Abstract
The balance between metabolism and biomass is very important in biological systems; however, to date there has been no quantitative method to characterize the balance. In this methodological study, we propose to use the distribution of amino acids in different domains to investigate this balance. It is well known that endogenous or exogenous amino acids in a biological system are either metabolized or incorporated into free amino acids (FAAs) or proteome amino acids (PAAs). Using glycine (Gly) as an example, we demonstrate a novel method to accurately determine the amounts of amino acids in various domains using serum, urine, and cell samples. As expected, serum and urine had very different distributions of FAA- and PAA-Gly. Using Tet21N human neuroblastoma cells, we also found that Myc(oncogene)-induced metabolic reprogramming included a higher rate of metabolizing Gly, which provides additional evidence that the metabolism of proliferating cells is adapted to facilitate producing new cells. It is therefore anticipated that our method will be very valuable for further studies of the metabolism and biomass balance that will lead to a better understanding of human cancers.
Keywords: biomass, cancer, glycine, metabolism, Warburg effect
An important question in biological systems is the balance between metabolism and biomass. For example, depending on the circumstance, cancer can either accumulate biomass in the tumor during proliferation, or sometimes reduce biomass of non-tumor tissues in the process of cachexia. Compared to normal differentiated cells, cancer cells often switch from producing most of their energy by the oxidation of pyruvate through the tricarboxylic acid (TCA) cycle in mitochondria, to lactate production in the cytosol (the Warburg effect).[1] One possible explanation of the Warburg effect is that the metabolism of proliferating cells adapts to facilitate the uptake and incorporation of nutrients to produce new cells.[1c]
Amino acids play one of the most important roles in biological processes, primarily because they are extensively involved in metabolism and constitute the basic building blocks of peptides and proteins. In fact, the aforementioned Warburg effect is being re-investigated due to new findings that show the importance of glutamine as an energy source to replace glucose-derived carbon diverted from the TCA cycle for proliferating cancer cells.[2] A recent study discovered that glycine (Gly) is strongly correlated with the rate of cancer cell proliferation and contributes directly to de novo purine biosynthesis.[3] In addition, we showed that N-Myc-oncogene can change the amino acid profiles and their relative distribution in the domains of free amino acids (FAAs) and proteome amino acids (PAAs).[4]
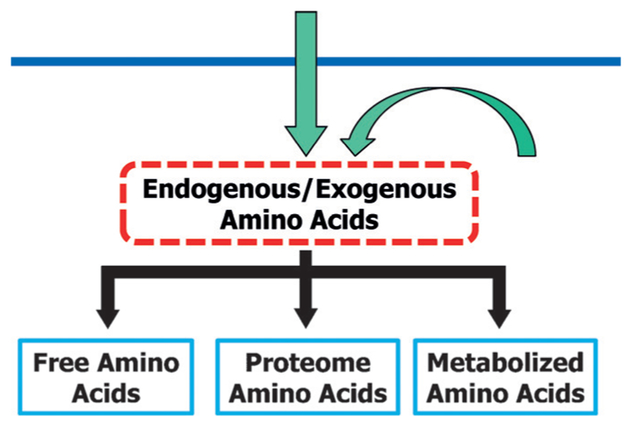
To date, the balance between metabolism and proteome biomass has been overlooked, and there has been no model developed to characterize their connections quantitatively. In this communication, we develop a method to explore the direct links between metabolism and the proteome biomass, and we propose to use amino acids as the probe to investigate this balance. As shown in Figure 1, endogenous or exogenous amino acids in a biological system are either metabolized or incorporated into the domains of FAAs or PAAs (including peptides). As an example, we measure the distribution of Gly in different domains to derive a quantitative model for the metabolism/proteome-biomass balance. In principle, metabolic flux analysis can also be used; however, flux analysis calculations are pathway dependent and often have multiple assumptions.[5]
Figure 1.
The concept of using amino acids as a probe to investigate the balance between metabolism and proteome biomass.
In this study, we used stable isotope labeled internal standards (iSTDs) for the accurate liquid chromatography tandem mass spectrometry (LC-MS/MS) measurements of FAAs and PAAs, using Gly as an example. To measure PAAs (including peptides that can produce Gly during hydrolysis), we improved a previously reported nonlinear least-squares extrapolation method,[6] and adapted it to our approach using iSTDs (Supporting Information, Section 1). Briefly, biological samples were spiked with iSTDs, and the mixtures were hydrolyzed with 6N hydrochloric acid (HCl) at 110 °C for t = 6, 12, 24, 48, and 96 h. Figure 2 illustrates the compartment model describing the simultaneous yield and decay processes of PAAs during hydrolysis. A is the level of PAAs in proteins, B is the level of MS-detectable amino acids, h is the hydrolysis rate, and l is the degradation rate.
Figure 2.
Schematic illustration of the compartment model describing the processes of hydrolysis and degradation of PAAs occurring simultaneously.
The MS-detectable amino acids at time t can be calculated from Equation (1), where Ap is the original level of PAAs in proteins prior to hydrolysis, and Ao is the amount of MS-detectable amino acids at t = 0 (FAAs).
| (1) |
We used the Monte Carlo method[7] to obtain the numerical solutions of Ap in Equation (2) (Supporting Information, Section 2). Io is the spiked level of iSTDs; k and b are the slope and intercept of the calibration curve, respectively, derived from the labeled and unlabeled standards.
| (2) |
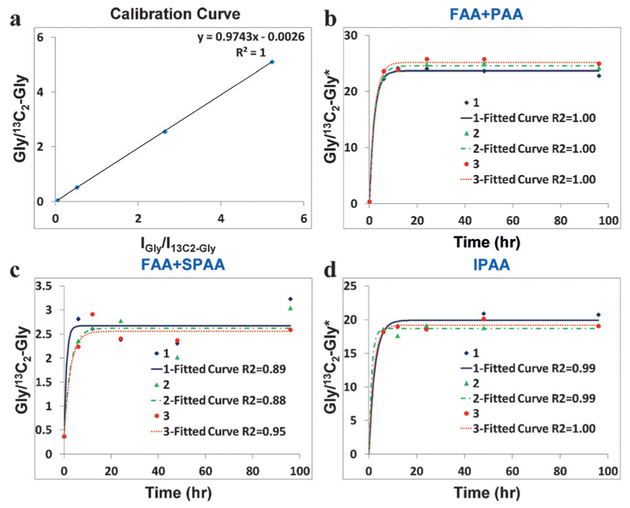
Figure 3a shows the calibration curve between Gly and 13C2-Gly, which converts the ratios of MS intensities to the ratios of amounts. Table 1 summarizes the concentrations of Gly in different domains of the serum and urine samples used in this study. From 5 μL serum, the amount of FAA-Gly was determined to be 1.87 × 10−9 mol (concentration: 0.373 ± 0.002 mM, standard error, n = 3), which fits well with the reported value from the HMDB database.[8] To obtain the amount of PAA-Gly, 5 μL serum was mixed with 13C2-Gly, and then the mixture was directly hydrolyzed with different time intervals. Figure 3b shows the Monte Carlo fitting of experimental data to Equation (2) (A0 = 1.87 × 10−9 mol). It can be clearly seen that our approach has a high reproducibility, and that the model of Equation (2) fits well with the collected data with high R2 values. The concentration of Gly in PAAs in the 5 μL serum sample was measured to be 23.9 ± 0.2 mM (Table 1).
Figure 3.
a) Calibration curve between Gly and 13C2-Gly; b) Monte Carlo fitting of experimental data to Equation (2) to determine the amount of PAAs (using Gly as an example) in 5 μL serum; c) Monte Carlo fitting of Equation (2) to determine the amount of SPAAs in 5 μL serum; d) fitting of Equation (2) to determine the amount of IPAAs in 5 μL serum. 1, 2, and 3 represent three replicates. *The ratio values were multiplied by 20 fold, except at t = 0, because more iSTD was spiked to approximately match the MS intensities of compounds (the intensity ratios were included in Figure 3 a).
Table 1:
The concentrations of Gly in different domains of the serum and urine samples analyzed in this study.
| Serum [mM] | Urine [mM] | |
|---|---|---|
| FAAs | 0.373 ± 0.002 | 0.812 ± 0.009 |
| PAAs | 23.9 ± 0.2 | 1.62 ± 0.01 |
| SPAAs | 2.25 ± 0.04 | 1.64 ± 0.02 |
| IPAAs | 19.2 ± 0.3 | 0.0679 ± 0.0008 |
| SPAAs/IPAAs[a] | 0.117 | 24.1 |
| PAAs/FAAs[a] | 64.0 | 1.98 |
Average values were used in calculations; no unit.
To further investigate the distribution of Gly in proteins, after methanol extraction we hydrolyzed the aqueous layer and the precipitated protein pellet to obtain Gly in the soluble protein amino acid (SPAA) and insoluble protein amino acid (IPAA) domains, respectively. For the same 5 μL serum, Figure 3c presents the Monte Carlo fitting curves used to determine the amount of SPAA-Gly. High R2 values were obtained, although they were lower than those in Figure 3b. The concentration of Gly in SPAAs was found to be 2.25 ± 0.04 mM (Table 1) using Equation (2). In addition, from the fitting curves in Figure 3d (Ao = 0), Gly in IPAAs in the serum sample was measured to be 19.2 ± 0.3 mM. The total amount of Gly in SPAAs and IPAAs should equal to that in PAAs. In this study, the sum of Gly in SPAAs and IPAAs was found to be 107.3 × 10−9 mol, which indicated that our approach had a recovery rate of 90% for this healthy human serum (PAAs: 119 × 10−9 mol).
Similarly, we measured Gly in different domains using a 50 μL urine sample from a healthy human volunteer, and the results are shown in Table 1 and the Supporting Information, Section 3. Notably, the recovery rate was determined to be 105 % for this healthy human urine, providing additional evidence that the protein hydrolysis method in this study is appropriate. Our results showed that the FAA-Gly concentration in the urine sample was much higher than that in the serum sample, and that urine contained much less PAA-Gly than serum (as anticipated, since in general urine has less total protein). Serum had a higher amount of Gly in the IPAA domain than the SPAA domain (SPAAs/IPAAs = 0.117); however, this was opposite for the urine sample (SPAAs/IPAAs = 24.1). Interestingly, the PAAs/FAAs ratio from the urine sample was 1.98, which was much lower than that from the serum sample (64.0).
To further demonstrate the concept of the balance between metabolism and biomass, we used 15N-13C2-Gly as the iSTD to construct a quantitative model describing the distribution of spiked 13C2-Gly in all sample domains of the Tet21N human neuroblastoma cells. We compared untreated cells (Myc-On) and those with N-Myc repressed (Myc-Off) by doxycycline. As is well known, Myc can cause significant alterations in global gene and protein expression, and activation of the oncogene Myc facilitates proliferation and biomass accumulation required for cell division through metabolic reprogramming.[9]
Figure 4 shows the experimental design and results. Analytically, it is very challenging to quantify the amount of metabolized amino acids. However, in this study we first quantify the amounts of extracellular FAAs (B), cellular FAAs (C), and cellular PAAs (D); if a known amount of 13C2-Gly (A) is spiked into the cells, consumed (E) and metabolized 13C2-Gly (F) can be easily calculated from the simple equation in Figure 4a (E = A−B=C+D+F), according to the law of conservation of matter. Notably, the D term is calculated using the Monte Carlo fitting of Equation (2). The amount of extracellular PAA 13C2-Gly was very small and was ignored in the calculation.
Figure 4.
The experimental design and results of a quantitative model describing the distribution of spiked 13C2-Gly in every domain of the Myc-On (a) and Myc-Off (b) Tet21N human neuroblastoma cells.
In Figures 4a and 4b, the amount of spiked 13C2-Gly was scaled to 100.0 (A), with a standard error of 0.4 (n = 2). After 72 hrs of incubation, extracellular FAA 13C2-Gly (B) was measured to be 82 ± 1 for the Myc-On Tet21N cells. Cellular FAA (C) and PAA (D) 13Q-Gly into the Myc-On Tet21N cells were determined to be 0.8 ± 0.1 and 3.1 ± 0.1, respectively. Therefore, consumed (E) and metabolized 13C2-Gly (F) in Myc-On cells were calculated to be 18 ± 1 and 14 ± 1, respectively. Similarly, extracellular FAA 13C2-Gly was 92 ± 4, cellular FAA 13C2-Gly was 0.50 ± 0.02, cellular PAA 13C2- Gly was 2.3 ± 0.2, consumed 13C2-Gly was 8 ± 4, and metabolized 13C2-Gly was 6 ± 4 for the Myc-Off Tet21N cells.
As expected, Myc-On Tet21N cells consumed more 13C2-Gly than Myc-Off cells. This fits well with the observation that Myc-On cells have a faster (≈2 ×) proliferation rate than Myc-Off cells during the 72-hr incubation. This could also indicate that Myc drives Gly uptake and metabolism in a similar manner as it increases glucose and glutamine uptake and metabolism.[9c] Myc-Off cells consumed less 13C2-Gly, which is also an important reason for the larger relative variation in the E and F terms than those for Myc-On cells. In future studies, using medium without Gly or a longer incubation time can further increase 13C2-Gly consumption in Myc-Off cells.
It can be clearly seen in Figure 4c that our method generated different metabolism-biomass balance models for the Myc-On and Myc-Off cells, which could be characteristic for tumorigenesis cells. The C/E and D/E values were lower and a higher proportion (F/E) of consumed 13C2-Gly (E) was metabolized (F) in Myc-On cells compared to that in Myc-Off cells. The major domain for consumed 13C2-Gly was the metabolized domain (F/C and F/D > 1) for both Myc-On and Myc-Off cells. The second major domain for cellular 13C2-Gly was the proteome (D). In addition, Myc-On cells had larger F/C and F/D ratio values than Myc-Off cells, indicating that Myc enhanced the conversion of incorporated glycine into other metabolites, instead of proteome- or FAA-Gly.
Although detailed biological analysis is beyond the scope of this study, we used 13C2-Gly as the tracer to examine major metabolic pathways that could contribute to the different behaviour of metabolizing Gly. As shown in Figure S4 (Supporting Information, Section 5), the enrichment of 13C2-Gly into lactate and TCA metabolites was very low. The average enrichment rate into serine was 7.9 % and 10 % for Myc-On and Myc-Off cells, respectively, indicating that serine synthesis is not mainly responsible for (in fact, opposite to) the different Gly metabolism.
Gly contributes to multiple other metabolic pathways, including the de novo synthesis of purine nucleotides and glutathione. Figure 5a shows that 13C2-Gly in Myc-On cells had a higher incorporation rate into AMP than that in Myc-Off cells. Direct incorporation into the purine backbone (M + 2) was a more important mechanism for Gly to be converted into AMP than the Gly cleavage system (GCS, M +1). Consistent with Figure 5a, the gene expression data in Figure 5b shows that N-Myc in Tet21N cells causes robust activation of all the enzymes in purine biosynthesis pathway. This fits well with the dependency of these cells upon the enzyme PFAS, as using siRNA against this enzyme would selectively kill Myc-On cells.[9c] Compared to AMP, there was no significant change induced by Myc, in terms of the reduced and oxidized glutathione ratio (GSH/GSSG, Figure S5a), or enrichment of 13C2-Gly in either GSH (Figure S5b) or GSSG (Figure S5c). Consistently, our gene expression analysis (Figure S5d) indicated no significant change in the expression of the glutathione synthesis pathway.[9c] Taken together, our results suggest that incorporation into the purine backbone to facilitate DNA synthesis could be an important mechanism for Myc-On cells to have a higher rate of metabolizing Gly than Myc-Off cells.
Figure 5.
a) The incorporation of 13C2-Gly into AMP in Myc-On and Myc-Off cells, M + n (n = 0, 1, and 2) means that n carbons are 13C-labled in AMP; b) the N-Myc-induced changes of de novo purine biosynthesis enzymes as measured using transcriptomic analysis.
Our approach derives a quantitative model describing the balance between metabolism and biomass, while at the current stage it cannot identify specific proteins/peptides. We improved the compartment model[6] using iSTDs and Monte Carlo fitting to measure proteome amino acids, since in principle at any given time point the MS measured levels of amino acids are not true hydrolysis yields (degradation occurs simultaneously). For PAA-Gly measurements, the reproducibility of h and l is relatively poor, which is similar to the results of previous studies.[6] To accurately measure h and l, multiple time points are required both before and after the levels of hydrolyzed amino acids reach their respective maxima. However, this is challenging because sample preparation includes a drying step and non-hydrolyzed proteins can interfere with LC-MS/MS detection. Fortunately, in most cases Ap is a more important contributor to the model than either h or l.
In summary, we have proposed a method to use the distribution of amino acids in different domains to characterize the balance between metabolism and biomass. Using Gly as an example, we demonstrated how to accurately determine the amounts of FAAs and PAAs (including peptides that can produce Gly during hydrolysis) in serum, urine, and cell samples. It was shown that our approach using iSTDs and the compartment model fits well with the collected data, and that it has high reliability for measuring the amounts of amino acids in various domains. The Gly models from serum and urine were very different. Furthermore, this work reports the first quantitative model describing the distribution of Gly (and amino acids more generally) in all sample domains of the Myc-On and Myc-Off Tet21N cells. Myc-induced metabolic reprogramming included a higher rate of metabolizing Gly, compared to FAA- and PAA-Gly, which supports the observation that the metabolism of fast proliferating cells is adapted to producing new cells. Based on our results, we expect that the relative distribution of amino acids in different domains could be a characteristic index of biological status, and that our method could be routinely used in biological and medical sciences.
Supplementary Material
Acknowledgements
This work was supported by the NIH (2R01 GM085291, RO1 CA57138, and T32 CA009657), FAPESP (FCN grant 2012/20672–0), and the NSFC (21365001 and 2011YQ170067).
Footnotes
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under http://dx.doi.org/10.1002/anie.201609236.
Contributor Information
Dr. Haiwei Gu, Northwest Metabolomics Research Center, Department of Anesthesiology and Pain Medicine, University of Washington Seattle, WA 98109 (USA) Jiangxi Key Laboratory for Mass Spectrometry and Instrumentation, East China University of Technology Nanchang, Jiangxi Province 330013 (China).
Dr. Jianhai Du, Department of Biochemistry, University of Washington Seattle, WA 98109 (USA)
Dr. Daniel Raftery, Northwest Metabolomics Research Center, Department of Anesthesiology and Pain Medicine, University of Washington Seattle, WA 98109 (USA); Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA 98109 (USA).
References
- [1].a) Warburg O, Science 1956,123,309–314; [DOI] [PubMed] [Google Scholar]; b) Kim J, Dang CV, Cancer Res. 2006, 66, 8927–8930; [DOI] [PubMed] [Google Scholar]; c) Vander Heiden MG, Cantley LC, Thompson CB, Science 2009, 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB, Proc. Natl. Acad. Sci. USA 2007,104, 19345–19350; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB, Proc. Natl. Acad. Sci. USA 2008,105, 18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK, Science 2012, 336, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gu H, Du J, Carnevale Neto F, Carroll PA, Turner SJ, Chiorean EG, Eisenman RN, Raftery D, Analyst 2015,140, 2726–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Fan TWM, Lane AN, J. Biomol. NMR 2011, 49, 267–280; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zamboni N, Fendt SM, Ruhl M, Sauer U, Nat. Protoc 2009, 4, 878–892; [DOI] [PubMed] [Google Scholar]; c) Fischer E, Sauer U, J. Biol. Chem 2003, 278, 46446–46451. [DOI] [PubMed] [Google Scholar]
- [6].a) Darragh AJ, Garrick DJ, Moughan PJ, Hendriks WH, Anal. Biochem 1996, 236, 199–207; [DOI] [PubMed] [Google Scholar]; b) Robel EJ, Crane AB, Anal Biochem. 1972, 48, 233–246. [DOI] [PubMed] [Google Scholar]
- [7].Han XL, Pozdin V, Haridass C, Misra P, J. Inf. Comput. Sci 2006, 3, 1–7. [Google Scholar]
- [8]. http://www.hmdb.ca/.
- [9].a) Dang CV, Cold Spring Harbor Symp. Quant. Biol 2011, 76, 369–374; [DOI] [PubMed] [Google Scholar]; b) Sloan EJ, Ayer DE, Genes Cancer 2010,1, 587–596; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Carroll PA, Diolaiti D, McFerrin L, Gu H, Djukovic D, Du J, Cheng PF, Anderson S, Ulrich M, Hurley JB, Raftery D, Ayer DE, Eisenman RN, Cancer Cell 2015, 27, 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.