Abstract
Patients with severe, treatment-refractory asthma are at risk for death from acute exacerbations. The cytokine IL17A has been associated with airway inflammation in severe asthma, and novel therapeutic targets within this pathway are urgently needed. We recently showed that IL17A increases airway contractility by activating the procontractile GTPase RhoA. Here, we explore the therapeutic potential of targeting the RhoA pathway activated by IL17A by inhibiting RhoA guanine nucleotide exchange factors (RhoGEFs), intracellular activators of RhoA. We first used a ribosomal pulldown approach to profile mouse airway smooth muscle by qPCR and identified Arhgef12 as highly expressed among a panel of RhoGEFs. ARHGEF12 was also the most highly expressed RhoGEF in patients with asthma, as found by RNA sequencing. Tracheal rings from Arhgef12-KO mice and WT rings treated with a RhoGEF inhibitor had evidence of decreased contractility and RhoA activation in response to IL17A treatment. In a house dust mite model of allergic sensitization, Arhgef12-KO mice had decreased airway hyperresponsiveness without effects on airway inflammation. Taken together, our results show that Arhgef12 is necessary for IL17A-induced airway contractility and identify a therapeutic target for severe asthma.
Keywords: Muscle Biology, Pulmonology
Keywords: Asthma
The Rho guanine nucleotide exchange factor 12 (Arhgef12) is necessary for IL17A-induced airway contractility and could represent a therapeutic target for severe asthma.
Introduction
Ten percent of patients with asthma have severe disease (1, 2). These patients account for the majority of morbidity and mortality associated with asthma and are exacerbation-prone despite maximal therapy. Furthermore, despite recent advances in inflammatory profiling and targeted treatments for mild-to-moderate asthmatics, progress has been limited for patients with severe disease. Therefore, a major goal of current translational research is to identify therapeutic targets that are relevant to severe asthma (1, 3–5).
A core challenge of treating patients with severe asthma is their limited treatment response to corticosteroids. Th17 cells producing the cytokine IL17A have been implicated in steroid-refractory asthma by several preclinical and clinical studies (6–12). Furthermore, there is evidence that IL17A may drive disease activity most directly in patients with baseline airway hyperresponsiveness (AHR): in a 12-week clinical trial, anti-IL17A antibody treatment improved symptom scores only in the subgroup of patients who, in baseline pulmonary function testing, exhibited reversibility of airway obstruction with bronchodilator treatment (13). Since bronchodilator reversibility suggests airway contractility as a root cause of poor control in this subgroup, we reasoned that finding therapeutic targets downstream of IL17A, specifically in airway smooth muscle, should yield therapeutic targets relevant to this subgroup of severe asthmatics.
One such downstream target is the small GTPase RhoA. We recently showed that IL17A secreted by Th17 cells activates and increases airway contractility in mouse models via activation of the small GTPase RhoA (12). RhoA is a positive regulator of airway smooth muscle contraction via multiple pathways, including the effector Rho kinase, which augments myosin phosphorylation, and actin cytoskeletal effects that enable increased force transmission across the tissue (12, 14–17). Given these multiple procontractile effects of RhoA activation, we reasoned that RhoA guanine nucleotide exchange factors (RhoGEFs) (18, 19), activators of RhoA that catalyze GTP loading, should represent an upstream chokepoint for broad-based targeting of the contractile response to IL17A.
While vascular smooth muscle RhoGEF activity has been identified as a potential target for treatment of hypertension (20), the relative expression in airway smooth muscle of individual genes among the large family of RhoGEFs is unknown. Our approach was to employ a smooth muscle–specific ribosomal pulldown to profile airway smooth muscle RhoGEF expression. This screen led to the identification of Arhgef12, which we found was necessary for IL17A-mediated RhoA activation and airway contractility as well as allergic AHR in vivo. Taken together, our findings encourage the development of ARHGEF12 inhibition as a therapy we believe to be novel for severe asthma.
Results
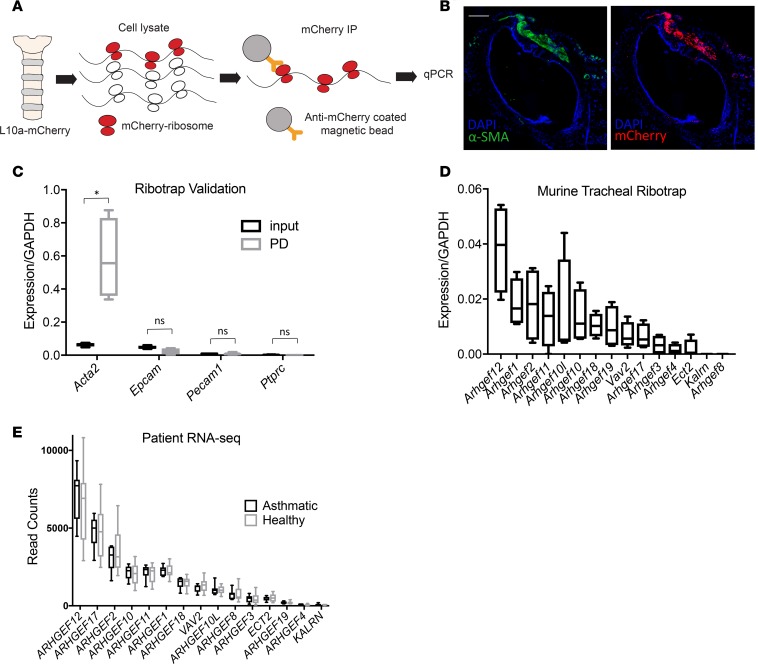
A major challenge to analysis of gene expression of airway smooth muscle is its poor suitability for flow cytometry–based sorting, given a lack of established cell surface markers. To overcome this limitation, we performed smooth muscle riboprofiling by pulldown of smooth muscle polysomes from fresh tracheal lysates using α–smooth muscle actin-CreERT2 (21) mice crossed with a ribotag allele, Rosa26-loxp-STOP-loxp-Rpl10a-mcherry (22). The mRNA thus isolated represents the actively translating mRNA in airway smooth muscle (Figure 1, A and B). Comparison by qPCR of expression of smooth muscle and non–smooth muscle genes before and after pulldown demonstrated the specificity of the pulldown (Figure 1C).
Figure 1. Riboprofiling identifies Arhgef12 as the most highly expressed RhoA guanine exchange factor in airway smooth muscle.
(A) Schematic of smooth muscle RNA riboprofiling of trachea from acta2-CreERT2/Rosa26loxpSTOPloxpRpl10a-mCherry mice. (B) Immunofluorescence of trachea from acta2-CreERT2/Rosa26loxpSTOPloxpRpl10a-mCherry mice. Scale bar: 200 μm. (C) Validation of smooth muscle specificity of riboprofiling by qPCR of smooth muscle and non–smooth muscle genes (n = 4 mice). PD, pulldown. (D) Riboprofiling of mouse tracheal RhoGEFs in healthy mice (n = 4 mice). (E) RhoGEF expression in airway smooth muscle from asthma patients (n = 7) and healthy controls (n = 12). Where 2 samples were available (4 of 7 asthma patients), counts were averaged. Box plots show minimum, maximum, and median, with 25th to 75th percentile range. *P < 0.01 by 1-tailed Student’s t test.
We then performed a qPCR screen of tracheal mRNA isolated by smooth muscle ribotag pulldown to determine highly expressed RhoGEFs in airway smooth muscle in healthy mice. To compile a list of candidate RhoGEFs, we searched gene identity and functional data for RhoGEFs in the Universal Protein Resource (https://www.uniprot.org/) and consulted published reviews, deriving a list of 15 RhoA-specific GEFs that have been studied across multiple tissue types (18, 19). SYBR qPCR for these targets identified Arhgef12 as the most highly expressed GEF in mouse tracheal smooth muscle by riboprofiling (Figure 1D). Of note, neither allergic inflammation nor IL17A increased Arhgef12 expression (Supplemental Figure 1). In order to measure relative ARHGEF12 expression in human airway smooth muscle, we collected surgical lung samples from asthmatics (Table 1) as well as healthy controls and isolated bronchial airway smooth muscle for RNA sequencing (RNA-seq). In both patients and controls, ARHGEF12 was again more highly expressed than any other detected RhoGEF from our panel (Figure 1E).
Table 1. Patients with a clinical diagnosis of asthma.
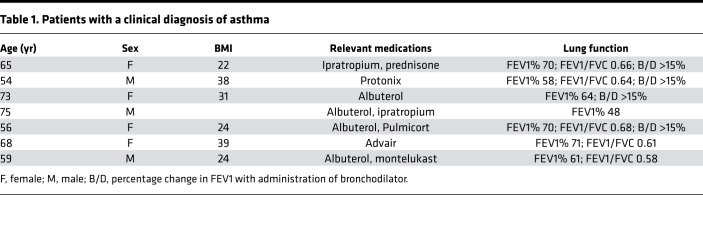
To explore the physiologic relevance of Arhgef12 to airway smooth muscle function, we measured the biochemical effects of IL17A in posterior trachealis muscle lysates from Arhgef12-KO mice. Absence of Arhgef12 or treatment of WT rings with the small-molecule RhoGEF inhibitor Y16 (23) decreased IL17A-induced airway smooth muscle myosin light-chain phosphorylation and RhoA activity in tracheal smooth muscle (Figure 2, A–C). Given the importance of RhoA activation to airway smooth muscle contraction downstream of IL17A, we then performed tracheal ring contraction assays using tracheal rings from Arhgef12-KO mice or WT rings treated with Y16. In both cases, the hypercontractility induced by IL17A was inhibited (Figure 2, D and E).
Figure 2. Arhgef12 is necessary for IL17A-induced RhoA activation and airway hypercontractility.
(A and B) Western blot for phosphorylated myosin light chain in Arhgef12–/– and WT mouse tracheal lysates and WT mouse tracheal lysates treated with or without Arhgef12 inhibitor Y16, with densitometry (n = 4 mice per condition; veh, DMSO; *P < 0.05, **P < 0.01 by 1-tailed Student’s t test; box plots show minimum, maximum, and median with 25th to 75th percentile range). (C) ELISA for Rhotekin-rho–binding domain affinity-captured active RhoA (n = 5 mice per condition; *P < 0.05 by 1-tailed Student’s t test; box plots show minimum, maximum, and median with 25th to 75th percentile range).). (D and E) Methacholine-induced (Mch-induced) contractile force of tracheal rings treated with IL17A (n = 4 rings per group; *P < 0.05, **P < 0.001, ***P < 0.0001 by Tukey’s multiple comparisons test following 2-way ANOVA for Il17A-treated comparing WT versus Arhgef12–/– or vehicle versus Y16; data are mean ± SEM).
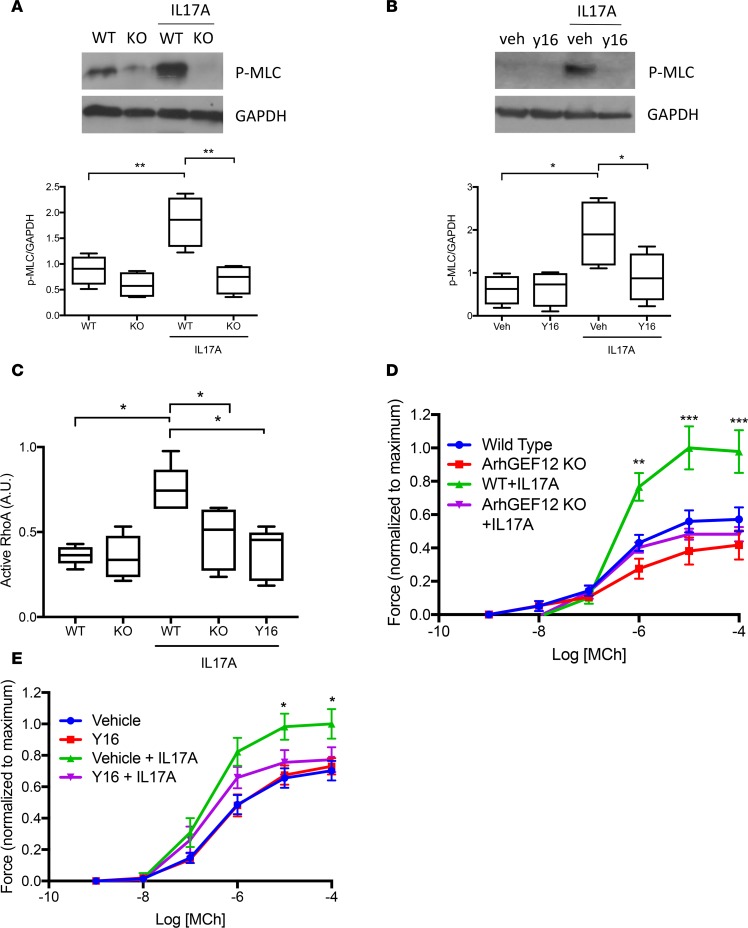
Having established Arhgef12’s role in IL17A-induced RhoA activation and hypercontractility, we next tested its pathologic relevance with an in vivo model of allergic AHR. We selected house dust mite extract as the allergic sensitizer for AHR testing, because AHR has been shown to be IL17A-dependent in this model (24). After airway sensitization with house dust mite, Arhgef12-KO mice had significantly decreased pulmonary resistance compared with WT mice. No difference was seen in WT or KO mice treated with saline (Figure 3A). Importantly, Arhgef12-KO mice had intact inflammatory responses in the face of suppressed AHR, supporting a smooth muscle–specific effect (Figure 3, B–D).
Figure 3. Arhgef12 is necessary for allergic airway hyperresponsiveness.
(A) Airway responsiveness to intravenous methacholine in Arhgef12–/– and WT mice following sensitization with house dust mite extract (n = 10 animals per group; mean ± SEM). (B–D) Serum IgE (n = 10 mice per condition; data are mean ± SEM), airway histologic inflammation scoring (n = 6 mice per condition; box plots show minimum, maximum, and median with 25th to 75th percentile range), and airway lavage cell counts (n = 10 mice per condition; box plots show minimum, maximum, and median with 25th to 75th percentile range and median). LC, lymphocytes. *P < 0.01, **P < 0.0001 for WT+HDM versus KO+HDM by Tukey multiple comparisons test following 2-way ANOVA.
Discussion
Severe asthma remains an unaddressed therapeutic need, and new molecular targets within pathways thought to contribute to severity are urgently needed. One such pathway is inflammation induced by IL17A, a cytokine that has been associated with treatment-refractory asthma. In this report, we extend our previous finding that IL17A activates the procontractile GTPase RhoA by testing the effect of Arhgef12 in airway contraction and hyperresponsiveness models (12). Taken together, our results provide proof of principle for a therapeutic strategy for severe asthma focused on RhoGEFs.
Starting with a qPCR screen, we identified Arhgef12 as much more highly expressed than other GEFs within murine airway smooth muscle. RNA-seq data from patients and healthy controls confirmed a high expression of ARHGEF12. Interestingly, we did not find an effect of allergic inflammation or IL17A on its expression, nor was ARHGEF12 expressed more highly in patients with asthma than healthy controls. However, it will be interesting in the future to determine whether ARHGEF12 is more highly expressed — and thus might explain refractory bronchospasm — in a larger, more severe cohort. We then explored the functional role of Arhgef12 with studies in Arhgef12-KO mice. Our results show that IL17A-induced RhoA activation and hypercontractility are Arhgef12 dependent. In vivo, Arhgef12-KO mice had decreased allergic AHR compared with WT mice, an effect that is attributable to blunted airway smooth muscle contractility, given that the absence of Arhgef12 did not have any effect on airway inflammation.
Recent studies have implicated GEFs in both airway inflammation and contractility (25, 26). This study adds to this growing literature by revealing Arhgef12 to be dominantly expressed among GEFs in the smooth muscle compartment and by showing its relevance to the IL17A pathway, a driver of severe asthma. The results for airway contractility with Y16 provide proof of concept for targeting Arhgef12, but given its inhibition of Arhgef1 and Arhgef11 in addition (23), future studies should develop more specific inhibitors to parse the relative benefit of focused inhibition of Arhgef12 versus broader GEF targeting. In the era of precision medicine for asthma, therapeutic development within the RhoA pathway holds promise for patients with severe, steroid refractory asthma in whom a Th17 inflammatory signature is predominant. Our data provide impetus for developing the targeting of Arhgef12 as a bronchodilator target for this urgent, unmet therapeutic indication.
Methods
Antibodies.
Rabbit anti-phospho-myosin light-chain (3671S) and GAPDH (2118S) antibodies were purchased from Cell Signaling Technology. Mouse anti–smooth muscle actin antibody was purchased from MilliporeSigma (clone 1A4). ARHGEF12 antibody was purchased from Abcam (ab136072).
Mice.
For ribotag pulldown experiments, L10a-mCherry mice, as previously described (22), were crossed with mice expressing α–smooth muscle actin promoter-driven CreERT2 (21) for tamoxifen-inducible L10a-mCherry in smooth muscle. To achieve recombination of floxed alleles in adult α-SMA-creERT2 lox-STOP-lox L10aRFP mice, tamoxifen dissolved in corn oil at a concentration of 20 mg/ml was administered via intraperitoneal injection at a dose of 2 mg every other day for 5 days. Mice deficient in Arhgef12 have been previously characterized (20), and WT littermates were used as controls. Adult WT 129S1 mice were purchased from The Jackson Laboratory. Mice used for all experiments were sex matched, 6–8 weeks old, and housed under specific pathogen–free conditions in the Animal Barrier Facility at University of California, San Francisco.
Isolation of translating mRNA.
We used tamoxifen-inducible α–smooth muscle actin-CreERT2 (21) mice crossed with an allele for expression of an mcherry-tagged ribosome, Rosa26-loxp-STOP-loxp-L10a-mcherry (22), followed by mcherry affinity purification for pulldown of cell-type-specific, actively translating mRNA, as previously described (27). In brief, whole trachea was excised, homogenized in 1 ml supplemented homogenization buffer (10 mM HEPES, pH 7.4, 150 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 100 μg/ml cycloheximide, protease inhibitors, RNAsin, calyculin), and centrifuged at 16,000 g for 4 minutes. Supernatants were incubated at 4°C for 1 hour with anti-RFP magnetic beads (MBL anti-RFP mAb) and then washed with a high-salt buffer (10 mM HEPES, pH 7.4, 350 mM KCl, 5 mM MgCl2, 1% NP40, 0.5 mM DTT, 100 μg/ml cycloheximide, RNAsin, calyculin).
Immunofluorescence.
Tracheas from tamoxifen-induced RiboTRAP mice were excised and embedded in OCT. 10-μM sections were prepared using Leica Cryostat CM1850. The sections were fixed with 1% paraformaldehyde, permeabilized with 0.3% Triton X-100, blocked with 10% serum, and incubated with primary antibody overnight. This was followed by PBS washes, incubation with secondary antibody for 1 hour, and PBS washes. Sections were then mounted in mounting medium containing DAPI and visualized with a Zeiss LSM 780 NLO microscope.
Quantitative PCR.
Input or TRAP RNA was purified using RNeasy Kit (Qiagen) with on-column DNaseI digestion. iScript reverse transcription super mix (Bio-Rad) was used for first-strand synthesis, followed by qPCR using SYBR Green super mix (Thermo Fisher Scientific). Gene expression was normalized to Gapdh or ribosomal 16S RNA. To validate isolation of smooth muscle–specific RNA, we interrogated our TRAP RNA for smooth muscle–specific gene Acta2 and non–smooth muscle–specific genes Epcam, Pecam1, and Ptprc (CD45). Table 2 lists primer sequences used for qRT-PCR.
Table 2. Primer sequences used for qRT-PCR.
Measurement of tracheal smooth muscle contractility ex vivo.
Mouse tracheal ring contraction studies were performed in a muscle bath system as described previously (12). Briefly, after incubation with or without 100 ng/ml IL17A for 16 hours at 37°C with 5% CO2, tracheal rings from WT or Arhgef12–/– mice were treated with increasing doses of methacholine (MilliporeSigma) for measurement of contractile force. In some case, rings were also incubated with the Arhgef12 inhibitor Y16 at 15 μM Y16 (MilliporeSigma) or vehicle control (DMSO) for 16 hours at 37°C with 5% CO2.
Allergic asthma model.
Allergic asthma was induced in mice using a total house dust mite (Greer) extract as previously described (24). Mice were sensitized by intranasal application of 150 μg house dust mite in 30 μl saline once per week for 4 weeks. Control mice were sensitized with saline.
Airways responsiveness measurement in vivo.
Twenty-two hours after the last challenge, pulmonary resistance was measured in response to a range of concentrations of intravenous acetylcholine using the forced oscillation technique with the FlexiVent system (SCRIREQ) as previously described (28).
Determination of HDM-specific IgE.
Serum samples were analyzed for house dust mite-specific IgE by ELISA. Wells were coated with HDM, and a biotinylated IgE antibody (BD Biosciences, clone R35-72) was used to analyze blood samples for IgE.
Assessment of pulmonary inflammation.
Total and differential cell percentages were determined by hemocytometer and by light microscopic evaluation of more than 300 cells per slide as previously described (28). After lavage, lungs were inflated with 10% buffered formalin to 25-cm H2O and transferred to 10% buffered formalin. 5-μm sections were stained with H&E for semiquantitative assessment of inflammation. To quantify inflammation, H&E-stained lung sections were deidentified for blinding and scored for peribronchial and perivascular inflammatory cell infiltration: grade 0, no infiltration; grade 1, <25% of examined area; grade 2, 25%–50%; grade 3, 51%–75%; and grade 4, >75%.
Immunoblots.
Smooth muscle dissected from mouse trachea and treated overnight with murine IL17A (Peprotech) or early passage human airway smooth muscle cells (Lonza) treated overnight with human IL17A were homogenized in lysis buffer (MilliporeSigma RIPA buffer) with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Lysates were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (MilliporeSigma). Membranes were blocked for 1 hour with 5% BSA in Tris-buffered saline with Tween-20, incubated at 4°C overnight or for 1 hour with primary antibodies, washed in Tris-buffered saline with Tween-20, incubated for 1 hour with peroxidase-conjugated secondary antibody, washed in Tris-buffered saline with Tween-20, and developed with ECL plus (Perkin-Elmer) prior to exposure to film (Denville Scientific).
Measurement of RhoA activation.
Affinity capture of active RhoA (RhoA-GTP) was performed by RhoA G-LISA (Cytoskeleton) according to the manufacturer’s instructions. Briefly, smooth muscle dissected from posterior mouse trachea was homogenized in supplied lysis buffer. After measurement of protein concentration, equal quantities were loaded in supplied RhoA-GTP affinity plates (Cytoskeleton) for 30 minutes. After wash, captured active RhoA was detected by ELISA with supplied anti-RhoA antibody (Cytoskeleton) for colorimetric detection at 490 nm.
Patient airway smooth muscle cells.
Airway smooth muscle cells were isolated from specimens obtained at surgery at the Mayo Clinic. Indications for surgery included focal, noninfectious causes, such as focal cancers. Based on history and chart review, patients were identified as asthmatic (medications for asthma and lung function are listed at Table 1) or nonasthmatic, age-matched controls. Third- to sixth-generation bronchi from normal lung regions were isolated and denuded of epithelium, and the remaining airway smooth muscle tissue was enzymatically dissociated to generate cells used for experiments. First- or second- cultures were maintained under standard conditions of 37°C (5% CO2, 95% air) and serum starved for 24 hours prior to experimentation.
RNA-seq of human airway samples.
RNA libraries were prepared using 200 ng total RNA per manufacturer’s instructions for the TruSeq RNA Sample Prep Kit (v2, Illumina). Concentration and size distribution of completed libraries was determined using an Agilent Bioanalyzer DNA 1000 chip and Qubit fluorometry (Invitrogen). Libraries were sequenced at 8 samples per lane to generate approximately 30–45 million reads per sample following standard protocols using the Illumina cBot and cBot Paired end cluster kit (version 3). Flow cells were sequenced as 51 paired-end reads on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit (version 3) and HCS (v2.0.12) data collection software. Base calling was performed using Illumina’s RTA version 1.17.21.3.
Allergen exposure and laser capture microdissection.
Mice were intranasally challenged with 10 mg Alternaria alternata (Greer), 10 mg Aspergillus fumigatus (Greer), 10 mg house dust mite (Greer), and 10 mg ovalbumin (MilliporeSigma) 3 times per week for 4 weeks. Control mice were given sterile PBS 3 times per week for 4 weeks. Twenty-four hours after the last challenge, mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (20 mg/kg). Under RNase-free conditions, lungs were harvested, frozen in liquid nitrogen, and stored at –80°C. Frozen lungs were cut into 10-μm sections and processed as previously described (29, 30). In brief, laser capture microdissection of airway smooth muscle was performed using an Arcturus XT microdissection system (Molecular Devices). Small airways (300- to 350-μm diameter) were visualized by light microscopy (×200 magnification). Airway epithelial and smooth muscle layers were identified. Using an infrared laser, layers were microdissected and captured onto CapSure Macro LCM caps. Total airway smooth muscle RNA was isolated from caps using the ARCTURUS PicoPure RNA Isolation Kit (Applied Biosystems). cDNA for qRT-PCR was synthesized (Roche LightCycler 96 System).
Data availability.
RNA-seq data for the human airway smooth muscle samples have been deposited at the NCBI’s Gene Expression Omnibus database (GEO GSE119580).
Statistics.
Data were evaluated by 2-way ANOVA with post hoc pairwise comparison Tukey’s test and 1-tailed Student’s t test. Differences with a P value less than or equal to 0.05 were considered statistically significant.
Study approval.
For patients studies, written informed consent was obtained from all subjects, and the study was approved by the Mayo Clinic Institutional Review Board. For animal studies, all experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
Author contributions
VF and AH equally conducted riboprofiling, airway contractility, and biochemical experiments; analyzed data; and contributed toward writing the manuscript. EW performed biochemical experiments. APL performed experiments involving fluorescence microscopy. PG performed riboprofiling. XR and DS performed AHR experiments. SAW, MAT, RDB, and YSP performed RNA-seq analysis of patient airway smooth muscle samples. MB designed the research studies, oversaw all experiments, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by a National Heart, Lung, And Blood Institute award (U54HL119893) and NIH/National Center for Advancing Translational Sciences–University of California, San Francisco–Clinical and Translational Science Institute grant (UL1 TR001872-01) to MB; NIH grant R01 HL088029 to YSP, and NIH grant HL131560 to MB. The authors thank Pierre Chambon (Strasbourg, France) for sharing α-SMACreERT2 mice.
Version 1. 11/02/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: JCI Insight. 2018;3(21):e123578. https://doi.org/10.1172/jci.insight.123578.
Contributor Information
Valerie Fong, Email: valfong8@gmail.com.
Austin Hsu, Email: austin.hsu@ucsf.edu.
Esther Wu, Email: esther.wu@ucsf.edu.
Previn Ganesan, Email: previn.ganesan@ucsf.edu.
Xin Ren, Email: Xin.Ren@ucsf.edu.
Dean Sheppard, Email: dean.sheppard@ucsf.edu.
Sarah A. Wicher, Email: Wicher.Sarah@mayo.edu.
Y.S. Prakash, Email: prakash.ys@mayo.edu.
Mallar Bhattacharya, Email: mallar.bhattacharya@ucsf.edu.
References
- 1.Wenzel S. Severe asthma in adults. Am J Respir Crit Care Med. 2005;172(2):149–160. doi: 10.1164/rccm.200409-1181PP. [DOI] [PubMed] [Google Scholar]
- 2.Fahy JV. Type 2 inflammation in asthma--present in most, absent in many. Nat Rev Immunol. 2015;15(1):57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wenzel SE, Busse WW, National Heart, Lung, Blood Institute’s Severe Asthma Research Program Severe asthma: lessons from the Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(1):14–21. doi: 10.1016/j.jaci.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel S. Severe asthma: from characteristics to phenotypes to endotypes. Clin Exp Allergy. 2012;42(5):650–658. doi: 10.1111/j.1365-2222.2011.03929.x. [DOI] [PubMed] [Google Scholar]
- 5.Fajt ML, Wenzel SE. Development of new therapies for severe asthma. Allergy Asthma Immunol Res. 2017;9(1):3–14. doi: 10.4168/aair.2017.9.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullens DM, et al. IL-17 mRNA in sputum of asthmatic patients: linking T cell driven inflammation and granulocytic influx? Respir Res. 2006;7:135. doi: 10.1186/1465-9921-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers ES, et al. Distinct endotypes of steroid-resistant asthma characterized by IL-17A(high) and IFN-γ(high) immunophenotypes: Potential benefits of calcitriol. J Allergy Clin Immunol. 2015;136(3):628–637.e4. doi: 10.1016/j.jaci.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKinley L, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008;181(6):4089–4097. doi: 10.4049/jimmunol.181.6.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang YH, et al. A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J Exp Med. 2010;207(11):2479–2491. doi: 10.1084/jem.20101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogli LK, et al. T cell-derived IL-17 mediates epithelial changes in the airway and drives pulmonary neutrophilia. J Immunol. 2013;191(6):3100–3111. doi: 10.4049/jimmunol.1301360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Lloyd CM, Noble A. Th17 responses in chronic allergic airway inflammation abrogate regulatory T-cell-mediated tolerance and contribute to airway remodeling. Mucosal Immunol. 2013;6(2):335–346. doi: 10.1038/mi.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudo M, et al. IL-17A produced by αβ T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat Med. 2012;18(4):547–554. doi: 10.1038/nm.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busse WW, et al. Randomized, double-blind, placebo-controlled study of brodalumab, a human anti-IL-17 receptor monoclonal antibody, in moderate to severe asthma. Am J Respir Crit Care Med. 2013;188(11):1294–1302. doi: 10.1164/rccm.201212-2318OC. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Huang Y, Gunst SJ. The small GTPase RhoA regulates the contraction of smooth muscle tissues by catalyzing the assembly of cytoskeletal signaling complexes at membrane adhesion sites. J Biol Chem. 2012;287(41):33996–34008. doi: 10.1074/jbc.M112.369603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudo M, et al. Mfge8 suppresses airway hyperresponsiveness in asthma by regulating smooth muscle contraction. Proc Natl Acad Sci USA. 2013;110(2):660–665. doi: 10.1073/pnas.1216673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter I, Cobban HJ, Vandenabeele P, MacEwan DJ, Nixon GF. Tumor necrosis factor-alpha-induced activation of RhoA in airway smooth muscle cells: role in the Ca2+ sensitization of myosin light chain20 phosphorylation. Mol Pharmacol. 2003;63(3):714–721. doi: 10.1124/mol.63.3.714. [DOI] [PubMed] [Google Scholar]
- 17.Chiba Y, Matsusue K, Misawa M. RhoA, a possible target for treatment of airway hyperresponsiveness in bronchial asthma. J Pharmacol Sci. 2010;114(3):239–247. doi: 10.1254/jphs.10R03CR. [DOI] [PubMed] [Google Scholar]
- 18.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 19.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33(31):4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirth A, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14(1):64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 21.Wendling O, Bornert JM, Chambon P, Metzger D. Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis. 2009;47(1):14–18. doi: 10.1002/dvg.20448. [DOI] [PubMed] [Google Scholar]
- 22.Hupe M, Li MX, Gertow Gillner K, Adams RH, Stenman JM. Evaluation of TRAP-sequencing technology with a versatile conditional mouse model. Nucleic Acids Res. 2014;42(2):e14. doi: 10.1093/nar/gkt995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang X, et al. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc Natl Acad Sci USA. 2013;110(8):3155–3160. doi: 10.1073/pnas.1212324110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chesné J, et al. Prime role of IL-17A in neutrophilia and airway smooth muscle contraction in a house dust mite-induced allergic asthma model. J Allergy Clin Immunol. 2015;135(6):1643–1643.e3. doi: 10.1016/j.jaci.2014.12.1872. [DOI] [PubMed] [Google Scholar]
- 25.Brown JP, et al. Arhgef1 is required by T cells for the development of airway hyperreactivity and inflammation. Am J Respir Crit Care Med. 2007;176(1):10–19. doi: 10.1164/rccm.200702-270OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaifta Y, et al. Transforming growth factor-β enhances Rho-kinase activity and contraction in airway smooth muscle via the nucleotide exchange factor ARHGEF1. J Physiol (Lond) 2018;596(1):47–66. doi: 10.1113/JP275033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106(33):13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Huang X, Sheppard D. ADAM33 is not essential for growth and development and does not modulate allergic asthma in mice. Mol Cell Biol. 2006;26(18):6950–6956. doi: 10.1128/MCB.00646-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faksh A, et al. Effects of antenatal lipopolysaccharide and postnatal hyperoxia on airway reactivity and remodeling in a neonatal mouse model. Pediatr Res. 2016;79(3):391–400. doi: 10.1038/pr.2015.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravamudan B, et al. Caveolin-1 knockout mice exhibit airway hyperreactivity. Am J Physiol Lung Cell Mol Physiol. 2012;303(8):L669–L681. doi: 10.1152/ajplung.00018.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data for the human airway smooth muscle samples have been deposited at the NCBI’s Gene Expression Omnibus database (GEO GSE119580).