Abstract
Splicing is an essential step in eukaryotic gene expression. While the majority of introns is excised by the U2-dependent, or major class, spliceosome, the appropriate expression of a very small subset of genes depends on U12-dependent, or minor class, splicing. The U11/U12 65K protein (hereafter 65K), encoded by RNPC3, is one of seven proteins that are unique to the U12-dependent spliceosome, and previous studies including our own have established that it plays a role in plant and vertebrate development. To pinpoint the impact of 65K loss during mammalian development and in adulthood, we generated germline and conditional Rnpc3-deficient mice. Homozygous Rnpc3−/− embryos died prior to blastocyst implantation, whereas Rnpc3+/− mice were born at the expected frequency, achieved sexual maturity, and exhibited a completely normal lifespan. Systemic recombination of conditional Rnpc3 alleles in adult (Rnpc3lox/lox) mice caused rapid weight loss, leukopenia, and degeneration of the epithelial lining of the entire gastrointestinal tract, the latter due to increased cell death and a reduction in cell proliferation. Accompanying this, we observed a loss of both 65K and the pro-proliferative phospho-ERK1/2 proteins from the stem/progenitor cells at the base of intestinal crypts. RT-PCR analysis of RNA extracted from purified preparations of intestinal epithelial cells with recombined Rnpc3lox alleles revealed increased frequency of U12-type intron retention in all transcripts tested. Our study, using a novel conditional mouse model of Rnpc3 deficiency, establishes that U12-dependent splicing is not only important during development but is indispensable throughout life.
Keywords: RNPC3, minor class splicing, U12 intron, development, gastrointestinal epithelium
INTRODUCTION
Splicing, the excision of introns from pre-mRNA, is an essential step in gene expression. Catalyzing this process are the spliceosomes, which consist of small nuclear ribonucleoproteins (snRNPs), each one comprising a different small nuclear RNA (snRNA) and several proteins (Wahl et al. 2009). Most multicellular eukaryotic cells require two types of spliceosome: the major class or U2-dependent spliceosomes that remove U2-type introns, and the minor class or U12-dependent spliceosomes that remove U12-type introns, which constitute approximately 0.35% of introns (∼822 and ∼647 in the human and mouse genomes, respectively) (Merico et al. 2015; Horiuchi et al. 2018). Even though they are rare, U12-type introns are highly conserved across distantly related eukaryotic taxa, including most plants, fungi, animals, and even some single cell eukaryotes (Burge et al. 1998), indicating an early evolutionary origin and important function (Russell et al. 2006). Indeed, genes that contain U12-type introns are over-represented in functions and pathways related to development, such as the cell cycle, DNA replication and repair, RNA processing, as well as voltage gated ion channels and MAPK signaling (Supplemental Fig. S1A; Merico et al. 2015).
U12-type introns are distinguished from U2-type introns by two highly conserved motifs: a 7 bp sequence adjacent to the 5′ splice site and the branch point sequence (BPS), just upstream of the 3′ splice site. Like U2-dependent spliceosomes, U12-dependent spliceosomes utilize 5 snRNPS for splicing, including the U5 snRNP, which is shared by the two spliceosomes. The other four snRNPs are exclusive to the U12-dependent spliceosome and contain the U11, U12, U4ATAC, and U6ATAC snRNAs, respectively. Unlike the U1 and U2 snRNPs of the major class spliceosome, which recognize and bind to U2-type introns as monomers, the corresponding U11 and U12 snRNPs form a stable di-snRNP prior to assembling on U12-type introns. The U11/U12 di-snRNP contains 7 unique proteins: 20K, 25K, 31K, 35K, 48K, 59K, and 65K (Will et al. 2004), with the latter three forming a “molecular bridge” structure (Supplemental Fig. S1B) that brings the two ends of the intron together (Benecke et al. 2005), ready for the catalytic steps of splicing.
Impaired minor class splicing generally leads to intron retention and, less frequently, exon skipping and alternative splicing (Chang et al. 2007). These perturbations often result in frame-shifts, the creation of premature stop codons (PTCs) and reduced expression and function of affected genes (Patel et al. 2002; Niemelä and Frilander 2014). Although only directly affecting a small subset of genes (Markmiller et al. 2014), impaired minor class splicing has the potential to impact on many downstream pathways, leading to catastrophic outcomes. This is best exemplified by the human developmental disease microcephalic osteodysplastic primordial dwarfism 1 (MOPD1), also known as Taybi-Linder syndrome (TALS), which is caused by biallelic autosomal recessive mutations in the RNU4ATAC gene (Edery et al. 2011; He et al. 2011). The clinical features of this genetic disease are severe growth retardation with brain and skeletal abnormalities, usually culminating in death during infancy. Roifman syndrome is a less severe minor class splicing syndrome caused by the inheritance of compound heterozygous mutations in less detrimental nucleotides in RNU4ATAC similarly presenting with growth retardation and cognitive delay, more pronounced retinal dystrophy and immunodeficiency (Merico et al. 2015; Heremans et al. 2018).
The N-terminal portion of the 65K protein binds to the 59K protein and its C-terminal RNA binding domain binds to U12 snRNA (Will et al. 2004; Singh et al. 2016) (Supplemental Fig. S1B). Loss of 65K function in developing Arabidopsis and zebrafish larvae causes inefficient U12-dependent splicing and a failure to thrive (Jung and Kang 2014; Markmiller et al. 2014; Park et al. 2016). In our zebrafish study, the digestive organs and several other tissues that exhibit high rates of proliferation during larval development were the most severely affected by 65K loss. In contrast, the inheritance of two different RNPC3 alleles in human revealed an important but discrete role for 65K in pituitary gland development (Argente et al. 2014; Norppa et al. 2018), which could be managed effectively by administration of growth hormone. These dissimilar outcomes from 65K deficiency during zebrafish and human development suggest the presence of tissue-specific contexts during development when critical thresholds of minor class splicing efficiency must be met.
To explore this question in the setting of mammalian development and adulthood, we generated germline and conditional mouse models of Rnpc3 deficiency. We found that Rnpc3-null mice die very early in development, prior to implantation. Meanwhile, induced 65K deficiency in adults impacted most severely on the highly proliferative epithelial lining of the gastrointestinal tract and the constantly renewing hematopoietic compartment of the bone marrow. In this study, we focused our analysis on the intestinal epithelium since this is the most vigorously renewing tissue in adult mammals. It comprises an elaborately folded monolayer of cells that is organized into spatially distinct compartments dedicated to stem/progenitor cell proliferation, lineage commitment, terminal differentiation, and cell death (Barker et al. 2008). Notably, we found the 65K protein localized to the nuclei of stem cells and their progenitors situated at the base of goblet-shaped invaginations called crypts. RT-PCR analysis of RNA extracted from purified, 65K-deficient, intestinal epithelium revealed increased levels of incorrectly spliced transcripts of U12-type intron-containing genes, including Mapk8 and Mapk11, which encode the pro-proliferative proteins, JNK1 and p38β, respectively. Collectively our data show that 65K is required for efficient U12-dependent splicing and is indispensable for rapid cell renewal, not only during development but throughout life.
RESULTS
Generation of constitutive and conditional alleles of Rnpc3 deficiency
To generate constitutive and conditional alleles of Rnpc3 deficiency in mice, we purchased ES cells (C57BL6/N) containing a targeted knockout (KO)-first, conditional ready, mutant Rnpc3 allele (Rnpc3neo; Fig. 1) from the European Conditional Mouse Mutagenesis Program (EUCOMM). These cells also harbor a LacZ reporter gene introduced into the endogenous Rnpc3 locus to monitor Rnpc3 transcription. Upon injection into C57BL6 blastocysts, these ES cells contributed to the generation of germline chimeric offspring that bred to produce heterozygous Rnpc3neo/+ mice (Fig. 1A). To remove the neomycin and LacZ cassettes, we catalyzed recombination between the FRT sites in the KO-first construct by crossing Rnpc3neo/+ mice with an embryonic Flp deleter transgenic line, EIIA-Flp, which reconstituted a functionally wild-type Rnpc3 allele with loxP sites flanking exons 4 and 5 (Rnpc3lox; Fig. 1B). To generate a pure null allele lacking exons 4 and 5 (Rnpc3−; Fig. 1C), we crossed Rnpc3neo mice with mice carrying a Cre deleter transgene, EIIa-Cre (Lakso et al. 1996).
FIGURE 1.

Heterozygous Rnpc3-deficient mice exhibit a normal lifespan but complete loss of Rnpc3 is indispensable for preimplantation development. (A) The initial targeted allele (Rnpc3neo) used in this study contained an IRES:LacZ and a loxP-flanked promoter-driven neo cassette inserted into intron 3 of the Rnpc3 gene, thereby disrupting its function. (B) Mice harboring conditional alleles in which exons 4 and 5 are flanked by loxP sites were generated by crossing Rnpc3neo mice with an embryonic Flp deleter transgenic line. (C) A germline null allele (Rnpc3−) was generated by crossing Rnpc3neo mice with mice carrying a Cre deleter transgene. Diagram modified from Skarnes et al. (2011) with permission from Springer Nature. (D) Longevity of Rnpc3+/− mice is the same as WT littermate controls: n = 58–62, Student's t-test P = 0.1131. (E) Pituitary gland weights of Rnpc3+/− mice (60–70 wk) are comparable to WT littermate controls (n = 11–17, Student's t-test P = 0.4822). (F) Several litters of mice were generated from in-crossing Rnpc3+/− parents and no Rnpc3−/− mice were born or hatched in culture from E3.5 blastocysts. (G) Genotyping of E3.5 blastocysts that had been cultured for 24 h identified all potential genotypes. Rnpc3+/+ and Rnpc3+/− blastocysts appeared normal while Rnpc3−/− exhibited an arrested morula phenotype (right panel). Scale bar in G = 100 µm.
65K is required for preimplantation murine development
Adult Rnpc+/− mice are morphologically indistinguishable from Rnpc3+/+ mice, are fertile, have a similar lifespan to Rnpc3+/+ littermates (n = 58–62, Student's t-test P = 0.1131; Fig. 1D) and exhibit no gross skeletal or growth abnormalities. Heterozygous Rnpc3neo/+ mice are also completely normal, fertile, and have the same lifespan as wild-type C57Bl/6 littermates (not shown). Because the inheritance of compound heterozygous RNPC3 mutations in three out of four human siblings resulted in pituitary hypoplasia during development (Argente et al. 2014), we compared pituitary gland weight between adult (60–70 wk) Rnpc3+/− and Rnpc3+/+ mice and found no significant difference (n = 11–17, Student's t-test P = 0.4822; Fig. 1E).
In contrast, Rnpc3−/− mouse embryos are not born. Of the 166 pups born from in-crossing Rnpc3+/− parents none of them was Rnpc3−/− (χ2 statistic 56.783, P < 0.0001; Fig. 1F). We determined the stage at which Rnpc3−/− embryos are lost by crossing Rnpc3+/− males and females and collecting 89 E3.5 blastocysts from the uteri of Rnpc3+/− females for culture and in vitro analysis. Of this cohort, 23 (26%) arrested at a very early stage in development and did not hatch (Fig. 1F). When we genotyped hatched blastocysts (n = 66) that had been cultured for 5 d (E3.5 + 5), no Rnpc3−/− blastocysts were detected (χ2 statistic 22.818, P < 0.0001; Fig. 1F), indicating that the ∼25% of blastocysts that failed to hatch were likely to include Rnpc3−/− embryos. To investigate this further, we collected E3.5 blastocysts from a single mother, cultured them for 24 h (E3.5 + 1) and genotyped them all (n = 11). Four embryos were retarded at the morula stage, and all were found to be Rnpc3−/− (Fig. 1G), demonstrating that Rnpc3−/− embryos fail to develop beyond the compacted cell morula stage and do not hatch from the zona pellucida. Thus, Rnpc3 is indispensable for preimplantation murine development.
To determine sites of Rnpc3 transcription in adult mice, we performed β-galactosidase staining of histological sections of tissues harvested from heterozygous Rnpcneo/+ mice (Supplemental Fig. S2). We observed strong β-galactosidase activity in the epithelial stem/progenitor cell compartments of the gastrointestinal tract (stomach, small intestine, and colon; Supplemental Fig. S2A–C) and in the epithelial stem cells of the hair follicle bulge (Supplemental Fig. S2D), bronchiolar epithelial cells lining the lung (Supplemental Fig. S2E), and the Purkinje cells of the cerebellum (Supplemental Fig. S2F). The particularly robust β-galactosidase staining in the stem cell/progenitor cells of the stomach, small and large intestines, Purkinje cells and hair follicle suggests that Rnpc3 transcription is likely to be important for the correct functioning of these tissues. Much weaker β-galactosidase activity was also detected in several other tissues and organs, including the thyroid, heart, liver, and kidney (data not shown) indicating that some degree of activation of the Rnpc3 locus is widespread in adult mice.
The 65K protein is required for the integrity of multiple tissues in adult mice
To investigate the impact of depleting 65K from all tissues of adult mice, we circumvented the embryonic lethality of Rnpc3−/− mice by using UBC-CreERT2, encoding a tamoxifen-inducible, systemic Cre recombinase. In these experiments, mice of all genotypes examined were treated with tamoxifen. UBC-CreERT2;Rnpc3lox/+ mice were phenotypically normal throughout the 8 d experiment, indicating that induced heterozygous loss of one Rnpc3 allele in adults has no acute toxicity (Fig. 2A). In contrast, UBC-CreERT2;Rnpc3lox/lox mice exhibited significantly reduced body weight 6 d after tamoxifen treatment, which became more severe with time (Fig. 2A). Peripheral blood collected at 8 d post-tamoxifen treatment revealed significant leukopenia (Fig. 2B), reduced numbers of monocytes (Fig. 2B), anemia (Fig. 2C), and thrombocytopenia (Fig. 2D). At this stage, the mice showed signs of malnutrition and stress (scruffy coat, docile, and hunched behavior) and were euthanized. Following this, a comprehensive histological analysis was performed by a pathologist in the Department of Pathology, University of Melbourne node of the Australian Phenomics Network (APN). This revealed many morphological defects, including a markedly smaller thymus (Fig. 2E) with no discernible medullary (black arrow) or cortical regions (white arrow) (Fig. 2F,G). In addition, the skin showed subtle abnormalities with a paucity of sebaceous glands and an attenuated epidermis (data not shown). However, the most patently affected tissue was the epithelial lining of the gastrointestinal tract.
FIGURE 2.
65K is an essential protein in adult mice and is necessary for the integrity of self-renewing tissues. The effect of acute, global 65K deficiency was studied in Rnpc3lox/lox and Rnpc3lox/+ adult mice (8–12 wk) harboring an inducible UBC-CreERT2 allele. These mice were weighed prior to tamoxifen treatment and daily thereafter. At the end of the experiment, tissues and peripheral blood were collected for analysis. (A) UBC-CreERT2;Rnpc3lox/lox (green line, circles) mice had lost weight irreversibly by 6 d post-tamoxifen treatment and reached the ethical end-point requiring euthanasia at 8 d. In contrast, no significant weight loss was experienced by tamoxifen-treated UBC-CreERT2;Rnpc3lox/+ mice (blue line, diamonds) or the three control strains: Rnpc3lox/+ (blue line, inverted triangles), Rnpc3lox/lox (red line, squares), and UBC-CreERT2 (red line, triangles). Data are expressed as mean + SEM (n = 3–9). (*) P < 0.05, two-way ANOVA with Tukey's multiple comparison testing. (B) Analysis of peripheral blood at 8 d post-tamoxifen treatment revealed a significant decrease in the numbers of white blood cells (WBC), lymphocytes, and monocytes in UBC-CreERT2;Rnpc3lox/lox (green) compared to controls (red). (C) Red blood cell (RBC) numbers were also significantly decreased (D) and mice exhibited severe thrombocytopenia. (E) Systemic deletion of 65K induces thymic atrophy. Thymi were collected from control and UBC-CreERT2;Rnpc3lox/lox mice at 8 d post-tamoxifen treatment and weighed. Data are expressed as mean + SEM (n = 7–9). (****) P < 0.0001, (***) P < 0.0002, (**) P < 0.01 Student's t-test. (F) Histological analysis of the thymus displays characteristic cortex (white arrow) and medulla (black arrow) regions in control animals (Rnpc3lox/lox) at 8 d post-tamoxifen treatment. (G) This organization is absent in thymi taken from UBC-CreERT2;Rnpc3lox/lox mice. Scale bar in F–G = 100 µm.
Systemic recombination of conditional alleles in Rnpc3lox/lox mice causes severe atrophy of the gastrointestinal epithelium
Histological analysis of the entire gastrointestinal tract showed a severely disrupted mucosa from the esophagus to the rectum (Fig. 3). The organization of the squamous epithelium of the esophagus was disrupted with signs of erosive esophagitis (cf. Fig. 3A and E). The columnar mucous epithelium of the glandular stomach was also damaged (cf. Fig. 3B and F). The monolayer of epithelium lining the small intestine and colon contained vacuolated epithelial cells scattered throughout the submucosa, and dispersed, dilated glandular structures (arrowheads), reminiscent of crypts, were observed in the attenuated epithelium suggesting attempted regeneration (cf. Fig. 3C,D with Fig. 3G,H, respectively).
FIGURE 3.
Systemic Rnpc3 deficiency causes severe atrophy of the entire gastrointestinal epithelium. Histological analysis of the epithelial lining of the esophagus, stomach, small intestine, and colon in tamoxifen-treated control (Rnpc3lox/lox) (A–D) and experimental (UBC-CreERT2;Rnpc3lox/lox) mice (E–H) 8 d post-tamoxifen treatment. Whereas the organization and structural integrity of the gastrointestinal epithelium is intact in control organs (A–D), recombination of both Rnpc3lox alleles causes large-scale degeneration of the epithelial layer interspersed with discrete pockets of regenerating crypt-like structures (arrowheads in F, G, and H). Scale bar in A = 50 µm and applies to all images.
The 65K protein is expressed in the epithelial cells of intestinal crypts and is depleted following Rnpc3 deletion
We used immunocytochemistry to follow the fate of cells lacking the 65K protein at 4 d and 6 d post-tamoxifen treatment. In control Rnpc3lox/lox (no UBC-Cre) mice, we found nuclear localization of the 65K protein in the crypt cells of both the small intestine (Fig. 4A,A′) and colon (Fig. 4B,B′), consistent with our LacZ reporter analysis (Supplemental Fig. S2B,C). As the cells migrated up the crypt-villus axis, the 65K staining became progressively weaker or was absent, indicating a greater requirement for 65K in the stem/progenitor cell compartment of the crypts (lower bracket), than the terminally differentiated cells of the villus (upper bracket). Next, we carried out immunohistochemical analysis of U1A, a 32 kDa major class (U1/U2-dependent) spliceosome component that binds directly to stem-loop II of U1 snRNA. In contrast to 65K expression, we observed robust U1A staining in the nuclei of all cells throughout the crypt-villus axis of the intestinal epithelium, including in the differentiated cells (Supplemental Fig. S3).
FIGURE 4.
65K is expressed in the nuclei of cells in intestinal crypts and is lost following induced Rnpc3 deletion. (A,A′) Immunostaining with 65K antibody highlights positive nuclei at the base of the crypts of the small intestine (A, lower bracket) and colon (B,B′) with weak to absent expression in cells of the small intestinal villi (A, upper bracket) and in the lamina propria. Black arrows denote cells with 65K-positive nuclei in the stem cell compartment of the crypts. (C,D) Four days after tamoxifen (TMX) treatment of UBC-CreERT2;Rnpc3lox/lox mice, 65K protein expression is decreased in the small intestine (C, dashed bracket; magnified in C′) and colon (D,D′). The small intestine epithelial structure is disorganized with shorter villi. (E,F) Six days after TMX treatment, some areas of the small intestinal (E, bracket; magnified in E′) and colonic epithelium (F,F′) in UBC-CreERT2;Rnpc3lox/lox mice have completely lost 65K expression, coinciding with widespread epithelial disruption. Meanwhile, foci of regenerating crypt-like structures arise from cells in which complete recombination has not occurred and 65K protein is still evident (E′, arrows). A′–F′ show regions of interest at higher magnification. Scale bar in A, B, C, D, E, and F = 100 µm. Scale bar in A′, B′, C′, D′, E′, and F′ = 50 µm.
After 4 d of tamoxifen treatment of UBC-CreERT2;Rnpc3lox/lox mice, there was a marked reduction in 65K staining in the crypt cells of both the small intestine (Fig. 4C,C′, dashed bracket) and colon (Fig. 4D,D′). This was accompanied by loss of crypt structure and epithelial integrity with nuclei rounding up and cells losing apico-basal polarity. Two days later, the epithelium of the small intestine was highly disorganized. Large areas of epithelial disruption were devoid of 65K expression, though a few scattered cells retained 65K expression (Fig. 4E,E′, arrows), most likely corresponding to stem/progenitor cells that escaped recombination. Similarly, in the colon, large vacuolated spaces appeared in the crypts. The epithelial cells were thinner and flatter and 65K staining was largely weak or absent (Fig. 4F,F′).
65K depletion is accompanied by a marked increase in cell death and reduced intestinal cell proliferation
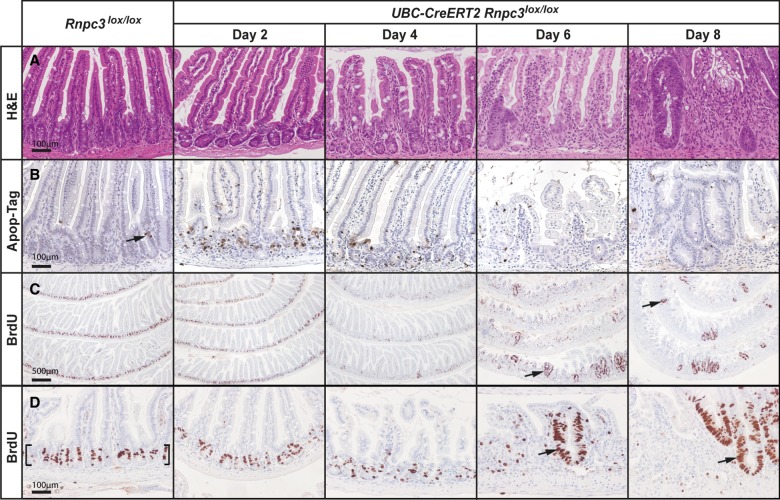
Histological analysis of the monolayer of small intestinal epithelium over an 8 d time-course after tamoxifen treatment, revealed a progressive deterioration of the intestinal epithelium that was clearly evident at 6 d post-tamoxifen treatment (Fig. 5A). Morphological abnormalities included crypt loss, reduced villus height, infiltration of immune cells, and ulceration (Fig. 5A). We analyzed the role of cell death in producing these morphological defects using Apop-Tag. We observed an early wave of brown punctate staining, indicative of apoptotic cells, from 2- to 4-d post-tamoxifen treatment. Brown staining was restricted to the highly proliferative, stem/progenitor compartment of the small intestinal crypts (Fig. 5B), essentially overlapping the region where cells expressing 65K were observed (Fig. 4A,A′). Apoptosis is rare or undetectable in this region of wild-type mice. We also assessed the impact of 65K loss on cell proliferation using bromodeoxyuridine (BrdU) incorporation to identify cells in the S-phase of the cell cycle (Fig. 5C,D). In sections of small intestinal epithelium from tamoxifen-treated, control (Rnpc3lox/lox) mice (shown in Fig. 5C,D at low and high magnifications, respectively), intense brown nuclear staining was present in the crypts (bracketed region), whereas staining in the villi was negligible. In contrast, 4 d after tamoxifen treatment, there was a marked reduction in BrdU-positive cells in corresponding sections of small intestinal epithelium from tamoxifen-treated, UBC-CreERT2;Rnpc3lox/lox mice, indicating fewer cells progressing through the cell cycle (Fig. 5C,D). There was no difference in the frequency of BrdU-positive cells scattered throughout the differentiated cell compartment of the small intestinal villi (Fig. 5C,D). At 6 d and 8 d after tamoxifen treatment, we observed the discontinuous appearance of new, BrdU-positive crypt-like structures (arrows), corresponding to sites of regeneration, akin to a localized wound healing response (Ashton et al. 2010). In summary, loss of 65K expression produced an increase in apoptosis and a decrease in cell proliferation in the proliferative compartment of the small intestinal epithelium, which together contributed to a severe and progressive deterioration of this tissue.
FIGURE 5.
Loss of 65K induces apoptosis of epithelial cells in small intestinal crypts. The impact of acute global 65K deficiency on the integrity of the small intestine was studied in tamoxifen-treated Rnpc3lox/lox adult mice (8–12 wk) harboring an inducible UBC-CreERT2 allele. (A) Imaging of hematoxylin and eosin (H&E)-stained sections of small intestine from UBC-CreERT2;Rnpc3lox/lox mice revealed progressive epithelial damage from 4 d post-tamoxifen treatment, characterized by loss of crypt structure, shortening of villi, and invasion of inflammatory cells. Enlarged, regenerative crypts 6 d and 8 d post-tamoxifen treatment are indicative of a wound healing response. (B) Apo-tag (brown staining) identifies cells undergoing apoptosis. Such cells are rare in the unrecombined epithelium (arrow), but increase markedly in number in UBC-CreERT2;Rnpc3lox/lox mice 2 d and 4 d post-tamoxifen treatment. The apoptotic cells are found mainly in the crypts. (C) At 2, 4, 6, and 8 d post-tamoxifen treatment, mice were injected with BrdU to mark cells in the S-phase of the cell cycle, prior to being euthanized 2 h later. Low magnification images of BrdU-positive cells (brown nuclei) throughout the small intestine highlight the location of proliferative cells in the crypts; such cells are absent from the villi. Scale bar = 500 µm. Upon recombination, BrdU-positive cells are fewer in number and more dispersed, particularly 4 d after tamoxifen treatment. Areas of regenerating proliferating cells are observed from 6- to 8-d post-tamoxifen treatment (arrows). (D) Higher magnification views highlighting proliferative cells in control crypts (brackets) and at sites of regeneration in Rnpc3-depleted intestinal epithelium (arrows). Scale bars in A, B, D = 100 µm (applies to all images in the row).
The MAPK signaling proteins, pERK1/2, are expressed in the same cells as 65K in regenerating crypts
Extracellular signal-regulated protein kinases 1 and 2 (ERK1/2) are members of the mitogen-activated protein kinase (MAPK) superfamily that can mediate cell proliferation and apoptosis. To compare the localization of the 65K protein with these regulators of cell proliferation, we compared the localization of the 65K protein with the phosphorylated (activated) forms of ERK1/2 (pERK1/2) in adjacent histological sections of the small intestine (Fig. 6). In control, tamoxifen-treated Rnpc3lox/lox mice, we observed robust nuclear and cytoplasmic pERK1/2 staining, which was highly restricted to cells at the base of the crypts (Fig. 6A). At higher magnifications (Fig. 6A′,A″), it was clear that the pERK1/2 staining was present in the crypt base columnar (CBC) cells, tightly wedged between the terminally differentiated Paneth cells, which were pERK-negative. These CBC cells have been shown to be Lrg5-positive (Barker et al. 2007) and to correspond, based on multiple strict criteria, to definitive intestinal stem cells, capable of repopulating entire crypts (Barker et al. 2007). This expression pattern was disrupted in sections of small intestine taken from UBC-CreERT2;Rnpc3lox/lox mice 6 d after tamoxifen treatment. The ensuing highly disorganized epithelium was punctuated with discontinuous regions of 65K (Fig. 6B,B′,B″) and pERK1/2 staining (Fig. 6C,C′,C″), which were largely coincident and glandular in morphology.
FIGURE 6.
pERK expression is concordant with 65K expression in the proliferative cells of intestinal crypts and in recovering regions of the epithelium. (A) Immunohistochemistry of pERK1/2 (pERK) reveals staining in the BrdU-positive compartment (cf. Fig. 5C,D) of crypts (boxed region in A is shown at higher magnification in A′ and A″). Meanwhile, pERK staining is absent from the terminally differentiated cells of the villi (bracket A′). The crypt base columnar (CBC) cells (arrowheads A″) strongly express pERK and can be clearly seen wedged between the large, granular, terminally differentiated Paneth cells, which are pERK negative. (B) Six days following tamoxifen treatment to induce recombination of Rnpc3lox alleles, the structural organization of the small intestinal epithelium is lost, save for discontinuous patches of cells that still express 65K due to incomplete recombination of Rnpc3lox alleles (B′ and arrows B″). (C) These same cells also show robust nuclear and cytoplasmic pERK expression (C is a serial section of B). The left box in B is magnified in panels B′ and C′. Regions predominantly lacking 65K (right box in B) also fail to express pERK (see brackets in magnified panels B″ and C″). Arrows denote individual cells in B and C that are both 65K- and pERK-positive (note that the 65K staining is nuclear only, whereas the pERK staining is found in the nucleus and cytoplasm). Scale bars in A, B, and C = 500 µm. Scale bars in A′, B′, C′, B″, and C″ = 50 µm. Scale bar in A″ = 20 µm.
Collectively, our data demonstrate that the crypts of the small intestine and colon are the main sites of Rnpc3 transcription and 65K synthesis in the intestinal epithelium. Induced loss of 65K protein leads to an increase in cell death, a decrease in cell proliferation and disrupted pERK signaling in cells at the base of the crypts, including the stem cells. The overall deficit in cell production produced by 65K loss resulted in a comprehensive breakdown in the organized structure of the intestinal epithelium, thereby compromising its integrity and function.
Loss of 65K in the intestinal epithelium results in impaired minor class splicing
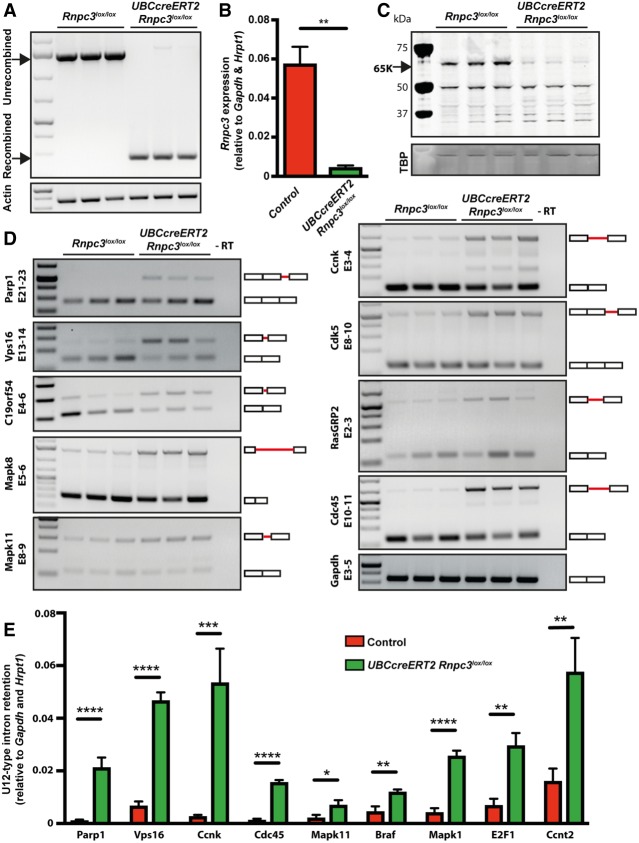
To investigate whether the impact of 65K loss on intestinal integrity was likely to be due to a reduction in the efficiency of U12-dependent splicing, we prepared genomic DNA (gDNA), mRNA, and protein from purified epithelial cells harvested from the small intestine and colon of three independent tamoxifen-treated Rnpc3lox/lox and UBC-CreERT2;Rnpc3lox/lox mice. By 4 d post-tamoxifen treatment, efficient recombination of Rnpc3lox alleles in UBC-CreERT2;Rnpc3lox/lox mice was observed (Fig. 7A, arrow). Using RT-qPCR and western blot analysis, we showed that this was accompanied by a significant decrease in Rnpc3 transcripts (Fig. 7B) and 65K protein (Fig. 7C, arrow), respectively. Using a combination of previously validated (Lotti et al. 2012; Doktor et al. 2017) and newly designed primers, we used semi-quantitative RT-PCR to detect spliced and un-spliced transcripts from a subset of U12-type intron-containing genes. We observed an increase in transcripts retaining the U12-type intron (upper bands in all cases), and a corresponding decrease in correctly spliced transcripts, specifically in intestinal cells taken from tamoxifen-treated UBC-CreERT2;Rnpc3lox/lox mice, compared to cells taken from tamoxifen-treated Rnpc3lox/lox mice (Fig. 7D). There was little effect on U2-type intron retention in the same subset of transcripts (Supplemental Fig. S4). Using RT-qPCR we quantified U12-type intron retention in these transcripts along with others selected from the U12 database (http://genome.crg.es/datasets/U12) (Alioto 2007). This analysis revealed a significant increase in U12-type intron retention for all the genes tested in the 4 d post tamoxifen-treated UBC-CreERT2;Rnpc3lox/lox mice, compared to controls (Fig. 7E). Thus, our data show that loss of 65K significantly impairs U12-dependent splicing in purified intestinal epithelial cells, consistent with the notion that efficient minor class splicing is required for the normal homeostasis and integrity of the gastrointestinal epithelium of adult mice.
FIGURE 7.
Loss of 65K impairs minor class splicing in colonic epithelial cells. Preparations of gDNA, total RNA, and protein were extracted from purified colon crypt epithelial cells 4 d post-tamoxifen treatment. (A) Allele-specific PCR-based analysis demonstrates almost complete recombination of the Rnpc3 locus in UBC-CreERT2;Rnpc3lox/lox samples, but not in Rnpc3lox/lox samples. (B) RT-qPCR confirms that rnpc3 mRNA levels are significantly reduced in UBC-CreERT2;Rnpc3lox/lox samples (green bars) compared to control (red bars; combination of tamoxifen treated UBC-CreERT2;Rnpc3+/+ and Rnpc3lox/lox samples). (C) Western blot analysis shows a marked reduction in 65K protein in UBC-CreERT2;Rnpc3lox/lox compared to Rnpc3lox/lox controls (relative to TBP loading control). (D) RT-PCR and gel electrophoresis analysis of U12-type intron containing genes in colon crypt epithelial cells of mice shown in A. Schematic representation of spliced and intron-retaining mRNAs is shown on the right, with retained U12-type introns represented by red solid lines between exons (white boxes). (E) RT-qPCR using primers designed to amplify transcripts retaining their U12-type intron in several candidate U12-type intron-containing genes (Parp1, Vps16, Ccnk, Cdc45, Mapk11, Braf, Mapk1, E2F1, and Ccnt2) shows that U12-type intron retention is significantly increased in recombined UBC-CreERT2;Rnpc3lox/lox samples (green bars) compared to controls (red bars; combination of tamoxifen-treated UBC-CreERT2;Rnpc3+/+ and Rnpc3lox/lox samples). Data are expressed as mean + SEM relative to expression of housekeeping genes Gapdh and Hrpt1; n = 3 for each genotype. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001 Student's t-test.
DISCUSSION
65K is indispensable for early embryonic development
Our laboratory previously reported the first in vivo vertebrate model of minor class splicing deficiency when we showed that 65K is essential for the normal growth of organs during zebrafish development (Markmiller et al. 2014). Herein we show that 65K is a critical protein component of the U12-dependent spliceosome in mice. It is indispensable for early embryogenesis and for homeostasis of self-renewing/proliferative tissues in later life.
Homozygous mutant mice were never born from heterozygous in-crosses. Harvesting of E3.5 blastocysts for in vitro analysis revealed that whereas Rnpc3+/+ and Rnpc3+/− blastocysts progressed normally through early developmental stages, Rnpc3−/− embryos arrested after the cleavage stage resulting in compacted morulas. Maternally encoded proteins typically support embryonic development until the blastocyst stage; thus, the failure of embryos lacking Rnpc3 to develop beyond the morula stage points to an essential function for minor class splicing in preimplantation mouse development. Interestingly, the Rnpc3 gene is expressed transiently in E3.5 primitive endoderm (Gerovska and Araúzo-Bravo 2016), further supporting a role for 65K in early mouse embryogenesis.
From our study, we cannot say whether the early developmental lethality caused by 65K loss is brought about by the dysregulation of one or many U12-type intron-containing gene(s). There are several possible candidates among the 647 mouse U12-type intron-containing genes (Horiuchi et al. 2018) whose deficiency causes early embryonic lethality in mice, including Mapk1, Plc3b, GAK, and Cyclin K. However, we did not investigate whether the phenotype we observed is due to the disrupted expression of a single U12-type intron-containing gene, or the combined effect of disrupting many such genes.
Robust activation of the Rnpc3 locus is evident in the self-renewing tissues of adult mice
In the adult mouse, Rnpc3 transcription is activated in the stem/progenitor cell compartments of several self-renewing tissues. It is particularly striking in the intestinal epithelial stem cells located at the base of crypts (Barker et al. 2007) and in the progenitor cells occupying the transit-amplifying zone immediately above them. In contrast, Rnpc3 transcription is not conspicuous in the more differentiated cells of the small intestinal villi, where cells progressing through the cell cycle are rare. That activation of transcription was accompanied by expression of 65K protein in the same cells was confirmed by immunohistochemistry, which showed that the 65K protein was present in the nucleus of stem/progenitor cells in the small intestine and colon crypts. We also demonstrated stem cell-specific Rnpc3 transcription in the skin bulge and hair germ region, the locations of the hair follicle and melanocyte stem cells, respectively (Hsu et al. 2014). A subset of lung epithelial cells, located at the bronchio-alveolar duct junction (BADJ), the putative location of lung stem cells that help to maintain homeostasis of the lung epithelia, also showed active Rnpc3 transcription. Much lower levels of expression were observed in several other, but not all, tissues of adult mice. Of note, it has been reported that 65K expression is not only tightly controlled at the transcriptional level but also by an elaborate post-transcriptional mechanism (Verbeeren et al. 2010, 2017). Through this mechanism, the length of the 3′ untranslated region (3′UTR) of human RNPC3 mRNA can be adjusted to produce either a translation-competent mRNA isoform with a short 3′UTR or a nonproductive isoform with a long 3′UTR, which is retained in the nucleus (Verbeeren et al. 2017). The authors of this study speculated that switching to the long 3′UTR splicing isoform may provide a mechanism to keep the levels of the 65K protein balanced in cells undergoing differentiation (Verbeeren et al. 2017).
65K impairment impacts most severely on self-renewing tissues
Systemic deletion of 65K in the adult mouse impacted most severely on tissues that undergo continuous self-renewal to maintain homeostasis, such as the hematopoietic compartment, thymus and, most particularly, the gastrointestinal epithelium, where the progenitor cells of intestinal crypts divide every 12–16 h in order to replace the entire small intestinal epithelium every 2–6 d (Sancho et al. 2004). Likewise, the significant anemia, drop in peripheral blood lymphocytes and platelets we observed are most likely due to an effect on the continually renewing hematopoietic stem cells of the bone marrow. The leukopenia/immunodeficiency phenotype we observed is reminiscent of that seen in Roifman syndrome patients where impaired U12-dependent splicing of MAPK1 (critical for B-cell differentiation) and DIAPH1 (known to regulate platelet formation) has been detected in clinical samples (Heremans et al. 2018). Similarly, analysis of Zrsr1 mutant mice indicates a requirement for efficient minor class splicing in RBC production (Horiuchi et al. 2018).
Meanwhile, defects in other tissues with active Rnpc3 transcription, such as the skin and lung, were less evident, perhaps due to the more quiescent nature of the progenitor cells in these tissues. For example, the turnover time of the tracheal–bronchial epithelium of adult rodents is estimated to be more than 100 d (Blenkinsopp 1967), compared to 2–4 d in the intestinal epithelium. Indeed, we think that tissues with lower mitotic indices may ultimately have been negatively impacted by 65K loss in our experiments. However, the rapid weight loss of UBC-CreERT2;Rnpc3lox/lox mice upon induced 65K deficiency meant that all mice reached the ethical end-point of the experiment at 8 d post-tamoxifen and were euthanized, thereby precluding observation of any delayed aspects of the phenotype.
Apop-Tag staining of histological sections of the small intestine demonstrated that following deletion of 65K, the transit-amplifying compartment was depleted of proliferating cells via apoptosis. Intestinal stem cells are known to be extremely susceptible to apoptosis when placed under cellular stress (Potten and Grant 1998). However, 6 d after tamoxifen treatment, discrete pockets of large, BrdU-positive crypts emerged in a typical wound healing response to epithelial damage (Ashton et al. 2010). A likely explanation for the appearance of regenerative crypts is that they originated from a small proportion of stem cells in which complete recombination of Rnpc3lox alleles did not occur. Immunohistochemical analysis supports this interpretation because we observed expression of 65K in regenerating crypt cells that was coincident with activation of the pro-proliferative MAPK family proteins, ERK1/2.
Our data show that 65K is expressed in the pluripotent stem cells of intestinal crypts where it is required for efficient minor class splicing and intestinal integrity. In contrast, the quiescent cells that have migrated onto the villi survive longer with only residual/negligible amounts of 65K protein present. Thus, in the intestinal epithelium, the terminally differentiated cells appear not to require efficient minor class splicing to survive. In contrast, our observation that the major class spliceosome-specific component, U1A is robustly synthesized by all intestinal epithelial cells suggests that major class splicing is required in both the proliferative and differentiated compartments of this tissue.
While our observations suggest that mouse Rnpc3 expression may not be essential in all cells, one context in which RNPC3 may be an essential gene is cancer. This is indicated by the results of a large number of genome-wide CRISPR/Cas9 targeting screens designed to identify genes that are essential for the proliferation and viability of human cancer cell lines in culture (Blomen et al. 2015; Hart et al. 2015; Wang et al. 2015; Meyers et al. 2017). We determined that RNPC3 is an essential gene in this context by interrogating the GenomeCRISPR database (Rauscher et al. 2017), which comprises data from ∼500 different CRISPR/Cas9 targeting experiments performed in 421 different human cancer cell lines. We discovered that RNPC3 is almost invariably classified as an essential gene in these studies (http://genomecrispr.dkfz.de/#!/results/RNPC3), thereby leading us to hypothesize that RNPC3 may represent a genetic vulnerability of cancer cells that could be targeted therapeutically.
Relevance of Rnpc3 mutations to minor class splicing clinical disorders
A small number of human developmental disorders that are caused by a mutation in a minor class splicing component have been identified and these are characterized by a broad range of severity and distinct, tissue-specific effects. In contrast to the severity of MOPD1, familial isolated growth hormone deficiency is a treatable growth retardation phenotype that was observed in offspring inheriting compound heterozygous mutations in RNPC3 (Argente et al. 2014). This was the first time a gene encoding a minor class splicing-specific protein had been implicated in a genetically inherited human disease (Argente et al. 2014). In the single family affected, the RNPC3 mutations occurred in the part of the gene encoding the more C-terminal of its two RNA binding domains. This has been shown to disrupt binding of the protein to the U12 and U6atac snRNAs in the U11/U12 di-snRNP intron recognition complex (Norppa et al. 2018). One sequence variant causes an amino acid change at codon 474 (P474T) and the second introduces a stop codon at position 502 (R502X) of the 517 aa/65kDa protein, resulting in impaired expression of the corresponding mRNA (Argente et al. 2014). Comparing the disease outcomes in this family (severe isolated growth hormone deficiency and pituitary hypoplasia) with the severe phenotypes of Rnpc3-null zebrafish and mice, suggests that the missense (P474T) mutation is hypomorphic. Based on our work, we would expect the inheritance of two truly null RNPC3 alleles in human to be embryonic lethal.
There are several instances where defects in minor class splicing impact on the developing and adult central nervous system. For example, mutations in the RNU12 gene cause congenital cerebellar ataxia characterized by delayed motor milestones in development, mild learning difficulties and hypotonia in infancy (Elsaid et al. 2017). Interestingly, the impaired expression of the CULIN1, ATAXIN10, and SLC9A6 genes, which all contain a U12-type intron, has been implicated in several cerebellar ataxia-related phenotypes. A mouse model of SLC9A6 deficiency causes a marked and progressive loss of Purkinje cells and a discrete clinical phenotype attributable to motor hyperactivity and cerebellar dysfunction (Strømme et al. 2011). Our observation that Purkinje neurons exhibit strong Rnpc3 transcription is consistent with the notion that efficient minor class splicing is required for the integrity of these cells and may be indispensable for their fundamental role in controlling motor movement. Finally, in a mouse model of conditional Rnu11 deficiency, recombination of Rnu11 alleles in the developing neuroepithelium of the pallium from E9.5 caused the death of the self-amplifying radial glial cells, leading to severe microcephaly at birth (Baumgartner et al. 2018). Less sensitive to U11 snRNA loss were the intermediate progenitor cells and neurons, demonstrating cell type-specific requirements for minor class spliceosome activity in the developing brain.
Widespread versus tissue-specific impact of impaired minor class splicing
An unresolved question regarding the pathological consequences of impaired minor class splicing is whether the observed clinical phenotypes are caused by global defects in the splicing of U12-type intron-containing genes, or tissue-specific effects produced by a small subset of these genes. Another layer of influence stems from the participation of genes containing U12-type introns in essential pathways and processes such as DNA repair (PARP1, DDB1, EXO1, MSH3), transcription (E2F1-6), general splicing (ESRP1, SRPK1-3) and translation (EIF3I, EIF3K, EIF4G3, EIF2B4). Using RNA-seq in zebrafish, we showed that impaired minor class splicing has the potential to impact indirectly on the expression of all genes, not just those containing U12-type introns (Markmiller et al. 2014). Given this far-reaching capacity to alter the expression of the entire genome, it is hardly surprising that disruption of this “minor” process is incompatible with life.
Gene-set enrichment analysis reveals that U12-type introns are over-represented in genes encoding components of signaling pathways relevant to cancer such as ERK2, MAPK11/P38β, JNK1, BRAF (Supplemental Fig. S1A; Markmiller et al. 2014; Merico et al. 2015). Moreover, our RT-PCR analysis showed splicing defects in several MAPK genes following 65K protein deletion. This, together with the results of the cancer gene essentiality studies described above, leads us to hypothesize that impairing the efficiency of minor class splicing may also arrest the growth of tumors, particularly those reliant on RAS-MAPK signaling (Dhillon et al. 2007; McCormick 2011). Now that we have generated conditional alleles of Rnpc3 deficiency, we will test our hypothesis using models of genetically engineered, tumor-prone mice.
MATERIALS AND METHODS
Gene-set enrichment analysis
Seven hundred and forty-four human U12-type intron containing genes (Merico et al. 2015) were analyzed using the Broad GSEA tool to identify enriched GO terms for biological pathways, processes, and molecular functions. Significant enrichment was assessed with an FDR-q of <0.05 (Mootha et al. 2003; Subramanian et al. 2005).
Mice
All procedures performed on mice were conducted with the approval of the Animal Ethics Committees of the Walter and Eliza Hall Institute of Medical Research, and the Parkville Branch of the Ludwig Institute for Cancer Research, Melbourne, Victoria, Australia.
We purchased an ES cell clone containing a targeted Rnpc3 “knockout-first” allele (EPD0441_1_B10) comprising a LacZ-tagged (Rnpc3neo) null allele (Fig. 1A), from the EUCOMM (European Conditional Mouse Mutagenesis) consortium (Skarnes et al. 2011). ES cells were transplanted into blastocysts at the Monash University node of the Australian Phenomics Network (APN). We tested the resulting chimeras for germline transmission using a copulatory plug screening protocol (Wilson and Sheardown 2011). Heterozygous offspring of positive chimeras were identified by genotyping (for PCR primers, see Supplemental Table S1). All mice except those used to harvest E3.5 blastocysts (see below) were maintained on a pure C57BL/6 genetic background.
We obtained UBC-CreERT2 (Soriano 1999; Ruzankina et al. 2007) mice from Prof Wayne Phillips at the Peter MacCallum Cancer Centre, Melbourne, Australia. Recombination of floxed alleles was achieved by crossing Rnpc3lox/+ or Rnpc3lox/lox mice (Fig. 1B) with mice carrying a single UBC-CreERT2 allele. Tamoxifen (Sigma; 30 mg/mL) was administered by oral gavage in two consecutive daily doses (150 µL) to bring about ubiquitous recombination. After tamoxifen treatment, mouse weight was monitored daily. To assess Cre-mediated deletion of Rnpc3, gDNA was extracted from tissues of interest at timed intervals following tamoxifen treatment. Primers were designed to detect un-recombined and recombined Rnpc3 alleles (see Supplemental Table S1). At the end of the experiment, histopathology and organ pathology were performed by the University of Melbourne Department of Pathology node of the APN.
β-galactosidase staining
To detect cells with transcriptionally active Rnpc3 loci, dissected tissues from Rnpc3neo/+ mice were washed thoroughly with PBS. To allow scanning of the entire intestine, 10 cm segments were slit open longitudinally, cleaned with PBS and rolled up longitudinally (“Swiss rolls”), mucosa inwards, using a 21G syringe needle. Tissues were fixed in 4% PFA in PBS for 1 h on a roller at 4°C. Each sample was then rinsed three times for 20 min in 2 mL wash buffer (23 mM NaH2P4, 77 mM Na2H2P4, 2 mM MgCl2, 0.02% NP40, 0.02% sodium deoxycholate, 7.5 µM BSA in water) at RT. Each tissue was stained overnight in 1 mL X-gal buffer [1 mg/mL X-gal, 5 mM K3Fe(CN)6, 6 mM K4Fe(CN)6, 2 mM MgCl2 in wash buffer] at 37°C. Stained tissues were washed twice in PBS and post-fixed in 2% PFA overnight at 4°C. Tissues were transferred to 70% ethanol prior to embedding in paraffin for sectioning and light counterstaining with nuclear fast red.
Blastocyst experiments
For blastocyst studies, Rnpc3+/− mice were crossed onto a FVB background to introduce hybrid vigor and generate larger litter sizes. For genotyping, gDNA was extracted from individual blastocysts in DirectPCR Lysis Reagent (Viagen Biotech) with 1 mg/mL proteinase K (4 h at 55°C followed by inactivation at 85°C for 45 min). Where indicated, blastocysts were cultured first in ES medium (Voss et al. 1997) on 0.1% gelatin-coated plates and photographed every 24 h using a Diaphot 300 phase-contrast microscope (Nikon) to capture progress through ex vivo development.
Blood analysis
Blood was collected from the retro-orbital plexus of adult mice and cell counts were obtained with an Advia 2120 hematological analyzer (Bayer).
Histology and immunohistochemistry
Histological sections (4 µm) of various tissues, including Swiss roll preparations of the small and large intestines, were stained with hematoxylin and eosin (H&E). Visualization of apoptosis was performed using the Apop-Tag Peroxidase In Situ Apoptosis Detection Kit (Merck Millipore) according to the manufacturer's instructions. To detect cells in S-phase of the cell cycle, mice were given an intraperitoneal injection of bromodeoxyuridine (BrdU; 100 mg/kg; Sigma-Aldrich), 2 h prior to collection of tissues. For immunohistochemistry, the following antibodies were used: anti-65K antibody (ab90090 1:250; Abcam, Cambridge, UK), anti-U1A antibody (ab155054 1:1000; Abcam), anti-pERK (#4370 1:500; Cell Signaling Technologies), and anti-BrdU antibody (BD555624; Life Technologies). Antigen retrieval was performed by heating to 100°C in 10 mM pH 6 sodium citrate buffer, endogenous peroxidase activity was inhibited with 3% H2O2 (v/v; BioLab) and nonspecific antibody binding was blocked using 10% goat serum (w/v; Sigma-Aldrich). Primary antibody binding was performed overnight at 4°C in a humidified box, followed by exposure to anti-rabbit biotinylated secondary antibodies and Vectastain Elite ABC HRP reagent (Vector laboratories) for 30 min at room temperature (RT). Signals were detected with the Liquid DAB+ Substrate Chromogen System (Dako) prior to counterstaining with haematoxylin.
All images were captured using an upright Nikon Eclipse 90i microscope or a Panoramic Scan II (3DHISTECH).
Western blotting
Preparations of pure intestinal epithelial cells harvested from freshly dissected small and large intestines of 8- to 13-wk-old mice (Whitehead et al. 1987) were lysed in RIPA buffer (150 mM NaCl, 0.1% NP40, 0.5% sodium deoxycholate, 50 mM Tris [pH 8.0]) supplemented with Complete Protease Inhibitor Cocktail and PHOSTOP phosphatase inhibitors (Roche). Lysates were incubated on ice for 30 min and the soluble fraction was obtained following centrifugation for 20 min at 13,000 rpm. The protein concentration of samples was determined by BCA protein assay (Pierce).
Protein lysate (50 µg of per lane) was loaded onto NuPAGE Novex Bis-Tris 4–12% polyacrylamide gels and transferred onto Immobilon-FL PVDF membranes (Millipore). Membranes were blocked overnight at 4°C in Odyssey Blocking Buffer (Li-Cor) and incubated with primary antibodies either overnight at 4°C for anti-65K (25820-1-AP, 1;1000; Proteintech) or for 1 h at RT for anti-TATA binding protein (TBP; loading control) (ab51841, 1:1000; Abcam). Both secondary antibodies: donkey anti-rabbit 680 (Odyssey IRDye 92632221) and donkey anti-mouse 800cw (Odyssey IRDye 923632212) were used at 1:10,000 and incubated with membranes for 1 h at RT. An Odyssey infrared imaging system (Licor) was used to scan membranes.
Splicing analysis using reverse transcriptase-PCR (RT-PCR)
Total RNA from purified intestinal epithelial cells was extracted using TRIsure reagent (Bioline) and the Qiagen RNeasy mini kit (QIAGEN). RNA integrity was analyzed on a 2100 Bioanalyzer (Agilent) and 1 µg aliquots were used to generate cDNA with the Superscript III First Strand Synthesis System (Invitrogen) according to the manufacturer's instructions. Splicing and intron retention were visualized by electrophoresis of RT-PCR products generated using primer pairs in exons spanning either U12- or U2-type introns (see Supplemental Table S1 for PCR primer sequences). To determine the efficiency of U12-type intron removal in response to Rnpc3 deficiency, we used a primer within the U12-type intron to amplify un-spliced (intron-retaining) mRNA. RT-quantitative PCR (RT-qPCR) was performed using a SensiMix SYBR kit (Bioline) on an Applied Biosystems ViiA™7 Real-Time PCR machine. Expression data were normalized by reference to Gapdh and Hprt1 expression. LinRegPCR V11.0 was used for baseline correction, PCR efficiency calculation, and transcript quantification analysis (Ruijter et al. 2009).
Statistical methods
Pairwise comparisons were made using two-tailed Student's t-tests assuming equal variances in Prism (Version 6; GraphPad Software). Error bars represent the mean ± SEM, n ≥ 3 independent mice of each genotype. Two-way ANOVA with Tukey's multiple comparison testing was used to define significance in normalized mouse weight across genotype and over time. A P-value <0.05 was used to define statistical significance. Animal survival data were plotted as Kaplan-Meier curves.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Andrew Naughton, Melanie Asquith, Mel Pritchard, Emily Sutherland, and Faye Dobrowski (mouse husbandry), Cary Tsui (histology), Qian Du, Lotta Burstroem, Julia Griesbach, and Samantha Eccles (technical assistance), and Anne Voss and Michael Buchert (helpful discussions). We also thank Drs. Stephen Scherer and Daniele Merico for providing us with their list of 822 human U12-type introns distributed across 744 genes. Rnpc3-targeted mice were generated and analyzed in collaboration with the European Conditional Mouse Mutagenesis Consortium (EUCOMM) and the Monash and Melbourne University nodes of the Australian Phenomics Network (APN). This work was supported by the National Health and Medical Research Council of Australia (grant 1024878 and Senior Research Fellowship 1022870 to J.K.H.), Ludwig Cancer Research and a Victorian State Government Operational Infrastructure Support grant.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.068221.118.
Freely available online through the RNA Open Access option.
REFERENCES
- Alioto TS. 2007. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res 35: D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argente J, Flores R, Gutiérrez-Arumí A, Verma B, Martos-Moreno GÁ, Cuscó I, Oghabian A, Chowen JA, Frilander MJ, Pérez-Jurado LA. 2014. Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Mol Med 6: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton GH, Morton JP, Myant KK, Phesse TJ, Ridgway RA, Marsh VV, Wilkins JA, Athineos D, Muncan V, Kemp R, et al. 2010. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev Cell 19: 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Barker N, van de Wetering M, Clevers H. 2008. The intestinal stem cell. Genes Dev 22: 1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner M, Olthof AM, Aquino GS, Hyatt KC, Lemoine C, Drake K, Sturrock N, Nguyen N, Al Seesi S, Kanadia RN. 2018. Minor spliceosome inactivation causes microcephaly, owing to cell cycle defects and death of self-amplifying radial glial cells. Development 145: dev166322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benecke H, Lührmann R, Will CL. 2005. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11–59K protein. EMBO J 24: 3057–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkinsopp WK. 1967. Proliferation of respiratory tract epithelium in the rat. Exp Cell Res 46: 144–154. [DOI] [PubMed] [Google Scholar]
- Blomen VA, Májek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science 350: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Burge CB, Padgett RA, Sharp PA. 1998. Evolutionary fates and origins of U12-type introns. Mol Cell 2: 773–785. [DOI] [PubMed] [Google Scholar]
- Chang WC, Chen YC, Lee KM, Tarn WY. 2007. Alternative splicing and bioinformatic analysis of human U12-type introns. Nucleic Acids Res 35: 1833–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W. 2007. MAP kinase signalling pathways in cancer. Oncogene 26: 3279–3290. [DOI] [PubMed] [Google Scholar]
- Doktor TK, Hua Y, Andersen HS, Brøner S, Liu YH, Wieckowska A, Dembic M, Bruun GH, Krainer AR, Andresen BS. 2017. RNA-sequencing of a mouse-model of spinal muscular atrophy reveals tissue-wide changes in splicing of U12-dependent introns. Nucleic Acids Res 45: 395–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al. 2011. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332: 240–243. [DOI] [PubMed] [Google Scholar]
- Elsaid MF, Chalhoub N, Ben-Omran T, Kumar P, Kamel H, Ibrahim K, Mohamoud Y, Al-Dous E, Al-Azwani I, Malek JA, et al. 2017. Mutation in noncoding RNA RNU12 causes early onset cerebellar ataxia. Ann Neurol 81: 68–78. [DOI] [PubMed] [Google Scholar]
- Gerovska D, Araúzo-Bravo MJ. 2016. Does mouse embryo primordial germ cell activation start before implantation as suggested by single-cell transcriptomics dynamics? Mol Hum Reprod 22: 208–225. [DOI] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163: 1515–1526. [DOI] [PubMed] [Google Scholar]
- He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, et al. 2011. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 332: 238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heremans J, Garcia-Perez JE, Turro E, Schlenner SM, Casteels I, Collin R, de Zegher F, Greene D, Humblet-Baron S, Lesage S, et al. 2018. Abnormal differentiation of B cells and megakaryocytes in patients with Roifman syndrome. J Allergy Clin Immunol 142: 630–646. [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Perez-Cerezales S, Papasaikas P, Ramos-Ibeas P, López-Cardona AP, Laguna-Barraza R, Fonseca Balvís N, Pericuesta E, Fernández-González R, Planells B, et al. 2018. Impaired spermatogenesis, muscle, and erythrocyte function in U12 intron splicing-defective Zrsr1 mutant mice. Cell Rep 23: 143–155. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Li L, Fuchs E. 2014. Emerging interactions between skin stem cells and their niches. Nat Med 20: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HJ, Kang H. 2014. The Arabidopsis U11/U12-65K is an indispensible component of minor spliceosome and plays a crucial role in U12 intron splicing and plant development. Plant J 78: 799–810. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci 93: 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti F, Imlach WL, Saieva L, Beck ES, Hao le T, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, et al. 2012. An SMN-dependent U12 splicing event essential for motor circuit function. Cell 151: 440–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Cloonan N, Lardelli RM, Doggett K, Keightley MC, Boglev Y, Trotter AJ, Ng AY, Wilkins SJ, Verkade H, et al. 2014. Minor class splicing shapes the zebrafish transcriptome during development. Proc Natl Acad Sci 111: 3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. 2011. Cancer therapy based on oncogene addiction. J Surg Oncol 103: 464–467. [DOI] [PubMed] [Google Scholar]
- Merico D, Roifman M, Braunschweig U, Yuen RK, Alexandrova R, Bates A, Reid B, Nalpathamkalam T, Wang Z, Thiruvahindrapuram B, et al. 2015. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman syndrome by disrupting minor intron splicing. Nat Commun 6: 8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. 2017. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49: 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273. [DOI] [PubMed] [Google Scholar]
- Niemelä EH, Frilander MJ. 2014. Regulation of gene expression through inefficient splicing of U12-type introns. RNA Biol 11: 1325–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norppa AJ, Kauppala TM, Heikkinen HA, Verma B, Iwaï H, Frilander MJ. 2018. Mutations in the U11/U12-65K protein associated with isolated growth hormone deficiency lead to structural destabilization and impaired binding of U12 snRNA. RNA 24: 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Jung HJ, Nguyen Dinh S, Kang H. 2016. Structural features important for the U12 snRNA binding and minor spliceosome assembly of Arabidopsis U11/U12-small nuclear ribonucleoproteins. RNA Biol 13: 670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, McCarthy M, Steitz JA. 2002. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J 21: 3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Grant HK. 1998. The relationship between ionizing radiation-induced apoptosis and stem cells in the small and large intestine. Br J Cancer 78: 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher B, Heigwer F, Breinig M, Winter J, Boutros M. 2017. GenomeCRISPR–a database for high-throughput CRISPR/Cas9 screens. Nucleic Acids Res 45: D679–D686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AG, Charette JM, Spencer DF, Gray MW. 2006. An early evolutionary origin for the minor spliceosome. Nature 443: 863–866. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho E, Batlle E, Clevers H. 2004. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol 20: 695–723. [DOI] [PubMed] [Google Scholar]
- Singh J, Sikand K, Conrad H, Will CL, Komar AA, Shukla GC. 2016. U6atac snRNA stem-loop interacts with U12 p65 RNA binding protein and is functionally interchangeable with the U12 apical stem-loop III. Sci Rep 6: 31393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71. [DOI] [PubMed] [Google Scholar]
- Strømme P, Dobrenis K, Sillitoe RV, Gulinello M, Ali NF, Davidson C, Micsenyi MC, Stephney G, Ellevog L, Klungland A, et al. 2011. X-linked Angelman-like syndrome caused by Slc9a6 knockout in mice exhibits evidence of endosomal-lysosomal dysfunction. Brain 134: 3369–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeeren J, Niemelä EH, Turunen JJ, Will CL, Ravantti JJ, Lührmann R, Frilander MJ. 2010. An ancient mechanism for splicing control: U11 snRNP as an activator of alternative splicing. Mol Cell 37: 821–833. [DOI] [PubMed] [Google Scholar]
- Verbeeren J, Verma B, Niemelä EH, Yap K, Makeyev EV, Frilander MJ. 2017. Alternative exon definition events control the choice between nuclear retention and cytoplasmic export of U11/U12-65K mRNA. PLoS Genet 13: e1006824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Gruss P. 1997. Germ line chimeras from female ES cells. Exp Cell Res 230: 45–49. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead RH, Brown A, Bhathal PS. 1987. A method for the isolation and culture of human colonic crypts in collagen gels. In Vitro Cell Dev Biol 23: 436–442. [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Lührmann R. 2004. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10: 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S, Sheardown SA. 2011. Identification of germline competent chimaeras by copulatory plug genotyping. Transgenic Res 20: 429–433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.