Abstract
The most severe manifestation of peripheral arterial disease (PAD) is critical limb ischemia (CLI). CLI patients suffer high rates of amputation and mortality; accordingly, there remains a clear need both to better understand CLI and to develop more effective treatments. Gastrocnemius muscle was obtained from 32 older (51–84 years) non-PAD controls, 27 claudicating PAD patients (ankle-brachial index [ABI] 0.65 ± 0.21 SD), and 19 CLI patients (ABI 0.35 ± 0.30 SD) for whole transcriptome sequencing and comprehensive mitochondrial phenotyping. Comparable permeabilized myofiber mitochondrial function was paralleled by both similar mitochondrial content and related mRNA expression profiles in non-PAD control and claudicating patient tissues. Tissues from CLI patients, despite being histologically intact and harboring equivalent mitochondrial content, presented a unique bioenergetic signature. This signature was defined by deficits in permeabilized myofiber mitochondrial function and a unique pattern of both nuclear and mitochondrial encoded gene suppression. Moreover, isolated muscle progenitor cells retained both mitochondrial functional deficits and gene suppression observed in the tissue. These findings indicate that muscle tissues from claudicating patients and non-PAD controls were similar in both their bioenergetics profile and mitochondrial phenotypes. In contrast, CLI patient limb skeletal muscles harbor a unique skeletal muscle mitochondriopathy that represents a potentially novel therapeutic site for intervention.
Keywords: Metabolism
Keywords: Atherosclerosis, Cardiovascular disease, Skeletal muscle
Critical limb ischemia is characterized by consistent and pervasive deficits in skeletal muscle oxidative metabolism.
Introduction
Peripheral arterial disease (PAD) has historically been viewed as a progressive continuum from intermittent claudication (IC) to critical limb ischemia (CLI). Despite the presence of overlapping indices of disease severity (i.e., ankle-brachial index [ABI], associated comorbidities, and occlusive plaque anatomical location; see ref. 1), 70%–80% of IC patients do not progress to CLI (2–6). This suggests that CLI may be a unique manifestation of PAD with distinct etiology.
In a series of recent preclinical studies, we found that the onset of the muscle myopathy increases susceptibility to CLI and is defined by decreased muscle mitochondrial function (7–12). Moreover, therapeutically targeting mitochondria protects the skeletal muscle against ischemia-induced mitochondrial dysfunction, reducing the onset of the myopathy and preventing limb necrosis (8, 10). Mitochondria are well recognized to serve as gatekeepers for the mortality of ischemic cells (13, 14) and, as such, these findings raise the intriguing possibility that the etiology of CLI may originate in skeletal muscle from disruptions in cellular and/or mitochondrial bioenergetics. To begin to test this hypothesis, we obtained skeletal muscle from non-PAD control, IC, and CLI patients and performed whole transcriptome sequencing and comprehensive mitochondrial phenotyping. Our data provide, regardless of risk-factor influence, a yet unseen characterization of CLI that includes a consistent and pervasive deficit in the skeletal muscle oxidative metabolism profile. This represents a critical first step in better understanding the unique local microenvironment present in CLI limb muscles and provides a promising direction for therapeutic development.
Results
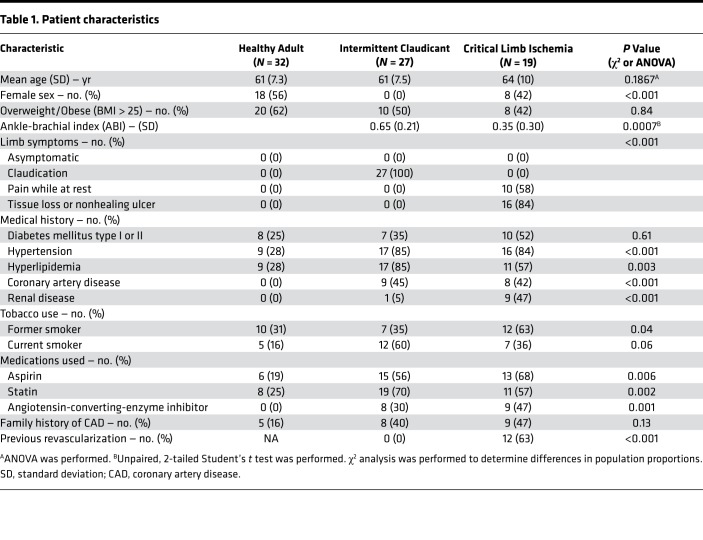
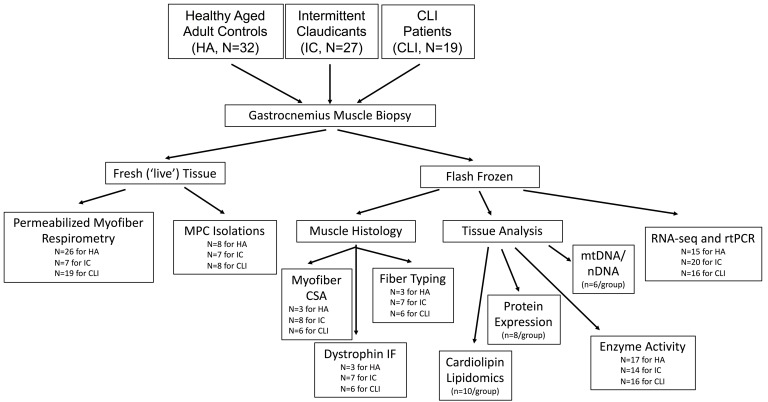
This study was designed to be an observational, cross-sectional comparison of gene expression in patients with claudicating PAD (IC), critical limb ischemia (CLI), and non-PAD control participants (healthy adults, HAs). Patient characteristics are reported in Table 1. Non-PAD controls were diagnosis- and symptom-free by self-report. Claudicating patients enrolled had a clinical diagnosis of occlusive limb disease secondary to pain with exertion relieved by rest. All CLI patients presented with ischemic rest pain with or without tissue necrosis/nonhealing ulcers. ABI values were significantly lower in CLI compared with IC (0.35 ± 0.30 vs. 0.65 ± 0.21 SD; P = 0.0007). Seven CLI patients had undetectable ABIs (noncompressible arteries). Nine CLI patients had chronic or end-stage renal disease requiring hemodialysis. Twelve CLI patients underwent revascularization procedures that failed to prevent amputation. Two of 9 bypass grafts were patent at the time of amputation. Of the enrolled patients, 20% were African American, 31% were women, and 52% were overweight/obese (BMI > 25). Forty-three percent were current or former tobacco users and 35% were diabetic. A flowchart describing how tissue samples were processed including experiments performed and sample sizes used is shown in Figure 1.
Table 1. Patient characteristics.
Figure 1. Flowchart of sample processing for experimental procedures.
Descriptive flowchart of how human specimens were processed for data collection including analyses and sample sizes for each experiment performed.
Unique bioenergetics gene expression profile in CLI tissues.
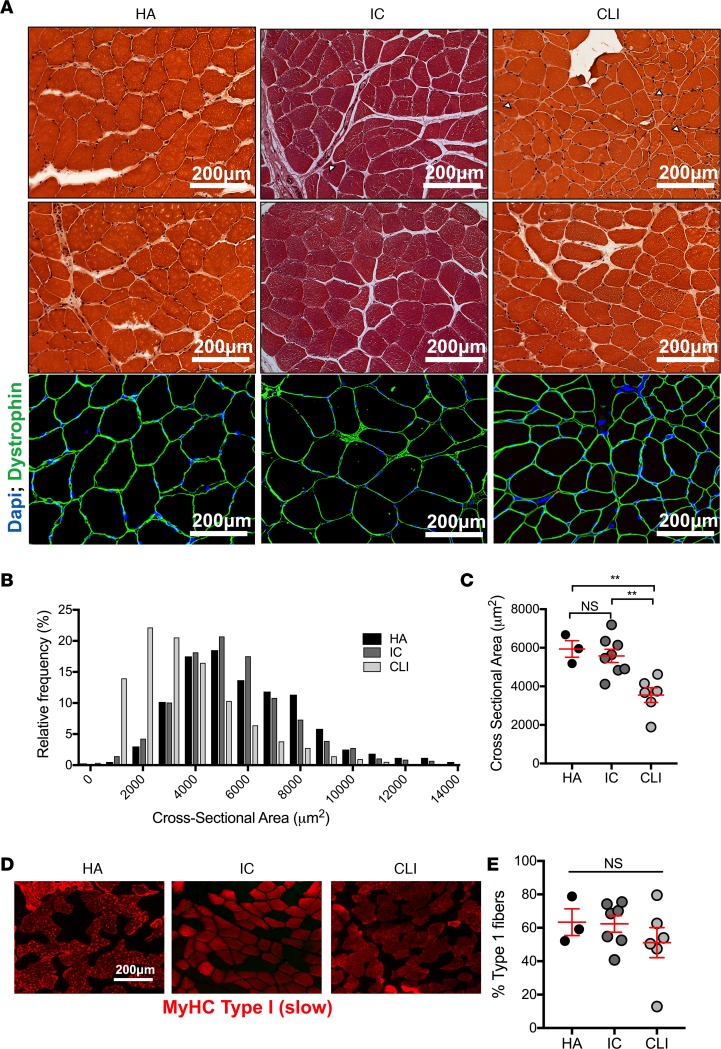
Muscle biopsy samples were uniformly collected from the gastrocnemius muscle (10 cm distal to the tibial tuberosity) of HA controls, IC patients, and CLI patients undergoing limb amputation. Histological and dystrophin staining confirmed intact muscle architecture (Figure 2A) and ensured that downstream experiments would not be artificially influenced by overt necrosis in the biopsy specimens. CLI specimens displayed morphological indicators of skeletal myopathy including smaller and nonuniform myofiber sizes (Figure 2, B and C). Muscle fiber typing analysis indicated a similar proportion of type I (slow twitch) fibers across all groups (Figure 2, D and E). Transcriptome (mRNA) sequencing was then performed (Figure 3A). Principal component analysis revealed a unique gene expression file in CLI patients (Figure 3B) defined by the following: 397 genes expressed differently between HA and IC (Figure 3, C and D); 3,627 genes expressed differently between CLI and HA patients; and 3,999 genes expressed differently between CLI and IC patients (Figure 3, E–H). Differential gene expression results (both uncorrected and FDR-corrected P values) can be found in Supplemental Dataset 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.123235DS1). Gene ontology (GO) enrichment analysis revealed a unique alteration in the transcriptional program of mitochondrial metabolism in CLI patients: the most enriched pathways being cellular respiration, mitochondrial inner membrane, and NADH dehydrogenase activity (Figure 4, A and C). Heatmaps of mitochondrial metabolism mRNA expressions highlight the uniform and striking differences observed in CLI patients (Figure 4B). Quantitative reverse transcription PCR (qRT-PCR) was performed on selected gene targets as a means of validating RNA-sequencing. Consistent with RNA-sequencing results, CLI patients displayed significant decreased mRNA expression of Cox6a2, ATP5a1, NDUFA1, MRPL15, and UQCRFS1 (Figure 4C). Importantly, Cox6a1 (a gene in which expression was similar across groups in the RNA-sequencing analysis) was not significantly different between the 3 groups by qRT-PCR (Figure 4C).
Figure 2. Histological assessment of skeletal muscle specimens.
Skeletal muscle biopsy specimens were obtained from the gastrocnemius of healthy adults (HA), intermittent claudicants (IC), and critical limb ischemia (CLI) patients. (A) Histological (H&E staining) assessment and immunofluorescent staining for dystrophin confirms that samples obtained were not from necrotic regions within the limb (note: color differences in IC samples are due to paraffin embedding). Small white arrows indicate evidence of small, irregularly shaped myofibers in IC and CLI patients. (B) Distribution plots of myofiber cross-sectional area from each patient group (N = 3 for HA, N = 8 for IC, N = 6 for CLI). (C) Quantification of mean myofiber cross-sectional area (N = 3 for HA, N = 8 for IC, N = 6 for CLI). (D) Representative immunofluorescence images stained for myosin heavy chain (MyHC) type I (slow twitch myofibers). (E) Quantification of the percentage of type I myofibers in each group (N = 3 for HA, N = 7 for IC, N = 6 for CLI). **P < 0.01 using ANOVA with Tukey’s multiple comparison test. NS, not significant. Data are presented as the mean ± SEM.
Figure 3. RNA-sequencing in muscle biopsy specimens.
(A) Schematic diagram of tissue acquisition and RNA-sequencing analysis. (B) Principal component analysis (PCA) of the samples. (C) Volcano plot representing differential gene expression between HA and IC. (D) Heatmap of all differentially expressed genes from panel C. (E) Volcano plot representing differential gene expression between HA and IC. (F) Heatmap of all differentially expressed genes from panel E. (G) Volcano plot representing differential gene expression between HA and IC. (H) Heatmap of all differentially expressed genes from panel G. N = 15 for HA, N = 20 for IC, N = 16 for CLI.
Figure 4. Gene expression signature is unique to CLI.
Gene expression profiles were determined by whole genome sequencing of RNA isolated from muscle biopsy samples of the gastrocnemius. (A) Gene ontology (GO) enrichment analysis indicating the most significant gene expression changes between groups. (B) A heatmap of gene expression differences (log2[fold change from HA]) for the GO terms and genes associated with mitochondrial function. (C) mRNA changes were validated by qRT-PCR of selected genes. ***P < 0.001, ****P < 0.0001 using ANOVA with Tukey’s multiple comparison test. NS, not significant. Sample sizes for RNA-sequencing and qRT-PCR were N = 15 for HA, N = 20 for IC, N = 16 for CLI. Data are presented as the mean ± SEM.
Mitochondrial respiratory function, content, and enzyme kinetics in CLI tissues.
To determine the functional impact of the unique bioenergetics gene expression signature in CLI, comprehensive mitochondrial phenotyping experiments were performed using permeabilized myofibers and multiple substrate high-resolution respirometry (15). This approach retains the inherent myofiber mitochondrial structure and avoids artificially influencing mitochondrial respiratory function by isolating mitochondria. Mitochondrial respiratory capacity in myofiber bundles from IC patients was largely not statistically different from HA muscle, the exception being complex I–supported state 3 respiration (235.12 ± 34.51 vs. 359.51 ± 18.91 pmol/s/mg dry weight for IC and HA, respectively; P = 0.0097). Compared with myofibers from either HA or IC patient limbs and regardless of substrate used, an approximately 40%–60% lower mitochondrial respiratory capacity was observed in CLI muscle myofibers (Figure 5A). Patient limb muscle mitochondrial number/density was then assessed using citrate synthase activity, cardiolipin content (a lipid species only present in mitochondrial membranes), and mitochondrial DNA/nuclear DNA (mtDNA/nDNA) ratios. Citrate synthase enzyme activity assays revealed similar mitochondrial content across patients (Figure 5B). Monolyso-, tetralinoleoyl-, and trilinoleoyl-palmitoleic cardiolipin species were quantified by mass spectrometry. Both IC and CLI limb muscles displayed decreased monolyso- and tetralinoleoyl-cardiolipin species when compared with muscles from HA patients. Limb muscle cardiolipin abundance was not different between IC and CLI patient tissues (Figure 5C). Assessment of the mtDNA/nDNA ratio also indicated similar mitochondrial content across all groups (Figure 5D). Collectively, the citrate synthase, cardiolipin, and mtDNA/nDNA analyses demonstrate that decreases in skeletal muscle mitochondrial respiratory function in CLI muscle myofibers occurred independent of overt changes in mitochondrial content/density. Specific activities of the respiratory complexes I, II, III, and IV were next determined spectrophotometrically in gastrocnemius homogenates to further characterize changes in mitochondrial respiration. Statistically significant changes in relative specific activity were not detected in IC patients (all P > 0.08). The relative specific activities of complexes I, III, and IV were reduced by approximately 60%–80% in muscle homogenates from CLI patients (Figure 5E).
Figure 5. Comprehensive mitochondrial phenotyping.
Skeletal muscle mitochondrial respiratory function was measured in permeabilized myofiber samples (representative image in panel A). (A) CLI patients displayed decreased complex I3– (state 3), I+II–, II–, and IV–supported oxygen consumption compared with both IC and HA patients (N = 26 for HA, N = 7 for IC, N = 19 for CLI). Mitochondrial content was assessed by citrate synthase activity (B) (N = 17 for HA, N = 14 for IC, N = 16 for CLI), cardiolipin content (C) (N = 10/group), and mtDNA/nDNA ratio (D) (N = 6/group), indicating that mitochondrial content was not different between IC and CLI. (E) Biochemical enzyme assays of muscle lysates were performed to further dissect changes in the mitochondrial electron transport system. These assays indicate decreased specific activities (normalized for citrate synthase activity) in complexes I, III, and IV in CLI patients (N = 17 for HA, N = 14 for IC, N = 16 for CLI). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 using ANOVA with Tukey’s multiple comparison test. NS, not significant. Data are presented as the mean ± SEM (note: error bars in panel B are 95% CI).
Mitochondrial protein expression.
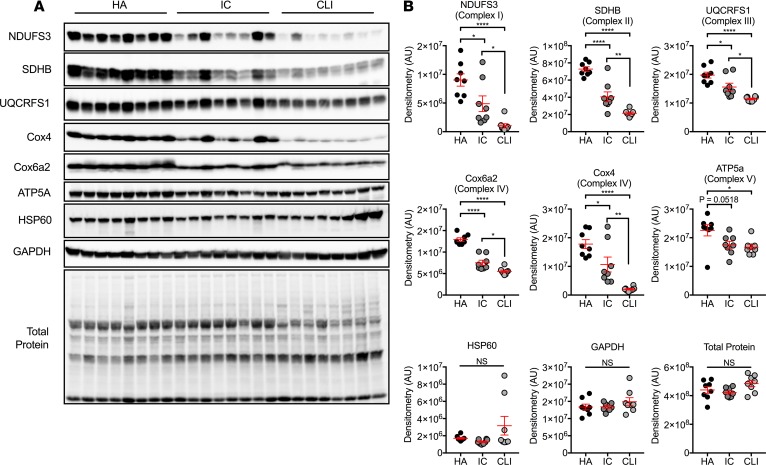
Western blotting analysis was performed for selected mitochondrial proteins to examine the impact of gene expression differences. Muscle homogenates from both IC and CLI patients displayed lower expression of oxidative phosphorylation proteins compared with the HAs, including NDUFS3, SDHB, UQCRFS1, Cox4, Cox6a2, and ATP5a (Figure 6; see complete unedited blots in the supplemental material). When compared with IC patient homogenates, NDUFS3, SDHB, UQCRFS1, Cox6a2, and Cox4 proteins were reduced in CLI patient muscles (all P < 0.05). Interestingly, protein abundances related to complexes I and IV (NFUFS3, Cox4, and Cox6a2) were affected the most: approximately 40%–45% reductions in IC vs. approximately 60%–90% reductions in CLI. Importantly, no differences were observed in any protein control measure, including HSP60 (a nonmetabolic mitochondrial loading control), GAPDH (cytosolic loading control), and total membrane protein (confirming even protein loading).
Figure 6. Mitochondrial protein changes in CLI.
Patient muscle specimens were analyzed by Western blotting. (A) Whole blot images are shown using chemiluminescence. (B) Selected proteins (panel A) were quantified by standard densitometry. *P < 0.05, **P < 0.01, ****P < 0.0001 using ANOVA with Tukey’s multiple comparison test (N = 8/group). NS, not significant. Data are presented as the mean ± SEM.
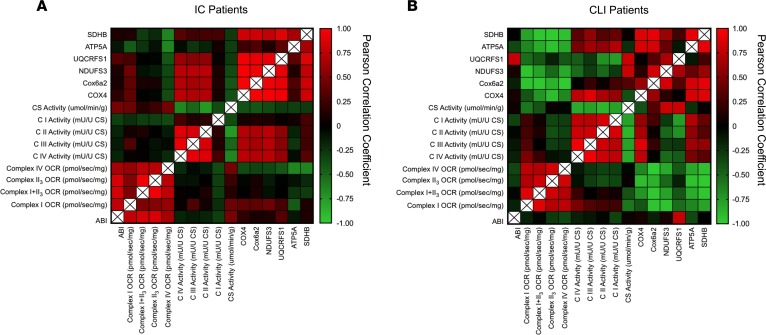
Correlational relationship between patient mitochondrial function and ABI.
Pearson correlation coefficients were calculated within each patient population to examine potential relationships between mitochondrial function and hemodynamics (i.e., ABI). Heatmaps generated are shown for IC and CLI patients in Figure 7. Strong correlations between permeabilized myofiber respiration and ABI were observed in IC, but not CLI myofibers. The lack of correlation in CLI may be related to the narrow range of ABIs in these patients (all <0.5). Complex specific enzyme activities exhibited moderately strong correlations (approximately 0.4–0.6) with protein abundance in both groups. Patients enrolled in this study had diverse clinical characteristics including diabetes, hypertension, hyperlipidemia, renal failure, CAD, and smoking. Preliminary multiple regression analyses failed to find any of these clinical characteristics as a significant predictor of the mitochondrial phenotype. It is noteworthy that although this study is one of the larger mechanistic studies on CLI patients, the present sample size does not provide adequate levels of power for strong multiple regression analyses.
Figure 7. Correlational analyses of mitochondrial outcomes and ABI.
Pearson correlation coefficients were performed for mitochondrial function outcomes and ABI in both IC and CLI patients. Permeabilized myofiber oxygen consumption rate (OCR, pmol/s/mg), complex-specific enzyme activity (mU/U citrate synthase [CS]), protein abundance by Western blotting, and ABI were used. Heatmaps were generated from these Pearson correlation matrixes for both IC (A) and CLI (B) patients.
Mitochondriopathy is retained in muscle progenitor cells from CLI patients.
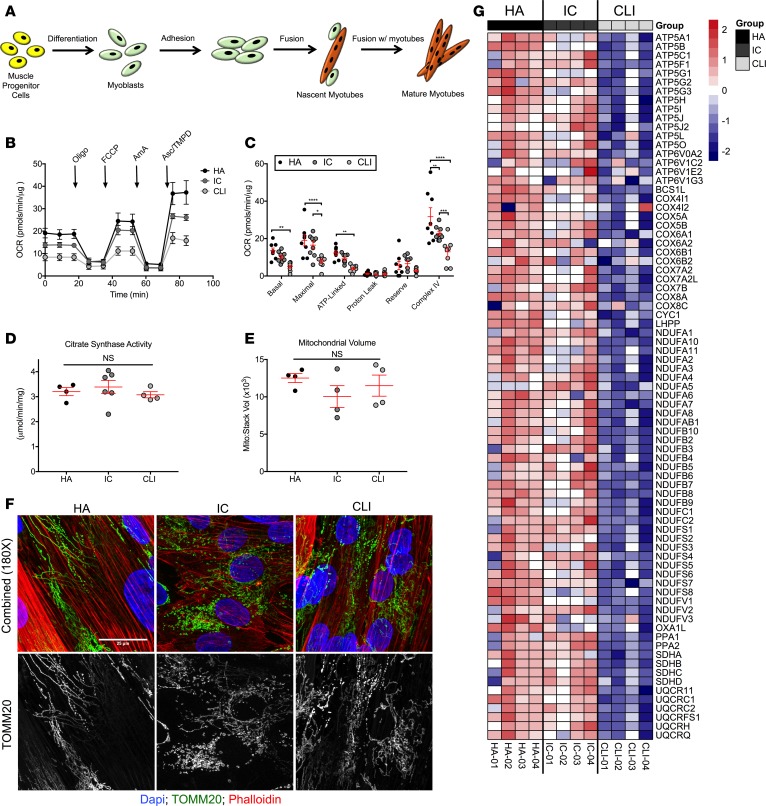
To investigate whether the programmatic CLI limb muscle mitochondriopathy described might impact the health and function of patient muscle progenitor/satellite cells (MPCs), we isolated cells from patient gastrocnemius muscles. MPCs were isolated from 8 HA patients (age = 62.5 ± 6.5 years, N = 3 females/5 males), 7 IC patients (age = 64.8 ± 2.9 years, N = 7 males, ABI = 0.81 ± 0.11), and 8 CLI patients (age = 61.7 ± 10.8 years, N = 4 females/4 males, ABI = 0.43 ± 0.28). MPCs were differentiated into myotubes in standard normoxic culture conditions prior to experimentation (Figure 8A). Basal mitochondrial respiration was decreased in CLI MPCs compared with HA MPCs. Proton leak was similar across all patient MPCs. We found an approximately 60% decrease in both maximal (7.75 ± 1.13 pmol/min/g for CLI vs. 19.06 ± 2.96 pmol/min/μg for HA and 16.50 ± 1.92 pmol/min/μg for HA; P < 0.05) and complex IV–linked (12.94 ± 2.47 pmol/min/μg for CLI vs. 31.69 ± 4.81 pmol/min/μg for HA and 22.72 1.21 pmol/min/μg for HA; P < 0.01) oxygen consumption compared with either HA or IC MPCs (Figure 8, B and C). Citrate synthase activity (Figure 8D) and confocal Z-stack imaging of mitochondrial volume (Alexa Fluor 488–conjugated TOMM20, Figure 8, E and F) both confirmed similar patient MPC mitochondrial contents. Interestingly, mitochondrial oxygen consumption in MPCs isolated from HA patients were not affected by a 12-hour hypoxia/nutrient deprivation treatment (Supplemental Figure 1), suggesting that the observed deficit in CLI MPCs may be unique to the ischemic microenvironment they were derived from.
Figure 8. Skeletal muscle stem cells from CLI patients display a unique bioenergetics gene expression signature.
Primary muscle progenitor cells (satellite cells) were isolated from muscle biopsies. Isolated myoblasts were differentiated into myotubes by serum withdrawal (A). (B) Cellular respiration was assessed using a Seahorse XF analyzer. Oligo, oligomycin; AmA, antimycin A; Asc, ascorbate; OCR, oxygen consumption rate. (C) Quantification of cellular respiration under different substrate/inhibitor combinations indicates impaired basal, maximal, and complex IV–linked respiration in cells from CLI patients (N = 8 for HA, N = 7 for IC, and N = 8 for CLI). (D) Citrate synthase activity was not different in cells (N = 4 for HA, N = 6 for IC, and N = 4 for CLI). (E) Quantification of mitochondrial volume from Z-stack confocal imaging of fluorescently labeled mitochondria in MPCs (N = 4 for HA, N = 4 for IC, and N = 4 for CLI). (F) Representative images of labeled mitochondria (TOMM20, green). Counterstaining was performed with DAPI (blue, nuclei) and phalloidin (red, actin). (G) Gene expression analysis by qRT-PCR indicated a unique gene signature related to mitochondrial metabolism. All sample sizes indicate independent MPC isolations from different patients. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 using ANOVA with Tukey’s multiple comparison test. NS, not significant. Data are presented as the mean ± SEM.
We next performed a qRT-PCR gene array focused on mitochondrial metabolism to determine whether the myotube mitochondriopathy was paralleled by alterations in the bioenergetics transcriptional program, similar to that observed in the patient limb muscle tissues. Relative gene expression (ΔΔCT) was calculated first by correcting each gene CT by the average CT value for housekeeping genes that were unchanged across groups (B2M, HPRT1, RPL13A, GAPDH, and ACTB), and the log2 fold change was calculated relative to the myotubes differentiated from HA MPCs. Gene expression defects observed in CLI patient muscle tissues were mirrored in the differentiated CLI patient myotubes, despite normoxic and nutrient-rich culture conditions (Figure 8G). The reduced mitochondrial gene expression program was surprisingly uniform across myotubes from 4 different CLI patient MPC cultures.
Discussion
In this study, we discovered what we believe is a unique and severe mitochondriopathy in human patients diagnosed with CLI. This mitochondriopathy is defined by the following: reduced mitochondrial oxidative capacity, reduced electron transport system enzyme function, and decreased abundance of mitochondria-associated mRNAs and proteins. Mitochondriopathy occurred in muscle tissue devoid of necrotic lesions and similar in mitochondrial content to IC muscle tissue. Prior to this study, clinical medicine lacked detailed biological information to adequately differentiate CLI limb muscle pathology as anything other than progressively worse than IC tissues. Our findings reveal that mitochondriopathy is a potentially novel target for the development of stand-alone or adjuvant therapies for the myopathy of CLI.
Vascular medicine/science has largely failed to advance our understanding of CLI. As previously discussed, this may be due to both a lack of interest from pharmaceutical industries in the smaller CLI population and the risks/difficulties associated with working with these patients (16). Preclinical mouse models of CLI demonstrate that skeletal muscle myopathy develops within hours of ischemia initiation. This is well before the onset of necrosis (11) and temporally precedes a concomitant regeneration from myopathy and blood flow recovery (7, 8, 10, 12, 17). The primary clinical treatments for CLI, surgical bypass or endovascular intervention, have limited effectiveness in preventing mortality or amputation (18); 30-day hospital readmission rates for CLI patients who have undergone a vascular procedure are greater than 40% (19). Furthermore, investigational approaches derived from preclinical studies (e.g., proangiogenic and stem cell–based therapies) have been ineffective at promoting CLI limb salvage (20–22). We contend that the stagnant nature of biomedical advances for CLI patients warrants new approaches to identify targets for investigation. Metabolism has been extensively studied in IC patients, likely a direct result of the known beneficial effects of physical activity in relieving symptomatology in these patients. Here, we reveal that the limb-muscle mitochondria of CLI patients possess a unique bioenergetic signature. This presents a unique opportunity and biological target to develop focused therapeutics for the CLI patient. While our results do not directly define preexisting mitochondriopathy as a mechanism for disease presentation, they present a defining limb-tissue signature. This signature could also be used in conjunction with clinical classification systems (23) to accurately identify CLI patients at greater risk for negative outcomes.
Limb tissue mitochondrial content is a commonly used indicator of mitochondrial health in PAD patients. Studies report both stable citrate synthase activity (24, 25) and increases in mtDNA copy number (26) in IC patient limb muscles. We report no alterations in mitochondrial content between IC and CLI patients as measured by either standard (citrate synthase activity and mtDNA/nDNA ratios) or refined (cardiolipin lipidomics) approaches. The maintenance of limb skeletal muscle mitochondrial content in CLI patients is somewhat surprising considering that the following occurred: substantial functional deficits, globally reduced bioenergetic mRNA expression, and lower mitochondrial protein content. Additionally, and despite similar mitochondrial content, IC and CLI patient muscle myofiber mitochondrial function was markedly different. CLI-associated reductions in mitochondrial protein levels likely negatively impacted respiratory function in a manner not accurately reflected by measures of mitochondrial content. This disconnect highlights the importance of parallel measurements of both mitochondrial content and function in studies of PAD patients, particularly those with CLI.
Whether genetically based or risk-factor induced, a muscle cell–specific compromise in mitochondrial respiratory function is catastrophic to the health of the ischemic limb. Mitochondria function as continuously operating batteries, creating the energy and redox charges essential for establishing and maintaining life for cells. For skeletal muscle myofibers to survive and respond to the environment created by critical levels of ischemia, there is a significant demand for ATP that must be met by properly functioning resident mitochondria (8, 10, 27). This ATP is absolutely necessary for the muscle degenerative and regenerative processes that occur in both chronic and acute phases of the CLI pathophysiology. Muscles’ regeneration from ischemic pathophysiology temporally occurs in parallel with blood flow recovery (28–41). Physiologically, this temporal association makes perfect sense; the return of arterial flow to a metabolically fragile or incompetent limb microenvironment is unlikely to yield a sustainable reversal or halting of pathophysiology. Proliferating and differentiating muscle cells and regenerating myofibers are both sources of and targets for traditional vascular growth factors (7, 9, 42–45), providing critical proangiogenic factors to support existing vessels and promote neovascularization in the ischemic limb. A logical explanation for the failure of previous trials may be that these therapies fail to stimulate the survival and metabolic viability of the limb muscle, which would nurture the local environment and provide support for therapeutics specifically aimed at the vasculature.
Isolated muscle progenitor/satellite cells (i.e., MPCs) provide an excellent tool for studying muscle cell bioenergetics outside of the local environment of the critically ischemic limb. These cells are necessary for recovery from an acute injury (46). MPCs, when activated, fuse into existing damaged myofibers undergoing a profound and necessary metabolic change that includes a switch from glycolytic to oxidative metabolism (47–50). This switch is replicated in vitro during myoblast differentiation into muscle myotubes. Isolating these cells from PAD patients provided us an opportunity to examine oxidative metabolism under tightly controlled normoxic and nutrient-rich conditions, effectively minimizing the influence of differing local environments created by PAD clinical manifestations. MPCs appear to be uniquely prone to mirroring the local metabolic environment from which they were isolated (51, 52). CLI patient MPCs retained the following characteristics: stable myotube mitochondrial content, attenuated oxygen consumption, and a suppressed bioenergetic mRNA profile. This finding is of paramount importance to ischemic limb muscle, which undergoes cyclical periods of degeneration/regeneration depending on the fusion of these cells to ensure function of the muscle. The presence of a progenitor cell memory lends insight into the biology behind amputation outcomes despite successful interventional revascularization. Based on our results, targeted therapies to either induce metabolic sufficiency in endogenous MPCs or exogenously provide metabolically potent MPCs represent intriguing therapeutic options for CLI myopathy that warrant further exploration.
The study population we utilized is considered small from a clinical perspective (46 PAD patients across IC and CLI populations); it includes patients of both sexes, multiple race/ethnicities, and multiple risk factors/comorbidities, and CLI tissue acquisition occurred at the terminal point in the patient’s disease process (i.e., amputation). A larger study population would allow both an independent assessment of the influence of each risk factor on mitochondrial physiology and the ability to further segregate patients into distinct categories of disease severity. The homogeneity of our findings within the CLI clinical presentation indicates that, despite our patients’ diverse background genetics and additional diseases/risk factors, the mitochondrial profiles are fairly uniform within PAD clinical presentations. Future clinical trials are needed to determine the chronology of skeletal muscle mitochondrial dysfunction in CLI patients.
In summary, CLI patients suffer from a unique limb skeletal muscle mitochondriopathy that distinguishes them from their claudicating counterparts. Our data do not provide direct evidence of whether the suppression of oxidative metabolism is adaptive or maladaptive in these patients. In either case, there are significant implications for the therapeutic rehabilitation of CLI patients. This is the first time to our knowledge that a defining characteristic of CLI patient limb muscle has been reported. A suppressed bioenergetic gene expression signature uniformly occurred in CLI patients, contributing to an inadequate ability to consume oxygen and provide ATP in support of critical cellular functions. Mitochondria are richly abundant in limb skeletal muscles and play pivotal roles in both energy production and initiation of necrotic/apoptotic cell death for cells exposed to ischemic conditions (13, 14). Given the known negative independent influences of PAD risk factors on limb muscle mitochondria, this research also leads us to suggest that limb muscle mitochondrial bioenergetics may serve as a key site of convergence in the pathophysiology of risk factors for CLI, such as diabetes. This represents a critical step forward in targeted therapeutic development for a patient population with limited options.
Methods
Study approval and participants.
Thirty-two HAs without PAD, 27 PAD patients with IC, and 19 patients with CLI were recruited through print advertising or identified by vascular surgeons at East Carolina University Brody Medical Center and the University of Nebraska Medical Center (UNMC) and Veterans Affairs Nebraska-Western Iowa under approved IRB protocols. Inclusion criteria comprised patients representative of all clinical classifications of PAD, as indicated in the patient physical and clinical characteristics shown in Table 1. Exclusion criteria consisted only of CLI amputation patients who previously provided biological specimens from the contralateral limb. All data collection was carried out by blinded investigators at ECU or UNMC.
Tissue acquisition.
Percutaneous muscle biopsy samples were taken from the lateral gastrocnemius muscle of age-matched volunteer HAs and identified IC patients. The skin of the muscle biopsy sight was first cleansed with a providone-iodine swab and then anesthetized with 5 ml of lidocaine. A small incision was made and the muscle biopsy sample was aspirated through a 5-mm Bergstrom needle. Tissue obtained from CLI patients occurred inside the operating room immediately following limb amputation performed without the use of a tourniquet. Muscle specimens from CLI patients were collected form the same anatomical location of the gastrocnemius muscle as the percutaneous muscle biopsies performed in HA and IC patients. Part of the muscle biopsy sample was immediately flash frozen in liquid nitrogen and stored at –80°C until subsequent analysis. Other portions of the muscle biopsy (approximately 80–150 mg) were used for human primary muscle cell isolation and permeabilized myofiber mitochondrial experiments.
RNA isolation and transcriptome sequencing.
Total RNA was extracted using Qiagen RNeasy Midi kits per manufacturer instructions. RNA-sequencing was performed by Quick Biology Inc. RNA integrity was checked using an Agilent Bioanalyzer 2100; only samples with clean rRNA peaks were used. The library for RNA-sequencing was prepared according to a KAPA Stranded mRNA-Seq poly(A) selected kit with 201–300 bp insert size (KAPA Biosystems) using 250 ng total RNA as input. Final library quality and quantity were analyzed by Agilent Bioanalyzer 2100 and Life Technologies Qubit3.0 Fluorometer. Paired-end (150-bp) reads were sequenced on an Illumina HighSeq 4000. The reads were first mapped to the latest UCSC transcript set using Bowtie2 version 2.1.0 (53) and the gene expression level was estimated using RSEM v1.2.15 (54). Trimmed mean of M values (TMM) was used to normalize the gene expression. Differentially expressed genes were identified using the edgeR program (55). Genes showing altered expression with P less than 0.05 and more than 1.5-fold changes were considered differentially expressed. Adjusted P values were also calculated assuming FDR = 0.05. Goseq was used to perform the GO enrichment analysis and Kobas was used to performed the pathway analysis. Heatmaps were generated with Pheatmap program using log2(fold change from non-PAD). To further validate RNA-sequencing findings, RNA was reverse transcribed using Superscript IV reverse transcriptase according to manufacturer instructions (Invitrogen). Real-time PCR on selected gene targets was performed using a Quantstudio 3 Real-Time PCR system (Applied Biosystems). Relative quantification of mRNA levels was determined using the comparative threshold cycle (ΔΔCT) method using FAM-labeled Taqman gene expression assays (Applied Biosystems) specific to the given gene run in multiplex with a VIC-labeled 18S control primer. The RNA-sequencing data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and can be accessed using GEO Series accession number GSE120642.
Preparation of permeabilized muscle fibers.
A portion of each muscle sample was immediately placed in ice-cold buffer X (50 mM K-MES, 7.23 mM K2EGTA, 2.77 mM CaK2EGTA, 20 mM imidazole, 20 mM taurine, 5.7 mM ATP, 14.3 mM phosphocreatine, and 6.56 mM MgCl2-6H2O, pH 7.1) for preparation of permeabilized fiber bundles (PmFBs) as previously described (15, 56). Fiber bundles were separated along their longitudinal axis using needle-tipped forceps under magnification (MX6 Stereoscope, Leica Microsystems), permeabilized with saponin (30 μg/ml) for 30 minutes at 4°C on a nutating mixer, and subsequently washed in cold buffer Z (105 mM K-MES, 30 mM KCl, 1 mM EGTA, 10 mM K2HPO4, 5 mM MgCl2-6H2O, 0.5 mg/ml BSA, pH 7.1) for 15 minutes until analysis. At the conclusion of each experiment, PmFBs were washed in double-distilled H2O to remove salts, freeze-dried (Labconco), and weighed. Typical fiber bundle sizes were 0.2–0.6 mg dry weight.
Mitochondrial respiration measurements.
High-resolution O2 consumption measurements (57) were conducted at 37°C in buffer Z, supplemented with creatine monohydrate (20 mM), using the OROBOROS O2K Oxygraph. A substrate inhibitor titration protocol was performed as follows: 2 mM malate plus 10 mM glutamate (state 2 respiration), followed by the addition of 4 mM ADP to initiate state 3 respiration supported by complex I substrates. Convergent electron flow through complexes I and II was initiated with the addition of 10 mM succinate. Rotenone (10 μM) was subsequently added to inhibit complex I, followed by 10 μM cytochrome c to test the integrity of the mitochondrial membrane. Complex IV–supported respiration was examined using the electron donor N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) at 0.4 mM in the presence of 2 mM ascorbate (to limit auto-oxidation of TMPD) and 5 μM of antimycin A (to prevent reverse electron flow through complex III). The rate of respiration was expressed as pmol/s/mg fiber dry weight. All respiration measurements were conducted at 37°C and a working range [O2] of approximately 350 to 200 μM.
Citrate synthase activity.
Activity assays were performed using a citrate synthase activity assay kit (Sigma-Aldrich, CS0720) per the manufacturer’s instructions. Briefly, skeletal muscles or primary myotube lysates were generated by glass pestle homogenization by hand in RIPA buffer. Protein concentrations were determined using a Pierce BCA protein assay (Thermo Fisher Scientific, 23225). Activity assays were performed in assay buffer containing 100 mM Tris, 1 mM EDTA, 1 mM EGTA, 10 mM DTNB (Sigma-Aldrich, D8130), and 30 mM acetyl CoA at pH 8.35. All samples were measured in duplicate and the average absorbance was used in final calculations of activity. Background absorbance was measured prior to addition of 10 mM oxaloacetate (Sigma-Aldrich, O4126) and final activity rates were corrected for those values.
Relative specific activity of mitochondrial OXPHOS complexes.
Frozen skeletal muscle from the muscle biopsy procedure was homogenized in 0.3 M sucrose, 10 mM HEPES, and 1 mM EGTA on ice (adapted from ref. 58), centrifuged at 800 g for 10 minutes, and protein in the supernatant was quantified using a BCA protein assay kit (Life Technologies). Specific activities of all individual ETS complexes were determined spectrophotometrically as previously described (59). Briefly, aliquots of skeletal muscle lysates were diluted in hypotonic medium (25 mM K2HPO4, 5.3 mM MgCl2, pH 7.2) and further subjected to 3–4 freeze-thaw cycles. Complex I activity was determined in 5 mM Tris, 0.5 mg/ml BSA, 24 μM KCN, 0.4 μM antimycin A, pH 8, following the oxidation of NADH (0.8 mM) at 340 nm (ε340 = 6,220 M–1cm–1) for 3 minutes using oxidized decyl-ubiquinone (50 μM DCUox) as the electron acceptor. Rotenone (4 μM) was added to measure rotenone-sensitive NADH-DCU oxidoreductase activity. Complex II activity was measured in SQR medium (10 mM KH2PO4, 2 mM EDTA, 1 mg/ml BSA) in the presence of 10 mM succinate, following the reduction of dichlorophenolindophenol (80 μM DCPIP) at 600 nm (ε340 = 19,100 M–1cm–1) for 3 minutes, using DCUox as the electron acceptor. The reaction was inhibited by the addition of the competitive substrate malonate (10 mM). Complex III activity was measured in SQR medium supplemented with 200 μM ATP and 240 μM KCN, using DCUred (80 mM) as electron donor and oxidized cytochrome c as the acceptor (40 μM). The reaction was followed measuring the reduction of cytochrome c at 550 nm (ε340 = 18,500 M–1cm–1) for 3 minutes, and finally inhibited by the addition of 0.5 μM myxothiazol. Cytochrome c oxidase (complex IV) was determined with the addition of 1 mM reduced cytochrome c by following the decrease in absorbance at 550 nm. Potassium cyanide was added to check for the specificity of complex IV activity.
Cardiolipin quantification.
Cardiolipin was quantified using previously published methods with liquid chromatography coupled to electrospray ionization mass spectrometry in an API 4000 mass spectrometer (Sciex) (60). For mass spectrometry sample preparation, muscle was homogenized using a glass-on-glass homogenizer in PBS and lipids were extracted from 0.25 mg of protein per sample using a modified Bligh and Dyer method with 1,000 nmoles tetramyristal cardiolipin added as an internal standard (Avanti Polar Lipids) (60, 61).
Analysis of mtDNA/nDNA ratio.
For a third measurement of mitochondrial content/density, mtDNA and nDNA were quantified and expressed as a ratio as previously described (62). Genomic DNA was isolated from 10–15 mg of the muscle biopsy specimen using a Nucleospin tissue kit (Takara). qPCR was performed using 100 ng of gDNA. mtDNA was assessed using the FAM-labeled Taqman probe Hs02596864_g1 for the MT-CO1 gene. Multiplex analysis of nuclear DNA was assessed using the VIC-labeled Taqman probe Hs02758991_g1 for GAPDH. The mtDNA/nDNA ratio was calculated by dividing MT-CO1 expression by GAPDH expression.
Immunoblotting.
Frozen skeletal muscle from the muscle biopsy procedure was homogenized in RIPA buffer supplemented with protease inhibitors (Roche). Protein concentrations were determined using the Pierce BCA protein assay. Proteins were then separated using an SDS-PAGE gel (Midi-Protean Stain-Free TGX, Bio-Rad). Separated proteins were transferred to PVDF membranes using the Bio-Rad Turbo Transfer System, followed imaging for total protein using the stain-free labeling on a Bio-Rad Chemidoc Imaging system. PVDF membranes were then blocked for 90 minutes in nonfat dry milk in Tris-buffered saline with 0.1% Tween. Membranes were incubated overnight at 4°C with the following primary antibodies: Total Oxphos Cocktail (Abcam, MS601), Cox6a2 (Thermo Fisher Scientific/Invitrogen, PA5-26622), HSP60 (Proteintech, 15282), NDUFS3 (Proteintech, 15066), UQCRFS1 (Proteintech, 18443), Cox4 (Abcam, ab14744), and GAPDH (Cell Signaling Technology, 5174). Following incubation with primary antibodies, membranes were washed in Tris-buffered saline with 0.1% Tween (3 times 10 minutes) and incubated with HRP-conjugated secondary antibodies of the appropriate host for 1 hour. Total blots were visualized with chemiluminescence (Pierce ECL Substrate, Thermo Fisher Scientific, 32209) using a Bio-Rad Chemidoc imaging system. Densitometry was performed using ImageLab Software v5.2.1 (Bio-Rad).
Skeletal muscle histology.
Skeletal muscle morphology was assessed by standard light microscopy. Transverse sections (10 μm thick) from muscle biopsy samples of the gastrocnemius muscle frozen in liquid-nitrogen-cooled isopentane in OCT were cut using a Leica 3050S cryotome and collected on charged slides for staining. IC patient gastrocnemius muscle samples were fixed in methacarn and paraffin embedded, resulting in the observed color difference with H&E staining. Sections were deparaffinized in xylene and an ethanol gradient prior to staining. For morphological analyses, standard methods for H&E histological staining were performed and images were acquired using an Evos FL auto wide-field microscope (Life Technologies) with a plan-fluorite 20× coverslip-corrected objective (NA 0.5, air).
Immunofluorescence.
HA and CLI tissues were prepared as described above. HA and CLI sections were fixed in 1:1 acetone/methanol for 10 minutes at –20°C and rehydrated in 1× PBS. IC sections were deparaffinized and rehydrated in 1× PBS. All sections were blocked in 5% goat serum plus 1× PBS for 1 hour at room temperature. For dystrophin immunofluorescence, sections were incubated with rabbit anti–human dystrophin primary antibody at 4°C overnight (1:100 dilution, Thermo Fisher Scientific, Rb-9024; targeted to the protein C-terminus). Sections were washed 3 times for 10 minutes each with cold 1× PBS and incubated for 1 hour with Alexa Fluor 488–conjugated anti-rabbit IgG secondary antibody (1:250, Invitrogen). For fiber typing analysis, sections were incubated with mouse anti–bovine myosin heavy chain type I (slow) (DSHB, BA-D5) primary antibody at 4°C overnight (1:100 dilution). Sections were washed 3 times for 10 minutes each with cold 1× PBS. For dystrophin immunofluorescence, sections were incubated for 1 hour with Alexa Fluor 488–conjugated anti-rabbi IgG1 secondary antibody (1:250, Invitrogen). For fiber typing, sections were incubated for 1 hour with Alexa Fluor 594–conjugated goat anti-mouse IgG2b secondary antibodies (1:250, Invitrogen). Sections were mounted using Vectashield hard mount medium with DAPI (Vector Labs). Images were obtained as described above.
Image processing.
Image processing was performed using ImageJ (NIH, v1.49). All processing was performed uniformly over the entire image, and processing parameters were made constant for the entire image set. H&E-stained images were dark-field corrected using a correction image collected at the time of acquisition. Corrections were performed using the Image Calculator Plus plugin. Images were contrast enhanced with a 0.3% pixel saturation threshold. In order to facilitate feature enhancement in representative image panels where features may be obscured due to the small size of the images, an unsharp mask filter was applied to the dark-field-corrected/contrast-enhanced H&E images (mask weight = 0.6; pixel radius = 1). No images used for quantification were altered from their original format prior to analysis. Muscle fiber cross-sectional area was measured by a blinded investigator. Two hundred fibers were measured from each ×4 magnification image and samples were pooled for frequency distributions. The analysis of 200 fibers was determined from a test set of each type of patient sample as the minimum number of measurements required to achieve a stable variance among the measurements. Fiber type proportions were counted from two ×4 magnification images from each sample.
Primary MPC isolation and culture.
Primary human MPCs (human myoblasts) were derived from fresh muscle biopsy samples. Briefly, skeletal muscle specimens were trimmed of connective tissue, and placed in 10-cm dishes containing ice-cold sterile DMEM with 4.5 g/l glucose, supplemented with 1% penicillin/streptomycin (isolation medium, IM). Muscle was further cleaned of connective tissue and fat inside a sterile culture hood and then transferred to a third 10-cm dish containing 5 ml of cold IM and minced for 2 minutes using sterile razor blades. The minced slurry was transferred to 15-ml tubes, 5 ml additional MPC IM was added, tubes were inverted several times and centrifuged at 4°C for 3 minutes at 1,000 g to remove contaminants. IM was subsequently aspirated and the pellet was resuspended in 10 ml of IM and inverted 5–10 times to loosen the pellet and mix. Plates were subsequently rinsed in IM to ensure that all tissue was collected and then 0.1% Pronase (Calbiochem, 53702) was added. Tubes were placed on rotator at 37°C and 5% CO2 for 1 hour. The digested tissue slurry was then transferred to 50-ml conical tubes and centrifuged for 5 minutes at 1,000 g at room temperature. Supernatant was aspirated and the digested pellet was resuspended in purification medium (PM: DMEM with 4.5 g/l glucose, supplemented with 20% fetal bovine serum [FBS] and 1% penicillin/streptomycin). The suspension was then triturated approximately 20 times through a blunt-end pipetting needle attached to a sterile 30-cc syringe. The suspension was then passed through a 100-μm disposable Steriflip vacuum filter into a 50-ml tube, including 3 successive 8-ml washes of the sieve with prewarmed PM, and subsequently centrifuged at room temperature for 5 minutes at 1,000 g. The cell pellet was then resuspended in 1 ml FBS before addition to primary MPC growth medium (GM: Ham’s F10, supplemented with 20% FBS and 1% penicillin/streptomycin, and supplemented immediately prior to use with 5 ng/ml basic FGF). Cells were plated on collagen-coated flasks or dishes and allowed to adhere and proliferate for 3 days. After day 3, cells were trypsinized in 0.25% trypsin/EDTA and preplated at 37°C and 5% CO2 for 1 hour and 15 minutes on an uncoated flask to allow for fibroblast purification, and the supernatant containing MPCs was removed and centrifuged at 800 g for 5 minutes at room temperature prior to replating in MPC GM on collagen-coated flasks. After reaching approximately 70% confluence, MPCs were then plated on entactin/collagen/laminin plates or dishes and differentiated into mature myotubes by serum withdrawal in differentiation media (DM: DMEM with 4.5 g/l glucose, supplemented with 2% horse serum and 1% penicillin/streptomycin and supplemented with 0.1% insulin/transferrin/selenium immediately prior to use). DM was changed every 24 hours.
Primary cell respiration measurements.
Primary muscle cell respiration was assessed using the Seahorse XF24 Analyzer after 96 hours of differentiation. Cells were placed in assay media containing 2.5 mM glucose, 10 mM pyruvate, and 2 mM GlutaMax approximately 30 minutes prior to the start of the experiment, and allowed to equilibrate. Respiration was measured with sequential additions of 1 μg/ml oligomycin (to inhibit ATP synthase), 1 μM FCCP (to uncouple respiration), 2.5 μM antimycin A (to inhibit electron flow by blocking complex III), and 0.5 mM ascorbate plus 0.1 mM TMPD. All cell respiration experiments were performed using identical cell seeding densities, corrected for background rates of oxygen consumption, and normalized to total protein in each well using BCA assay following the addition of RIPA lysis buffer.
Primary muscle cell mitochondrial volume.
Differentiated myotubes were fixed in 4% paraformaldehyde in 1× PBS for 15 minutes at 25°C, permeabilized for 10 minutes in 1× PBS plus 30 μg/ml saponin, then blocked at 25°C for 1 hour in 5% goat serum/1% BSA in 1× PBS. Cells were then incubated for 1 hour at 37°C with 1:500 TOMM20 Alexa Fluor 488–conjugated primary antibody (Abcam, ab205486). Cells were washed and incubated with 1:40 phalloidin Alexa Fluor 594 conjugate (Thermo Fisher Scientific, A12381) and DAPI reagent (Thermo Fisher Scientific, R37606). Images were acquired with an Olympus FV1000 laser scanning confocal microscope with Fluoview acquisition software. A 60× plan apochromatic oil immersion objective lens was used for all imaging (NA 1.35). DAPI and Alexa Fluor 488 excitation was achieved using the 405 nm and 488 nm line of a multiline argon laser, respectively. Alexa Fluor 594 (phalloidin) excitation was achieved using a 559 nm laser diode. Images were sequentially scanned at 2 μs/pixel using Kalman line filtering (5 times). For Z-stacked image acquisition, 0.46 μm slice thickness was selected for optimum sampling with a pinhole diameter of 110 μm. Image processing was performed using ImageJ (NIH, v1.49). All processing was performed uniformly over the entire image, and processing parameters were made constant for the entire image set. Representative maximal intensity Z projections from myotube imaging were contrast enhanced with a 0.3% pixel saturation threshold and unsharp mask filtered to facilitate feature enhancement in representative images only. No images used for quantification were altered from their original format prior to analysis. Mitochondrial morphology was assessed using ImageJ with the 3D object counter plugin. Stacks were split into RBG channels and thresholded through the stack by a blinded investigator. Mitochondrial volume was summed for each stack and normalized to the total stack volume.
Primary muscle cell mitochondrial metabolism gene array.
Total RNA was isolated from differentiated human myotubes using TRIzol reagent (Thermo Fisher Scientific). RNA quality and quantity were determined using a Nanodrop 2000 (Thermo Fisher Scientific). RNA (500 ng) was reverse transcribed using RT2 First Strand kits (Qiagen, 330401). Gene expression arrays for Human Mitochondrial Energy Metabolism (Qiagen, PAHS-008ZA) were run with RT2 SYBR Green/ROX PCR master mix on a Quantstudio 3 Real-Time PCR system (Applied Biosystems). Gene expression was normalized to the average CT value of 5 loading control genes on each plate (B2M, HPRT1, RPL13A, GAPDH, and ACTB). Relative quantification of mRNA levels was determined using the comparative threshold cycle (ΔΔCT) method and fold change from non-PAD was calculated using 2–ΔΔCT. Heatmaps were generated in Pheatmap program using the log2(fold change vs. HA).
Statistics.
Data are presented as mean ± SEM unless otherwise specified. Comparisons of data with more than 2 groups were performed using ANOVA with Tukey’s post hoc test for multiple comparisons. In all cases, P < 0.05 was considered statistically significant.
Study approval.
This study was approved by the IRBs at East Carolina University (Greenville, NC), the University of Nebraska Medical Center (Omaha, NE), and Veterans Affairs Nebraska-Western Iowa (Omaha, NE) and carried out in accordance with the Declaration of Helsinki. All participants gave written informed consent prior to enrollment.
Author contributions
JMM, TER, and DJY conceived and designed the study. JMM, TER, DJY, CAS, TNZ, SRS, PB, TDG, MDT, RK, EJG, GCS, MJT, PDN, BHA, and EES acquired, analyzed, or interpreted data. JMM, TER, and DJY drafted the manuscript. JMM, TER, DJY, BHA, PDN, and EES revised/edited the manuscript. JMM, TER, DJY, CAS, TNZ, SRS, PB, TDG, MDT, RK, GCS, MJT, PDN, BHA, and EES provided administrative, technical, or material support. JMM obtained funding and had full access to the data presented and takes responsibility for the integrity and accuracy of the data.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants R00HL103797 and R01HL125695 (to JMM), R01AR066660 (to EES), R01HL116455 and R01HL121635 (to BHA), R01HL123647 (to SRS), and F32HL129632 (to TER).
The authors would also like to acknowledge Iraklis I. Pipinos, Julian K. Kim, and George P. Casale from the University of Nebraska Medical Center for their contributions to the manuscript, including tissue acquisition.
Version 1. 11/02/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: JCI Insight. 2018;3(21):e123235. https://doi.org/10.1172/jci.insight.123235.
Contributor Information
Terence E. Ryan, Email: ryant@ufl.edu.
Dean J. Yamaguchi, Email: yamaguchid@ecu.edu.
Cameron A. Schmidt, Email: schmidtc14@students.ecu.edu.
Patricia Brophy, Email: brophyp@ecu.edu.
Thomas D. Green, Email: GREENT@ecu.edu.
Michael D. Tarpey, Email: TARPEYM15@ECU.EDU.
Reema Karnekar, Email: karnekarr@ecu.edu.
Emma J. Goldberg, Email: goldberge17@students.ecu.edu.
Genevieve C. Sparagna, Email: GENEVIEVE.SPARAGNA@UCDENVER.EDU.
Maria J. Torres, Email: maria.j.torres@duke.edu.
Brian H. Annex, Email: BHA4N@virginia.edu.
References
- 1.Sukul D, Grey SF, Henke PK, Gurm HS, Grossman PM. Heterogeneity of ankle-brachial indices in patients undergoing revascularization for critical limb ischemia. JACC Cardiovasc Interv. 2017;10(22):2307–2316. doi: 10.1016/j.jcin.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumakura H, Kanai H, Hojo Y, Iwasaki T, Ichikawa S. Long-term survival and fate of the leg in de novo intermittent claudication. Eur Heart J Qual Care Clin Outcomes. 2017;3(3):208–215. doi: 10.1093/ehjqcco/qcw057. [DOI] [PubMed] [Google Scholar]
- 3.Aquino R, et al. Natural history of claudication: long-term serial follow-up study of 1244 claudicants. J Vasc Surg. 2001;34(6):962–970. doi: 10.1067/mva.2001.119749. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbloom MS, et al. Risk factors affecting the natural history of intermittent claudication. Arch Surg. 1988;123(7):867–870. doi: 10.1001/archsurg.1988.01400310081013. [DOI] [PubMed] [Google Scholar]
- 5.Sigvant B, Lundin F, Wahlberg E. The risk of disease progression in peripheral arterial disease is higher than expected: A meta-analysis of mortality and disease progression in peripheral arterial disease. Eur J Vasc Endovasc Surg. 2016;51(3):395–403. doi: 10.1016/j.ejvs.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Mätzke S, Lepäntalo M. Claudication does not always precede critical leg ischemia. Vasc Med. 2001;6(2):77–80. [PubMed] [Google Scholar]
- 7.McClung JM, et al. BAG3 (Bcl-2-associated athanogene-3) coding variant in mice determines susceptibility to ischemic limb muscle myopathy by directing autophagy. Circulation. 2017;136(3):281–296. doi: 10.1161/CIRCULATIONAHA.116.024873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan TE, et al. Mitochondrial therapy improves limb perfusion and myopathy following hindlimb ischemia. J Mol Cell Cardiol. 2016;97:191–196. doi: 10.1016/j.yjmcc.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan TE, Schmidt CA, Green TD, Brown DA, Neufer PD, McClung JM. Mitochondrial regulation of the muscle microenvironment in critical limb ischemia. Front Physiol. 2015;6:336. doi: 10.3389/fphys.2015.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan TE, Schmidt CA, Green TD, Spangenburg EE, Neufer PD, McClung JM. Targeted expression of catalase to mitochondria protects against ischemic myopathy in high-fat diet-fed mice. Diabetes. 2016;65(9):2553–2568. doi: 10.2337/db16-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt CA, et al. Strain-dependent variation in acute ischemic muscle injury. Am J Pathol. 2018;188(5):1246–1262. doi: 10.1016/j.ajpath.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt CA, et al. Diminished force production and mitochondrial respiratory deficits are strain-dependent myopathies of subacute limb ischemia. J Vasc Surg. 2017;65(5):1504–1514.e11. doi: 10.1016/j.jvs.2016.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karch J, Molkentin JD. Regulated necrotic cell death: the passive aggressive side of Bax and Bak. Circ Res. 2015;116(11):1800–1809. doi: 10.1161/CIRCRESAHA.116.305421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirihai OS, Song M, Dorn GW. How mitochondrial dynamism orchestrates mitophagy. Circ Res. 2015;116(11):1835–1849. doi: 10.1161/CIRCRESAHA.116.306374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan TE, Brophy P, Lin CT, Hickner RC, Neufer PD. Assessment of in vivo skeletal muscle mitochondrial respiratory capacity in humans by near-infrared spectroscopy: a comparison with in situ measurements. J Physiol (Lond) 2014;592(15):3231–3241. doi: 10.1113/jphysiol.2014.274456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teraa M, Conte MS, Moll FL, Verhaar MC. Critical limb ischemia: current trends and future directions. J Am Heart Assoc. 2016;5(2) doi: 10.1161/JAHA.115.002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClung JM, et al. Skeletal muscle-specific genetic determinants contribute to the differential strain-dependent effects of hindlimb ischemia in mice. Am J Pathol. 2012;180(5):2156–2169. doi: 10.1016/j.ajpath.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abu Dabrh AM, et al. Bypass surgery versus endovascular interventions in severe or critical limb ischemia. J Vasc Surg. 2016;63(1):244–53.e11. doi: 10.1016/j.jvs.2015.07.068. [DOI] [PubMed] [Google Scholar]
- 19.Martinez RA, et al. Nationally representative readmission factors in patients with claudication and critical limb ischemia. Ann Vasc Surg. 2018;52:96–107. doi: 10.1016/j.avsg.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Lederman RJ, et al. Therapeutic angiogenesis with recombinant fibroblast growth factor-2 for intermittent claudication (the TRAFFIC study): a randomised trial. Lancet. 2002;359(9323):2053–2058. doi: 10.1016/S0140-6736(02)08937-7. [DOI] [PubMed] [Google Scholar]
- 21.Rajagopalan S, et al. Regional angiogenesis with vascular endothelial growth factor in peripheral arterial disease: a phase II randomized, double-blind, controlled study of adenoviral delivery of vascular endothelial growth factor 121 in patients with disabling intermittent claudication. Circulation. 2003;108(16):1933–1938. doi: 10.1161/01.CIR.0000093398.16124.29. [DOI] [PubMed] [Google Scholar]
- 22.Fadini GP, Agostini C, Avogaro A. Autologous stem cell therapy for peripheral arterial disease meta-analysis and systematic review of the literature. Atherosclerosis. 2010;209(1):10–17. doi: 10.1016/j.atherosclerosis.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Mills JL, et al. The Society for Vascular Surgery Lower Extremity Threatened Limb Classification System: risk stratification based on wound, ischemia, and foot infection (WIfI) J Vasc Surg. 2014;59(1):220–34.e1. doi: 10.1016/j.jvs.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Regensteiner JG, et al. Chronic changes in skeletal muscle histology and function in peripheral arterial disease. Circulation. 1993;87(2):413–421. doi: 10.1161/01.CIR.87.2.413. [DOI] [PubMed] [Google Scholar]
- 25.Hiatt WR, Regensteiner JG, Wolfel EE, Carry MR, Brass EP. Effect of exercise training on skeletal muscle histology and metabolism in peripheral arterial disease. J Appl Physiol. 1996;81(2):780–788. doi: 10.1152/jappl.1996.81.2.780. [DOI] [PubMed] [Google Scholar]
- 26.McDermott MM, et al. Peripheral artery disease, calf skeletal muscle mitochondrial DNA copy number, and functional performance. Vasc Med. 2018;23(4):340–348. doi: 10.1177/1358863X18765667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koopman R, Ly CH, Ryall JG. A metabolic link to skeletal muscle wasting and regeneration. Front Physiol. 2014;5:32. doi: 10.3389/fphys.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brass EP, Hiatt WR. Acquired skeletal muscle metabolic myopathy in atherosclerotic peripheral arterial disease. Vasc Med. 2000;5(1):55–59. doi: 10.1177/1358836X0000500109. [DOI] [PubMed] [Google Scholar]
- 29.Jain A, et al. The Walking Impairment Questionnaire stair-climbing score predicts mortality in men and women with peripheral arterial disease. J Vasc Surg. 2012;55(6):1662–73.e2. doi: 10.1016/j.jvs.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain A, et al. Declining walking impairment questionnaire scores are associated with subsequent increased mortality in peripheral artery disease. J Am Coll Cardiol. 2013;61(17):1820–1829. doi: 10.1016/j.jacc.2013.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leeper NJ, et al. Exercise capacity is the strongest predictor of mortality in patients with peripheral arterial disease. J Vasc Surg. 2013;57(3):728–733. doi: 10.1016/j.jvs.2012.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsubara Y, et al. Sarcopenia is a prognostic factor for overall survival in patients with critical limb ischemia. J Vasc Surg. 2015;61(4):945–950. doi: 10.1016/j.jvs.2014.10.094. [DOI] [PubMed] [Google Scholar]
- 33.McDermott MM, et al. Decline in functional performance predicts later increased mobility loss and mortality in peripheral arterial disease. J Am Coll Cardiol. 2011;57(8):962–970. doi: 10.1016/j.jacc.2010.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDermott MM, et al. Calf muscle characteristics, strength measures, and mortality in peripheral arterial disease: a longitudinal study. J Am Coll Cardiol. 2012;59(13):1159–1167. doi: 10.1016/j.jacc.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pipinos, et al. The myopathy of peripheral arterial occlusive disease: Part 1. Functional and histomorphological changes and evidence for mitochondrial dysfunction. Vasc Endovascular Surg. 2007;41(6):481–489. doi: 10.1177/1538574407311106. [DOI] [PubMed] [Google Scholar]
- 36.Pipinos, et al. The myopathy of peripheral arterial occlusive disease: Part 2. Oxidative stress, neuropathy, and shift in muscle fiber type. Vasc Endovascular Surg. 2008;42(2):101–112. doi: 10.1177/1538574408315995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pipinos, et al. Mitochondrial defects and oxidative damage in patients with peripheral arterial disease. Free Radic Biol Med. 2006;41(2):262–269. doi: 10.1016/j.freeradbiomed.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Pipinos, et al. Abnormal mitochondrial respiration in skeletal muscle in patients with peripheral arterial disease. J Vasc Surg. 2003;38(4):827–832. doi: 10.1016/S0741-5214(03)00602-5. [DOI] [PubMed] [Google Scholar]
- 39.Singh N, et al. Leg strength predicts mortality in men but not in women with peripheral arterial disease. J Vasc Surg. 2010;52(3):624–631. doi: 10.1016/j.jvs.2010.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JR, et al. Protein concentration and mitochondrial content in the gastrocnemius predicts mortality rates in patients with peripheral arterial disease. Ann Surg. 2015;261(3):605–610. doi: 10.1097/SLA.0000000000000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss DJ, et al. Oxidative damage and myofiber degeneration in the gastrocnemius of patients with peripheral arterial disease. J Transl Med. 2013;11:230. doi: 10.1186/1479-5876-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClung JM, et al. Subacute limb ischemia induces skeletal muscle injury in genetically susceptible mice independent of vascular density. J Vasc Surg. 2016;64(4):1101–1111.e2. doi: 10.1016/j.jvs.2015.06.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClung JM, et al. Muscle cell derived angiopoietin-1 contributes to both myogenesis and angiogenesis in the ischemic environment. Front Physiol. 2015;6:161. doi: 10.3389/fphys.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mofarrahi M, et al. Angiopoietin-1 enhances skeletal muscle regeneration in mice. Am J Physiol Regul Integr Comp Physiol. 2015;308(7):R576–R589. doi: 10.1152/ajpregu.00267.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rissanen TT, et al. Expression of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 (KDR/Flk-1) in ischemic skeletal muscle and its regeneration. Am J Pathol. 2002;160(4):1393–1403. doi: 10.1016/S0002-9440(10)62566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dumont NA, Bentzinger CF, Sincennes MC, Rudnicki MA. Satellite cells and skeletal muscle regeneration. Compr Physiol. 2015;5(3):1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- 47.Fortini P, Iorio E, Dogliotti E, Isidoro C. Coordinated metabolic changes and modulation of autophagy during myogenesis. Front Physiol. 2016;7:237. doi: 10.3389/fphys.2016.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duguez S, Féasson L, Denis C, Freyssenet D. Mitochondrial biogenesis during skeletal muscle regeneration. Am J Physiol Endocrinol Metab. 2002;282(4):E802–E809. doi: 10.1152/ajpendo.00343.2001. [DOI] [PubMed] [Google Scholar]
- 49.Sharma N, et al. Use of quantitative membrane proteomics identifies a novel role of mitochondria in healing injured muscles. J Biol Chem. 2012;287(36):30455–30467. doi: 10.1074/jbc.M112.354415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichenko AS, et al. Mitochondrial maintenance via autophagy contributes to functional skeletal muscle regeneration and remodeling. Am J Physiol, Cell Physiol. 2016;311(2):C190–C200. doi: 10.1152/ajpcell.00066.2016. [DOI] [PubMed] [Google Scholar]
- 51.Houmard JA, Pories WJ, Dohm GL. Severe obesity: evidence for a deranged metabolic program in skeletal muscle? Exerc Sport Sci Rev. 2012;40(4):204–210. doi: 10.1097/JES.0b013e31825d53fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heden TD, et al. Greater oxidative capacity in primary myotubes from endurance-trained women. Med Sci Sports Exerc. 2017;49(11):2151–2157. doi: 10.1249/MSS.0000000000001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perry CG, et al. Inhibiting myosin-ATPase reveals a dynamic range of mitochondrial respiratory control in skeletal muscle. Biochem J. 2011;437(2):215–222. doi: 10.1042/BJ20110366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fisher-Wellman KH, et al. Pyruvate dehydrogenase complex and nicotinamide nucleotide transhydrogenase constitute an energy-consuming redox circuit. Biochem J. 2015;467(2):271–280. doi: 10.1042/BJ20141447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2(2):287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 59.Barrientos A. In vivo and in organello assessment of OXPHOS activities. Methods. 2002;26(4):307–316. doi: 10.1016/S1046-2023(02)00036-1. [DOI] [PubMed] [Google Scholar]
- 60.Sparagna GC, Johnson CA, McCune SA, Moore RL, Murphy RC. Quantitation of cardiolipin molecular species in spontaneously hypertensive heart failure rats using electrospray ionization mass spectrometry. J Lipid Res. 2005;46(6):1196–1204. doi: 10.1194/jlr.M500031-JLR200. [DOI] [PubMed] [Google Scholar]
- 61.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 62.Andres AM, et al. Mitophagy and mitochondrial biogenesis in atrial tissue of patients undergoing heart surgery with cardiopulmonary bypass. JCI Insight. 2017;2(4):e89303. doi: 10.1172/jci.insight.89303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.