Abstract
Automated measurements of the human cone mosaic requires the identification of individual cone photoreceptors. The current gold standard, manual labeling, is a tedious process and can not be done in a clinically useful timeframe. As such, we present an automated algorithm for identifying cone photoreceptors in adaptive optics optical coherence tomography (AO-OCT) images. Our approach fine-tunes a pre-trained convolutional neural network originally trained on AO scanning laser ophthalmoscope (AO-SLO) images, to work on previously unseen data from a different imaging modality. On average, the automated method correctly identified 94% of manually labeled cones when compared to manual raters, from twenty different AO-OCT images acquired from five normal subjects. Voronoi analysis confirmed the general hexagonal-packing structure of the cone mosaic as well as the general cone density variability across portions of the retina. The consistency of our measurements demonstrates the high reliability and practical utility of having an automated solution to this problem.
OCIS codes: (100.0100) Image processing, (170.4470) Ophthalmology, (110.1080) Active or adaptive optics
1. Introduction
Adaptive optics (AO) techniques have been used to facilitate the visualization of the retinal photoreceptor mosaic in ocular imaging systems by improving the lateral resolution [1–3]. AO techniques have been combined with scanning light ophthalmoscope (AO-SLO) [4], flood illumination ophthalmoscopy (AO-FIO) [2, 5] and optical coherence tomography (AO-OCT) [6]. Similar to AO-SLO and AO-FIO, AO-OCT systems can provide high-resolution en face images of the photoreceptor mosaic in which the cones appear as bright circles surrounded by dark regions [7–9], but with the added benefit of high axial resolution which allows for cross-sectional tomography images in which the different outer retinal layers can be clearly delineated.
High-resolution images acquired using AO-assisted imaging systems have been used to investigate various changes in the appearance of the photoreceptor mosaic in both normal eyes [10–13] and eyes with degenerative retinal diseases, such as cone-rod dystrophy [14–16], retinitis pigmentosa [17–19] and occult macular dystrophy [20, 21]. Although these images can be qualitatively useful, quantitative analysis of different morphometric properties of the mosaic is generally preferred and requires identification of individual cones.
As manual segmentations are both subjective and laborious, several automated methods for detecting cones in AO images using a variety of traditional image processing techniques such as local intensity maxima detection [22–25], graph-theory and dynamic programming (GTDP) [26], and estimation of cone spatial frequency [27–29] have been developed. Although these mathematical methods work for the specific data for which they were designed, reliance on ad hoc rules and specific algorithmic parameters does not allow for alternative imaging conditions, such as different resolutions, areas within the retina, and imaging modalities.
A new alternative approach is using deep convolution neural networks (CNNs) where features of interest are learned directly from data. This allows for a higher degree of adaptability as the same machine learning algorithm can be re-purposed by using different training images given a sufficiently large training set [30]. Several of these networks have shown high performance for many different image analysis tasks, including ophthalmic applications [31]. For example, CNNs have been used for the segmentation of retinal blood vessels [32, 33], and detection of diabetic retinopathy [34] in retinal fundus images, and classification of pathology [35] or segmentation of retinal layers [36] and microvasculature [37] in optical coherence tomography (OCT) images. More recently, a CNN using a large dataset of manually marked images for training, has been developed to identify cones in AO-SLO images [38].
Using supervised deep learning approaches for quantification of the photoreceptors requires manually marked images. Unfortunately, a large dataset of manually marked images from an AO-OCT system does not currently exist and the construction of a model on an inadequate amount of data can have a negative impact on performance by causing overfitting. One method of addressing a small dataset is to use data from a similar domain, a technique known as transfer learning. Transfer learning has proven to be highly effective when faced with domains with limited data [39–41]. Instead of training a new network, by fixing the weights in certain layers of a network already optimized to recognize general structures from a larger dataset, and retraining the weights of the non-fixed layers, the model can recognize features with appreciably fewer examples [42]. In this study, we present an effective transfer learning algorithm for retraining a CNN originally trained on manually segmented confocal AO-SLO images in order to detect cones in AO-OCT images with a different field-of-view (FOV). Three different transfer learning techniques were applied and compared against manual raters.
2. Methods
2.1. AO-OCT dataset
All AO-OCT subject recruitment and imaging was performed at the Eye Care Centre of Vancouver General Hospital. The project protocol was approved by the Research Ethics Boards at the University of British Columbia and Vancouver General Hospital, and the experiment was performed in accordance with the tenets of the Declaration of Helsinki. Written informed consent was obtained by all subjects. The 1060nm, 200 kHz swept source AO-OCT system used in this study is similar to a previously described prototype [8], but a fixed collimator was used in place of a variable collimator. The axial resolution defined by the −6 dB width was measured to be 8.5μm in air (corresponding to a resolution of 6.2μm in tissue (n = 1.38)), and the transverse resolution with a 5.18mm beam diameter incident on the cornea was estimated to be 3.6μm assuming a 22.2mm focal length of the eye and refractive index of 1.33 for water at 1.06μm. Images were acquired with the system focus placed at the photoreceptor layer using GPU-based real-time OCT B-scan images. Wavefront distortion correction was realized by optimizing the shape of a deformable mirror in the system using a Sensorless AO (SAO) technique.
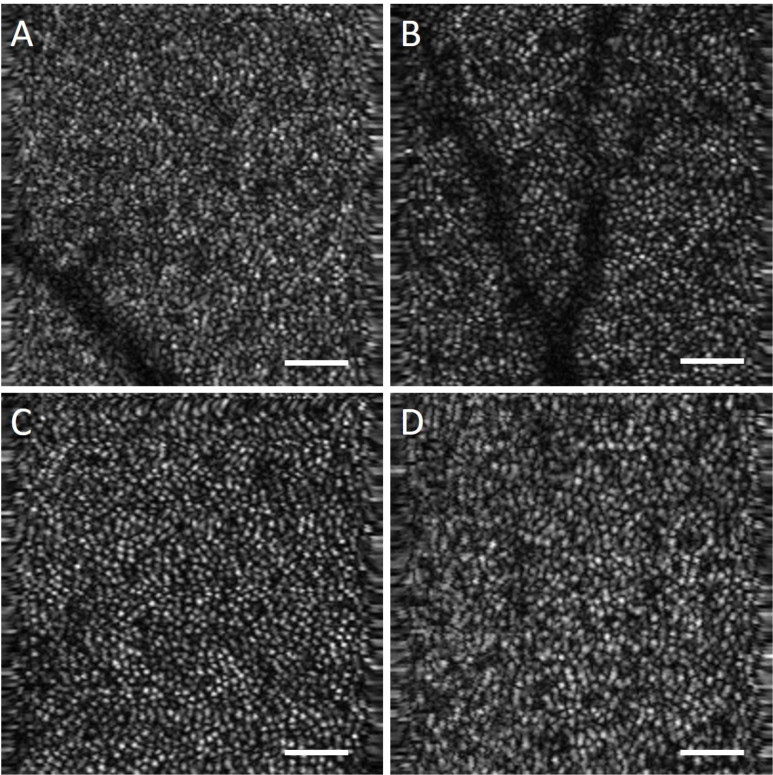
Four different locations (centered at ∼3.5°, ∼5°, ∼6.5°, and ∼8°) temporal to the fovea were imaged in five subjects. For each retinal location, five AO-OCT volumes (1.25° × 1.25° FOV) were acquired with 200 × 200 sampling density in a second. From the acquired volumes, a single volume with the least motion artifact was chosen and used for the rest of the analysis. Each AO-OCT image was resized to 400 × 400 pixels using bicubic interpolation to allow for the ’non-cone’ space to be at least 1 pixel. An example of the data acquired from one subject is shown in Fig. 1.
Fig. 1.
Data acquired from a 26 year old female control subject at retinal eccentricities of (a–d) ∼3.5°, ∼5°, ∼6.5°, and ∼8° respectively. Scalebar 50 μm.
The center of each cone photoreceptor was manually segmented using a Wacom Intuos 4 tablet and free image processing software [GNU image manipulation program (GIMP)] in all 20 AO-OCT images. One manual rater (Rater A) segmented data from three subjects (12 images) and another manual rater (Rater B) segmented the data from the remaining two subjects (8 images). For analysis of inter-rater agreement, two AO-OCT images from a normal subject not included in retraining the network were segmented by both raters and compared to the CNN output.
2.2. AO-SLO dataset
The AO-SLO dataset used to implement the initial conditions for the convolutional neural network was obtained from Ref. [38], and consisted of 840 confocal AO-SLO images acquired at 0.65° from the center of fixation, as well as the corresponding manual segmentations for the center of each cone. Each of the images within this dataset were extracted from a 0.96° × 0.96° FOV image, resulting in a FOV ranging from ∼0.20° × 0.20° to ∼0.25° × 0.25° from 21 subjects (including 20 normal subjects and 1 subject with deuteranopia) [38].
2.3. Data pre-processing
The image acquisition protocols and processing strategies of the AO-SLO and AO-OCT systems are quite different. In particular, the data from the AO-OCT system has a 5–6.25 times larger FOV. Therefore, data augmentation was performed on the AO-SLO data used for training the base network in order to improve the similarity between the two datasets. The data was sub-sampled at a rate of 1.5, 2, 2.5 and 3 to be a closer representation of the resolutions used for AO-OCT imaging.
Image patches were then extracted to use as inputs for training the network. As the manual segmentation protocol did not include non-cone locations, these were extracted using the protocol in [38, 43].In brief, a Voronoi diagram was constructed using the manually labeled cones as the center of each Voronoi cell. The boundaries were then assumed to be non-cone pixels, and a single point along each boundary was randomly chosen and placed in the non-cone set. A 33×33 window was then placed over each cone and non-cone location and used as input to the network. Locations closer than 16 pixels to the edge were discarded and not used.
2.4. Network training methods
The convolutional neural network used in this experiment is a slightly modified Cifar network taken from the AO-SLO cone segmentation paper [38]. The details of the network architecture are given in Table 1. Inputs to this network are 33×33×1 feature maps centered on a cone or non-cone pixel with the corresponding binary label. The final fully connected layer provides a score for each class (cone and non-cone), which are input into a soft-max layer that outputs the probability of the original center pixel belonging to each class.
Table 1.
CNN Architecture
| Layer | Type | Input Size | Filter Size | Stride |
|---|---|---|---|---|
| 1 | Convolutional | 33×33×1 | 5×5×32 | 1 |
| 2 | Batch Normalization | 33×33×32 | - | - |
| 3 | Max Pooling | 33×33×32 | 3×3 | 2 |
| 4 | ReLu Activation | 16×16×32 | - | - |
| 5 | Convolutional | 16×16×32 | 5×5×32 | 1 |
| 6 | Batch Normalization | 16×16×32 | - | - |
| 7 | ReLu Activation | 16×16×32 | - | - |
| 8 | Average Pooling | 16×16×32 | 3×3 | 2 |
| 9 | Convolutional | 8×8×32 | 5×5×32 | 1 |
| 10 | Batch Normalization | 8×8×64 | - | - |
| 11 | ReLu Activation | 8×8×64 | - | - |
| 12 | Average Pooling | 8×8×64 | 3×3 | 2 |
| 13 | Fully connected | 4×4×64 | 4×4×64 | - |
| 14 | Batch Normalization | 1×1×64 | - | - |
| 15 | ReLu Activation | 1×1×64 | - | - |
| 16 | Fully connected | 1×1×64 | 1×1×64 | - |
| 17 | Soft Max | 1×1×2 | - | - |
A similar network was also used in [43] to incorporate confocal AO-SLO and split detector AO-SLO image pairs. In brief, separate paths were used for layers 1–15 for the confocal and the split detector images, after which a concatenation layer combined the two 1 × 1 × 64 vectors output from the confocal and split detector paths into a single 1 × 1 × 128 vector that was fed into the rest of the network.
The experiments reported in this paper use two different methods to modify the AO-SLO network to segment cone photorecepters in AO-OCT images: transfer learning and fine-tuning. In both methods, certain layers of the base network trained on AO-SLO data were set to be non-trainable and the rest of the layers were then retrained using the AO-OCT data. In the first experiment, transfer learning was used so that only the classifier would be retrained and the first 15 layers of the base network were set to be non-trainable. The other two experiments used fine-tuning, where the base network was frozen before the second and third convolutional layers (layer 5 and 9, respectively) and the remaining trainable weights were subsequently retrained using the AO-OCT data. There was an average of 35,494 cone AO-OCT patches and 82,867 non-cone AO-OCT patches used on average for fine-tuning, and 336,280 AO-SLO patches used for the initial training. The batch size for training the base network was set to 100, and the maximum number of epochs was set to be 50 with an early stopping parameter set to when the validation loss hadn’t decreased in 4 epochs. For the transfer learning techniques, the batch size was decreased to 32. The learning rate was 0.001 and the weight decay was set at 0.0001 for both initial training and fine tuning.
Five-fold cross-validation on all manually segmented AO-OCT images was performed. The 20 original images were divided into 5 sets, so that all images from the same subject were placed into the same set. Images from four of the subjects were used to train the network, and images from the remaining subject were used to test the network. This procedure was repeated five times with a different test subject each time.
The CNN based detection method was implemented in TensorFlow and the Keras API [44] using Python 3.5.4. We ran the algorithm on a desktop PC with an i7-6700K CPU at 4.0 GHz, 16 GB of RAM, and a GeForce GTX 780 Ti X GPU. The average run time for segmenting a new image after training was 20 seconds.
2.5. Performance evaluation methods
A number of quantitative metrics were used to determine the effectiveness of the convolutional neural network. Probability maps were generated by extracting 33x33 pixel patches from each pixel location in the image as inputs to the trained network. The outputs of the network, the probabilities that each pixel was centered on a cone, were then arranged to generate a probability map the same size as the original image. These probability maps were binarized using Otsu’s method [45] and the centroid of any 4-connected components were taken to be the centers of cones. Any pixels within half the input size (16 pixels) to the edge of the input image were discarded from analysis. Within this implementation, True Positive (TP) results will indicate that the data was located within 0.5 of the median spacing between manually marked cones to a manually marked cone, False Positive (FP) results will indicate that the automatically detected cones were not matched to a manually detected cone, and False Negative (FN) results will indicate that manually marked cones did not automatically match detected cones. Given these definitions, Dice’s coefficient, Sensitivity, and False Discovery Rate are defined in Equations (1–3) respectively.
| (1) |
| (2) |
| (3) |
2.6. Cone mosaic analysis methods
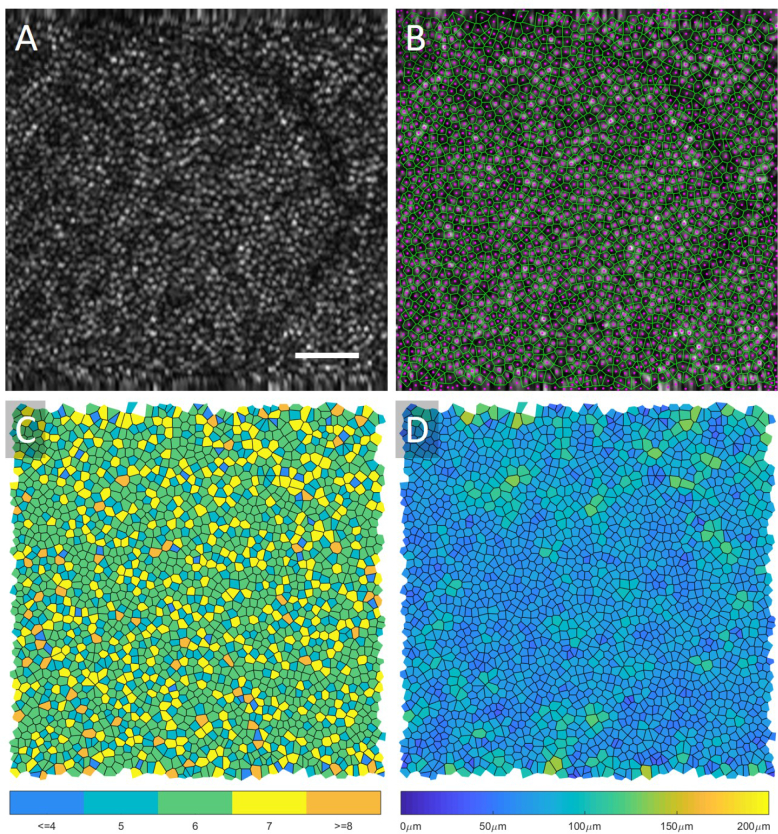
Voronoi diagrams were automatically constructed from each automated cone mosaic to calculate density, area, and proportion of hexagonal Voronoi domains, which are indicators of the regularity of the cone packing arrangement [46–48]. To analyze the regularity of the cone mosaics, the images were grouped together by their retinal eccentricities where Area 1 was closest to the fovea (∼3.5°), and Area 4 was furthest (∼8°). Cone density was defined as the ratio of the number of bound Voronoi cells in an image to the summed area of the bound Voronoi cells. To calculate the proportion of 6-sided Voronoi domains, the number of Voronoi cells with six sides was divided by the total number of bound Voronoi cells within an image. The number of neighbours was calculated as the mean number of sides of all bound Voronoi cells in an image. Similarly, the Voronoi cell area was calculated as the mean area of the bound Voronoi cells in an image. An example summary of this analysis is shown in Fig. 2 along with the original AO-OCT image. The Voronoi boundary map (green) and the automated centres of each cone (magenta) are shown in Fig. 2B, the number of neighbours map where each Voronoi cell is shaded depending on the number of neighbours is shown in Fig. 2C and the Voronoi cell area map where each Voronoi cell is shaded based on the cell area is shown in Fig. 2D.
Fig. 2.
An original AO-OCT image taken at ∼6.5° retinal eccentricity is displayed in (a), and the center of the cones (magenta) and Voronoi map (green) is overlaid onto the image in (b). In (c) the Voronoi cells are shaded based on the number of neighbours, and in (d) the cells are shaded based on their area. Scalebar 50 μm.
3. Results
3.1. Performance evaluation
The performances of the automated algorithms in comparison to manual grading are summarized in Table 2, including training on a network with randomly initialized weights. For the case of the randomly initialized weights, only AO-OCT images were used to train the network. The results from all four methods are within the standard deviations across all three quantitative measurements. A trend of over-segmenting in the methods which retrained more weights (Fine-Tuning Layer 5 and Layer 9) can be seen in the lower resultant sensitivities and slightly better false discovery rates. These numbers are slightly worse than the original network trained only on confocal AO-SLO data, where the sensitivity was 0.989 ± 0.012, the false discovery rate was 0.008 ± 0.014, and the Dice’s Coefficient was 0.990 ± 0.010 [38].
Table 2.
Average performance of the automated methods with respect to manual marking
| Sensitivity | False Discovery Rate | Dice’s Coefficient | |
|---|---|---|---|
| Transfer Learning | 0.940 ± 0.041 | 0.079 ± 0.037 | 0.929 ± 0.018 |
| Fine-Tuning (Layer 5) | 0.936 ± 0.046 | 0.062 ± 0.038 | 0.935 ± 0.017 |
| Fine-Tuning (Layer 9) | 0.936 ± 0.049 | 0.073 ± 0.043 | 0.930 ± 0.018 |
| Random Initialization | 0.942 ± 0.034 | 0.093 ± 0.038 | 0.923 ± 0.017 |
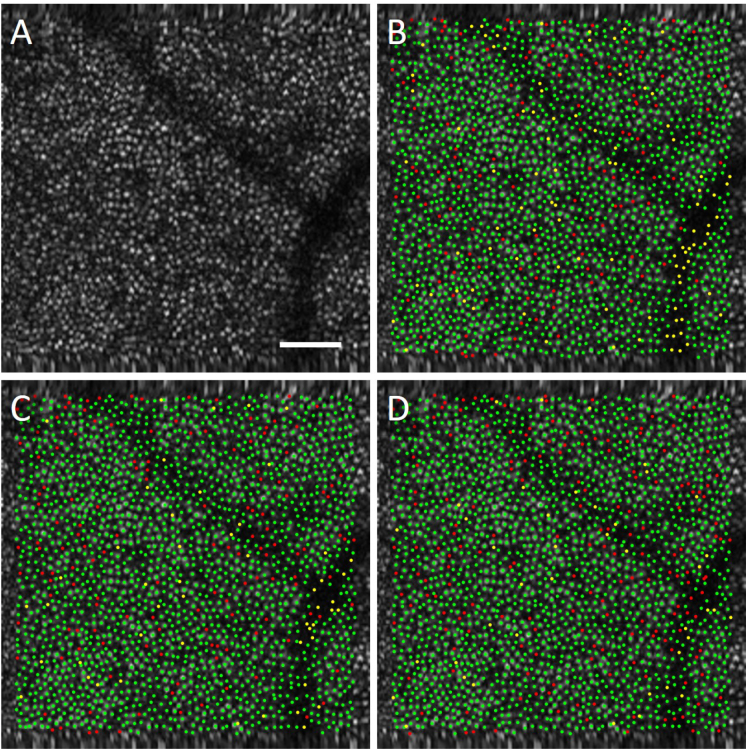
Figure 3 displays the results of the automated algorithms in comparison to manual grading for one of the AO-OCT datasets. In the marked images, a green point indicates an automatically detected cone that was matched to a manually marked cone (true positive), a yellow point indicates a cone missed by the automatic algorithm (false negative), and a red point indicates an automatic marking with no corresponding manually marked cone (false positive). From the images we can see that the methods performed quite well and that the methods that retrained more weights in the network produced more automated cone locations, resulting in fewer missed cones (hence less yellow locations) and more red locations, than the transfer learning method which only retrained the classifier.
Fig. 3.
Comparison of automated results in (a) an AO-OCT image from the (b) transfer learning, (c) fine-tuning (Layer 5) and (d) fine-tuning (Layer 9) methods. In the marked images, a green point indicates an automatically detected cone that was matched to a manually marked cone (true positive), a yellow point indicates a cone missed by the automatic algorithm (false negative), and a red point indicates an automatic marking with no corresponding manually marked cone (false positive). Scalebar 50 μm.
3.2. Inter-rater agreement
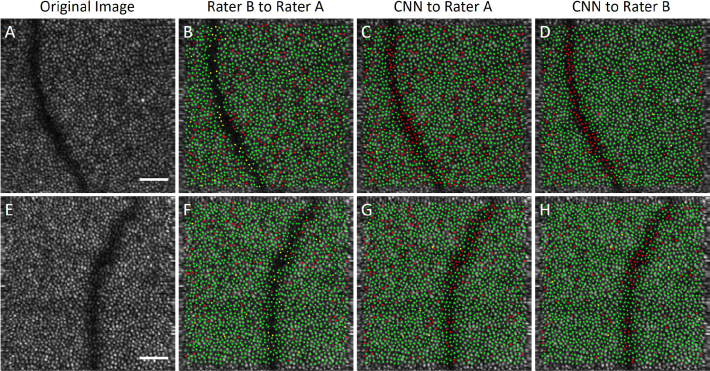
As previously mentioned in Section 2.1, two AO-OCT images from a normal subject not included in training the network were segmented by both raters for inter-rater analysis. For the AO-SLO images in [38], the manual rater segmentation quality was high because the FOV was small, the images highly sampled, and the cones hence had a high contrast. The AO-OCT images used in this report had a larger FOV, and hence a lower sampling density. Consequently, the contrast of the cones was poorer, leading to minor disagreement in segmentation even between the manual graders. The inter-rater performance can be seen in Fig. 4 along with the original AO-OCT image and a comparison of Rater A to the CNN output. As can be seen in Fig. 4b,e, Rater B found more cones than Rater A did in the majority of the image area, with the exception of areas in the blood vessel shadow where they were markedly more conservative. Similarly, the CNN found more cones than Rater A and in fact was more similar to Rater B as shown in Table 3. All inter-rater measurements are within the standard deviation of the automated results, suggesting that the automated segmentation is comparable to that of a human rater.
Fig. 4.
Comparison of manual results in two AO-OCT images (a) and (e) to a second rater (b,f) and the CNN (c–d,g–h). In the manually marked comparison images (b,f), a green point indicates a cone marked by Rater A that was matched to a cone marked by Rater B (true positive), a yellow point indicates a cone missed by Rater B (false negative), and a red point indicates a marking by Rater B with no corresponding cone marked by Rater A (false positive). In the comparison images to the CNN (c–d,g–h), a green point indicates a cone marked by a Rater that was matched to a cone marked by the CNN (true positive), a yellow point indicates a cone missed by the CNN (false negative), and a red point indicates a marking by the CNN with no corresponding cone marked by the Rater (false positive). Scalebar 50 μm.
Table 3.
Average performance of both raters and the automated methods with respect to manual marking
| Sensitivity | False Discovery Rate | Dice’s Coefficient | |
|---|---|---|---|
| Inter-rater | 0.978 ± 0.007 | 0.109 ± 0.043 | 0.932 ± 0.027 |
| CNN to Rater A | 0.998 ± 0.002 | 0.177 ± 0.066 | 0.902 ± 0.039 |
| CNN to Rater B | 0.997 ± 0.001 | 0.099 ± 0.033 | 0.946 ± 0.018 |
3.3. Cone mosaic analysis
From the performance evaluation of the three different transfer learning methods, we chose the results from the Fine-Tuning (Layer 5) method to further analyze as it had the highest Dice’s Coefficient. The results are summarized in Table 4 for all cone analysis parameters. The proportion of hexagonal cells and mean number of neighbours remained relatively constant over the areas imaged, while a general trend of the mosaic becoming less dense further from the fovea can be observed.
Table 4.
Cone Mosaic Measurements by Area
| Area 1 (∼3.5°) | Area 2 (∼5°) | Area 3 (∼6.5°) | Area 4 (∼8°) | |
|---|---|---|---|---|
| Automated Results | ||||
| Cone Density (cones/mm2(×1000)) | 15.49 ± 1.02 | 13.15 ± 0.88 | 11.95 ± 0.53 | 10.96 ± 0.39 |
| Percent 6-Sided (%) | 47.71 ± 2.56 | 47.42 ± 3.30 | 48.40 ± 1.36 | 46.89 ± 1.33 |
| Number of Neighbors | 5.87 ± 0.34 | 5.89 ± 0.33 | 5.87 ± 0.34 | 5.87 ± 0.36 |
| Voronoi Cell Area (μm2) | 52.50 ± 11.22 | 61.91 ± 12.82 | 67.94 ± 13.84 | 73.98 ± 14.72 |
| Manual Results | ||||
| Cone Density (cones/mm2(×1000)) | 16.12 ± 1.31 | 12.86 ± 0.88 | 11.77 ± 0.89 | 10.71 ± 1.09 |
| Percent 6-Sided (%) | 47.39 ± 1.58 | 44.01 ± 1.35 | 45.87 ± 1.71 | 43.77 ± 0.92 |
| Number of Neighbors | 5.87 ± 0.35 | 5.86 ± 0.35 | 5.86 ± 0.32 | 5.85 ± 0.36 |
| Voronoi Cell Area (μm2) | 50.66 ± 18.05 | 63.34 ± 18.02 | 69.20 ± 21.61 | 76.54 ± 23.80 |
4. Discussion
In this work, we investigated the use of transfer learning techniques using an automatic CNN based method for detecting cone photoreceptors in AO-OCT images. Using manually marked images from a confocal AO-SLO system to initialize the weights of our network, we have demonstrated retraining a CNN to extract features of interest and classify cones in previously unseen images from an AO-OCT imaging system. We tested our method on images of various retinal eccentricities and showed that our method had good agreement with the current gold standard of manual marking. In addition, we used various morphometric cone mosaic measurements to show quantitative agreement with measurements from AO-SLO systems.
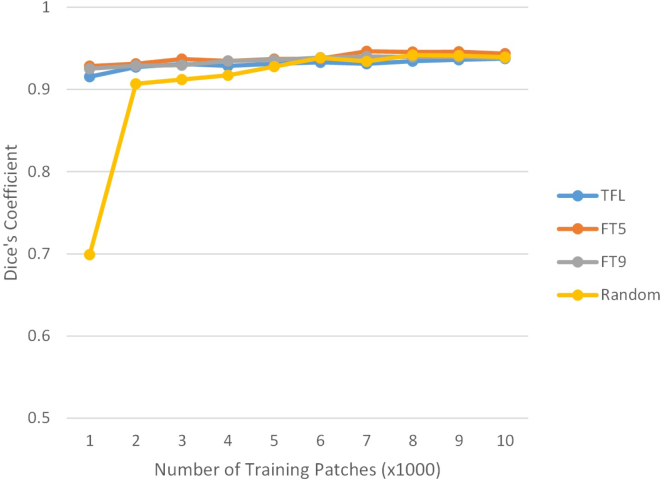
As shown in Table 2, performance of the CNN based algorithms were comparable to the current gold standard of manual grading which suggests that the automated segmentation is comparable to that of a human rater. This is highly encouraging, because traditional methods of photoreceptor cone segmentation heavily utilize modality and FOV specific ad hoc rules, which limit their application to other imaging protocols and require further algorithm modification and development for new imaging protocols. All that was needed to adapt the algorithm from confocal AO-SLO to AO-OCT was the corresponding training dataset. As we can see from the Table, the results from only using AO-OCT images and random weight initialization are comparable to using fine-tuning and transfer learning methods. We postulate that this is due to the large number of AO-OCT training data (118,361 patches) and that for a CNN, recognizing simple shapes like high contrast cones is a relatively straightforward task. As such, Fig. 5 shows how the number of training patches affects the performance of the different CNN algorithms. Random initialization performed poorly for 1000 patches, was close to the transfer learning methods at 2000 patches, but ultimately fit within the standard deviation at 3000 patches and above, whereas the transfer learning methods were stable from 1000 patches. This has important implications as we look to use CNNs for pathological images where the signal-to-noise ratio is lower and there are much fewer datasets from which to train.
Fig. 5.
How the number of training patches affects the performance of the different CNN algorithms. Random weight initialization (Random) is comparable to the Transfer Learning (TFL), Fine-Tuning Layer 5 (FT5), and Fine-Tuning Layer 9 (FT9) methods at 3,000 training patches.
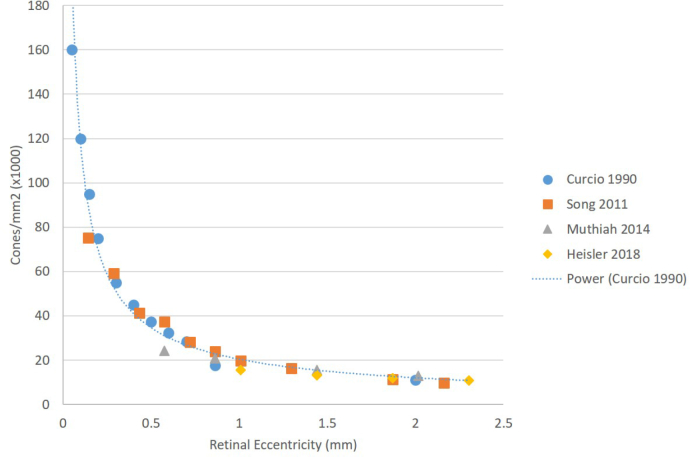
Additionally, the algorithm’s output measurements are congruent with AO-SLO data from the Literature [48]. The proportion of hexagonal cells remained relatively constant over the areas imaged, which is consistent with the Literature although the overall values are slightly lower than reported for AO-SLO data (52.6 ± 6.56 at 3.5 ° and 50.9 ± 7.32 at 8 °) [48]. Similarly, the mean number of neighbours also remained relatively constant over the areas imaged, which is consistent with the Literature although the overall values are slightly lower than reported for AO-SLO data (52.6 ± 6.56 at 3.5 ° and 50.9 ± 7.32 at 8 °) [48]. The general trend of the mosaic becoming less dense further from the fovea was also observed and is shown in Fig. 6 which compares our cone density measurements to measurements found in the Literature [49–51] for histology and AO-SLO data. Datapoints from Ref. [49] were extracted from the figures in the paper. As shown, our data follows the general trend for the eccentricities imaged.
Fig. 6.
Comparison of cone density measurements from the Literature to the cone density measurements from the AO-OCT system. The gold standard of histology [49] is shown with a trendline, as well as measurements from two AO-SLO systems [50, 51].
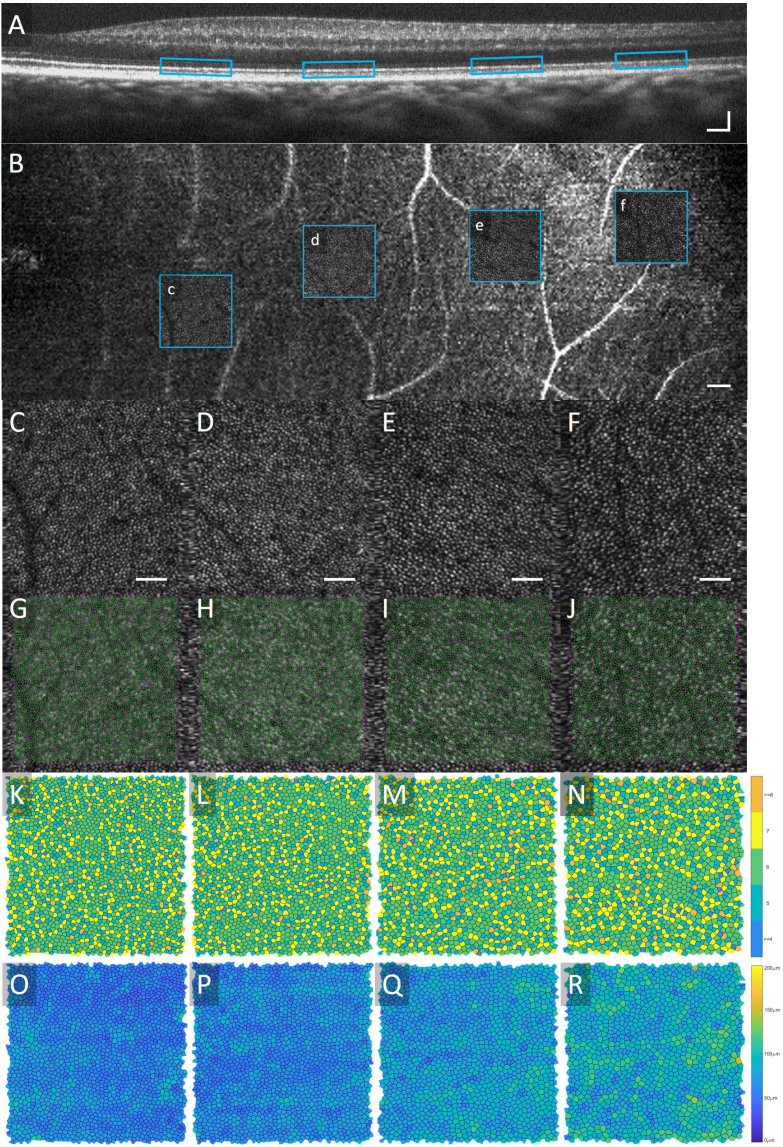
There is a trend towards overestimating cones in the algorithms where more layers were frozen with the AO-SLO initialized weights (Fine-Tuning (Layer 9) and Transfer Learning) as shown in the higher false discovery rate in Table 2 although the true positive rate was better. This is also reflected in Fig. 3, where the Fine-Tuning (Layer 5) results show less false positive cones. The majority of false positives was in regions below blood vessels. This could be improved by including more training data specifically around regions of vessels. This could either be done by acquiring more data around blood vessels, or using data augmentation techniques to modify the current dataset. Alternatively, the CNN could be combined with other pre-processing steps, such as blood vessel segmentation (for example, using OCT-A as shown in Fig. 7) to identify and remove the regions below vessels from the quantitative analysis. Moreover, poor inter-observer agreement can negatively affect the performance of learning based methods such as CNN. Utilization of datasets graded by multiple observers, for example both Rater A and Rater B, could further improve performance.
Fig. 7.
Results from a 22 year old male subject. Colocalization of the Areas 1 and 2 to a (a) widefield OCT Bscan [scalebar 100 μm] and (b) widefield AO-OCT-A en face view [scalebar 100 μm] are shown. The original AO-OCT images from Areas 1–4 are shown in (c–f) [scalebar 50 μm], automated segmentation results and the Voronoi diagrams are in (g–j), the Voronoi cells in (k–n) are shaded based on the number of neighbours, and in (o–r) the cells are shaded based on their area.
There is also space for improvement in all of the proposed algorithms mentioned in this work. The network architecture was chosen as it was the one published with the open-source confocal AO-SLO dataset, and was not optimized for the AO-OCT data. Additionally, the hyper-parameters were empirically chosen to provide good performance for both the fine-tuning and transfer learning methods. It is possible these parameters could be further optimized to provide better performance. Additionally, applying further custom pre-processing and post-processing steps such as multi-acquisition registration and averaging [52] may improve the results presented here.
Previously demonstrated in the Literature was confocal AO-SLO to split aperture AO-SLO; although different, there were similarities in field of view. In this report, transfer to a completely different imaging modality was demonstrated. Furthermore, the differences in scale of the PRs across retinal eccentricities while maintaining good sensitivity and false discovery rate is a significant achievement and demonstration of CNN usefulness. In general, for the cone photoreceptor mosaic, the features have a relatively well defined underlying structure, and a regularity for which CNNs are well suited. This implies that this transfer learning technique could be used to analyze images from commercial flood fundus photoreceptor images as well. There exists potential for CNNs to help unite images from different modalities with or without AO enabled imaging techniques for understanding photoreceptor changes in disease.
An example of this kind of multi modality imaging is shown in Fig. 7 where structural OCT, OCT-A and AO-OCT were used. Though the individual AO-OCT images used in this report are currently considered to be large field of view when imaging photoreceptors, being able to view wide field structural images is necessary for locating areas of interest. Further studies looking at various pathologies using wide field structural OCT cross sections and OCT-A to observe microvasculature changes and locate smaller regions of interest to then image with the higher resolution AO-OCT, using the CNN for quantification of the cone mosaic and vasculature [37], would be pertinent for proving clinical utility.
5. Conclusion
CNNs provide the opportunity to adapt to changing conditions without having to adjust ad hoc rules, but instead retraining the network on a sufficiently large database. In this paper, we experimentally demonstrated three different transfer learning methods to identify the cones in a small set of AO-OCT images using a base network trained on AO-SLO images which all obtained results similar to that of a manual rater. Using the results from the Fine-Tuning (Layer 5) method, we calculated four different cone mosaic parameters which were similar to results found in AO-SLO images showing the utility of our method.
Acknowledgments
The authors would like to express their sincerest gratitude to Profs. Sina Farsiu, Joseph Carroll and Alfredo Dubra and their groups for making the AO-SLO datasets and corresponding software open source and available for research purposes.
Funding
Brain Canada; National Sciences and Engineering Research Council of Canada; Canadian Institutes of Health Research; Alzheimer Society Canada; Michael Smith Foundation for Health Research; Genome British Columbia.
Disclosures
MJJ: Seymour Vision, Inc. (E), MVS: Seymour Vision, Inc. (I).
References and links
- 1.Williams D. R., “Imaging single cells in the living retina,” Vis. Res. 51, 1379–1396 (2011). Vision Research 50th Anniversary Issue: Part 2. 10.1016/j.visres.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang J., Williams D. R., Miller D. T., “Supernormal vision and high-resolution retinal imaging through adaptive optics,” J. Opt. Soc. Am. A 14, 2884–2892 (1997). 10.1364/JOSAA.14.002884 [DOI] [PubMed] [Google Scholar]
- 3.Pircher M., Zawadzki R. J., “Review of adaptive optics oct (ao-oct): principles and applications for retinal imaging [invited],” Biomed. Opt. Express 8, 2536–2562 (2017). 10.1364/BOE.8.002536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roorda A., Romero-Borja F., Da W. J., III, Queener H., Hebert T. J., Campbell M. C., “Adaptive optics scanning laser ophthalmoscopy,” Opt. Express 10, 405–412 (2002). 10.1364/OE.10.000405 [DOI] [PubMed] [Google Scholar]
- 5.Merino D., Duncan J. L., Tiruveedhula P., Roorda A., “Observation of cone and rod photoreceptors in normal subjects and patients using a new generation adaptive optics scanning laser ophthalmoscope,” Biomed. Opt. Express 2, 2189–2201 (2011). 10.1364/BOE.2.002189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermann B., Fernández E. J., Unterhuber A., Sattmann H., Fercher A. F., Drexler W., Prieto P. M., Artal P., “Adaptive-optics ultrahigh-resolution optical coherence tomography,” Opt. Lett. 29, 2142–2144 (2004). 10.1364/OL.29.002142 [DOI] [PubMed] [Google Scholar]
- 7.Kocaoglu O. P., Liu Z., Zhang F., Kurokawa K., Jonnal R. S., Miller D. T., “Photoreceptor disc shedding in the living human eye,” Biomed. Opt. Express 7, 4554 (2016). 10.1364/BOE.7.004554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ju M. J., Heisler M., Wahl D., Jian Y., Sarunic M. V., “Multiscale sensorless adaptive optics OCT angiography system for in vivo human retinal imaging,” J. Biomed. Opt. 22, 1 (2017). 10.1117/1.JBO.22.12.121703 [DOI] [PubMed] [Google Scholar]
- 9.Salas M., Augustin M., Felberer F., Wartak A., Laslandes M., Ginner L., Niederleithner M., Ensher J., Minneman M. P., Leitgeb R. A., Drexler W., Levecq X., Schmidt-Erfurth U., Pircher M., “Compact akinetic swept source optical coherence tomography angiography at 1060 nm supporting a wide field of view and adaptive optics imaging modes of the posterior eye,” Biomed. Opt. Express 9, 1871 (2018). 10.1364/BOE.9.001871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubra A., Sulai Y., Norris J. L., Cooper R. F., Dubis A. M., Williams D. R., Carroll J., “Noninvasive imaging of the human rod photoreceptor mosaic using a confocal adaptive optics scanning ophthalmoscope,” Biomed. Opt. Express 2, 1864 (2011). 10.1364/BOE.2.001864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chui T. Y., Song H., Burns S. a., “Adaptive-optics imaging of human cone photoreceptor distribution,” J. Opt. Soc. Am. A, Opt. image science, vision 25, 3021–3029 (2008). 10.1364/JOSAA.25.003021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kocaoglu O. P., Lee S., Jonnal R. S., Wang Q., Herde a. E., Derby J. C., Gao W. H., Miller D. T., “Imaging cone photoreceptors in three dimensions and in time using ultrahigh resolution optical coherence tomography with adaptive optics,” Biomed. Opt. Express 2, 748–763 (2011). 10.1364/BOE.2.000748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pircher M., Zawadzki R. J., Evans J. W., Werner J. S., Hitzenberger C. K., “Simultaneous imaging of human cone mosaic with adaptive optics enhanced scanning laser ophthalmoscopy and high-speed transversal scanning optical coherence tomography,” Opt. letters 33, 22–24 (2008). 10.1364/OL.33.000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi S. S., Doble N., Hardy J. L., Jones S. M., Keltner J. L., Olivier S. S., Werner J. S., “In vivo imaging of the photoreceptor mosaic in retinal dystrophies and correlations with visual function,” Investig. Ophthalmol. & Vis. Sci. 47, 2080 (2006). 10.1167/iovs.05-0997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfing J. I., Chung M., Carroll J., Roorda A., Williams D. R., “High-Resolution Retinal Imaging of Cone-Rod Dystrophy,” Ophthalmology. 113 1014 (2006). 10.1016/j.ophtha.2006.01.056 [DOI] [PubMed] [Google Scholar]
- 16.Song H., Rossi E. A., Stone E., Latchney L., Williams D., Dubra A., Chung M., “Phenotypic diversity in autosomal-dominant cone-rod dystrophy elucidated by adaptive optics retinal imaging,” The Br. journal ophthalmology 102, 136–141 (2018). 10.1136/bjophthalmol-2017-310498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan J. L., Zhang Y., Gandhi J., Nakanishi C., Othman M., Branham K. E., Swaroop A., Roorda A., “High-resolution imaging with adaptive optics in patients with inherited retinal degeneration,” Investig. Ophthalmol. Vis. Sci. 48, 3283–3291 (2007). 10.1167/iovs.06-1422 [DOI] [PubMed] [Google Scholar]
- 18.Talcott K. E., Ratnam K., Sundquist S. M., Lucero A. S., Lujan B. J., Tao W., Porco T. C., Roorda A., Duncan J. L., “Longitudinal study of cone photoreceptors during retinal degeneration and in response to ciliary neurotrophic factor treatment,” Investig. ophthalmology & visual science 52, 2219–2226 (2011). 10.1167/iovs.10-6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makiyama Y., Ooto S., Hangai M., Takayama K., Uji A., Oishi A., Ogino K., Nakagawa S., Yoshimura N., “Macular cone abnormalities in retinitis pigmentosa with preserved central vision using adaptive optics scanning laser ophthalmoscopy,” PLoS ONE 8 e79447 (2013). 10.1371/journal.pone.0079447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitaguchi Y., Kusaka S., Yamaguchi T., Mihashi T., Fujikado T., “Detection of photoreceptor disruption by adaptive optics fundus imaging and fourier-domain optical coherence tomography in eyes with occult macular dystrophy,” Clin. Ophthalmol. 5, 345–351 (2011). 10.2147/OPTH.S17335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakanishi A., Ueno S., Kawano K., Ito Y., Kominami T., Yasuda S., Kondo M., Tsunoda K., Iwata T., Terasaki H., “Pathologic changes of cone photoreceptors in eyes with occult macular dystrophy,” Investig. Ophthalmol. Vis. Sci. 56, 7243–7249 (2015). 10.1167/iovs.15-16742 [DOI] [PubMed] [Google Scholar]
- 22.Li K. Y., Roorda A., “Automated identification of cone photoreceptors in adaptive optics retinal images,” J. Opt. Soc. Am. A, Opt. image science, vision 24, 1358–1363 (2007). 10.1364/JOSAA.24.001358 [DOI] [PubMed] [Google Scholar]
- 23.Xue B., Choi S. S., Doble N., Werner J. S., “Photoreceptor counting and montaging of en-face retinal images from an adaptive optics fundus camera,” J. Opt. Soc. Am. A, Opt. image science, vision 24, 1364–1372 (2007). 10.1364/JOSAA.24.001364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wojtas D. H., Wu B., Ahnelt P. K., Bones P. J., Millane R. P., “Automated analysis of differential interference contrast microscopy images of the foveal cone mosaic,” J. Opt. Soc. Am. A 25, 1181–1189 (2008). 10.1364/JOSAA.25.001181 [DOI] [PubMed] [Google Scholar]
- 25.Garrioch R., Langlo C., Dubis A. M., Cooper R. F., Dubra A., Carroll J., “Repeatability of in vivo parafoveal cone density and spacing measurements,” Optom. Vis. Sci. 89, 632–643 (2012). 10.1097/OPX.0b013e3182540562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu S. J., Lokhnygina Y., Dubis A. M., Dubra A., Carroll J., Izatt J. a., Farsiu S., “Automatic cone photoreceptor segmentation using graph theory and dynamic programming,” Biomed. optics express 4, 924–937 (2013). 10.1364/BOE.4.000924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohammad F., Ansari R., Wanek J., Shahidi M., “Frequency-based local content adaptive filtering algorithm for automated photoreceptor cell density quantification,” in 2012 19th IEEE International Conference on Image Processing, (2012), pp. 2325–2328. 10.1109/ICIP.2012.6467362 [DOI] [Google Scholar]
- 28.Cooper R. F., Langlo C. S., Dubra A., Carroll J., “Automatic detection of modal spacing (Yellott’s ring) in adaptive optics scanning light ophthalmoscope images,” Ophthalmic Physiol. Opt. 33, 540–549 (2013). 10.1111/opo.12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunefare D., Cooper R. F., Higgins B., Katz D. F., Dubra A., Carroll J., Farsiu S., “Automatic detection of cone photoreceptors in split detector adaptive optics scanning light ophthalmoscope images,” Biomed. Opt. Express 7, 2036 (2016). 10.1364/BOE.7.002036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan M. I., Mitchell T. M., “Machine learning: Trends, perspectives, and prospects,” Science 349, 255–260 (2015). 10.1126/science.aaa8415 [DOI] [PubMed] [Google Scholar]
- 31.Lee A., Taylor P., Kalpathy-Cramer J., Tufail A., “Machine learning has arrived!” Ophthalmology 124, 1726–1728 (2017). 10.1016/j.ophtha.2017.08.046 [DOI] [PubMed] [Google Scholar]
- 32.Liskowski P., Krawiec K., “Segmenting retinal blood vessels with deep neural networks,” IEEE Transactions on Med. Imaging 35, 2369–2380 (2016). 10.1109/TMI.2016.2546227 [DOI] [PubMed] [Google Scholar]
- 33.Li Q., Feng B., Xie L., Liang P., Zhang H., Wang T., “A cross-modality learning approach for vessel segmentation in retinal images,” IEEE Transactions on Med. Imaging 35, 109–118 (2016). 10.1109/TMI.2015.2457891 [DOI] [PubMed] [Google Scholar]
- 34.Ga V, Pa L, Ca M, et al. , “Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs,” JAMA 316, 2402–2410 (2016). 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 35.Karri S. P. K., Chakraborty D., Chatterjee J., “Transfer learning based classification of optical coherence tomography images with diabetic macular edema and dry age-related macular degeneration,” Biomed. Opt. Express 8, 579–592 (2017). 10.1364/BOE.8.000579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang L., Cunefare D., Wang C., Guymer R. H., Li S., Farsiu S., “Automatic segmentation of nine retinal layer boundaries in oct images of non-exudative amd patients using deep learning and graph search,” Biomed. Opt. Express 8, 2732–2744 (2017). 10.1364/BOE.8.002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prentašic P., Heisler M., Mammo Z., Lee S., Merkur A., Navajas E., Beg M. F., Šarunic M., Loncaric S., “Segmentation of the foveal microvasculature using deep learning networks,” J. Biomed. Opt. 21, 075008 (2016). 10.1117/1.JBO.21.7.075008 [DOI] [PubMed] [Google Scholar]
- 38.Cunefare D., Fang L., Cooper R. F., Dubra A., Carroll J., “Open source software for automatic detection of cone photoreceptors in adaptive optics ophthalmoscopy using convolutional neural networks,” Sci. Reports 7 6620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donahue J., Jia Y., Vinyals O., Hoffman J., Zhang N., Tzeng E., Darrell T., “Decaf: A deep convolutional activation feature for generic visual recognition,” CoRR abs/1310.1531 (2013).
- 40.Razavian A. S., Azizpour H., Sullivan J., Carlsson S., “Cnn features off-the-shelf: An astounding baseline for recognition,” in 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops, (2014), pp. 512–519. 10.1109/CVPRW.2014.131 [DOI] [Google Scholar]
- 41.Yosinski J., Clune J., Bengio Y., Lipson H., “How transferable are features in deep neural networks?” in Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2, (MIT Press, Cambridge, MA, USA, 2014), NIPS’14, pp. 3320–3328. [Google Scholar]
- 42.Kermany D. S., Goldbaum M., Cai W., Valentim C. C., Liang H., Baxter S. L., McKeown A., Yang G., Wu X., Yan F., Dong J., Prasadha M. K., Pei J., Ting M., Zhu J., Li C., Hewett S., Dong J., Ziyar I., Shi A., Zhang R., Zheng L., Hou R., Shi W., Fu X., Duan Y., Huu V. A., Wen C., Zhang E. D., Zhang C. L., Li O., Wang X., Singer M. A., Sun X., Xu J., Tafreshi A., Lewis M. A., Xia H., Zhang K., “Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning,” Cell 172, 1122–1124 (2018). 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 43.Cunefare D., Langlo C. S., Patterson E. J., Blau S., Dubra A., Carroll J., Farsiu S., “Deep learning based detection of cone photoreceptors with multimodal adaptive optics scanning light ophthalmoscope images of achromatopsia,” Biomed. Opt. Express 9, 3740–3756 (2018). 10.1364/BOE.9.003740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chollet F., et al. , “Keras,” https://keras.io (2015).
- 45.Otsu N., “A threshold selection method from gray-level histograms,” IEEE Transactions on Syst. Man, Cybern. 9, 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- 46.Ooto S., Hangai M., Takayama K., Sakamoto A., Tsujikawa A., Oshima S., Inoue T., Yoshimura N., “High-resolution imaging of the photoreceptor layer in epiretinal membrane using adaptive optics scanning laser ophthal-moscopy,” Ophthalmology. 118, 873–881 (2011). 10.1016/j.ophtha.2010.08.032 [DOI] [PubMed] [Google Scholar]
- 47.Li K. Y., Roorda A., “Automated identification of cone photoreceptors in adaptive optics retinal images,” J. Opt. Soc. Am. A 24, 1358–1363 (2007). 10.1364/JOSAA.24.001358 [DOI] [PubMed] [Google Scholar]
- 48.Cooper R. F., Wilk M. A., Tarima S., Carroll J., “Evaluating descriptive metrics of the human cone mosaic,” Investig. Ophthalmol. & Vis. Sci. 57, 2992 (2016). 10.1167/iovs.16-19072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curcio C. A., Sloan K. R., Kalina R. E., Hendrickson A. E., “Human photoreceptor topography,” J. Comp. Neurol. 292, 497–523 (1990). [DOI] [PubMed] [Google Scholar]
- 50.Song H., Chui T. Y. P., Zhong Z., Elsner A. E., Burns S. A., “Variation of cone photoreceptor packing density with retinal eccentricity and age,” Investig. Ophthalmol. & Vis. Sci. 52, 7376 (2011). 10.1167/iovs.11-7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muthiah M. N., Gias C., Chen F. K., Zhong J., McClelland Z., Sallo F. B., Peto T., Coffey P. J., Cruz L. Da, “Cone photoreceptor definition on adaptive optics retinal imaging,” Br. J. Ophthalmol. 98, 1073–1079 (2014). 10.1136/bjophthalmol-2013-304615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heisler M., Lee S., Mammo Z., Jian Y., Ju M., Merkur A., Navajas E., Balaratnasingam C., Beg M. F., Sarunic M. V., “Strip-based registration of serially acquired optical coherence tomography angiography Strip-based registration of serially acquired optical,” J. Biomed. Opt. 22, 036007 (2017). 10.1117/1.JBO.22.3.036007 [DOI] [PubMed] [Google Scholar]