Abstract
BACKGROUND. Commercial gene expression assays are guiding clinical decision making in patients with prostate cancer, particularly when considering active surveillance. Given heterogeneity and multifocality of primary prostate cancer, such assays should ideally be robust to the coexistence of unsampled higher grade disease elsewhere in the prostate in order to have clinical utility. Herein, we comprehensively evaluated transcriptomic profiles of primary multifocal prostate cancer to assess robustness to clinically relevant multifocality.
METHODS. We designed a comprehensive, multiplexed targeted RNA-sequencing assay capable of assessing multiple transcriptional classes and deriving commercially available prognostic signatures, including the Myriad Prolaris Cell Cycle Progression score, the Oncotype DX Genomic Prostate Score, and the GenomeDX Decipher Genomic Classifier. We applied this assay to a retrospective, multi-institutional cohort of 156 prostate cancer samples. Derived commercial biomarker scores for 120 informative primary prostate cancer samples from 44 cases were determined and compared.
RESULTS. Derived expression scores were positively correlated with tumor grade (rS = 0.53–0.73; all P < 0.001), both within the same case and across the entire cohort. In cases of extreme grade-discordant multifocality (co-occurrence of grade group 1 [GG1] and ≥GG4 foci], gene expression scores were significantly lower in low- (GG1) versus high-grade (≥GG4) foci (all P < 0.001). No significant differences in expression scores, however, were observed between GG1 foci from prostates with and without coexisting higher grade cancer (all P > 0.05).
CONCLUSIONS. Multifocal, low-grade and high-grade prostate cancer foci exhibit distinct prognostic expression signatures. These findings demonstrate that prognostic RNA expression assays performed on low-grade prostate cancer biopsy tissue may not provide meaningful information on the presence of coexisting unsampled aggressive disease.
FUNDING. Prostate Cancer Foundation, National Institutes of Health (U01 CA214170, R01 CA183857, University of Michigan Prostate Specialized Program of Research Excellence [S.P.O.R.E.] P50 CA186786-05, Weill Cornell Medicine S.P.O.R.E. P50 CA211024-01A1), Men of Michigan Prostate Cancer Research Fund, University of Michigan Comprehensive Cancer Center core grant (2-P30-CA-046592-24), A. Alfred Taubman Biomedical Research Institute, and Department of Defense.
Keywords: Oncology
Keywords: Cancer, Molecular diagnosis, Prostate cancer
Gene expression prognostic tests performed on low-grade prostate cancer may not provide meaningful information on the presence of coexisting, unsampled high-grade disease.
Introduction
Widespread adoption of serum prostate-specific antigen (PSA) screening has led to the overdiagnosis and overtreatment of low-grade (Gleason score 6; grade group 1 [GG1]) prostate cancer (1–3). To better balance the harms and benefits of PSA screening, a number of active surveillance strategies have been introduced for low-grade favorable-risk prostate cancer. Active surveillance is composed of multimodal serial monitoring, with an increasing reliance on the utilization of a number of biopsy tissue-based prognostic gene expression tests performed in commercial laboratories (e.g., Myriad Prolaris Cell Cycle Progression [CCP] score, Oncotype DX Genomic Prostate Score [GPS], and GenomeDX Decipher Genomic Classifier [GC]) to provide information on active surveillance suitability versus definitive therapy (4–7). Tissue-based prostate cancer biomarkers, an expenditure in aggregate estimated to be more than $250 million annually, have been aggressively marketed by industry and rapidly adopted by physicians. Additionally, these assays have been included in the 2018 National Comprehensive Cancer Network prostate cancer guidelines and received Centers for Medicare and Medicaid Services approval for use in risk stratification of prostate cancer. Of the estimated 180,000 men diagnosed with prostate cancer annually in the United States, approximately 70% with favorable-risk prostate cancer are thought to be potential candidates for these tests that cost up to $4,500 each (8, 9).
The histologic, spatial, and temporal genomic heterogeneity of solid organ cancers continues to be an impediment for the development of single biopsy–based prognostic biomarkers as well as the implementation of precision medicine approaches that rely on tissue sampling (10, 11). While biopsy-based RNA expression signatures have shown clinical utility in several cancers, understanding the biology of all clinically important foci, within a given organ, can be challenging when the multiplicity as well as the histologic and genomic diversity of cancer foci is high. Prostate cancer is heterogeneous and is multifocal, with multiple, genomically independent tumors identified in up to 80% of men undergoing radical prostatectomy for clinically localized disease (12–15). Hence, there has been enormous interest in identifying prostate cancer prognostic biomarkers that are unaffected by tissue sampling bias. Critically, for maximum clinical utility, effective prognostic biomarkers must be able to determine which men with low-grade cancer on diagnostic biopsy have (a) an undersampled high-grade component of the same tumor focus and/or (b) an unsampled, spatially and genomically distinct high-grade tumor focus missed by the original biopsy.
To date, a comprehensive transcriptomic analysis of grade discordant primary multifocal prostate cancer has not been reported. Notably, the advent and refinement of technologies compatible with formalin-fixed, paraffin-embedded (FFPE) tissue samples, such as qRT-PCR, microarray-based expression profiling, NanoString, and RNA-based next-generation sequencing (NGS), has enabled the rapid clinical translation of transcriptomic biomarkers (16–19). Application of these methodologies to low-grade prostate cancer, however, is particularly challenging due to the minute volume of cancerous biopsy tissue often available for analysis (20). Importantly, of the above described prognostic expression tests, one directly claims to be robust to both undersampling of high-grade components of the same tumor focus as well as the presence of unsampled high-grade tumor foci elsewhere in the prostate (6). Although these assays have been analytically and clinically validated to predict a variety of clinically relevant endpoints, recent evidence supports transcriptional differences between multifocal low- and high-grade prostate cancer, challenging the potential robustness of these assays to tumor multifocality and genomic heterogeneity (21). In addition, it is important to note that CCP score, GPS, and GC have yet to be evaluated in a head-to-head manner.
Here, we developed a multiplex prostate cancer–specific RT-PCR–based RNA-sequencing (mxRNAseq) assay compatible with minute FFPE specimens in order to directly interrogate and compare commercially available prognostic transcriptomic signatures and relevant individual candidate biomarkers in primary, clinically relevant multifocal prostate cancer.
Results
Cohort composition.
Our cohort included 193 FFPE tissue specimens chosen to represent the complete spectrum of prostate cancer progression (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.123468DS1). The complete sample information, including tissue type (e.g., primary prostate cancer [n = 156]; hormonal therapy naive metastases [n = 18]; metastatic castration resistant prostate cancer [n = 7]; benign prostate tissue [n = 11]; and benign lymph node [n = 1]), specimen type, prior treatment, and previous molecular characterization is provided in Supplemental Table 1. Prior to exclusion of samples that did not pass stringent quality control (QC) metrics, as described in the Supplemental Methods, our FFPE tissue cohort included 156 primary prostate cancer samples from 50 cases, including 21 multifocal cases with at least 1 sample from a GG1 focus and at least 1 sample from a GG2–5 focus; 13 of these multifocal cases had at least 1 sample from a GG1 focus and at least 1 sample from a ≥GG4 focus, representing the extremes of GG difference in multifocality (Supplemental Table 1). For comparison, prior to exclusion of samples not passing QC metrics, our cohort also included 9 GG1 samples (from 6 cases) from prostates with exclusively GG1 prostate cancer.
After exclusion of samples that did not pass QC metrics, our informative cohort contained 120 primary prostate cancer samples from 44 cases. This cohort included 15 multifocal cases with at least 1 sample each from a GG1 focus and a higher grade (GG2–5) focus; 8 of these multifocal cases had at least 1 sample each from a GG1 focus and a ≥GG4 focus, representing the extremes of GG discordance in multifocal disease. For comparison, our cohort included 8 GG1 informative samples from 6 cases with organ-confined GG1 prostate cancer.
mxRNAseq assay assessment and validation.
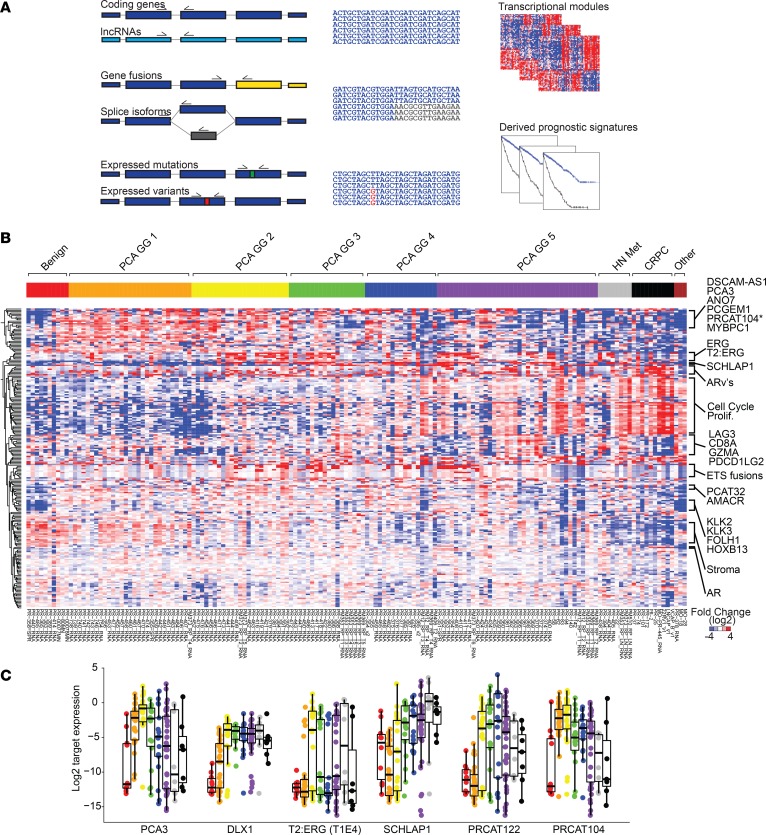
The details of our mxRNAseq assay assessment and validation are as described in the Supplemental Methods. Briefly, to detect and/or quantify multiple potentially relevant prostate cancer transcriptomic alterations, we designed a custom 306-amplicon mxRNAseq panel (Supplemental Table 2) that targeted genes required to derive the 3 commercially available prognostic signatures as well as diagnostic/subtyping relevant genes, transcriptional modules (e.g., AR signaling, activated T cell infiltrate, neuroendocrine gene expression), recurrent 5′ and 3′ gene fusion partners (e.g., ETS genes, BRAF and RAF1, and their common 5′ partners), lncRNAs (e.g., SChLAP1), expressed recurrent somatic mutations (e.g., hotspot residues in SPOP, BRAF and IDH1), and expressed relevant germline variants (e.g., HOXB13 G84E) (Figure 1A). Raw read counts for all amplicons in all samples are provided in Supplemental Table 2, and normalized reads from amplicons/samples passing QC are provided in Supplemental Table 3.
Figure 1. Targeted multiplexed RNA sequencing enables robust transcriptional profiling of prostate cancer.
(A) Prostate cancer multiplexed RNA-sequencing (mxRNAseq) panel. To interrogate multiple classes of prostate cancer relevant transcriptional biomarkers, we designed a custom 306-amplicon mxRNAseq assay targeting coding genes, lncRNAs, gene fusions (e.g., TMPRSS2:ERG [T2:ERG]), splice isoforms (e.g., AR variants), expressed somatic mutations (e.g., SPOP hotspot mutations), and expressed germline risk variants (e.g., HOXB13 p.G84E). Targets to derive commercially available prognostic assays, including Prolaris Cell Cycle Progression (CCP) score, Oncotype Dx Genomic Prostate Score (GPS), and Decipher Genomic Classifier (GC) score were also included. (B) mxRNAseq confirms expected transcriptional deregulation across prostate cancer progression. Unsupervised, centroid linkage hierarchical clustering of log2-normalized expression for 223 informative amplicons from our cohort of FFPE samples representing the full spectrum of prostate cancer progression (n = 156) is shown. Samples are sorted from benign prostate tissue, ascending grade group (GG) of localized prostate cancer (GG1–5, equivalent to Gleason scores 6, 3+4 = 7, 4+3 = 7, 8, and >8), hormone therapy naive metastases (HN Met), castration-resistant prostate cancer (CRPC), and nonprostate cancer specimens included as assay controls (far right). Individual transcripts and modules (e.g., cell cycle/proliferation [Prolif.], stroma, etc.) are indicated to the right. (C) Transcriptional deregulation of individual prostate cancer transcriptional biomarker. Log2-normalized expression for prostate cancer–specific markers (PCA3, DLX1, T2:ERG [isoform T1E4]), overexpressed in aggressive prostate cancer–specific markers (SCHLaP1 and PRCAT122) and underexpressed in aggressive prostate cancer–specific markers (PRCAT104), is shown across prostate cancer progression (same order/color legend as in B).
Unsupervised hierarchical clustering of target gene/transcript expression across the 156–informative sample compendium of FFPE tissue and cell line samples meeting high-stringency QC parameters (see Supplemental Methods) is shown in Figure 1B. Multiple analyses support the validity of our approach and observed expression profiles, as described in the Supplemental Methods and Supplemental Figures 1 and 2. Importantly, single-coding genes and lncRNAs known to be dysregulated in prostate cancer displayed expected differential expression patterns, including PCA3, DLX1, and TMPRSS2:ERG, as biomarkers of prostate cancer versus benign prostate tissue (Figure 1C). Similarly, SChLAP1, a lncRNA consistently identified as one of the most prognostic single transcripts across prostate cancer profiling studies (22, 23), was confirmed to be overexpressed in aggressive prostate cancer (Figure 1C). Furthermore, lncRNAs reported as prostate cancer specific (e.g., PRCAT122) (24) or prostate cancer specific but decreased in high-grade disease (e.g., PCAT-14 [also known as PRCAT104] (24, 25), showed expected tissue expression patterns (Figure 1C). Detection of prostate cancer relevant gene fusions, expressed somatic mutations, and germline risk variant profiles, and androgen receptor (AR) full-length and AR splice variants by mxRNAseq are as described in the Supplemental Methods and Supplemental Figures 3–5. Taken together, these findings support the ability of our mxRNAseq assay to characterize a diverse set of transcriptional events (essential coding genes and gene fusions, lncRNAs, somatic mutations, germline variants, and splice variants) important for prostate cancer molecular subtyping, multiclonality assessment, risk stratification, and potential treatment guidance.
Assessment of single-gene and clinically available prognostic biomarkers.
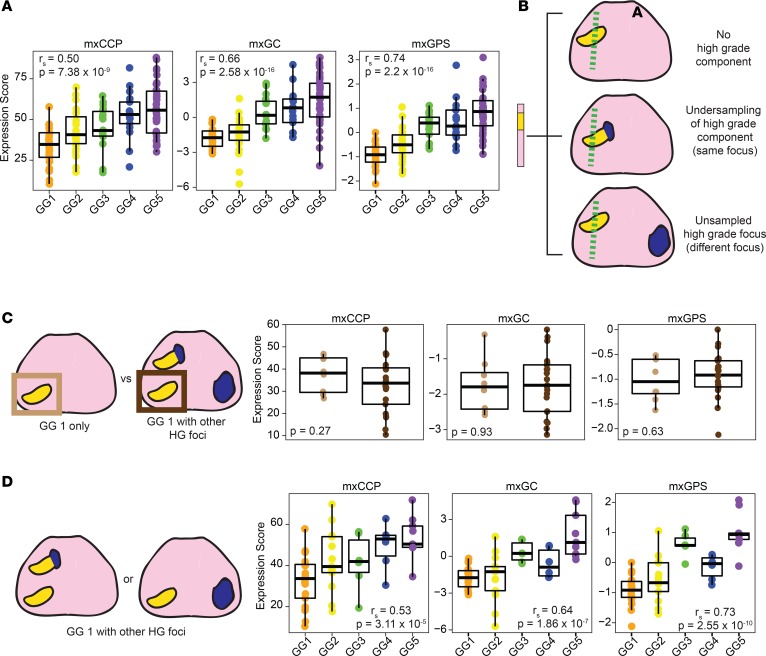
We designed our assay to interrogate both single-transcript prognostic biomarkers, such as SCHLAP1 and PRCAT104, as well as the target genes/transcripts used to derive the commercially available CCP, GPS, and GC scores. Hence, as described in detail in the Supplemental Methods, we derived CCP (mxCCP), GPS (mxGPS), and GC (mxGC) scores, and as expected, mxCCP (rS = 0.50, P = 7.38 × 10–9), mxGPS (rS = 0.74, P = 2.2 × 10–16), and mxGC (rS = 0.66, P = 2.58 × 10–16) scores significantly increased across prostate cancer GGs (Figure 2A) and showed expected expression patterns between benign prostate tissue and metastatic samples (Supplemental Figure 6A). Supporting these derived scores, cell cycle/proliferation–related genes form one of the most correlated transcriptional modules in our assay (Figure 1B and Supplemental Figure 2), regardless of their inclusion in CCP (all target genes), GPS (TPX2), or GC (e.g., UBE2C and NUSAP1). Likewise, the individual components of the mxGPS score were evaluated and showed expected expression across GGs and in benign prostate tissue and metastatic samples (Supplemental Figure 6B). Finally, clustering of GC target gene/transcript expression from our mxRNAseq data resulted in clustered expression patterns across prostate cancer progression (Supplemental Figure 6C). Hence, despite the obvious limitations inherent in deriving prognostic signatures developed on other platforms (particularly for GC given the included target transcripts and analytical approach), these results support the ability and internal validity of our mxRNAseq assay to derive multiple commercially available, expression-based prognostic biomarkers.
Figure 2. Derivation and assessment of clinically available prognostic signatures.
(A) mxRNAseq-derived CCP (mxCCP), Decipher Genomic Classifier (mxGC), and Oncotype Dx GPS (mxGPS) scores are plotted stratified by grade group (GG1–5) for 125 FFPE primary prostate cancer tissue samples. Derived scores significantly increase with ascending GG (Spearman rank [rs] correlation, and 1-sided ANOVA P values are shown). (B) Due to both intratumoral grade heterogeneity as well as true multifocality, diagnostic prostate biopsy sampling of only low-grade prostate cancer (GG1, yellow) may reflect a lack of high-grade (HG) tumor component (top), undersampling of a high-grade (blue) component (GG2 or GG3 tumor focus, middle), or unsampling of a separate high-grade prostate cancer focus (bottom). As the most concerning scenario for a patient considering active surveillance (AS) is the latter, we tested the robustness of derived prognostic scores from GG1 to multifocality through several analyses. (C) Derived prognostic scores do not differ between GG1 prostate cancer when present in isolation or when other HG tumor foci are present. Derived prognostic mxCCP, mxGC, and mxGPS scores were plotted from samples of pT2 GG1 prostate cancer without HG foci (clinically indolent, light brown, n = 8 from 6 cases) versus scores from GG1 prostate cancer foci (n = 21 from 15 cases) where other HG foci were present. No significant differences between the 2 groups for any derived prognostic score were observed (2-sample, unpaired 2-sided t test P values are shown). (D) Derived prognostic scores differ between GG1 prostate cancer foci and concurrent HG foci. Derived prognostic scores were plotted for all samples from the 15 cases from C where samples were taken from both GG1 prostate cancer foci as well as other concurrent higher grade foci. Samples were stratified by the GG of the profiled component. Spearman rank (rs) correlation and 1-sided ANOVA P values for scores stratified by GG are shown, demonstrating significantly increased derived prognostic scores by GG of the profiled component (similar to findings from our entire cohort, including these samples as shown in Supplemental Figure 7C).
Assessment of prognostic biomarkers in the context of tumor multifocality.
For maximum clinical utility, prognostic prostate cancer biomarkers used at biopsy should be robust to both undersampling of high-grade cancer in the same tumor focus as well as to an unsampled spatially distinct high-grade focus (Figure 2B). Thus, we comprehensively assessed the robustness of individual candidate prognostic transcripts and mxRNAseq-derived signatures to clinically relevant tumor multifocality using a multistep approach on prostatectomy specimens.
First, we hypothesized that if prognostic signatures in low-grade tumors could predict unsampled high-grade tumor foci elsewhere in the prostate, such signatures should differ between GG1 foci from prostates with only organ-confined GG1 tumor foci (no metastatic potential) (26) and prostates with GG1 and concomitant higher-grade (GG2–5) foci. As shown in Figure 2C, no significant difference in mxCCP (2-sample unpaired t test; P = 0.27), mxGPS (P = 0.63), or mxGC scores (P = 0.93) was observed between GG1 samples from GG1-only prostates (n = 8 samples from 6 cases) and GG1 samples with coexisting multifocal GG2–5 foci (n = 21 samples from 15 cases). Likewise, PRCAT104 showed no significant difference between GG1 samples from GG1-only and GG1 with multifocal GG2–5 foci (P = 0.78; Supplemental Figure 7A), while SChLAP1 showed significantly lower expression in GG1 with multifocal GG2–5 foci versus GG1 only (P = 0.006; Supplemental Figure 7A).
Next, limiting the cohort to grade-discordant multifocal cases only (n = 15 cases), we determined whether commercial prognostic signatures were equivalent in GG1 samples (n = 21) versus all coexisting GG2–5 samples (n = 34) from the same set of cases, as would be expected if such biomarkers were truly robust to multifocality. As shown in Figure 2D and Supplemental Figure 7C, in these patients, mxCCP (1-sided ANOVA for mxCCP score by GG, P = 0.002), mxGPS (P = 1.2 × 10–10), and mxGC scores (P = 7.9 × 10–7), as well as SCHLAP1 expression (P = 4.9 × 10–4), were significantly different across GG, with significant direct correlations observed between SChLAP1 (rS = 0.52, P = 4.87 × 10–5) and all prognostic scores (mxCCP [rS = 0.53, P = 3.11 × 10–5], mxGPS [rS = 0.73, P = 2.55 × 10–10], mxGC (rS = 0.64, P = 1.86 × 10–7]) with increasing GG. Likewise, PRCAT104 expression was significantly inversely correlated with GG (rS = –0.34, P = 0.011). Taken together, these results do not support equivalent prognostic expression profiles in low-grade foci and concomitant multifocal high-grade foci.
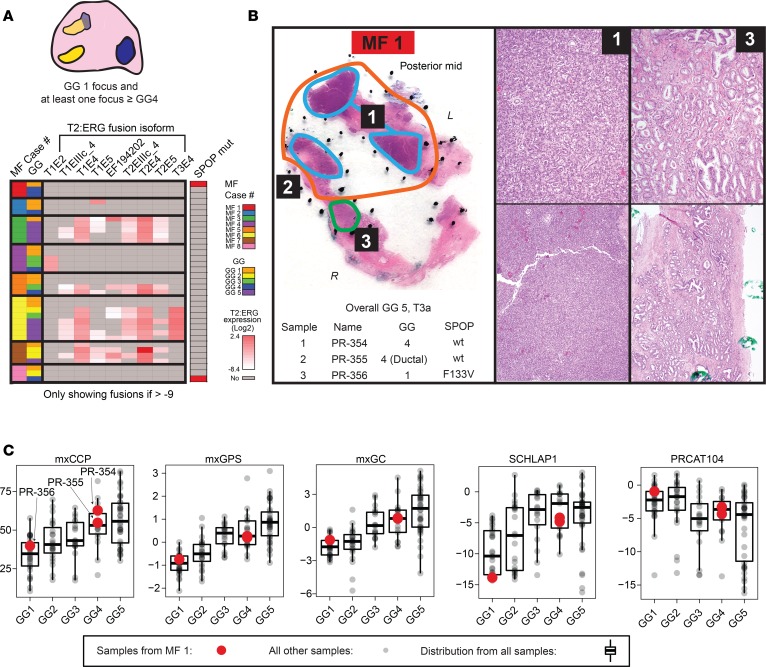
The most concerning false-negative potential for prognostic tests given multifocality is when only a GG1 focus is sampled by diagnostic biopsy, yet an unsampled, very aggressive prostate cancer focus (≥GG4) is present elsewhere in the prostate (Figure 2B). Hence, our cohort specifically included 8 informative cases with at least 1 sample from a GG1 focus and at least 1 sample from a ≥GG4 focus, to represent extremes of histologic aggressiveness in multifocality (MF cases 1–8; Figure 3A and Figure 4A). In addition to appearing multifocal by histopathology, molecular subtyping by our mxRNAseq assay (gene fusion and SPOP mutation status) supports clear multiclonality between the GG1 and ≥GG4 foci in 6 of 8 cases (MF cases 1, 2, 4–6, and 8), as shown in the integrative heatmap in Figure 3A. For example, in MF cases 4–6, samples from the GG1 tumor foci were T2:ERG-negative/SPOPwt, while samples from the GG4 foci (including samples from GG2 and GG3 morphology) were all T2:ERG-positive/SPOPwt. In the other 2 cases (MF cases 3 and 7), although samples from both the GG1 and ≥GG4 foci were T2:ERG-positive/SPOPwt, unique T2:ERG fusion isoforms were expressed exclusively in the low- or high-grade foci, consistent with multiclonality, with MF 3 showing definitive multiclonality by global genomic profiling (see below).
Figure 3. Derived prognostic signatures in the context of prostate cancer multifocality with extreme grade differences.
(A) Our cohort included 8 informative multifocal cases (MF cases 1–8) with “extreme” grade differences (e.g., at least 1 low-grade [GG1] focus and at least 1 spatially independent high-grade [≥GG4] focus), as shown in the diagram on top. An integrative heatmap summarizes mxRNAseq profiling data supporting multifocality of these cases below. Individual T2:ERG fusion isoform expression and SPOP mutation status are shown for the 35 samples from these cases (case and GG of each sample shown according to the legend). T2:ERG/SPOP status (and unique T2:ERG isoform expression) support multifocality in these cases as described in the text). (B) Histology and mxRNAseq support extreme multifocality in case MF1, which had a large pT3a GG5 index tumor focus (orange). The posterior capsule section additionally contained a small, nearby but spatially distinct GG1 focus (green). Two samples were taken from the GG5 index focus (cyan 1 and 2), and 1 sample was taken from the GG1 focus (green, 3). Sample name, profiled morphologic GG, and SPOP mutation status is shown, along with high- and low-power histopathology of the indicated samples (original magnification, ×10 [top]; ×4 [bottom]). The morphology and SPOP p.F133V mutation only in the GG1 focus support clear multifocality. (C) Derived prognostic scores (mxCCP, mxGPS, mxGC), as well as expression levels of candidate prognostic long noncoding RNA (lncRNA) biomarkers (SCHLAP1 and PRCAT104) from MF 1 samples, are indicated as red points (stratified by profiled GG) overlying the distribution of all 125 localized prostate cancer samples from our cohort (see Figure 2A).
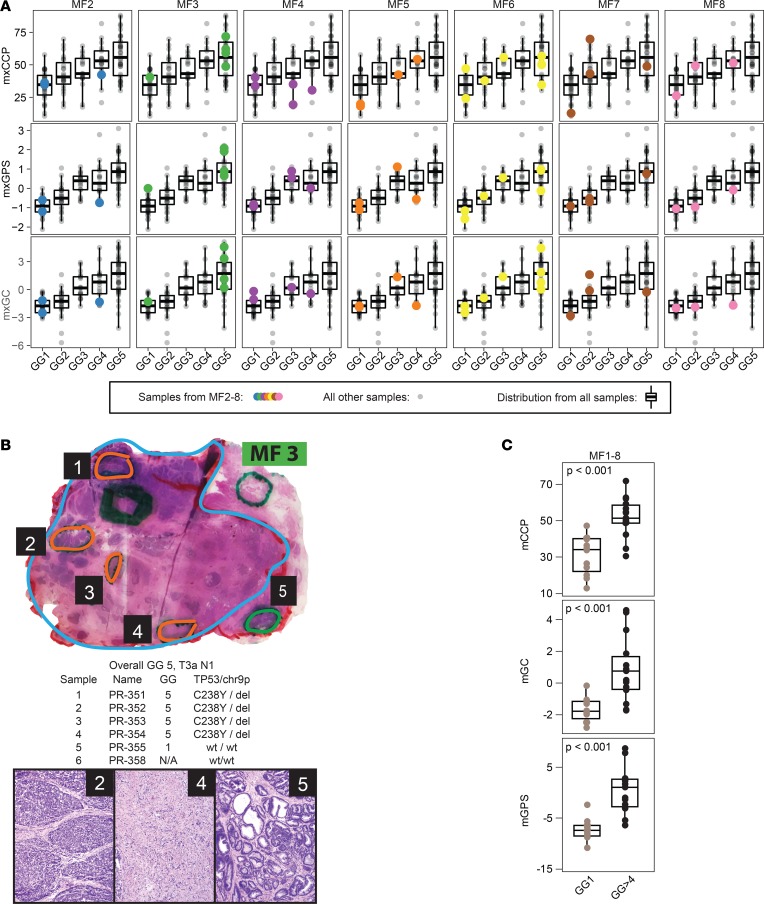
Figure 4. Derived prognostic scores are not robust to multifocal prostate cancer with extreme grade differences.
(A) Derived prognostic scores (mxCCP, mxGPS, mxGC) from all profiled multifocal cases (MF2–8; MF1 in Figure 3) with extreme grade differences (i.e., at least 1 sample each from GG1 and ≥GG4 tumor foci). For each case, profiled samples are indicated by colored points overlying the overall cohort distribution, as in Figure 3C. (B) Histology and mxRNAseq support extreme multifocality in case MF3, which had a large pT3a GG5 index tumor focus (cyan) and a spatially distinct small GG1 focus (focus 5, green). Informative samples from the GG5 (orange) and GG1 (focus 5, green) foci are indicated (as well as a sampled area of normal prostate stroma in gray), with the chart showing sample name, profiled morphologic GG, and TP53 mutation/chromosome [chr] 9p deletion status, along with histopathology of the indicated samples (original magnification, ×4). The morphology and distinct TP53 p.C238Y/9p deletion status support clear multifocality, in addition to the unique T2:ERG isoform expression seen only in the GG1 focus (Figure 3A). (C) Derived prognostic scores are not robust to multifocal prostate cancer with extreme grade differences. Derived prognostic scores from all GG1 samples versus ≥GG4 tumor foci from the 8 multifocal cases with extreme grade group differences shown in Figure 3, A and C, are plotted (2-sample, unpaired 2-sided t test P values are shown).
Next, we assessed whether derived prognostic signatures and single-gene prognostic biomarkers were robust to extreme grade-discordant multifocality. Histology from MF case 1 (overall GG5, pT3a) is shown in Figure 3B. In this case, posterior capsule sections in the same block showed tumor with distinct morphology from two separate foci. Two sampled areas of GG4 morphology (samples PR-354 and PR-355) were taken from the larger GG5 index tumor focus, along with a sample from a distinct focus of GG1 (PR-356). Of note, the GG1 sample harbored a SPOP p.F133V mutation detected by both mxRNAseq and mxDNAseq, while the GG4 samples were SPOPwt, consistent with multiclonality despite the spatial proximity of the tumor foci. In this case of extreme multifocality, we would expect that if transcriptional biomarkers and signatures were robust to multifocality, the multifocal GG1 and GG4 samples should show similar profiles. However, as shown in Figure 3C, the GG1 and GG4 samples from this case display distinct derived prognostic signatures and individual transcript biomarker expression more in keeping with their morphology, based on comparison to all other samples in our compendium.
MF case 3 (overall GG5, pT3a, N1), showed a GG5 index focus involving nearly the entire prostate (Figure 4B, cyan), with a minute GG1 focus (with HGPIN-like morphology) that was separate from the index focus on all levels (Figure 4B, focus 5, green). mxDNAseq demonstrated a TP53 p.C238Y somatic mutation and chr 9p deletion in all 4 samples from the index GG 5 focus (orange) and 2 samples from the lymph node metastasis (data not shown) but not in the GG1 focus (focus 5, green). By mxRNAseq, although the GG5 and GG1 samples expressed multiple T2:ERG gene fusions at similar levels, the GG1 focus uniquely expressed the T2:ERG.EF194202 isoform (Figure 3A). Collectively, these results demonstrate that the low- and high-grade tumor foci in this case represent true multifocality at the extremes of aggressiveness, making this case ideal to assess the robustness of transcriptomic biomarkers to multifocality.
Assessment of the transcriptional profile of other MF cases (see Supplemental Results and Supplemental Figures 8A and 9 for additional case descriptions) consistently demonstrated similar findings in nearly all cases (Figure 4C and Supplemental Figure 8B). Across the 8 extreme grade-discordant MF cases, we observed a significantly higher mxGPS score (P = 6.2 × 10–6), mxCCP score (P = 3.5 × 10–5), mxGC score (P = 0.0004), and SCHLAP1 expression (P = 4.2 × 10–5) in ≥GG4 versus GG1 samples (n = 12 and 15, respectively; Figure 4C and Supplemental Figure 8B). Likewise, PRCAT104 was significantly lower in the ≥GG4 versus GG1 samples (P = 0.008; Supplemental Figure 8B). Taken together, our data demonstrate that, in the context of true multifocality, derived prognostic expression signatures and single-gene biomarkers are not robust across concomitant tumor foci with extremes of histologic grade.
Discussion
We developed a targeted mxRNAseq assay to enable comprehensive profiling of prostate cancer–specific transcriptomic events, including derived commercially available prognostic tests, across routine prostate cancer FFPE specimens. Applying this assay to prostate cancer tissue samples — including cases of extreme grade discordant multifocality (distinct foci of GG1 and ≥GG4) — we demonstrate that derived commercially available prognostic signatures and candidate single-transcript biomarkers obtained from a low-grade cancer focus do not predict the presence of coexisting high-grade foci.
An important feature of localized prostate cancer is the nearly ubiquitous nature of multifocal disease, with most men having more than one genomically distinct tumor focus in the prostate at radical prostatectomy. Such multifocality has the potential to profoundly influence multiple aspects of prostate cancer clinical management, particularly a recommendation to pursue active surveillance. A common question faced by physicians and patients alike when confronted with a new diagnosis of apparent low-grade/low-risk prostate cancer is could there be more aggressive disease elsewhere in the prostate? To be sure, inter/intrafocal genomic heterogeneity is not unique to prostate cancer; it has been reported in several other malignancies, including kidney cancer, lung cancer, and medulloblastoma, and has been shown to affect cancer outcomes (10, 27, 28). Notably, in a recent genomic characterization of localized prostate cancer, multiple subclones of cancer were found in 59% of patients(29). The authors reported that multiclonality was associated with a higher risk of relapse among cases of localized prostate cancer, suggesting that biomarkers that can capture tumor heterogeneity should be developed for low-risk tumors in patients thought to be suitable for active surveillance (29).
Current tissue-based prognostic assays applicable to patients considering active surveillance after biopsy, such as Prolaris, Oncotype DX, and Decipher, were constructed on the premise that risk stratification can be significantly improved through molecular assessment of the biopsy detected prostate cancer (4–7). In validation studies, these tests were shown to improve on the performance of multivariate clinicopathological models for predicting adverse surgical pathology as well as longer term oncologic outcomes (6, 30–32). However, in many of these studies, the initial development and validation was performed using prostatectomy tissue, enabling a priori capture of the index/highest grade lesion (6, 30–32). Importantly, one test directly claims that its signature is capable of capturing the biological profile of all prostate cancer foci within a given prostate, even if the biopsy missed the higher grade focus (6). Tests with such robustness to multifocality would be of tremendous clinical value, especially when one considers the large numbers of men diagnosed with low-risk prostate cancer every year in the United States. Although these tests were not validated on low-grade cancers in the context of multifocality, the 2018 National Comprehensive Cancer Network guidelines include Oncotype Dx, Decipher, and Prolaris as methods of molecular risk stratification for men with very-low- and low-risk disease on biopsy. Additionally, while these tests are not recommended by the American Urological Association and the European Association of Urology for use in this setting, Oncotype Dx and Prolaris have received positive coverage decisions from Centers for Medicare and Medicaid Services and many urologists may be using these tests in this manner.
Without accurate assessment, patients who initially have an unsampled higher grade focus may appear to “rapidly progress” on active surveillance when subsequent biopsy samples identify a preexisting high-grade focus or unexpected aggressive pathology at radical prostatectomy in those who elect definitive management (33). Our study, which also included profiling of coisolated DNA to inform on true multifocality, clearly demonstrates that the molecular profile of a single cancer focus does not capture the multifocal genomic and transcriptomic landscape of primary prostate cancer, particularly when the sampled focus is low grade. These findings are supported by smaller scale studies of multifocal prostate cancer that found significant interfocal heterogeneity (21, 34). Notably, Wei et al. also observed discordant RNA expression profiles among distinct foci from 4 radical prostatectomy samples; however, this study did not use precisely isolated FFPE samples and no correlation between GG and prognostic scores was determined (21). Likewise, Radtke et al. demonstrated that although GC scores were generally consistent between MRI/ultrasound fusion-guided biopsies containing the highest grade prostate cancer and the index foci at prostatectomy in a cohort of 7 patients, 1 patient showed discordant GC profiles in high- and low-grade biopsy cores (34). Additional large-scale studies are needed to examine the gene expression profiles associated with cancers identified by MRI-guided biopsy.
Our study has several limitations. First, we did not precisely assess for robustness to intrafocal tumor heterogeneity. Several tissue-based prognostic tests have directly assessed this, and orthogonal protein and genomic data support this concept (6, 35). Second, we did not use the actual commercially available tests (Oncotype Dx, Prolaris, or Decipher, including their direct analytical methods) or perform our assay in a CLIA-accredited laboratory environment. Specific to the challenges of deriving scores on an orthogonal platform, we were only able to design amplicons for 18 of the 22 included transcripts in the GC score, and we used the housekeeping genes of GPS to normalize CCP scores. Nevertheless, numerous lines of evidence described in the Supplemental Methods support the validity of our results and observed expression profiling. It is unlikely that the commercial assays would perform any differently in assessing multifocal disease, given that our results demonstrated clear differences in the overall gene expression profile of low- and high-grade prostate cancer. Third, we utilized prostatectomy tissue samples for multifocality robustness assessment instead of biopsy tissue samples as would be typically assessed by these commercially available assays. This approach was necessary to ensure profiling of true multifocal disease. Studies are ongoing to perform a similar assessment on biopsy tissues (e.g., assessing concordance of signatures from biopsies with small volume, GG1 prostate cancer on one side of the prostate versus biopsies with ≥GG4 prostate cancer on the contralateral side).
Both tumor multifocality and heterogeneity represent significant challenges in the evaluation and management of men with primary prostate cancer. In aggregate, our findings suggest that without clear validation in patients with extremes of GG discordant multifocal disease, caution should be exercised when using expression-based genomic tests obtained from a single low-grade cancer focus to make treatment decisions. Development of blood, urine, tissue, and imaging-based biomarkers that can overcome obstacles imposed by prostate cancer multifocality and heterogeneity are urgently needed to better inform initial management in the thousands of men with apparent low-grade prostate cancer considering active surveillance each year.
Methods
Patients and specimens.
We assembled a retrospective, multi-institution, nonconsecutive cohort of 193 FFPE tissue specimens chosen to represent the complete spectrum of prostate cancer progression (Supplemental Table 1). Additionally, we included melanoma samples (n = 2) and FFPE prostate cancer cell line pellets (n = 3; 22RV1, LNCAP, and VCAP) as controls for specific amplicons. Relevant demographic, clinical, and pathologic data were extracted from each patient’s medical chart. All profiled prostate cancer samples were evaluated for multifocality, assigned a Gleason score and GG, and outlined for isolation by a board certified anatomic pathologist with experience in prostate and molecular pathology (36). We classified samples from regions of Gleason score 4+3 cancer with tertiary pattern 5 as GG4 given equivalently aggressive behavior (37).
mxRNAseq assay design.
Relevant prostate cancer transcripts and prognostic signatures were identified from internal and external comprehensive expression and mutational profiling efforts, clinically available prognostic tests, and potentially useful biomarkers. Using the Ampliseq White Glove service, we designed a custom 306-amplicon multiplex Ampliseq RNA-sequencing (mxRNAseq) panel targeting multiple classes of relevant transcriptional alterations (Supplemental Table 2). Amplicons were included to assess 3 previously assessed candidate housekeeping genes (19), all housekeeping (n = 5) and target genes (n = 12) from Oncotype Dx GPS (6), and all target genes (n = 31) from Myriad’s Prolaris assay [CCP score] (4). We were able to design amplicons for 18 of the 22 included transcripts in GenomeDX’s Decipher prostate cancer test (which generates a GC score, with 3 distinct amplicons each targeted against different regions of the MYBPC1, ANO7, and UBE2C loci [for a total of 24 amplicons]) (7).
RNA and DNA NGS and bioinformatics methods.
RNA and DNA isolation and NGS were performed as detailed in the Supplemental Methods and as previously described (18, 38, 39). Detailed bioinformatics methods, fusion isoform- and partner-level analyses, AR and AR splice variant analysis, and DNA variant calls and copy number analyses are as previously reported and described in the Supplemental Methods (18, 38, 40).
Derivation of commercially available prognostic scores.
For each sample, we derived CCP, GPS, and GC (mxCCP, mxGPS and mxGC scores, respectively) based on the normalized expression of the specific target genes included in each respective assay as described in detail in the Supplemental Methods (5, 7, 20). Briefly, for mxCCP scores, we used the previously published formula for calculating CCP scores for nonreplicate experiments using GPS housekeeping genes (5). For mxGPS, scores were derived by adding log2-normalized gene expression values for each score module and adding the weighted modules as previously published (20). Given that the Decipher GC score was built using a random forest-based classifier and not all target transcript locations have been published, we calculated mxGC scores as previously reported (7) and as follows: (average of the log2-normalized values of overexpressed in prostate cancer progression target genes) – (average of the log2-normalized values of underexpressed in prostate cancer progression target genes).
Statistics.
One-way ANOVA tests were used to compare differences in mean score among GG1–5 for derived prognostic scores (mxCCP, mxGPS, and mxGC) and normalized expression of single candidate prognostic targets. Spearman rank-order correlations (rs) were determined for derived prognostic scores and normalized expression of single candidate prognostic targets versus GG. Two-sample, unpaired, two-tailed t tests were performed to compare derived prognostic scores and single prognostic targets between groups of samples. All statistical analyses were performed using R. P < 0.05 was considered statistically significant.
Study approval.
The study was approved by the institutional review boards of the University of Michigan Medical School, Office of Research, University of Michigan Medical School. This is a retrospective assessment of archived specimen with minimal risks, thus informed consent was not required for the conduct of this study.
Author contributions
SSS, SFS, SAT, and GSP designed research studies. SSS, KRP, and LLDLV conducted experiments. SSS, DHH, JBK, RM, AMU, NEC, ML, KRP, LL, MS, NRL, JC, MAR, SFS, and SAT acquired data. SSS, DHH, JBK, and SAT analyzed data. SSS, DHH, JBK, SAT, and GSP wrote the first draft of manuscript. SSS, DHH, JBK, RM, AMU, NEC, ML, KRP, LLDLV, MS, NRL, DES, TMM, MSD, AMC, JC, MAR, SFS, SAT, and GSP critically reviewed the manuscript.
Supplementary Material
Acknowledgments
The authors acknowledge Elai Davicioni, Nicholas Erho, and Mohammed Alshalalfa (GenomeDX, Vancouver, British Columbia, Canada) for their assistance in designing amplicons to target the Decipher assay genes. The custom RNAseq panel was designed through the Ampliseq White Glove service of Thermo Fisher Scientific. This work was supported in part by the Prostate Cancer Foundation (to DES, JC, MAR, SAT, SSS, and TMM), the National Institutes of Health (U01 CA214170 to AMC and SAT and R01 CA183857 to SAT; the University of Michigan Prostate Specialized Program of Research Excellence [S.P.O.R.E.] P50 CA186786-05; Weill Cornell Medicine S.P.O.R.E. P50 CA211024-01A1). It was also supported by the Men of Michigan Prostate Cancer Research Fund and the University of Michigan Comprehensive Cancer Center (core grant 2-P30-CA-046592-24). TMM and SAT have been supported by the A. Alfred Taubman Biomedical Research Institute. DES has been supported by the Department of Defense. JC was supported by the Nuovo-Soldati Foundation.
Version 1. 11/02/2018
Electronic publication
Footnotes
Conflict of interest: SAT has received travel support from and had a sponsored research agreement with Compendia Bioscience/Life Technologies/Thermo Fisher Scientific, which provided access to a DNA-sequencing panel used herein. The University of Michigan and Brigham and Women’s Hospital have been issued patents (10,041,123; 9,745,635; 9,719,143; 9,303,291; 9,284,609; 8,969,527; 8,580,509; 8,211,645; and 7,718,369) on ETS gene fusions in prostate cancer, on which AMC, MAR, and SAT are coinventors. The diagnostic field of use was licensed to Hologic/Gen-Probe Inc., which has sublicensed rights to Roche/Ventana Medical Systems. SAT has served as a consultant for and received honoraria from Janssen, AbbVie, Sanofi, Almac Diagnostics, and Astellas/Medivation. SAT has sponsored research agreements with Astellas/Medivation and GenomeDX. SAT is a cofounder and consultant for Strata Oncology. TMM has received research funding from MDxHealth, Myriad Genetics, and GenomeDX. TMM has served as a consultant for Myriad Genetics.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: JCI Insight. 2018;3(21):e123468. https://doi.org/10.1172/jci.insight.123468.
Contributor Information
Daniel H. Hovelson, Email: hovelson@med.umich.edu.
Romain Mathieu, Email: romainmath@yahoo.fr.
Nicole E. Curci, Email: ncurci@med.umich.edu.
Matthew Lee, Email: lematthe@med.umich.edu.
Lorena Lazo de la Vega, Email: llazodel@med.umich.edu.
Martin Susani, Email: martin.susani@me.com.
Nathalie Rioux-Leclercq, Email: Nathalie.RIOUX-LECLERCQ@chu-rennes.fr.
Todd M. Morgan, Email: tomorgan@med.umich.edu.
Joanna Cyrta, Email: joc2071@med.cornell.edu.
Mark A. Rubin, Email: rubinma@med.cornell.edu.
References
- 1.Draisma G, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95(12):868–878. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 2.Andriole GL, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360(13):1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schröder FH, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366(11):981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuzick J, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106(6):1095–1099. doi: 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuzick J, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12(3):245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein EA, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66(3):550–560. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Erho N, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner AB, Patel SG, Etzioni R, Eggener SE. National trends in the management of low and intermediate risk prostate cancer in the United States. J Urol. 2015;193(1):95–102. doi: 10.1016/j.juro.2014.07.111. [DOI] [PubMed] [Google Scholar]
- 9.Clinton TN, Bagrodia A, Lotan Y, Margulis V, Raj GV, Woldu SL. Tissue-based biomarkers in prostate cancer. Expert Rev Precis Med Drug Dev. 2017;2(5):249–260. doi: 10.1080/23808993.2017.1372687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlinger M, Catto JW, Orntoft TF, Real FX, Zwarthoff EC, Swanton C. Intratumour heterogeneity in urologic cancers: from molecular evidence to clinical implications. Eur Urol. 2015;67(4):729–737. doi: 10.1016/j.eururo.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Meiers I, Waters DJ, Bostwick DG. Preoperative prediction of multifocal prostate cancer and application of focal therapy: review 2007. Urology. 2007;70(6 Suppl):3–8. doi: 10.1016/j.urology.2007.06.1129. [DOI] [PubMed] [Google Scholar]
- 13.Wise AM, Stamey TA, McNeal JE, Clayton JL. Morphologic and clinical significance of multifocal prostate cancers in radical prostatectomy specimens. Urology. 2002;60(2):264–269. doi: 10.1016/S0090-4295(02)01728-4. [DOI] [PubMed] [Google Scholar]
- 14.Rubin MA, Demichelis F. The Genomics of Prostate Cancer: emerging understanding with technologic advances. Mod Pathol. 2018;31(S1):S1–11. doi: 10.1038/modpathol.2017.166. [DOI] [PubMed] [Google Scholar]
- 15.Boutros PC, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47(7):736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 16.Van Allen EM, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20(6):682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoshida Y, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359(19):1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovelson DH, et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17(4):385–399. doi: 10.1016/j.neo.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grasso CS, et al. Integrative molecular profiling of routine clinical prostate cancer specimens. Ann Oncol. 2015;26(6):1110–1118. doi: 10.1093/annonc/mdv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knezevic D, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. doi: 10.1186/1471-2164-14-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei L, et al. Intratumoral and intertumoral genomic heterogeneity of multifocal localized prostate cancer impacts molecular classifications and genomic prognosticators. Eur Urol. 2017;71(2):183–192. doi: 10.1016/j.eururo.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chua MLK, et al. A prostate cancer “nimbosus”: genomic instability and SChLAP1 dysregulation underpin aggression of intraductal and cribriform subpathologies. Eur Urol. 2017;72(5):665–674. doi: 10.1016/j.eururo.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White NM, et al. Multi-institutional analysis shows that low PCAT-14 expression associates with poor outcomes in prostate cancer. Eur Urol. 2017;71(2):257–266. doi: 10.1016/j.eururo.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Ross HM, Kryvenko ON, Cowan JE, Simko JP, Wheeler TM, Epstein JI. Do adenocarcinomas of the prostate with Gleason score (GS) ≤6 have the potential to metastasize to lymph nodes? Am J Surg Pathol. 2012;36(9):1346–1352. doi: 10.1097/PAS.0b013e3182556dcd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrissy AS, et al. Spatial heterogeneity in medulloblastoma. Nat Genet. 2017;49(5):780–788. doi: 10.1038/ng.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamal-Hanjani M, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 29.Espiritu SMG, et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell. 2018;173(4):1003–1013.e15. doi: 10.1016/j.cell.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 30.Bishoff JT, et al. Prognostic utility of the cell cycle progression score generated from biopsy in men treated with prostatectomy. J Urol. 2014;192(2):409–414. doi: 10.1016/j.juro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Freedland SJ, et al. Prognostic utility of cell cycle progression score in men with prostate cancer after primary external beam radiation therapy. Int J Radiat Oncol Biol Phys. 2013;86(5):848–853. doi: 10.1016/j.ijrobp.2013.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooperberg MR, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol. 2013;31(11):1428–1434. doi: 10.1200/JCO.2012.46.4396. [DOI] [PubMed] [Google Scholar]
- 33.Haffner MC, De Marzo AM, Yegnasubramanian S, Epstein JI, Carter HB. Diagnostic challenges of clonal heterogeneity in prostate cancer. J Clin Oncol. 2015;33(7):e38–e40. doi: 10.1200/JCO.2013.50.3540. [DOI] [PubMed] [Google Scholar]
- 34.Radtke JP, et al. Transcriptome Wide Analysis of Magnetic Resonance Imaging-targeted Biopsy and Matching Surgical Specimens from High-risk Prostate Cancer Patients Treated with Radical Prostatectomy: The Target Must Be Hit. Eur Urol Focus. doi: 10.1016/j.euf.2017.01. https://doi.org/10.1016/j.euf.2017.01.005 [published online ahead of print January 28, 2017]. [DOI] [PubMed] [Google Scholar]
- 35.Trock BJ, et al. PTEN loss and chromosome 8 alterations in Gleason grade 3 prostate cancer cores predicts the presence of un-sampled grade 4 tumor: implications for active surveillance. Mod Pathol. 2016;29(7):764–771. doi: 10.1038/modpathol.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Epstein JI, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol. 2016;40(2):244–252. [DOI] [PubMed] [Google Scholar]
- 37.Sauter G, et al. Clinical utility of quantitative Gleason grading in prostate biopsies and prostatectomy specimens. Eur Urol. 2016;69(4):592–598. doi: 10.1016/j.eururo.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 38.Palapattu GS, et al. Molecular profiling to determine clonality of serial magnetic resonance imaging/ultrasound fusion biopsies from men on active surveillance for low-risk prostate cancer. Clin Cancer Res. 2017;23(4):985–991. doi: 10.1158/1078-0432.CCR-16-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cani AK, et al. Next-gen sequencing exposes frequent MED12 mutations and actionable therapeutic targets in phyllodes tumors. Mol Cancer Res. 2015;13(4):613–619. doi: 10.1158/1541-7786.MCR-14-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warrick JI, et al. Tumor evolution and progression in multifocal and paired non-invasive/invasive urothelial carcinoma. Virchows Arch. 2015;466(3):297–311. doi: 10.1007/s00428-014-1699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.