Abstract
The IMP family of RNA binding proteins, also named as insulin-like growth factor 2 (IGF2) mRNA-binding proteins (IGF2BPs), are highly conserved RNA regulators that are involved in many RNA processing stages, including mRNA stability, localization, and translation. There are three paralogs in the IMP family, IMP1-3, in mammals that all adopt the same domain arrangement with two RNA recognition motifs (RRM) in the N terminus and four KH domains in the C terminus. Here, we report the structure and biochemical characterization of IMP3 RRM12 and its complex with two short RNAs. These structures show that both RRM domains of IMP3 adopt the canonical RRM topology with two α-helices packed on an anti-parallel four stranded β-sheet. The spatial orientation of RRM1 to RRM2 is unique compared with other known tandem RRM structures. In the IMP3 RRM12 complex with RNA, only RRM1 is involved in RNA binding and recognizes a dinucleotide sequence.
Keywords: RNA-binding protein, RRM, X-ray crystallography
INTRODUCTION
RNA-binding proteins (RBPs) participate in all levels of gene expression by orchestrating the assembly and disassembly of distinct mRNA–protein complexes (mRNPs) that function at specific stages of a transcript's life to regulate transcription, splicing, export, localization, translation, and degradation (Müller-McNicoll and Neugebauer 2013; Singh et al. 2015). While the repertoire of domains that have been found to interact with RNA continues to increase, many RBPs contain a few well-characterized domains (Gerstberger et al. 2014; Castello et al. 2016). Among the most abundant of these domains are the RNA recognition motif (RRM) and hnRNP K-homology (KH) domain, which both predominantly recognize short RNA motifs (Query et al. 1989; Siomi et al. 1993). To compensate for this limited sequence specificity, the cooperative interaction of multiple RBDs within an RBP or multiple RBPs within an mRNP enables recognition of specific RNAs (Lunde et al. 2007; Hennig and Sattler 2015). Understanding how the higher order assembly of RNA–protein complexes integrates both sequence and structural information remains a central challenge in post-transcriptional regulation (Gronland and Ramos 2017).
The IMP RNA-binding proteins (IMP1, IMP2, and IMP3) are a conserved family of multidomain RBPs (two RRMs and four KH domains) that have been found to regulate RNA localization, translation, and stability (Degrauwe et al. 2016b). Their diversity of function is exemplified by the multiple independent discoveries (Vg1RBP/Vera, IMP1-3, CRD-BP, KOC, ZBP1, and IGF2BP1-3) of these RBPs in a variety of experimental systems (Deshler et al. 1997; Müeller-Pillasch et al. 1997; Ross et al. 1997; Doyle et al. 1998; Havin et al. 1998; Nielsen et al. 1999). The IMPs are highly expressed during development and are down-regulated postnatally, though IMP2 expression is maintained in adult tissues, but are often reexpressed or amplified in numerous cancers and correlate with poor survival (Bell et al. 2013; Degrauwe et al. 2016b). Recent work has also demonstrated a direct role for the IMP family in promoting oncogenic transformation (JnBaptiste et al. 2017).
The six RBDs of the IMP proteins are arranged into three pairs RRM1 + RRM2 (RRM12), KH1 + KH2 (KH12), and KH3 + KH4 (KH34) that are separated by flexible linkers. The interaction between IMP1 (ZBP1) KH34 and the zipcode cis-acting element of the β-actin 3′UTR has been extensively biochemically and structurally characterized (Chao et al. 2010; Patel et al. 2012; Nicastro et al. 2017). While the isolated KH34 domain is capable of high affinity and specific RNA recognition, less is known concerning how the RRM12 and KH12 domains contribute to the function of the full-length proteins. Several genome-wide studies of IMP family members have identified RNAs that are bound in vivo; however, a complete understanding of how IMP family members recognize and regulate their respective target mRNAs has not yet been achieved (Hafner et al. 2010; Conway et al. 2016; Degrauwe et al. 2016a; Ennajdaoui et al. 2016).
Here, we report the crystal structures of unbound IMP3 RRM12 domain and its complex with RNA. These structures show that both of the RRM1 and RRM2 domains adopt canonical RRM folds (β1α1β2β3α2β4); however, only the RNA-binding surface of RRM1 is accessible. Biochemical experiments show that IMP3 RRM12 recognizes short cytosine and adenine sequences with modest affinity. The structure of IMP3 RRM12 bound to ACAC and CCCC oligonucleotides confirmed that the RNA-binding interface is exclusively comprised of residues within RRM1, and RNA-binding does not induce a conformational change in the orientation of RRM1 to RRM2. These findings provide insight into the function of the N-terminal RRM domains of the IMP family of RBPs.
RESULTS AND DISCUSSION
Crystal structure of IMP3 RRM12
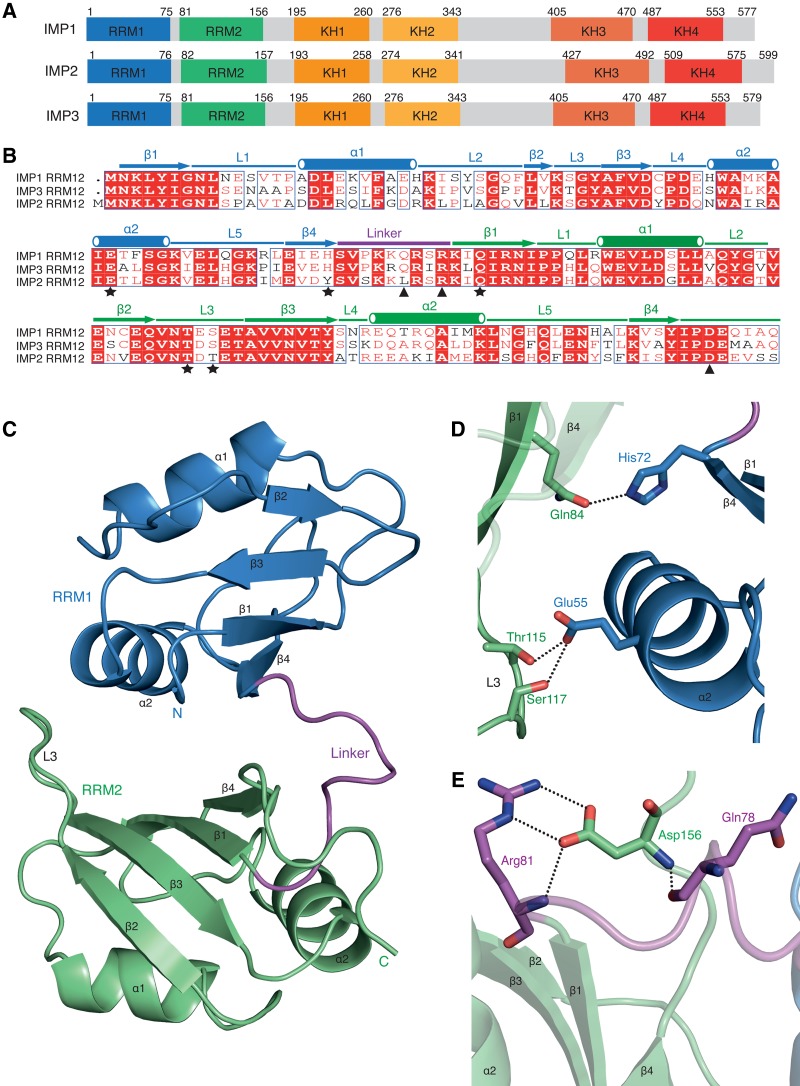
While the role of the C-terminal KH domains (KH34) of ZBP1/IMP1 in recognition of RNA has been extensively biochemically and structurally characterized, much less is known about the function of N-terminal RRM domains (RRM12) and the middle KH domains (KH12) (Fig. 1A). To better understand the function of RRM12, we expressed and purified IMP3 RRM12 (residues 1–161) from bacteria with an N-terminal maltose binding protein (MBP) tag and a C-terminal His6 tag. After removal of the MBP-tag by TEV protease, IMP3 RRM12 was found to be a monomer in solution (Supplemental Fig. S1A). Crystals of IMP3 RRM12 were obtained that belonged to the space group P1 and diffracted to 1.3 Å resolution (Supplemental Table 1). The structure was solved by molecular replacement using an NMR model of the IMP3 RRM2 domain (PDB 2E44). There are two copies of IMP3 RRM12 in the crystallographic asymmetric unit and the electron density allowed complete modeling of IMP3 RRM12 (residues 1–161, chain A and residues 1–159, chain B) as well as an N-terminal serine residue, which remained after TEV cleavage, and part of the C-terminal His-tag (chain A). Both structures of IMP3 RRM12 are nearly identical with a root-mean-square deviation (RMSD) of 0.47 Å for 159 Cα atoms.
FIGURE 1.
Structure of IMP3 RRM12 domain. (A) Schematic diagram showing conserved domain arrangement of IMP-family members. (B) Sequence alignment of RRM12 domains of IMP1-3. Identical residues (white), highly conserved (red), and not conserved (black) are depicted. Residues involved in the RRM1-RRM2 interface (star) and the linker-RRM2 (C terminus) interactions (triangle) are shown. (C) Domain orientation of IMP3 RRM12. RRM1 (blue), RRM2 (green), and linker (purple) with secondary structures labeled. The β-sheet surface of RRM1 is exposed to solvent, while the β-sheet surface of RRM2 is occluded by RRM1. (D) The hydrogen bonds formed in the interface of RRM1 and RRM2. (E) Hydrogen bonds and salt bridges formed between residues of linker and RRM2 (C terminus).
In the IMP3 RRM12 structure, both of the individual RRM1 and RRM2 domains adopt the canonical RRM-fold with β1α1β2β3α2β4 topology, which positions the two α-helices over an anti-parallel four-stranded β-sheet (Fig. 1C; Afroz et al. 2015). The tandem RRM domains form a compact structure that is stabilized by an interface of ∼700 Å2 of buried surface area. In RRM1, the canonical RNA-binding surface formed by β1 (RNP2 motif) and β3 (RNP1 motif) are solvent accessible (Fig. 1C). The N terminus of RRM1 and RRM1 α2 pack against the surface formed by the β1 and β3 strands of RRM2, which occludes the potential RNA-binding interface of RRM2 (Fig. 1C, Supplemental Fig. S1B). Gln84 within RRM2 β1, which is usually a conserved phenylalanine or tyrosine residue within the RNP2 motif, makes a hydrogen bond with His72 in RRM1 β4 (Fig. 1B,D). Also, in the interface of RRM1 and RRM2, hydrogen bonds formed between Glu55 on RRM1 α2 and Thr115, Ser117 on Loop3 of RRM2 contribute to the stabilization of this conformation (Fig. 1D). The orientation of RRM2 relative to RRM1 is further stabilized by the C terminus of RRM2 that is threaded through a pocket formed by the linker that connects RRM1 and RRM2. Hydrogen bonds formed between Gln78, Arg81, and Asp156 as well as two salt bridges between the sidechains of Arg81 and Asp156 also stabilize this conformation (Fig. 1C,E). In addition, a dense network of interactions among the linker residues, which are conserved within the IMPs, contributes to the conformation of the IMP3 RRM12 domain (Supplemental Figs. S1C, S2).
The arrangement of the tandem IMP3 RRM domains is unique compared to previously determined structures of proteins containing multiple RRMs (Supplemental Fig. S3). The orientation of RRM1 to RRM2 in FBP-interacting repressor (FIR) has the highest structural similarity (RMSD 3.07 Å) and also closely packs the α2 helix of RRM1 onto the β-sheet surface of RRM2, though it is rotated by ∼45° (Supplemental Fig. S3; Cukier et al. 2010). Interestingly, the structure of FIR RRM12 does not change upon nucleic acid binding and, consequently, only RRM1 is involved in DNA recognition.
RNA-binding affinity and sequence specificity of IMP3 RRM12
In order to identify the RNA-binding sequence specificity of IMP3 RRM12, we performed an in vitro SELEX (systematic evolution of ligands by exponential enrichment) experiment using an RNA library containing 14 randomized nucleotides. RNA sequences were selected for by binding to MBP-IMP3 RRM12 that was immobilized on amylose resin. While the structure of IMP3 RRM12 suggested that only RRM1 is accessible for RNA-binding, a 14-nucleotide random sequence was used in the SELEX experiment. This length RNA would potentially be able to interact with RRM2, if the conformation we observed in the crystal structure was dynamic in solution, or identify any noncanonical RNA-binding interfaces within IMP3 RRM12. After nine rounds of selection, the enriched RNA population was determined by sequencing (Supplemental Table 2). A majority (80%) of the sequences contained a stretch of pyrimidine nucleotides of at least three with CCC being the most common. The other sequences all contained a CC dinucleotide within the randomized nucleotides; however, this sequence was also present in the constant regions used for PCR amplification. IMP3 RRM12 recognition of only a short RNA sequence is consistent with electrophoretic mobility shift assays that demonstrated only weak binding affinity for the selected RNA population during the course of the selection (data not shown).
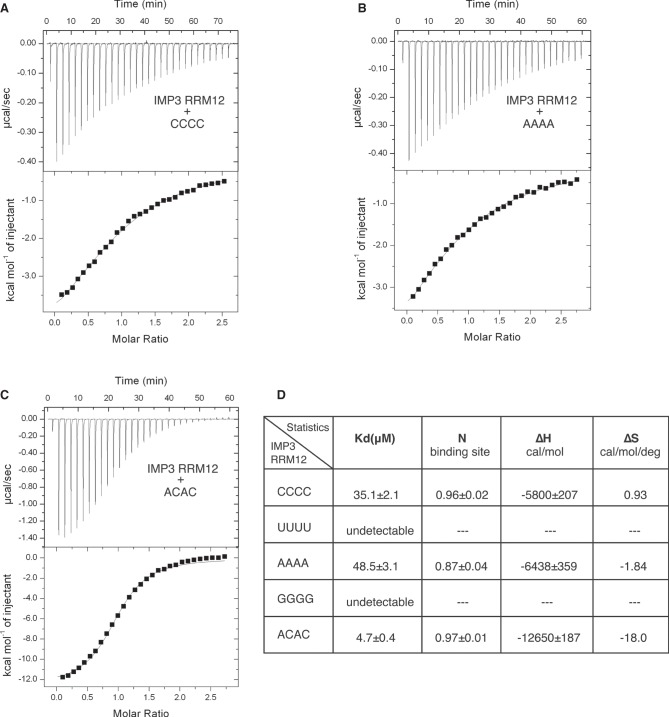
Since our SELEX experiments suggested that IMP3 RRM12 recognizes only short RNA sequences, we measured the binding affinity of IMP3 RRM12 to a short RNA oligonucleotide UUCCCG (∼30 µM), one of the most abundant SELEX sequences (Supplemental Table 2), using isothermal titration calorimetry (ITC) (Supplemental Fig. S4A). To further characterize its RNA-binding specificity, we measured the affinity of IMP3 RRM12 to RNA oligonucleotides containing all four homopolymers (CCCC, UUUU, AAAA, GGGG). IMP3 RRM12 bound to CCCC and AAAA with similar, but, weak affinity (∼40 µM) (Fig. 2A,B,D). We did not detect the binding, however, to either UUUU or GGGG indicating modest sequence specificity (Supplemental Fig. S4B,C). IMP3 RRM12 bound to ACAC with almost an order of magnitude higher affinity (∼5 µM) indicating its preference for this dinucleotide sequence (Fig. 2C,D). Multiple CA dinucleotides were pres-ent in the constant regions of the SELEX RNAs, which may have prevented the selection from enriching for this sequence within the randomized regions. We also measured IMP3 RRM12 binding to longer RNA sequences that revealed similar sequence preferences but could also potentially provide multiple protein binding sites (Supplemental Fig. S5).
FIGURE 2.
Binding affinity determination of IMP3 RRM12 with RNA. Titration of IMP3 RRM12 into CCCC (A), AAAA (B), and ACAC (C), monitored by ITC. The top panel shows titration experiment plotted as heat (µcal sec−1) versus time (min), and the lower panel shows binding isotherm with the integrated heat (kcal mol−1) of IMP3 RRM12 versus the molar ratio of IMP3 RRM12 to RNA. The continuous line shows the fit of the data to a one-site binding model. (D) Summary of ITC measurements of IMP3 RRM12 binding to above RNAs.
Crystal structures of IMP3 RRM12 in complex with ACAC and CCCC RNA
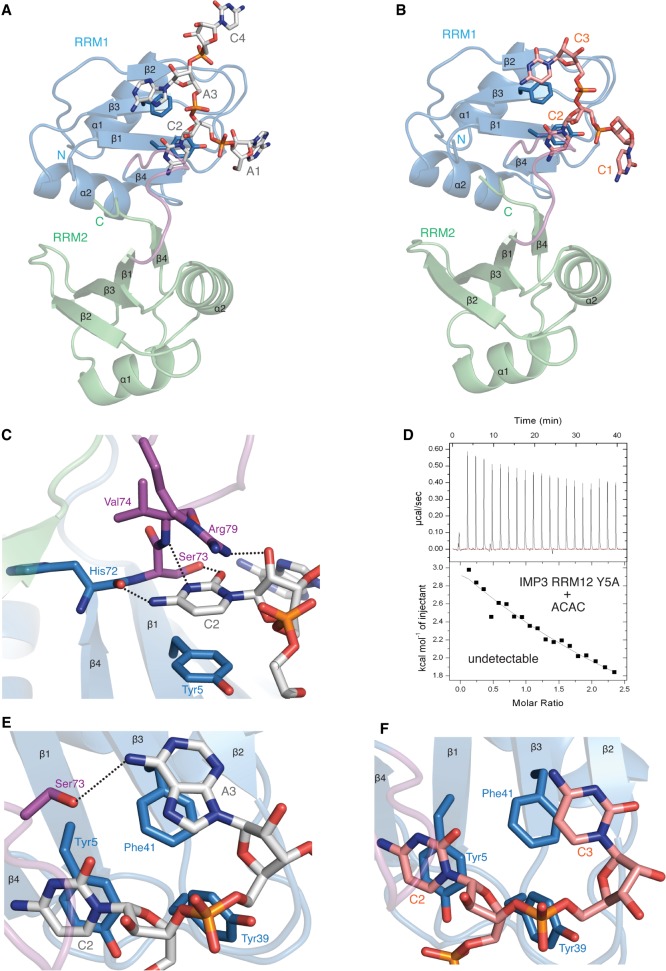
To understand the structural basis of IMP3 RRM12 recognition of RNA, we performed cocrystallization trials with short RNA sequences. Crystals were obtained of IMP3 RRM12 in complex with either ACAC or CCCC that diffracted to 2.0 Å and 2.1 Å resolution, respectively. The IMP3 RRM12 + ACAC crystals belonged to a trigonal space group and had one copy of the complex in the asymmetric unit, while the IMP3 RRM12 + CCCC crystals had the same space group and unit cell dimensions as the unbound IMP3 RRM12 crystals (Supplemental Table 1). For the IMP3 RRM12 complex with ACAC, the RNA electron density allowed building of the first three nucleotides and the backbone of C4, while for the CCCC structure, only three cytosine nucleotides were visible in only one of the two copies of IMP3 RRM12 in the asymmetric unit (Fig. 3A,B; Supplemental Fig. S6). Crystal lattice contacts prevent RNA from binding to the other copy of IMP3 RRM12. IMP3 RRM12 in the free (residues 1–159) and RNA-bound (residues 1–157) structures are very similar (RMSD of 0.57 Å with CCCC and 0.78 Å with ACAC, for 157 Cα atoms) indicating that RNA-binding does not induce a conformational change between the RRM1 and RRM2 domains.
FIGURE 3.
The structure of IMP3 RRM12 in complex with ACAC and CCCC RNAs. (A) Overall structure of IMP3 RRM12 and RNA ACAC. RRM1 (blue), RRM2 (green), linker (purple), and RNA (in sticks) are shown. Only RRM1 is involved in RNA binding. (B) Overall structure of IMP3 RRM12 and RNA CCCC. (C) Contact details between RRM1 β-sheet and cytosine C2 in IMP3 RRM12 and ACAC complex. (D) IMP3 RRM12 Y5A mutant disrupts the interaction between IMP3 RRM12 and RNA ACAC. (E) In the complex of IMP3 RRM12 with ACAC, the base of A3 stacks on the sidechain of Phe41 and forms a hydrogen bond with Ser73 on IMP3 RRM12 linker. (F) In the complex of IMP3 RRM12 with CCCC, the base of C3 stacks on the sidechain of Phe41.
The canonical RNA-binding interface of RRM1 formed by β1 (RNP2) and β3 (RNP1) is involved in recognition of the two central nucleotides, either C2A3 or C2C3 (Fig. 3A,B). The cytosine nucleotide (C2) is involved in most of the RNA–protein contacts and interacts with two conserved tyrosine residues (Tyr5 in RNP2 and Tyr39 in RNP1) in both structures. The cytosine base stacks on the aromatic ring of Tyr5, similar to the binding of other RRM domains to pyrimidines in this position of the RNP2 motif (Fig. 3C; Afroz et al. 2015). Mutation of Tyr5 to an alanine abolishes binding to the ACAC RNA indicating that this residue is critical for RNA-binding (Fig. 3D). The aromatic side chain of Tyr39 (RNP1) also makes van der Waals contacts to the C2 backbone, as well as inserting into the space between the sugars of the second and third nucleotides (Fig. 3E,F). The position of C2 is further stabilized by a network of hydrogen bonds. His72, Ser73, and Val74, which are located in the linker between RRM1 and RRM2, make hydrogen bonds to the base of C2 (Fig. 3C). Additionally, the guanidinium group of the Arg79 sidechain makes a hydrogen bond to the 2′OH of the C2 sugar (Fig. 3C). Almost all of these interacting residues are conserved among IMP family members suggesting that this RNA-binding specificity may also be shared (Supplemental Fig. S2). The only exception is His72, which is a tyrosine in IMP2; however, it is the backbone carbonyl of this amino acid that hydrogen bonds to the C2 base (Fig. 3C; Supplemental Fig. S2). The bases of the third nucleotides (A3 and C3) both stack upon the aromatic ring of Phe41 within the RNP1 motif; however, A3 forms an additional hydrogen bond with the hydroxyl group of Ser73 (Fig. 3E,F). The increased stacking interaction of A3 and its additional hydrogen bond likely account for the increased affinity measured for ACAC. The A1 and C1 nucleotides in the two structures both make contacts with residues from symmetry-related copies of IMP3 RRM12, and these interactions are artifacts of crystal packing (data not shown).
Our structural and biochemical characterization of IMP3 RRM12 contributes to the understanding of how multidomain RBPs function in recognition of specific transcripts. While the isolated IMP3 tandem RRM domains recognize RNA with only modest sequence specificity and affinity, these interactions may be functionally important in the context of the full-length protein bound to its RNA targets. Recently, the importance of RNA conformation and dynamics in ZBP1 (IMP1) binding to the β-actin 3′-UTR was previously described (Woods et al. 2017). The interactions of RRM12 and KH12 with RNA may alter or stabilize specific RNA structures when coupled to the high affinity and sequence-specific interactions of KH34, which could nucleate the assembly of a larger mRNP. In the yeast ASH1 transport mRNP, the cooperative assembly of She2p and She3p to the ASH1 transcript is necessary for transport, which highlights the importance of multiple low-affinity interactions in stabilizing specific RNA–protein complexes (Edelmann et al. 2017). Interestingly, IMP1 RRM12 has been shown to bind the motor KIF11 while IMP3 can interact with KIF20A (Taniuchi et al. 2014; Song et al. 2015). An important future goal will be to understand how RNA-binding of the RRM12 domain may influence the interaction of IMP family members with other proteins.
MATERIALS AND METHODS
Protein preparation
IMP3 RRM12 (residues 1–161) and the Y5A mutant were cloned by PCR into a derivative of pMalc (New England BioLabs) that contains a Tobacco Etch virus (TEV) protease site after the maltose-binding protein (MBP) tag. A C-terminal His6 tag was added by PCR to ensure purification of full-length fusion proteins. The constructs were transformed into Escherichia coli strain Rosetta2 (EMD Bio-sciences), and recombinant protein was induced with 1 mM IPTG for 4 h at 37°C. Cell pellets were resuspended in lysis buffer (50 mM Tris pH 7.5, 1.5 M NaCl, 1 mM EDTA, 1 mM DTT) supplemented with one Complete EDTA-free protease inhibitor tablet (Roche), and were lysed by sonication. Cell debris was removed by centrifugation, and the soluble fusion protein was purified by amylose affinity chromatography (New England BioLabs) followed by Ni-NTA affinity chromatography and size exclusion column (SEC) S200 (GE Healthcare). The protein concentrations were calculated by measuring the absorbance at 280 nm and using extinction coefficients determined by ProtParam (Gasteiger et al. 2005).
Crystallization and structure determination of IMP3 RRM12 and IMP3 RRM12 + RNA complex
IMP3 RRM12 (0.75 mM) was crystallized using sitting-drop vapor diffusion at 20°C by mixing equal volumes of the protein (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT) and reservoir solution (0.2 M KNO3, 20% PEG3350). The crystals were cryoprotected by soaking in reservoir solution supplemented with 25% ethylene glycol before flash-cooling in liquid nitrogen. Data were collected to 1.3 Å resolution from a single crystal at the Swiss Light Source X06DA beamline at a wavelength of 0.9795 Å. The diffraction data were indexed, integrated, and scaled using MOSFLM and the CCP4 suite of programs (Battye et al. 2011; Winn et al. 2011).
IMP3 RRM12 (0.75 mM) in complex with RNA, CCCC, or ACAC (1.12 mM), were also crystallized using sitting-drop vapor diffusion at 20°C by mixing equal volumes of the complex (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT) and reservoir solution (100 mM Bis–Tris pH 6.2, 0.2 M MgCl2, 25% PEG3350 for RNA CCCC, and 0.2 M KH2PO4, 20% PEG3350 for RNA ACAC). Crystals were cryoprotected by soaking in reservoir solution supplemented with 20% ethylene glycol before flash-cooling in liquid nitrogen. Data were collected to 2.1 Å resolution for complex with CCCC and 2.0 Å resolution for complex with ACAC from a single crystal at the Swiss Light Source X06DA beamline at a wavelength of 0.9999 Å. The diffraction data of IMP3 RRM12 + CCCC were indexed, integrated, and scaled using XDS (Kabsch 2010), while the complex data of IMP3 RRM12 + ACAC is processed by MOSFLM.
All of the structures of IMP3 RRM12 in free and RNA-bound forms were determined by molecular replacement with Phaser using a truncated version of the NMR model of IMP3 RRM2 (PDB 2E44) (McCoy et al. 2007). Rounds of refinement and model building were carried out with Phenix and Coot (Emsley and Cowtan 2004; Adams et al. 2010). TLS-refinement was performed in the initial rounds of refinement with domains corresponding to RRM1 and RRM2.
Isothermal titration calorimetry
ITC measurements were performed on an ITC200 Micro calorimeter (MicroCal) at 25°C. IMP3 RRM12, IMP3 RRM12 Y5A mutant and the synthesized RNAs (four nucleotide containing oligos were ordered from Dharmacon, eight and seven nucleotide containing oligos were ordered from IDT) were dialyzed in 20 mM HEPES pH 8.0, 150 mM NaCl, and 1 mM TCEP buffer. The titrations were carried out by injecting 35.2 µL aliquots of the protein (0.9 mM) into RNA (0.065 mM for all four and eight nucleotide oligos, 0.045 mM for RNA ACACACA) at time intervals of 2 min to ensure that the titration peak returned to the baseline. The titration data were analyzed using the program Origin7.0 (MicroCal).
RNA selection by SELEX using a random library
A 14-random nucleotides library with the constant sequence for PCR amplification 5′-GGGAATGGATCCACATCTACGA-(N14)-TTCACTGCAGACTTGACGAAGCTT-3′ was prepared by chemical synthesis (Microsynth). Fourteen nucleotides, positions 23–36, were randomized to 25%A, 25%T, 25%G, and 25%C. The dsDNA library preparation and the random RNA pool used for initial selection were performed as described previously (Chao et al. 2010). The concentration of MBP-IMP3 RRM12 for the first three rounds was 1 µM, and the RNA pool concentration was also 1 µM. From round four to nine, the protein concentration decreased to 500 nM, while the RNA concentration kept 1 µM. RNA from the third, sixth and ninth round of selection and the random RNA pool were fluorescein-labeled, and their affinity for MBP–IMP3 RRM12 was quantified by EMSA (data not shown).
SEC-MALS (multi-angle light scattering)
Purified IMP3 RRM12 (in buffer 20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT) was concentrated to 5 mg ml−1 and loaded to a Superdex 200 10/300 GL gel-filtration column (GE Healthcare) equilibrated in 20 mM Tris pH 7.5, 200 mM NaCl, 0.02% w/v NaN3, and 1 mM DTT. Light scattering was recorded with an in-line miniDAWN TREOS three angle light scattering detector (Wyatt Technology), and protein concentration was detected using an in-line Optilab T-rEX refractive index detector (Wyatt Technology). The molecular mass of IMP3 RRM12 was calculated using ASTRA 6 software (Wyatt Technology).
DATA DEPOSITION
The coordinates of IMP3 RRM 12 (6FQ1) and IMP3 RRM12 + RNA complex (6FQR and 6GX6) have been deposited in the Protein Data Bank.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Novartis Research Foundation (J.A.C.), Swiss National Science Foundation (SNF) grant 31003A_156477 (J.A.C.), and the EMBO Young Investigator Program (J.A.C.). We thank J. Keusch (FMI) for crystallography support, Nicolas Thomä (FMI) for the valuable input, and the staff of the Swiss Light Source X06DA beamline, Paul Scherrer Institute (Villingen) for assistance with data collection.
Author contributions: M.J. performed all experiments. H.G. assisted with structural biology. M.J. and J.A.C wrote the manuscript with input from H.G.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.065649.118.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afroz T, Cienikova Z, Cléry A, Allain FH. 2015. One, two, three, four! How multiple RRMs read the genome sequence. Methods Enzymol 558: 235–278. [DOI] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. 2013. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci 70: 2657–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW. 2016. Comprehensive identification of RNA-binding domains in human cells. Mol Cell 63: 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. 2010. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev 24: 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AE, Van Nostrand EL, Pratt GA, Aigner S, Wilbert ML, Sundararaman B, Freese P, Lambert NJ, Sathe S, Liang TY, et al. 2016. Enhanced CLIP uncovers IMP protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Rep 15: 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukier CD, Hollingworth D, Martin SR, Kelly G, Díaz-Moreno I, Ramos A. 2010. Molecular basis of FIR-mediated c-myc transcriptional control. Nat Struct Mol Biol 17: 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrauwe N, Schlumpf TB, Janiszewska M, Martin P, Cauderay A, Provero P, Riggi N, Suvà ML, Paro R, Stamenkovic I. 2016a. The RNA binding protein IMP2 preserves glioblastoma stem cells by preventing let-7 target gene silencing. Cell Rep 15: 1634–1647. [DOI] [PubMed] [Google Scholar]
- Degrauwe N, Suvà ML, Janiszewska M, Riggi N, Stamenkovic I. 2016b. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev 30: 2459–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Schnapp BJ. 1997. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276: 1128–1131. [DOI] [PubMed] [Google Scholar]
- Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J. 1998. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res 26: 5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann FT, Schlundt A, Heym RG, Jenner A, Niedner-Boblenz A, Syed MI, Paillart JC, Stehle R, Janowski R, Sattler M, et al. 2017. Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex. Nat Struct Mol Biol 24: 152–161. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Ennajdaoui H, Howard JM, Sterne-Weiler T, Jahanbani F, Coyne DJ, Uren PJ, Dargyte M, Katzman S, Draper JM, Wallace A, et al. 2016. IGF2BP3 modulates the interaction of invasion-associated transcripts with RISC. Cell Rep 15: 1876–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook (ed. Walker J). pp. 571–607. Humana Press, NY. [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronland GR, Ramos A. 2017. The devil is in the domain: understanding protein recognition of multiple RNA targets. Biochem Soc Trans 45: 1305–1311. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK. 1998. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev 12: 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig J, Sattler M. 2015. Deciphering the protein-RNA recognition code: combining large-scale quantitative methods with structural biology. Bioessays 37: 899–908. [DOI] [PubMed] [Google Scholar]
- JnBaptiste CK, Gurtan AM, Thai KK, Lu V, Bhutkar A, Su MJ, Rotem A, Jacks T, Sharp PA. 2017. Dicer loss and recovery induce an oncogenic switch driven by transcriptional activation of the oncofetal Imp1-3 family. Genes Dev 31: 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müeller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, Varga G, Friess H, Büchler M, Beger HG, et al. 1997. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene 14: 2729–2733. [DOI] [PubMed] [Google Scholar]
- Müller-McNicoll M, Neugebauer KM. 2013. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 14: 275–287. [DOI] [PubMed] [Google Scholar]
- Nicastro G, Candel AM, Uhl M, Oregioni A, Hollingworth D, Backofen R, Martin SR, Ramos A. 2017. Mechanism of β-actin mRNA recognition by ZBP1. Cell Rep 18: 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19: 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, et al. 2012. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 26: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Bentley RC, Keene JD. 1989. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell 57: 89–101. [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Pratt G, Yeo GW, Moore MJ. 2015. The clothes make the mRNA: past and present trends in mRNP fashion. Annu Rev Biochem 84: 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res 21: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Zheng Y, Wang Y, Katz Z, Liu X, Chen S, Singer RH, Gu W. 2015. Specific interaction of KIF11 with ZBP1 regulates the transport of β-actin mRNA and cell motility. J Cell Sci 128: 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi K, Furihata M, Saibara T. 2014. KIF20A-mediated RNA granule transport system promotes the invasiveness of pancreatic cancer cells. Neoplasia 16: 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CT, Lackey L, Williams B, Dokholyan NV, Gotz D, Laederach A. 2017. Comparative visualization of the RNA suboptimal conformational ensemble in vivo. Biophys J 113: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.