Abstract
Flagellar motility of Campylobacter jejuni and Helicobacter pylori influences host colonization by promoting migration through viscous milieus such as gastrointestinal mucus. This review explores mechanisms C. jejuni and H. pylori employ to control flagellar biosynthesis and chemotactic responses. These microbes tightly control the activities of σ54 and σ28 to mediate ordered flagellar gene expression. In addition to phase-variable and posttranslational mechanisms, flagellar biosynthesis is regulated spatially and numerically so that only a certain number of organelles are placed at polar sites. To mediate chemotaxis, C. jejuni and H. pylori combine basic chemotaxis signal transduction components with several accessory proteins. H. pylori is unusual in that it lacks a methylation-based adaptation system and produces multiple CheV coupling proteins. Chemoreceptors in these bacteria contain nonconserved ligand binding domains, with several chemoreceptors matched to environmental signals. Together, these mechanisms allow for swimming motility that is essential for colonization.
Keywords: flagella, Epsilonproteobacteria, gene expression, CheZ, FlgS, FlgR
INTRODUCTION
Campylobacter and Helicobacter species are representative Epsilonproteobacteria that frequently associate with animal and human hosts. Campylobacter jejuni is a leading worldwide cause of diarrheal disease in humans, but it is also a harmless commensal organism of the intestinal tracts of many wild and agriculturally important animals, especially poultry (14, 79). Helicobacter pylori primarily infects humans, residing in the gastric mucosa in roughly half of the world’s population. This infection can lead to many gastric diseases, including gastritis, gastric ulcers, and gastric carcinoma (21).
During infection of hosts, C. jejuni and H. pylori reside primarily within the thick mucus layer lining the intestinal and gastric epithelium, respectively (6, 41, 65). Flagellar motility is an important colonization determinant for these organisms, presumably enabling migration to and movement within mucus to reach microenvironments conducive to growth. As such, nonmotile C. jejuni or H. pylori mutants are attenuated for colonization of human or animal hosts (46, 83, 84). Like other motile bacteria, motility in Campylobacter and Helicobacter species is regulated by a chemotactic signaling system that allows the organisms to follow favorable chemical gradients in environments. Nonchemotactic (Che−) C. jejuni and H. pylori mutants are reduced for host colonization, indicating that recognition of proper environmental parameters via chemosensory systems is critical for in vivo growth (32, 40, 46, 111, 128). This review highlights the critical features of the flagellar and chemotactic systems of Campylobacter and Helicobacter species, with emphasis on those characterized in C. jejuni and H. pylori, that allow for the regulated construction of polar flagella and the proper chemosensory network to mediate rotational movement of the flagellar nanomachine.
VISCOSITY-INFLUENCED FLAGELLAR MOTILITY
Owing to possible evolutionary pressures from residing in viscous mucus milieus, Campylobacter and Helicobacter species demonstrate unusually high velocities and movements in viscous substances compared with many motile bacteria. In the typically studied peritrichous, rodshaped bacteria (such as Salmonella species and Escherichia coli), highest swimming velocities occur in low to slightly viscous substances, with severe velocity reductions as viscosity increases (31, 41, 102). In contrast, C. jejuni and H. pylori are helical bacteria with exclusively polar flagella. Many of these bacterial strains demonstrate roughly twice the velocity of rod-shaped bacteria in substances of low viscosity (ranging from ~22 to 40 μm s−1) and can retain these velocities as the viscosity increases 40- to 80fold (31, 41, 60, 102, 122). Proportionally, the high velocity of C. jejuni and H. pylori in viscous substances is roughly equivalent to a human male of average height swimming in corn oil at 50 m s−1!
The unusual motility of Campylobacter and Helicobacter species in viscous substances is hypothesized to be due to a helical cell shape and the presence of polar flagella. The flagella provide propulsive torque as well as a rotary movement of the cell body; a helical cell shape creates a corkscrew-like rotation, which may allow the bacteria to push against a rigid substance to promote motility in the viscous mucus (8, 31). However, H. pylori mutants with less helical cell bodies retain a high swimming velocity in viscous liquids, suggesting that helical shape may not be critical for movement through viscous substances (9, 105). An alternative idea is that the bacteria facilitate movement in mucus by altering the physical nature of the mucus immediately surrounding the organism. Through the activity of its urease, H. pylori increases the pH of the mucus, which lowers the viscosity to promote greater movement (13).
C. jejuni exhibits two different viscosity-dependent modes of motility in cecal mucosal scrapings from infected mice (65). At low viscosity, C. jejuni predominantly swims via rather straight vectoral movements with few direction changes (31, 102). In contrast, at high viscosity a darting motility characterized by oscillation along a relatively short and straight path is observed (102, 108). These different types of motility may be relevant in different regions of the gastrointestinal mucus layer, as viscosity is thought to increase from the lumen to the epithelial surface. Darting motility may increase the time and number of contacts between C. jejuni and the epithelium, thereby augmenting adherence to or invasion of these cells (108).
REGULATION OF FLAGELLAR GENE EXPRESSION AND BIOSYNTHESIS
Overview of Flagellar Structure
A fully functional motility system includes the flagellum and the chemosensory system that influences flagellar rotation based on environmental conditions. Approximately 25 to 30 relatively conserved proteins compose a flagellum, with additional factors required for synthesis or secretion of these components. The collection of chemosensory components is less conserved and can involve either a few to dozens of chemoreceptors and signaling proteins in a bacterium. In all, 40 to 100 different proteins are required to form a rotating, chemoresponsive flagellum.
Based on bioinformatics, genetic screens, and functional studies, many proteins of Campylobacter and Helicobacter species are known to be required for motility, although exact roles for some have not yet been assigned. Combining these studies with information from the well-studied systems of Salmonella species and E. coli (see References 17, 73, and 77 for extensive reviews), a description of flagellar structure and biosynthesis in Campylobacter and Helicobacter species follows.
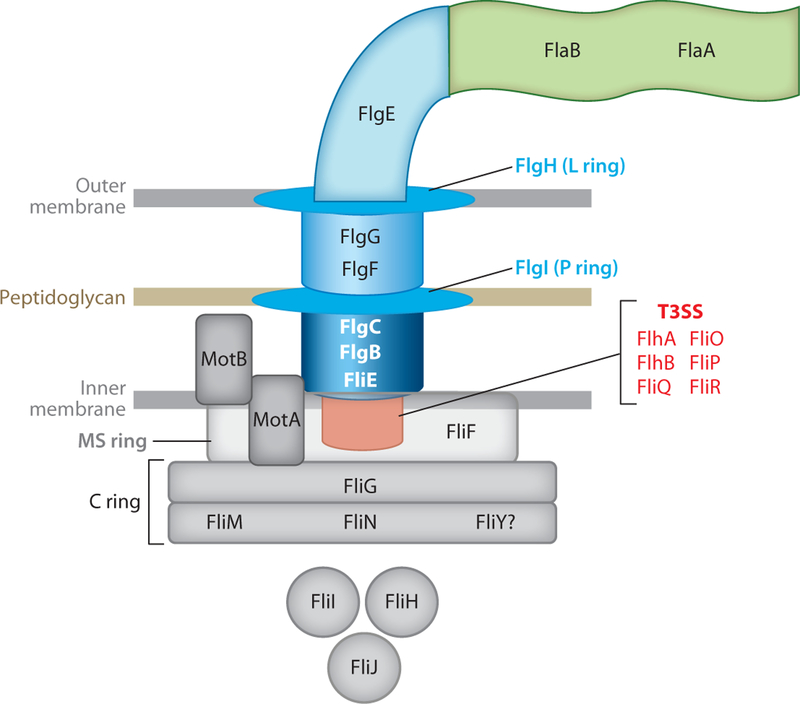
A flagellum can be divided into two broad substructures, the hook-basal body (HBB) complex and the extracellular filament (Figure 1). The HBB complex is further divided into three structures: (a) the base located in the cytoplasm and inner membrane; (b) the periplasmic rod and associated ring structures; and (c) the surface-localized hook.
Figure 1.
Structure of the flagellar organelle of Campylobacter jejuni and Helicobacter pylori. The major components common to the flagella of C. jejuni and H. pylori are shown. Other relatively less-characterized proteins specific to each bacterium are not shown for simplicity. Flagellar components are color-coded based on the classification of the respective genes in the transcriptional regulatory cascade for the organisms: red (class 1), blue (class 2), and green (class 3). An exception includes flaB, which is a σ54-dependent class 2 gene. Proteins in gray include those encoded by genes whose transcription has not been analyzed or is known to be outside the flagellar transcriptional hierarchy. Other class 1 proteins that are required for expression of σ54-dependent class 2 genes but are not part of the flagellar structure are not shown. Abbreviation: T3SS, type III secretion system.
The flagellar base consists of the MS ring, the flagellar type III secretion system (T3SS), the cytoplasmic C ring or switch complex, and the motor. The inner membrane MS ring is a homomultimer of FliF with a functional flagellar T3SS (composed of FlhA, FlhB, FliO, FliP, FliQ, and FliR) within the central membrane patch of the ring (Figure 1). The flagellar T3SS exports many proteins that are part of the flagellum beyond the inner membrane. The C ring, located at the cytoplasmic face of the MS ring, functions as the flagellar switch and also aids in secretion. This complex is composed of FliG, FliM, and FliN or FliY, although Epsilonproteobacteria contain both FliN and FliY (71; discussed below). FliG is responsible largely for the rotary component of the switch, turning the flagellum either clockwise or counterclockwise depending on input from the chemotaxis system. FliM and FliN bind the phosphory-lated form of the CheY (CheY-P) chemotaxis response regulator (95). As described below, CheY-P binding to the switch results in clockwise flagellar rotation; without CheY-P bound, counterclockwise rotation occurs. FliN additionally interacts with protein complexes involved in export such as substrate-chaperone partners or with the FliH-FliI complex that assists in efficient export through the T3SS (113). FliY shares homology with FliN and may have a partially redundant function in H. pylori (71). Motor components of the base include MotA and MotB, which form a proton channel and the stator component of the flagellum; these components function to link proton flow and generation of torque for flagellar rotation (77).
Most flagellar proteins located beyond the inner membrane are dependent on the T3SS, the MS ring, and the switch complex for export. The flagellum is made from the inside out, beginning with the MS and switch complex, continuing with the HBB complex, and terminating with the extracellular filament. Ordered secretion of proteins to promote proper flagellar biosynthesis is an inherent property of the T3SS. FliE, FlgB, and FlgC are the first rod proteins secreted, which polymerize on the periplasmic surface of the MS ring to form a hollow tube. Additional nascently secreted flagellar proteins (such as the distal FlgF and FlgG rod proteins) are deposited into this conduit and polymerize on the tip of the sprouting flagellar structure. This pattern of secretion and polymerization on the tip extends throughout the biosynthesis of the hook and filament.
FlgI and FlgH form the P ring in the pep-tidoglycan and the L ring in the outer membrane, respectively. These rings form around the rod, likely providing support and passage through these cell envelope components. After completion of the rod, FlgE and minor hook proteins are secreted. FliK determines the correct number of FlgE proteins needed to form a proper hook length in Campylobacter and Helicobacter species (57, 94). As shown in Salmonella species (29), FliK likely functions as a temporal molecular ruler to measure the hook length as it is synthesized. FliK is secreted intermittently through the forming HBB complex in a fairly linear fashion. Simultaneous contact of the N and C termini of FliK with FlgD, which is present at the tip of the growing hook, and with FlhB of the T3SS at the flagellar base, signifies that a hook of an ideal length has formed. An autoproteolytic cleavage event then occurs in FlhB that alters the substrate specificity of secretion by the T3SS so that proteins for filament biosynthesis, such as the flagellins, are then exported.
Flagellar biosynthesis terminates with filament biosynthesis. In C. jejuni and H. pylori, the filament is composed primarily of the major flagellin, FlaA, with the minor flagellin, FlaB, less abundant in the structure (67, 69). Whereas FlaA is essential for flagellation and motility, the requirement of FlaB varies between species (52, 104, 118, 119).
Regulatory Cascade for Flagellar Gene Expression
Expression of flagellar genes is controlled in a hierarchical manner that allows flagellar proteins to be produced in stages, facilitating their ordered secretion and proper interactions so that flagellar biosynthesis occurs correctly. This regulated mechanism of gene expression is accomplished by linking transcription of classes of flagellar genes to structural steps in flagellar biosynthesis.
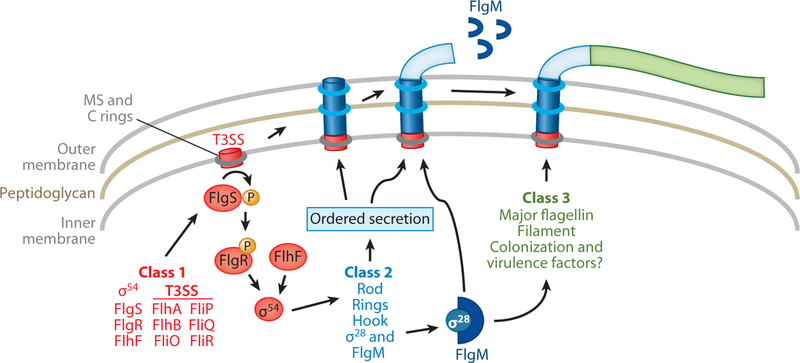
In motile bacteria, flagellar genes can be organized into classes on the basis of their temporal order of expression. Class 1 genes are transcribed first and usually encode master transcriptional regulators that control expression of class 2 genes, which include those encoding HBB proteins. Genes for these canonical flagellar master regulators appear to be absent from C. jejuni and H. pylori (87, 114). Instead, C. jejuni and H. pylori class 1 genes include those encoding the flagellar T3SS, the FlgSR two-component system (TCS), the FlhF GT-Pase, and σ54 (12, 45, 82, 103, 124) (Figure 2). Unlike genes encoding master regulators in other motile bacteria, expression of class 1 genes in C. jejuni and H. pylori appears to be constitutive.
Figure 2.
The flagellar transcriptional regulatory cascade for Campylobacter jejuni and Helicobacter pylori. The flagellar genes and respective proteins included in the flagellar transcriptional cascade are shown. These factors are color-coded on the basis of their classification in the transcriptional regulatory cascade: red (class 1 genes), blue (class 2 genes), and green (class 3 genes). fliA (encoding σ28) and flgM are included as class 2 genes in C. jejuni, but these genes are dependent only partially on σ54 for expression. Formation of the flagellar T3SS (which is composed of FlhA, FlhB, FliO, FliP, FliQ, and FliR) has been proposed to create a signal detected by the FlgS sensor kinase, resulting in autophosphorylation of the protein. Phosphotransfer to the FlgR response regulator activates the protein, allowing for interactions with and stimulation of σ54. The FlhF GTPase is likely involved in a separate pathway that converges with or functions downstream of the FlgSR-T3SS pathway to fully activate expression of σ54-dependent class 2 genes. The T3SS facilitates the ordered secretion of the class 2 rod, ring, and hook proteins to complete the HBB complex. Secretion of the antisigma factor FlgM through the HBB complex relieves repression of σ28 activity, resulting in expression of class 3 genes. The class 3 genes include flaA, which encodes the major flagellin, and those for other minor filament proteins. Other class 3 genes of C. jejuni appear to encode proteins not involved in motility and, instead, may be involved in virulence or colonization of C. jejuni. Abbreviations: HBB, hook-basal body; T3SS, type III secretion system.
Through genetic and biochemical analyses in C. jejuni, a regulatory pathway has been proposed for how class 1 proteins likely influence class 2 gene expression through σ54 (Figure 2) (5, 45, 56). Many aspects of this model appear to be valid for H. pylori (82, 103). In the C. jejuni model, two pathways either converge or work sequentially to influence activity of σ54 and, consequently, expression of class 2 genes. The first pathway involves communication between the FlgSR TCS and the flagellar T3SS (56). The FlgSR TCS is composed of the cytoplasmic FlgS histidine kinase and the FlgR response regulator, an NtrC-like transcriptional regulator. Members of the NtrC family of regulators commonly activate σ54 in bacteria (55, 56, 124). Activation of the FlgSR TCS is dependent on the flagellar T3SS components, as any T3SS mutant appears to disrupt phosphotransfer through FlgSR (56). FlgSR appears to function downstream of the T3SS on the basis of the finding that FlgR mutants that function independently of FlgS suppress the phenotypes of flagellar T3SS mutants for decreased expression of σ54-dependent flagellar genes (56). Because many class 2 proteins are dependent on the T3SS for secretion, the T3SS ultimately controls expression of its own secretion substrates.
It is not yet known how the flagellar T3SS activates FlgSR. Mutations in the T3SS that selectively impair secretion of the FlaA flagellin but not formation of the system do not cause defects in σ54-dependent flagellar gene expression (56). These findings suggest thatT3SS formation, rather than its secretory function, activates FlgS. FlgS could possibly detect creation of a T3SS by interacting directly with one or more components of the fully formed secretory system.
The second pathway required for expression of σ54-dependent class 2 genes involves the FlhF GTPase, but the mechanism is unclear. In a GTPase-independent manner, FlhF appears to function in a pathway that is separate from FlgSR and the flagellar T3SS (5) (Figure 2). It is not yet known whether these two pathways converge or whether the FlhF pathway functions downstream of the FlgSR-T3SS pathway to activate σ54.
Similar to other bacterial systems, the C. jejuni and H. pylori σ28 (encoded by fliA) is required for expression of class 3 genes such as flaA and other filament genes and is negatively controlled by the antisigma factor FlgM (12, 19, 45, 53, 123, 124) (Figure 2). As in the Salmonella system, expression of C. jejuni and H. pylori σ28-dependent class 3 genes is repressed until the HBB is completed because of FlgM (19, 53, 123). Repression is mediated by FlgM binding to σ28, which prevents the sigma factor from associating with RNA polymerase. Once the HBB complex is complete in C. jejuni and H. pylori, FlgM is secreted by the flagellar T3SS, which relieves σ28 from repression and allows it to interact with RNA polymerase to express class 3 genes (92, 123) (Figure 2). FlaA and other filament proteins are produced and secreted through the HBB complex for filament biosynthesis.
Curiously, FlgM-mediated repression of σ28 activity in C. jejuni is temperature sensitive (123). FlgM interacts with σ28 at 37° C but not at 42° C, which allows σ28-dependent class 3 gene expression to occur at 42° C regardless of the status of the HBB complex. It is not yet clear whether an advantage exists in expressing class 3 filament genes at 42 °C before a HBB is complete. However, σ28-dependent genes encoding proteins with functions not obviously involved in motility have been identified (12, 35, 90). Considering that avian species are natural hosts for C. jejuni and have body temperatures around 42° C, these proteins maybe expressed via a σ28-dependent, flagellum-independent mechanism to possibly assist in colonization of these hosts.
In other flagellar systems, mot genes encoding the stator components of the motor and genes encoding many chemosensory proteins are dependent on σ28 for expression (reviewed in Reference 18). Generally, these genes in C. jejuni or H. pylori are controlled neither by σ54 nor by σ28, with the exception of one H. pylori chemotaxis gene (53, 101). Expression of these genes appears to be constitutive and outside the flagellar transcriptional cascade, although there is one identified regulator for a chemoreceptor gene (25).
Phase-Variable Control of Motility
In addition to the complex transcriptional regulatory cascade described above, flagellar gene expression and biosynthesis are also controlled by phase variation in Campylobacter and Helicobacter species. Phase variation is a random and reversible genetic process that can affect transcription or translation of a target gene. One common mechanism of phase variation involves alteration in the number of nucleotide repeats in the coding sequences or promoters of genes. These alterations may affect the level of transcription of a gene or lead to frameshifting of the coding sequence to truncate translation of the encoded protein. These mechanisms allow transcription or translation to switch reversibly between phase “on” and phase “off” states.
In Campylobacter coli, a phase-variable homopolymeric tract of thymines is present in flhA, which encodes a flagellar T3SS component (86). Alteration of this polyT tract truncates FlhA to presumably affect both σ28- and σ54-dependent flagellar gene expression and motility. Motility in C. jejuni is affected by phase variation of the FlgSR TCS (42, 43). FlgSR phase variation occurs in homopolymeric nucleotide tracts consisting of adenine or thymine residues within the coding sequences offlgR or flgS, with additional variation of heteropolymeric nucleotide tracts occurring in flgS (42, 43). Phase variation of these repeats affect translation ofboth FlgS and FlgR, providing an additional level of control of σ54-dependent flagellar gene expression outside of the requirement of these proteins for signal transduction to activate σ54. To date, the FlgSR TCS is unique in that both regulatory proteins are controlled by phase-variable mechanisms. In H. pylori, phase variation of a homopolymeric tract of cytosines in fliP leads to a truncated T3SS component that does not alter expression of class 3 genes but prevents flagellar biosynthesis (51).
Posttranscriptional and Posttranslational Mechanisms Affecting Flagellar Gene Expression and Biosynthesis
Both posttranscriptional and posttranslational regulatory mechanisms influence flagellar biosynthesis in C. jejuni and H. pylori.
HP0958.
HP0958 is a cytoplasmic protein of H. pylori that binds both protein and mRNA substrates to influence transcription or translation of different flagellar components. HP0958 interacts with σ54, FliH (a regulator of the FliI ATPase that assists in dissociating flagellar proteins from chaperones for secretion), and flaA mRNA (27, 88, 91, 93). Binding flaA mRNA promotes translation of the major flagellin, whereas interactions with σ54 likely increase stability of the sigma factor (11, 88). An interesting hypothesis has been proposed that after completion of the HBB complex, newly transcribed class 3 flaA mRNAs may compete with σ54 for binding to HP0958. As a result, flaA mRNAs are translated with a concomitant decrease in σ54 levels due to instability after release from HP0958. Thus, a reduction in σ54 levels would result in decreased class 2 gene expression (27). If true, this mechanism would provide a possible explanation for cessation ofclass 2 flagellar gene expression after completion of the HBB complex in H. pylori.
Flagellin glycosylation influences flagellar biosynthesis.
Campylobacter and Helicobacter flagellins must be glycosylated posttranslationally for flagellar filament biosynthesis (26, 36, 38, 54, 97). FlaA flagellins of both species and the H. pylori FlaB flagellin are modified by an O-linked protein glycosylation system, which adds a glycan to specific serines or threonines of the flagellins (96, 97, 112). Pseudaminic acid, legionaminic acid, and modified versions of each glycan are the major glycan additions to the Campylobacter flagellins (70, 76, 96, 97, 112). In contrast, flagellin glycosylation in H. pylori is fairly homogeneous, with only pseudaminic acid modifying the proteins (97).
Flagellin glycosylation in Campylobacter and Helicobacter spp. contributes approximately 4%−10% of the protein mass (96, 97, 112). Sixteen to 19 different serines or threonines of C. jejuni FlaA and 7 serines or threonines of H. pylori FlaA are glycosylated (97, 112). These glycosylated residues are limited to the D2 and D3 subdomains of the flagellins, which in Salmonella species are the surface-exposed regions of the folded filament-incorporated flagellins (97, 112, 129). Curiously, only three specific residues of C. jejuni FlaA need to be glycosylated for normal filament biosynthesis and motility (30).
Although the function is not entirely clear, flagellin glycosylation likely provides additional subunit-subunit interactions for stacking the flagellins during filament biosynthesis. The Campylobacter and Helicobacter flagellins lack the D1 subdomain, which in the prototypical Salmonella flagellin aids in flagellin subunit interactions during filament biosynthesis (4,129). Glycosylation of the Campylobacter and Helicobacter flagellins may compensate for the lack of the D1 domain and provide structural alterations to the flagellins to promote filament biosynthesis.
SPATIAL AND NUMERICAL CONTROL OF POLAR FLAGELLAR BIOSYNTHESIS
Unlike peritrichous bacteria, flagellar biosynthesis in polarly flagellated bacteria is spatially and numerically restricted. Campylobacter species commonly produce a single flagellum at one or both bacterial poles, and H. pylori normally produces up to six flagella at a single pole. The FlhF GTPase and FlhG (annotated as FleN in Pseudomonas species) influence spatial and numerical parameters of flagellar biosynthesis in other polarly flagellated bacteria (20, 37, 63, 81, 85). In Vibrio cholerae flhF mutants, ff54-dependent flagellar gene expression is reduced, with only a few bacteria able to synthesize a flagellum; these flagella form at lateral sites on the cell body rather than at polar sites (20, 37). Pseudomonas flhF mutants also produce lateral rather than polar flagella, indicating that FlhF is involved in spatial control of flagellar biosynthesis (81, 85). FlhG/FleN of both Vibrio and Pseudomonas species regulate flagellar numbers by repressing the expression or activity of respective master transcriptional regulators that control flagellar gene expression (20, 23, 24). Increased flagellar gene expression in flhG/fleN mutants likely drives an increase in polar flagellar numbers. FlhG is encoded in Campylobacter and Helicobacter species but has not been well characterized in either bacterium.
The FlhF GTPases of C. jejuni and H. pylori are required for flagellar gene expression and biosynthesis, but each mechanism differs by the dependency on the GTPase activity of FlhF (5, 45, 82). As described above, C. jejuni FlhF is required for expression of ff54-dependent flagellar genes in a GTPase-independent manner. However, C. jejuni strains producing FlhF mutant proteins locked in a GTP-bound state (and with reduced GTP hydrolysis) generate populations of bacteria with diverse flagellar biosynthesis phenotypes, including production of multiple flagella at a single pole or flagella at lateral sites along the cell body (5). Thus, GTP hydrolysis is not required for flagellar gene expression but is required for controlling proper spatial and numerical parameters of flagellar biosynthesis.
The mechanism by which the GTPase activity of FlhF influences flagellar biosynthesis is currently unknown. One hypothesis for how FlhF may regulate spatial parameters of flagellar biosynthesis is that the protein may function to correctly position components of the flagellar base to the poles. Division of a C. jejuni cell with a flagellum at each pole results in daughter cells without a flagellum at the new pole. FlhF, through its GTPase activity, may specifically recognize newpoles of a daughter cells and assist in the synthesis of the MS ring, T3SS, and switch complex at the new pole. Placement of these initial components dictates where a flagella will be formed. In support of this hypothesis, FlhF is unipolarly localized in C. jejuni, but it is unclear if these poles are new or old poles after division (30). Continued exploration is required for understanding how the GTPase activity of FlhF is regulated and how this activity functions to precisely place a single flagellum only at poles.
A SHEATH ENCASES HELICOBACTER FLAGELLA
Each flagella of Helicobacter species is encased in a membranous sheath, a relatively atypical property of bacterial flagella (33, 34). The sheaths are contiguous with the outer membrane, but the mechanism of sheath biosynthesis is unknown. Mutations in flagellin or filament genes that reduce filament biosynthesis generally result in shorter sheaths equal to the length of the filament, suggesting that the sheath may form around the filament as it is synthesized (52). However, some mutants occasionally produce what were considered empty sheaths lacking internal flagella or short filaments with empty sheath extensions (52, 61). If the sheaths are indeed empty, it is unclear how a membrane would spontaneously form a sheath-like structure without an internal proteinaceous protrusion. In terms of function, one possibility is that the sheath prevents recognition by host antibodies or innate immune receptors, a suggestion that is supported by the observation thatflagellin-specific antibodies do not bind to H. pylori flagellins in sheaths (62). However, some nonpathogenic bacteria have sheathed flagella, indicating that the sheath likely has functions outside of promoting immune evasion or persistence in a host.
CHEMOTAXIS
Overview of Chemotaxis
Chemotactic signaling systems allow bacteria to follow favorable chemical gradients (117). The E. coli system represents the paradigm for chemotaxis, utilizing chemoreceptors to sense particular environmental cues and transmit this information to a signal transduction cascade that affects flagellar rotation (reviewed in Reference 117) (Figure 3). The core signal transduction proteins include chemoreceptors, the CheA kinase, the CheY response regulator, and the CheW coupling protein, which physically couples receptors to CheA (126). The direction of flagellar rotation is influenced by the phosphorylation status of CheY. The amount of CheY-P is regulated by ligand binding to the chemoreceptors, such that an attractant ligand squelches CheA kinase activity and non-phosphorylated CheY predominates. In this form, CheY fails to interact with the flagellar switch, the flagella rotate counterclockwise, and the bacteria swim. In the absence of attractant ligand, CheA phosphorylates CheY. CheY-Pbinds the flagellar switch to turn the flagellum clockwise, which promotes bacterial tumbling and direction changes.
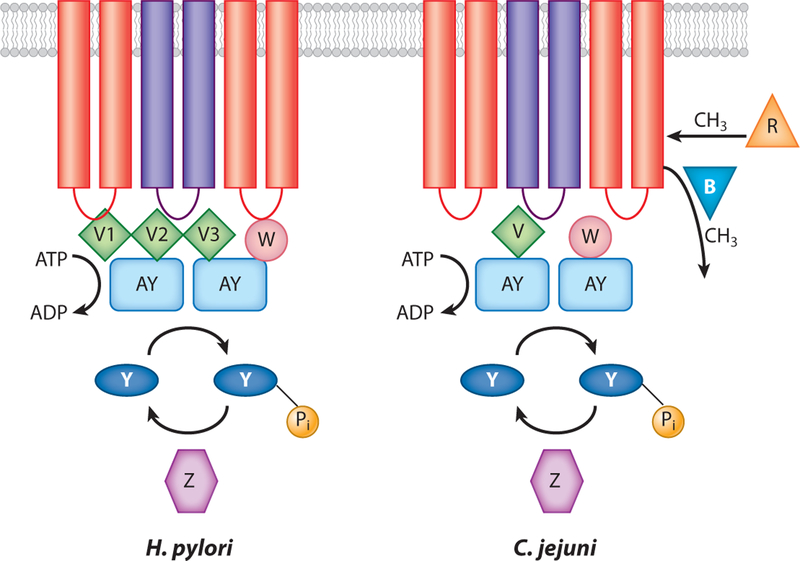
Figure 3.
Chemotaxis signal transduction systems of Campylobacter jejuni and Helicobacter pylori. Transmembrane chemoreceptors are shown as purple and red bars interacting with the coupling proteins CheW (W) or CheV (V). H. pylori contains three CheV proteins, termed CheV1 (V1), CheV2 (V2), and CheV3 (V3). The coupling proteins in turn interact with CheA, also called CheAY (AY), which catalyzes a kinase reaction using ATP as the phosphodonor. The phosphoryl group is passed to CheY (Y) and removed by CheZ (Z). C. jejuni contains CheR (R) and CheB (B), which methylate and demethylate, respectively, the chemoreceptors in other microbes. H. pylori and C. jejuni also contain cytoplasmic chemoreceptors that are not shown but are presumed to participate in the same protein-protein interactions.
Several chemotaxis proteins outside of the core chemotaxis set are commonly found in many but not all motile bacteria (126). The first set are phosphatases, such as CheZ and FliY, that accelerate the intrinsic dephosphorylation activity of CheY. The second set are the CheR and CheB enzymes that methylate and demethylate, respectively, chemoreceptor glutamyl residues to control adaptation responses to sustained ligand levels. The third set are the alternative coupling proteins such as CheV, which are homologs of CheW proteins with an additional receiver (REC) domain. H. pylori and C. jejuni collectively contain all of the above chemotaxis proteins and respond chemotactically to several different compounds (see sidebar, Chemotactic Signals for H. pylori and C. jejuni).
Chemotaxis During Infections
The role of chemotaxis in infection has been dissected using mutants that are Che− due to loss of core signal transduction proteins or mutants that lack individual chemoreceptors. Generally speaking, nonmotile mutants demonstrate the most severe in vivo growth defects, followed by Che− mutants, and lastly mutants lacking a particular chemoreceptor (3, 83, 111). This trend has been most well studied in H. pylori; in contrast, C. jejuni Che− or nonmotile mutants demonstrate similar colonization defects in hosts, with chemoreceptor mutants displaying more subtle defects (16, 40, 46, 84). In H. pylori, motility and chemotaxis play key roles in infection establishment. For example, H. pylori Che− mutants have a 50% infectious dose (ID50) 250-fold higher than wild-type bacteria, and isogenic nonmotile mutants have an ID50 25,000-fold higher than wild-type strains (83, 111). In some studies, H. pylori Che− mutants do not colonize stomachs of hosts (32, 75). This outcome may be due to differences in infectious dose between different rodent species and strains, such that infectious doses were not adequate to achieve colonization by Che− mutants. The colonization defects of H. pylori chemotaxis and motility mutants are exacerbated greatly in a competitive infection with wild-type strains; typically in these coinfections little or no mutant is recovered (83, 111).
It has been more difficult to assess whether chemotaxis plays a role in persistent infection. At a high dose, H. pylori Che− mutants establish colonization and persist for months similar to wild-type strains (111, 120). Che− mutants are found at roughly fivefold-lower levels than wild-type bacteria during the first month of infection, but the numbers of wild-type bacteria and Che− mutants equalize over time (111, 120). In addition, wild-type H. pylori can displace some Che− mutants from a preestablished infection (111). When two strains of equal colonization capacity are used in such a superinfection strategy, the first strain colonizing is not affected by the second strain (111). The finding that Che− mutants can be displaced from a persistent infection by wild-type bacteria suggests that chemotactic motility is beneficial in a persistent infection.
Chemotaxis furthermore influences H. pylori localization within the stomach. First, wild-type bacteria occupy both the corpus (acid-producing) and antrum regions of the stomach, whereas Che− mutants are restricted to the corpus (111). This finding suggests that chemotaxis either guides H. pylori to the antrum or helps it survive in this niche. Chemotaxis also promotes closer contact of H. pylori with the gastric epithelial cells (120). H. pylori Che− mutants elicit less host inflammation in mouse and gerbil models, possibly because of the observed decrease in host cell interactions as H. pylori adherence enhances the inflammatory response (39, 75, 120).
C. jejuni and H. pylori Contain Multiple Chemoreceptors
In H. pylori and C. jejuni, chemotactic signals are sensed by chemoreceptors that exist as either integral membrane or soluble cytoplasmic proteins. Chemoreceptors are multidomain proteins, bearing a sensory domain and a signaling domain (125). The signaling domain, also called the methyl-accepting (MA) domain, is highly conserved and interacts with either the CheW or the CheV coupling proteins (Figure 3). The chemoreceptor sensory domains are highly variable, residing extracytoplasmically in transmembrane chemoreceptors, but cytoplasmically in soluble ones (125). Sensory domains can sense ligands directly or indirectly via a protein-protein interaction with a ligand-bound periplasmic binding protein. It is not possible to predict the ligand for a given receptor or whether the receptor even binds a ligand directly (125). Several chemoreceptor sensory domains are well characterized (117), although none of the chemoreceptor sensory domains of the Epsilonproteobacteria is characterized to date.
H. pylori has three integral membrane chemoreceptors, TlpA, TlpB, and TlpC, and one soluble chemoreceptor, TlpD (3, 22, 99, 120). TlpA, TlpB, and TlpD have specific responses ascribed to them. TlpA has been proposed to sense arginine (15). TlpB mediates a response to low pH in a noncanonical chemotaxis assay (22). In this assay, wild-type H. pylori displays arcing swimming behavior and forms a wall of bacteria in response to a drop of 0.1 N HCl introduced under a coverslip; mutants lacking tlpB do not display this behavior. Movement away from low pH has been noted for many bacterial species and suggested to account for the observed localization of H. pylori within the stomach (98, 100). It will be exciting to see if additional pH taxis assays support these findings, as well as to determine the mechanistic details for TlpB-dependent pH sensing.
The soluble chemoreceptor TlpD mediates a tactic response to bacterial energy levels (99). Specifically, treating H. pylori with chemicals that disrupt electron transport and deplete cellular ATP levels causes the bacteria to swim without direction changes, a response that matches the predicted attractant response extrapolated from mutant phenotypes (72, 110). Adding back nutrients or electron donors boosted the frequency of directional switches for wild-type H. pylori but not for strains lacking tlpD or cheA. Further experiments showed that using 10-fold-lower concentrations of one of the electron transport disruption chemicals triggered a repellent response and prevented wild-type H. pylori from migrating into a compartment containing this chemical (99). These results thus suggest that TlpD senses something about cellular energy status. Future studies to examine how TlpD senses these cellular alterations, and the exact nature of what is altered, are exciting avenues for future work.
Three of the four H. pylori chemoreceptors promote stomach colonization in wild-type animal models. tlpA, tlpC, or tlpD mutants demonstrate colonization defects in competition studies with wild-type strains, although they colonize mice normally when any is the sole infecting bacterium (3, 22, 120; K.M. Ottemann, unpublished data). The role of TlpB in colonization has been difficult to interpret. Two studies report that H. pylori tlpB mutants have no colonization defect in either mice or gerbils and actually slightly outcompete their wild-type parental strain (75, 120). One study, however, found that H. pylori tlpB mutants have a defect in single-strain colonization assays in transgenic IL-12-deficient mice, which are typically more permissive for colonization (22, 47). Future experiments that analyze the phenotype of Che− mutants in this model would help scientists to understand the reported tlpB mutant defect.
C. jejuni contains seven integral membrane chemoreceptors and three soluble chemoreceptors. Three of these have been paired to input signals. The first is the chemoreceptor composed of the CetA and CetB proteins (44). CetA bears the MA domain plus a transmembrane domain, and CetB is a cytoplasmic protein with similarity to a critical region of the Aer oxygen-sensing chemoreceptor. These two proteins presumably function together and trigger a response to pyruvate, a respiratory electron donor, and fumarate, a respiratory electron acceptor (28, 44). This observation coupled with homology of CetA to domains of other energy-sensing chemoreceptors suggests that this system responds to some aspect of respiratory electron flow. The chemoreceptor Tlp7 is also encoded by two genes (cj0952c and cj0951c) and senses formate (109). However, it is unclear if Cj0952c and Cj0951c function together to sense this compound. The other signal-paired chemoreceptor is the integral membrane protein Tlp1, which senses aspartate directly as a chemoattractant (40).
Several of the C. jejuni chemoreceptors are required for in vivo colonization. Mutant strains lacking one of two chemoreceptors, Cj0019c/DocB or Cj0262c, resulted in lower numbers of C. jejuni in the chick ceca, while removal of the other chemoreceptors had no affect on colonization (46). Later work found that Cj1506c/tlp1 mutants in a different C. jejuni strain had a defect in chick intestinal colonization, suggesting that the importance of chemoreceptors in colonization may vary with strains (40).
Core Signaling Proteins: CheA, CheW, and CheY
Chemoreceptors communicate their ligand binding information to the flagellar motor via the chemotaxis system, utilizing the core signal transduction proteins CheA, CheW, and CheY. Discussion of each protein in Campylobacter and Helicobacter species is presented below.
Campylobacter and Helicobacter species contain CheA kinases with additional REC domains.
CheA plays a pivotal role in chemotaxis by promoting CheY phosphorylation (Figure 3). Both C. jejuni and H. pylori cheA mutants are nonchemotactic in vitro and show defects in colonization of mice, confirming a role for CheA in chemotaxis and also for chemotaxis in infection (16, 32, 111).
CheA proteins of C. jejuni and H. pylori differ somewhat from canonical CheA proteins primarily in two ways. First, C. jejuni and H. pylori CheA proteins have the typical domains that autophosphorylate, function in phosphorelay to downstream targets, and bind CheW, but these CheA proteins lack sequence conservation in the P2 CheY binding domain (125). Despite the lack of conservation in this region, the H. pylori CheA P2 portion does bind CheY and even prefers it to other H. pylori REC-domain-containing chemotaxis proteins such as CheA or CheV (64). Across the Epsilonproteobacteria, the P2 region generally is not well conserved. This lack of conservation is perhaps not surprising because the P2 domain of E. coli CheA can be removed with minimal slowing of phosphorelay to substrates and only modest decreases in chemotaxis (49). The variability of this region suggests that the rate of phosphotransfer from CheA to CheY might differ between strains.
The second modification to Campylobacter and Helicobacter CheA proteins is the addition of a REC domain in their C termini (32, 87, 74). REC domains are found in TCS sensor proteins and are typically phosphorylated to allow phosphorylation-based control of themselves or proteins to which they are attached. Many bacteria as well as most members of the Epsilonproteobacteria have CheA with an added REC domain, called CheAY. Wuichet et al. (125) observed that CheA proteins that lack a well-conserved P2 domain are typically CheAY proteins. It is not yet known, however, what role this REC domain plays in the function of any CheA. Several experiments in H. pylori have shown that CheA can phosphorylate the CheA REC domain but CheY is the preferred substrate (50). In addition, CheY can retrophosphorylate CheA and the phosphoryl group is transferred to the CheAREC domain (50). One possible function of the CheA REC domain is to act to remove phosphoryl groups from CheY, as a so-called phosphate sink, although such a model has not been proven.
The coupling protein CheW and response regulatory CheY.
CheW is a coupling or scaffolding protein that links chemoreceptors with CheA to modulate the kinase activity of CheA (Figure 3). CheW is essential for chemotaxis as C. jejuni and H. pylori cheW mutants are Che− presumably because CheA is not coupled to the receptors and is not active (89, 111, 115). Consistent with their Che− nature, H. pylori cheW mutants have decreased ability to colonize mice (111). Many critical residues are conserved between well-studied CheW proteins and those of H. pylori and C. jejuni, suggesting preservation of CheW function (1, 72).
CheY-P plays a key role in the chemotaxis system by binding to the flagellar switch proteins, FliM and FliN, and causing the motor to rotate in the clockwise, or nondefault, direction, which spurs direction changes (95, 117). As in model systems, H. pylori and C. jejuni cheY null mutants are Che− and colonize animals poorly (7, 32, 75, 84, 111).
C. jejuni and H. pylori CheY proteins are highly homologous to each other and maintain the conserved phosphorylation site at Asp53. As expected, H. pylori CheY can be phosphorylated and it interacts with FliM in a phosphorylation-dependent manner (50, 71). The crystal structure of H. pylori CheY-P shows that it has the basic structure of a typical REC domain, although it differs somewhat from E. coli CheY (64). These results suggest that H. pylori and E. coli CheY proteins may interact with their respective FliM proteins differently. This possibility was supported by the inability of H. pylori CheY to complement an E. coli cheYnull mutant even though H. pylori CheY is phosphorylated by E. coli CheA (64). However, the functional consequence of a possibly different CheY-FliM interaction remains to be established.
Accessory Chemotaxis Proteins
The function of the core chemotaxis proteins may be augmented by additional proteins that are not found in all chemotactic bacteria (126). These proteins fall into three classes: CheY-P phosphatases, adaptation-related CheR methyltransferases and CheB methylesterases, and CheV CheW-REC hybrid coupling proteins.
CheY-P phosphatases: CheZ and FliY.
CheY dephosphorylation is critical for efficiently controlling the direction of flagellar rotation to promote chemotaxis. Bacteria utilize several types of phosphatases for this reaction (107). H. pylori and C. jejuni produce the CheZ and FliY phosphatases which dephosphorylate CheY-P by enhancing the endogenous CheY autodephosphorylation activity (107, 130) (Figure 3). H. pylori and C. jejuni CheZ proteins are very distant from other CheZ proteins and only conserve residues corresponding to the active site and CheY-P binding regions (66, 110).
Biochemical analysis showed that the putative H. pylori CheZ (HP0170, CheZHP) employs the same mechanism as E. coli CheZ to de-phosphorylate CheY-P, including both a similar active site and a C-terminal binding region (66). CheZHP unexpectedly has an expanded substrate range and is able to dephosphorylate CheV2 and the REC domain of CheA. This CheZ is the first to have substrates identified beyond CheY, which suggests it can affect phosphate flow at multiple levels (66).
H. pylori and C. jejuni also produce the FliY phosphatase (71, 107). FliY is a component of the flagellar switch, along with FliM, FliG, and FliN. Bacteria typically encode only three flagellar switch proteins (FliM, FliG, and either FliY or FliN), but a common feature of Epsilonproteobacteria is that they encode all four, as discussed below (71). The well-studied B. sub-tilis FliY contains a CheY-P binding sequence and has phosphatase activity toward CheY-P (106). Neither C. jejuni nor H. pylori FliY contains an apparent CheY-P binding domain, but the FliY proteins harbor the active site consensus sequence of the CheC phosphatase family ((D/S-X3-E-X2-N-X22-P) (71, 80; P. Lertsethtakarn & K.M. Ottemann, unpublished data). It is unknown whether these proteins have phosphatase activity. Analysis of fliY mutants for in vivo phosphatase activity is complicated by the fact that H. pylori fliY mutants are nonmotile, therefore preventing analysis of chemotaxis phenotypes (71).
Methylation adaptation system proteins: CheB and CheR.
Chemotaxis systems respond to a wide range of chemical concentrations that extend from nanomolar to millimolar amounts because of a process known as adaptation (10). Chemotaxis adaptation is typically performed by the CheB methylesterase and the CheR methyltransferase. Another adaptation mechanism involves CheV, hybrids of CheW, and a REC domain (discussed below). Most of the Epsilonproteobacteria (other than H. pylori) encode CheB, CheR, and CheV; H. pylori lacks CheB and CheR and instead has three CheV proteins (Figure 3).
CheB and CheR are functionally antagonistic, demethylating and methylating chemoreceptors as part of the adaptation process. CheR in Epsilonproteobacteria contains the canonical CheR domain and thus is predicted to function similarly to other studied CheR proteins (125). CheB proteins typically contain both a CheB methylesterase domain and a REC domain that contributes phosphorylation-mediated control of the demethylation activity. CheB in C. jejuni and Helicobacter species, however, lacks the REC domain, suggesting it uses a different regulatory mechanism (74, 59). Recent studies have shown that these proteins function as expected to methylate/demethylate at least DocC and Tlp1, but apparently not CetA (59).
CheV coupling proteins.
CheV has been best studied in B. subtilis, where it has a dual function as a CheW-like coupling protein and in mediating adaptation to attractants (1). As noted above, H. pylori has three CheV proteins, CheV1, CheV2, and CheV3, whereas other Epsilonproteobacteria have only one protein, simply annotated as CheV, and it is most closely related to H. pylori CheV3 (Figure 3). All three CheV proteins of H. pylori have roles in chemotaxis but their exact functions have not been determined (72, 89). H. pylori cheV1 mutants have the most severe chemotaxis phenotype, although they are not as impaired as Che− strains, whereas mutants lacking cheV2 or cheV3 show only very modest chemotaxis defects. cheV1 and cheV2 mutant strains display predominantly smoothswimming behavior akin to cheW, cheY, or cheA mutants; cheV3 mutants have opposite hyperswitching behavior (72). C. jejuni cheVmutants have a chemotaxis defect similar to that of a cheW mutant, suggesting CheV plays a role equal to that of CheW in this microbe (40).
CheV phosphorylation is required for adaptation in B. subtilis, but the phosphorylation status of Campylobacter and Helicobacter CheV proteins is not yet clear (1). H. pylori CheV proteins catalyze the removal of phosphate from CheA, but stable phosphorylated CheV proteins have not been detected, suggesting possibly that CheV-P has a very short half-life (50, 66). Alternative methods using smallmolecule phosphodonors suggest that CheV proteins are poorly phosphorylated, so the phosphorylation status of these proteins in Epsilonproteobacteria is not yet clear (66, 89).
It is not yet known in general whether particular chemoreceptors prefer CheV or CheW for CheA coupling (1). The C. jejuni chemoreceptor Tlp1/Cj1506 interacted more strongly with CheV than with CheW, supporting the importance of CheV in this system and possibly suggesting some chemoreceptors preferentially interact with CheV versus CheW (40).
Flagellar Switch Proteins: FliG, FliM, FliN, and FliY
The signal from the chemoreceptor complex is relayed to the flagellar motor via the switch complex that consists of FliG, FliM, and FliN (77). As noted above, C. jejuni, H. pylori, and all other Epsilonproteobacteria encode an additional switch protein, FliY, which is homologous to FliN at its C terminus (71). Mutation of any one of these genes in H. pylori results in nonmotile strains. H. pylorifliG orfliMmutants are aflagellated, whereas fliNorfliYmutants retain partial flagellation (2, 71). fliN and fliY appear to be redundant in H. pylori, as double fliN/fliYmutants are aflagellated (71). Although the role of each protein is not yet fully known in C. jejuni or H. pylori, it seems that all four switch proteins are essential for full flagellation and motility (71).
In the E. coli system, CheY-P binds to FliM and FliN to control rotational direction of the flagellar rotor FliG (95, 117). H. pylori CheY-P seems to interact similarly with FliM, but interactions with FliN have not yet been tested (71). One spontaneous fliM mutation has been analyzed, with results supporting the finding that H. pylori FliM functions similarly to that of E. coli (71).
CONCLUDING REMARKS
Understanding mechanisms of flagellar gene expression, flagellar biosynthesis, and chemotactic responses in Campylobacter and Helicobacter species is still in the relatively early stages of scientific exploration. However, within this relatively short time frame, significant advances have been made in these species to identify key mechanisms that have so far shown noteworthy significant deviations from paradigmatic models of E. coli and Salmonella species. Thus, continued exploration into regulatory mechanisms of flagellar gene transcription and biosynthesis and the chemotactic signaling systems of C. jejuni and H. pylori will undoubtedly open new paradigms of how bacteria achieve a functional, chemotactic rotary nanomachine to promote swimming behavior necessary for infectious processes.
CHEMOTACTIC SIGNALS FOR H. PYLORI AND C. JEJUNI.
H. pylori and C. jejuni respond chemotactically to several compounds. Recent findings suggest that assays that work well for analyzing chemotaxis in E. coli are not conducive to other bacteria (58, 68). Specifically, the chemical-in-plug assay gives falsepositive responses for both H. pylori and C. jejuni (58, 68). In addition, H. pylori is notmotile in buffers that lack complex nutrients (K.M. Ottemann, unpublished data), although several studies utilize such solutions. With these caveats in mind, reported attractants and repellents are noted below.
H. pylori responds to urea (15, 78, 121), bicarbonate (15, 78), arginine and several other amino acids (15, 121), and cholesterol (127) as attractants. H. pylori is repelled by low pH (22) andNiCl2 (15); alterations in protonmotive force trigger both attractant and repellent responses (99).
C. jejuni was initially reported to be attracted to multiple amino acids, carbohydrates, and organic acids (48). Several of these responses, however, were subsequently found to occur in nonchemotactic mutants and thus represent false-positive responses (58). Several compounds have withstood additional chemotaxis assays, however, with fucose, pyruvate, fumarate, aspartate, and formate being attractants for C. jejuni (40, 44, 48, 109, 116).
SUMMARY POINTS.
Relative to many other motile bacteria, C. jejuni and H. pylori are capable of a characteristically high velocity of flagellar motility in substances of moderately high viscosity.
A transcriptional regulatory cascade involving the flagellar T3SS, the FlgSR TCS, the FlhF GTPase, and σ54 is essential for expression of many genes required to form the flagellar HBB complex in C. jejuni and H. pylori.
Highly regulatedmechanisms exist to tightly control the spatial and numerical parameters of flagellar biosynthesis in the polarly flagellated Campylobacter and Helicobacter species.
C. jejuni and H. pylori contain the basic core chemotaxis components—chemoreceptors, CheA, CheW, and CheY—that all microbes have. These appear to function similarly to orthologs in model systems, although CheA in particular has some atypical modifications.
The chemotaxis system of H. pylori is unusual in that it contains no methylationbased adaptation systems and produces multiple copies of the CheV coupling/adaptation protein.
Chemoreceptors of C. jejuni and H. pylori contain nonconserved ligand binding domains, and only Tlp1 of C. jejuni has been shown to bind a particular ligand directly.
FUTURE ISSUES.
How do the flagellar T3SS and the FlgS sensor kinase communicate to initiate phosphotransfer to FlgR for full activation of σ54-dependent flagellar gene expression?
How does the GTPase activity of FlhF contribute to regulating the spatial and numerical parameters of flagellar biosynthesis?
Are other factors such as FlhG involved in influencing parameters of polar flagellar biosynthesis?
How does the flagellar sheath form around each filament in H. pylori?
What ligands are sensed by each chemoreceptor and are these direct or indirect?
Does H. pylori adapt to sustained ligand, and if so, are the CheV proteins involved?
Do motility and/or chemotaxis serve functions in vivo beyond nutrient acquisition, such as immune avoidance or transmission?
ACKNOWLEDGMENTS
Research in the laboratory of David R. Hendrixson is supported by NIH grant R01 AI065539 and by National Research Initiative grant 2009-35201-05039 from the USDA Cooperative State Research, Education, and Extension Service Food Safety Program. Research in the laboratory of Karen M. Ottemann was made possible by grant number AI050000 from the National Institutes of Allergy and Infectious Diseases (NIAID) of the NIH. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or any other funding agency.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. 2010. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol 18:494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan E, Dorrell N, Foynes S, Anyim M, Wren BW. 2000. Mutational analysis of genes encoding the early flagellar components of Helicobacter pylori: evidence for transcriptional regulation of flagellin A biosynthesis. J. Bacteriol 182:5274–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andermann TM, Chen YT, Ottemann KM. 2002. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect. Immun 70:5877–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, et al. 2005. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. USA 102:9247–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balaban M, Joslin SN, Hendrixson DR. 2009. FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J. Bacteriol 191:6602–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beery JT, Hugdahl MB, Doyle MP. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol 54:2365–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beier D, Spohn G, Rappuoli R, Scarlato V. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol 179:4676–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg HC, Turner L. 1979. Movement of microorganisms in viscous environments. Nature 278:349–51 [DOI] [PubMed] [Google Scholar]
- 9.Bonis M, Ecobichon C, Guadagnini S, Prevost MC, Boneca IG. 2010. AM23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol. Microbiol 78:809–19 [DOI] [PubMed] [Google Scholar]
- 10.Bray D 2002. Bacterial chemotaxis and the question of gain. Proc. Natl. Acad. Sci. USA 99:7–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caly DL, O’Toole PW, Moore SA. 2010. The 2.2-A structure of the HP0958 protein from Helicobacter pylori reveals a kinked anti-parallel coiled-coil hairpin domain and a highly conserved ZN-ribbon domain. J. Mol. Biol 403:405–19 [DOI] [PubMed] [Google Scholar]
- 12.Carrillo CD, Taboada E, Nash JH, Lanthier P, Kelly J, et al. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem 279:20327–38 [DOI] [PubMed] [Google Scholar]
- 13.Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I,et al. 2009. Helicobacterpylori moves through mucus by reducing mucin viscoelasticity. Proc. Natl. Acad. Sci. USA 106:14321–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Center for Disease Control. 2010. Preliminary FoodNet data on the incidence ofinfectionwith pathogens transmitted commonly through food - 10 states, 2009. MMWR Morb. Mortal. Wkly Rep 59:418–22 [PubMed] [Google Scholar]
- 15.Cerda O, Rivas A, Toledo H. 2003. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMSMicrobiol. Lett 224:175–81 [DOI] [PubMed] [Google Scholar]
- 16.Chang C, Miller JF. 2006. Campylobacter jejuni colonization of mice with limited enteric flora. Infect. Immun 74:5261–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol 6:455–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev 64:694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colland F, Rain JC, Gounon P, Labigne A, Legrain P, De Reuse H.2001. Identification of the Helicobacter pylori anti-a28 factor. Mol. Microbiol 41:477–87 [DOI] [PubMed] [Google Scholar]
- 20.Correa NE, Peng F, Klose KE. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol 187:6324–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cover TL, Blaser MJ. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croxen MA, Sisson G, Melano R, Hoffman PS. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol 188:2656–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dasgupta N, Arora SK, Ramphal R. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol 182:357–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol 183:6636–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delany I, Spohn G, Rappuoli R, Scarlato V. 2002. Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacterpylori. J. Bacteriol 184:4800–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doig P, Kinsella N, Guerry P, Trust TJ. 1996. Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol. Microbiol 19:379–87 [DOI] [PubMed] [Google Scholar]
- 27.Douillard FP, Ryan KA, Caly DL, Hinds J, Witney AA, et al. 2008. Posttranscriptional regulation of flagellin synthesis in Helicobacter pylori by the RpoN chaperone HP0958. J. Bacteriol 190:7975–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott KT, Dirita VJ 2008. Characterization of CetA and CetB, a bipartite energy taxis system in Campylobacter jejuni. Mol. Microbiol 69:1091–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erhardt M, Hirano T, Su Y, Paul K, Wee DH, et al. 2010. The role of the FliK molecular ruler in hook-length control in Salmonella enterica. Mol. Microbiol 75:1272–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewing CP, Andreishcheva E, Guerry P. 2009. Functional characterization of flagellin glycosylation in Campylobacter jejuni 81–176. J. Bacteriol 191:7086–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrero RL, Lee A. 1988. Motility of Campylobacter jejuni in a viscous environment: comparison with conventional rod-shaped bacteria. J. Gen. Microbiol 134:53–59 [DOI] [PubMed] [Google Scholar]
- 32.Foynes S, Dorrell N, Ward SJ, Stabler RA, McColm AA, et al. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun 68:2016–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geis G, Leying H, Suerbaum S, Mai U, Opferkuch W. 1989. Ultrastructure and chemical analysis of Campylobacter pylori flagella. J. Clin. Microbiol 27:436–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodwin CS,McCulloch RK, Armstrong JA, Wee SH. 1985. Unusual cellular fatty acids and distinctive ultrastructure in a new spiral bacterium (Campylobacter pyloridis) from the human gastric mucosa. J. Med. Microbiol 19:257–67 [DOI] [PubMed] [Google Scholar]
- 35.Goon S, Ewing CP, Lorenzo M, Pattarini D, Majam G, Guerry P. 2006. A σ28-regulated nonflagella gene contributes to virulence of Campylobacter jejuni 81–176. Infect. Immun 74:769–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol 50:659–71 [DOI] [PubMed] [Google Scholar]
- 37.Green JC, Kahramanoglou C, Rahman A, Pender AM, Charbonnel N, Fraser GM. 2009. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J. Mol. Biol 391:679–90 [DOI] [PubMed] [Google Scholar]
- 38.Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, et al. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol 60:299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guruge JL, Falk PG, Lorenz RG, Dans M, Wirth HP, et al. 1998. Epithelial attachment alters the outcome ofHelicobacterpylori infection. Proc. Natl. Acad. Sci. USA 95:3925–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartley-Tassell LE, Shewell LK, Day CJ, Wilson JC, Sandhu R, et al. 2010. Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol. Microbiol 75:710–30 [DOI] [PubMed] [Google Scholar]
- 41.Hazell SL, Lee A, Brady L, Hennessy W. 1986. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J. Infect. Dis 153:658–63 [DOI] [PubMed] [Google Scholar]
- 42.Hendrixson DR. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol 61:1646–59 [DOI] [PubMed] [Google Scholar]
- 43.Hendrixson DR. 2008. Restoration of flagellar biosynthesis byvaried mutational events in Campylobacter jejuni. Mol. Microbiol 70:519–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendrixson DR, Akerley BJ, DiRita VJ. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol 40:214–24 [DOI] [PubMed] [Google Scholar]
- 45.Hendrixson DR, DiRita VJ. 2003. Transcription of a54-dependent but not a28-dependent flagellar genes in Campylobacterjejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol 50:687–702 [DOI] [PubMed] [Google Scholar]
- 46.Hendrixson DR, DiRita VJ. 2004. Identification of Campylobacterjejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol 52:471–84 [DOI] [PubMed] [Google Scholar]
- 47.Hoffman PS, Vats N, Hutchison D, Butler J, Chisholm K, et al. 2003. Development of an interleukin-12-deficient mouse model that is permissive for colonization by a motile KE26695 strain ofHelicobacter pylori. Infect. Immun 71:2534–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hugdahl MB, Beery JT, Doyle MP. 1988. Chemotactic behavior of Campylobacterjejuni. Infect. Immun 56:1560–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jahreis K, Morrison TB, Garzon A, Parkinson JS. 2004. Chemotactic signaling by an Escherichia coli CheA mutant that lacks the binding domain for phosphoacceptor partners. J. Bacteriol 186:2664–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jimenez-Pearson MA, Delany I, Scarlato V, Beier D. 2005. Phosphate flow in the chemotactic response system ofHelicobacterpylori. Microbiology 151:3299–311 [DOI] [PubMed] [Google Scholar]
- 51.Josenhans C, Eaton KA, Thevenot T, Suerbaum S. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun 68:4598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Josenhans C, Labigne A, Suerbaum S. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol 177:3010–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Josenhans C, Niehus E, Amersbach S, Horster A, Betz C, et al. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol 43:307–22 [DOI] [PubMed] [Google Scholar]
- 54.Josenhans C, Vossebein L, Friedrich S, Suerbaum S. 2002. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol. Lett 210:165–72 [DOI] [PubMed] [Google Scholar]
- 55.Joslin SN, Hendrixson DR. 2008. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J. Bacteriol 190:2422–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joslin SN, Hendrixson DR. 2009. Activation of the Campylobacterjejuni FlgSR two component system is linked to the flagellar export apparatus. J. Bacteriol 191:2656–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamal N, Dorrell N,Jagannathan A, Turner SM, Constantinidou C,et al. 2007. Deletion of a previously uncharacterized flagellar-hook-length control gene fliK modulates the sigma54 -dependent regulon in Campylobacter jejuni. Microbiology 153:3099–111 [DOI] [PubMed] [Google Scholar]
- 58.Kanungpean D, Kakuda T, Takai S. 2011. False positive responses of Campylobacter jejuni when using the chemical-in-plug chemotaxis assay. J. Vet. Med. Sci 73:389–91 [DOI] [PubMed] [Google Scholar]
- 59.Kanungpean D, Kakuda T,Takai S. 2011Participationof CheRand CheB in the chemosensoryresponse of Campylobacter jejuni. Microbiology 157:1279–89 [DOI] [PubMed] [Google Scholar]
- 60.Karim QN, Logan RP, Puels J, Karnholz A, Worku ML. 1998. Measurement of motility of Helicobacter pylori, Campylobacter jejuni, and Escherichia coli by real time computer tracking using the Hobson BacTracker. J. Clin. Pathol 51:623–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim JS, Chang JH, Chung SI, Yum JS. 1999. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J. Bacteriol 181:6969–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kostrzynska M, Betts JD, Austin JW, Trust TJ. 1991. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol 173:937–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kusumoto A, Shinohara A, Terashima H, Kojima S, Yakushi T, Homma M. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390–99 [DOI] [PubMed] [Google Scholar]
- 64.Lam KH, Ling TK, Au SW. 2010. Crystal structure of activated CheY1 from Helicobacter pylori. J. Bacteriol 192:2324–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee A, O’Rourke JL, Barrington PJ, Trust TJ. 1986. Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacterjejuni: a mouse cecal model. Infect. Immun 51:536–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lertsethtakarn P, Ottemann KM. 2010. A remote CheZ orthologue retains phosphatase function. Mol. Microbiol 77:225–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leying H, Suerbaum S, Geis G, Haas R. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol 6:2863–74 [DOI] [PubMed] [Google Scholar]
- 68.Li J, Go AC, Ward MJ, Ottemann KM. 2010. The chemical-in-plug bacterial chemotaxis assay is prone to false positive responses. BMCRes. Notes 3:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Logan SM, Harris LA, Trust TJ. 1987. Isolation and characterization of Campylobacter flagellins. J. Bacteriol 169:5072–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Logan SM, Kelly JF, Thibault P, Ewing CP, Guerry P. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol 46:587–97 [DOI] [PubMed] [Google Scholar]
- 71.Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. 2009. Functional analysis of the Helicobacter pylori flagellar switch proteins. J. Bacteriol 191:7147–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lowenthal AC, Simon C, Fair AS, Mehmood K, Terry K, et al. 2009. A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology 155:1181–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol 57:77–100 [DOI] [PubMed] [Google Scholar]
- 74.Marchant J, Wren B, Ketley J. 2002. Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol 10:155–59 [DOI] [PubMed] [Google Scholar]
- 75.McGee DJ, Langford ML, Watson EL, Carter JE, Chen YT, Ottemann KM. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun 73:1820–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McNally DJ, Aubry AJ, Hui JP, Khieu NH, Whitfield D, et al. 2007. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J. Biol. Chem 282:14463–75 [DOI] [PubMed] [Google Scholar]
- 77.Minamino T, Imada K, Namba K. 2008. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol 18:693–701 [DOI] [PubMed] [Google Scholar]
- 78.Mizote T, Yoshiyama H, Nakazawa T. 1997. Urease-independent chemotactic responses of Helicobacter pylori to urea, urease inhibitors, and sodium bicarbonate. Infect. Immun 65:1519–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molbak K, Havelaar A. 2008. Burden ofillness ofcampylobacteriosis and sequelae In Campylobacter, ed. Nachamkin I, Szymanski CM, Blaser MJ, pp. 151–62. Washington, DC: ASM Press [Google Scholar]
- 80.Muff TJ, Ordal GW. 2008. The diverse CheC-type phosphatases: chemotaxis and beyond. Mol. Microbiol 70:1054–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murray TS, Kazmierczak BI. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol 188:6995–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, et al. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol 52:947–61 [DOI] [PubMed] [Google Scholar]
- 83.Ottemann KM, Lowenthal AC. 2002. Helicobacterpylori uses motility for initial colonization and to attain robust infection. Infect. Immun 70:1984–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ottemann KM, Miller JF. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol 24:1109–17 [DOI] [PubMed] [Google Scholar]
- 85.Pandza S, Baetens M, Park CH, Au T, Keyhan M, Matin A. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol 36:414–23 [DOI] [PubMed] [Google Scholar]
- 86.Park SF, Purdy D, Leach S. 2000. Localized reversible frameshift mutation in the flhA gene confers phase variability to flagellin gene expression in Campylobacter coli. J. Bacteriol 182:207–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–68 [DOI] [PubMed] [Google Scholar]
- 88.Pereira L, Hoover TR. 2005. Stable accumulation of sigma54 in Helicobacter pylori requires the novel protein HP0958. J. Bacteriol 187:4463–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pittman MS, Goodwin M, Kelly DJ. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493–504 [DOI] [PubMed] [Google Scholar]
- 90.Poly F, Ewing C, Goon S, Hickey TE, Rockabrand D, et al. 2007. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect. Immun 75:3859–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, et al. 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409:211–15 [DOI] [PubMed] [Google Scholar]
- 92.Rust M, Borchert S, Niehus E, Kuehne SA, Gripp E, et al. 2009. The Helicobacter pylori anti-sigma factor FlgM is predominantly cytoplasmic and cooperates with the flagellar basal body protein FlhA. J. Bacteriol 191:4824–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ryan KA, Karim N, Worku M, Moore SA, Penn CW, O’Toole PW. 2005. HP0958 is an essential motility gene in Helicobacter pylori. FEMS Microbiol. Lett 248:47–55 [DOI] [PubMed] [Google Scholar]
- 94.Ryan KA, Karim N, Worku M, Penn CW, O’Toole PW. 2005. Helicobacterpylori flagellar hook-filament transition is controlled by a FliK functional homolog encoded by the gene HP0906. J. Bacteriol 187:5742–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sarkar MK, Paul K, Blair D. 2010. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. USA 107:9370–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schirm M, Schoenhofen IC, Logan SM, Waldron KC, Thibault P. 2005. Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal. Chem 77:7774–82 [DOI] [PubMed] [Google Scholar]
- 97.Schirm M, Soo EC, Aubry AJ, Austin J, Thibault P, Logan SM. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol 48:1579–92 [DOI] [PubMed] [Google Scholar]
- 98.Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, et al. 2004. The spatial orientation of Helicobacterpylori in the gastric mucus. Proc. Natl. Acad. Sci. USA 101:5024–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schweinitzer T, Mizote T, Ishikawa N, Dudnik A, Inatsu S, et al. 2008. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J. Bacteriol 190:3244–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seymour FW, Doetsch RN. 1973. Chemotactic responses by motile bacteria. J. Gen. Microbiol 78:287–96 [DOI] [PubMed] [Google Scholar]
- 101.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–55 [DOI] [PubMed] [Google Scholar]
- 102.Shigematsu M, Umeda A, Fujimoto S, Amako K. 1998. Spirochaete-like swimming mode of Campylobacter jejuni in a viscous environment. J. Med. Microbiol 47:521–26 [DOI] [PubMed] [Google Scholar]
- 103.Spohn G, Scarlato V. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol 181:593–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suerbaum S, Josenhans C, Labigne A. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol 175:3278–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, et al. 2010. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell 141:822–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szurmant H, Muff TJ, Ordal GW. 2004. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J. Biol. Chem 279:21787–92 [DOI] [PubMed] [Google Scholar]
- 107.Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev 68:301–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Szymanski CM, King M, Haardt M, Armstrong GD. 1995. Campylobacterjejuni motility and invasion of Caco-2 cells. Infect. Immun 63:4295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tareen AM, Dasti JI, Zautner AE, Gross U, Lugert R. 2010. Campylobacter jejuni proteins Cj0952c and Cj0951c affect chemotactic behavior towards formic acid and are important for invasion of host cells. Microbiology 156:3123–35 [DOI] [PubMed] [Google Scholar]
- 110.Terry K, Go AC, Ottemann KM. 2006. Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol. Microbiol 61:871–82 [DOI] [PubMed] [Google Scholar]
- 111.Terry K, Williams SM, Connolly L, Ottemann KM. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun 73:803–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thibault P, Logan SM, Kelly JF, Brisson JR, Ewing CP, et al. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem 276:34862–70 [DOI] [PubMed] [Google Scholar]
- 113.Thomas J, Stafford GP, Hughes C. 2004. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. USA 101:3945–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–47 [DOI] [PubMed] [Google Scholar]
- 115.van Alphen LB, Bleumink-Pluym NM, Rochat KD, van Balkom BW, Wosten MM, van Putten JP. 2008. Active migration into the subcellular space precedes Campylobacter jejuni invasion of epithelial cells. Cell Microbiol 10:53–66 [DOI] [PubMed] [Google Scholar]
- 116.Vegge CS, Brondsted L, Li YP, Bang DD, Ingmer H. 2009. Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl. Environ. Microbiol 75:5308–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol 5:1024–37 [DOI] [PubMed] [Google Scholar]
- 118.Wassenaar TM, Bleumink-Pluym NMC, Newell DG, Nuijten PJ, van der Zeijst BAM. 1994. Differential flagellin expression in a flaAflaB+ mutant of Campylobacter jejuni. Infect. Immun 62:3901–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wassenaar TM, Bleumink-Pluym NMC, van der Zeijst BAM. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBOJ 10:2055–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun 75:3747–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Worku ML, Karim QN, Spencer J, Sidebotham RL. 2004. Chemotactic response of Helicobacter pylori to human plasma and bile. J. Med. Microbiol 53:807–11 [DOI] [PubMed] [Google Scholar]
- 122.Worku ML, Sidebotham RL, Baron JH, Misiewicz JJ, Logan RP, et al. 1999. Motility of Helicobacter pylori in a viscous environment. Eur. J. Gastroenterol. Hepatol 11:1143–50 [DOI] [PubMed] [Google Scholar]
- 123.Wosten MM, van Dijk L, Veenendaal AK, de Zoete MR, Bleumink-Pluijm NM, van Putten JP. 2010. Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol. Microbiol 75:1577–91 [DOI] [PubMed] [Google Scholar]
- 124.Wosten MMSM, Wagenaar JA, van Putten JPM. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem 279:16214–22 [DOI] [PubMed] [Google Scholar]
- 125.Wuichet K, Alexander RP, Zhulin IB. 2007. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol 422:1–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal 3:ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wunder C, Churin Y, Winau F, Warnecke D, Vieth M, et al. 2006. Cholesterol glucosylation promotes immune evasion by Helicobacter pylori. Nat. Med 12:1030–38 [DOI] [PubMed] [Google Scholar]
- 128.Yao R, Burr DH, Guerry P. 1997. CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol 23:1021–31 [DOI] [PubMed] [Google Scholar]
- 129.Yonekura K, Maki-Yonekura S, Namba K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643–50 [DOI] [PubMed] [Google Scholar]
- 130.Zhao R, Collins EJ, Bourret RB, Silversmith RE. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol 9:570–75 [DOI] [PubMed] [Google Scholar]