Abstract
Stem–loop binding protein (SLBP) is required for replication-dependent histone mRNA metabolism in mammals. Zebrafish possesses two slbps, and slbp1 is necessary for retinal neurogenesis. However, the detailed expression and function of slbp2 in zebrafish are still unknown. In this study, we first identified zebrafish slbp2 as an oocyte-specific maternal factor and then generated a maternal-zygotic slbp2 F3 homozygous mutant (MZslbp2Δ4−/−) using CRISPR/Cas9. The depletion of maternal Slbp2 disrupted early nuclear cleavage, which resulted in developmental arrest at the MBT stage. The developmental defects could be rescued in slbp2 transgenic MZslbp2Δ4−/− embryos. However, homozygous mutant MZslbp1Δ1−/− developed normally, indicating slbp1 is dispensable for zebrafish early embryogenesis. Through comparative proteome and transcriptome profiling between WT and MZslbp2Δ4−/− embryos, we identified many differentially expressed proteins and genes. In comparison with those in WT embryos, four replication-dependent histones, including H2a, H2b, H3, and H4, all reduced their expression, while histone variant h2afx significantly increased in MZslbp2Δ4−/− embryos at the 256-cell stage and high stage. Zebrafish Slbp2 can bind histone mRNA stem–loop in vitro, and the defects of MZslbp2Δ4−/− embryos can be partially rescued by overexpression of H2b. The current data indicate that maternal Slbp2 plays a pivotal role in the storage of replication-dependent histone mRNAs and proteins during zebrafish oogenesis.
Keywords: slbp2, replication-dependent histones, ribosomal proteins, maternal effect, oogenesis, early embryogenesis
INTRODUCTION
Replication-dependent histones are tightly regulated in the cell cycle and are necessary for packaging the newly replicated chromosomal DNA (Gunjan et al. 2006; Marzluff et al. 2008; Marzluff and Koreski 2017). In metazoan, replication-dependent histone mRNAs are not polyadenylated but instead terminated with a conserved stem–loop (SL), which possesses similar functions of poly(A) tail in translational efficiency and mRNA stability (Hentschel and Birnstiel 1981; Jackson and Standart 1990; Marzluff 1992). Stem–loop binding protein (SLBP) plays a central role at all steps of histone mRNA metabolism (Bernstein and Ross 1989; Gallie et al. 1996) via interactions between the SL and the RNA binding domain (RBD) of SLBP (Dominski et al. 2001).
SLBP, also known as hairpin-binding protein (HBP), was initially and partially purified from Xenopus oocyte nuclear extracts (Hanson et al. 1996), then identified from human, mouse, and frog by using yeast three-hybrid systems (Wang et al. 1996; Martin et al. 1997). During mouse oocyte growth and maturation, SLBP accumulates and regulates the synthesis of replication-dependent histones (Allard et al. 2002, 2005), which are the only known targets (Townley-Tilson et al. 2006). Due to insufficient histones H3 and H4, most of the mouse embryos derived from SLBP-depleted oocytes arrested at the two-cell stage (Arnold et al. 2008). The conserved functions of SLBP in histone mRNA metabolism and early embryo development were also revealed in Drosophila (Sullivan et al. 2001) and Caenorhabditis elegans (Pettitt et al. 2002).
Two SLBPs, named as xSLBP1 and xSLBP2, were identified from Xenopus oocytes and showed differential expression during oogenesis and early embryogenesis. xSLBP1 is the mammalian ortholog of SLBP and participates in pre-mRNA processing, while xSLBP2 is oocyte-specific and is supposed to be involved in storage (Wang et al. 1999). xSLBP2 has no effect on translation (Sánchez and Marzluff 2002) and is degraded, paralleling the activation of histone mRNA translation after oocyte maturation (Sánchez and Marzluff 2004). Considering that only the SLBP2 pseudogene is found in mouse, human, and rabbit genomes (Thelie et al. 2012), the sole SLBP appears to regulate replication-dependent histone synthesis in these mammalian animals (Allard et al. 2002, 2005). However, the SLBP2 transcripts were isolated from some other mammals, such as bovine, rat, dog, horse, and pig. Additionally, bovine SLBP2 and xSLBP2 display differences in their expression and localization during oogenesis (Thelie et al. 2012). Although the functions of SLBP in histone mRNA metabolism have been well revealed (Marzluff and Koreski 2017), the reports about SLBP2 are very scarce (Wang et al. 1999; Thelie et al. 2012; Liu et al. 2015) and the function of SLBP2 is still mostly unknown.
Zebrafish (Danio rerio) is an important vertebrate model animal for research on development and disease (Grunwald and Eisen 2002). In zebrafish slbp1 mutant, retinal neurogenesis is severely delayed (Imai et al. 2014). Recently, we identified oocyte-specific slbp2 from gibel carp (Carassius gibelio) (Gui and Zhou 2010; Zhou and Gui 2017) and analyzed its dynamic expression pattern during oogenesis and embryogenesis (Liu et al. 2015). Imai et al. (2014) also detected maternal slbp2 in early zebrafish embryos. However, the detailed expression pattern and function of slbp2 in early embryogenesis have not been revealed. In this study, we first characterized zebrafish Slbp2 as an oocyte-specific maternal protein. Then its role in zebrafish oogenesis and early embryogenesis was revealed by knockout using CRISPR/Cas9. Finally, we confirmed the expression changes of histone proteins influenced by Slbp2 deficiency and explored the interaction between replication-dependent histones and Slbp2.
RESULTS
Oocyte-specific and maternal expression pattern of zebrafish slbp2
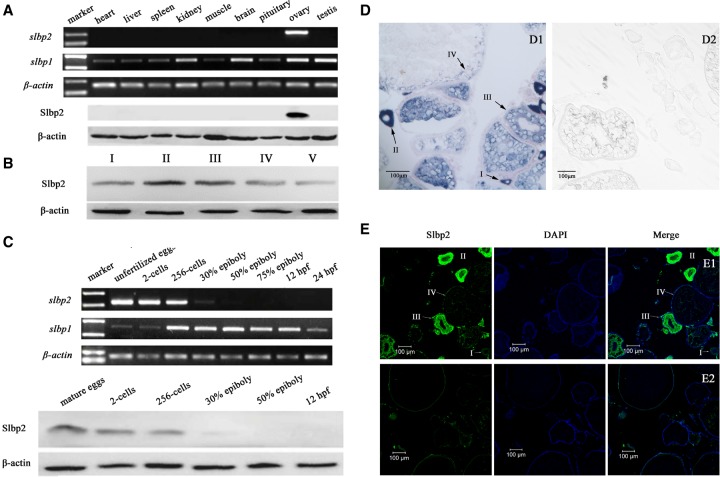
Both zebrafish slbp2 transcripts and proteins were specifically detected in the ovary, not in other analyzed tissues (Fig. 1A). During oogenesis, Slbp2 was expressed in primary growth oocytes (stage I), reached a peak in cortical alveolar oocytes (stage II), and then decreased from vitellogenic stage oocytes (stage III). After oocyte maturation, the Slbp2 protein was slightly reduced in mature eggs (stage V) (Fig. 1B). Consistent with Cgslbp2 in gibel carp (Liu et al. 2015), zebrafish slbp2 transcripts and proteins were detected only in unfertilized eggs and early embryos before the 50% epiboly stage (Fig. 1C), indicating that zebrafish Slbp2 is a maternal factor and no zygotic product is synthesized during embryogenesis. The distribution of zebrafish slbp2 in the ovary is also the same as that of Cgslbp2 in gibel carp (Liu et al. 2015). Zebrafish slbp2 was specifically expressed in the oocytes, not in somatic cells surrounding oocytes (Fig. 1D,E). The slbp2 transcript signals were intensively observed in the cytoplasm of primary growth oocytes (I), cortical alveolus oocytes (II), and vitellogenic oocytes (III), and subsequently reduced and concentrated in peripheral cytoplasm of maturing oocytes (IV) (Fig. 1D). The protein signals were weak in primary growth oocytes (I), and distributed strongly and uniformly in the cytoplasm of cortical alveolar oocytes (II). Along with the oocyte growth, Slbp2 began to reduce its abundance in vitellogenic oocytes (III). In maturing oocytes (IV), the signal was faint owing to the reduction and diffusion of Slbp2 protein along with oocyte growth (Fig. 1E). The data indicate that zebrafish Slbp2 is a maternal factor expressed specifically in oocytes.
FIGURE 1.
Dynamic expression pattern of zebrafish slbp2 during oogenesis and embryogenesis. (A) slbp2 and slbp1 expression in adult tissue detected by RT-PCR and Slbp2 protein detected by western blot. (B) Slbp2 expression in oocytes during oogenesis detected by western blot. (C) slbp2 and slbp1 expression in embryos during embryogenes is detected by RT-PCR and Slbp2 protein detected by western blot. (D) slbp2 mRNA localization during oogenesis. Ovarian cross sections after in situ hybridization with antisense (D1) or sense (D2) slbp2 probe. (E) Slbp2 protein localization in zebrafish mature ovary. Ovarian cross-section was stained with DAPI and anti-Slbp2 antibody (E1) or preimmune serum (E2). I, primary growth stage; II, cortical alveolus stage; III, vitellogenic stage; IV, maturing oocyte stage; V, mature eggs.
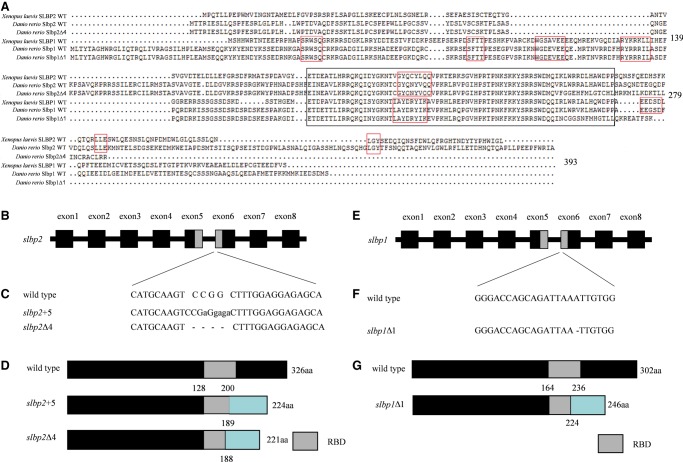
Although multiple protein sequence alignments showed that zebrafish Slbp2 possessed very low identities to zebrafish Slbp1 (16.9%), xSLBP1 (18.1%), and xSLBP2 (19.0%), the RBD domain of zebrafish Slbp2 showed 64.4% to 71.23% identities to RBD domains of zebrafish Slbp1, xSLBP1, and xSLBP2 (Fig. 2A). The region “CGYQNYVQQ” in the RBD domain responsible for Slbp2 being inactive in processing is conserved between zebrafish Slbp2 and xSLBP2, while the same region in zebrafish Slbp1 (LAYDKYIKA) distinctly differs from zebrafish Slbp2, which is conserved in xSLBP1 and critical for processing (Dominski et al. 2001). In comparison with xSLBP1, zebrafish Slbp1 also contains several conserved elements, including “SFTT” necessary for cell cycle regulation of xSLBP1 (Zheng et al. 2003), “WGDEVEE” required for translation of histone mRNA (Sánchez and Marzluff 2002), “SRWSQ,” and “RYRRRIL.” In addition, we also identified the conserved motifs “LLE” and “LGY” between zebrafish Slbp2 and xSLBP2.
FIGURE 2.
The establishment of zebrafish slbp2 and slbp1 knockout mutant lines by CRISPR/Cas9. (A) The multiple amino sequence alignments of Xenopus laevis SLBPs (xSLBP1 and xSLBP2) and zebrafish Slbps (Slbp1 WT, Slbp1Δ1, Slbp2 WT, and Slbp2Δ4). The RBD domain of the SLBPs is shown by a black rectangle, and other conserved motifs are shown by a red rectangle. (B,E) The target sites of zebrafish slbps. The exons and introns are depicted as rectangular boxes and thick lines. (C,F) Sequences of WT slbps and slbps mutations. (D,G) Schematic representation of WT Slbps proteins and mutated Slbps proteins. The RNA binding domain (RBD) is indicated by the gray box. The different protein sequences between WT Slbps and mutated Slbps are shown by the blue rectangular box.
Different from the oocyte-specific expression pattern of zebrafish slbp2, zebrafish slbp1 was ubiquitously expressed in adult tissues, and abundant transcripts were detected in ovary, testis, kidney, and brain (Fig. 1A). In addition, a few of maternal slbp1 transcripts were detected in unfertilized eggs, and the zygotic mRNA slbp1 level was strikingly activated at the mid-blastula transition and maintained a relatively stable expression level during the following embryogenesis embryos (Fig. 1C).
Establishment of zebrafish slbp2 knockout mutant lines
To explore the function of slbp2, we generated slbp2 gene-disrupted zebrafish using CRISPR/Cas9. The targeting site was chosen in the sixth exon to disrupt the RBD domain (Fig. 2B). The sequencing results showed several types of mutations (data not shown), of which two mutated types modified at the target site; a 4-bp deletion (slbp2Δ4) and a 5-bp insertion (slbp2+5) (Fig. 2C) were chosen to construct mutant lines. The mutations in slbp2Δ4 and slbp2+5 caused a reading frame shift and created a premature stop codon. The two slbp2 knockout mutant lines both altered the RBD domain (Fig. 2A,D).
Considering that slbp2 is a maternal factor, three different kinds of mutants, including zygotic slbp2 F2 homozygous mutants (Zslbp2Δ4−/− and Zslbp2+5−/−), maternal slbp2 F3 heterozygous mutants (Mslbp2Δ4+/− and Mslbp2+5+/−), and maternal-zygotic slbp2 F3 homozygous mutants (MZslbp2Δ4−/− and MZslbp2+5−/−), were generated (Fig. 3). Owing to the same phenotypes observed in slbp2Δ4 and slbp2+5 mutant lines, we chose slbp2Δ4 to perform the following experiments. We first examined Zslbp2Δ4−/− phenotypes by crossing heterozygous F1 female (slbp2Δ4+/−) and male (slbp2Δ4+/−) (Fig. 3A). Zebrafish Zslbp2Δ4−/− developed normally and grew to fertile adults. Subsequently, Mslbp2Δ4+/− and MZslbp2Δ4−/− were obtained by crossing female Zslbp2Δ4−/− with WT male or female Zslbp2Δ4−/− with male Zslbp2Δ4−/− (Fig. 3A). In contrast to WT and Zslbp2Δ4−/−, both Mslbp2Δ4+/− and MZslbp2Δ4−/− arrested embryo development at mid-blastula transition (MBT, 3.5 hpf) and finally died. Additionally, the transcripts of zebrafish slbp2 were hardly detected in MZslbp2Δ4−/− at the 256-cell stage and high stage by qPCR (Fig. 3B), which might be due to nonsense mediated decay. Moreover, embryos of three slbp1 mutants (Zslbp1Δ1−/−, Mslbp1Δ1+/−, and MZslbp1Δ1−/−) constructed by the similar strategy of CRISPR/Cas9 developed normally (Fig. 2E–G). The results indicate that maternal Slbp2, not Slbp1, is indispensable for the very early zebrafish embryogenesis. Thus, we focused our study on the comparison between MZslbp2Δ4−/− and WT before MBT.
FIGURE 3.
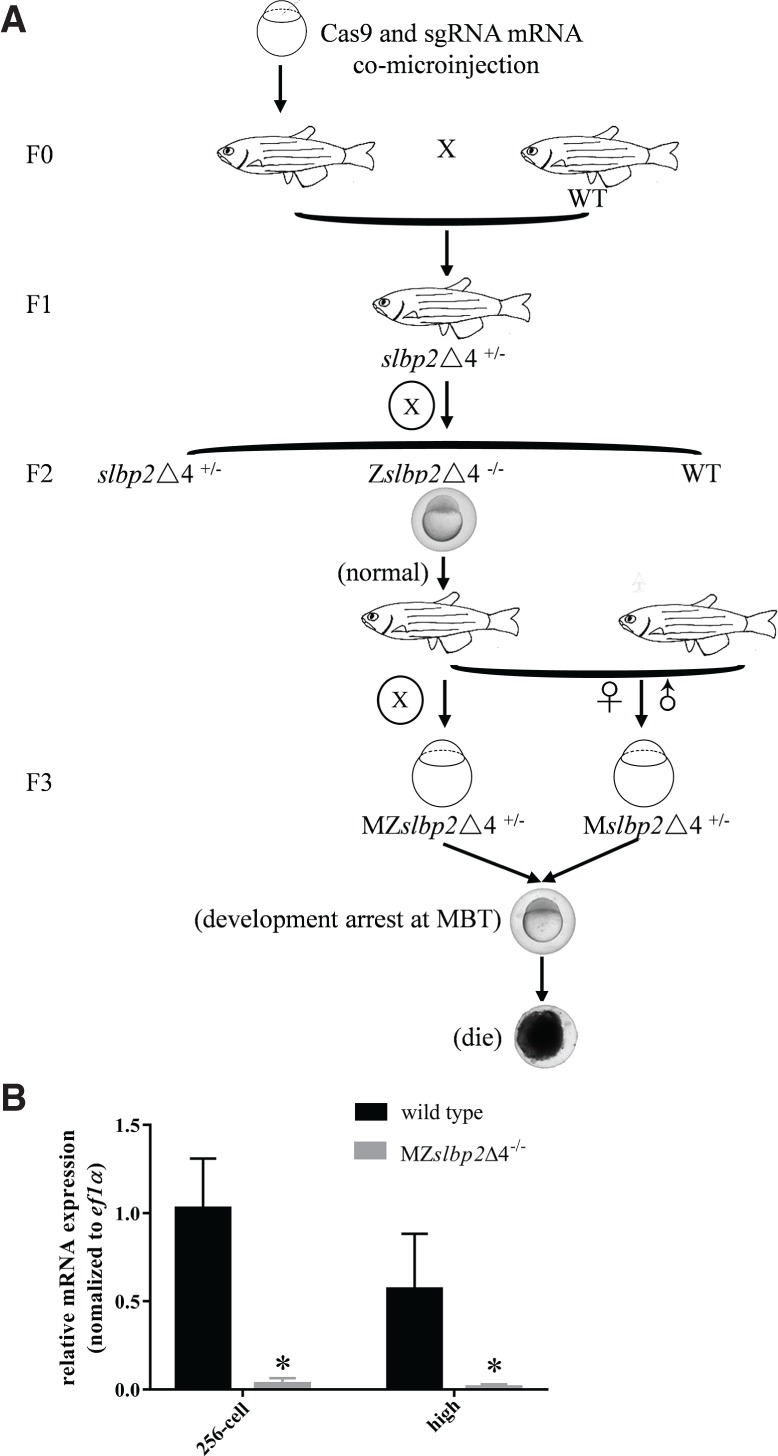
Flowchart and establishment of three zebrafish slbp2 mutant lines. (A) Flowchart and phenotypes of three zebrafish slbp2 mutant lines. (B) The relative expression of slbp2 in MZslbp2Δ4−/− and wild-type embryos at the 256-cell and high stages detected by qPCR. Zslbp2Δ4−/−: zygotic slbp2 F2 homozygous mutant; Mslbp2Δ4+/−: maternal slbp2 F3 heterozygous mutant; MZslbp2Δ4−/−: maternal-zygotic slbp2 F3 homozygous mutant.
Maternal Slbp2 depletion disrupts early nuclear cleavage
In comparison with WT zebrafish (Fig. 4A), MZslbp2Δ4−/− embryos appeared to develop normally during cleavage stage and blastula stage, but they did not initiate epiboly and arrested at the MBT stage (Fig. 4B), and all embryos died after 8 hpf. In order to visualize the dynamic cell behavior, in vitro transcribed H2B-GFP mRNAs were injected into the one-cell WT and MZslbp2Δ4−/− embryos, respectively. The first and second cycles of MZslbp2Δ4−/− embryos appeared somewhat normal, but they showed obvious disruptions of nuclear cleavage from the eight-cell stage. The mitotic nuclei in WT and MZslbp1Δ1−/− embryos condensed at anaphase (Fig. 4F,H), while the nuclei in MZslbp2Δ4−/− embryos had aberrant morphologies, forming a bridge connecting the two mitotic daughter nuclei (Fig. 4G).
FIGURE 4.
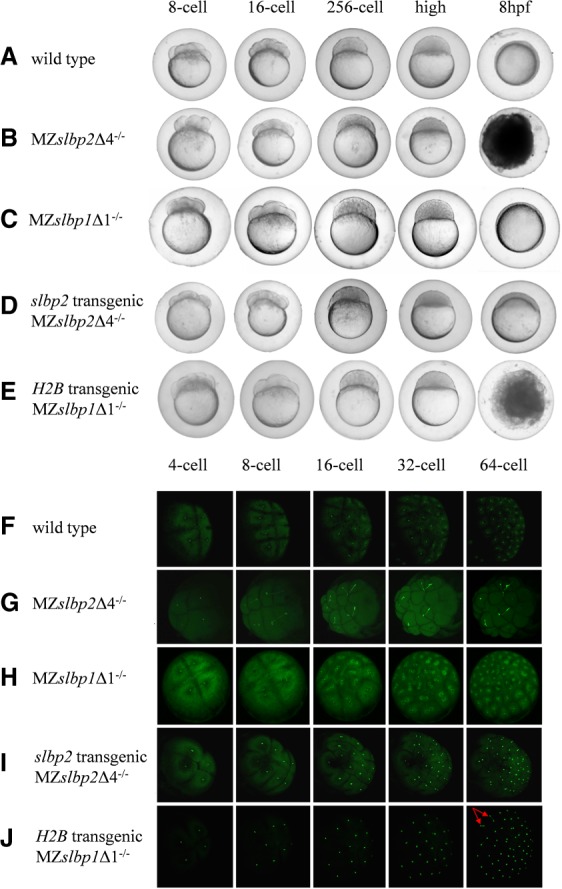
Comparisons of embryogenesis (A–E) and nuclear cleavage (F–J) among WT (A,F), MZslbp2Δ4−/− (B,G), MZslbp1Δ1−/− (C,H), slbp2 transgenic MZslbp2Δ4−/− embryos (D,I), and H2B-GFP transgenic MZslbp2Δ4−/− embryos (E,J). In vitro transcribed H2B-GFP mRNAs were injected into the one-cell embryos to visualize the dynamic nuclear cleavage (F–J). The developmental stages of embryos are marked at the top.
To confirm the defects in MZslbp2Δ4−/− embryos caused by the deficiency of maternal Slbp2, plasmid slbp2-GFP containing slbp2 ORF driven by β-actin promoter (Wang et al. 2001) was injected into the one-cell embryos produced by crossing heterozygous F1 female and male (slbp2Δ4+/−), and the female Zslbp2Δ4−/− mutants were screened by PCR. As described above, Zslbp2Δ4−/− developed and normally spawned (Fig. 3A). Subsequently, 10 females of slbp2 transgenic Zslbp2Δ4−/− fish were randomly selected to cross with males of Zslbp2Δ4−/−. Owing to the germline mosaic pattern in F0 transgenic fish, only ∼2%–15% of one-cell embryos exhibited the GFP fluorescent, indicating that these embryos expressed Slbp2. Compared to the MZslbp2Δ4−/− embryos, all of slbp2 transgenic MZslbp2Δ4−/− embryos completed epiboly (Fig. 4D), which suggested that the defects of early developmental arrest might be rescued by the expression of Slbp2. However, only 30%–55% of these embryos developed normally and grew to fertile adults. The others grew abnormally and showed a series of morphological deformations, including yolk sac deformity, head or tail hypoplasia, or growth retardation (data not shown). Additionally, the nuclear cleavage of slbp2 transgenic MZslbp2Δ4−/− exhibited normally (Fig. 4I). The injection of in vitro transcribed slbp2-GFP mRNAs or plasmid slbp2-GFP into MZslbp2Δ4−/− could not rescue the defects (data not shown). Therefore, the very early developmental arrest and the disruption of early nuclear cleavage in MZslbp2Δ4−/− embryos may have resulted from the specific depletion of maternal Slbp2.
Comparative proteomics and transcriptome profiling between WT and MZslbp2Δ4−/− embryos
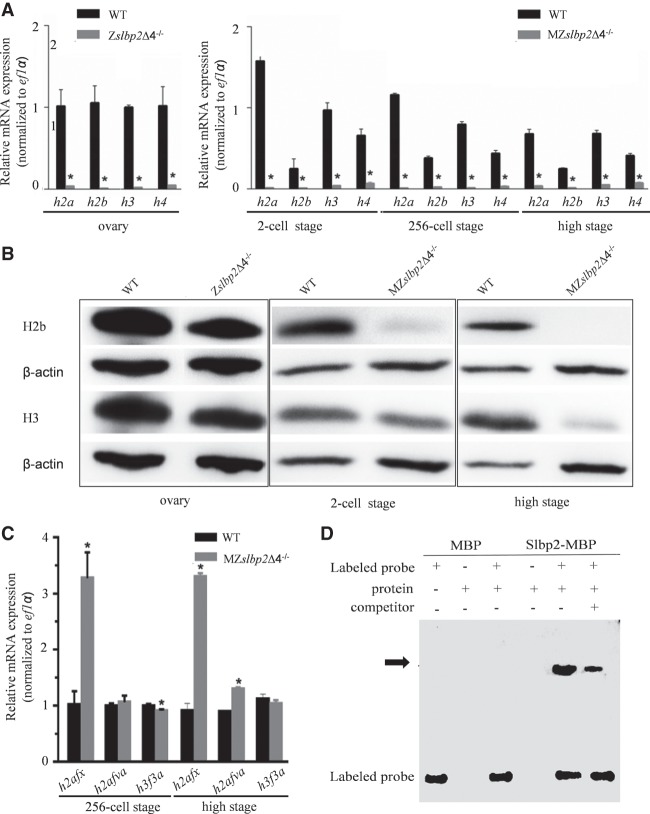
To reveal the proteins influenced by slbp2 deficiency, iTRAQ was used to assess proteome changes between WT and MZslbp2Δ4−/− embryos at the 256-cell stage (MZslbp2Δ4−/−-256 versus WT-256) and high stage (MZslbp2Δ4−/− -high versus WT-high), respectively. A total of 2944 proteins were identified and annotated in six public databases (Supplemental Table S1). We applied a threshold of >1.2-fold change and a P-value of <0.05 to identify differentially expressed proteins (DEPs). Additionally, only DEPs that were detected in at least two replicates were defined as significant DEPs. Based on these three criteria, 38 and 41 DEPs were identified from MZslbp2Δ4−/−-256 versus WT-256 and MZslbp2Δ4−/−-high versus WT-high, respectively (Supplemental Table S2). A fish-egg, lectin-like and histone H2a were the most highly down-regulated proteins in MZslbp2Δ4−/−-256 versus WT-256 and MZslbp2Δ4−/−-high versus WT-high, respectively. Thirteen commonly down-regulated DEPs were screened, including H2a, H4, vitellogenin 3, tubulin, alpha 4–like, and others. Compared to their contents in WT embryos, H2a reduced by two-thirds and H4 decreased by one-third, while H3 slightly reduced expression. Additionally, H2b was also detected only in one of three biological replicates and displayed the reduction by 60%. Eosinophil chemotactic cytokine, the glycyl-tRNA synthetase, and acyl-CoA dehydrogenase medium chain were commonly up-regulated in MZslbp2Δ4−/− embryos at the 256-cell and high stages compared to WT embryos.
Subsequently, we performed comparative transcriptome analysis of MZslbp2Δ4−/−-256 versus WT-256 and MZslbp2Δ4−/−-high versus WT-high, respectively. Three biological replicates were carried out. In comparison with WT embryos at the 256-cell and high stages, a total of 165 and 142 differentially expressed genes (DEGs) were respectively revealed in MZslbp2Δ4−/− embryos at corresponding stages (Supplemental Table S3). Zebrafish slbp1 kept similar expression levels between WT and MZslbp2Δ4−/− embryos at the 256-cell stage (Supplemental Table S4). Owing to the absence of poly(A) tails, replication-dependent histones were not credibly and repeatedly detected from transcriptomic profiles obtained by using the Illumina HiSeq 4000 platform. Four transcripts of h2a variants and five transcripts of h3 variants were screened from transcriptomic profiles. Compared to WT embryos, only h2afx expression increased up to 7.36- and 9.13-fold in MZslbp2Δ4−/− embryos at the 256-cell stage and high stage, respectively (Supplemental Table S4).
Dynamic expression changes of histones between WT and MZslbp2Δ4−/− embryos
To confirm dynamic expression changes of histones between WT and Zslbp2Δ4−/− ovary, and WT and MZslbp2Δ4−/− embryos, the replication-dependent histones and histone variants identified from proteomic and transcriptomic profiles were selected to perform qPCR analyses. Compared to their expression levels in wild-type ovary, zebrafish h2a, h2b, h3, and h4 mRNAs all largely decreased, while many histone proteins were detected in Zslbp2Δ4−/− ovary (Fig. 5A). In comparison with their expression levels in wild-type embryos, traces of zebrafish h2a, h2b, h3, and h4 mRNA were detected in MZslbp2Δ4−/− embryos at the two-cell, 256-cell, and high stages by qPCR (Fig. 5A). The results of western blot confirmed the large reduction of H2b and H3 proteins in MZslbp2Δ4−/− embryos at two-cell and high stages (Fig. 5B). In contrast, the expression levels of h2afx mRNA in MZslbp2Δ4−/− embryos increased up to 3.2- and 3.6-fold at the 256-cell stage and high stage, respectively. Additionally, other histone variants, such as h2afva and h3f3a, kept similar expression levels between WT and MZslbp2Δ4−/− embryos (Fig. 5C).
FIGURE 5.
Zebrafish Slbp2 regulates the storage of histone mRNAs and proteins during zebrafish oogenesis. (A) Relative expression by qPCR detection of four replication-dependent histones in WT and Zslbp2Δ4−/− ovaries, or WT and MZslbp2Δ4−/− embryos at the two-cell, 256-cell, and high stages. ef1α was used as the control. Each bar represents mean ± SD (n = 3). Asterisks (*) indicate significant differences (P < 0.05) between WT and Zslbp2Δ4−/− ovaries, or WT and MZslbp2Δ4−/− embryos. Data were performed from three independent experiments. (B) Western blot detection of H2b and H3 proteins in WT and Zslbp2Δ4−/− ovaries, and WT and MZslbp2Δ4−/− embryos at two-cell stage and high stage. The developmental stages of embryos are marked at the bottom. (C) Relative expression by qPCR detection of three histone variants in WT and MZslbp2Δ4−/− embryos at the 256-cell stage and high stage. ef1α was used as the control. Each bar represents mean ± SD (n = 3). Asterisks (*) indicate significant differences (P < 0.05) between WT and MZslbp2Δ4−/− embryos. (D) EMSA analysis of the binding ability between Slbp2 and SL of H2a mRNA.
Maternal Slbp2 is required for replication-dependent histone storage in zebrafish oogenesis
The significant reduction of replication-dependent histones (Fig. 5A,B) suggests that maternal Slbp2 might be required for replication-dependent histone mRNA and protein metabolism during zebrafish oogenesis and early embryogenesis. To confirm the assumption, the binding ability of zebrafish Slbp2 with the SL of replication-dependent histone mRNAs was assessed. Replication-dependent histone mRNAs share a conserved SL (Marzluff et al. 2008). Therefore, the SL of zebrafish h2a mRNA was selected to perform an electrophoretic mobility shift assay (EMSA) to determine its binding ability with a recombinant Slbp2 protein with MBP tag in vitro. As shown in Figure 5D, the Slbp2-MBP fusion protein could bind to the biotin-labeled h2a mRNA SL, and the excessive amount of unlabeled competitors effectively competed for this binding. Under the same condition, MBP protein did not bind to h2a mRNA SL. These results indicate that zebrafish Slbp2 can interact with the SL of replication-dependent histone mRNAs.
To verify whether or not the defects of MZslbp2Δ4−/− embryos can be rescued by overexpression of replication-dependent histones, the plasmid H2B-GFP containing human H2B ORF driven by β-actin promoter was injected into the one-cell embryos produced by crossing heterozygous F1 female and male (slbp2Δ4+/−). The H2B transgenic Zslbp2Δ4−/− and MZslbp2Δ4−/− fish were screened by the similar procedure used in the establishment of slbp2 transgenic Zslbp2Δ4−/− and MZslbp2Δ4−/− fish. Compared to the WT embryos (Fig. 4A), all of the H2B transgenic MZslbp2Δ4−/− embryos exhibited similar defects to MZslbp2Δ4−/− embryos (Fig. 4B) and also did not initiate epiboly (Fig. 4E). However, the defects of nuclear cleavage in H2B transgenic MZslbp2Δ4−/− embryos started at the 64-cell stage (Fig. 4J), which is later than that observed in MZslbp2Δ4−/− embryos at the eight-cell stage (Fig. 4G). The nuclei bridges were formed in a few mitotic daughter cells of H2B transgenic MZslbp2Δ4−/− embryos at the 64-cell stage (Fig. 4J). The injection of in vitro transcribed replication-dependent histone mRNAs into MZslbp2Δ4−/− could not rescue the defects (data not shown), indicating that maternal H2b might partially rescue the defects of MZslbp2Δ4−/− embryos. The results confirm the role of Slbp2 in the storage of replication-dependent histone mRNAs and proteins during zebrafish oogenesis.
DISCUSSION
The replication-dependent histone synthesis is cell cycle-regulated and is tightly controlled by the regulation of the SLBP level during the cell cycle in mammals (Marzluff and Koreski 2017). In this study, we first characterized zebrafish slbp2 as an oocyte-specific maternal factor by analyzing its dynamic expression pattern during oogenesis and embryogenesis (Fig. 1). Then, three slbp2 mutants, Zslbp2Δ4−/−, Mslbp2Δ4+/−, and MZslbp2Δ4−/−, were constructed by CRISPR/Cas9 (Figs. 2B–D, 3). To reveal the function of maternal Slbp2, the detailed comparison between WT and MZslbp2Δ4−/− embryos, such as early nuclear cleavage, proteome and transcriptome profiling, and dynamic expression of replication-dependent histones and histone variants were carried out. Similar to the defects in C. elegans, Drosophila, and mice SLBP-depleted embryos (Sullivan et al. 2001; Pettitt et al. 2002; Arnold et al. 2008), MZslbp2Δ4−/− embryos arrested development at MBT, which was caused by the abnormalities of nuclear morphology and behavior in early cleavage cells (Fig. 4). Subsequently, we observed a dramatic reduction of H2a and H2b, a moderate decrease of H3 and H4, and a significant increase of h2afx expression in Zslbp2Δ4−/− ovary and MZslbp2Δ4−/− embryos (Fig. 5A–C). The partial rescue by maternal H2b in MZslbp2Δ4−/− embryos might be due to the compensation of some histone variants, such as H2afx, and the residual H3 and H4 protein in MZslbp2Δ4−/− embryos. Combined with the confirmation of the binding ability between Slbp2 and h2a SL (Fig. 5D), we suggest that maternal Slbp2 should be required for early nuclear cleavage and replication-dependent histone expression in zebrafish embryogenesis.
The knockout of maternal Slbp1 had no effect on zebrafish early embryogenesis and nuclear cleavage (Fig. 4C,G). In the zebrafish rw440 homozygous mutant, a nonsense mutation occurred at the third amino acid of Slbp1. At 3 dpf, the rw440 mutant showed slow retinal cell proliferation and delay of retinal neurogenesis (Imai et al. 2014). The normal early embryogenesis of rw440 mutant and MZslbp1Δ1−/− indicate that they might be hypomorphs. Interestingly, zebrafish slbp1 and slbp2 showed obvious differences in their expression (Fig. 1), and the zygotic mRNA slbp1 level was strikingly activated at the MBT (Fig. 1C). In zebrafish, zygotic genome activation (ZGA) coincides with the MBT (Vastenhouw et al. 2010; Lindeman et al. 2011). However, the maternal factors that regulate ZGA remain largely unknown in vertebrates (Tadros and Lipshitz 2009; Andersen et al. 2013). The function of slbp1 in ZGA awaits further investigation. Nonetheless, our results indicate that slbp2, not slbp1, plays a conserved and pivotal role in maintaining normal early nuclear cleavage (proper chromatin assembly).
Replication-dependent histones play an essential role in mitotic chromosome condensation (de la Barre et al. 2000). The defect of early nuclear cleavage and chromosome condensation in zebrafish MZslbp2Δ4−/− embryos might be due to deficiency of replication-dependent histones. In Zslbp2Δ4−/− ovary, mRNAs of replication-dependent histones all largely decreased, while a lot of histone proteins were detected (Fig. 5A,B). Since they are not dividing, the other cell types in the ovary will not produce histone mRNAs but have normal levels of histone protein. This accounts for the large drop in histone mRNAs in the Zslbp2Δ4−/− ovary, while there is still a lot of histone proteins present. qPCR and western blot confirmed the large reduction of replication-dependent histones in MZslbp2Δ4−/− embryos (Fig. 5A,B). The results indicate that the reduction of histone proteins in the Zslbp2Δ4−/− ovary is likely due to failure of the oocyte to produce and store histone proteins. A small quantity of H3 proteins detected by western blot might be the H3.3 variant histones. Because of the replication-dependent histone deficiency, the DNA in cleavage cells could not be appropriately packaged into normal chromatin. Consequently, this probably led to the failure of chromosome condensation and impaired nuclear division (Fig. 4G). Compared to WT embryos, the C. elegans and mouse SLBP-depleted embryos had much less H3 and H4 (Pettitt et al. 2002; Arnold et al. 2008), and the protein level of H3 also decreased in the zebrafish slbp1 mutant rw440 embryos at 2 and 3 dpf (Imai et al. 2014). The Drosophila dSLBP mutant embryos showed similar defects observed in MZslbp2Δ4−/− and accumulated abnormal polyadenylated H3 and H4 mRNAs (Sullivan et al. 2001). Surprisingly, the total amount of H2a and H2b in mouse SLBP-depleted embryos was similar to WT embryos (Arnold et al. 2008), which indicates that SLBP is not necessary for storage of H2a and H2b in mouse oocytes (Arnold et al. 2008). In Xenopus, the storage of replication-dependent histone mRNAs and proteins during oogenesis and its subsequent uses during embryogenesis have been extensively studied (Woodland 1980). Xenopus SLBP2 is oocyte-specific and is supposed to be involved in storage and translational repression of histone mRNA (Wang et al. 1999; Sánchez and Marzluff 2002, 2004). Our results suggest that Slbp2 should be required for the accumulation of four replication-dependent histones in zebrafish oocytes (Fig. 5A,B), which may reflect the Slbp regulatory mechanism differences in histone metabolism among mammal, amphibian, and teleost.
Interestingly, a total of nine histone variants were screened from transcriptomic profiles, and the only h2afx was up-regulated expression in MZslbp2Δ4−/− embryos at the 256-cell stage and high stage, respectively (Fig. 5C). H2afx, also known as H2ax, is a variant of histone H2a and represents 2%–25% of total H2a (Rogakou et al. 1998). The phosphorylated form of H2AFX (γ-H2AFX) plays a critical role in DNA repair and maintenance of genomic integrity (Rogakou et al. 1998; Bassing et al. 2002; Celeste et al. 2002, 2003a,b; Yuan et al. 2010; Testa et al. 2018). In addition, H2afx is also required for the chromatin remodeling of sex chromosomes and meiotic silencing in mouse (Fernandez-Capetillo et al. 2003; Cloutier et al. 2015). The accumulation of γ-H2afx was observed in XO female mice (Baarends et al. 2005), and the ablation of H2afx could restore oocyte numbers in XO females to XX WT levels (Cloutier et al. 2015). In our previous study, an oocyte-specific histone H2a variant H2af1o was identified in gibel carp and showed significantly higher mobility in nucleosomes than ubiquitous H2afx (Wu et al. 2009). Moreover, H2af1o deficiency disturbed cell synchrony in early cleavages before MBT (Yue et al. 2013). Dramatic global chromatin remodeling events occur in oocyte maturation and early embryogenesis, including changes in histone biosynthesis, modification, and exchange (Becker et al. 2005). The reason why h2afx increases expression levels in zebrafish MZslbp2Δ4−/− embryos is unclear. The increased expression of h2afx in MZslbp2Δ4−/− embryos indicated that a more polyadenylated form of this mRNA was produced and might function in the absence of Slbp2.
In summary, this work confirms slbp2 as the key player in teleost histone storage in oogenesis, and establishes a fundamental groundwork for future investigations toward elucidating the regulative mechanisms behind Slbp2 and histone storage.
MATERIALS AND METHODS
Zebrafish maintenance and samples collection
Wild-type zebrafish of strain AB was used to produce mutant lines. All fishes maintained at 28.5°C under a reproduction regime (14 h light/10 h dark cycle) (Westerfield 2007). The samples, including adult tissues, oocytes, and embryos at different stages, were collected according to previous reports (Xie et al. 2003; Guan et al. 2008; Liu et al. 2015). Zebrafish embryos at two-cell (0.75 hpf), four-cell (1 hpf), eight-cell (1.25 hpf), 16-cell (1.5 hpf), 32-cell (1.75 hpf), 64-cell (2 hpf), 256-cell (2.5 hpf), high (3.3 hpf), 30% epiboly (4.7 hpf), 50% epiboly (5.3 hpf), 75% epiboly (8 hpf), 12 hpf, and 24 hpf were collected. The zebrafish MBT begins at cleavage cycle 10 (Kane and Kimmel 1993). Zebrafish oocyte development has been divided into five stages, including primary growth stage (I), cortical alveolus stage (II), vitellogenesis (III), oocyte maturation (IV), and mature egg (V) (Selman et al. 1993). All samples were quick frozen by liquid nitrogen and stored at −80°C or used directly. All procedures were performed with the approval of the Animal Care and Use Committee of the Institute of Hydrobiology, Chinese Academy of Sciences.
RNA isolation, reverse transcription PCR (RT-PCR), and quantitative real time PCR (qPCR)
Total RNAs were extracted by RNeasy Plus Mini Kit (Qiagen). One microgram of DNase-treated total RNA was transcribed reversely using Goldscript cDNA Synthesis Kit (Invitrogen), as described by the manufacturer. The cDNA templates of replication dependent histones were transcribed by random primer, and others were transcribed by oligo(dT) primer. RT-PCR and qPCR were performed as described previously (Huang et al. 2009; Zhong et al. 2014). β-actin or ef1α was selected as housekeeping gene (Livak and Schmittgen 2001). The primers are listed in Supplemental Table S5.
Probe synthesis and in situ hybridization
A 618 bp cDNA fragment of zebrafish slbp2 was amplified by specific primers (Supplemental Table S5). Antisense or sense DIG-labeled RNA probes were synthesized using T7 polymerase by in vitro transcription (Roche). The ovarian tissues were sampled for in situ hybridization, which was performed according to a previous report (Liu et al. 2015). Images were acquired by Zeiss Axio Observer A1 inverted microscope.
Polyclonal antibody preparation, western blot detection, and immunofluorescence localization
The 15 amino acid peptide (CLEKMNTELSDGSEK) that locates at the 222–235 amino acid site in the C terminus of zebrafish Slbp2 was chosen to produce the polyclonal antibody, which is different from zebrafish Slbp1 or other proteins. The peptide was conjugated to the KLH (Keyhole Limpet Hemocyanin) peptide and then used to immunize rabbits to get the polyclonal antibody. Anti-H2b (ab1790) and anti-H3 (ab1791) antibodies were purchased from Abcam Company. Western blot detection and immunofluorescence localization were also performed as described previously (Liu et al. 2015). The results of western blot and immunofluorescence were acquired by ImageQuant LAS 4000mini (GE) and Leica confocal laser scanning microscope, respectively.
Establishment of slbp2/1 mutant zebrafish lines by CRISPR/Cas9
Gene knockout of zebrafish slbp2 and slbp1 by CRISPR/Cas9 was performed as previously described (Lin et al. 2017; Xiong et al. 2017a,b). Due to the importance and conservation of RNA binding domains, the slbp2 sgRNA target site was designed on the sixth exon with the online service website (http://zifit.partners.org/ZiFiT/CSquare9Nuclease.aspx), and gRNA was transcribed with the TranscriptAid T7 High-Yield Transcription Kit (Thermo Fisher Scientific). The zebrafish-Codon-Optimized Cas9 plasmid pCS2-Cas9 (Liu et al. 2014) was digested with XbaI (NEB). The linearized pCS2-Cas9 was purified and transcribed using the T7 mMessage mMachine Kit (Ambion). Twenty picograms of sgRNA and 300 pg of Cas9 mRNA were coinjected into the one-cell stage zebrafish WT embryos. The mutations were detected by genomic PCR and sequenced as previously described (Supplemental Table S5; Yang et al. 2017). The three kinds of slbp2 mutant zebrafish lines, including Zslbp2Δ4−/−, Mslbp2Δ4+/−, and MZslbp2Δ4−/−, were generated by crossing combinations described in Figure 2A. Through the same strategy, the three kinds of slbp1 mutant zebrafish lines, including Zslbp1Δ1−/−, Mslbp1Δ1+/−, and MZslbp1Δ1−/−, were also constructed.
Generation of slbp2 or H2B transgenic MZslbp2Δ4−/− embryos
Zebrafish transgenic lines Tg(actb:slbp2-GFP) and Tg(actb:HIST1H2BJ-GFP) were constructed as follows. The full-length open reading frame (ORF) of zebrafish slbp2 (zgc:158856) was amplified by specific primers with the SmaI/KpnI restriction site (Supplemental Table S5) and inserted into the β-actin promoter construct pCAGcGH (Wang et al. 2001) digested by SmaI/KpnI. Similarly, the full-length ORF of human H2B (AK311849.1) was synthesized and inserted into the pMD-18T vector, which was used as a template to amplify H2B ORF by specific primers with XmalI/KpnI restriction enzyme sites (Supplemental Table S5). The PCR production was ligated to the β-actin promoter construct pCAGcGH (Wang et al. 2001) digested by XmalI/KpnI. One hundred picograms of slbp2-GFP or H2B-GPF plasmid DNA and 100 pg of Tol2 transposase mRNA were coinjected into one-cell embryos produced by crossing heterozygous F1 female and male (slbp2Δ4+/−). The female Zslbp2Δ4−/− mutants were screened by PCR (Supplemental Table S5), and then females of slbp2 transgenic Zslbp2Δ4−/− fish were randomly selected to cross with Zslbp2Δ4−/−. The slbp2 or H2B transgenic MZslbp2Δ4−/− embryos were screened by GFP fluorescence.
In vitro transcribed H2B-GFP mRNAs and microinjection
The H2B-GFP was amplified from pCAG-H2B-GFP and inserted into the PCS2+ vector. The plasmid PCS2+-H2B-GFP was linearized by NotI, which was used as a template to in vitro transcribe H2B-GFP mRNAs by Message Machine Kit (Ambion). To visualize the dynamic nuclear cleavage, about 120 pg H2B-GFP mRNA was injected into the zebrafish WT, MZslbp2Δ4−/−, and slbp2 transgenic MZslbp2Δ4−/− embryos at the one-cell stage.
Proteome and transcriptome analysis
Zebrafish WT and MZslbp2Δ4−/− at the 256-cell stage and high stage were sampled to perform iTRAQ and RNA-seq analysis, respectively. Three biological replicates were analyzed per group. The detection of peptides, library construction, sequencing, and bioinformatics analysis was accomplished by the Beijing Genomics Institute (BGI) according to the standard procedures. The raw data of transcriptomes have been submitted to the NCBI database (accession no. SRP126712), and the mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (Vizcaino et al. 2016) partner repository, with the data set identifier PXD008614.
Recombinant protein production and electrophoretic mobility shift assay (EMSA)
The optimized 981 bp ORF of slbp2 was synthesized and cloned into pMAL-c5x (NEB). Plasmid pMAL-c5x-slbp2 was transformed into BL21(DE3) Condon Plus (Stratagene). Single colonies were screened and cultured in LB medium until OD600 = 0.6, and then 0.2 mM isopropyl thio-β-d-galactoside (IPTG) was added for induction at 20°C for 16 h. E. coli cells were collected and disrupted by ultrasonic. The fused Slbp2-MBP protein was purified from the supernatant by dextran gel separation and ion exchange and detected by SDS-PAGE.
EMSA analysis was performed as described previously (Qiu et al. 2015). In brief, the conserved H2a SL (AAAAAGGUUCUUUUAAGAACCACCCAUUUUU) was synthesized for the RNA probe labeled with 5′-biotin (Thermo Scientific) (Williams et al. 1994; Wang et al. 1999). Five microgams of purified Slbp2-MBP protein was incubated with 1 nmol 5′-biotin-labeled probe RNA and 10 µmol mimic negative control (5′-biotin-unlabeled competitor) in binding buffer for 30 min. Binding reactions were resolved by 6% PAGE and electrophoretically transferred to nylon membrane. The transferred RNA was cross-linked to the nylon membrane by using CL-1000 Ultraviolet Crosslinker (UVP). Biotin-labeled RNA was detected by chemiluminescence and exposed with ChemiDoc XRS+ chemiluminescent imaging analysis system (Bio-Rad).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Key Program of Frontier Sciences of the Chinese Academy of Sciences (QYZDY-SSW-SMC025), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA08030201), the Earmarked Fund for Modern Agro-industry Technology Research System (NYCYTX-49), the Autonomous Project of the State Key Laboratory of Freshwater Ecology and Biotechnology (2016FBZ01). We want to thank Fang Zhou for providing confocal services (Analytical and Testing Center, IHB, CAS) and Da-wei Zhang in the Wu-han Xiao Laboratory (IHB, CAS) for kindly providing CRISPR/cas9 plasmids.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067090.118.
REFERENCES
- Allard P, Champigny MJ, Skoggard S, Erkmann JA, Whitfield ML, Marzluff WF, Clarke HJ. 2002. Stem-loop binding protein accumulates during oocyte maturation and is not cell-cycle-regulated in the early mouse embryo. J Cell Sci 115: 4577–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard P, Yang Q, Marzluff WF, Clarke HJ. 2005. The stem-loop binding protein regulates translation of histone mRNA during mammalian oogenesis. Dev Biol 286: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen IS, Lindeman LC, Reiner AH, Østrup O, Aanes H, Aleström P, Collas P. 2013. Epigenetic marking of the zebrafish developmental program. Development 104: 85–112. [DOI] [PubMed] [Google Scholar]
- Arnold DR, Francon P, Zhang J, Martin K, Clarke HJ. 2008. Stem-loop binding protein expressed in growing oocytes is required for accumulation of mRNAs encoding histones H3 and H4 and for early embryonic development in the mouse. Dev Biol 313: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA. 2005. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol 25: 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, et al. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci 99: 8173–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Miyara F, Han ZM, Kihara M, Brown DT, Hager GL, Latham K, Adashi EY, Misteli T. 2005. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol Biol Cell 16: 3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P, Ross J. 1989. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem Sci 14: 373–377. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. 2002. Genomic instability in mice lacking histone H2AX. Science 296: 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. 2003a. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114: 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. 2003b. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679. [DOI] [PubMed] [Google Scholar]
- Cloutier JM, Mahadevaiah SK, ElInati E, Nussenzweig A, Toth A, Turner JMA. 2015. Histone H2AFX links meiotic chromosome asynapsis to prophase I oocyte loss in mammals. PLoS Genet 11: e1005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Barre AE, Gerson V, Gout S, Creaven M, Allis CD, Dimitrov S. 2000. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J 19: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Erkmann JA, Greenland JA, Marzluff WF. 2001. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and for histone pre-mRNA processing. Mol Cell Biol 21: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A. 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4: 497–508. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Lewis NJ, Marzluff WF. 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res 24: 1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald DJ, Eisen JS. 2002. Headwaters of the zebrafish emergence of a new model vertebrate. Nat Rev Genet 3: 717–724. [DOI] [PubMed] [Google Scholar]
- Guan M, Rawson DM, Zhang T. 2008. Development of a new method for isolating zebrafish oocytes (Danio rerio) from ovary tissue masses. Theriogenology 69: 269–275. [DOI] [PubMed] [Google Scholar]
- Gui JF, Zhou L. 2010. Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploid Carassius auratus gibelio (review). Sci China Life Sci 53: 409–415. [DOI] [PubMed] [Google Scholar]
- Gunjan A, Paik J, Verreault A. 2006. The emergence of regulated histone proteolysis. Curr Opin Genet Dev 16: 112–118. [DOI] [PubMed] [Google Scholar]
- Hanson RJ, Sun JH, Willis DG, Marzluff WF. 1996. Efficient extraction and partial purification of the polyribosome-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry 35: 2146–2156. [DOI] [PubMed] [Google Scholar]
- Hentschel CC, Birnstiel ML. 1981. The organization and expression of histone gene families. Cell 25: 301–313. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhou L, Li Z, Gui JF. 2009. Expression pattern, cellular localization and promoter activity analysis of ovarian aromatase (Cyp19a1a) in protogynous hermaphrodite red-spotted grouper. Mol Cell Endocrinol 307: 224–236. [DOI] [PubMed] [Google Scholar]
- Imai F, Yoshizawa A, Matsuzaki A, Oguri E, Araragi M, Nishiwaki Y, Masai I. 2014. Stem-loop binding protein is required for retinal cell proliferation, neurogenesis, and intraretinal axon pathfinding in zebrafish. Dev Biol 394: 94–109. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Standart N. 1990. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell 62: 15–24. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. 1993. The zebrafish mid-blastula transition. Development 119: 447–456. [DOI] [PubMed] [Google Scholar]
- Lin QH, Mei J, Li Z, Zhang XM, Zhou L, Gui JF. 2017. Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics 207: 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, Winata C, Mathavan S, Müller F, Aleström P, et al. 2011. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell 21: 993–1004. [DOI] [PubMed] [Google Scholar]
- Liu D, Wang ZX, Xiao A, Zhang YT, Li WY, Zu Y, Yao SH, Lin S, Zhang B. 2014. Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized Cas9 and evaluation of off-targeting effect. J Genet Genomics 41: 43–46. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang XJ, Wang W, Zhang J, Li Z, Gui JF. 2015. Molecular characterization and expression of an oocyte-specific histone stem-loop binding protein in Carassius gibelio. Comp Biochem Physiol B Biochem Mol Biol 190: 46–53. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Martin F, Schaller A, Eglite S, Schumperli D, Muller B. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J 16: 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. 1992. Histone 3′ ends: essential and regulatory functions. Gene Expression 2: 93–97. [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Koreski KP. 2017. Birth and death of histone mRNAs. Trends Genet 33: 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt J, Crombie C, Schumperli D, Muller B. 2002. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone gene expression and is essential for embryonic and postembryonic cell division. J Cell Sci 115: 857–866. [DOI] [PubMed] [Google Scholar]
- Qiu K, Li ZP, Yang Z, Chen JY, Wu SX, Zhu XY, Gao S, Gao J, Ren GD, Kuai BK, et al. 2015. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyII catabolic genes in Arabidopsis. PLoS Genet 11: e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Sánchez R, Marzluff WF. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol 22: 7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R, Marzluff WF. 2004. The oligo(A) tail on histone mRNA plays an active role in translational silencing of histone mRNA during Xenopus oogenesis. Mol Cell Biol 24: 2513–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman K, Wallace RA, Sarka A, Qi X. 1993. Stages of oocyte development in the zebrafish, Brachydanio rerio. J Morphol 218: 203–224. [DOI] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang XC, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev 15: 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Testa E, Nardozi D, Antinozzi C, Faieta M, Di Cecca S, Caggiano C, Fukuda T, Bonanno E, Zhenkun L, Maldonado A, et al. 2018. H2AFX and MDC1 promote maintenance of genomic integrity in male germ cells. J Cell Sci 131: jcs214411. [DOI] [PubMed] [Google Scholar]
- Thelie A, Pascal G, Angulo L, Perreau C, Papillier P, Dalbies-Tran R. 2012. An oocyte-preferential histone mRNA stem-loop-binding protein like is expressed in several mammalian species. Mol Reprod Dev 79: 380–391. [DOI] [PubMed] [Google Scholar]
- Townley-Tilson WH, Pendergrass SA, Marzluff WF, Whitfield ML. 2006. Genome-wide analysis of mRNAs bound to the histone stem-loop binding protein. RNA 12: 1853–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZF, Whitfield ML, Ingledue TC, Dominski Z, Marzluff WF. 1996. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Gene Dev 10: 3028–3040. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Ingledue TC, Dominski Z, Sánchez R, Marzluff WF. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol Cell Biol 19: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Hu W, Wu G, Sun YH, Chen SP, Zhang FY, Zhu ZY, Feng JX, Zhang XR. 2001. Genetic analysis of “all-fish” growth hormone gene transferred carp (Cyprinus carpio L) and its F1 generation. Chinese Sci Bull 46: 1174–1178. [Google Scholar]
- Westerfield M. 2007. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio), 5th ed University of Oregon Press, Eugene, OR. [Google Scholar]
- Williams AS, Ingledue TC, Kay BK, Marzluff WF. 1994. Changes in the stem-loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res 22: 4660–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland HR. 1980. Histone synthesis during the development of Xenopus. FEBS Lett 121: 1–10. [DOI] [PubMed] [Google Scholar]
- Wu N, Yue HM, Chen B, Gui JF. 2009. Histone H2A has a novel variant in fish oocytes. Biol Reprod 81: 275–283. [DOI] [PubMed] [Google Scholar]
- Xie J, Wen JJ, Yang ZA, Wang HY, Gui JF. 2003. Cyclin A2 is differentially expressed during oocyte maturation between gynogenetic silver crucian carp and gonochoristic color crucian carp. J Exp Zool Part A 295a: 1–16. [DOI] [PubMed] [Google Scholar]
- Xiong ST, Wu JJ, Jing J, Huang PP, Li Z, Mei J, Gui JF. 2017a. Loss of stat3 function leads to spine malformation and immune disorder in zebrafish. Sci Bull 62: 185–196. [DOI] [PubMed] [Google Scholar]
- Xiong ST, Mei J, Huang PP, Jing J, Li Z, Kang JL, Gui JF. 2017b. Essential roles of stat5.1/stat5b in controlling fish somatic growth. J Genet Genomics 44: 577–585. [DOI] [PubMed] [Google Scholar]
- Yang YJ, Wang Y, Li Z, Zhou L, Gui JF. 2017. Sequential, divergent, and cooperative requirements of Foxl2a and Foxl2b in ovary development and maintenance of zebrafish. Genetics 205: 1551–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Adamski R, Chen JJ. 2010. Focus on histone variant H2AX: to be or not to be. FEBS Lett 584: 3717–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue HM, Li Z, Wu N, Liu Z, Wang Y, Gui JF. 2013. Oocyte-specific H2A variant H2af1o is required for cell synchrony before mid-blastula transition in early zebrafish embryos. Biol Reprod 89: 82. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dominski Z, Yang XC, Elms P, Raska CS, Borchers CH, Marzluff WF. 2003. Phosphorylation of SLBP on two threonines triggers degradation of SLBP, the sole cell-cycle regulated factor required for regulation of histone mRNA processing, at the end of S-phase. Mol Cell Biol 23: 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong JX, Zhou L, Li Z, Gui JF. 2014. Zebrafish Noxa promotes mitosis in early embryonic development and regulates apoptosis in subsequent embryogenesis. Cell Death Differ 21: 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou L, Gui JF. 2017. Natural and artificial polyploids in aquaculture. Aquac Fish 2: 103–111. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.