Abstract
Riboswitches contain structured aptamer domains that, upon ligand binding, facilitate helical switching in their downstream expression platforms to alter gene expression. To fully dissect how riboswitches function requires a better understanding of the energetic landscape for helical switching. Here, we report a sequencing-based high-throughput assay for monitoring in vitro transcription termination and use it to simultaneously characterize the functional effects of all 522 single point mutants of a glycine riboswitch type-1 singlet. Mutations throughout the riboswitch cause ligand-dependent defects, but only mutations within the terminator hairpin alter readthrough efficiencies in the absence of ligand. A comprehensive analysis of the expression platform reveals that ligand binding stabilizes the antiterminator by just 2–3 kcal/mol, indicating that the competing expression platform helices must be extremely close in energy to elicit a significant ligand-dependent response. These results demonstrate that gene regulation by this riboswitch is highly constrained by the energetics of ligand binding and conformational switching. These findings exemplify the energetic parameters of RNA conformational rearrangements driven by binding events.
Keywords: riboswitch, expression platform, glycine, singlet, transcription termination
INTRODUCTION
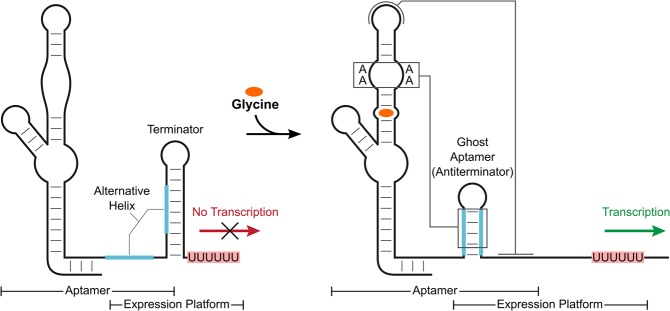
Riboswitches are noncoding RNA elements that alter gene expression in response to changing levels of specific effector molecules (Mandal and Breaker 2004b; Nudler and Mironov 2004). They typically contain a structured aptamer domain that, upon ligand-binding, stabilizes one of two or more competing helices in a downstream expression platform (Barrick and Breaker 2007). This promotes gene expression in “ON” switches and reduces expression in “OFF” switches by presenting or sequestering key elements, often at the levels of transcription or translation (Sherwood and Henkin 2016). For instance, ligand-binding by a transcriptional ON switch stabilizes an antiterminator helix that is mutually exclusive with the formation of a terminator hairpin. This leads to production of more full-length mRNA and increased levels of gene expression (Fig. 1).
FIGURE 1.
Gene regulation by a transcriptional ON switch. Depicted is an example of a glycine riboswitch type-1 singlet transcriptional ON switch. A terminator hairpin forms in the absence of ligand, which leads to termination of the mRNA upstream of the gene of interest. Ligand binding induces a structural rearrangement and stabilizes an antiterminator helix that is mutually exclusive with the terminator hairpin. This causes an increase in production of full-length mRNA and gene expression.
The molecular basis of ligand recognition by the conserved aptamer domain is well understood for many riboswitches. X-ray crystal structures of isolated aptamers bound to ligand have elucidated details of the aptamer folds and revealed which functional groups directly contact the ligand (Serganov and Patel 2012). Mutagenesis and structure-activity relationship analyses have subsequently established the energetic contributions of specific RNA-ligand interactions (Gilbert et al. 2007; Shanahan et al. 2011; Meehan et al. 2016). These studies form the framework for our current understanding of how aptamers achieve physiologically sufficient specificity and affinity.
However, the role of the aptamer extends beyond ligand binding. Aptamers must facilitate helical switching in the associated expression platform to alter gene expression. The nucleotides in these helices are typically not conserved, yet the sequence identity affects riboswitch function. Multiple studies have demonstrated that the dynamic range of a riboswitch can be modified by altering the strength of these helices (Heppell et al. 2011; Ceres et al. 2013; Marcano-Velázquez and Batey 2015), indicating that helical switching depends on their relative stabilities. Yet, it is not known how close in energy these competing helices need to be or how deviations from the consensus aptamer sequence affect the energy supplied to the expression platform. Resolving these questions would provide important insights about the energetic constraints riboswitches face. Therefore, we set out to systematically analyze how mutations throughout the aptamer and expression-platform domains of a transcriptionally controlled riboswitch affect termination efficiency. We selected the glycine riboswitch type-1 singlet as a model system.
The glycine riboswitch is widespread across bacteria and controls genes related to glycine metabolism and transport (Mandal et al. 2004; Kazanov et al. 2007; McCown et al. 2017). There are three subtypes of glycine riboswitches: two single-aptamer variants, referred to as singlets, and a tandem dual-aptamer version (Mandal et al. 2004; Ruff et al. 2016). The tandem system is the most common and uses two consecutive and homologous aptamer domains to control a single expression platform (Barrick and Breaker 2007; Ruff and Strobel 2014; Ruff et al. 2016). Singlets contain just one aptamer that is either followed (type-1) or preceded (type-2) by a short hairpin termed the “ghost aptamer” (Ruff et al. 2016). Bioinformatic information for these ghost aptamers is limited, but the biochemical evidence indicates that both singlets form an A-minor motif between their ghost aptamer and conserved adenines in the P3 helix. For type-1 singlets, the ghost aptamer is expected to be part of the expression platform and become stabilized upon ligand binding, likely through the formation of this A-minor motif (Ruff et al. 2016). Meanwhile, the ghost aptamer is expected to play a structural role for type-2 singlets with ligand binding predicted to stabilize the P1 stem, similar to tandem systems (Mandal et al. 2004; Ruff et al. 2016). All three subtypes of the glycine riboswitch have been shown to bind glycine (Mandal et al. 2004; Ruff et al. 2016). However, the ability to regulate gene expression has only been demonstrated for the tandem system (Mandal et al. 2004; Babina et al. 2017).
Here, we report consensus alignments for the type-1 and type-2 singlets and demonstrate that both singlet subtypes can functionally modulate transcription termination in vitro. We also present a high-throughput method for performing in vitro transcription termination that we have termed SMARTT (Sequencing-based Mutational Analysis of RNA Transcription Termination). Using SMARTT, we quantitatively and simultaneously analyzed all 522 single point mutants and several combinations of double mutants of a glycine riboswitch type-1 singlet. The effects of these mutations provide a quantitative measure of the functional importance of individual structural elements. These data demonstrate an exemplary case of riboswitch gene regulation that is highly constrained by the energetics of ligand binding and conformational switching.
RESULTS
Bioinformatic analysis of the type-1 and type-2 singlets
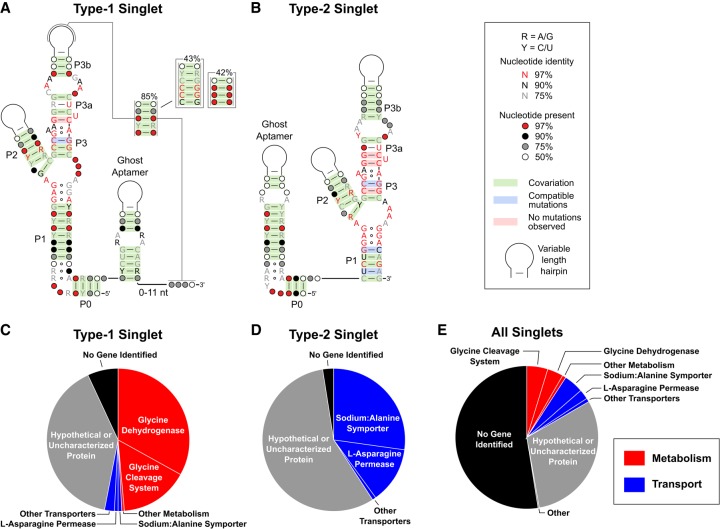
We generated consensus alignments for the type-1 and type-2 singlet sequences to determine if there are any systematic differences between these variants (Fig. 2). We found 15,847 unique glycine riboswitch sequences, of which 4221 (27%) contain a single aptamer. After applying stringent filters to limit the number of false positives (Materials and Methods), 269 and 380 of these single-aptamer variants could be unambiguously classified as type-1 and type-2 singlets, respectively.
FIGURE 2.
Bioinformatic analysis of glycine riboswitch singlets. Consensus sequences are shown for the type-1 (A) and type-2 (B) glycine riboswitch singlets. Pie charts depict the relative abundance of the genes located directly downstream from the type-1 singlets (C), the type-2 singlets (D), and all identified singlets (E).
The consensus sequences for these two classes of singlets are highly homologous with two main exceptions. First, a putative pseudoknot was identified between the P3b stem–loop and the nucleotides following the ghost aptamer in 85% of type-1 singlets. This pseudoknot displays covariation and frequently contains 3–4 consecutive cytosine nucleotides within the P3b stem–loop and 3–4 guanosine nucleotides following the ghost aptamer. The location of this G-rich sequence downstream from the ghost aptamer tends to roughly correlate with the length of the P3b stem–loop and often overlaps with portions of the alternative expression platform helix. Similar pseudoknots have been implicated to play key roles in the mechanism of gene regulation in other riboswitches (Corbino et al. 2005; Klein et al. 2009; Johnson et al. 2012; Nelson et al. 2013) and an analogous pseudoknot was previously described for the tandem glycine system (Kladwang et al. 2012), but was not experimentally verified. Pseudoknots were not identified in any of the type-2 singlets. A second difference between the two singlet subtypes is that the type-1 singlet typically contains a longer P1 helix in its aptamer domain and a shorter ghost aptamer helix compared to the type-2 singlet. This is similar to tandem systems, where the P1 stem tends to be longer in the first aptamer than the second (Mandal et al. 2004; Barrick and Breaker 2007).
The consensus sequences additionally revealed that the ghost aptamer helix of both singlets frequently contains a set of purine–purine mispairs—depicted as an internal loop in Figure 2A. This motif resembles the stretch of purine–purine pairs that stack on top of the P1 helix of glycine aptamers (Mandal et al. 2004; Barrick and Breaker 2007; Butler et al. 2011). In the crystal structure of the Fusobacterium nucleatum tandem glycine riboswitch, these purines form two sheared GA base pairs that stack with an intercalating adenine from the P3 helix of the same aptamer (Supplemental Fig. S1). Singlets do not have this adenine, so it is unclear if a similar structure is formed within the ghost aptamers. These observations suggest that the ghost aptamer may not be just a generic helix, but contains specific sequence elements that may be important for function.
The genes located downstream from a riboswitch often provide clues about its function. The genetic context of type-1 and type-2 singlets was analyzed to determine whether the two subtypes control similar sets of genes. Consistent with previous bioinformatic analyses of glycine riboswitches, the majority of characterized proteins following the 4221 singlets were either involved in the metabolism of glycine and other amino acids (55%) or annotated as various types of amino acid transporters (44%; Fig. 2E; Mandal et al. 2004; Kazanov et al. 2007; Ruff et al. 2016). However, a clear bias was observed regarding genes controlled by type-1 singlets versus type-2 singlets. 93% of the characterized genes located downstream from type-1 singlets are involved in glycine catabolism, whereas 99% of the characterized genes controlled by type-2 singlets encode amino acid transporters (Fig. 2C,D). Tandem glycine riboswitches that regulate gcvT (glycine cleavage system aminomethlytransferase T) and alsT (annotated as a sodium:alanine symporter) have been shown to act as ON and OFF switches, respectively (Mandal et al. 2004; Babina et al. 2017; Khani et al. 2018). This means that at high concentrations of glycine, gcvT would be up-regulated and used to degrade excess glycine, while alsT would be down-regulated and limit the import of glycine and/or alanine. Altogether, these data suggest that type-1 singlets primarily function as ON switches whereas type-2 singlets predominantly act as OFF switches.
Type-1 and type-2 singlets are functional in transcription termination assays
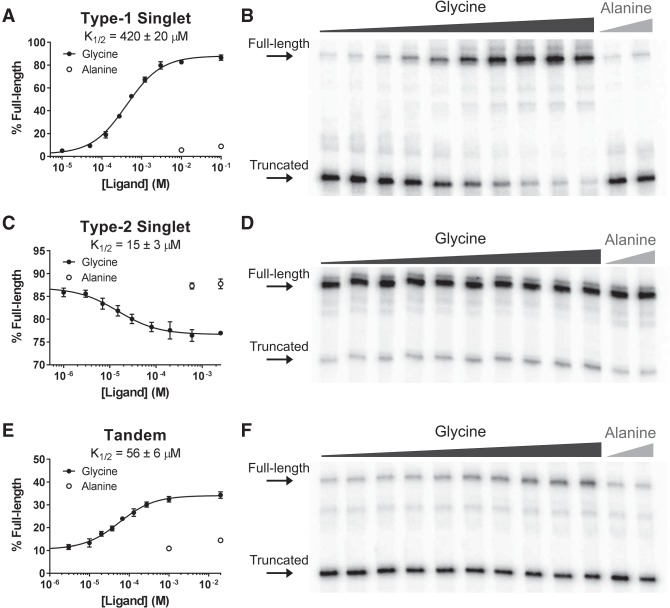
Ruff et al. previously reported that isolated aptamers from type-1 and type-2 singlets bind glycine with affinities similar to tandem variants (Ruff et al. 2016). However, the ability to bind ligand does not always translate to a functional role in gene control (Li et al. 2016). We set out to determine whether both singlet subtypes are capable of regulating gene expression using an in vitro transcription termination assay. This assay monitors termination efficiency across a range of ligand concentrations. Transcription products are separated by gel electrophoresis and the relative intensities of the full-length and truncated transcripts are quantified.
We selected a type-1 singlet from Clostridium tetani and a type-2 singlet from Desulfitobacterium hafniense that were both predicted to contain a Rho-independent terminator helix in their expression platforms. The type-1 singlet is located upstream of rhaT, a gene encoding a permease from the drug/metabolite transporter superfamily. The type-2 singlet is located upstream of an uncharacterized gene.
Both singlet constructs successfully modulate transcription termination in a glycine-dependent manner in vitro (Fig. 3). Consistent with our bioinformatics observations, the type-1 singlet is an ON switch and the type-2 singlet is an OFF switch. The type-1 singlet had a robust amplitude of 86 ± 1% and half-maximal termination (K1/2) occurred at 420 ± 20 µM. The type-2 singlet had a modest amplitude of −10 ± 1% and a K1/2 of 15 ± 3 µM. For additional comparison, we tested a tandem variant from Bacillus subtilis. This construct is an ON switch and displayed an amplitude of 24 ± 1% and a K1/2 of 56 ± 6 µM. It had a Hill coefficient of 1.0 ± 0.1, indicating that this riboswitch is not cooperative under these conditions. These results demonstrate that both type-1 and type-2 singlets function as genetic regulators and produce similar response profiles to tandem constructs.
FIGURE 3.
Ligand-dependent modulation of transcription termination in vitro. Ligand-dependent response profiles and representative gels are shown for prototypical examples of the type-1 singlet (A,B), type-2 singlet (C,D), and tandem (E,F) glycine riboswitches. All three examples functionally modulate transcription termination in vitro. Error bars represent the standard deviation of three replicates.
Mutational analysis of all type-1 singlet point mutants highlights conserved sequence elements
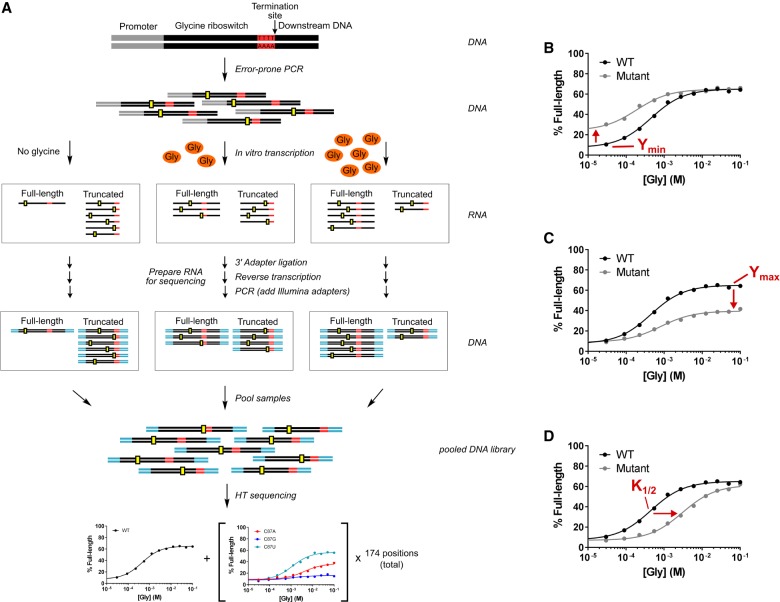
We set out to systematically analyze the functional effect of every point mutant within the singlet aptamer and expression-platform domains to determine the sequence and energetic requirements for gene control. To achieve this, we developed Sequencing-based Mutational Analysis of RNA Transcription Termination (SMARTT), a high-throughput method for generating ligand-dependent transcription termination profiles. SMARTT is a modified version of the gel-based assay for in vitro transcription termination (Fig. 4A). It is similar to the sequencing-based approach that was reported by the Yokobayashi Laboratory to study the activity of ribozyme variants (Kobori et al. 2015) and adds to the growing list of sequencing-based high-throughput approaches for studying mutational effects (Pitt and Ferré-D'Amaré 2010; Kladwang et al. 2011; Fowler et al. 2014). We chose to analyze the C. tetani type-1 singlet due to the large amplitude we observed for its wild-type (WT) response profile in the gel-based assay.
FIGURE 4.
Sequencing-based mutational analysis of RNA transcription termination (SMARTT). (A) Schematic of the strategy used for the high-throughput mutational analysis of transcription termination by sequencing. Mutant libraries were made by error-prone PCR (hypothetical mutations shown in yellow) and used as DNA template for a series of in vitro transcription reactions containing varying concentrations of glycine. RNA obtained from these reactions was prepared and sent for high-throughput sequencing (sequencing adapters are shown in cyan). The resulting sequences were computationally analyzed to produce response profiles for all variants with 0–2 mutations simultaneously. Representative response profiles are shown for mutants that alter the parameters Ymin (B), Ymax (C), and K1/2 (D). Error bars representing the standard deviation do not appear as they would be smaller than the size of each point.
Riboswitches that control gene expression transcriptionally produce RNAs of distinct lengths based on whether a terminator or antiterminator hairpin forms in the expression platform. SMARTT leverages high-throughput sequencing technology to quantify the relative abundance of full-length and truncated transcripts produced by these riboswitches as a function of ligand concentration and RNA sequence. Because the exact sequence and length is determined for each read, thousands of mutants can be quantitatively analyzed simultaneously.
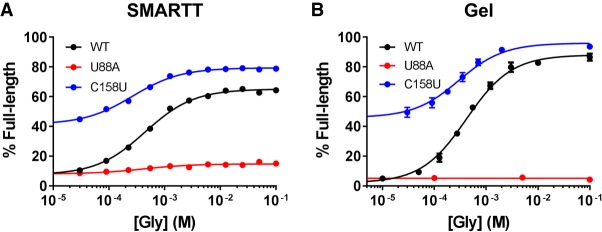
A library containing all possible single point mutants within the aptamer and expression-platform domains of the C. tetani type-1 singlet was created by error-prone PCR (Supplemental Tables S1, S2 and Supplemental Text). This library was used as template for in vitro transcription reactions with varying concentrations of glycine, which resulted in full-length and truncated RNAs. The RNAs were subsequently prepared for high-throughput sequencing. Full response profiles were generated for WT and all 522 singlet point mutants by counting the number of full-length and truncated sequences observed at each glycine concentration for all variants. These SMARTT-generated profiles are consistent with the conventional gel-based assay (Fig. 5; see also Supplemental Table S3 and Supplemental Text). A comparison of these data with those obtained in a preliminary evaluation of the assay illustrates that the results are reproducible between independent trials (Supplemental Fig. S2). Together, these results indicate these SMARTT-generated profiles can be relied upon to draw inferences about the riboswitch.
FIGURE 5.
Validation of SMARTT-generated response profiles. In vitro transcription termination profiles generated using SMARTT (A) and the conventional gel-based (B) approach are shown. Comparable response profiles are produced with both methods. Error bars represent the standard deviation.
Many of the individual mutants in the high-throughput data set affected one or more of the three fit parameters: Ymin, Ymax, and K1/2 (Fig. 4B–D). Ymin and Ymax are the percentages of full-length RNA produced either in the absence of ligand (Ymin) or at saturating concentrations (Ymax). These values report how “OFF” the riboswitch is when no ligand is bound and how “ON” it is when fully bound. K1/2 is the concentration of ligand needed to produce half-maximal modulation. It provides a measure of the responsiveness of the riboswitch to ligand concentration. Changes to these parameters reveal which states of the system mutations affect.
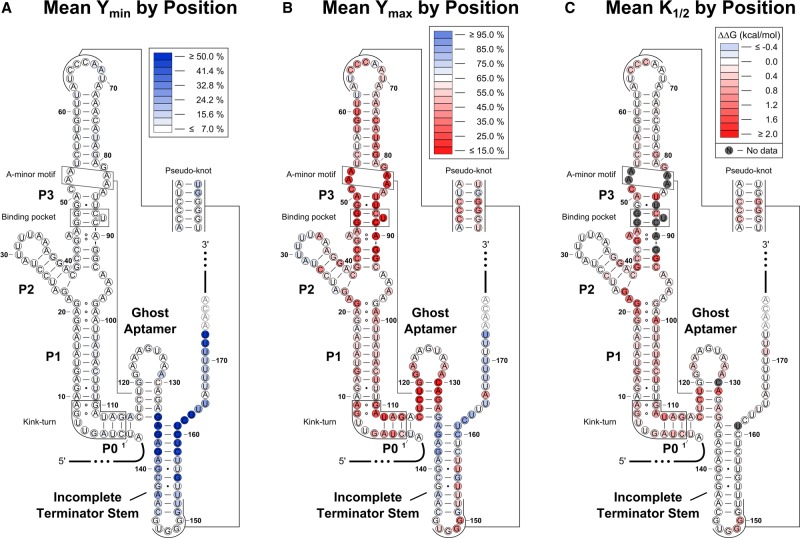
To visualize the impact of each mutation on these parameters, we mapped the mean value observed at each position for Ymin, Ymax, and K1/2 onto the predicted RNA secondary structure (Fig. 6). The most common effects were an increase in both Ymin and Ymax or a decrease in Ymax and/or an increase in K1/2. The only mutations that significantly altered the readthrough efficiency in the absence of ligand (Ymin) were located within the terminator hairpin (Fig. 6A). An increase in Ymin signifies that these mutants start in a partially or fully ON state. Most of these variants appear to weaken the overall stability of the terminator hairpin. In contrast, mutations that alter Ymax and/or K1/2 were located throughout the riboswitch (Fig. 6B,C). Changes to K1/2 suggest an altered binding affinity, which is caused by mutating structurally important regions in the aptamer domain. Notably, any mutation resulting in more than a ∼1000-fold defect to K1/2 will not show significant modulation over the concentrations tested and will instead have a decreased Ymax. Ymax can also be reduced if the communication pathway between the aptamer and expression-platform domains is disrupted or if the helix that becomes stabilized upon ligand binding is weakened.
FIGURE 6.
Heat maps. The mean Ymin (A), Ymax (B), and K1/2 (C) values for the three variants at each nucleotide position were mapped onto the predicted secondary structure. Deleterious mutations highlight functionally relevant regions of the riboswitch. Variants with an amplitude of <15% were excluded when computing the K1/2 mean values shown in C, as K1/2 values for these mutants may be influenced by minor amounts of WT contamination (see Supplemental Text).
Important structural elements of the riboswitch are highlighted by these heat maps. As described previously, ligand binding is proposed to stabilize the ghost aptamer through the formation of an A-minor motif between the minor groove of the ghost aptamer and conserved adenines in the P3 helix (Ruff et al. 2016). Consistent with this hypothesis, mutations to the binding pocket (located in the P3a helix), the conserved P3 adenines, and the ghost aptamer all cause substantial effects to Ymax and/or K1/2. Mutations to a kink-turn predicted to form in the area between P0 and the start of P1 (Ruff et al. 2016) also negatively affect these parameters. This is consistent with the known importance of an analogous kink-turn found in tandem systems, where it is proposed to help organize the quaternary structure (Baird and Ferré-D'Amaré 2013). These data suggest that this motif plays a similar role in singlet systems.
The high-throughput data set also provides evidence for the importance of the two conserved sequence elements identified by bioinformatics. A purine-rich region was identified within the ghost aptamer of both singlet subtypes. In the C. tetani system, this region consists of putative GA and AA pairs at the top of this helix. Mutations to the GA pair and, to a lesser extent, the following AA pair caused significant defects, mainly to Ymax (Supplemental Fig. S3). As predicted by the consensus sequence, the GA-to-AA conversion was the least disruptive GA mutation. Previous data showed that truncating the ghost aptamer loop of the Listeria monocytogenes type-1 singlet to a GAGA tetraloop results in significant binding defects, but a GAUAA pentaloop restores ligand binding (Ruff et al. 2016). The pentaloop, but not the tetraloop, would allow for at least the bottom GA pair to form. Altogether, these data indicate that these noncanonical pairs in the ghost aptamer have an important functional role.
The bioinformatic search also identified a pseudoknot in 85% of type-1 singlets. In the C. tetani system, this pseudoknot is predicted to form between the P3b stem–loop and the apex of the terminator hairpin. Single mutations to the CG base pairs in this motif display reduced values for Ymax and increased values for K1/2. Changes to both Ymax and K1/2 within the terminator hairpin indicate these nucleotides are involved in glycine-dependent long-range interactions. These results provide additional evidence that the pseudoknot identified in the consensus alignment is an authentic motif and has functional importance.
Ligand binding stabilizes the ghost aptamer by 2–3 kcal/mol
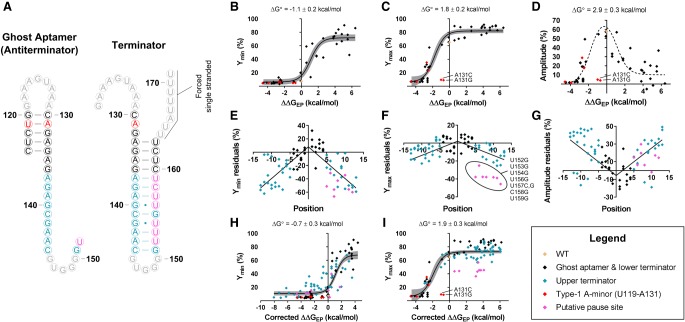
The standard model of gene regulation by riboswitches indicates that ligand binding by the aptamer domain promotes a conformational rearrangement that stabilizes one of two or more competing helices in a downstream expression platform. Based on this model, we hypothesized that the relative populations of these expression platform helices would be dependent on the difference in their free energies. This would be expected for riboswitches that are thermodynamically controlled. It should also hold for riboswitches that are kinetically controlled if a linear relationship exists between the free energy of the competing expression platform helices and their rates of formation and/or interconversion. Such linear free energy relationships have been observed for small perturbations in other contexts (Wells 1963; Kingery and Strobel 2012). To test this hypothesis, the readthrough efficiencies observed in the absence of ligand (Ymin) and at saturating levels (Ymax) for the mutations located within base-pairing regions of the type-1 singlet expression platform were compared with the predicted net free energy change to the expression platform (ΔΔGEP) caused by these same mutations (Fig. 7). Here, ΔGEP is defined as the difference in the predicted stability of the terminator and antiterminator (ghost aptamer) helices and ΔΔGEP is the change to this value upon mutation (Equation 7, Materials and Methods).
FIGURE 7.
Dependence of readthrough efficiencies on ΔΔGEP. (A) Predicted secondary structures for the antiterminator and terminator sequences used in the free energy calculations of ΔΔGEP. Nucleotides are colored according to plots B–I. Correlations are shown between ΔΔGEP and Ymin (B), Ymax (C), and amplitude (D). Only data associated with WT and mutations within the base-pairing regions of the ghost aptamer or the first four base pairs of the terminator hairpin were considered for the fits displayed in B–D. Red diamonds indicate nucleotides that are expected to participate in a type-1 A-minor interaction. These data were excluded from all fits as energetic contributions from tertiary interactions are not accounted for in the free energy calculations of ΔΔGEP. Correlations between the positional distance of a mutation from the base of the terminator stem and the associated residuals (based on the fits depicted in B–D) are shown for Ymin (E), Ymax (F), and amplitude (G). A linear dependence was observed and a correction term was added to the model to account for this dependence. Fits obtained with the updated model are shown for Ymin (H) and Ymax (I). The gray bands in B, C, H, and I are the 95% confidence intervals for the displayed fits. The dotted line shown in D depicts the difference between the Ymin and Ymax fits depicted in B and C.
We started by only considering mutations to base pairs in the ghost aptamer and the first four base pairs of the terminator hairpin. This subset was chosen because previous work indicates that the formation of these base pairs within the terminator provides the energy required for disruption of the RNA:DNA hybrid in the elongation complex and because mutations distal to the base of a terminator hairpin have been observed to have minimal effects on termination efficiencies (Wilson and von Hippel 1995; Larson et al. 2008). As hypothesized, mutations at these positions exhibit a dependence on ΔΔGEP and fit well to a standard two-state model (Equation 10, Materials and Methods; Fig. 7B–D).
However, two variants located within the ghost aptamer significantly deviate from the overall observed trend. A131G and A131C are completely nonfunctional despite predicted net free energy changes of just −0.7 and −1.0 kcal/mol, respectively. The predicted values for ΔΔGEP used in our analysis only take into account energetic contributions from secondary structure (Materials and Methods). Deviations from the overall trend could therefore indicate that important tertiary interactions are being disrupted. Significant biochemical and structural evidence indicates that the minor groove of type-1 singlet ghost aptamers is involved in a series of important A-minor interactions (Butler et al. 2011; Ruff et al. 2016). Type-1 A-minor interactions are known to discriminate against noncanonical base pairs due to steric clashes (Battle and Doudna 2002). The A131G mutation replaces the canonical U119–A131 base pair located in the ghost aptamer with a noncanonical UG wobble pair. This suggests that a type-1 A-minor interaction may form in the minor groove of this base pair in the WT construct, but is disrupted by the UG wobble pair present in this variant. Because of possible tertiary interactions with the U119–A131 pair, all mutations at these two positions were excluded from the fits displayed in Figure 7.
Although this subset of the data fit well to Equation 10 (Materials and Methods), the model breaks down when more terminator stem variants are included (Supplemental Fig. S4). The effects of mutations located farther from the base of the terminator hairpin deviate by increasing amounts from the values predicted by the fits described above. The residuals for these variants display a linear dependence on the distance between the location of the mutation and the base of the terminator helix (Fig. 7E–G). This indicates that it is largely the stability of the base of the terminator that determines the probability of termination. This is consistent with the observations made by Larson et al. (2008) that the energy required for disrupting the RNA:DNA hybrid of the elongation complex is obtained from the formation of the final 3–4 bp of the terminator hairpin.
To incorporate this position-dependence into the model, Equation 10 was modified to include an additional variable to be fit, a correction factor for ΔΔGEP, that is multiplied by a preset distance term equal to the number of base pairs separating the mutation from the base of the terminator helix (Equation 11, Materials and Methods; Fig. 7H,I). Upon fitting the data to Equation 11, optimal correction factors of −0.66 ± 0.05 kcal/(bp mol) and −0.08 ± 0.05 kcal/(bp mol) were determined for the Ymin and Ymax values of terminator stem variants, respectively. The near-zero correction applied to Ymax values indicates that the terminator helix is already largely destabilized in the WT construct under high glycine conditions. A difference of 2.6 ± 0.4 kcal/mol was observed between the values obtained for ΔG° (the additive inverse of the midpoint of the fit) in the absence of ligand (Ymin; −0.7 ± 0.3 kcal/mol) and at saturating concentrations (Ymax; 1.9 ± 0.3 kcal/mol). ΔG° provides a measure of the overall stability of the WT expression platform under the condition analyzed. Therefore, these data indicate that the addition of saturating ligand concentrations stabilizes the ghost aptamer by just 2–3 kcal/mol in this construct.
A few mutations in the terminator hairpin deviate from the model even after this correction term is added, particularly at saturating concentrations of glycine (Ymax). This includes five U-to-G mutations, one U-to-C mutation, and one C-to-G mutation all between positions 152 and 159. This is a uracil-rich region in the helix, which is a sequence motif predicted to induce polymerase pausing (Gusarov and Nudler 1999; Yarnell and Roberts 1999; Shundrovsky et al. 2004). In the wild-type construct, low levels of termination are observed immediately following positions C158 and U159 at all glycine concentrations tested (Supplemental Fig. S5A), which is consistent with this being a pause site (Larson et al. 2014). Thus, one explanation for this observation is that these mutations disrupt a critical pausing event that provides additional time for the aptamer domain to bind glycine and undergo a conformational rearrangement before the rest of the terminator is transcribed. This would explain why these mutants all significantly deviate from the model under conditions of saturating ligand, but follow the trend in its absence.
Targeted analysis of double mutant data reveals a mutant with a substantially improved K1/2
The C. tetani riboswitch displayed the weakest K1/2 of the three glycine riboswitch variants we analyzed. However, an alternate type-1 singlet from L. monocytogenes was previously shown to bind glycine with an affinity similar to the D. hafniense type-2 singlet tested here (Ruff et al. 2016). Therefore, we predicted a more responsive C. tetani mutant may exist. No single mutation in the SMARTT data set was sufficient to substantially improve the K1/2, indicating that such a variant would require multiple mutations if it exists at all.
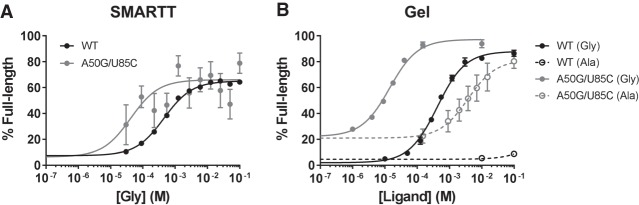
We compared the C. tetani construct to the consensus alignment and identified an AU base pair between positions 50 and 85 that is conserved as a GC base pair in 89% of type-1 singlets. Although the double mutant A50G/U85C had low sequence coverage in the SMARTT data set—an average of just 22.8 reads per glycine concentration—its fit indicated a substantial improvement to the K1/2 (40 ± 30 µM; Fig. 8A). This was confirmed by the gel-based assay (14 ± 1 µM; Fig. 8B). This K1/2 is 30-fold better than WT and comparable to the values obtained for the type-2 singlet and tandem constructs tested here. Unexpectedly, the variant also showed an improved affinity for alanine with a K1/2 of 4 ± 1 mM. This corresponds to a decrease in selectivity from ∼5000-fold for WT to 300-fold and is significantly lower than what was observed for the D. hafniense type-2 singlet (>2500-fold) and B. subtilis tandem construct (∼2000-fold). This result suggests that a sacrifice of binding affinity for higher selectivity may be advantageous in this system.
FIGURE 8.
Targeted analysis of double mutant data. (A) In vitro transcription termination response profiles generated by SMARTT for WT and A50G/U85C. Although a mean of just 22.8 reads per glycine concentration was obtained for A50G/U85C, the data suggest this double mutant has an improved K1/2 compared to WT. This was verified by gel. (B) In vitro transcription termination response profiles obtained by the conventional gel-based approach for WT and A50G/U85C. Filled circles indicate that glycine was used as the ligand; open circles indicate alanine. Error bars represent the standard deviation.
DISCUSSION
For a riboswitch to provide functional utility, it must bind specifically to its ligand at a physiologically relevant concentration and facilitate helical switching in an associated expression platform. These requirements must be satisfied using only the free energy provided from interactions between the RNA and the ligand. However, any energy that is devoted to stabilizing conformational rearrangements is not available to improve the binding affinity, and vice versa (Gartenberg and Crothers 1988; Williamson 2000). This means that the free energy available to the riboswitch for promoting helical switching is constrained by the number and type of contacts that can be made between the RNA and ligand and the binding affinity that is physiologically required.
In the case of the C. tetani type-1 singlet, we have demonstrated that ligand binding stabilizes its ghost aptamer by just 2–3 kcal/mol. This is roughly equivalent to making a single GU-to-GC mutation in this region and indicates that its expression platform helices have to be extremely close in energy for a significant ligand-dependent change in gene expression to be possible. Although riboswitch expression platforms typically display little to no sequence conservation, this result suggests that these helices are finely tuned based on the amount of energy that is available for helical switching.
As a transcriptional riboswitch, the C. tetani type-1 singlet must additionally respond to glycine within a limited timeframe. Previous work has demonstrated that many riboswitches under similar time constraints are unable to reach equilibrium before a genetic decision must be made (Wickiser et al. 2005). The data presented here cannot distinguish whether this type-1 singlet is kinetically controlled or if it reaches equilibrium over the time course of transcription. However, it is clear from our results that its readthrough efficiency is dependent on the stabilities of both the terminator and antiterminator, indicating both helices must be fine-tuned for function. This fine-tuning includes preserving a critical pausing element located within its expression platform. Similar pause sites are frequently observed within the expression platform of many riboswitches. They serve to extend the time allotted for ligand binding and conformational rearrangements and can also coordinate folding (Wickiser et al. 2005; Perdrizet et al. 2012).
The particular constraints felt by individual riboswitches are likely to vary based on several factors, including the ligand recognized, the range of ligand concentrations that are physiologically relevant, and the type of expression platform utilized (transcriptional versus translational, isolated riboswitch versus logic gate, etc.). For example, the C. tetani type-1 singlet displays a significantly higher K1/2 compared to the D. hafniense type-2 singlet and B. subtilis tandem constructs. Yet, a double mutation within its P3 helix was shown here to provide a 30-fold improvement to its K1/2. This indicates that these differences in responsiveness may be functionally important—likely based on the unique physiological and genetic contexts of these constructs. Continuing these comparisons, tandem glycine riboswitches bind two molecules of glycine to stabilize a single expression platform, while singlets only bind one. As such, tandem systems have access to additional energy for driving helical switching compared to singlets. This could allow for more complex and diverse expression platforms in these systems.
Other riboswitches (e.g., the thiamine pyrophosphate (TPP), cyclic-di-GMP, and adenosylcobalamine riboswitches) respond to ligands that are much larger than glycine. As the size and complexity of these ligands increases, so too does the potential for these molecules to more extensively contact the aptamer binding pocket and contribute more free energy to the system (Kuntz et al. 1999; Reynolds et al. 2007). Consistent with this concept, optical trapping experiments performed on examples of the adenine and TPP riboswitches report that ligand binding stabilizes the aptamer domains of these constructs by 4 ± 1 kcal/mol, 8 ± 2 kcal/mol, and 17 ± 5 kcal/mol for the pbuE adenine, add adenine, and thiC TPP riboswitches, respectively (Greenleaf et al. 2008; Neupane et al. 2011; Anthony et al. 2012). Thus, glycine, which is smaller and forms fewer contacts than either of these ligands, contributes the least amount of energy toward stabilizing the aptamer domain.
The fraction of energy that can be devoted toward conformational changes is also dependent on the observed binding affinity. Glycine, adenine, and TPP riboswitches all respond to their ligands across similar ranges of concentrations: the low- to mid-micromolar range for glycine riboswitches (Sherman et al. 2012; Ruff and Strobel 2014; Ruff et al. 2016), the mid-nanomolar to low-micromolar range for adenine riboswitches (Mandal and Breaker 2004a; Gilbert et al. 2006; Rieder et al. 2007), and the low- to mid-nanomolar range for TPP riboswitches (Sudarsan et al. 2003; Kulshina et al. 2010). Yet, some riboswitches must respond to their ligands at even lower concentrations than these. Such riboswitches will necessarily devote more of the total energy provided from ligand binding toward the affinity. For instance, cyclic-di-GMP riboswitches have affinities as low as 10 pM (Smith et al. 2009). This is roughly 105-fold tighter than the affinities observed for glycine riboswitches and is equivalent to devoting an extra ∼7 kcal/mol toward the affinity (Ruff et al. 2016). In the case of the c-di-GMP riboswitch, there is still likely to be sufficient energy to promote conformational rearrangements given the number of contacts it makes with its ligand. Yet, riboswitches that bind much smaller ligands with fewer potential contacts are likely to find such a binding affinity to be a limiting factor. The ability of RNA to finely tune helical energy and other parameters through small sequence alterations is therefore an important property that allows it to successfully regulate gene expression in a variety of contexts for a diverse set of ligands.
The concept of RNA undergoing functionally important conformational changes in response to the formation of intermolecular interactions is prevalent in biology. RNA commonly associates with proteins and other binding partners. The interactions formed upon binding often result in RNA structural rearrangements that are critical to the function of either the RNA or the associated protein. For example, the RNA portions of telomerase, the spliceosome, and the ribosome all undergo conformational rearrangements driven by interactions with their protein binding partners (Berman et al. 2010; Singh et al. 2012; Fica and Nagai 2017; Noller et al. 2017). As with riboswitches, the interactions formed contribute the energy necessary to stabilize functionally relevant states of the RNA. Therefore, the findings presented here regarding the energetics of ligand-induced conformational switching by the C. tetani riboswitch expand our understanding of this general principle.
MATERIALS AND METHODS
Bioinformatic analysis of RNA sequences
Updated seed alignments of the type-1 singlet, type-2 singlet, and the tandem version of the glycine riboswitch were produced by manually modifying existing alignments from previous bioinformatic searches (Barrick and Breaker 2007; Ruff et al. 2016) to include the kink-turn motif that was identified more recently (Kladwang et al. 2012; Sherman et al. 2012) on the 5′ end. Bioinformatic searches based on these seed alignments, as well as a minimal glycine riboswitch aptamer, were performed using Infernal 1.1 (Nawrocki and Eddy 2013) on the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) Database (O'Leary et al. 2016) release 76, as well as several metagenomics environmental data sets described previously (Weinberg et al. 2015). An E-value cutoff of 0.1 was applied. Sequences were removed if they contained an aptamer that lacked P1 and/or P3a (the binding pocket) helices. This left a total of 15,847 unique glycine riboswitch sequences. A list of all single-aptamer glycine riboswitches (4221 unique sequences) was created by removing sequences with an aptamer that was within 150 nt of another aptamer identified in the search.
To limit the number of false positives in the single-aptamer list, an additional search for glycine aptamers was performed within a region of 200 nt on either side of the identified singlet aptamers. Constructs were removed if an additional aptamer was identified or if full sequence data was not available in this region. Sequences in this subset that were also uniquely identified in the initial type-1 and type-2 singlet searches (excluding all type-1 and type-2 singlets found in the list lacking a reasonable ghost aptamer) were manually aligned. Using these as seed alignments, a final bioinformatic search was performed on the filtered subset of all single-aptamer glycine riboswitches (potential false positives removed) with Infernal 1.1 (E-value cutoff of 0.1). The search space for this final run was limited to the aptamer plus 5 nt on the side lacking the ghost aptamer and 150 nt on the side containing the ghost aptamer. Constructs without a reasonable ghost aptamer were again removed and the remaining sequences that were uniquely identified by each seed alignment were aligned manually. Consensus sequence figures based on these final alignments were created with R2R and edited with Inkscape.
Downstream gene associations were determined for the sequences in these final alignments as described previously (Weinberg et al. 2015). The locations of genes were retrieved from RefSeq (O'Leary et al. 2016) or IMG/M (Markowitz et al. 2008) annotations, or predicted with MetaGene (Noguchi et al. 2006) or MetaGeneMark (Zhu et al. 2010). Conserved domains were predicted using the Conserved Domain Database (Marchler-Bauer et al. 2015) version 2.25. A custom MATLAB script was used to combine and count equivalent genes. Additional manual curation was done on the list of genes to further combine and count equivalent genes. Downstream genes identified as RNAs or ambiguous annotations were discarded and counted as “No gene identified.” This occurred rarely and usually no other gene was identified downstream. Corresponding gene-association pie charts were created with Adobe Illustrator and further edited with Inkscape. Only the first downstream gene or domain was considered for these pie charts.
Design of plasmids and template DNA for gel-based in vitro transcription termination assays
A plasmid containing the B. subtilis riboswitch was created using standard molecular biology techniques. Plasmids containing the C. tetani and D. hafniense riboswitches were ordered from GeneArt (Thermo Fisher Scientific). For the B. subtilis construct, 30 nt upstream of its WT promoter through ∼30 nt downstream from its terminator hairpin (NC_000964.3/2549731-2549344) were cloned into a pUC19 plasmid. This region, plus an additional 50 nt downstream, was PCR amplified with Phusion HF Polymerase (NEB or Thermo Fisher Scientific) and used as the template DNA for the gel-based in vitro transcription termination assays. For the C. tetani and D. hafniense singlet constructs, the aptamer region of the B. subtilis construct (starting at P0) through the ∼30 nt downstream from the expression platform was replaced by the corresponding singlet aptamer through ∼40 nt downstream from their associated terminator hairpins (C. tetani: NC_004557.1/1925407-1925619; D. hafniense: CP001336.1/1219348-1219627). These regions, plus an additional 40 nt downstream, were PCR amplified with Phusion HF Polymerase and used as the template DNA in the associated gel-based transcription termination assays (see Supplemental Table S4 for the exact sequences used for each construct).
Preparation of mutagenized DNA library
A mutagenized library was made by error-prone PCR using the GeneMorph II Random Mutagenesis Kit from Agilent Technologies. Primers were chosen to flank the aptamer and expression-platform domains of the C. tetani type-1 singlet and mutations were introduced using a 16 cycle error-prone PCR reaction. Additional DNA was added upstream of the riboswitch through a two-step PCR reaction using two forward primers with large overhangs (∼80 nt) on their 5′ ends and Phusion HF Polymerase (NEB or Thermo Fisher Scientific). This additional DNA included a B. subtilis promoter (also recognizable by E. coli RNA polymerase) and matches the DNA template used in the gel-based transcription termination assay. This region, plus ∼25 nt downstream from the terminator hairpin, was further amplified by PCR with Phusion HF Polymerase (NEB or Thermo Fisher Scientific) and used as the template DNA in the SMARTT transcription termination assays (see Supplemental Table S4 for the specific sequence used).
Gel-based in vitro transcription termination assays
Sets of 20 μl in vitro transcription reaction mixtures were prepared on ice. Mixtures contained 40 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 150 mM KCl, 1 mM DTT, 10 µg/mL BSA, 1% Glycerol, 25 nM DNA template, 50 µM NTPs, approximately 2.5 µCi [α-32P] GTP, 0.5 units E. coli RNA Polymerase Holoenzyme (NEB), and varying concentrations of ligand (glycine or alanine). Reactions were incubated at 37°C for 1 h and then placed on ice. 2 volumes of stop/loading buffer (25 mM EDTA, <0.1% Xylene Cyanol, <0.1% Bromophenol, ∼93% formamide) were added to each reaction. Full-length and truncated RNA transcripts were separated using a 6% PAGE gel and visualized with a Typhoon FLA 9500 (GE Healthcare). Individual band intensities were determined using ImageQuant (GE Healthcare) and normalized based on the expected number of guanosine nucleotides contained in each band. Data from individual glycine titrations were fit to Equation 1:
| (1) |
where X is the ligand concentration, Ymax is the percentage of full-length RNA at saturating concentrations of ligand, Ymin is the percentage of full-length RNA in the absence of ligand, and K1/2 is the point of half-maximal termination. Ligand titrations were performed in triplicate. When testing the response of the riboswitch to alanine, little modulation was often observed over the range of concentrations tested. In these cases, estimates of the K1/2 were obtained by constraining the amplitude to be equal to the value obtained with glycine as the ligand.
Preparation of RNA for high-throughput sequencing
A set of 100 μl in vitro transcription reactions was prepared on ice. Reaction mixtures contained the same reagents as in the gel-based transcription termination assay described above, except no radiolabeled GTP was added. A single titration was performed. Reactions were incubated at 37°C for 1 h and then placed back on ice for >1 min. Template DNA was removed through DNase treatment. Ninety microliters of the transcription sample were combined with 10 µL of 10× TURBO DNase Buffer (Thermo Fisher Scientific) and 1 µL TURBO DNase (2 units; Thermo Fisher Scientific) and incubated at 37°C for 20 min. The RNA was then purified using an RNeasy Minelute Cleanup Kit (QIAGEN).
A preadenylated DNA adapter was ligated onto the 3′ end of the RNA (/5rApp/NNNNNCTGTAGGCACCATCAAT/3ddC/; ordered from IDT). Ligation reaction mixtures were incubated at 16°C overnight (final concentrations: 1× T4 RNase Ligase II Buffer [NEB], 100 µM DNA adapter, 15% PEG8000, 10 U/µL T4 RNA Ligase II, Trunc. K227Q [NEB], and ∼70% of the purified RNA). The RNA was again purified using an RNeasy Minelute Cleanup Kit (QIAGEN).
The ligated adapter provides a handle for reverse-transcription into cDNA. A primer (36.2 nM; 5′GATTGATGGTGCCTACAG) was annealed to the RNA for 5 min at 65°C in the presence of dNTPs (725 µM) and then cooled to 4°C for 2 min before addition of the rest of the reagents. The full reaction mixture (final concentrations: 1× FS Buffer [Thermo Fisher Scientific], 500 µM dNTPs, 25 nM primer, 5 mM DTT, 0.2 U/µL RNaseOUT [Invitrogen], 10 U/µL SuperScript III [Thermo Fisher Scientific]) was incubated at 55°C for 45 min and then heated to 85°C for 5 min to heat inactivate the SuperScript III enzyme. The RNA was then degraded by incubation with RNase A (0.5 µg/µL; Thermo Fisher Scientific) and RNase H (0.1 U/µL; Invitrogen) at 37°C for 1 h. The resulting cDNA was purified using Agencourt Ampure XP magnetic beads (Beckman Coulter). Relative concentrations of cDNA were then determined by qPCR.
Illumina sequencing adapters and a unique index were added to the cDNA from each sample condition through two rounds of PCR using Phusion HF Polymerase (NEB or Thermo Fisher Scientific) and primers with large 5′ overhangs (Supplemental Table S5). Five cycles were used in the first round of PCR and 4–6 cycles were used in the second round (based on cDNA qPCR results). Relative final DNA concentrations were estimated for each sample by qPCR following the second round of PCR. All samples were combined at approximately equal ratios before being submitted for sequencing at the Yale Center for Genome Analysis (YCGA). A final purification was performed by the YCGA using solid phase reversible immobilization (SPRI) beads to remove any residual short DNA impurities (e.g., primer dimers) prior to sequencing.
High-throughput sequencing
Sequencing was performed at the YCGA on an Illumina HiSeq 4000 (2 × 150). Samples were combined and mixed with human genome libraries by the YCGA to prevent sequencing issues stemming from the low sequence diversity of the type-1 singlet samples. The 13 samples produced in the full glycine titration were sequenced across two half lanes (∼7.7% of a lane per sample in total). The three samples described in Supplemental Figure S2 (Replicate B) occupied ∼2.5% of a lane per sample. Demultiplexing was done by the YCGA.
Analysis of sequencing results
The region upstream of the aptamer and the 3′ adapter were removed from the sequencing reads using the program Cutadapt (Martin 2011). Flags were applied to require a minimum adapter overlap length of 10 bases, a minimum sequence length of 80 nt after adapter removal, and a minimum quality score of five. Sequences missing adapters were discarded. The remaining sequences were aligned to a reference file containing the full-length WT sequence using Bowtie2 (Langmead and Salzberg 2012). Discordant alignments were not permitted.
Custom Python scripts adapted from the RTEventsCounter software by the Simon Laboratory (Sexton et al. 2017) were used to analyze SAM files to determine the fraction of full-length RNA for all variants with 0–2 mutations at each ligand concentration. Reads containing any base call quality scores below 20 within the mutagenized region of the RNA were discarded. The remaining reads were classified according to the mutations contained through position 173. Reads were labeled as terminated if the last nucleotide was between positions 165 and 173 (inclusive). This window was determined manually based on an observed glycine-dependent increase in the frequency of termination in this region (see Supplemental Fig. S5A). Reads were labeled as full-length if the last nucleotide fell after this window and discarded if it fell before. The number of full-length and truncated reads were then counted for each variant to determine the fraction of full-length RNA at each ligand concentration.
Graphpad Prism 7 or custom R scripts were used to perform nonlinear regression with inverse-variance weighting to fit the data obtained for all variants with 0–2 mutations to Equation 1 (see above). Individual reads were treated as independent Bernoulli trials. Two alternative methods were used to approximate the mean and variance of the percentage of full-length reads based on whether or not the normal approximation was reasonable. When both np and n(1-p) were greater than five at all concentrations of a given variant, the normal approximation was used and the mean and variance were determined by solving Equations 2 and 3:
| (2) |
| (3) |
where µ is the mean, σ2 is the variance, p is the fraction of full-length reads, and n is total the number of full-length and truncated reads. If these criteria were not met for any concentration, Bayesian inference was used. The mean and variance of the posterior distribution function obtained using the Jeffreys prior [Beta(0.5,0.5)] were calculated by solving Equations 4 and 5:
| (4) |
| (5) |
where s is the number of full-length reads and n is the total number of full-length and truncated reads.
Heat maps
A vector image of the predicted secondary structure of the C. tetani riboswitch was created using Inkscape in SVG format. Objects corresponding to each position were manually relabeled with unique object IDs based on the WT sequence. A custom Python script was then used to edit the fill colors of these objects in the associated SVG file. The assigned object IDs were used to look up the parameter values for all three mutants at each position and the mean value was calculated. The color of each object was determined based on this value using linear interpolation in RGB color space and the SVG file was updated accordingly. Additional minor editing of heat maps was done manually using Inkscape.
Due to the close relationship between binding affinity and K1/2, heat maps related to K1/2 report the apparent free energy (ΔΔGapp). This is done because this relationship suggests that cumulative effects for this parameter are likely to be multiplicative instead of additive and therefore are better represented on a log scale. ΔΔGapp was determined by solving Equation 6:
| (6) |
where R is the ideal gas constant (0.001987 kcal K−1 mol−1), T is the temperature of the transcription reaction (310 K), and the point of half-maximal termination for a particular variant and WT are denoted as K1/2,variant and K1/2,WT, respectively. K1/2 values for mutants with an amplitude under 15% were not included in the mean values reported (see Supplemental Text).
RNA secondary structure free energy calculations
The enf2 function of RNAStructure (Reuter and Mathews 2010) version 6.0 was used to calculate the free energy of the terminator and antiterminator helices in the expression platform for WT and all single point mutants in the expression platform. The WT sequences used are: 5′-CUCUGGAAAGUAAACAGAGAGAGAGCGAACGUGGGGU (ghost aptamer) and 5′-GAAAGUAAACAGAGAGAGAGCGAACGUGGGGUUUGUUCUCUCUuuauuuuu (terminator hairpin). Underlined nucleotides were predicted to base pair in the WT construct. Lowercase nucleotides were forced to be single stranded when searching for the minimum free energy secondary structure. The exact length of the terminator hairpin used was determined based on the site of termination and other factors (see Supplemental Text). Nucleotides outside of the WT base-pairing regions were kept for these two helices if they were located within the expression platform and remained single stranded in the lowest free energy WT structure determined by RNAStructure 6.0. This was done to allow for as many alternate secondary structures of mutant constructs as possible. Predicted net free energy changes to the expression platform (ΔΔGEP) were determined by solving Equation 7:
| (7) |
where ΔG is the calculated free energy of a given helix and the subscripts/superscripts T, AT, Mut, and WT correspond to the terminator, antiterminator (ghost aptamer), mutant construct, and WT construct, respectively.
The data were initially fit to the two-state model described in Equation 10. This model is based on the hypothesis that readthrough efficiencies depend on the fraction of antiterminator formed at a given ligand concentration and can be derived from Equations 8 and 9:
| (8) |
| (9) |
| (10) |
where Bmax is the readthrough efficiency when terminator hairpin formation is fully disrupted, Bmin is the readthrough efficiency when terminator hairpin formation is fully stabilized, KFL is the equilibrium constant for the conversion between the terminator and antiterminator helices, ΔG° is the additive inverse of the free energy of the WT expression platform (the midpoint of the fit), and ΔΔGEP is the predicted net free energy change to the expression platform.
As noted in the text, only a subset of the data fit well to Equation 10. However, a linear dependence was observed between the positional distance of a variant from the start of the terminator hairpin and the effect of that mutation. A correction term was added to account for this dependence and the data were fit again to Equation 11:
| (11) |
where C is a correction factor variable (determined in the fit) and D is the distance from the base of the terminator hairpin. Variants located within the first base pair of the terminator hairpin or outside of the terminator hairpin were assigned a distance D of zero.
DNA oligonucleotides and chemicals
All DNA oligos were ordered from the W.M. Keck Oligonucleotide Synthesis Facility at Yale University unless specified otherwise. Glycine and alanine were obtained from Sigma.
Code availability
Custom Python and R scripts used in the SMARTT analyses and for the generation of heat maps are available on GitHub at https://github.com/strobel-lab/SMARTT. All other scripts related to this work are available from the corresponding author upon request.
DATA DEPOSITION
Raw sequencing data used for the analyses presented in this manuscript have been deposited in the Sequence Read Archive under accession number SRP150789. An Excel file containing the number of reads observed at each concentration, the observed percentage of full-length RNA, and the fit parameter values generated for all single and double mutants are available in the Supplemental Material. Sequence alignments (in Stockholm format) for the type-1 singlet, type-2 singlet, and all single-aptamer constructs identified in the bioinformatic searches are also available in the Supplemental Material. All other data that support the findings presented in this work are available from the corresponding author upon request.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Caroline Reiss, Andrew Knappenberger, Karen Ruff, Dan Spakowicz, and Lydia Herzel for valuable advice and discussions. We thank Matt Simon, Alec Sexton, and Michael Rutenberg Shoenberg for helpful discussions regarding data processing. We thank Zasha Weinberg and Glenn Gaffield for help and advice concerning the bioinformatic analyses. We also thank the Yale Center for Genome Analysis, particularly Chris Castaldi, and the Yale Center for Research Computing for support and use of their infrastructure. C.D.T. was supported by the National Institutes of Health Biophysics Training grant (T32GM008283). This work was also supported by National Institutes of Health grant GM022778 (S.A.S. and Ronald R. Breaker).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067884.118.
REFERENCES
- Anthony PC, Perez CF, García-García C, Block SM. 2012. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci 109: 1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babina AM, Lea NE, Meyer MM. 2017. In vivo behavior of the tandem glycine riboswitch in Bacillus subtilis. mBio 8: e01602-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Ferré-D'Amaré AR. 2013. Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches. RNA 19: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8: R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle DJ, Doudna JA. 2002. Specificity of RNA–RNA helix recognition. Proc Natl Acad Sci 99: 11676–11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AJ, Gooding AR, Cech TR. 2010. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol Cell Biol 30: 4965–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA. 2011. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol 18: 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceres P, Garst AD, Marcano-Velázquez JG, Batey RT. 2013. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth Biol 2: 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. 2005. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in α-proteobacteria. Genome Biol 6: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Nagai K. 2017. Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat Struct Mol Biol 24: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Stephany JJ, Fields S. 2014. Measuring the activity of protein variants on a large scale using deep mutational scanning. Nat Protoc 9: 2267–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR, Crothers DM. 1988. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature 333: 824–829. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Mediatore SJ, Batey RT. 2006. Modified pyrimidines specifically bind the purine riboswitch. J Am Chem Soc 128: 14214–14215. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Love CE, Edwards AL, Batey RT. 2007. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry 46: 13297–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. 2008. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319: 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Nudler E. 1999. The mechanism of intrinsic transcription termination. Mol Cell 3: 495–504. [DOI] [PubMed] [Google Scholar]
- Heppell B, Blouin S, Dussault A-M, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA. 2011. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol 7: 384–392. [DOI] [PubMed] [Google Scholar]
- Johnson JE Jr, Reyes FE, Polaski JT, Batey RT. 2012. B12 cofactors directly stabilize an mRNA regulatory switch. Nature 492: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanov MD, Vitreschak AG, Gelfand MS. 2007. Abundance and functional diversity of riboswitches in microbial communities. BMC Genomics 8: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khani A, Popp N, Kreikemeyer B, Patenge N. 2018. A glycine riboswitch in Streptococcus pyogenes controls expression of a sodium:alanine symporter family protein gene. Front Microbiol 9: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingery DA, Strobel SA. 2012. Analysis of enzymatic transacylase Brønsted studies with application to the ribosome. Acc Chem Res 45: 495–503. [DOI] [PubMed] [Google Scholar]
- Kladwang W, VanLang CC, Cordero P, Das R. 2011. A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat Chem 3: 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, Chou F-C, Das R. 2012. Automated RNA structure prediction uncovers a kink-turn linker in double glycine riboswitches. J Am Chem Soc 134: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Edwards TE, Ferré-D'Amaré AR. 2009. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol 16: 343–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori S, Nomura Y, Miu A, Yokobayashi Y. 2015. High-throughput assay and engineering of self-cleaving ribozymes by sequencing. Nucleic Acids Res 43: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Edwards TE, Ferré-D'Amaré AR. 2010. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA 16: 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz ID, Chen K, Sharp KA, Kollman PA. 1999. The maximal affinity of ligands. Proc Natl Acad Sci 96: 9997–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Greenleaf WJ, Landick R, Block SM. 2008. Applied force reveals mechanistic and energetic details of transcription termination. Cell 132: 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. 2014. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344: 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hwang XY, Stav S, Breaker RR. 2016. The yjdF riboswitch candidate regulates gene expression by binding diverse azaaromatic compounds. RNA 22: 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. 2004a. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 11: 29–35. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. 2004b. Gene regulation by riboswitches. Nat Rev Mol Cell Biol 5: 451–463. [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. 2004. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306: 275–279. [DOI] [PubMed] [Google Scholar]
- Marcano-Velázquez JG, Batey RT. 2015. Structure-guided mutational analysis of gene regulation by the Bacillus subtilis pbuE adenine-responsive riboswitch in a cellular context. J Biol Chem 290: 4464–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D, Chen I-MA, Grechkin Y, Dubchak I, Anderson I, et al. 2008. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res 36: D534–D538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. 2017. Riboswitch diversity and distribution. RNA 23: 995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan RE, Torgerson CD, Gaffney BL, Jones RA, Strobel SA. 2016. Nuclease-resistant c-di-AMP derivatives that differentially recognize RNA and protein receptors. Biochemistry 55: 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. 2013. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol 9: 834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane K, Yu H, Foster DAN, Wang F, Woodside MT. 2011. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res 39: 7677–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. 2006. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34: 5623–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Lancaster L, Mohan S, Zhou J. 2017. Ribosome structural dynamics in translocation: yet another functional role for ribosomal RNA. Q Rev Biophys 50: e12. [DOI] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. 2004. The riboswitch control of bacterial metabolism. Trends Biochem Sci 29: 11–17. [DOI] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdrizet GA, Artsimovitch I, Furman R, Sosnick TR, Pan T. 2012. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc Natl Acad Sci 109: 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JN, Ferré-D'Amaré AR. 2010. Rapid construction of empirical RNA fitness landscapes. Science 330: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CH, Bembenek SD, Tounge BA. 2007. The role of molecular size in ligand efficiency. Bioorg Med Chem Lett 17: 4258–4261. [DOI] [PubMed] [Google Scholar]
- Rieder R, Lang K, Graber D, Micura R. 2007. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem 8: 896–902. [DOI] [PubMed] [Google Scholar]
- Ruff KM, Strobel SA. 2014. Ligand binding by the tandem glycine riboswitch depends on aptamer dimerization but not double ligand occupancy. RNA 20: 1775–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff KM, Muhammad A, McCown PJ, Breaker RR, Strobel SA. 2016. Singlet glycine riboswitches bind ligand as well as tandem riboswitches. RNA 22: 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. 2012. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys 41: 343–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Wang PY, Rutenberg-Schoenberg M, Simon MD. 2017. Interpreting reverse transcriptase termination and mutation events for greater insight into the chemical probing of RNA. Biochemistry 56: 4713–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan CA, Gaffney BL, Jones RA, Strobel SA. 2011. Differential analogue binding by two classes of c-di-GMP riboswitches. J Am Chem Soc 133: 15578–15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman EM, Esquiaqui J, Elsayed G, Ye J-D. 2012. An energetically beneficial leader–linker interaction abolishes ligand-binding cooperativity in glycine riboswitches. RNA 18: 496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood AV, Henkin TM. 2016. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu Rev Microbiol 70: 361–374. [DOI] [PubMed] [Google Scholar]
- Shundrovsky A, Santangelo TJ, Roberts JW, Wang MD. 2004. A single-molecule technique to study sequence-dependent transcription pausing. Biophys J 87: 3945–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Wang Z, Koo B-K, Patel A, Cascio D, Collins K, Feigon J. 2012. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell 47: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16: 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR. 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9: 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR. 2015. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 11: 606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells PR. 1963. Linear free energy relationships. Chem Rev 63: 171–219. [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. 2005. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18: 49–60. [DOI] [PubMed] [Google Scholar]
- Williamson JR. 2000. Induced fit in RNA–protein recognition. Nat Struct Mol Biol 7: 834–837. [DOI] [PubMed] [Google Scholar]
- Wilson KS, von Hippel PH. 1995. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci 92: 8793–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284: 611–615. [DOI] [PubMed] [Google Scholar]
- Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.