Abstract
Virulence-associated genes have been recognised and detected in Campylobacter species. The majority of them have been proven to be associated with pathogenicity. This study aimed to detect the presence of virulence genes associated with pathogenicity and responsible for invasion, expression of adherence, colonisation and production of the cytolethal distending toxin (cdt) in Campylobacter jejuni and Campylobacter coli. Commercial chicken faecal samples were randomly sampled from chicken farms within the Durban metropolitan area in South Africa. Furthermore, human clinical Campylobacter spp. isolates were randomly sampled from a private pathology laboratory in South Africa. Out of a total of 100 chicken faecal samples, 78% (n = 78) were positive for Campylobacter growth on modified charcoal cefoperazone deoxycholate and from the random laboratory collection of 100 human clinical isolates, 83% (n = 83) demonstrated positive Campylobacter spp. growth following culturing methods. These samples were screened for the presence of the following virulence genes: cadF, hipO, asp, ciaB, dnaJ, pldA, cdtA, cdtB and cdtC. As expected, the cadF gene was present in 100% of poultry (n = 78) and human clinical isolates (n = 83). Campylobacter jejuni was the main species detected in both poultry and human clinical isolates, whilst C. coli were detected at a significantly lower percentage (p < 0.05). Eight per cent of the C. jejuni from human clinical isolates had all virulence genes that were investigated. Only one C. coli isolate demonstrated the presence of all the virulence genes investigated; however, the pldA virulence gene was detected in 100% of the C. coli isolates in poultry and a high percentage (71%) in human clinical C. coli isolates as well. The detection of cdt genes was found at higher frequency in poultry than human clinical isolates. The high prevalence rates of virulence genes detected in poultry and human clinical isolates demonstrate their significance in the pathogenicity of Campylobacter species.
Introduction
Campylobacteriosis is a significant public health concern worldwide with most infections in humans caused by Campylobacter jejuni subsp. jejuni (from here on referred to as C. jejuni) and Campylobacter coli (Thakur et al. 2010). Sources of infection of Campylobacter spp. in humans include house flies, migratory birds, wildlife, companion animals, livestock, milk, water and other environmental sources (Epps et al. 2013). However, poultry and retail meat products have extensively been reported and implicated as the major sources of infection in human cases of campylobacteriosis (Komba et al. 2013). Campylobacter jejuni and C. coli have for a long time been the focus of studies within the genus compared to other Campylobacter species (Iraola et al. 2014). Research over the years has demonstrated that C. jejuni is the primary cause of approximately 80% – 95% of Campylobacter infections in human cases and the remaining cause is usually because of C. coli (Ragimbeau et al. 2014).
A range of gastrointestinal conditions such as colorectal cancer and inflammatory bowel disease are caused by Campylobacter infections. Guillain–Barré syndrome is a rare but serious consequence of Campylobacter infections and occurs in 1 out of 1000 cases. Guillain–Barré syndrome is an immune-mediated polyneuropathy of the peripheral nervous system which can result in neuromuscular paralysis (Van den Berg et al. 2014) and Barrett’s oesophagus in humans has also been associated with infection by Campylobacter species. Studies have also reported Campylobacter spp. in cases of extra-gastrointestinal manifestations such as reactive arthritis, bacteraemia, brain abscesses and lung infections (Broaders, Gahan & Marchesi 2013; Iraola et al. 2014; Nielsen et al. 2010; Swartz 2002). However, the specific role of Campylobacter species in the development of all these clinical manifestations remains unclear and further investigation is therefore required (Kaakoush et al. 2015). The epidemiology of Campylobacter is complex because of the wide distribution of the bacterium, its genetic variability and its interaction with the host (Khoshbakht et al. 2013). These factors regarding Campylobacter spp. have made studying its pathogenicity particularly challenging (Caro-Quintero, Rodriguez-Castaño & Konstantinidis 2009).
Campylobacteriosis is mainly confined to the very young, usually children under the age of 5 years or to elderly patients and patients with immunosuppression (Silva et al. 2011). This presents as a major threat, especially in South Africa because of the extremely high incidence of individuals who are infected with immune-compromising conditions such as HIV and AIDS (Kaakoush et al. 2015), which makes these hosts more susceptible to infection by numerous pathogens including Campylobacter spp. Patients infected with C. jejuni or C. coli or both experience severe watery or bloody diarrhoea, extremely high fevers, major weight loss and severe abdominal cramping which lasts on average for about 6 days. Symptoms usually begin in the period within 24 and 72 h after ingestion of the bacterium depending on the dosage of the organism present in the contaminated food or liquid ingested (Zaidi et al. 2012). Avian species are important reservoirs for the transmission of Campylobacter species and their high body temperature provides an optimal environment for the growth of the organism (Noormohamed & Fakhr 2014).
Campylobacter are able to colonise the caecum of chicken in extremely high numbers of up to 109 CFU/g of faecal matter, even though this pathogen is present in such high quantities the chicken rarely exhibits symptoms of disease (Thibodeau et al. 2015). The illness in humans can last for a few days, depending on the individual, and the abdominal pain is usually mistaken for appendicitis. Understanding the molecular basis of the virulence-associated genes with disease and knowledge of the genetic diversity amongst the different strains of Campylobacter is of paramount importance in the control of diseases and syndromes associated with this organism (Fonseca et al. 2014). Studies have also demonstrated that infections caused by C. jejuni and C. coli are more common during the summer months of the year (Chansiripornchai & Sasipreeyajan 2009; Fonseca et al. 2014; Garénaux et al. 2008; Hermans et al. 2011). Although C. coli is less prevalent than C. jejuni in many geographical regions, C. coli infections are responsible for as many as 25% of all gastroenteritis clinical cases caused by Campylobacter species (Kaakoush et al. 2015).
Gastroenteritis induced by C. coli is clinically impossible to differentiate from that by C. jejuni because of these organisms being fastidious in nature and their need for nutrient rich-based medium as well as anaerobic conditions for growth. This is the main reason why Campylobacter spp. are not applied in routine diagnostic programmes of clinical laboratories in most developing countries (Ghorbanalizadgan et al. 2014). The majority of Campylobacter infections are sporadic, and unlike for other food-borne pathogens, huge outbreaks are not very typical. Nevertheless, it is likely that outbreaks or small-case clusters occur far more frequently than previously suggested because of cases of Campylobacter infection going unreported and as such the true epidemiological statistics are under-reported (Taboada et al. 2013).
Successful invasion and organisation in host cells depends on the adhesin and fibronectin-binding protein which is the product of the cadF gene that plays a significant role during the infection process of Campylobacter binding to the extracellular matrix of human intestinal cells (Khoshbakht et al. 2013). The presence of the ciaB gene plays a very significant role because of the secretion of a CiaB 73 kDa protein which is important for the invasion of epithelial cells as well as colonisation of intestines of avian species, and CiaB proteins are also secreted in the presence of poultry serum and mucus (Biswas et al. 2011). The pldA gene is also related to cell invasion and is responsible for the synthesis of an outer membrane phospholipase that is important for caecal colonisation (O Cróinín & Backert 2012).
The dnaJ virulence gene enables Campylobacter species to cope with diverse physiological stresses and is also considered to be a chaperone protein (Chansiripornchai & Sasipreeyajan 2009); the ciaB, pldA and dnaJ that are recognised as heat shock protein genes are important for caecal colonisation and mutations in these genes limit the ability to achieve this (Jakee et al. 2015). The cytolethal distending toxin (cdt) genes, cdtA, cdtB and cdtC, form polycistronic cdt operons that are responsible for the expression of cytotoxins and lethal for host enterocytes (Carvalho et al. 2014). Against this background, this study aimed to isolate and identify C. jejuni and C. coli from chicken faeces as well and human clinical isolates as well as to determine the prevalence of virulence genes related to adherence, invasion and cytotoxicity.
Materials and methods
Ethical considerations
Human and animal studies have been approved by the appropriate ethics committee of the University of KwaZulu-Natal; therefore, they have been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Origin of samples and processing procedures
One hundred human Campylobacter isolates cryopreserved in Brucella broth (Oxoid) with 15% glycerol were randomly sampled from a collection that was received from a private laboratory in Durban, KwaZulu-Natal, during the year 2014 as described by Reddy and Zishiri (2017). In summary, the cultures were revived on modified charcoal cefoperazone deoxycholate (mCCDA) agar (Oxoid, England) containing Campylobacter-selective supplement SR0155 (Oxoid, England). A sterile loop was then streaked across the area of inoculation several times to achieve isolated colonies and plates were incubated at 37 °C for 48 h under microaerobic conditions created by CampyGen (Oxoid, UK) gas generating packs in an anaerobic jar.
One hundred chicken faecal samples were randomly sampled between March and September 2016 from commercial farms within the Durban metropolitan area as described by Reddy and Zishiri (2017). Fresh broiler faeces were aseptically collected with sterile swabs then directly inoculated into charcoal broth (Sigma-Aldrich, USA) and then transported to the laboratory for incubation at 37 °C for 48 h, under microaerophilic conditions (5% O2, 10% CO2 and 85% N2) created by CampyGen (Oxoid, UK) gas generating packs in an anaerobic jar.
Following incubation, the faecal samples in charcoal broth (Sigma-Aldrich) were filtered through a 0.65-μm cellulose nitrate filter (Sartorius Stedim Biotech, Germany) onto mCCDA (blood-free agar) (Oxoid, England). Approximately 500 μL of the incubated charcoal broth was evenly distributed over the filter aseptically; once the liquid had been filtered through, forceps were used to aseptically remove the filter. The culture plates were then set in an inverted position in an anaerobic jar containing an atmosphere generation system (CampyGen sachet, Oxoid) and then incubated at 37 °C for 48 h. Following incubation, species identity was confirmed, after Deoxyribonucleic acid (DNA) isolation, by polymerase chain reaction (PCR) targeting of the hipO gene specific for C. jejuni (Marinou et al. 2012) and the asp gene specific for C. coli (Al Amri et al. 2013).
Template DNA for PCR was extracted via the conventional boiling method. In summary, characteristic colonies of Campylobacter spp. were isolated from plates and suspended in 300 μL TE buffer then vortexed for homogenisation of cells. The suspensions were boiled at 100 °C for 10 min and then immediately cooled on ice. After centrifugation at 14 000 × g for 5 min, the supernatants were transferred to a new tube and stored at -20 °C until use in PCR (Datta et al. 2003). A positive Campylobacter spp. control was also prepared by isolating DNA from a reference strain of C. jejuni ATCC 29428 that was incubated under the same conditions and subjected to the same DNA extraction methods. The Thermo Scientific Nanodrop 2000, UV-Vis spectrophotometer (Wilmington, Delaware, USA) was used to check the concentration and quality of the isolated DNA (Reddy & Zishiri 2017). Following analysis of the extracted DNA, results within the range of 1.8–1.9 at the ratio of 260/280 were regarded as pure DNA and used in PCR. Concentrations of the DNA were also adjusted accordingly with sterile water for subsequent PCR reactions.
Detection of virulence genes using polymerase chain reaction
The DNA from samples demonstrating positive Campylobacter growth was amplified using PCR. In order to differentiate between the species responsible for infection in human clinical cases as well as in poultry, two species-specific genes were used. The hipO gene region is the hippuricase gene specific for C. jejuni (Marinou et al. 2012) and the asp gene region, the aspartokinase gene specific for C. coli (Al Amri et al. 2013). The PCR was used to detect nine virulence genes in the total DNA of Campylobacter isolates: cadF, hipO, asp, dnaJ, ciaB, pldA, cdtA, cdtB and cdtC. Polymerase chain reaction primers were sourced from Inqaba Biotechnologies, South Africa. Forward and reverse primers specific for the virulence genes under investigation were designed based on the gene sequence information in the GenBank database and in previously published studies (Al Amri et al. 2007; Chansiripornchai & Sasipreeyajan 2009; Rizal, Kumar & Vidyarthi 2010). Target genes, primer sequences, product sizes and annealing temperatures are presented in Table 1.
TABLE 1.
Target virulence genes, primer sequences, amplicon sizes and annealing temperatures.
| Target gene | Primer sequence (5’–3’) | Product size (bp) | Annealing temperature (°C) | References |
|---|---|---|---|---|
| cadF | F-TTGAAGGTAATTTAGATATG R-CTAATACCTAAAGTTGAAAC |
400 | 43 | Chansiripornchai and Sasipreeyajan (2009) |
| asp | F-GGTATGATTTCTACAAAGCGAGA R-ATAAAAGACTATCGTCGCGTG |
500 | 53 | Al Amri et al. (2007) |
| hipO | F-GAAGAGGGTTTGGGTGGT R-AGCTAGCTTCGCATAATAACTTG |
735 | 53 | Al Amri et al. (2007) |
| ciaB | F-TGCGAGATTTTTCGAGAATG R-TGCCCGCCTTAGAACTTACA |
527 | 54 | Chansiripornchai and Sasipreeyajan (2009) |
| dnaJ | F-ATTGATTTTGCTGCGGGTAG R-ATCCGCAAAAGCTTCAAAAA |
177 | 50 | Chansiripornchai and Sasipreeyajan (2009) |
| pldA | F-AAGAGTGAGGCGAAATTCCA R-GCAAGATGGCAGGATTATCA |
385 | 46 | Chansiripornchai and Sasipreeyajan (2009) |
| cdtA | F-CCTTGTGATGCAAGCAATC R-ACACTCCATTTGCTTTCTG |
370 | 49 | Rizal et al. (2010) |
| cdtB | F-GTTAAAATCCCCTGCTATCAACCA R-GTTGGCACTTGGAATTTGCAAGGC |
495 | 51 | Rizal et al. (2010) |
| cdtC | F-CGATGAGTTAAAACAAAAAGATA R-TTGGCATTATAGAAAATACAGTT |
182 | 48 | Rizal et al. (2010) |
Note: Please see the full reference list of the article, Reddy, S. & Zishiri, O.T., 2018, ‘Genetic characterisation of virulence genes associated with adherence, invasion and cytotoxicity in Campylobacter spp. isolated from commercial chickens and human clinical cases’, Onderstepoort Journal of Veterinary Research 85(1), a1507. https://doi.org/10.4102/ojvr.v85i1.1507, for more information.
Polymerase chain reactions were carried out in the BIO-RAD, T100™ Thermal Cycler (Singapore) for a 25 μL reaction using ThermoScientific DreamTaq Green PCR Master Mix (2X). A total of 12.5 μL DreamTaq Green PCR Master Mix was used with 1.5 μL of each primer of a 10 μM primer concentration, 5 μL template DNA and 4.5 μL nuclease-free water making a total volume of 25 μL. The amplification conditions for cadF, hipO, asp, dnaJ, ciaB and pldA consisted of an initial denaturalisation at 95 °C for 3 min, 45 cycles at 94 °C for 30 s, specific Tm for each primer for 30 s, and 72 °C for 1 min, followed by a final extension at 72 °C for 5 min. The cdt genes (cdtA, cdtB and cdtC) were run using different amplification conditions according to Rizal et al. (2010). The conditions consisted of an initial denaturation at 94 °C for 15 min, 45 cycles at 94 °C for 1 min, specific Tm for each cdt primer for 1 min, and 72 °C for 1 min, followed by a final extension at 72 °C for 7 min. Polymerase chain reaction products were then electrophoresed on a 1.5% agarose gel run at 60 V for 60 min, stained with ethidium bromide and then visualised using the BIO-RAD, ChemiDoc™ MP Imaging System.
Statistical analyses
Virulence genes detected in C. jejuni and C. coli were analysed using IBM SPSS Statistics (version 23). Pearson’s correlation analyses were implemented in order to establish the strength and direction of relationships between the individual virulence-associated genes in an effort to ascertain whether the presence of one virulence gene was linked to the presence of the other. Fisher’s exact tests, chi-square tests and logistic regression analysis were implemented to test for significance of whether the presence of virulence genes detected using PCR was determined by whether the isolates emanated from either human clinical cases or chicken faecal samples. In each model that was fitted, the dependent variable was whether a virulence gene was either present or absent (0 = absent; 1 = present) and the explanatory variables were whether the isolates emanated from human clinical cases or chicken faecal samples. The statistics were considered significant when the probability value was less than 0.05 (p < 0.05).
Results
Prevalence of virulence genes
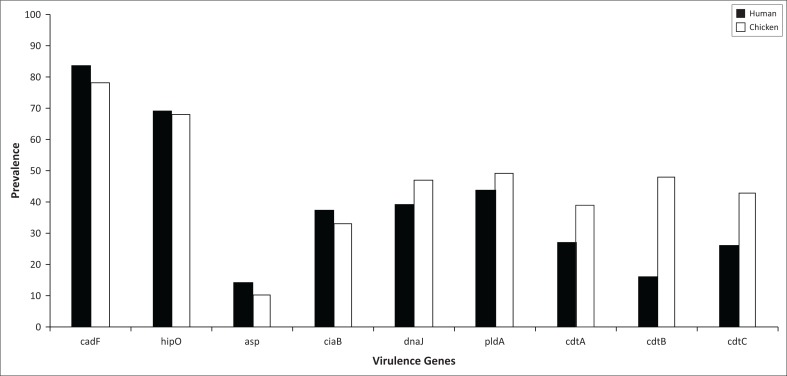
Of the 100 chicken faecal samples, 78% (n = 78) were presumptive for Campylobacter spp. growth on mCCDA and from the collection of a 100 human clinical isolates, 83% (n = 83) demonstrated positive Campylobacter spp. growth following culturing methods. Virulence genes investigated in this study are shown in Figure 1 and species differentiation was confirmed by detection of hipO (lane 1: 735 bp) and asp gene (lane 2: 500 bp). Campylobacter jejuni is the only species known to have the hippuricase (hipO) gene because it has not been detected in any other Campylobacter species. Furthermore, the aspartokinase (asp) gene is specific only in C. coli. All isolates (100%), irrespective of the species, were positive for the cadF gene based on PCR detection of a 400 bp amplicon (lane 3). This gene encodes Campylobacter species adhesion to fibronectin that is an important virulence factor for colonisation of epithelial cells. The dnaJ gene is depicted in lane 4 at 177 bp. Lane 5 depicts the pldA virulence gene with an amplicon of 385 bp. Lane 6 is the ciaB gene at 527 bp, lane 7 is the cdtA gene (370 bp), lane 8 is the cdtB gene (495 bp) that is a catalytic subunit in the CDT cluster and lastly lane 9 is the cdtC gene (182 bp), and cdtA and cdtC are binding proteins responsible for delivering cdtB into target cells. The number of isolates that were positive of each virulence gene is depicted in Figure 2.
FIGURE 1.
Representative gel picture of species identification and virulence genes investigated from Campylobacter spp.
FIGURE 2.
Prevalence of virulence genes in Campylobacter spp. in human clinical isolates and chicken faeces.
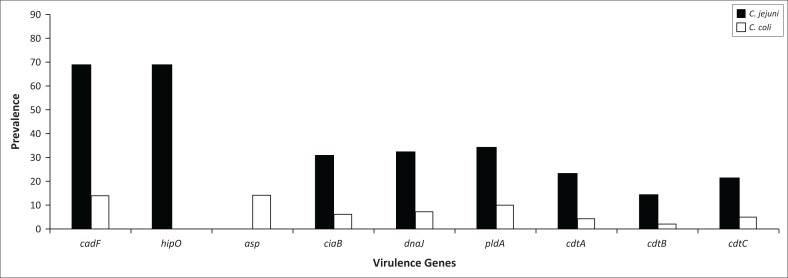
Results in Figure 3 indicate that C. jejuni is responsible for the majority of infection (83%; n = 69) in human gastroenteritis cases compared to the low detection of C. coli found only in 17% (n = 14) of cases. Of the C. jejuni isolated from human clinical cases, 45%, 46%, 49%, 33%, 20% and 30% were positive for ciaB, dnaJ, pldA, cdtA, cdtB and cdtC, respectively, and C. coli revealed 43%, 50%, 71%, 29%, 14% and 36%, respectively.
FIGURE 3.
Prevalence of virulence genes in Campylobacter jejuni and Campylobacter coli isolated from human clinical isolates.
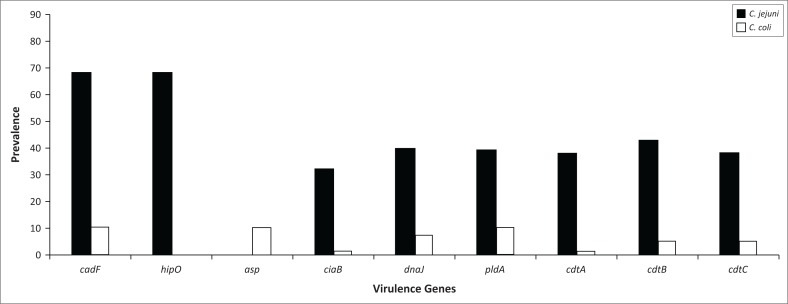
The isolates from chicken faeces (Figure 4) demonstrated a higher percentage of C. jejuni (87%) (n = 68) present as compared to only 13% (n = 10) of C. coli. The poultry isolates for C. jejuni demonstrated 47%, 59%, 57%, 56%, 63% and 56% for the presence of ciaB, dnaJ, pldA, cdtA, cdtB and cdtC, respectively, and C. coli isolates revealed 10%, 70%, 100%, 10%, 50% and 50% for the presence of ciaB, dnaJ, pldA, cdtA, cdtB and cdtC, respectively.
FIGURE 4.
Prevalence of virulence genes in Campylobacter jejuni and Campylobacter coli isolated from chicken faeces.
Statistical analyses
Pearson’s correlations (Table 2) demonstrated significant (p < 0.05) positive correlations between some virulence genes. The asp gene that identifies C. coli was the only gene which was not statistically significantly (p > 0.05) correlated with some of the virulence genes such as ciaB, dnaJ and all the cdt genes screened. Generally, most of the correlations amongst the virulence genes were statistically significant (p < 0.05), as depicted in Table 2. The presence of the dnaJ and cdt genes was positively correlated (p < 0.05). Furthermore, the presence of the hipO that confirms the presence of C. jejuni was positively correlated (p < 0.05) with the presence of the dnaJ gene and the cdt genes. There was also evidence of a positive correlation (p < 0.05) between the ciaB gene, dnaJ, pldA and cdt genes.
TABLE 2.
Comparison of Pearson’s correlations for virulence genes detected in Campylobacter species from human clinical isolates and chicken faeces.
| Genes | Statistical tests | cadF | hipO | asp | ciaB | dnaJ | pldA | cdtA | cdtB | cdtC |
|---|---|---|---|---|---|---|---|---|---|---|
| cadF | Pearson’s correlation | 1 | 0.630** | 0.141 | 0.286** | 0.342** | 0.369** | 0.272** | 0.266** | 0.282** |
| Sig. (2-tailed) | - | 0.000 | 0.058 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| hipO | Pearson’s correlation | 0.630** | 1 | −0.680** | 0.270** | 0.185* | 0.076 | 0.300** | 0.235** | 0.185* |
| Sig. (2-tailed) | 0.000 | - | 0.000 | 0.000 | 0.012 | 0.306 | 0.000 | 0.001 | 0.012 | |
| asp | Pearson’s correlation | 0.141 | −0.680** | 1 | −0.074 | 0.087 | 0.251** | −0.125 | −0.049 | 0.030 |
| Sig. (2-tailed) | 0.058 | 0.000 | - | 0.318 | 0.246 | 0.001 | 0.092 | 0.512 | 0.686 | |
| ciaB | Pearson’s correlation | 0.286** | 0.270** | −0.074 | 1 | 0.157* | 0.209** | 0.484** | 0.340** | 0.406** |
| Sig. (2-tailed) | 0.000 | 0.000 | 0.318 | - | 0.035 | 0.005 | 0.000 | 0.000 | 0.000 | |
| dnaJ | Pearson’s correlation | 0.342** | 0.185* | 0.087 | 0.157* | 1 | 0.442** | 0.339** | 0.340** | 0.327** |
| Sig. (2-tailed) | 0.000 | 0.012 | 0.246 | 0.035 | - | 0.000 | 0.000 | 0.000 | 0.000 | |
| pldA | Pearson’s correlation | 0.369** | 0.076 | 0.251** | 0.209** | 0.442** | 1 | 0.349** | 0.329** | 0.334** |
| Sig. (2-tailed) | 0.000 | 0.306 | 0.001 | 0.005 | 0.000 | - | 0.000 | 0.000 | 0.000 | |
| cdtA | Pearson’s correlation | 0.272** | 0.300** | −0.125 | 0.484** | 0.339** | 0.349** | 1 | 0.354** | 0.400** |
| Sig. (2-tailed) | 0.000 | 0.000 | 0.092 | 0.000 | 0.000 | 0.000 | - | 0.000 | 0.000 | |
| cdtB | Pearson’s correlation | 0.266** | 0.235** | −0.049 | 0.340** | 0.340** | 0.329** | 0.354** | 1 | 0.421** |
| Sig. (2-tailed) | 0.000 | 0.001 | 0.512 | 0.000 | 0.000 | 0.000 | 0.000 | - | 0.000 | |
| cdtC | Pearson’s correlation | 0.282** | 0.185* | 0.030 | 0.406** | 0.327** | 0.334** | 0.400** | 0.421** | 1 |
| Sig. (2-tailed) | 0.000 | 0.012 | 0.686 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | - |
Correlation is significant at the 0.05 level (2-tailed)
correlation is significant at the 0.01 level (2-tailed).
There were statistically significant (p < 0.05) relationships between the cadF, hipO, dnaJ, pldA, cdtA, cdtB and cdtC genes because of their presence in Campylobacter human clinical cases and chicken faeces investigated (Table 3). The virulence genes asp and ciaB were not statistically significant from the results observed because the results demonstrated a p > 0.05 for chi-square and Fisher’s exact statistical tests when compared to the other genes investigated.
TABLE 3.
Chi-square test and Fisher’s exact test for virulence genes investigated.
| Statistical tests | Asymptotic significance (2-sided) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| cadF | hipO | asp | ciaB | dnaJ | pldA | cdtA | cdtB | cdtC | |
| Pearson’s chi-square test | 0.011 | 0.030 | 0.720 | 0.654 | 0.014 | 0.034 | 0.004 | 0.000 | 0.000 |
| Fisher’s exact test | 0.011 | 0.038 | 0.827 | 0.760 | 0.017 | 0.038 | 0.005 | 0.000 | 0.000 |
Logistic regression analyses were conducted to predict the presence of virulence genes in chicken faeces and human clinical isolates using the source of the isolates as a predictor (Table 4). A test of the full model against a constant only model indicated that genes ciaB, dnaJ and pldA were not statistically significant (p > 0.05). The Wald criterion demonstrated that cdtA, cdtB and cdtC virulence genes made a significant contribution to prediction of the presence of these genes in human clinical isolates and poultry with p = 0.025, < 0.001 and 0.003, respectively (p < 0.05).
TABLE 4.
Logistic regression analysis results demonstrating the significance of virulence genes found in human clinical isolates and chicken faeces.
| Virulence genes | -2 Log likelihood | B | SE | Wald p-value | OR | 95% CI |
|---|---|---|---|---|---|---|
| ciaB | 220.362 | 0.092 | 0.318 | 0.771 | 1.097 | 0.588–2.047 |
| dnaJ | 219.587 | −0.537 | 0.319 | 0.093 | 0.585 | 0.313–1.093 |
| pldA | 217.706 | −0.404 | 0.321 | 0.209 | 0.668 | 0.356–1.253 |
| cdtA | 212.844 | −0.730 | 0.326 | 0.025 | 0.482 | 0.255–0.913 |
| cdtB | 185.315 | −1.902 | 0.363 | 0.000 | 0.149 | 0.073–0.304 |
| cdtC | 210.508 | −0.991 | 0.328 | 0.003 | 0.371 | 0.195–0.707 |
Discussion
Campylobacter jejuni and Campylobacter coli are the most prevalent Campylobacter spp. that are responsible for gastroenteritis infections in humans. The pathogenicity of Campylobacter spp. depends on the virulence factors that differ amongst strains of different origin. Several virulence-associated genes have been recognised and detected in Campylobacter, and the majority of them are associated with pathogenicity. The information regarding Campylobacter virulence genes and genotypes of Campylobacter spp. is unclear in developing countries such as South Africa; therefore, research is essential to characterise pathogenic markers in this organism and to, furthermore, initiate subsequent preparation of strategies for its proper control and prevention.
All isolates that were presumptively identified as Campylobacter, irrespective of the bacterial species, were positive for the presence of the cadF gene, which facilitates adherence to fibronectin in contact regions (Flanagan et al. 2009). These findings are in concordance with earlier studies with regard to the presence of the cadF gene in Campylobacter spp. isolated from human as well as chicken (Al Amri et al. 2007; Datta et al. 2003; Rozynek et al. 2005). The presence of gene products from cadF (Campylobacter adhesin to fibronectin) has clearly demonstrated involvement in Campylobacter colonisation and this has been shown by in vivo colonisation using a chicken model (Thibodeau et al. 2015). This gene encodes a protein that interacts with a host extracellular matrix protein fibronectin, and is required for Campylobacter adherence to and colonisation of the host cell surface (Ghorbanalizadgan et al. 2014). The high prevalence (100%) of the cadF gene in the present study demonstrates that many isolates originating from poultry faeces have pathogenic potential properties for humans. The ubiquitous existence of the highly conserved cadF gene in 100% of Campylobacter spp. was previously reported by Konkel et al. (1999) and was subsequently used by other investigators for successful detection of Campylobacter spp. (Rizal et al. 2010; Wieczorek, Szewczyk & Osek 2012).
The hippuricase gene (hipO) is specific for C. jejuni and is not detected in any other Campylobacter species. The Pearson’s correlation analyses demonstrated that there was a significant (p < 0.05) strong positive correlation (63%) between the cadF and the hipO genes. Our study utilised the positive PCR amplification of asp gene in order to identify which samples belonged to the species C. coli. Furthermore, the correlation analysis demonstrated that the presence of the asp gene was negatively correlated (-0.63%) to the presence of the hipO gene (p < 0.05). This is in agreement with other studies because C. coli is not as prevalent as C. jejuni as the cause of infection in humans (Feodoroff et al. 2011; Khoshbakht et al. 2013; Samie et al. 2007).
Many factors play a significant role in the varying isolation or recovery rates of Campylobacter species that include whether the samples are fresh or frozen, the type of sampling procedure used and the isolation protocol followed as well as the time of year for collection of samples. The ciaB (Campylobacter invasive antigen B) gene is known to be involved in the translocation of Campylobacter into host cells for the purpose of host cell invasion and also plays a significant role in caecal colonisation in chicken (O Cróinín & Backert 2012). The ciaB gene was detected in 45% and 42% in human and chicken samples, respectively, and had a significant low positive correlation (15.7%) with the dnaJ gene (p < 0.05). With regard to the dnaJ gene that enables Campylobacter species to cope with diverse physiological stresses, there was a significant positive correlation when there was presence of the hipO and ciaB gene found in samples (p < 0.05).
Another important factor for colonisation of Campylobacter species in the intestine of chickens is the pldA gene (an outer membrane phospholipase A) that encodes proteins associated with increased bacterial invasion on cultured epithelial cells (Ghorbanalizadgan et al. 2014). The distribution of this gene was dissimilar amongst the two species investigated as C. jejuni showed a higher presence of this gene compared to C. coli in both human and chicken isolates. The percentage of pldA in chicken and human isolates was 53% and 63%, respectively.
There is an increase ranging from 88% to 100% in the presence of the pldA gene in C. jejuni isolates from chicken with age of broilers as a major contributing factor (Datta, Niwa & Itoh 2009). The collection of samples recovered from broilers at the end of the breeding period increases the chances of poultry serovars having better colonisation and making them more virulent for their other hosts. The results from this study indicate that Campylobacter spp. recovered from chicken faeces may have potential virulence consequences in humans. The study also demonstrated that C. jejuni and C. coli in chicken faecal samples from the slaughterhouse are indicative of a public health hazard because of the spread of this emerging food-borne pathogen. The contamination of carcasses may occur from intestinal contents during slaughtering and or post-slaughtering processes and if these products are not cleaned, stored or cooked properly, this could lead to outbreaks of infection in the public.
Damage to nuclear DNA resulting in the inhibition of the cell cycle in G2 or M phase is the cytopathic effect of the cytotoxin released by cdtA, cdtB and cdtC (Carvalho et al. 2014). The low percentage of the cdtB gene in humans found in this study (19%) is in agreement with a study conducted in India (Rizal et al. 2010), which reported a prevalence of 28%. However, chickens also demonstrated a low percentage of this gene (20%) in contrast to this study which found 62% that could be because of genetic factors or variation in the isolates from different geographic areas. Translocation of cdtB to the nucleus of cells induces the genotoxic effects on host DNA; triggering DNA repair mechanisms that could lead to cell cycle arrest and eventually cause cell death (Ge et al. 2008). In addition, it has also been suggested that the CDT cluster could also play a role in adhesion and invasion of the pathogen in a host (Ge et al. 2008). Cell invasion could result in cellular injury, leading to reduced absorptive capacity of the intestine, whereas cdt production is important for interleukin-8 release by intestinal cells in vitro which plays an important role in the host mucosal inflammatory response caused by Campylobacter species (Hermans et al. 2011).
Cell invasion of epithelial cells and cdt production are important bacterial virulence mechanisms that play a significant role for inducing enterocolitis. The presence of a single cdt gene does not have any effect on the bacterium virulence; however, if all three cdt genes are present together in a cluster, then they are responsible for the release of a functional cytotoxin (Lapierre et al. 2016). Our study revealed that 10 of the human clinical isolates had all three cdt genes compared to 18 of the chicken isolates. The low percentage of cdt genes needs to be compared with cytolethal distending toxin production levels and tested on a variety of different cell lines in order to study the phenotypic characteristics of the isolates for a better understanding (Wieczorek, Szewczyk & Osek 2012).
Conclusion
The current study revealed that isolates of C. jejuni and C. coli from chicken faeces and human clinical cases possess a variety of different virulence genes associated with vital processes such as invasion, from the genes investigated in this study: cadF, ciaB and pldA play significant roles in successful invasion within a host. Expression of adherence in host cells depends on the adhesin and fibronectin-binding protein which is the product of the cadF gene and this gene was present at 100% in both chicken faecal samples (n = 78) and human clinical isolates (n = 83). Colonisation within a host also plays an important role for Campylobacter survival and ciaB, pldA and dnaJ are important for caecal colonisation in chickens; however, mutations in these genes can limit the ability to achieve this. The production of cytolethal distending toxins cdtA, cdtB and cdtC forms a polycistronic cdt operon that is responsible for the expression of cytotoxins, from these genes, and lethal for host enterocytes. Findings from this study demonstrate the pathogenic potential of the isolates recovered and this is a major public health concern. This necessitates the need for proper preventive measures to prevent contamination of food with Campylobacter spp. at the food production level in South Africa. This is one of the first studies emanating from South Africa in which the virulence genes in Campylobacter spp. have been screened in both human and animal samples and is therefore a good foundation for more intensive studies.
Acknowledgements
The authors would like to acknowledge the College of Agriculture, Engineering and Science as well as the School of Life Sciences at the University of KwaZulu-Natal (Westville campus) for financial support and the anonymous (for confidentiality reasons) private pathology laboratory in Durban which provided the human clinical isolates. They would also like to express their most sincere gratitude to the anonymous (for confidentiality reasons) poultry farms within the Durban metropolitan area in South Africa for providing chicken faecal samples for this study.
Competing interests
The authors declare that they have no financial or personal relationships that may have inappropriately influenced them in writing this article.
Authors’ contributions
Both authors contributed equally to this work. Both authors read and approved the final manuscript.
Footnotes
How to cite this article: Reddy, S. & Zishiri, O.T., 2018, ‘Genetic characterisation of virulence genes associated with adherence, invasion and cytotoxicity in Campylobacter spp. isolated from commercial chickens and human clinical cases’, Onderstepoort Journal of Veterinary Research 85(1), a1507. https://doi.org/10.4102/ojvr.v85i1.1507
References
- Al Amri A., Senok A.C., Ismaeel A.Y., Al-Mahmeed A.E. & Botta G.A, 2007, ‘Multiplex PCR for direct identification of Campylobacter spp. in human and chicken stools’, Journal of Medical Microbiology 56, 1350–1355. https://doi.org/10.1099/jmm.0.47220-0 [DOI] [PubMed] [Google Scholar]
- Al Amri A., Senok A.C., Ismaeel A.Y., Al-Mahmeed A.E., Botta G.A., Lake R. et al. , 2013, ‘Human and animal Campylobacteriosis in Tanzania: A review’, BMC Microbiology 95, 169–174. [Google Scholar]
- Biswas D., Hannon S.J., Townsend H.G.G., Potter A. & Allan B.J, 2011, ‘Genes coding for virulence determinants of Campylobacter jejuni in human clinical and cattle isolates from Alberta, Canada, and their potential role in colonization of poultry’, International Microbiology 14(1), 25–32. [DOI] [PubMed] [Google Scholar]
- Broaders E., Gahan C.G.M. & Marchesi J.R, 2013, ‘Mobile genetic elements of the human gastrointestinal tract: Potential for spread of antibiotic resistance genes’, Gut Microbes 4, 271–280. https://doi.org/10.4161/gmic.24627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Quintero A., Rodriguez-Castaño G.P. & Konstantinidis K.T, 2009, ‘Genomic insights into the convergence and pathogenicity factors of Campylobacter jejuni and Campylobacter coli species’, Journal of Bacteriology 191, 5824–5831. https://doi.org/10.1128/JB.00519-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A.F. De, Martins D., Azevedo S.S., Piatti R.M., Genovez M.E. & Scarcelli E, 2014, ‘Detection of CDT toxin genes in Campylobacter spp. strains isolated from broiler carcasses and vegetables in São Paulo, Brazil’, Brazilian Journal of Microbiology 44(3), 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chansiripornchai N. & Sasipreeyajan J, 2009, ‘PCR detection of four virulence-associated genes of Campylobacter jejuni isolates from Thai broilers and their abilities of adhesion to and invasion of INT-407 cells’, The Journal of Veterinary Medical Science/The Japanese Society of Veterinary Science 71, 839–844. [DOI] [PubMed] [Google Scholar]
- Datta S., Niwa H. & Itoh K, 2009, ‘Age-dependent variation of virulence-associated genes retained in Campylobacter jejuni isolated from chickens in a poultry farm’, The Journal of Veterinary Medical Science/The Japanese Society of Veterinary Science 71(9), 1247–1249. [DOI] [PubMed] [Google Scholar]
- Datta S., Niwa H., Itoh K., Mazi W., Senok A., Al-Mahmeed A. et al. , 2003, ‘Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet (O) -mediated tetracycline resistance’, Society 23, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epps S.V.R., Harvey R.B., Hume M.E., Phillips T.D., Anderson R.C. & Nisbet D.J, 2013, ‘Foodborne campylobacter: Infections, metabolism, pathogenesis and reservoirs’, International Journal of Environmental Research and Public Health 10, 6292–6304. https://doi.org/10.3390/ijerph10126292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feodoroff B., Lauhio A., Ellström P. & Rautelin H, 2011, ‘A nationwide study of Campylobacter jejuni and Campylobacter coli bacteremia in Finland over a 10-year period, 1998–2007, with special reference to clinical characteristics and antimicrobial susceptibility’, Clinical Infectious Diseases 53(8), e99–e106. https://doi.org/10.1093/cid/cir509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan R.C., Neal-Mckinney J.M., Dhillon A.S., Miller W.G., Konkel M.E, 2009, ‘Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization’, Infection and Immunity 77, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca B.B., Rossi D.A., Maia C.A., Nalevaiko P.C., Melo R.T., Cuccato L.P. et al. , 2014, ‘Characterization of the virulence, growth temperature and antibiotic resistance of the Campylobacter jejuni IAL 2383 strain isolated from humans’, Brazilian Journal of Microbiology 274, 271–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garénaux A., Jugiau F., Rama F., de Jonge R., Denis M., Federighi M. et al. , 2008, ‘Survival of Campylobacter jejuni strains from different origins under oxidative stress conditions: Effect of temperature’, Current Microbiology 56, 293–297. https://doi.org/10.1007/s00284-007-9082-8 [DOI] [PubMed] [Google Scholar]
- Ge Z., Schauer D.B. & Fox J.G, 2008, ‘In vivo virulence properties of bacterial cytolethal-distending toxin’, Cellular Microbiology 10, 1599–1607. https://doi.org/10.1111/j.1462-5822.2008.01173.x [DOI] [PubMed] [Google Scholar]
- Ghorbanalizadgan M., Bakhshi B., Lili A.K., Najar-Peerayeh S. & Nikmanesh B, 2014, ‘A molecular survey of Campylobacter jejuni and Campylobacter coli virulence and diversity’, Iranian Biomedical Journal 18, 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans D., Van Deun K., Martel A., Van Immerseel F., Messens W., Heyndrickx M. et al. , 2011, ‘Colonization factors of Campylobacter jejuni in the chicken gut’, Veterinary Research 42, 82 https://doi.org/10.1186/1297-9716-42-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraola G., Perez R., Naya H., Paolicchi F., Pastor E., Valenzuela S. et al. , 2014, ‘Genomic evidence for the emergence and evolution of pathogenicity and niche preferences in the genus Campylobacter’, Genome Biology and Evolution 6, 2392–2405. https://doi.org/10.1093/gbe/evu195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakee J.E., Ata N.S., Hakim A.S., Syame S.M. & Omara S.T, 2015, ‘Prevalence of virulence genes and antimicrobial resistance patterns of Campylobacter species isolated from chicken in Egypt’, Asian Journal of Poultry Science 9, 250–261. https://doi.org/10.3923/ajpsaj.2015.250.261 [Google Scholar]
- Kaakoush N.O., Castano-Rodriguez N., Mitchell H.M. & Man S.M, 2015, ‘Global epidemiology of campylobacter infection’, Clinical Microbiology Reviews 28, 687–720. https://doi.org/10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshbakht R., Tabatabaei M., Hosseinzadeh S., Shekarforoush S.S. & Aski H.S, 2013, ‘Distribution of nine virulence-associated genes in Campylobacter jejuni and C. coli isolated from broiler feces in Shiraz, Southern Iran’, Foodborne Pathogens and Disease 10, 764–770. https://doi.org/10.1089/fpd.2013.1489 [DOI] [PubMed] [Google Scholar]
- Komba E.V.G., Mdegela R.H., Msoffe P.L.M. & Ingmer H, 2013, ‘Human and animal campylobacteriosis in Tanzania: A review’, Tanzania Journal of Health Research 15, 1–13. https://doi.org/10.4314/thrb.v15i1.6 [DOI] [PubMed] [Google Scholar]
- Lapierre L., Gatica M.D.L.A., Riquelme V., Vergara C., Yañez J.M., San Martín B. et al. , 2016, ‘Characterization of antimicrobial susceptibility and its association with virulence genes related to adherence, invasion, and cytotoxicity in Campylobacter jejuni and Campylobacter coli isolates from animals, meat, and humans’, Microbial Drug Resistance 22(5), 432–444. https://doi.org/10.1089/mdr.2015.0055 [DOI] [PubMed] [Google Scholar]
- Marinou I., Bersimis S., Ioannidis A., Nicolaou C., Mitroussia-Ziouva A., Legakis N.J. et al. , 2012, ‘Identification and antimicrobial resistance of Campylobacter species isolated from animal sources’, Frontiers in Microbiology 3, 1–6. https://doi.org/10.3389/fmicb.2012.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Hansen K.K., Gradel K.O., Kristensen B., Ejlertsen T., Østergaard C. et al. , 2010, ‘Bacteraemia as a result of Campylobacter species: A population-based study of epidemiology and clinical risk factors’, Clinical Microbiology and Infection 16, 57–61. https://doi.org/10.1111/j.1469-0691.2009.02900.x [DOI] [PubMed] [Google Scholar]
- Noormohamed A. & Fakhr M.K, 2014, ‘Prevalence and antimicrobial susceptibility of Campylobacter spp. in oklahoma conventional and organic retail poultry’, The Open Microbiology Journal 8, 130–137. https://doi.org/10.2174/1874285801408010130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O Cróinín T. & Backert S, 2012, ‘Host epithelial cell invasion by Campylobacter jejuni: Trigger or zipper mechanism?’, Frontiers in cellular and infection microbiology 2, 25 https://doi.org/10.3389/fcimb.2012.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragimbeau C., Colin S., Devaux A., Decruyenaere F., Cauchie H.-M., Losch S. et al. , 2014, ‘Investigating the host specificity of Campylobacter jejuni and Campylobacter coli by sequencing gyrase subunit A.’, BMC Microbiology 14, 205 https://doi.org/10.1186/s12866-014-0205-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S. & Zishiri O.T, 2017, ‘Detection and prevalence of antimicrobial resistance genes in Campylobacter spp. isolated from chickens and humans’, Onderstepoort Journal of Veterinary Research 84(1), a1411 https://doi.org/10.4102/ojvr.v84i1.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizal A., Kumar A. & Vidyarthi A.S, 2010, ‘Prevalence of pathogenic genes in Campylobacter jejuni isolated from poultry and human’, Internet Journal of Food Safety 12, 29–34. [Google Scholar]
- Rozynek E., Dzierzanowska-Fangrat K., Jozwiak P., Popowski J., Korsak D. & Dzierzanowska D, 2005, ‘Prevalence of potential virulence markers in Polish Campylobacter jejuni and Campylobacter coli isolates obtained from hospitalized children and from chicken carcasses’, Journal of Medical Microbiology 54, 615–619. https://doi.org/10.1099/jmm.0.45988-0 [DOI] [PubMed] [Google Scholar]
- Samie A., Ramalivhana J., Igumbor E.O. & Obi C.L, 2007, ‘Prevalence, haemolytic and haemagglutination activities and antibiotic susceptibility profiles of Campylobacter spp. isolated from human diarrhoeal stools in Vhembe District, South Africa’, Journal of Health, Population and Nutrition 25, 406–413. [PMC free article] [PubMed] [Google Scholar]
- Silva J., Leite D., Fernandes M., Mena C., Gibbs P.A. & Teixeira P, 2011, ‘Campylobacter spp. as a foodborne pathogen: A review’, Frontiers in Microbiology 2, 1–12. https://doi.org/10.3389/fmicb.2011.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz M.N, 2002, ‘Human diseases caused by foodborne pathogens of animal origin’, Clinical Infectious Diseases 34, S111–S122. https://doi.org/10.1086/340248 [DOI] [PubMed] [Google Scholar]
- Taboada E.N., Clark C.G., Sproston E.L. & Carrillo C.D, 2013, ‘Current methods for molecular typing of Campylobacter species’, Journal of Microbiological Methods 95, 24–31. https://doi.org/10.1016/j.mimet.2013.07.007 [DOI] [PubMed] [Google Scholar]
- Thakur S., Zhao S., McDermott P.F., Harbottle H., Abbott J., English L. et al. , 2010, ‘Antimicrobial resistance, virulence, and genotypic profile comparison of Campylobacter jejuni and Campylobacter coli isolated from humans and retail meats’, Foodborne Pathogens and Disease 7(7), 835–844. https://doi.org/10.1089/fpd.2009.0487 [DOI] [PubMed] [Google Scholar]
- Thibodeau A., Fravalo P., Taboada E.N., Laurent-Lewandowski S., Guévremont E., Quessy S. et al. , 2015, ‘Extensive characterization of Campylobacter jejuni chicken isolates to uncover genes involved in the ability to compete for gut colonization’, BMC Microbiology 15, 97 https://doi.org/10.1186/s12866-015-0433-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B., Walgaard C., Drenthen J., Fokke C., Jacobs B.C. & Van Doorn P.A, 2014, ‘Guillain–Barré syndrome: Pathogenesis, diagnosis, treatment and prognosis’, Nature Reviews Neurology 10(8), 469–482. https://doi.org/10.1038/nrneurol.2014.121 [DOI] [PubMed] [Google Scholar]
- Wieczorek K., Szewczyk R. & Osek J, 2012, ‘Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter jejuni and C. coli isolated from retail raw meat in Poland’, Veterinarni Medicina 57, 293–299. [Google Scholar]
- Zaidi M.B., Campos F.D., Estrada-García T., Gutierrez F., León M., Chim R. et al. , 2012, ‘Burden and transmission of zoonotic foodborne disease in a rural community in Mexico’, Clinical Infectious Diseases 55, 51–60. https://doi.org/10.1093/cid/cis300 [DOI] [PubMed] [Google Scholar]