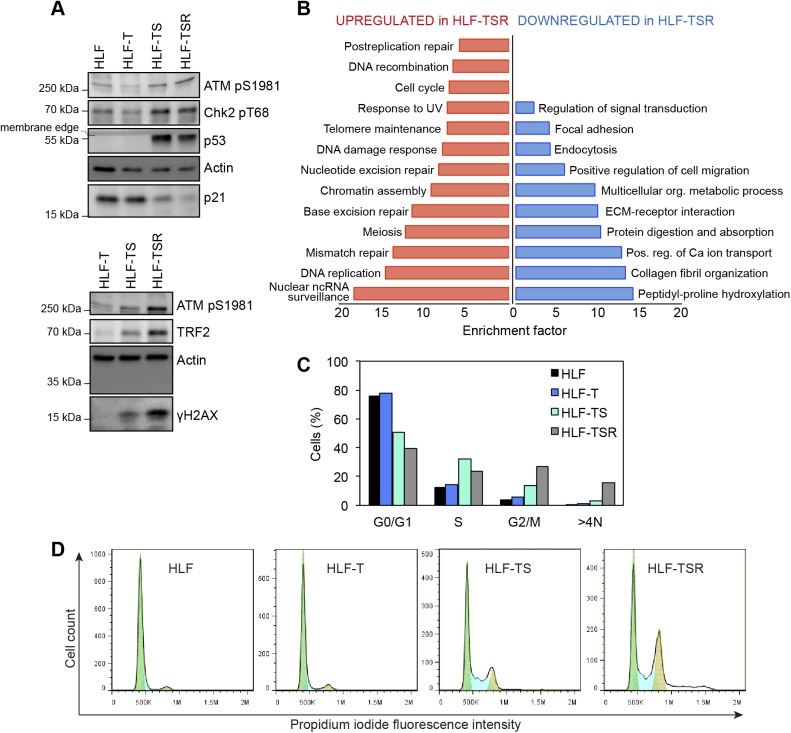
The four comparisons by QTIP (Fig 1B) identified 39 telomeric factors that became either up- or down-regulated during different steps of transformation (Figs 3 and S5, and Table 1). The most notable differences induced during transformation could be ascribed to expression of the SV40 early region. Several proteins involved in DNA replication, DDR, and DNA repair became up-regulated at telomeres upon expression of the T antigens. H-RasV12 expression was associated with milder alterations and contributed to down-regulation of a small subset of proteins of as-yet-unknown telomeric function. In some cases, H-RasV12 slightly counteracted the effect of SV40 T antigens (Fig 3B). The transformation-induced changes did not depend on telomere length because the comparison of HLF-TSR with either HLF or HLF-T cells yielded similar results. Cell transformation also increased the levels of shelterin components TRF1, TRF2, and its interacting protein RAP1, whereas TIN2, TPP1, and POT1 did not change (Figs 3 and S5). Up-regulation of TRF2 was confirmed on Western blots (Fig S6A) and is consistent with previous reports of increased TRF1 and TRF2 expression in human malignancies (Nakanishi et al, 2003; Diehl et al, 2011; Pal et al, 2015; Chen et al, 2017). Overall, our analysis indicated that factors that preserve telomere integrity became up-regulated during transformation. To further corroborate this notion, we analyzed the proteome of chromatin extracts of the four different HLF lines (Table S1). This analysis confirmed that factors which contribute to genome stability are up-regulated during transformation (Fig S6B), whereas the increase of phosphorylated ATM pS1981 and Chk2 pT68 indicated increased DNA damage (Fig S6A). The up-regulation of p53 with SV40 large T expression was expected as SV40 large T inactivates p53, preventing the up-regulation of the p53 target MDM2, which mediates p53 degradation in a negative feedback loop. Consistently, the p53 target p21 decreased upon transduction with SV40 large T, suggesting efficient suppression of p53 function (Fig S6A). Overall, the analysis suggested that the increased DNA damage at telomeres and elsewhere in the genome, which may result from oncogene-induced replication stress and hyperproliferation (Fig S6C and D), is counteracted through SV40 T antigen-induced up-regulation of proteins that contribute to DNA stability and repair.
Figure 3. Identification of transformation-responsive telomeric proteins.
(A) Scatter plots for QTIP2 showing differences in telomere recruitment between HLF-T and HLF-TSR cell lines. Plotted are mean values from the forward and reverse TRF IP replicates. See also Fig S5. (B) Heatmap of differentially regulated telomeric proteins. Top panel: shelterin proteins; bottom panel: putative telomeric proteins that were significantly up-/down-regulated in TRF IP in at least one QTIP (Significance B, both sides, P < 0.05). Missing values are displayed in gray.
Figure S5. Cellular transformation alters telomere protein composition (related to Fig 3).
Scatter plots for QTIPs showing differences in telomere recruitment between indicated cell lines. Plotted are mean values from the forward and reverse TRF IP replicates. A dataset of 134 telomeric proteins (Table S1, list B) was used for the plots.
Table 1.
Transformation-responsive telomeric proteins identified by QTIP, reported functions, and association with cancer.
| Gene name | Major pathway | Canonical function | Cancer association |
|---|---|---|---|
| APOA1 | Lipid metabolism | Participates in cholesterol transport from tissues to liver for excretion; roles in human sperm motility. | (Wang et al, 2009; Su et al, 2010; van Duijnhoven et al, 2011; Jiang et al, 2014; Zamanian-Daryoush & DiDonato, 2015) |
| ARHGAP1 | Rho GTPase signaling | GTPase activator for the Rho, Rac, and Cdc42 proteins. | (Ahn et al, 2012; Li et al, 2017) |
| CAT | Antioxidant activity | Decomposition of hydrogen peroxide. | (Bauer, 2012; Glorieux et al, 2015; Wang et al, 2016) |
| CEP170 | Microtubule dynamics | Role in microtubule organization and cell morphology. | Unknown |
| CLPX | Proteostasis | Component of the mitochondrial unfoldase–peptidase complex. | Unknown (Seo et al, 2016) |
| CLSTN1 | Intracellular transport | Regulates kinesin-mediated cargo transport and organizes microtubule polarity during axon development. | Unknown |
| CTNND1 | Cell adhesion, transcription | Transcription regulation, cell adhesion, Wnt signaling, and spatiotemporal control of small Rho-GTPases. | (Thoreson & Reynolds, 2002; Dann et al, 2014; Kourtidis et al, 2015; Li et al, 2015) |
| DCLRE1B (Apollo) | DNA repair, telomere maintenance | 5′-3′ exonuclease, control of DNA damage repair and topological stress, generation and maintenance of telomeric overhangs, and DNA replication. | Unknown (Natrajan et al, 2007; Karami et al, 2016) |
| GOPC | Proteostasis | Intracellular protein trafficking and degradation. | (Charest et al, 2003; Ohara et al, 2017) |
| HDGF | Transcription regulation | Heparin-binding protein with mitogenic activity. Regulates transcription. | (Hu et al, 2003; Chen et al, 2015; Lian et al, 2015; Wu et al, 2016; Yang et al, 2016) |
| HSD17B11 | Lipid metabolism | Converts 5α-androstan-3α, 17β-diol to androsterone. | (Nakamura et al, 2009) |
| HSPB6 | Proteostasis | Acts as a molecular chaperone. | (Noda et al, 2007; Matsushima-Nishiwaki et al, 2011, 2016; Nagasawa et al, 2014; Qiao et al, 2014; Ju et al, 2015) |
| KIF23 | Cytokinesis | Component of the centralspindlin complex, essential for cytokinesis in Rho-mediated signaling. | (Takahashi et al, 2012; Sun et al, 2015, 2016; Kato et al, 2016) |
| MAP4K4 | Protein kinase | Activates several mitogen-activated protein kinase pathways; involved in cancer cell growth, apoptosis, and migration. | (Qiu et al, 2012; Haas et al, 2013; Feng et al, 2016; Liu et al, 2016; Gao et al, 2017) |
| MCM4, 6, 7 | DNA replication | Components of the MCM2–7 DNA replicative helicase. | (Honeycutt et al, 2006; Shima et al, 2007; Bagley et al, 2012; Das et al, 2013; Kwok et al, 2015) |
| MPRIP | Cytoskeleton regulation | Targets myosin phosphatase to the actin cytoskeleton. Regulation of the actin cytoskeleton by RhoA and ROCK1. | (Ono et al, 2008) |
| NAV1 | Cytoskeleton regulation | Regulation of the microtubule cytoskeleton, important for neuronal development, and interacts with the RhoGEF TRIO. | Unknown |
| NPAT | Transcription, cell cycle | Required for progression through the G1 and S phases, for S phase entry, and for activation of histone gene transcription. | (Saarinen et al, 2011; Hamdi et al, 2017) |
| ORC1, 3, 5 | DNA replication | Components of the origin recognition complex; required for the assembly of pre-RC. | Unknown (Champeris Tsaniras et al, 2014) |
| PDLIM5 | Calcium signaling | Regulates intracellular calcium levels by linking calcium channel and PKC. | (Eeles et al, 2009; Kim et al, 2010) |
| PKP4 | Cell adhesion | Regulates RhoA signaling during cytokinesis. | Unknown |
| POLDIP2 | DNA replication and repair | DNA replication and damage repair, also implicated in mitochondrial function, extracellular matrix regulation, cell cycle progression, focal adhesion, and cell migration. | (Grinchuk et al, 2010; Chian et al, 2016) |
| RAI14 | Cytoskeleton regulation | May have roles in human testis development and spermatogenesis. | Unknown |
| RNMTL1 | RNA processing | Catalyses the formation of 2′-o-methylguanosine at position 1370 in the 16S mitochondrial large-subunit rRNA. | (Haiman et al, 2013) |
| SAMHD1 | dNTP and nucleic acid metabolism | dNTPase activity, has roles in DNA repair, innate immunity, cancer, and HIV-1 restriction; controversial nuclease activity. | (Clifford et al, 2014; Shi et al, 2014; Wang et al, 2014; de Silva et al, 2014; Merati et al, 2015; Rentoft et al, 2016) |
| SPTAN1 | Regulation of cytoskeleton | A scaffolding protein involved in cell adhesion and motility; implicated in repair of DNA interstrand cross-links. | (Tuominen et al, 1996; Gorman et al, 2007; Cunha et al, 2010; Wolgast et al, 2011; Hinrichsen et al, 2014) |
| SPTBN1 | Regulation of cytoskeleton | Scaffolding function in protein sorting, cell adhesion, and migration; implicated in TGF-β signaling. | (Gorman et al, 2007; Jiang et al, 2010; Yao et al, 2010; Zhi et al, 2015) |
| TCEA1 | Transcription | Required for efficient RNA polymerase II transcription elongation past template-encoded arresting sites. | (Hubbard et al, 2008; Shema et al, 2011) |
| TERF2 | Telomere maintenance | Required for telomere capping and protection. Inhibits nonhomologous end joining and ATM activation at telomeres. Required for t loop formation. | (Nakanishi et al, 2003; Bellon et al, 2006; Munoz et al, 2006; Diehl et al, 2011; Biroccio et al, 2013; Pal et al, 2015; Chen et al, 2017) |
| TMPO | Nuclear architecture | Involved in the structural organization of the nucleus and in the post-mitotic nuclear assembly. | (Brachner & Foisner, 2014; Zhang et al, 2016) |
| TP53 | DDR, cell cycle | Transcription factor inducing cell cycle arrest/senescence and apoptosis; induction of DNA repair genes. | (Muller et al, 2011; Muller & Vousden, 2013; Kaiser & Attardi, 2017) |
| TPR | Nucleocytoplasmic transport | Scaffolding element of the nuclear pore complex essential for normal nucleocytoplasmic transport of proteins and mRNAs. | (David-Watine, 2011) |
| TRIOBP | Cytoskeleton regulation | Might be involved in actin remodeling, directed cell movement, and cell cycle regulation. | (Bao et al, 2015; Thutkawkorapin et al, 2016) |
| TRMT6, TRMT61A | tRNA modification | Catalyses the formation of N1-methyladenine at position 58 in initiator methionyl-tRNA. | (Shi et al, 2015; Macari et al, 2016) |
Figure S6. Cellular transformation is associated with increased DNA damage burden and hyperproliferation.
(A) Immunoblot analysis of DDR, performed on whole-cell lysates. Actin is used as a loading control. (B) Gene annotation enrichment analysis of proteins that were significantly up- or down-regulated in chromatin-enriched extracts from HLF-TSR when compared with HLF-T. The extracts were analyzed in two biological and two technical replicates, and the mean HLF-TSR/HLF-T SILAC ratios were tested for outliers (Significance B, respective side, P < 0.05). This produced datasets of 85 up-regulated and 91 down-regulated proteins that were separately tested for enriched gene annotation terms when compared with the full dataset of 1,537 proteins (Fisher’s exact test, Benjamini–Hochberg FDR < 0.02). Plotted are overrepresented “KEGG pathway” and “GO Biological Process” categories. (C, D) Cell cycle profiles of HLF-derived cell lines. Cells were analyzed by flow cytometry after propidium iodide staining. The percentages of cells in G0/G1, S, and G2/M phases were determined using the Watson Pragmatic computational model in FlowJo.
TERRA is up-regulated during cellular transformation
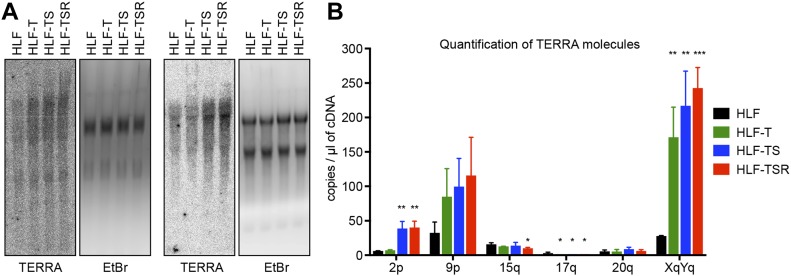
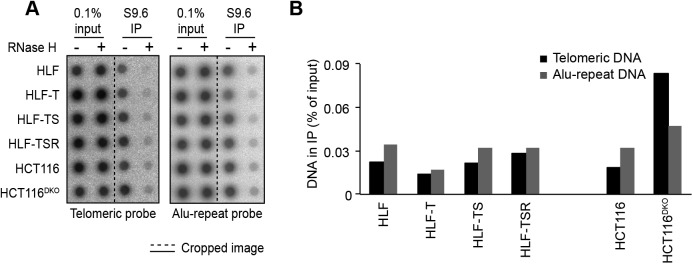
The telomeric long noncoding RNA TERRA has been implicated among others in the telomeric DDR and the regulation of telomerase (Azzalin & Lingner, 2015). In addition, TERRA can form RNA/DNA hybrid structures at telomeres, which represent obstacles for the replication machinery (Balk et al, 2013; Pfeiffer et al, 2013) while promoting homology-directed repair (Arora et al, 2014; Graf et al, 2017). We measured overall TERRA levels on Northern blots (Fig 4A) and TERRA molecules stemming from specific chromosome ends by quantitative RT-PCR (RT–qPCR) (Feretzaki & Lingner, 2017) (Fig 4B). This analysis revealed that the overall TERRA levels increased with every step of transformation. The analysis of individual TERRA molecules showed telomere end-specific regulation. TERRA stemming from telomeres 9p and XpYp increased most notably upon transduction with hTERT, whereas 2p TERRA increased upon transduction with the SV40 early region (Fig 4B). 15q, 17q, and 20q TERRA were not up-regulated during transformation. We also tested if the increased TERRA levels gave increased RNA/DNA hybrids at telomeres. The S9.6 monoclonal antibody, which recognizes RNA/DNA hybrids (Hu et al, 2006), was used to perform DNA–RNA immunoprecipitation (Fig S7). The immunoprecipitated nucleic acids contained telomeres, indicating the presence of RNA/DNA hybrid structures consistent with previous analyses (Arora et al, 2014; Sagie et al, 2017). The signal was abolished upon pretreatment with RNase H, which destroys the RNA part in RNA/DNA duplexes, demonstrating the specificity of the assay. However, the amounts of RNA/DNA hybrids at telomeres and at Alu repeats did not change notably during transformation, indicating that increased hybrids at telomeres are not responsible for increased replicative stress that may be induced during transformation.
Figure 4. TERRA levels are elevated during transformation.
(A) Northern blot analysis of total RNA from the HLF-derived cell lines. TERRA was detected using a telomeric DNA probe complementary to the UUAGGG repeats. Ethidium bromide (EtBr) staining is shown as a loading control. Two independent biological replicates are shown. (B) TERRA quantification by RT–qPCR with primers specific for the indicated subtelomeric sequences. Plotted is mean + SD from three biological and two technical replicates. Two-tailed unpaired t test, comparing all derived cell lines with primary HLFs (*P < 0.05; **P < 0.01; ***P < 0.001; bars lacking asterisks are not significant).
Figure S7. Cellular transformation is not accompanied by accumulation of R-loops.
(A) Detection of telomeric R-loops by immunoprecipitation using the S9.6 antibody that recognizes DNA/RNA hybrids. As a negative control, half of each nucleic acid preparation was treated with RNase H before the IP. The precipitated DNA was spotted on a membrane and hybridized either with a specific telomere probe (Telo) or a control Alu repeat probe (Alu). Accumulation of telomeric R-loops upon double knockout (DKO) of DNA methyltransferases DNMT1 and DNMT3b in HCT116 cells served as a positive control. (B) Quantification of the S9.6 DNA/RNA IP in (A). The amounts of precipitated DNA were quantified as compared with the total DNA in the inputs. The background RNAse H–resistant signals were subtracted.
Up-regulated proteins prevent telomere fragility
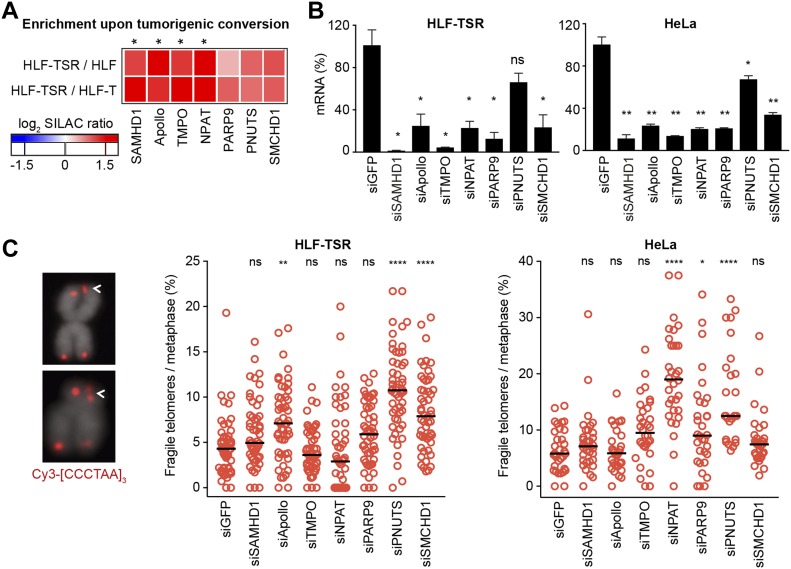
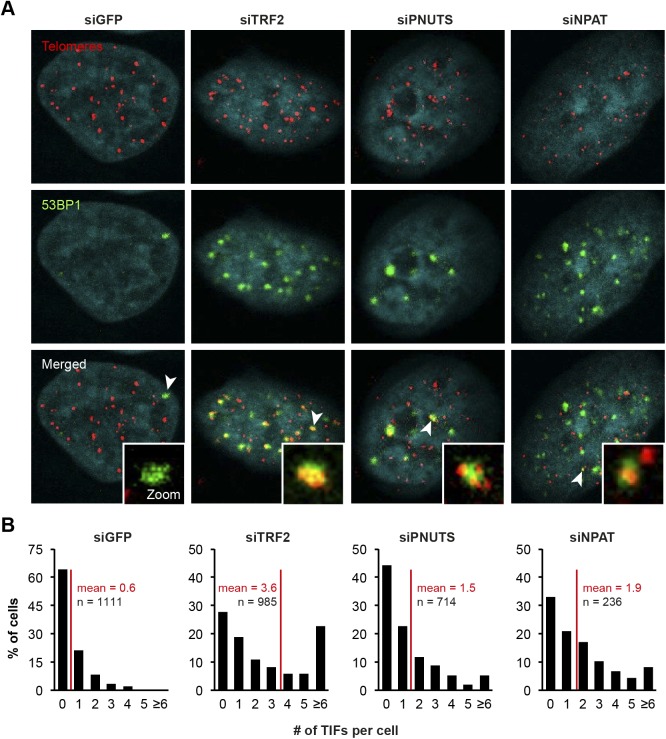
To test the hypothesis that during transformation, up-regulated proteins act against replicative stress at telomeres, we depleted candidate proteins (Fig 5A) using siRNA pools in HLF-TSR and HeLa cells. Measuring mRNA levels by RT–qPCR (Fig 5B) revealed efficient depletion of all candidates except for PNUTS, whose depletion strongly impacted on cell viability. Metaphase chromosomes were prepared and telomeric signals were detected by FISH. Replication stress at telomeres gives the so-called fragile telomeres, which are characterized by a smeary telomeric signal or multiple telomeric signals (Fig 5C). Telomere loss, apposition, and breakage (“outside”) events were also scored (Table S2). Quantification revealed increases in telomere fragility upon depletion of several candidates. Depletion of NPAT scored only positively in HeLa cells. NPAT has known roles in transcriptional activation of histones but was not known to be also telomeric (Zhao et al, 2000). Its depletion caused accumulation of genome-wide 53BP1 foci, a subset of which colocalized with telomeres (Fig S8). SAMHD1 plays, among others, roles in homology-directed repair (Daddacha et al, 2017). Its roles at telomeres are further characterized and discussed below. DCLRE1B/SNMB1/Apollo is a DNA 5′-3′ exonuclease with roles in telomere end processing (Lenain et al, 2006; van Overbeek & de Lange, 2006; Wu et al, 2012). Apollo also relieves topological stress during telomere replication (Ye et al, 2010). TMPO is a lamina-associated protein with roles in nuclear architecture and thus may contribute to telomere localization (Harris et al, 1995). PARP9 is a poly(ADP-ribose) polymerase involved in DNA repair (Yang et al, 2017). PNUTS binds and regulates protein phosphatase 1, which among others is involved in the telomeric DDR, DNA repair, and DNA replication origin licensing (Kim et al, 2009; Landsverk et al, 2010; Hiraga et al, 2017). PNUTS depletion caused accumulation of genome-wide 53BP1 foci, a subset of which colocalized with telomeres (Fig S8). SMCHD1 is required for X chromosome inactivation (Nozawa et al, 2013). It preferentially associates with long telomeres (Grolimund et al, 2013) and has reported roles in DNA repair (Coker & Brockdorff, 2014; Tang et al, 2014).
Figure 5. Cellular transformation up-regulates proteins that help preserve telomere integrity.
siRNA screens were performed to test the effect of candidate depletion on telomere structure visualized on metaphase chromosome spreads. (A) Heatmap showing behavior of the selected candidates in QTIPs. Reported are mean SILAC ratios from the forward and reverse TRF IP replicates. The asterisks indicate proteins that were classified as significantly “transformation responsive” in the QTIP screen (Table S1, list C). (B) RT–qPCR to verify candidate depletion using siRNA pools in HLF-TSR and HeLa cells. Knockdown efficiency was compared with the mRNA levels in cells transfected with control siRNA against GFP. Plotted is mean + SD from two biological and two technical replicates for HLF-TSR and from two technical replicates from one representative experiment for HeLa. Two-tailed unpaired t test (*P < 0.05; **P < 0.01; ns = not significant). (C) Analysis of fragile telomeres upon candidate depletion. Indicated telomere aberrations were scored on metaphase spreads with telomeres detected by FISH with a Cy3-[CCCTAA]3 probe (red) and DNA stained with DAPI (gray). HLF-TSR: >50 metaphases from two independent experiments (except for siGFP samples, for which data from three independent replicates) were analyzed for each condition. HeLa: >25 metaphases were analyzed per condition in a single experiment. The black line represents median. One-way ANOVA with Dunnett’s multiple comparisons test, comparing all conditions to siGFP (*P < 0.05; **P < 0.01; ****P < 0.0001; ns = not significant). See also Table S2.
Figure S8. Depletion of PNUTS and NPAT induces a global DDR.
(A) Induction of telomere dysfunction–induced foci (TIF) upon candidate knockdown in HeLa cells from Fig 5. Depletion of TRF2 was used as a positive control. 53BP1 was detected by indirect IF and telomeres by FISH with a Cy3-[CCCTAA]3 probe. (B) Frequency distribution of the number of TIFs per cell from (A). n indicates the number of cells analyzed for each condition and red lines mark the mean.
SAMHD1 counteracts telomere breakage in TRF1-depleted cells
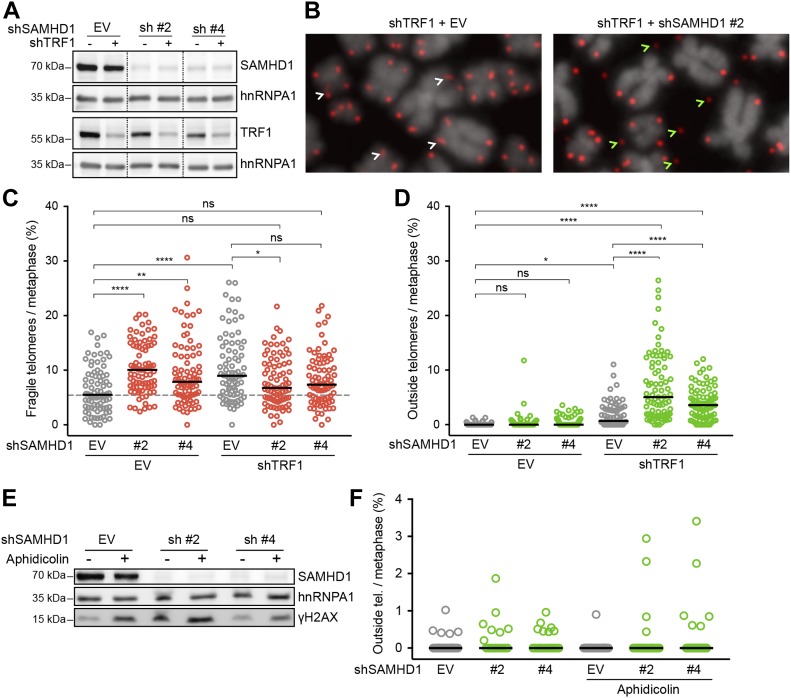
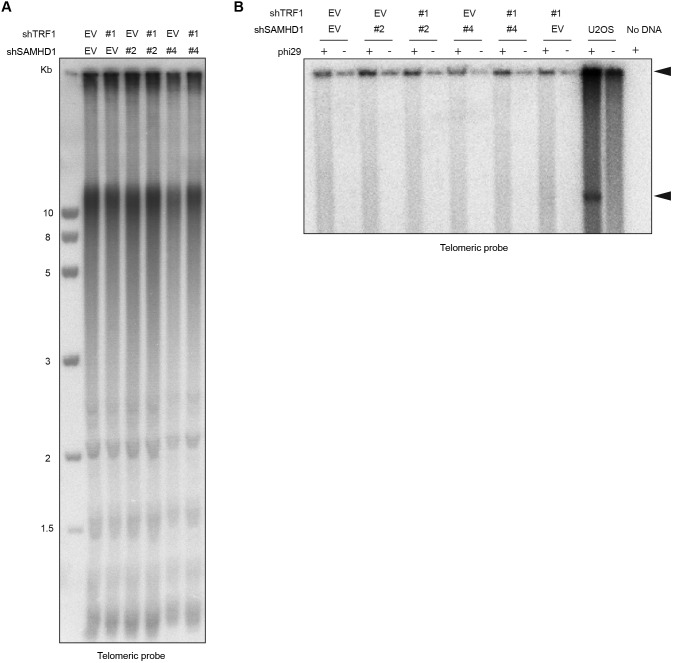
SAMHD1 has been implicated in homology-directed repair of DNA double-strand breaks, recruiting CtIP to promote DNA end resection (Daddacha et al, 2017). In addition, SAMHD1 has been recently shown to stimulate processing of stalled replication forks (Coquel et al, 2018). To assess the roles of SAMHD1 in curbing replication stress or maintenance of telomere intactness, we depleted it in HeLa cells using shRNAs (Fig 6A). As a control, we induced local replication stress at telomeres by depleting TRF1. TRF1 prevents telomere fragility presumably through the recruitment of the BLM and RTEL1 helicases (Sfeir et al, 2009). SAMHD1 depletion enhanced telomere fragility slightly (Figs 5 and 6), whereas TRF1 depletion showed a stronger effect (Fig 6B and C, and Table S3). Strikingly, we observed that the co-depletion of SAMHD1 and TRF1 caused a novel telomere phenotype we referred to as “outsider” in which the telomere signal was detached from the telomeric chromatin of metaphase chromosomes (Fig 6B and D). No notable effects on telomere length could be observed in a TRF analysis in this short-term experiment (Fig S9). Furthermore, the telomeres did not score positive for the presence of extrachromosomal DNA in the form of t-circles, which have been detected in cells that utilize the alternative lengthening of telomeres (ALT) mechanism (Cesare & Griffith, 2004). The most straightforward interpretation of these results is that telomeres break in the absence of TRF1 during replication and that SAMHD1 is required for their repair.
Figure 6. SAMHD1 is critical for telomere integrity in TRF1-depleted but not in aphidicolin-treated cells.
(A–D) Characterization of telomere aberrations on metaphase chromosomes induced by SAMHD1 knockdown in TRF1-proficient and TRF1-deficient HeLa cells. (A) Immunoblotting analysis of knockdown efficiency in cells treated with the indicated shRNAs for 6 d. SAMHD1 and TRF1 are shown on two separate membranes. hnRNPA1 is used as a loading control. Irrelevant lanes have been omitted from the image (dashed lines). (B) Representative metaphase chromosome spreads with telomeres detected by FISH with a Cy3-[CCCTAA]3 probe (red) and DNA stained with DAPI (gray). White arrows point to typical fragility (smears and multiple telomeric signals), whereas green arrows indicate outside telomeres (i.e., a telomeric signal positioned outside the DAPI signal). (C, D) Quantification of fragile and outside telomeres from cells in (A). 81 metaphases from three independent experiments were analyzed for each condition. The black line represents median. One-way ANOVA with Tukey’s multiple comparisons test (*P < 0.05; **P < 0.01; ****P < 0.0001; ns = not significant). (E, F) Effect of SAMHD1 depletion on telomere structure in aphidicolin-treated HeLa cells. (E) Immunoblot verification of SAMHD1 depletion and DDR induction. Where indicated, the cells were treated with 0.1 μg/ml aphidicolin for 20 h before harvest. (F) Quantification of outside telomeric FISH signal on metaphase chromosome spreads from cells in (E). Shown are data from a representative experiment with ≥17 metaphases and >3,000 telomeres analyzed per condition. Differences are not statistically significant. See also Table S3.
Figure S9. Co-depletion of TRF1 and SAMHD1 does not lead to rapid telomere shortening.
(A) TRF analysis of genomic DNA prepared from HeLa cells transfected with indicated pSuper plasmids. Genomic DNA was digested overnight with HinfI and RsaI and fractionated on an agarose gel. (B) Phi29-dependent telomeric circles (T-circles) amplification assay. Genomic DNA prepared from HeLa cells transfected with indicated pSuper plasmids was digested overnight with HinfI and RsaI, and 0.75 μg of DNA was used for phi29-dependent amplification reaction. Genomic DNA from U2OS cell line was used as a positive control. Arrows indicate T-circle amplification products.
We then tested if SAMHD1 depletion also triggers outsider telomeres in cells in which telomere fragility was induced by treatment with the DNA polymerase α and δ inhibitor aphidicolin (Figs 6E and F, and Table S3). However, the outsider phenotype was not triggered in this setting probably reflecting the different defects in TRF1-depleted versus DNA polymerase–inhibited cells.
Discussion
Changes in telomere length have been known for several decades to promote two key events during tumorigenesis, cellular senescence, and telomere crisis. Here, we discover that cellular transformation also involves crucial changes in telomere protein composition. We identify 134 proteins that are enriched in telomeric chromatin fractions of human fibroblasts. Importantly, 39 of these changed significantly in abundance at telomeres during oncogenic transformation. Many of the up-regulated proteins had been linked to cancer development in previous studies (Table 1); however, the link between their telomere recruitment and tumorigenesis remains unexplored. Most up-regulated telomeric factors play roles in either DNA replication, DNA repair, or telomere protection. The pre-replication complex (pre-RC) components ORC1, 3, and 5 and MCM4, 6, and 7 may be recruited to telomeres via TRF2, which becomes more abundant during transformation. It remains to be tested if the recruited pre-RC components form active origins. Origin firing within telomeres occurs rarely, but dormant origins within telomeres may be activated upon stalling of replication forks that enter the telomeres from subtelomeric regions (Drosopoulos et al, 2015). We also identify Apollo/DCLRE1B as a transformation up-regulated factor, which has been reported to counteract topological stress at telomeres (Ye et al, 2010). In support of a role in curbing replication stress, Apollo/DCLRE1B depletion increased telomere fragility. In addition, we identify six novel factors that were up-regulated during transformation and whose depletion caused telomere fragility.
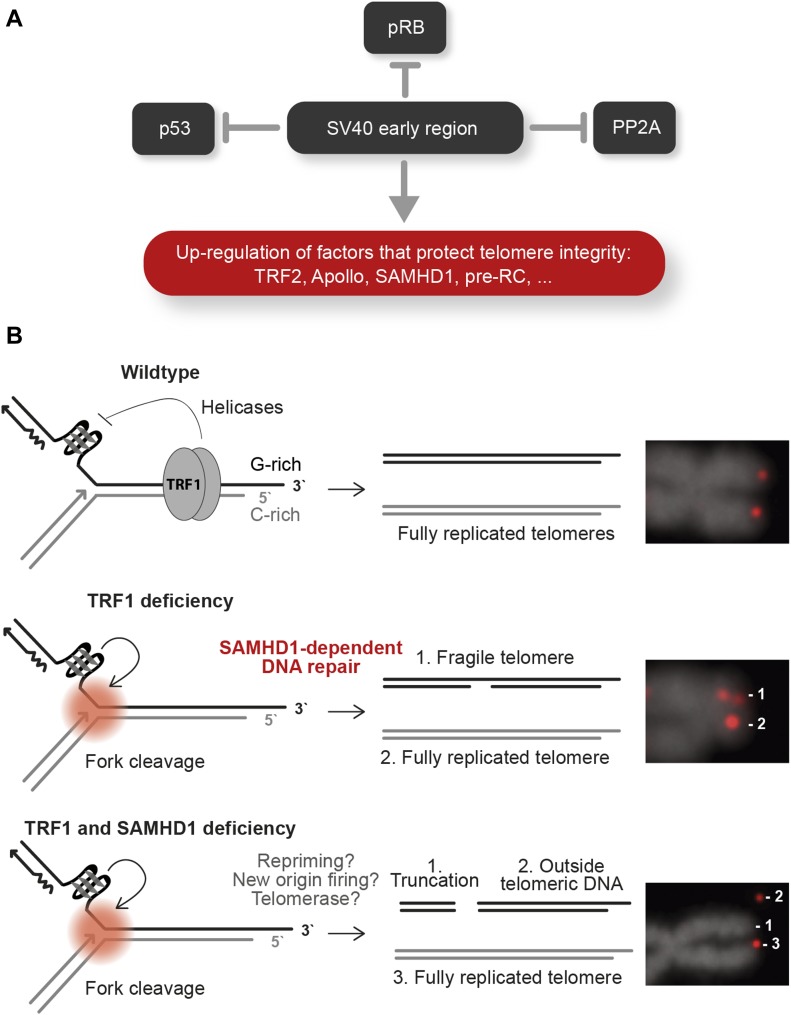
Oncogene-induced hyperproliferation leads to replication stress, possibly involving depletion of nucleotides or increased collisions with transcription complexes, although the exact mechanism remains unknown (Bartek et al, 2007; Hills & Diffley, 2014). During transformation of HLFs, TERRA levels but not DNA/RNA hybrids increased. Thus, increased telomere transcription may contribute to telomere fragility, although other mechanisms should not be excluded. We propose that the up-regulation of factors that suppress telomere fragility is critical to resist oncogene-induced replication stress at telomeres during transformation. The fragility-suppressing factors were induced upon expression of SV40 T antigens inactivating p53, RB, and protein phosphatase 2A. Expression of oncogenic H-RasV12, which was introduced as a last step in our transformation protocol, did not contribute to their up-regulation. During natural carcinogenesis, oncogene-induced hyperproliferation precedes inactivation of p53 and RB and therefore oncogene-induced senescence may be promoted (Halazonetis, 2004; Gorgoulis et al, 2005; Bartkova et al, 2006; Di Micco et al, 2006; Halazonetis et al, 2008; Suram et al, 2012). Our results suggest that targeting of p53, RB, and PP2A by SV40 T antigens not only alleviates checkpoint-induced cell cycle control but also triggers expression of factors that suppress DNA damage upon replication stress, which is prevalent at telomeres (Fig 7A).
Figure 7. Hypothetical model for transformation-associated changes at telomeres.
(A) The SV40 large T and small t antigens inhibit p53, RB, and PP2A. In addition, SV40 early gene expression leads to up-regulation of factors, which promote telomere protection and replication. (B) Telomere replication requires TRF1. In the absence of TRF1, G-quadruplex structures accumulate at telomeres, which become cleaved during replication and repaired by SAMHD1-dependent repair. In the absence of both TRF1 and SAMHD1, double-strand breaks are retained. Telomere synthesis proceeds downstream of the breaks mediated by replication fork restart, telomerase, or replication fork firing from within telomeric repeats.
One of the top candidates up-regulated upon transformation of HLFs was SAMHD1. Therefore, we decided to explore its functions at telomeres. SAMHD1 is a dNTPase, which restricts HIV-1 replication (Laguette et al, 2011). SAMHD1 is mutated in cancer (Clifford et al, 2014; Merati et al, 2015; Rentoft et al, 2016) and in the neurodegenerative autoimmune disorder Aicardi–Goutières syndrome in which aberrant nucleic acids accumulate, inducing innate immunity (Ballana & Este, 2015). More recently, SAMHD1 functions in homology-directed DNA repair (Daddacha et al, 2017) and DNA replication (Coquel et al, 2018) have been discovered. Specifically, SAMHD1 recruits CtIP to DNA double-strand breaks to facilitate end resection and homologous recombination (HR) (Daddacha et al, 2017). We find that double depletion of TRF1 and SAMHD1 gives outsider telomeres, which seem broken off from the chromosome arms. We suspect that this phenotype can be ascribed to the functions of SAMHD1 in HR and we propose the following model (Fig 7B). TRF1 depletion gives telomere fragility because of inefficient recruitment of the G-quadruplex unwinding helicases BLM and RTEL1. Therefore, replication forks may stall and are eventually processed into DNA double-strand breaks near the telomere–subtelomere boundary. In SAMHD1-proficient cells, the breaks are repaired by homology-directed repair. In SAMHD1-deficient cells, the broken forks remain unrepaired. However, the telomeric FISH signals of outsider telomeres indicate that telomere replication does proceed downstream of the break, possibly triggered by repriming events or activation of telomeric origins that converge toward the double-strand break (Pasero & Vindigni, 2017). A contribution of telomerase in promoting telomere synthesis downstream of the broken fork cannot be excluded. The latter was seen in fission yeast in which the TRF1-ortholog Taz1 was deleted (Miller et al, 2006). Loss of Taz1 led to stalled replication forks at telomeres, which could be maintained only in the presence of telomerase.
Overall, our study underlines the importance of telomere replication stress in cellular transformation. It identifies a number of novel factors that are induced by SV40 T antigens to counteract telomere fragility. Deregulation of these proteins at telomeres might play an unrecognized role in the development of cancer, as exemplified by SAMHD1, which has been found mutated in cancer cells and seems to have a pivotal function in the preservation of telomere integrity. This work will stimulate further studies to unravel the mechanisms that underlie telomere replication stress and oncogene-induced senescence during transformation.
Materials and Methods
Cell lines
The following cell lines were used: HeLa; HEK293T; HLF (Ducrest et al, 2001); HLF-T—HLF cells stably infected with hTERT expressing retroviruses; HLF-TS—HLF-T cells stably infected with SV40 early region expressing retroviruses; HLF-TSR—HLF-TS cells stably infected with H-RasV12 expressing retroviruses; HCT116; and HCT116DKO—HCT116 cells with genetic disruption of DNMT1 and DNMT3b (Rhee et al, 2002). Cells were maintained in DMEM (Gibco) containing 10% FBS (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified incubator containing 5% CO2. Conditions for SILAC labeling were reported previously (Grolimund et al, 2013; Majerska et al, 2017).
Plasmids
Recombinant retroviruses were produced in HEK293T cells transfected with a pBABE expression vector (pBABE-hygro-hTERT [Cristofari & Lingner, 2006], pBABE-puro-HRasV12 [9051; Addgene], pBABE-neo-largeTgenomic [10891; Addgene]), and pcl-Ampho packaging vector (Naviaux et al, 1996). For validation of telomeric localization of candidate proteins by anti-HA ChIP, cDNAs from HLF-TSR cells were amplified by PCR and introduced by InFusion cloning (Clontech) into pcDNA6-derived vectors (Invitrogen) for expression as 3xHA-tagged proteins. The shRNA-expressing vectors were generated by cloning the corresponding double-stranded DNA oligonucleotides into pSuper vectors digested with BglII and HindIII. The targeting sequences are as follows: pSuper-blast-shSAMHD1 #2 5′-CGCATGCTGAAGCTAAGTA-3′; pSuper-blast-shSAMHD1 #4 5′-GTATCGCATTTCTACAGCA-3′; and pSuper-puro-shTRF1 5′-GAATATTTGGTGATCCAAA-3′.
QTIP
QTIP experiments were carried out as described previously (Grolimund et al, 2013; Majerska et al, 2017) with the following modifications. 240–490 × 106 cells of each cell type were used per IP. Upon mixing the two cell types in a 1:1 ratio, cells in suspension were cross-linked for 10 min at 25°C using a combination of 1% formaldehyde and 2 mM ethylene glycol bis(succinimidyl succinate). Chromatin-enriched pellets were resuspended at 25 × 106 cells/ml in LB3 buffer (10 mM Tris–HCl, pH 8.0; 200 mM NaCl; 1 mM EDTA–NaOH, pH 8.0; 0.5 mM EGTA–NaOH, pH 8.0; 0.1% w/v sodium deoxycholate; 0.25% w/v sodium lauryl sarcosinate; and EDTA-free protease inhibitor complex [Roche]) and sonicated for 12 min at 4°C using the Focused-ultrasonicator (Covaris E220; 12 × 12-mm glass tubes with AFA fiber; duty: 5.0, peak incident power: 140, cycles: 200, amplitude: 0, velocity: 0, and dwell: 0). Immunoprecipitation was performed using beads covalently coupled with nonspecific IgGs (sc-2027; Santa Cruz Biotechnology) or beads coupled to affinity-purified anti-TRF1 (rabbit #714) and anti-TRF2 antibodies (rabbit #40) according to the previously published protocol (Aeby et al, 2016).
Mass spectrometry analysis and data processing
LC-MS/MS analysis was performed as described previously (Aeby et al, 2016). Raw MS data were analyzed using MaxQuant software (Cox & Mann, 2008) with FDR < 0.01 at the level of both proteins and peptides. Peak lists were searched against the human Uniprot database, using Arg10 and Lys8 as heavy labels (multiplicity of 2). The mass tolerance was set to 7 ppm for precursor ions and MS/MS accuracy was set at 0.5 D. Enzyme was set to trypsin with up to two missed cleavages. Proteins and peptides (minimum six amino acids) were identified using a target-decoy approach with a reversed database. At least two (unique + razor) peptides were required for protein identification. A cutoff was set to 0.1 for posterior error probability.
TRF/IgG enrichment was determined using MS/MS spectral counts, which were calculated from the evidences of protein groups. For each protein group, we summed up the MS/MS counts for each experiment. At least three MS/MS spectral counts were required per condition. The missing values were imputed with pseudo-counts of 0.5. The normalization of the spectral counts was performed following the normalization schema in Scaffold (http://www.proteomesoftware.com/products/scaffold/). Further statistical analyses and graphical displays were performed in Perseus software version 1.5.3.2 (http://www.coxdocs.org/), using mean values of the forward and reverse QTIP replicates. The expression values and ratios were log2-transformed and normalized by subtracting the median. Proteins enriched in TRF IP over control IgG IP (i.e., showing significant TRF/IgG ratios) were calculated separately for each QTIP as statistically significant outliers using a Significance A left-sided test (P < 0.05). Only proteins with significant TRF/IgG ratio in at least two of four QTIPs were considered “telomeric.” Upon manual inspection, proteins HRG and SERPINC1 were removed from the list of telomeric proteins because they were light labeled in both label-swap experiments, indicating they were external contaminants. Significant SILAC ratios were determined for each QTIP (both, for inputs and TRF IPs) using a Significance B two-sided test (both sides, P < 0.05). Proteins with a significant SILAC ratio in at least one of the four QTIPs were considered “transformation responsive.” To create heatmaps, hierarchical clustering was performed in Perseus based on average Euclidean distance of the log2 mean SILAC ratios. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al, 2016) partner repository with the dataset identifier PXD010088.
siRNA and plasmid transfection
The cells were transfected with siRNA pools (5 pmol of each siRNA per well of a six-well plate) by calcium phosphate precipitation twice with a 36-h interval and harvested 80–100 h after the first transfection. siRNA oligonucleotides were purchased from Qiagen: SAMHD1 (SI00710500, SI04137217, SI04189332, and SI04243673), DCLRE1B (SI00134757, SI02778692, and SI02778699), TMPO (SI03215730, SI04951156, and SI04951163), NPAT (SI00660814, SI04343157, and SI04355708), PARP9 (SI04136244, SI04196108, SI04212642, and SI04247285), PNUTS (SI00041811, SI00041832, SI03032666, and SI03088141), and SMCHD1 (SI00454664 and SI00454678). siGFP 5′-GCAGCACGACTTCTTCAAGTT-3′ was synthesized at Microsynth. For transient overexpression of 3xHA-tagged proteins, HEK293T cells in a 15-cm tissue culture dish were transfected with 30 μg of plasmid DNA using calcium phosphate precipitation. Cells were harvested for ChIP analysis 42–48 h post-transfection. HeLa cells were transfected with pSuper-Puro and pSuper-Blast constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. 1 μg/ml puromycin and 5 μg/ml blasticidin were added to the medium 1 d after the transfection, and selection was maintained for 5 d.
Chromatin immunoprecipitation
HEK293T cells were cross-linked with 1% formaldehyde for 25 min, resuspended in LB3 buffer (20-30 × 106 cells/ml), and sonicated for 30 min at 4°C using a Focused-ultrasonicator as described in the QTIP protocol. The sonicated extracts were centrifuged at 4°C for 15 min at 20,000 g and the supernatant was mixed with four volumes of ChIP dilution buffer and precleared for 1 h at 4°C with Sepharose 6B (Sigma-Aldrich) pre-blocked with yeast tRNA (0.5 mg/ml of beads). The precleared lysate corresponding to 2 × 106 cells was supplemented with 40 µl of yeast tRNA-blocked Protein G 50% bead slurry (GE Healthcare Life Sciences) and 3 μg of anti-HA antibody (ab9110; Abcam). After an overnight incubation at 4°C, the beads were washed as for QTIP. The DNA from input and IP samples was isolated and analyzed by dot blot hybridization as described previously (Grolimund et al, 2013).
Northern blotting analysis of TERRA
RNA extraction was performed using RNeasy Mini kit (Qiagen) according to manufacturer’s protocol. Northern blot TERRA analysis was performed as described previously (Azzalin et al, 2007). In brief, 10 μg of total RNA was separated by electrophoresis in a 1.2% formaldehyde agarose gel and blotted onto a nylon membrane (Hybond N+; GE Healthcare Life Sciences). Upon UV-cross-linking, the membrane was hybridized overnight with a [α32P]-dCTP–labeled telomeric probe at 50°C. After hybridization, the membrane was washed three times with a 1× SSC/0.5% SDS solution for 20 min at 60°C. Radioactive signals were detected using a phosphorimager (FLA-3000; Fujifilm).
Standard curves for TERRA copy number determination
To determine the copy number of TERRA in the qPCR reactions, we constructed a standard curve for each TERRA based on a recombinant plasmid containing the desired subtelomere sequence. The subtelomeres were amplified from HeLa genomic DNA using Phusion Green High-Fidelity DNA polymerase and the primers listed in Table S4. The PCR products were isolated from the gel, purified using the Qiagen gel extraction kit, and cloned in the pCR4 BLUNT-TOPO vector using the Zero Blunt TOPO PCR cloning kit for sequencing. All the plasmids were sequenced to confirm the subtelomere sequence. To construct the standard curves for each TERRA, we made a 10-fold serial dilution of each plasmid ranging from 3 × 108 to 3 × 10−1. The concentration of each plasmid was determined using Qubit 4 Fluorometer (Invitrogen). We calculated the copy number concentration using the following formula:
Table S4 List of primers for RT–qPCR of TERRA. (202.5KB, pdf)
Each standard curve was performed in duplicate. The CT values were plotted against the logarithm of the plasmid copy number and the standard curve was generated by a linear regression of the points. The PCR amplification efficiency was calculated as a percentage from the slope of the curve using the following formula:
RT–qPCR analysis of TERRA
RNA was isolated using RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. The RNA was reversed-transcribed using the Invitrogen SuperScript III Reverse Transcriptase as previously described (Feretzaki & Lingner, 2017). The cDNA was diluted 1/2 for routine relative quantification qPCR or 1/2 and 1/5 for absolute quantification. qPCR runs included both a no-template control and a no-reverse transcription control. All the experiments were performed in three biological replicates. qPCR was performed as previously described (Feretzaki & Lingner, 2017). To determine and analyze the relative changes in TERRA expression between the samples, we applied the 2−ΔΔCt method. The number of TERRA copies in the qPCR or the cDNA template was calculated based on the standard curve.
RT–qPCR analysis of depletion efficiency
3 μg of RNA was reverse-transcribed using SuperScript III RT (Invitrogen) with 150 ng random primers (Promega) and 500 ng oligo (dT)15 primers (Promega). The qPCR was performed as previously described (Feretzaki & Lingner, 2017) using primers listed in Table S5.
Table S5 List of primers for RT–qPCR to verify candidate depletion. (113.7KB, pdf)
Telomeric FISH on metaphase chromosomes
Cells were treated with 0.05 μg/ml demecolcine for 2 h, harvested, and incubated in 0.056 M KCl at 37°C for 7 min. Swollen cells were fixed in cold methanol/acetic acid (3:1) overnight and spread on glass slides. After ageing overnight, the slides were rehydrated in 1× PBS, fixed in 4% formaldehyde, and dehydrated in an ethanol series. The slides were allowed to air-dry before applying the hybridization solution (10 mM Tris–HCl, pH 7.4; 70% formamide; and 0.5% blocking reagent [Roche]) containing Cy3-OO-(CCCTAA)3 PNA probe. The spreads were denatured at 80°C for 3 min and hybridized at RT for 3 h. The slides were washed twice for 15 min with 10 mM Tris–HCl (pH 7.4)/70% formamide, and three times for 5 min with 0.1 M Tris–HCl (pH 7.4)/0.15 M NaCl/0.08% Tween-20 with DAPI in the second wash. The slides were dehydrated in an ethanol series and mounted in Vectashield embedding medium. Images were acquired using a Zeiss Axioplan II microscope.
Immunofluorescence FISH
Cells were grown on coverslips, fixed in 4% formaldehyde, and permeabilized for 5 min in 0.1% Triton X-100/0.02% SDS/1× PBS, and then pre-blocked for 10 min in 2% BSA/1× PBS and blocked for 45 min in 10% goat serum/2% BSA/1× PBS. Cells were then incubated with 0.5 μg/ml α-53BP1 antibody (NB100-304; Novus Biologicals) for 1 h, washed three times for 4 min in 2% BSA/1× PBS, and incubated with Alexa Fluor 633–conjugated secondary antibody (1:500, A-21070; Invitrogen) for 30 min. After three washes in PBS, stained cells were fixed in 4% formaldehyde for 5 min. Both antibodies were diluted in 10% goat serum/2% BSA/1× PBS, and all steps of the IF protocol were performed at RT. For telomeric FISH, the stained and fixed coverslips were dehydrated in an ethanol series, air-dried, and hybridized with the Cy3-OO-(CCCTAA)3 PNA probe as described above. Images were captured using a Zeiss LSM 700 confocal microscope.
Telomere Restriction Fragment (TRF) analysis
Genomic DNA was isolated using the Wizard Genomic DNA Purification kit (Promega) and subjected to restriction digestion with RsaI and HinfI, in the presence of RNase A, at 37°C overnight. Digested DNA was extracted using phenol–chloroform–isoamyl alcohol, followed by isopropanol precipitation. 1 μg of digested DNA was separated on 1% agarose gel by pulsed-field gel electrophoresis using a Bio-Rad CHEF DR-II apparatus or on 0.8% agarose gel by constant field gel electrophoresis. The gels were dried and hybridized with [α32P]-dCTP–labeled telomeric probe at 50°C as described (Grolimund et al, 2013). Radioactive signals corresponding to single-stranded telomeric DNA were detected using a phosphorimager (FLA-3000; Fujifilm). Thereafter, the gel was denatured in 0.5 M NaOH, 1.5 M NaCl for 30 min, neutralized for 15 min with 0.5 M Tris–HCl, pH 7.5, and 1.5 M NaCl; and hybridized again to detect total telomeric DNA.
Immunoblot analysis
Immunoblots were incubated separately with the following primary antibodies: anti-ATM pS1981 (1:1,000, ab81292; Abcam), anti-Chk2 pT68 (1:1,000, 2661; Cell Signaling), anti-p53 (1:1,000, sc-126; Santa Cruz), anti-p21 (1:5,000, ab109520; Abcam), anti-Actin (1:2,000, sc-1616; Santa Cruz), anti-TRF2 (1:1,000, 05-521; Millipore), and anti-γH2AX (1:1,000, 05-636; Millipore), followed by a corresponding horseradish peroxidase–conjugated secondary antibody.
Flow cytometry analysis
Following fixation in 70% ethanol, the pellet from 5 × 105 cells was resuspended in 250 µl of PBS containing 0.2 μg/ml RNAse A and incubated for 15 min at 37°C. The DNA was stained by adding 250 µl of 80 μg/ml of propidium iodide, followed by 10-min incubation at 4°C in the dark. 250 µl of PBS was added to each sample before FACS analysis on Accuri C6 (BD Biosciences). The percentage of cells in each phase of the cell cycle was determined using the Watson Pragmatic computational model in FlowJo software (TreeStar).
DNA/RNA hybrid immunoprecipitation
60–80 × 106 cells were harvested for each condition and lysed for 5 min in cold RLN buffer (50 mM Tris-Cl, pH 8.0; 140 mM NaCl; 1.5 mM MgCl2; 0.5% v/v Nonidet P-40; 175 µl buffer per 10 × 106 cells) supplemented with 1 mM DTT and 100 U/ml RNasin Plus (Promega). After centrifugation (300 g, 2 min, 4°C), the nuclei-enriched pellet was resuspended in RLT buffer (RNeasy Plus kit; [Qiagen]; 500 µl buffer per 20 × 106 cell equivalent) supplemented with βmercaptoethanol at 10 µl/ml and homogenized by passing through an insulin needle. Nucleic acids were thereafter isolated by a phenol:chloroform:isoamylalcohol (25:24:1; Biosolve BV) separation, followed by isopropanol precipitation. The nucleic acids were dissolved in H2O (400 µl per 20 × 106 cell equivalent) and sonicated using a Focused-ultrasonicator (Covaris E220) to obtain DNA fragments below 500 bp. 150 µl of sonicated nucleic acids were treated for 90 min at 37°C with either 10 µl of RNase H (1 U/µl; Roche) or H2O. The reaction was stopped by addition of 2 µl of 0.5 M EDTA. The samples were mixed with 1,738 µl of buffer 1 (10 mM Hepes-KOH, pH 7.5; 275 mM NaCl; 0.1% SDS; 1% Triton X-100; and 0.1% Na-deoxycholate) and precleared for 1 h at 4°C with 80 µl of 50% Protein G bead slurry (GE Healthcare Life Sciences) that had been pre-blocked for 1 h at 4°C with yeast tRNA (1 mg tRNA per 1 ml of 50% bead slurry). The IP reactions were set up containing 800 µl of precleared extract, 40 µl of tRNA-blocked Protein G beads, and 1 μg of S9.6 antibody (022715; Kerafast) or mouse IgG (sc-2025; Santa Cruz Biotechnology). The reactions were incubated for 90 min at 4°C on a rotating wheel and then washed consecutively for 5 min each time with 1 ml of buffer 2 (50 mM Hepes-KOH, pH 7.5; 140 mM NaCl; 1% Triton X-100; 0.1% Na-deoxycholate; and 1 mM EDTA), buffer 3 (50 mM Hepes-KOH, pH 7.5; 500 mM NaCl; 1% Triton X-100; 0.1% Na-deoxycholate; and 1 mM EDTA), buffer 4 (10 mM Tris–HCl, pH 8.0; 250 mM LiCl; 1% NP-40; 1% Na-deoxycholate; and 1 mM EDTA), and 1× TE buffer (10 mM Tris–HCl, pH 8.0, and 1 mM EDTA). The input and precipitated DNA was analyzed by dot blot hybridization.
T-circle assay
T-circle assay was modified from the work of Zellinger et al (2007). 2 μg of genomic DNA was digested with 10 U/μg (each) HinfI and MboI (New England Biolabs) in CutSmart buffer in the presence of 10 μg/ml RNase A overnight at 37°C. 1.5 μg of cut DNA was ethanol-precipitated and resuspended in an annealing buffer (0.2 M Tris, pH 7.5; 0.2 M KCl; and 1 mM EDTA) with 1 μM (CCCTAA)3 primer containing thiophosphate linkages between the three 3′ terminal nucleotides. The mix was denatured at 96°C for 5 min and cooled down to 25°C for 2 h. DNA was ethanol-precipitated and resuspended in 10 μl of H2O for the T-circle reaction (10 μl sample combined with 10 μl 0.2 mg/ml BSA, 0.1% Tween, 1 mM each dATP, dGTP, dTTP, dCTP, 1× ϕ29 buffer, and 7.5 U ϕ29 DNA polymerase [New England Biolabs]). Primer extension was carried out at 30°C for 12 h. The ϕ29 DNA polymerase was inactivated by incubation at 65°C for 20 min. The extension products were separated by agarose gel electrophoresis (0.6% agarose at 2 V/cm for 16 h). The gel was then dried at 50°C for 2 h, denatured and hybridized with a specific telomeric probe. U2OS genomic DNA of the ALT cell line was used as a positive control.
Supplementary Information
Supplementary Information is available at https://doi.org/10.26508/lsa.201800121.
Supplementary Material
Acknowledgements
We thank Patricia Renck Nunes for providing the plasmids for TERRA copy number determination. We gratefully acknowledge Patrick Reichenbach for immortalization of HLFs. The mass spectrometry analysis was performed by the Proteomics Core Facility and Technology Platform at Ecole Polytechnique Fédérale de Lausanne (EPFL) (Florence Armand, Romain Hamelin, and Marc Moniatte). Members of the Lingner laboratory are thanked for discussions, technical advice, and sharing reagents. Research in J Lingner’s laboratory was supported by the Swiss National Science Foundation (SNSF), the SNSF-funded NCCR RNA and disease network, an Initial Training Network grant (CodeAge) from the European Commission’s Seventh Framework Programme (grant agreement no. 316354), the Swiss Cancer League, and EPFL. M Feretzaki was supported by a Marie Curie postdoctoral fellowship.
Author Contributions
J Lingner: conceptualization, funding acquisition, and writing—original draft, review, and editing.
J Majerska: data curation, investigation, methodology, project administration, and writing—review and editing.
M Feretzaki: investigation.
G Glousker: investigation.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- Aeby E, Ahmed W, Redon S, Simanis V, Lingner J (2016) Peroxiredoxin 1 protects telomeres from oxidative damage and preserves telomeric DNA for extension by telomerase. Cell Rep 17: 3107–3114. 10.1016/j.celrep.2016.11.071 [DOI] [PubMed] [Google Scholar]
- Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, Pertsemlidis A, Gregory PA, Wright JA, Goodall GJ, et al. (2012) ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest 122: 3170–3183. 10.1172/jci63608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM (2014) RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 5: 5220 10.1038/ncomms6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J (2015) Telomere functions grounding on TERRA firma. Trends Cell Biol 25: 29–36. 10.1016/j.tcb.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801. 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Bagley BN, Keane TM, Maklakova VI, Marshall JG, Lester RA, Cancel MM, Paulsen AR, Bendzick LE, Been RA, Kogan SC, et al. (2012) A dominantly acting murine allele of Mcm4 causes chromosomal abnormalities and promotes tumorigenesis. PLoS Genet 8: e1003034 10.1371/journal.pgen.1003034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B (2013) Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 20: 1199–1205. 10.1038/nsmb.2662 [DOI] [PubMed] [Google Scholar]
- Ballana E, Este JA (2015) SAMHD1: At the crossroads of cell proliferation, immune responses, and virus restriction. Trends Microbiol 23: 680–692. 10.1016/j.tim.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Bao J, Wang S, Gunther LK, Kitajiri S, Li C, Sakamoto T (2015) The actin-bundling protein TRIOBP-4 and -5 promotes the motility of pancreatic cancer cells. Cancer Lett 356: 367–373. 10.1016/j.canlet.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J (2007) DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 26: 7773–7779. 10.1038/sj.onc.1210881 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637. 10.1038/nature05268 [DOI] [PubMed] [Google Scholar]
- Bartocci C, Diedrich JK, Ouzounov I, Li J, Piunti A, Pasini D, Yates JR 3rd, Lazzerini Denchi E (2014) Isolation of chromatin from dysfunctional telomeres reveals an important role for Ring1b in NHEJ-mediated chromosome fusions. Cell Rep 7: 1320–1332. 10.1016/j.celrep.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G. (2012) Tumor cell-protective catalase as a novel target for rational therapeutic approaches based on specific intercellular ROS signaling. Anticancer Res 32: 2599–2624. [PubMed] [Google Scholar]
- Bellon M, Datta A, Brown M, Pouliquen JF, Couppie P, Kazanji M, Nicot C (2006) Increased expression of telomere length regulating factors TRF1, TRF2 and TIN2 in patients with adult T-cell leukemia. Int J Cancer 119: 2090–2097. 10.1002/ijc.22026 [DOI] [PubMed] [Google Scholar]
- Biroccio A, Cherfils-Vicini J, Augereau A, Pinte S, Bauwens S, Ye J, Simonet T, Horard B, Jamet K, Cervera L, et al. (2013) TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat Cell Biol 15: 818–828. 10.1038/ncb2774 [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteinter S, Wright WE (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279: 349–352. 10.1126/science.279.5349.349 [DOI] [PubMed] [Google Scholar]
- Brachner A, Foisner R (2014) Lamina-associated polypeptide (LAP)2alpha and other LEM proteins in cancer biology. Adv Exp Med Biol 773: 143–163. 10.1007/978-1-4899-8032-8_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, Paumard-Hernandez B, Fernandez V, Dominguez F, Salas C, Romero-Laorden N, Garcia-Donas J, et al. (2015) A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nat Commun 6: 8383 10.1038/ncomms9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD (2004) Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol 24: 9948–9957. 10.1128/mcb.24.22.9948-9957.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champeris Tsaniras S, Kanellakis N, Symeonidou IE, Nikolopoulou P, Lygerou Z, Taraviras S (2014) Licensing of DNA replication, cancer, pluripotency and differentiation: An interlinked world? Semin Cel Dev Biol 30: 174–180. 10.1016/j.semcdb.2014.03.013 [DOI] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J (2007) Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev 21: 2485–2494. 10.1101/gad.1588807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest A, Lane K, McMahon K, Park J, Preisinger E, Conroy H, Housman D (2003) Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosomes Cancer 37: 58–71. 10.1002/gcc.10207 [DOI] [PubMed] [Google Scholar]
- Chen SC, Hu TH, Huang CC, Kung ML, Chu TH, Yi LN, Huang ST, Chan HH, Chuang JH, Liu LF, et al. (2015) Hepatoma-derived growth factor/nucleolin axis as a novel oncogenic pathway in liver carcinogenesis. Oncotarget 6: 16253–16270. 10.18632/oncotarget.3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Wang Y, Li F, Lin W, Liang Y, Ma Z (2017) Expression of telomere repeat binding factor 1 and TRF2 in prostate cancer and correlation with clinical parameters. Biomed Research Int 2017: 9764752 10.1155/2017/9764752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chian CF, Hwang YT, Terng HJ, Lee SC, Chao TY, Chang H, Ho CL, Wu YY, Perng WC (2016) Panels of tumor-derived RNA markers in peripheral blood of patients with non-small cell lung cancer: Their dependence on age, gender and clinical stages. Oncotarget 7: 50582–50595. 10.18632/oncotarget.10558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Louis T, Robbe P, Ackroyd S, Burns A, Timbs AT, Wright Colopy G, Dreau H, Sigaux F, Judde JG, et al. (2014) SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 123: 1021–1031. 10.1182/blood-2013-04-490847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker H, Brockdorff N (2014) SMCHD1 accumulates at DNA damage sites and facilitates the repair of DNA double-strand breaks. J Cell Sci 127: 1869–1874. 10.1242/jcs.140020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel F, Silva MJ, Techer H, Zadorozhny K, Sharma S, Nieminuszczy J, Mettling C, Dardillac E, Barthe A, Schmitz AL, et al. (2018) SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557: 57–61. 10.1038/s41586-018-0050-1 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J (2006) Telomere length homeostasis requires that telomerase levels are limiting. EMBO J 25: 565–574. 10.1038/sj.emboj.7600952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha IW, Carvalho KC, Martins WK, Marques SM, Muto NH, Falzoni R, Rocha RM, Aguiar S, Simoes AC, Fahham L, et al. (2010) Identification of genes associated with local aggressiveness and metastatic behavior in soft tissue tumors. Transl Oncol 3: 23–32. 10.1593/tlo.09166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198. 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Daddacha W, Koyen AE, Bastien AJ, Head PE, Dhere VR, Nabeta GN, Connolly EC, Werner E, Madden MZ, Daly MB, et al. (2017) SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep 20: 1921–1935. 10.1016/j.celrep.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann SG, Golas J, Miranda M, Shi C, Wu J, Jin G, Rosfjord E, Upeslacis E, Klippel A (2014) p120 catenin is a key effector of a Ras-PKCvarepsilon oncogenic signaling axis. Oncogene 33: 1385–1394. 10.1038/onc.2013.91 [DOI] [PubMed] [Google Scholar]
- Das M, Prasad SB, Yadav SS, Govardhan HB, Pandey LK, Singh S, Pradhan S, Narayan G (2013) Over expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesis. PLoS One 8: e69607 10.1371/journal.pone.0069607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Watine B. (2011) Silencing nuclear pore protein Tpr elicits a senescent-like phenotype in cancer cells. PloS one 6: e22423 10.1371/journal.pone.0022423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva S, Wang F, Hake TS, Porcu P, Wong HK, Wu L (2014) Downregulation of SAMHD1 expression correlates with promoter DNA methylation in Sezary syndrome patients. J Invest Dermatol 134: 562–565. 10.1038/jid.2013.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE (2009) Purification of proteins associated with specific genomic Loci. Cell 136: 175–186. 10.1016/j.cell.2008.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071. 10.1038/nature06065 [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642. 10.1038/nature05327 [DOI] [PubMed] [Google Scholar]
- Diehl MC, Idowu MO, Kimmelshue KN, York TP, Jackson-Cook CK, Turner KC, Holt SE, Elmore LW (2011) Elevated TRF2 in advanced breast cancers with short telomeres. Breast Cancer Res Treat 127: 623–630. 10.1007/s10549-010-0988-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X (2013) Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155: 345–356. 10.1016/j.cell.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL (2015) BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol 210: 191–208. 10.1083/jcb.201410061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA, Newbold RF, Nabholz M, Lingner J (2001) Regulation of human telomerase activity: Repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res 61: 7594–7602. [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, et al. (2009) Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet 41: 1116–1121. 10.1038/ng.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XJ, Pan Q, Wang SM, Pan YC, Wang Q, Zhang HH, Zhu MH, Zhang SH (2016) MAP4K4 promotes epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma. Tumour Biol 37: 11457–11467. 10.1007/s13277-016-5022-1 [DOI] [PubMed] [Google Scholar]
- Feretzaki M, Lingner J (2017) A practical qPCR approach to detect TERRA, the elusive telomeric repeat-containing RNA. Methods 114: 39–45. 10.1016/j.ymeth.2016.08.004 [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, et al. (2012) Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 14: 355–365. 10.1038/ncb2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Chen G, Gao C, Zhang DH, Kuan SF, Stabile LP, Liu G, Hu J (2017) MAP4K4 is a novel MAPK/ERK pathway regulator required for lung adenocarcinoma maintenance. Mol Oncol 11: 628–639. 10.1002/1878-0261.12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorieux C, Zamocky M, Sandoval JM, Verrax J, Calderon PB (2015) Regulation of catalase expression in healthy and cancerous cells. Free Radic Biol Med 87: 84–97. 10.1016/j.freeradbiomed.2015.06.017 [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, et al. (2005) Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913. 10.1038/nature03485 [DOI] [PubMed] [Google Scholar]
- Gorman EB, Chen L, Albanese J, Ratech H (2007) Patterns of spectrin expression in B-cell lymphomas: Loss of spectrin isoforms is associated with nodule-forming and germinal center-related lymphomas. Mod Pathol 20: 1245–1252. 10.1038/modpathol.3800962 [DOI] [PubMed] [Google Scholar]
- Graf M, Bonetti D, Lockhart A, Serhal K, Kellner V, Maicher A, Jolivet P, Teixeira MT, Luke B (2017) Telomere length determines TERRA and r-loop regulation through the cell cycle. Cell 170: 72–85 e14. 10.1016/j.cell.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Grinchuk OV, Motakis E, Kuznetsov VA (2010) Complex sense-antisense architecture of TNFAIP1/POLDIP2 on 17q11.2 represents a novel transcriptional structural-functional gene module involved in breast cancer progression. BMC Genomics 11 Suppl 1: S9 10.1186/1471-2164-11-s1-s9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolimund L, Aeby E, Hamelin R, Armand F, Chiappe D, Moniatte M, Lingner J (2013) A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat Commun 4: 2848 10.1038/ncomms3848 [DOI] [PubMed] [Google Scholar]
- Haas DA, Bala K, Busche G, Weidner-Glunde M, Santag S, Kati S, Gramolelli S, Damas M, Dittrich-Breiholz O, Kracht M, et al. (2013) The inflammatory kinase MAP4K4 promotes reactivation of Kaposi’s sarcoma herpesvirus and enhances the invasiveness of infected endothelial cells. PLoS Pathog 9: e1003737 10.1371/journal.ppat.1003737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA (1999) Creation of human tumour cells with defined genetic elements. Nature 400: 464–468. 10.1038/22780 [DOI] [PubMed] [Google Scholar]
- Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA (2002) Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol 22: 2111–2123. 10.1128/mcb.22.7.2111-2123.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Han Y, Feng Y, Xia L, Hsu C, Sheng X, Pooler LC, Patel Y, Kolonel LN, Carter E, et al. (2013) Genome-wide testing of putative functional exonic variants in relationship with breast and prostate cancer risk in a multiethnic population. PLoS Genet 9: e1003419 10.1371/journal.pgen.1003419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD. (2004) Constitutively active DNA damage checkpoint pathways as the driving force for the high frequency of p53 mutations in human cancer. DNA Repair 3: 1057–1062. 10.1016/j.dnarep.2004.03.036 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355. 10.1126/science.1140735 [DOI] [PubMed] [Google Scholar]
- Hamdi Y, Soucy P, Kuchenbaeker KB, Pastinen T, Droit A, Lemacon A, Adlard J, Aittomaki K, Andrulis IL, Arason A, et al. (2017) Association of breast cancer risk in BRCA1 and BRCA2 mutation carriers with genetic variants showing differential allelic expression: Identification of a modifier of breast cancer risk at locus 11q22.3. Breast Cancer Res Treat 161: 117–134. 10.1007/s10549-016-4018-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CA, Andryuk PJ, Cline SW, Mathew S, Siekierka JJ, Goldstein G (1995) Structure and mapping of the human thymopoietin (TMPO) gene and relationship of human TMPO beta to rat lamin-associated polypeptide 2. Genomics 28: 198–205. 10.1006/geno.1995.1131 [DOI] [PubMed] [Google Scholar]
- Hills SA, Diffley JF (2014) DNA replication and oncogene-induced replicative stress. Curr Biol 24: R435–R444. 10.1016/j.cub.2014.04.012 [DOI] [PubMed] [Google Scholar]
- Hinrichsen I, Ernst BP, Nuber F, Passmann S, Schafer D, Steinke V, Friedrichs N, Plotz G, Zeuzem S, Brieger A (2014) Reduced migration of MLH1 deficient colon cancer cells depends on SPTAN1. Mol Cancer 13: 11 10.1186/1476-4598-13-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga SI, Ly T, Garzon J, Horejsi Z, Ohkubo YN, Endo A, Obuse C, Boulton SJ, Lamond AI, Donaldson AD (2017) Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep 18: 403–419. 10.15252/embr.201641983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeycutt KA, Chen Z, Koster MI, Miers M, Nuchtern J, Hicks J, Roop DR, Shohet JM (2006) Deregulated minichromosomal maintenance protein MCM7 contributes to oncogene driven tumorigenesis. Oncogene 25: 4027–4032. 10.1038/sj.onc.1209435 [DOI] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. (2013) TERT promoter mutations in familial and sporadic melanoma. Science 339: 959–961. 10.1126/science.1230062 [DOI] [PubMed] [Google Scholar]
- Hu TH, Huang CC, Liu LF, Lin PR, Liu SY, Chang HW, Changchien CS, Lee CM, Chuang JH, Tai MH (2003) Expression of hepatoma-derived growth factor in hepatocellular carcinoma. Cancer 98: 1444–1456. 10.1002/cncr.11653 [DOI] [PubMed] [Google Scholar]
- Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH (2006) An antibody-based microarray assay for small RNA detection. Nucleic Acids Res 34: e52 10.1093/nar/gkl142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA (2013) Highly recurrent TERT promoter mutations in human melanoma. Science 339: 957–959. 10.1126/science.1229259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K, Catalano J, Puri RK, Gnatt A (2008) Knockdown of TFIIS by RNA silencing inhibits cancer cell proliferation and induces apoptosis. BMC Cancer 8: 133 10.1186/1471-2407-8-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Yang ZH, Luo DH, Guo L, Sun R, Chen QY, Huang PY, Qiu F, Zou X, Cao KJ, et al. (2014) Elevated apolipoprotein A-I levels are associated with favorable prognosis in metastatic nasopharyngeal carcinoma. Med Oncol 31: 80 10.1007/s12032-014-0080-y [DOI] [PubMed] [Google Scholar]
- Jiang X, Gillen S, Esposito I, Giese NA, Michalski CW, Friess H, Kleeff J (2010) Reduced expression of the membrane skeleton protein beta1-spectrin (SPTBN1) is associated with worsened prognosis in pancreatic cancer. Histol Histopathol 25: 1497–1506. 10.14670/HH-25.1497 [DOI] [PubMed] [Google Scholar]
- Jones RE, Oh S, Grimstead JW, Zimbric J, Roger L, Heppel NH, Ashelford KE, Liddiard K, Hendrickson EA, Baird DM (2014) Escape from telomere-driven crisis is DNA ligase III dependent. Cell Rep 8: 1063–1076. 10.1016/j.celrep.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Ju YT, Kwag SJ, Park HJ, Jung EJ, Jeong CY, Jeong SH, Lee YJ, Choi SK, Kang KR, Hah YS, et al. (2015) Decreased expression of heat shock protein 20 in colorectal cancer and its implication in tumorigenesis. J Cell Biochem 116: 277–286. 10.1002/jcb.24966 [DOI] [PubMed] [Google Scholar]
- Kaiser AM, Attardi LD (2017) Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ 25: 93–103. 10.1038/cdd.2017.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karami S, Han Y, Pande M, Cheng I, Rudd J, Pierce BL, Nutter EL, Schumacher FR, Kote-Jarai Z, Lindstrom S, et al. (2016) Telomere structure and maintenance gene variants and risk of five cancer types. Int J Cancer 139: 2655–2670. 10.1002/ijc.30288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Wada H, Patel P, Hu HP, Lee D, Ujiie H, Hirohashi K, Nakajima T, Sato M, Kaji M, et al. (2016) Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer 92: 53–61. 10.1016/j.lungcan.2015.11.018 [DOI] [PubMed] [Google Scholar]
- Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, Songyang Z (2009) TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat Struct Mol Biol 16: 372–379. 10.1038/nsmb.1575 [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. 10.1126/science.7605428 [DOI] [PubMed] [Google Scholar]
- Kim ST, Cheng Y, Hsu FC, Jin T, Kader AK, Zheng SL, Isaacs WB, Xu J, Sun J (2010) Prostate cancer risk-associated variants reported from genome-wide association studies: Meta-analysis and their contribution to genetic variation. Prostate 70: 1729–1738. 10.1002/pros.21208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtidis A, Yanagisawa M, Huveldt D, Copland JA, Anastasiadis PZ (2015) Pro-tumorigenic phosphorylation of p120 catenin in renal and breast cancer. PLoS One 10: e0129964 10.1371/journal.pone.0129964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok HF, Zhang SD, McCrudden CM, Yuen HF, Ting KP, Wen Q, Khoo US, Chan KY (2015) Prognostic significance of minichromosome maintenance proteins in breast cancer. Am J Cancer Res 5: 52–71. [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474: 654–657. 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk HB, Mora-Bermudez F, Landsverk OJ, Hasvold G, Naderi S, Bakke O, Ellenberg J, Collas P, Syljuasen RG, Kuntziger T (2010) The protein phosphatase 1 regulator PNUTS is a new component of the DNA damage response. EMBO Rep 11: 868–875. 10.1038/embor.2010.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E (2006) The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol 16: 1303–1310. 10.1016/j.cub.2006.05.021 [DOI] [PubMed] [Google Scholar]
- Li JP, Liu Y, Yin YH (2017) ARHGAP1 overexpression inhibits proliferation, migration and invasion of C-33A and SiHa cell lines. Onco Targets Ther 10: 691–701. 10.2147/ott.s112223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TF, Qin SH, Ruan XZ, Wang X (2015) p120-catenin participates in the progress of gastric cancer through regulating the Rac1 and Pak1 signaling pathway. Oncol Rep 34: 2357–2364. 10.3892/or.2015.4226 [DOI] [PubMed] [Google Scholar]
- Lian J, Tang J, Shi H, Li H, Zhen T, Xie W, Zhang F, Yang Y, Han A (2015) Positive feedback loop of hepatoma-derived growth factor and beta-catenin promotes carcinogenesis of colorectal cancer. Oncotarget 6: 29357–29374. 10.18632/oncotarget.4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Qu GQ, Lu YM, Kong WM, Liu Y, Chen WX, Liao XH (2016) Silencing of MAP4K4 by short hairpin RNA suppresses proliferation, induces G1 cell cycle arrest and induces apoptosis in gastric cancer cells. Mol Med Rep 13: 41–48. 10.3892/mmr.2015.4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macari F, El-Houfi Y, Boldina G, Xu H, Khoury-Hanna S, Ollier J, Yazdani L, Zheng G, Bieche I, Legrand N, et al. (2016) TRM6/61 connects PKCalpha with translational control through tRNAi(Met) stabilization: Impact on tumorigenesis. Oncogene 35: 1785–1796. 10.1038/onc.2015.244 [DOI] [PubMed] [Google Scholar]
- Maciejowski J, de Lange T (2017) Telomeres in cancer: Tumour suppression and genome instability. Nat Rev Mol Cell Biol 18: 175–186. 10.1038/nrm.2016.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerska J, Redon S, Lingner J (2017) Quantitative telomeric chromatin isolation protocol for human cells. Methods 114: 28–38. 10.1016/j.ymeth.2016.08.003 [DOI] [PubMed] [Google Scholar]
- Matsushima-Nishiwaki R, Adachi S, Yoshioka T, Yasuda E, Yamagishi Y, Matsuura J, Muko M, Iwamura R, Noda T, Toyoda H, et al. (2011) Suppression by heat shock protein 20 of hepatocellular carcinoma cell proliferation via inhibition of the mitogen-activated protein kinases and AKT pathways. J Cell Biochem 112: 3430–3439. 10.1002/jcb.23270 [DOI] [PubMed] [Google Scholar]
- Matsushima-Nishiwaki R, Toyoda H, Nagasawa T, Yasuda E, Chiba N, Okuda S, Maeda A, Kaneoka Y, Kumada T, Kozawa O (2016) Phosphorylated heat shock protein 20 (HSPB6) regulates transforming growth factor-alpha-induced migration and invasion of hepatocellular carcinoma cells. PLoS One 11: e0151907 10.1371/journal.pone.0151907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merati M, Buethe DJ, Cooper KD, Honda KS, Wang H, Gerstenblith MR (2015) Aggressive CD8(+) epidermotropic cutaneous T-cell lymphoma associated with homozygous mutation in SAMHD1. JAAD Case Rep 1: 227–229. 10.1016/j.jdcr.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP (2006) Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828. 10.1038/nature04638 [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH (2013) p53 mutations in cancer. Nat Cell Biol 15: 2–8. 10.1038/ncb2641 [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC (2011) p53 and its mutants in tumor cell migration and invasion. J Cell Biol 192: 209–218. 10.1083/jcb.201009059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz P, Blanco R, Blasco MA (2006) Role of the TRF2 telomeric protein in cancer and ageing. Cell Cycle 5: 718–721. 10.4161/cc.5.7.2636 [DOI] [PubMed] [Google Scholar]
- Nagasawa T, Matsushima-Nishiwaki R, Toyoda H, Matsuura J, Kumada T, Kozawa O (2014) Heat shock protein 20 (HSPB6) regulates apoptosis in human hepatocellular carcinoma cells: Direct association with Bax. Oncol Rep 32: 1291–1295. 10.3892/or.2014.3278 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Suzuki T, Arai Y, Sasano H (2009) 17beta-hydroxysteroid dehydrogenase type 11 (Pan1b) expression in human prostate cancer. Neoplasma 56: 317–320. 10.4149/neo_2009_04_317 [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Kawai T, Kumaki F, Hiroi S, Mukai M, Ikeda E, Koering CE, Gilson E (2003) Expression of mRNAs for telomeric repeat binding factor (TRF)-1 and TRF2 in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Clin Cancer Res 9: 1105–1111. [PubMed] [Google Scholar]
- Natrajan R, Williams RD, Grigoriadis A, Mackay A, Fenwick K, Ashworth A, Dome JS, Grundy PE, Pritchard-Jones K, Jones C (2007) Delineation of a 1Mb breakpoint region at 1p13 in Wilms tumors by fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer 46: 607–615. 10.1002/gcc.20446 [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM (1996) The pCL vector system: Rapid production of helper-free, high-titer, recombinant retroviruses. J Virol 70: 5701–5705. 0022-538X/96/$04.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kumada T, Takai S, Matsushima-Nishiwaki R, Yoshimi N, Yasuda E, Kato K, Toyoda H, Kaneoka Y, Yamaguchi A, et al. (2007) Expression levels of heat shock protein 20 decrease in parallel with tumor progression in patients with hepatocellular carcinoma. Oncol Rep 17: 1309–1314. 10.3892/or.17.6.1309 [DOI] [PubMed] [Google Scholar]
- Nozawa RS, Nagao K, Igami KT, Shibata S, Shirai N, Nozaki N, Sado T, Kimura H, Obuse C (2013) Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol 20: 566–573. 10.1038/nsmb.2532 [DOI] [PubMed] [Google Scholar]
- Ohara N, Haraguchi N, Koseki J, Nishizawa Y, Kawai K, Takahashi H, Nishimura J, Hata T, Mizushima T, Yamamoto H, et al. (2017) Low expression of the GOPC is a poor prognostic marker in colorectal cancer. Oncol Lett 14: 4483–4490. 10.3892/ol.2017.6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386. 10.1074/mcp.m200025-mcp200 [DOI] [PubMed] [Google Scholar]
- Ono R, Matsuoka J, Yamatsuji T, Naomoto Y, Tanaka N, Matsui H, Matsushita M (2008) M-RIP, a novel target of JNK signaling and a requirement for human cancer cell invasion. Int J Mol Med 22: 199–203. 10.3892/ijmm_00000009 [DOI] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691. 10.1016/j.cell.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal D, Sharma U, Singh SK, Kakkar N, Prasad R (2015) Over-expression of telomere binding factors (TRF1 & TRF2) in renal cell carcinoma and their inhibition by using SiRNA induce apoptosis, reduce cell proliferation and migration invitro. PLoS One 10: e0115651 10.1371/journal.pone.0115651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Vindigni A (2017) Nucleases acting at stalled forks: How to reboot the replication program with a few shortcuts. Annu Rev Genet 51: 477–499. 10.1146/annurev-genet-120116-024745 [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Crittin J, Grolimund L, Lingner J (2013) The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J 32: 2861–2871. 10.1038/emboj.2013.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao N, Zhu Y, Li H, Qu Z, Xiao Z (2014) Expression of heat shock protein 20 inversely correlated with tumor progression in patients with ovarian cancer. Eur J Gynaecol Oncol 35: 576–579. 10.4028/www.scientific.net/amr.886.576 [DOI] [PubMed] [Google Scholar]
- Qiu MH, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH (2012) Expression and prognostic significance of MAP4K4 in lung adenocarcinoma. Pathol Res Pract 208: 541–548. 10.1016/j.prp.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Quesada V, Ramsay AJ, Rodriguez D, Puente XS, Campo E, Lopez-Otin C (2013) The genomic landscape of chronic lymphocytic leukemia: Clinical implications. BMC Med 11: 124 10.1186/1741-7015-11-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay AJ, Quesada V, Foronda M, Conde L, Martinez-Trillos A, Villamor N, Rodriguez D, Kwarciak A, Garabaya C, Gallardo M, et al. (2013) POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet 45: 526–530. 10.1038/ng.2584 [DOI] [PubMed] [Google Scholar]
- Rentoft M, Lindell K, Tran P, Chabes AL, Buckland RJ, Watt DL, Marjavaara L, Nilsson AK, Melin B, Trygg J, et al. (2016) Heterozygous colon cancer-associated mutations of SAMHD1 have functional significance. Proc Natl Acad Sci USA 113: 4723–4728. 10.1073/pnas.1519128113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. (2002) DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416: 552–556. 10.1038/416552a [DOI] [PubMed] [Google Scholar]
- Robles-Espinoza CD, Harland M, Ramsay AJ, Aoude LG, Quesada V, Ding Z, Pooley KA, Pritchard AL, Tiffen JC, Petljak M, et al. (2014) POT1 loss-of-function variants predispose to familial melanoma. Nat Genet 46: 478–481. 10.1038/ng.2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarinen S, Aavikko M, Aittomaki K, Launonen V, Lehtonen R, Franssila K, Lehtonen HJ, Kaasinen E, Broderick P, Tarkkanen J, et al. (2011) Exome sequencing reveals germline NPAT mutation as a candidate risk factor for Hodgkin lymphoma. Blood 118: 493–498. 10.1182/blood-2011-03-341560 [DOI] [PubMed] [Google Scholar]
- Sagie S, Toubiana S, Hartono SR, Katzir H, Tzur-Gilat A, Havazelet S, Francastel C, Velasco G, Chedin F, Selig S (2017) Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA: RNA hybrids. Nat Commun 8: 14015 10.1038/ncomms14015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JH, Rivadeneira DB, Caino MC, Chae YC, Speicher DW, Tang HY, Vaira V, Bosari S, Palleschi A, Rampini P, et al. (2016) The mitochondrial unfoldase-peptidase complex ClpXP controls bioenergetics stress and metastasis. PLoS Biol 14: e1002507 10.1371/journal.pbio.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103. 10.1016/j.cell.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE (2005) Senescence and immortalization: Role of telomeres and telomerase. Carcinogenesis 26: 867–874. 10.1093/carcin/bgh296 [DOI] [PubMed] [Google Scholar]
- Shema E, Kim J, Roeder RG, Oren M (2011) RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol Cell 42: 477–488. 10.1016/j.molcel.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Yang XM, Tang DD, Liu G, Yuan P, Yang Y, Chang LS, Zhang LR, Song DK (2015) Expression and significance of m1A transmethylase, hTrm6p/hTrm61p and its related gene hTrm6/hTrm61 in bladder urothelial carcinoma. Am J Cancer Res 5: 2169–2179. [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lv G, Chu Z, Piao L, Liu X, Wang T, Jiang Y, Zhang P (2014) Identification of natural splice variants of SAMHD1 in virus-infected HCC. Oncol Rep 31: 687–692. 10.3892/or.2013.2895 [DOI] [PubMed] [Google Scholar]
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC (2007) A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet 39: 93–98. 10.1038/ng1936 [DOI] [PubMed] [Google Scholar]
- Soudet J, Jolivet P, Teixeira MT (2014) Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol Cell 53: 954–964. 10.1016/j.molcel.2014.02.030 [DOI] [PubMed] [Google Scholar]
- Su F, Kozak KR, Imaizumi S, Gao F, Amneus MW, Grijalva V, Ng C, Wagner A, Hough G, Farias-Eisner G, et al. (2010) Apolipoprotein A-I (apoA-I) and apoA-I mimetic peptides inhibit tumor development in a mouse model of ovarian cancer. Proc Natl Acad Sci USA 107: 19997–20002. 10.1073/pnas.1009010107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang C, Yang Z, Wu Y, Wang H, Bao Z, Jiang T (2016) KIF23 is an independent prognostic biomarker in glioma, transcriptionally regulated by TCF-4. Oncotarget 7: 24646–24655. 10.18632/oncotarget.8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jin Z, Song X, Wang J, Li Y, Qian X, Zhang Y, Yin Y (2015) Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma. BMC Cancer 15: 961 10.1186/s12885-015-1987-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suram A, Kaplunov J, Patel PL, Ruan H, Cerutti A, Boccardi V, Fumagalli M, Di Micco R, Mirani N, Gurung RL, et al. (2012) Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J 31: 2839–2851. 10.1038/emboj.2012.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Fusaki N, Ohta S, Iwahori Y, Iizuka Y, Inagawa K, Kawakami Y, Yoshida K, Toda M (2012) Downregulation of KIF23 suppresses glioma proliferation. J Neurooncol 106: 519–529. 10.1007/s11060-011-0706-2 [DOI] [PubMed] [Google Scholar]
- Tang M, Li Y, Zhang X, Deng T, Zhou Z, Ma W, Songyang Z (2014) Structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) promotes non-homologous end joining and inhibits homologous recombination repair upon DNA damage. J Biol Chem 289: 34024–34032. 10.1074/jbc.m114.601179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreson MA, Reynolds AB (2002) Altered expression of the catenin p120 in human cancer: Implications for tumor progression. Differ Res Biol Divers 70: 583–589. 10.1046/j.1432-0436.2002.700911.x [DOI] [PubMed] [Google Scholar]
- Thutkawkorapin J, Picelli S, Kontham V, Liu T, Nilsson D, Lindblom A (2016) Exome sequencing in one family with gastric- and rectal cancer. BMC Genet 17: 41 10.1186/s12863-016-0351-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H, Sormunen R, Kallioinen M (1996) Non-erythroid spectrin (fodrin) in cutaneous tumours: Diminished in cell membranes, increased in the cytoplasm. Br J Dermatol 135: 576–580. 10.1111/j.1365-2133.1996.tb03834.x [DOI] [PubMed] [Google Scholar]
- van Duijnhoven FJ, Bueno-De-Mesquita HB, Calligaro M, Jenab M, Pischon T, Jansen EH, Frohlich J, Ayyobi A, Overvad K, Toft-Petersen AP, et al. (2011) Blood lipid and lipoprotein concentrations and colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition. Gut 60: 1094–1102. 10.1136/gut.2010.225011 [DOI] [PubMed] [Google Scholar]
- van Overbeek M, de Lange T (2006) Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol 16: 1295–1302. 10.1016/j.cub.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ (2012) RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806. 10.1016/j.cell.2012.03.030 [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456. 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CD, Sun Y, Chen N, Huang L, Huang JW, Zhu M, Wang T, Ji YL (2016) The role of catalase C262T gene polymorphism in the susceptibility and survival of cancers. Scientific Rep 6: 26973 10.1038/srep26973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Lu FZ, Shen XY, Wu Y, Zhao LT (2014) SAMHD1 is down regulated in lung cancer by methylation and inhibits tumor cell proliferation. Biochem Biophys Res Commun 455: 229–233. 10.1016/j.bbrc.2014.10.153 [DOI] [PubMed] [Google Scholar]
- Wang X, Dai S, Zhang Z, Liu L, Wang J, Xiao X, He D, Liu B (2009) Characterization of apolipoprotein A-I as a potential biomarker for cholangiocarcinoma. Eur J Cancer Care 18: 625–635. 10.1111/j.1365-2354.2008.00965.x [DOI] [PubMed] [Google Scholar]
- Wolgast LR, Cannizzarro LA, Ramesh KH, Xue X, Wang D, Bhattacharyya PK, Gong JZ, McMahon C, Albanese JM, Sunkara JL, et al. (2011) Spectrin isoforms: Differential expression in normal hematopoiesis and alterations in neoplastic bone marrow disorders. Am J Clin Pathol 136: 300–308. 10.1309/ajcpsa5rnm9igfjf [DOI] [PubMed] [Google Scholar]
- Wu D, Niu X, Pan H, Zhang Z, Zhou Y, Qu P, Zhou J (2016) MicroRNA-497 targets hepatoma-derived growth factor and suppresses human prostate cancer cell motility. Mol Med Rep 13: 2287–2292. 10.3892/mmr.2016.4756 [DOI] [PubMed] [Google Scholar]
- Wu P, Takai H, de Lange T (2012) Telomeric 3′ overhangs derive from resection by Exo1 and apollo and fill-in by POT1b-associated CST. Cell 150: 39–52. 10.1016/j.cell.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Jividen K, Spencer A, Dworak N, Ni L, Oostdyk LT, Chatterjee M, Kusmider B, Reon B, Parlak M, et al. (2017) Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol Cell 66: 503–516.e5. 10.1016/j.molcel.2017.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GY, Zhang AQ, Wang J, Li CH, Wang XQ, Pan K, Zhou C, Dong JH (2016) Hepatoma-derived growth factor promotes growth and metastasis of hepatocellular carcinoma cells. Cell Biochem Funct 34: 274–285. 10.1002/cbf.3189 [DOI] [PubMed] [Google Scholar]
- Yao ZX, Jogunoori W, Choufani S, Rashid A, Blake T, Yao W, Kreishman P, Amin R, Sidawy AA, Evans SR, et al. (2010) Epigenetic silencing of beta-spectrin, a TGF-beta signaling/scaffolding protein in a human cancer stem cell disorder: Beckwith-Wiedemann syndrome. J Biol Chem 285: 36112–36120. 10.1074/jbc.m110.162347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Leger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, et al. (2010) TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142: 230–242. 10.1016/j.cell.2010.05.032 [DOI] [PubMed] [Google Scholar]
- Zamanian-Daryoush M, DiDonato JA (2015) Apolipoprotein A-I and cancer. Front Pharmacol 6: 265 10.3389/fphar.2015.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K (2007) Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell 27: 163–169. 10.1016/j.molcel.2007.05.025 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang G, Chen S, Ding J, Ju S, Cao H, Tian H (2016) Depletion of thymopoietin inhibits proliferation and induces cell cycle arrest/apoptosis in glioblastoma cells. World J Surg Oncol 14: 267 10.1186/s12957-016-1018-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E (2000) NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev 14: 2283–2297. 10.1101/gad.827700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X, Lin L, Yang S, Bhuvaneshwar K, Wang H, Gusev Y, Lee MH, Kallakury B, Shivapurkar N, Cahn K, et al. (2015) betaII-Spectrin (SPTBN1) suppresses progression of hepatocellular carcinoma and Wnt signaling by regulation of Wnt inhibitor kallistatin. Hepatology 61: 598–612. 10.1002/hep.27558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4 List of primers for RT–qPCR of TERRA. (202.5KB, pdf)
Table S5 List of primers for RT–qPCR to verify candidate depletion. (113.7KB, pdf)