Abstract
Our laboratory has recently proposed that the oxidation of guanine (G) to 8-oxo-7,8-dihydroguanine (OG) in G-rich promoter regions of DNA repair genes can serve as a regulatory mechanism of gene transcription. These regions also have the potential to fold into G-quadruplexes (G4). The human RAD17 promoter sequence has such a region in the template strand of the gene. In this work, the potential G-quadruplex sequence (PQS) of the RAD17 gene promoter was interrogated in different sequence contexts. With two extra nucleotides of the native sequence on either side of the G4, the structure was found to fold into a hybrid-like G4, similar to the hybrid-1 fold that the human telomere sequence can adopt. With only one nucleotide on either side of the PQS, the topology of the structure was observed to be mixed, and without extra nucleotides on the ends, the sequence adopted a parallel fold. Next, the sequence was studied with synthetic incorporation of the oxidative modification OG into specific sites and installed into the promoter of plasmids with a luciferase gene. These plasmids were transfected into a human cell line to observe the effect of the G4s on transcription. The RAD17 PQS was found to decrease luciferase expression with the presence of OG that is consistent with RAD17 expression under oxidative stress. This serves as an example of how oxidative modification could affect transcription in the context of a G4.
INTRODUCTION
Biomolecular chemical modifications resulting from oxidation by reactive oxygen species (ROS) have been implicated in the initiation and progression of aging, coronary disease, neurological diseases, and cancer.1 These disease states represent the deleterious effects of cellular ROS; on the other hand, ROS, such as H2O2, are also cellular signaling agents that oxidize specific proteins as part of the regulatory network for cells to respond to increasing ROS levels.2 All biomolecular components are susceptible to oxidation via ROS. In DNA, the electron-rich guanine (G) heterocycle is most susceptible to oxidation to yield many oxidatively modified products, in which 8-oxo-7,8-dihydroguanine (OG) is a key product identified in vivo.3 Further, a positive correlation between ROS and OG levels is well documented.4 Thus, our laboratory and others have proposed oxidation of G to OG in promoter regions of DNA can serve as a signaling agent to regulate gene transcription.5-7 This phenomenon of gene activation via G oxidation in a promoter region has been documented in the VEGF,8 SIRT1,9 TNF-α,10 and BCL211 genes in mammals. Under what circumstances and how globally does G oxidation in gene promoters modulate gene transcription are questions we have asked.
Our work on the topic of gene regulation by G oxidation to OG in a promoter region initially focused on the VEGF promoter because it contains a G-rich region in the coding (i.e., non-transcribed) strand that is responsible for gene regulation, and this region is prone to oxidation.12 More interestingly, the G-rich region is a potential G-quadruplex sequence (PQS) that can adopt a parallel-stranded G-quadruplex (G4) fold,13 and this feature has been proposed as part of the regulatory mechanism.14 Thus, we undertook studies to understand if G oxidation to OG in the VEGF PQS could activate transcription via favorable G4 formation.15 Briefly, the ability to synthesize OG site-specifically into the VEGF PQS for regulation of a luciferase reporter gene transfected into mammalian cells allowed the following observations to be made. When OG is present in the VEGF PQS in the coding strand, the reporter gene was turned on via a mechanism that involved base excision repair (BER) to remove the OG and yield a helix destabilizing abasic site (AP) that provided the thermodynamic drive to shift the structure to the G4 state. The VEGF PQS possesses five G tracks that allow flexibility in the structure to shift the damaged G-run into a large loop and stabilize the G4 structure with incorporation of the undamaged 5th G track.16 This new structure provides a platform for protein recruitment and gene activation. We followed this study with another that placed the VEGF promoter PQS into the template strand of the luciferase reporter gene.17 In the second study, we found transcription was turned off by the presence of OG in the G-rich context in the template (transcribed) strand. The work suggested OG and the G4 fold work together to block transcription preinitiation leading to the decrease in gene expression observed. In conclusion, G oxidation to OG in the G4 context in the coding strand of a gene promoter can turn transcription on, and in contrast, the same chemistry and sequence in the template strand of a gene promoter turns transcription off. These observations led us to question whether mechanisms like these could occur in other genes for transcriptional regulation.18
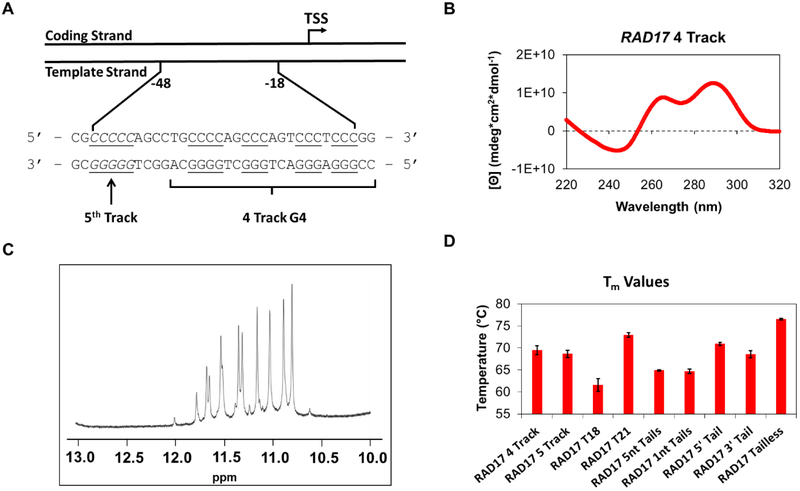
Thus, we conducted a bioinformatic analysis to identify hundreds of PQSs in human DNA repair gene promoters.18 We conducted initial structural studies on 30 of these sequences to establish G4 folding, and five were taken into the reporter gene studies to demonstrate the PQSs responded to G4 ligands by altering the transcription levels of a luciferase gene in glioblastoma cells. During the study, a PQS was found in the template strand of the promoter for the RAD17 gene (Figure 1A) that furnished a very interesting and unique circular dichroism (CD) spectrum for a promoter G4 (Figure 1B). The spectrum was nearly identical to that observed for the hybrid folds of the human telomere sequence, 19 this observation was fascinating because nearly all of the promoter G4s characterized adopt parallel or mixed folds and rarely hybrid folds.20 Thus, we elected to further interrogate the RAD17 PQS and the results are presented herein. Previous cellular studies have identified the RAD17 PQS to be possibly folded in the cellular context, 21,22and oxidative stress conditions result in down regulation of the gene.23 These two cellular observations suggest oxidation of G to OG in the RAD17 PQS in the template strand of the gene promoter may function to down regulate transcription. Herein, the structural results support the conclusion that this sequence can adopt a hybrid G4, as long as tails of two nucleotides each exist on the 5` and 3` ends of the sequence. When the tails were removed, the sequence was found to adopt a parallel-stranded G4. This observation provides a cautionary note to the G4 community that inclusion of the natural sequences flanking a promoter G4 may dramatically alter the fold observed. Lastly, studies were conducted to demonstrate that when an oxidation prone G in this sequence is modified to OG, the gene is down regulated, on the basis of a luciferase reporter assay in human glioblastoma cells.
Figure 1.
(A) Depiction of the location of the RAD17 PQS in the promoter region of the gene. (B) The CD spectrum obtained for the sequence with four domains of G runs. (C) Imino region (10–12 ppm) of the 1H NMR spectrum of the sequence. (D) Tm values obtained for sequences studied.
METHODS
Oligodeoxynucleotide Preparation
The oligodeoxynucleotides (ODNs) were synthesized by the DNA-Peptide Core Facility at the University of Utah using commercially available phosphoramidites. The ODNs were cleaved and deprotected following standard protocols and then purified using an anion-exchange semi-preparative HPLC column with a linear gradient from 25–100 %B over 30 min while monitoring the UV/vis absorbance at 260 nm. (A = 10% CH3CN/90% ddH2O, B = 1 M LiCl, 25 mM Tris in 10% CH3CN/90% ddH2O, pH 8, flow rate = 3 mL/min.) The purification salts were removed by dialysis against ddH2O for 36 h at 4 °C. The dialyzed DNA was lyophilized to dryness and resuspended in ddH2O followed by measurement of the absorbance at 260 nm and using the primary sequence to estimate the extinction coefficient to determine the DNA concentration. The DNA was then annealed at the specified concentration for each experiment via heating to 95 °C for 5 min and then slowly cooling to room temperature over 3 h followed by storage at 4 °C for 24 h prior to analysis.
NMR Studies
The 1H-NMR experiments were conducted on an 800-MHz NMR spectrometer running the Watergate solvent suppression pulse sequence. The spectra were recorded at a DNA concentration of 300 µM in 20 mM KPi (pH 7) with 50 mM KCl at 24 °C.
Circular Dichroism, Tm, and Thioflavin T Fluorescence Studies
The circular dichroism (CD) spectra were recorded using a 1-mm path length quartz cuvette on samples with a DNA concentration of 10 μM in a buffer containing 20 mM lithium cacodylate, 140 mM KCl, and 12 mM NaCl (pH 7, 20 °C) over a range of 200–320 nm. Thermal denaturation experiments were conducted to obtain Tm values on samples with a DNA concentration of 2 μM in the same buffer conditions by heating the samples at 1 °C/min and monitoring the absorbance changes at 295 nm. The thioflavin T fluorescence studies were measured on samples containing 1 μM DNA and 0.5 μM fluorophore, and the λem was scanned from 440–700 nm with a λex = 425 nm. The λem = 490 nm was monitored for each sequence studied.
Mapping Sites of Oxidation via Piperidine Cleavage
The annealed strands were 5’ radiolabeled and oxidized with the one-electron oxidant CO3•- following literature protocols.24 The sites of oxidation were revealed via treatment with freshly prepared 1 M piperidine at 90 °C for 2 h. This method was previously demonstrated to cleave most G oxidation products in a quantitative yield.24 After cleavage, the samples were lyophilized to dryness and resuspended in loading dye and then electrophoresed on a 20% denaturing PAGE at 45 W for 2 h. The sites of oxidation were visualized by storage phosphor autoradiography.
Plasmid Preparation
The plasmids were constructed with the psiCHECK2 plasmid (Promega) that contains the Renilla luciferase (Rluc) and firefly luciferase (luc) genes. Restriction-free cloning was utilized to insert the RAD17 promoter G4 sequences, followed by transformation of these plasmids to competent E. coli. The plasmids were isolated from these cells by a miniprep kit (Qiagen). Site-specific modification of OG was synthesized in short oligonucleotides and inserted into the plasmid via literature methods.15
Cell Culture Studies
Glioblastoma cells (U87-MG) were purchased from ATCC. The cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS, 20 µg/mL gentamicin, 1× glutamax, and 1× nonessential amino acids. Transfection experiments were conducted in white, 96-well plates with X-tremeGene HP DNA transfection agent (Roche) with 200–750 ng of plasmid following literature protocols.15 The Dual-Glo luciferase assay (Promega) was conducted following the manufacturer’s protocol to monitor Rluc and luc expression levels. Each experiment was conducted in four replicates as recommended by the manufacturer. The measured luminescence values were converted into normalized relative response ratios (RRR) that is the luminescence of Rluc divided by the luminescence of luc (i.e., RRR = Rluc/luc).
RESULTS AND DISCUSSION
Characterization of the RAD17 Promoter G-Quadruplex
The RAD17 promoter PQS was initially studied with four G tracks with two nucleotides of the natural sequence as tails on both the 5’ and 3’ ends (RAD17 4 Track, Table 1). Studies with the human telomere sequence found that omission of these natural sequence overhangs can impact the structures of the G4 fold.25 In the first set of studies, upon purification, the samples were annealed in NMR buffer (20 mM KPi pH 7.0, 50 mM KCl, 22 °C) at a concentration of 300 μM DNA to maximize signals in 1H-NMR studies. The diagnostic imino protons of G:G Hoogsteen base pairs (10–12 ppm)26,27 in G-tetrads of G4 structures were identified to support folding of the sequence to a G4 (Figure 1C). There exist 12 major peaks in this region of the spectrum consistent with one predominant G4 fold in solution. This observation was highly unusual compared to 1H-NMR spectra for other PQSs in promoter regions of various DNA repair genes that showed an array of broad signals in the imino proton region.18
Table 1.
The RAD17 promoter PQSs studied. A red X indicates a substitution from a G to an OG.
| Name | Sequence |
|---|---|
| RAD17 4 Track | 5′ – CC GGG A GGG ACT GGG CT GGGG CA – 3′ |
| RAD17 5 Track | 5′ – CC GGG A GGG ACT GGG CT GGGG CAG GCT GGGGG CG – 3′ |
| RAD17 T18 | 5′ – CC GGG A GGG ACT GGG CT TGGG CA – 3′ |
| RAD17 T21 | 5′ – CC GGG A GGG ACT GGG CT GGGT CA – 3′ |
| RAD17 5nt Tails | 5′ – CGGCC GGG A GGG ACT GGG CT GGGG CAGGC – 3′ |
| RAD17 1nt Tails | 5′ – C GGG A GGG ACT GGG CT GGGG C – 3′ |
| RAD17 5’ Tail | 5′ – C GGG A GGG ACT GGG CT GGGG – 3′ |
| RAD17 3’ Tail | 5′ – GGG A GGG ACT GGG CT GGGG C – 3′ |
| RAD17 Tailless | 5′ – GGG A GGG ACT GGG CT GGGG – 3′ |
| RAD17 5 OG13 | 5′ – CC GGG A GGG ACT XGG CT GGGG CAGGCT GGGGG CG – 3′ |
| RAD17 5 OG18 | 5′ – CC GGG A GGG ACT GGG CT XGGG CAGGCT GGGGG CG – 3′ |
| RAD17 4 OG13 | 5′ – CC GGG A GGG ACT XGG CT TGGG CA – 3′ |
| RAD17 4 OG18 | 5′ – CC GGG A GGG ACT GGG CT XGGT CA – 3′ |
The DNA sample was then diluted to be analyzed by other spectroscopic methods. The CD spectrum of the RAD17 PQS was recorded in a buffer to mimic the K+ and Na+ human intracellular concentrations (20 mM lithium cacodylate pH 7.0, 140 mM KCl, 12 mM NaCl) at 10 μM DNA concentration. The spectrum showed a λmax ≈ 292 nm, λshoulder ≈ 264 nm and λmin ≈ 245 nm that is nearly identical to the one observed for a hybrid fold which the human telomere sequence can adopt (Figure 1B).19 The Tm value (Figure 1D) for the sequence was measured by observing the decrease of absorbance at 295 nm with increasing temperature from 20 °C to 95 °C. The Tm value was found to be 69.5 ± 1.0 °C, higher than physiological temperature, and thus this sequence has the potential to fold in cellulo.28 In an additional experiment, the fluorescence emission enhancement (Figure S1) of the G4-specific fluorophore thioflavin T (ThT) was measured in the presence of the RAD17 PQS following a literature protocol that found enhancement of fluorescence of ThT of >20 Fl490nm/Flo in the presence of a folded G4.29 With a value of 18.9 ± 0.3 Fl490nm/Flo, the folded sequence produced results inconsistent with G4 formation; however, recent studies have identified ThT binding is more favorable with parallel-stranded G4s than hybrid-like G4s as the RAD17 sequence appears to adopt.30
The sequence with five G domains (RAD17 5 Track) also had signals in the imino proton range of the 1H-NMR spectrum (Figure S2). However, there were more peaks than observed for the RAD17 4 Track sequence. The peaks were also less defined, suggesting there exist many possible structures in solution, consistent with having five runs of Gs with one run of Gs having five nucleotides (Figure 1A).16 The CD spectrum for the RAD17 5 track (Figure S2) also had signals at λ ≈ 290 nm, λ ≈ 263 nm, and a λmin ≈ 245 nm. However, the signal at λ ≈ 263 nm was larger than that of λ ≈ 290 nm that is opposite of the spectrum for the four track sequence; the wavelengths at which λmax and λshoulder are found in this spectrum are switched compared to the four track sequence. This may indicate a different folding pattern or a mixture of folding topologies when compared to literature spectra.31 The Tm value was found to be 68.6 ± 0.8 °C (Figure 1D), similar to the value that was obtained for the four-track sequence. The sequence also gave a fluorescence enhancement value of 63.3 ± 4 Fl490nm/Flo (Figure S1). With these observations, the RAD17 5 Track sequence can fold at physiological conditions and is possibly more polymorphic with incorporation of the fifth run of Gs compared to the four-track sequence.
The sequence was then analyzed to elucidate the possible structure into which the quadruplex folds. The PQS has three runs of three Gs that could all participate in folding, and one run of four Gs of which only three Gs would participate in folding at a given time (Figure 1A). This would mean that one of the four Gs in the last run would be part of a loop. To find out which G this would be, two new sequences (RAD17 T18 and RAD17 T21) were studied (Table 1) with separate G-to-T substitutions that would prevent that nucleotide from participating in the formation of the G-tetrad core. Comparison of the new sequence and the wild-type sequence would help identify which G would be best for the folding of the G4.32 These DNA samples were obtained, purified, and prepared as above. In the 1H-NMR spectra (Figure S3), the RAD17 T18 showed defined peaks at the 10–12 ppm range while the RAD17 T21 showed peaks that were less defined. The CD spectrum (Figure S4) for T18 was similar to the one for the wild-type RAD17 sequence with a λmax ≈ 292 nm, λshoulder ≈ 264 nm and λmin ≈ 245 nm that is indicative of a hybrid-like fold. The RAD17 T21 sequence, however, had a λmax ≈ 266 nm and λmin ≈ 241 nm that is consistent with a parallel-stranded G4 on the basis of a comparison to literature sources.33-36 The Tm values for the RAD17 T18 and T21 sequences were 61.6 ± 1.4 °C and 73.0 ± 0.5 °C (Figure 1D), respectively, thus both sequences have the potential to fold in cellulo. The fluorescence enhancement assay with ThT found values of 26.0 ± 1.7 Fl490nm/Flo for RAD17 T18 and 74.2 ± 2.9 Fl490nm/Flo for RAD17 T21 (Figure S1), thus they have the capability to fold into G4s. Considering that the data for the sequence RAD17 T18 matched the data collected for the original RAD17 4 Track sequence, the G that was substituted to a T in that sequence (at position 18) does not participate in the core of the G4, but instead occupies a loop position in these contexts.
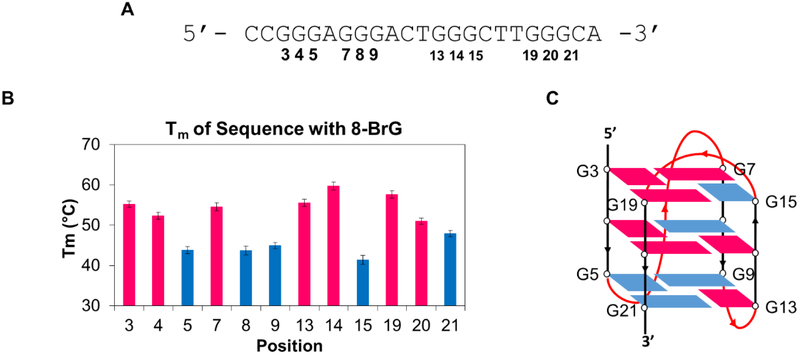
Using the RAD17 T18 sequence (Figure 2A) to decrease variability in folding of the G4, a study was then performed to support a structural model of the hybrid G4 fold. A key characteristic of hybrid G4s is that they possess both syn and anti G nucleotides. To probe which Gs were syn and which were anti, separate sequences (Table S1) were obtained with individual substitutions at each of the guanines to 8-bromoguanine (8-BrG). Because 8-BrG is known to strongly favor the syn conformation, its substitution for a G either stabilizes or destabilizes the structure depending on the preferred fold,37-40 i.e. the syn conformation is confirmed if the structure is stabilized and the opposite assignment is made (anti) if 8-BrG substitution is destabilizing. The sequences that gave CD spectra (Figure S5) similar to that of the one obtained for the RAD17 T18 and wild-type sequences were considered to have the substituted G in the syn conformation. The spectra that gave different signatures were considered to have the substituted G in the anti conformation. Assignments were further determined with Tm studies in which a lower melting temperature would correlate with lower stability, and thus the position was identified as being in the anti orientation (Figure 2B). With these data, the folding pattern of the sequence (Figure 2C) was found to be similar to that of the hybrid 1 structure that the human telomere sequence can adopt.19, 41,42
Figure 2.
Studies with 8-BrG substitution of Gs to elucidate folding pattern of RAD17 promoter PQS. Red represents a G in the syn position, blue represents a G in the anti position. (A) T18 Sequence used for studies with positions of the Gs marked. (B) Tm data obtained of sequences with 8-BrG substituted in different positions. (C) Depiction of the fold that the parent sequence adopts.
Effect of Tails on the RAD17 Promoter G-Quadruplex
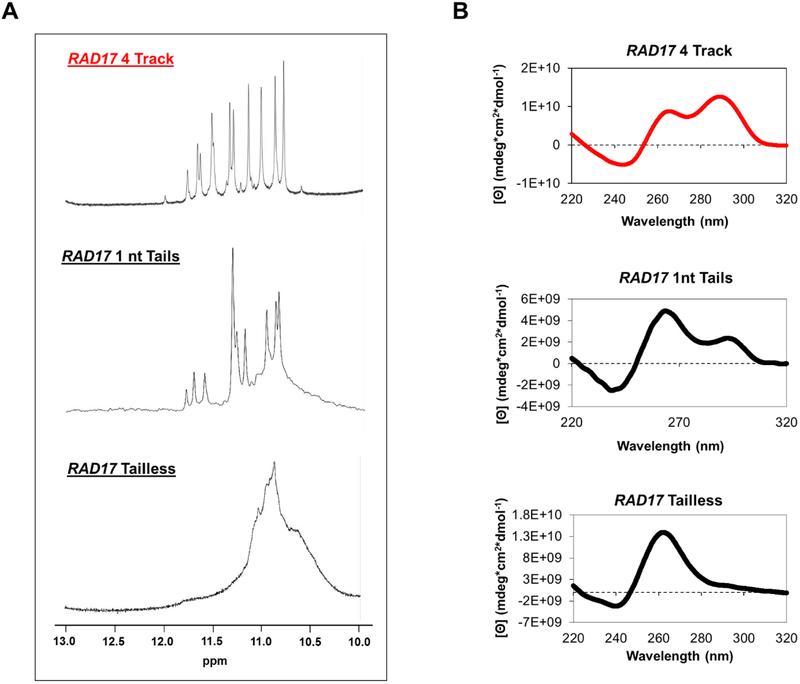
To learn more about the structure that the RAD17 PQS adopts, the absence and presence of tails was studied to find their effects on folding. Several sequences were interrogated, each varying in the number of nucleotides in the tails (Table 1). The molecularities of the different sequences were interrogated via size-exclusion chromatography (Figure S6) in which the results support intramolecular folding for the sequences under the conditions studied. With five nucleotides of the natural sequence on both the 5’ and 3’ ends (RAD17 5 nt), there were a few defined peaks in the 10–12 ppm region of the 1H-NMR spectrum, and the sequence adopted a hybrid-like fold based on CD spectroscopy (Figure 3A and 3B). The Tm was 64.9 ± 0.2 °C and fluorescence enhancement value with ThT of 21.6 ± 1 Fl490nm/Flo (Figures 1D and S1). The data collected for the RAD17 5 nt sequence concur with the original sequence and suggest that it folds into a hybrid G4; thus, adding more nucleotides at the ends of the sequence in addition to the two original nucleotides as single-stranded tails does not change the folding pattern.
Figure 3.
(A) 1H-NMR spectra and (B) CD spectra for the RAD17 promoter PQS with different numbers of nucleotides in the tails.
Because adding nucleotides does not change the folding pattern, the next sequences studied had a reduced number of nucleotides in the tails. With one-nucleotide tails on both ends (RAD17 1 nt), there were still some defined peaks in the 1H-NMR spectrum (Figure 3A), and the sequence adopted a similar fold to the wild-type RAD17 sequence. However, the signal at λ ≈ 292 nm decreased while the signal at λ ≈ 264 nm increased (Figure 3B). This may indicate that a fully parallel fold may be in equilibrium with a hybrid fold. The Tm value was measured to be 64.7 ± 0.5 °C, and the sequence showed a fluorescence enhancement value of 12.2 ± 0.3 Fl490nm/Flo (Figures 1D and S1) with thioflavin T. The fluorescence enhancement was lower than expected for a G4, but considering there was an enhancement observed, there is still the possibility that the sequence can fold to a parallel or anti-parallel structure.
With the interesting observation that mixed topologies could be formed with one nucleotide on both ends of the G4 forming sequence, more studies were performed on additional sequences to elucidate whether a single nucleotide on one end could give similar results. First, a sequence with one nucleotide at the 5’ end (RAD17 5’ Tail) was studied. For the CD spectrum (Figure S7), this sequence gave a λmax ≈ 263 nm and λmin ≈ 241 nm, which was similar to the spectrum of the RAD17 T21 sequence. Thus, the RAD17 5’ Tail sequence adopts only parallel folds, on the basis of the CD spectrum. The Tm value was found to be 71.0 ± 0.4 °C (Figure 1D), thus the G4 has the possibility of folding in physiological temperatures. As for the sequence with one nucleotide at the 3’ end (RAD17 3’ Tail), the CD spectrum was similar to that obtained for the RAD17 5’ Tail sequence (Figure S7) with a λmax ≈ 262 nm and λmin ≈ 240 nm. The Tm was found to be 68.6 ± 0.9 °C (Figure 1D). Thus, this sequence can adopt a parallel-stranded G4 with one nucleotide at either the 5’ or 3’ end of the sequence.
A final sequence without tails (RAD17 Tailless) was studied. Without nucleotides flanking the G4-forming sequence, there was a signal as a broad hump of undefined peaks between 10–12 ppm in the 1H-NMR spectrum (Figure 3A). The CD spectrum (Figure 3B) gave a λmax ≈ 262 nm and λmin ≈ 240 nm that indicates a parallel fold. The Tm value was found to be 76.6 ± 0.2 °C (Figure 1D), similar to the melting temperatures that have been obtained for the other sequences. The sequence also had a fluorescence enhancement value of 88.1 ± 7.1 Fl490nm/Flo (Figure S1). The results were expected considering that with one nucleotide on either end of the G4, the sequence adopted parallel folds. Without any nucleotides on either end, the sequence adopts a parallel fold.
Based on the results of the tail studies, we surmise that the tails have a major effect on the G4 folding pattern the PQSs can adopt. A change in tail length correlating with a change in folding of a G4 was previously observed for the human telomere sequence where a different number of nucleotides in overhanging tails would result in different hybrid folds.25 However, RAD17 is unique in a subtle change in the tail length can change the structure from a hybrid to a parallel-folded G4. This brings a cautionary note to those studying G4 structure in synthetic oligodeoxynucleotides; sequence truncation can potentially lead to erroneous interpretations.
Oxidation of the RAD17 promoter PQS
In addition to modifying the tail lengths of the RAD17 promoter PQS, studies to determine the impact that oxidation could have on the behavior of the sequence were conducted. The RAD17 4 Track sequence was exposed to the one-electron oxidant CO3•- and subsequent analysis via gel electrophoresis revealed hotspots for oxidation at G nucleotides at the 5’ face of the G runs as well as Gs in the loops. The data are consistent with previous studies finding similar hotspots of oxidation for G4 folds.16 A common G oxidation product from this reaction is 8-oxo-7,8-dihydroguanine (OG); 3,43,44 therefore, two sequence variations with oxidative modifications (Table 1) at one of these hotspots were selected to study: one with oxidation at the 5’-terminus (position 13) of a G run, and one with oxidation in a loop position (position 18). First, oxidation was site-specifically incorporated in the RAD17 PQS as OG in place of the guanine. After synthesis of the sequence containing OG, the DNA was purified and annealed in the same buffer previously described for collecting the CD spectra.
Initially, the CD spectra and Tm values (Figure S8) were obtained for the sequences with five domains. With oxidation at position 13 (core), there was a λmax ≈ 290 nm, λshoulder ≈ 262 nm and λmin ≈ 237 nm. Compared to the wild-type sequence, the signal at 290 nm increased, which may indicate a more hybrid-like structure in solution. On the other hand, the oxidation at position 18 (loop) gave a spectrum very similar to that of the five track sequence without damage with a λmax ≈ 264 nm, λshoulder ≈ 290 nm and λmin ≈ 238 nm. The CD spectra suggest that with the OG in a core position of the sequence, the folding is less polymorphic and a more defined fold is formed. Comparing the spectrum of position 13 to that of the four track sequence without damage, we found they were nearly identical. By introducing the damage at that position, the structure may adopt the same fold as the four-track sequence. Meanwhile, damage in a loop position (such as with position 18) does not appear to affect the folding pattern as the CD spectrum is similar to that of the five-track sequence without damage. These results concur with previous results found by our laboratory for the fifth track of the VEGF PQS in which we described how the presence of OG in a core position can cause a rearrangement of the G4 structure to include the fifth track instead of the track with the modification.16 The Tm values obtained for the sequences were >60 °C verifying that both sequences can fold into quadruplexes at physiological temperatures.
With the four-track sequence, modification at position 13 showed a CD spectrum (Figure S8) with λmax ≈ 288 nm, λshoulder ≈ 261 nm and λmin ≈ 238 nm. The modification at position 18 gave a similar spectrum (Figure S8) with a λmax ≈ 290 nm, λshoulder ≈ 266 nm and λmin ≈ 242 nm. Both of these spectra are similar to the one obtained for the wild-type four-track sequence without modification, indicating that OG at either position does not affect the folding pattern. However, the Tm values tell a different story. Both values are above physiological temperatures, but with an oxidative modification in the sequence, the values decreased compared to the original sequence. At position 13, the presence of OG decreased the Tm by almost 30 °C, while at position 18, the Tm decreased by about 6 °C (Figure S8). The large decrease in melting temperature at position 13 indicates that the presence of OG in the core position greatly destabilizes the G-quadruplex as the OG will not participate in the Hoogsteen base pairing of the tetrad.45 The small decrease at position 18 indicates that the damage in the loop position only marginally affects the fold considering the damage will not participate in the fold of the G4.
RAD17 Promoter PQS in a Human Cell Line
In the final set of studies, some of the G4-forming RAD17 sequences (Table 1) were inserted into the SV40 promoter sequence of the Renilla luciferase gene in a reporter plasmid to understand whether the folding of the G4 in human cells could have an effect on gene expression. The sequences were incorporated into the plasmid following a method that was previously reported by our laboratory. 15The firefly luciferase gene that was also present in the plasmid was not modified and was therefore used as an internal standard. The modified plasmids were then transfected into human glioblastoma cells (U87-MG). The luciferase expression levels were obtained and normalized to obtain relative response ratios (RRRs).
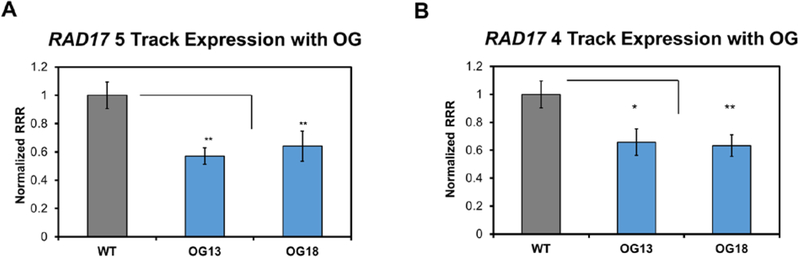
The RAD17 5 Track sequence was inserted into the plasmid, and to study the effect of oxidation on expression levels, RAD17 5 OG13 or RAD17 5 OG18-containing plasmids were similarly prepared. Compared to the normal wild-type (all guanine) G4 sequence, both positions of oxidative modification decreased expression of the Renilla luciferase gene (Figure 4A). Similar results were found for the RAD17 4 Track PQS when comparisons were made to sequences with OG at positions 13 and 18. Both positions decreased expression of the Renilla luciferase gene (Figure 4B) in the U87–MG cells. Our laboratory previously found that with the presence of a G-quadruplex containing OG or AP modifications in the template strand of a gene, such as with the RAD17 PQS that is natively in this strand, there was also a downregulation of expression of the gene.17 This concurred with previous reports that oxidation downregulates the expression of the RAD17 gene in U937 human lymphoma cancer cells.23 The magnitude of the downregulation is somewhat smaller with RAD17 construct compared to the previously studied VEGF promoter, possibly due to the less stable hybrid G4 fold of RAD17 compared to the parallel G4 of VEGF.17
Figure 4.
Expression levels of Renilla luciferase relative to firefly luciferase (RRR) measured for (A) the RAD17 5 Track sequence with and without oxidative modifications, and (B) the RAD17 4 Track sequence with and without oxidative modifications. Significance values for each comparison of data were calculated via a Student’s t test. Significance at *P < 0.05 or **P <0.01 is indicated.
CONCLUSIONS
In this report, we characterized the PQS that resides in the promoter region of the human RAD17 gene. The sequence was found to have the ability to fold to a G4 consistent with a hybrid-like topology under model physiological conditions. This was found to be different from other PQSs found in promoter regions that usually adopt parallel folds.20 Different nucleotide lengths of tails were also found to alter dramatically the folding pattern from hybrid-like to parallel topologies. With at least two nucleotides of the native sequence flanking both sides of the PQS, the G4 remained a hybrid-like fold. However, when less than two nucleotides were present on either the 5` or 3` sides, the folding of the G4 shifted to a parallel topology. The study with tail length serves as a cautionary note for any experiments performed on PQSs, including structural characterization by NMR, especially if conclusions are to be made based on the folding pattern of the G4 in physiological conditions.
Folding of the RAD17 sequence was also examined when either of two oxidation-prone Gs were synthetically substituted by OG, the common oxidation product of G. When OG was introduced into a core position (13), the G4 structure was hybrid-like and not very dynamic, while OG in a loop position (18) led to a less well-defined fold. Importantly, both points of modification still led to stably folded G-quadruplexes.
The RAD17 promoter PQS was then interrogated in human cells with or without the oxidatively modified base OG. The sequences were inserted into plasmids, transfected into human glioblastoma cells, and a luciferase assay was performed. The assay revealed that the sequences containing OG decreased the expression of luciferase compared to undamaged sequences, concurring with the cellular observation that in the presence of oxidative stress, the expression levels of RAD17 decreased.23 This work is an example of how a G-quadruplex in a promoter region of a gene coupled with the oxidative modification OG affects transcription, and this modification could possibly assist in regulation of gene expression via initiation of DNA repair.17
Supplementary Material
Acknowledgements
This work was supported by a National Cancer Institute Grant (R01 CA090689). The DNA strands were provided by the DNA/Peptide core facility at the University of Utah, which is supported in part by a NCI Cancer Center Support Grant (P30 CA042014). The authors are grateful to Y. Ding (University of Utah) for her helpful discussions regarding the cellular experiments.
Footnotes
Supporting Information
Copies of 1H-NMR spectra, CD spectra, ThT fluorescence, and Tm values for all RAD17 5 Track, G-to-T mutated sequences, and for sequences bearing 8-BrG and OG
Notes
The authors declare no competing financial interests in this work.
REFERENCES
- 1.Lonkar P, and Dedon PC (2011) Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int. J. Cancer 128, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wall SB, Oh J-Y, Diers AR, and Landar A (2012) Oxidative modification of proteins: an emerging mechanism of cell signaling. Front. Physiol 3, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadet J, Douki T, Gasparutto D, and Ravanat J-L (2003) Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res 531, 5–23. [DOI] [PubMed] [Google Scholar]
- 4.Gedik CM, and Collins A (2005) Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 19, 82–84. [DOI] [PubMed] [Google Scholar]
- 5.Fleming AM, and Burrows CJ (2017) 8-Oxo-7,8-dihydroguanine, friend and foe: epigenetic-like regulator versus initiator of mutagenesis. DNA Repair 56, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seifermann M, and Epe B (2017) Oxidatively generated base modifications in DNA: Not only carcinogenic risk factor but also regulatory mark? Free Radical Biol. Med 107, 258–265. [DOI] [PubMed] [Google Scholar]
- 7.Antoniali G, Malfatti MC, and Tell G (2017) Unveiling the non-repair face of the base excision repair pathway in RNA processing: A missing link between DNA repair and gene expression? DNA Repair 56, 65–74. [DOI] [PubMed] [Google Scholar]
- 8.Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, Borchert GM, and Gillespie MN (2015) An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell Mol. Physiol 309, L1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoniali G, Lirussi L, D’Ambrosio C, Dal Piaz F, Vascotto C, Casarano E, Marasco D, Scaloni A, Fogolari F, and Tell G (2014) SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol. Biol. Cell 25, 532–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba X, and Boldogh I (2016) Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase1-mediated epigenetic regulation of nuclear factor KappaB-driven gene expression. J. Biol. Chem 291, 25553–25566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, and Avvedimento EV (2008) DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206. [DOI] [PubMed] [Google Scholar]
- 12.Clark DW, Phang T, Edwards MG, Geraci MW, and Gillespie MN (2012) Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radical Biol. Med 53, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agrawal P, Hatzakis E, Guo K, Carver M, and Yang D (2013) Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 41, 10584–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, and Hurley LH (2008) The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 7, 880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming AM, Ding Y, and Burrows CJ (2017) Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A, 2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleming AM, Zhou J, Wallace SS, and Burrows CJ (2015) A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent. Sci 1, 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming AM, Zhu J, Ding Y, and Burrows CJ (2017) 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-quadruplex is an on-off switch for transcription. ACS Chem. Biol 12, 2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming AM, Zhu J, Ding Y, Visser JA, Zhu J, and Burrows CJ (2018) Human DNA repair genes possess potential G-quadruplex sequences in their promoters and 5’-untranslated regions. Biochemistry 57, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray RD, Buscaglia R, and Chaires JB (2012) Populated intermediates in the thermal unfolding of the human telomeric quadruplex. J. Am. Chem. Soc 134, 16834–16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kendrick S, and Hurley LH (2010) The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl. Chem 82, 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray LT, Vallur AC, Eddy J, and Maizels N (2014) G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol 10, 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, and Balasubramanian S (2016) G-quadruplex structures mark human regulatory chromatin. Nature Genetics 48, 1267. [DOI] [PubMed] [Google Scholar]
- 23.Islam MA, Thomas SD, Murty VV, Sedoris KJ, and Miller DM (2014) c-Myc quadruplex-forming sequence Pu-27 induces extensive damage in both telomeric and nontelomeric regions of DNA. J. Biol. Chem 289, 8521–8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming AM, and Burrows CJ (2013) G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol 26, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phan AT, Kuryavyi V, Luu KN, and Patel DJ (2007) Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 35, 6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feigon J, Koshlap KM, and Smith FW (1995) 1H NMR spectroscopy of DNA triplexes and quadruplexes, in Methods in Enzymology, pp 225–255. [DOI] [PubMed] [Google Scholar]
- 27.Adrian M, Heddi B, and Phan AT (2012) NMR spectroscopy of G-quadruplexes. Methods 57, 11–24. [DOI] [PubMed] [Google Scholar]
- 28.Piazza A, Adrian M, Samazan F, Heddi B, Hamon F, Serero A, Lopes J, Teulade-Fichou MP, Phan AT, and Nicolas A (2015) Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 34, 1718–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renaud de la Faverie A, Guédin A, Bedrat A, Yatsunyk LA, and Mergny J-L (2014) Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res. 42, e65-e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu S, Li Q, Xiang J, Yang Q, Sun H, Guan A, Wang L, Liu Y, Yu L, Shi Y, Chen H, and Tang Y (2016) Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Scientific Reports 6, 24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vorlíčková M, Kejnovská I, Sagi J, Renčiuk D, Bednářová K, Motlová J, and Kypr J (2012) Circular dichroism and guanine quadruplexes. Methods 57, 64–75. [DOI] [PubMed] [Google Scholar]
- 32.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, and Hurley LH (2004) The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc 126, 8702–8709. [DOI] [PubMed] [Google Scholar]
- 33.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, and Sasisekharan V (1992) Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 20, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Đapić V, Abdomerović V, Marrington R, Peberdy J, Rodger A, Trent JO, and Bates PJ (2003) Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 31, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kypr J, Kejnovská I, Renčiuk D, and Vorlíčková M (2009) Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karsisiotis AI, Hessari N. M. a., Novellino E, Spada GP, Randazzo A, and Silva M. W. d. (2011) Topological characterization of nucleic acid G‐quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed 50, 10645–10648. [DOI] [PubMed] [Google Scholar]
- 37.Lech Christopher J., Cheow Lim, Joefina K, Wen Lim, Jocelyn M, Amrane S, Heddi B, and Phan Anh T. (2011) Effects of site-specific guanine C8-modifications on an intramolecular DNA G-quadruplex. Biophys. J 101, 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dias E, Battiste JL, and Williamson JR (1994) Chemical probe for glycosidic conformation in telomeric DNAs. J. Am. Chem. Soc 116, 4479–4480. [Google Scholar]
- 39.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, and Katahira M (2007) Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J 274, 3545–3556. [DOI] [PubMed] [Google Scholar]
- 40.Karg B, and Weisz K (2018) Loop length affects syn-anti conformational rearrangements in parallel G-quadruplexes. Chemistry, 10.1002/chem.201801851. [DOI] [PubMed] [Google Scholar]
- 41.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, and Yang D (2006) Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luu KN, Phan AT, Kuryavyi V, Lacroix L, and Patel DJ (2006) Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc 128, 9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burrows CJ, and Muller JG (1998) Oxidative nucleobase modifications leading to strand scission. Chem. Rev 98, 1109–1152. [DOI] [PubMed] [Google Scholar]
- 44.Fleming AM, and Burrows CJ (2017) Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med 107, 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Fleming AM, Averill AM, Burrows CJ, and Wallace SS (2015) The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 43, 4039–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.