Abstract
The DEAD-box RNA helicase Dbp2p is highly conserved in eukaryotes and has been implicated in transcription, ribosome biogenesis, mRNP assembly, nuclear export, and long noncoding RNA (lncRNA) function. It is not understood how Dbp2p performs these seemingly unrelated biological roles. An important step toward addressing this question is the determination of cellular RNA binding sites of Dbp2p. Here, we identify transcriptome-wide RNA binding sites of Dbp2p from Saccharomyces cerevisiae using UV-crosslinking, denaturing tandem affinity purification, and next generation sequencing. We find that Dbp2p crosslinks to mRNAs and ribosomal RNAs, and markedly to noncoding RNAs, including snoRNA, snRNAs, and tRNAs. In snoRNAs, Dbp2p preferentially crosslinks at sites near the 3′ ends. These sites coincide with regions where RNA–DNA hybrids (R-loops) form and with binding sites of Sen1p, another RNA helicase that functions in transcription termination and 3′ processing of noncoding RNAs. We show that Dbp2p interacts in an RNA-independent manner with Sen1p in vivo. Dbp2p crosslinks to tRNAs and other RNAs also at sites where R-loops form. Collectively, our data link Dbp2p to noncoding RNAs, Sen1p, and R-loops. The transcriptome-wide connection to R-loops provides a unifying theme for diverse cellular roles of Dbp2p.
Keywords: helicase, snoRNA, nucleus, R-loops, NNS complex, noncoding RNA
INTRODUCTION
The DEAD-box RNA helicase Dbp2p from yeast is highly conserved in eukaryotes (Iggo et al. 1991; Fuller-Pace 2013). The human Dbp2p ortholog DDX5 (p68) has been linked to several cancers and is a therapeutic target for acute myeloid leukemia and potentially other tumors (Causevic et al. 2001; Shin et al. 2007; Clark et al. 2008; Wortham et al. 2009; Lin et al. 2013; Mazurek et al. 2014). In vitro, Dbp2p unwinds RNA duplexes in an ATP-dependent manner and hydrolyzes ATP in an RNA-dependent fashion (Ma et al. 2013). In the cell, Dbp2p has been implicated in transcription, ribosome biogenesis, mRNA decay, mRNP assembly, nuclear export, and nuclear RNA quality control (Barta and Iggo 1995; Bond et al. 2001; Bleichert and Baserga 2007; Cloutier et al. 2012; Ma et al. 2013, 2016). Recently, Dbp2p has been linked to the formation of hybrids between long noncoding RNA (lncRNA) and DNA (R-loops) at the GAL gene locus, which affects carbon source metabolism in S. cerevisiae (Cloutier et al. 2016).
The seemingly unrelated cellular roles of Dbp2p raise the question of which common functional themes underlie its biological functions. To address this problem, it is essential to define the cellular RNA binding sites of Dbp2p, which are currently not known. Here, we determine the RNA binding sites of Dbp2p in the cell, using a protocol that combines affinity purification of tagged, endogenous Dbp2p with the individual nucleotide resolution crosslinking (iCLIP) approach (König et al. 2010; Huppertz et al. 2014).
We find that Dbp2p binds rRNAs, mRNAs, and noncoding RNAs including snoRNAs, snRNAs, and tRNAs. Dbp2p interacts with snoRNAs and snRNAs predominantly near 3′ ends at positions that closely correspond to binding sites of Sen1p, another RNA helicase and component of the Nrd1-Nab3-Sen1 (NNS) complex, which functions in transcription termination and 3′ processing of noncoding RNAs in yeast. We show that Dbp2p and Sen1p interact in the cell in an RNA-independent manner. The link between Dbp2p and the NNS complex rationalizes the connection of Dbp2p to nuclear RNA quality control (Cloutier et al. 2012). We further find that Dbp2p binding sites on diverse RNAs correlate with sites where R-loops form (Wahba et al. 2016). These observations suggest that the link between Dbp2p and R-loops is not restricted to lncRNA at the GAL gene locus (Cloutier et al. 2016), but represents a common, transcriptome-wide theme for Dbp2p function on diverse RNA types.
RESULTS AND DISCUSSION
Dbp2p associates with noncoding RNAs
To determine the RNA binding sites of Dbp2p in S. cerevisiae, we combined denaturing purification of affinity-tagged, endogenous Dbp2p with the iCLIP approach. We implemented a stringent, denaturing affinity purification to prevent isolation of not crosslinked RNA fragments, which might be detected because Dbp2p, like other DEAD-box helicases, interacts with RNAs without known substrate preference (Ma et al. 2013). We generated a yeast strain with a histidine-TEV-biotin-histidine tag on the genomically encoded Dbp2p (DBP2-HTBH), thereby ensuring that the tagged protein was under the control of endogenous promoters. Growth characteristics for wild-type and DBP2-HTBH strains were virtually identical (Supplemental Fig. S1A). We performed UV-crosslinking (König et al. 2012) and isolated crosslinked Dbp2p-RNA fragments by dual affinity purification under denaturing conditions. Libraries for next generation sequencing were generated, sequenced, and analyzed according to the iCLIP protocol (Supplemental Fig. S1B; Huppertz et al. 2014).
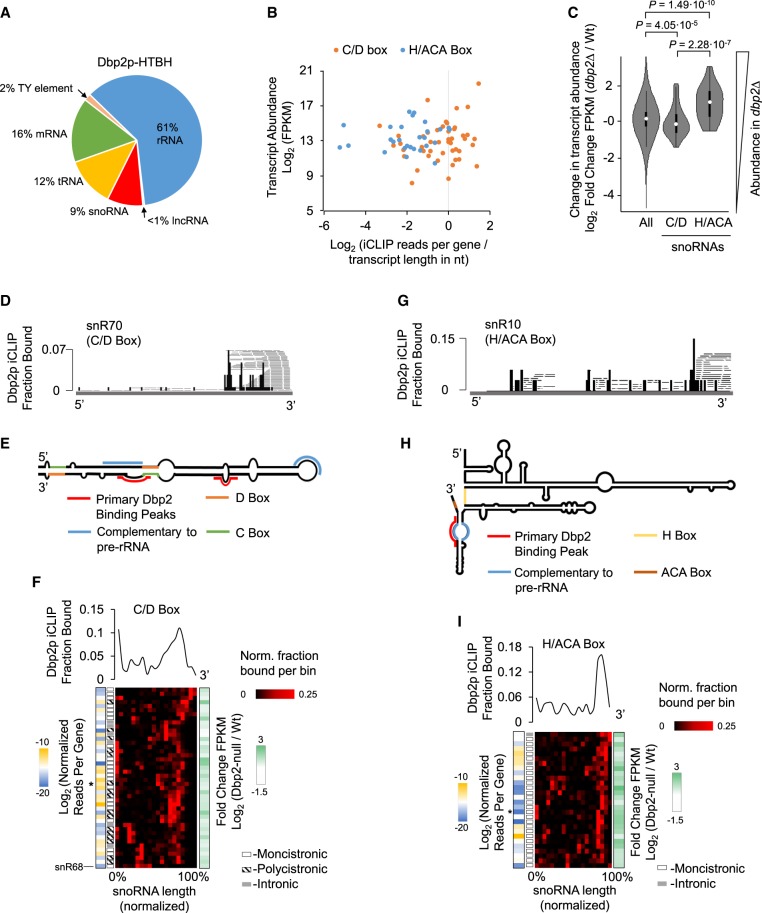
Dbp2p crosslinked to diverse RNA types (Fig. 1A). Most sequencing reads were found in rRNAs and mRNAs, consistent with reported roles of Dbp2p in mRNA and ribosome metabolism (Bond et al. 2001; Ma et al. 2013, 2016). Crosslinking of Dbp2p was also seen on noncoding RNAs, most notably to snoRNAs, tRNAs, and Ty-elements (Fig. 1A). Since snoRNAs comprise <1% of the cellular RNA mass, and Ty-elements <0.3%, the data indicated preferential association of Dbp2p to these RNA types. tRNAs comprise roughly 10% of the cellular RNA mass, but extensive interactions of proteins with tRNA are unusual, unless the proteins participate in tRNA metabolism (Gerstberger et al. 2014; Mukherjee et al. 2018). The data thus suggested preferential Dbp2p association with tRNAs as well. We did not detect significant sequence or structure signatures for Dbp2p crosslinking sites for any of the RNA types or for all RNAs combined (Supplemental Fig. S1C), suggesting that Dbp2p did not recognize specific RNA sequences or structures in the cell. The spectrum of RNAs crosslinked to Dbp2p differed markedly for the phylogenetically closely related DEAD-box helicase Ded1p (Guenther et al. 2018). Using an affinity-tag and denaturing purification protocol identical to that used for Dbp2p, <1% of Ded1p crosslinks mapped to tRNAs and <2% for snoRNAs (Guenther et al. 2018). Collectively, our observations indicated direct, physical links between Dbp2p and noncoding RNAs.
FIGURE 1.
Dbp2p binding to snoRNAs. (A) Distribution of unique, mapped crosslinking reads for Dbp2p. Values represent averages of two data sets from biological replicates. (B) Correlation between expression levels of C/D-box (orange) and H/ACA-box (blue) snoRNAs (determined by RNA-seq) and unique iCLIP reads per snoRNA, normalized for transcript length (nucleotides). Values for expression levels and normalized iCLIP reads represent averages from biological replicates (C/D-box snoRNA: n = 44. H/ACA-box snoRNA: n = 28). (C) Differences between wild-type and dbp2Δ strain expression levels for all RNAs, and C/D-box and H/ACA-box snoRNAs. Values represent averages from biological replicates. P-values were calculated according to Welch's two-tailed t-test (all RNAs: n = 6702. C/D-box snoRNA: n = 44. H/ACA-box snoRNA: n = 28). (D) Representative iCLIP trace for the C/D-box snR70. Black bars indicate 5′ ends of crosslink reads. Dbp2p iCLIP Fraction Bound refers to the black bars (RT stops) and indicates values normalized to the RNA. Gray lines show full sequencing reads. (E) Schematic secondary structure for snR70. Prominent Dbp2p binding sites are marked in red. C-box, D-box, and regions complementary to the ribosome are highlighted as noted in the legend. (F) Metagene analysis of Dbp2p binding to C/D-box snoRNAs (n = 44). (Upper panel) Plot of fractions of iCLIP reads per RNA bin (averaged from biological replicates), normalized for each C/D-box snoRNA (e.g., panel E) and averaged for all C/D-box snoRNAs. (Lower panel) Fraction of Dbp2p crosslinked (per bin) for all C/D-box snoRNAs, averaged from both biological replicates. Traces are arranged by hierarchal clustering. Blue and gold colored blocks on the left show the degree of Dbp2p crosslinked to a given transcript (log2 iCLIP reads per transcript, normalized to transcript length in nucleotides, divided by transcript abundance in FPKM). The grayscale blocks indicate whether the snoRNA is monocistronically, polycistronically, or intronically encoded. White and green colored blocks on the right show the changes in expression level for the respective snoRNA between wild-type and dbp2Δ strain (log2 fold change in FPKM). The asterisk marks snR70. (G) Representative iCLIP trace for the H/ACA-box snR10. Black bars indicate 5′ ends of crosslink reads. Fraction Bound refers to the black bars (RT stops) and indicates values normalized to the RNA. Gray lines show full sequencing reads. (H) Schematic secondary structure for snR30. Prominent Dbp2p binding sites are marked in red. H-box, ACA box, and regions complementary to the ribosome are highlighted as noted in the legend. (I) Metagene analysis of Dbp2p binding to H/ACA-box snoRNAs (n = 28). Data are presented as described for C/D-box snoRNAs in panel G. The asterisk marks snR10.
Dbp2p binds to 3′ proximal regions in snoRNAs
Because Dbp2p had not been physically linked to snoRNAs, we analyzed Dbp2p association with these noncoding RNAs in more detail. We first examined whether Dbp2p bound selectively to a subset of snoRNAs. To this end, we compared the expression level of each C/D-box and H/ACA-box snoRNA with the crosslinking reads of Dbp2p in each snoRNA, normalized to the transcript length (Fig. 1B). Expression levels of snoRNAs were determined by RNA-seq (Supplemental Fig. S1D). The number of Dbp2p crosslinking reads varied for both, C/D-box and H/ACA-box snoRNAs, but snoRNA expression levels showed no apparent correlation to Dbp2p crosslinking reads normalized to transcript length (Fig. 1B). These data indicated that Dbp2p association with snoRNAs is not primarily dictated by transcript levels, suggesting that other factors, processes, or RNA elements determine the degree of Dbp2p crosslinking. More Dbp2p reads were seen on C/D-box snoRNAs, compared to H/ACA-box snoRNAs, even though both snoRNA types were expressed over roughly similar ranges (Fig. 1B). The data revealed a preferential association of Dbp2p with C/D-box over H/ACA-box snoRNAs.
This notion prompted us to test whether Dbp2p differentially affected the expression of C/D- and H/ACA-box RNAs. Previous RNA-seq data had suggested that snoRNAs were among the genes whose expression levels were most affected by DBP2 deletion (Beck et al. 2014). However, stringent size selection of the RNA-seq libraries in these experiments precluded unbiased examination of the snoRNA genes. We therefore measured RNA levels by RNA-seq from libraries that explicitly accounted for RNAs in the length-range of snoRNAs (see Materials and Methods). To assess the impact of Dbp2p, we compared RNA levels in a wild-type and in a DBP2 deletion strain (dbp2Δ) (Supplemental Fig. S1D–F; Beck et al. 2014). Consistent with previous data, we observed relative differential expression of over 1000 RNAs by a factor of greater than 2. Both, up- and down-regulated RNAs also correlated broadly with the previous data (Supplemental Fig. S1G–I; Beck et al. 2014). In addition, we found that Dbp2p affects the relative expression levels of H/ACA- and C/D-box snoRNAs differently (Fig. 1C). Levels for most H/ACA-box snoRNAs increase in the dbp2Δ strain, whereas levels for most C/D-box snoRNAs decrease (Fig. 1C; Supplemental Fig. S1F). The data suggested a connection between Dbp2p-snoRNA association and the expression level of the snoRNA type.
We next analyzed the Dbp2p crosslinking patterns on snoRNAs (Fig. 1D–I). Crosslink sites of Dbp2p on C/D-box snoRNAs clustered toward the 3′ end, although less frequent crosslinking was also seen at other sites (Fig. 1D–F). Most crosslinked reads did not extend beyond the processed snoRNA terminus, suggesting that Dbp2p binds the snoRNAs after 3′ maturation (Fig. 1D; Supplemental Fig. S1J). Preferential crosslinking toward the 3′ end was seen for all C/D-box snoRNAs, except for snR68 (Fig. 1F), although it was not apparent why the Dbp2p binding pattern differed for this transcript.
The peak of crosslinked reads for C/D-box snoRNAs was located on average approximately 30 nt upstream of the 3′ terminus (Supplemental Fig. S2A). It is possible that actual binding of Dbp2p extends further toward the 3′ terminus, but due to the sequencing read-size bias inherent to the iCLIP procedure, shorter RNA fragments might be depleted during the library preparation.
A metagene analysis of the crosslink patterns of Dbp2p to all C/D-box snoRNAs revealed slight variations in the Dbp2p binding pattern mostly with respect to the distance of the major crosslink sites relative to the 3′ end (Fig. 1F). However, we could not detect correlations between these variations and the impact of Dbp2p on the expression level of the corresponding snoRNAs, or whether the snoRNAs were mono-cistronically, poly-cistronically, or intronically encoded (Fig. 1F). Nevertheless, our data revealed a preference of Dbp2p to associate with C/D-box snoRNAs proximal to processed 3′ ends.
Dbp2p crosslink sites on H/ACA-box snoRNAs also clustered toward the 3′ end (Fig. 1G–I). Crosslinked reads generally did not extend beyond the processed snoRNA terminus, suggesting that Dbp2p binds H/ACA-box snoRNAs after 3′-end maturation (Supplemental Fig. S1J). As seen for C/D-box snoRNAs, crosslinked reads peaked on average roughly 30 nt upstream of the 3′ terminus (Supplemental Fig. S2B). Dbp2p crosslinked to a subset of H/ACA-box snoRNAs to the region complementary to the rRNA (Fig. 1H). This observation raises the possibility that Dbp2p could impact the binding of these snoRNAs to rRNA, but these sites are also proximal to the 3′ end, which might render spurious the overlap between rRNA binding sites and Dbp2p crosslinks.
As for C/D-box snoRNAs, a metagene analysis of the crosslink patterns of Dbp2p to all H/ACA box snoRNAs revealed slight variations in the Dbp2p binding pattern (Fig. 1I). We did not detect differences in these variations for mono-cistronically or intronically encoded snoRNAs and no correlations between the binding pattern and impact of Dbp2p on the expression level of the given snoRNA (Fig. 1I). Thus, as seen for C/D-box snoRNAs, our data revealed Dbp2p association with H/ACA-box snoRNAs proximal to processed 3′ ends.
Dbp2p binding to snoRNAs reveals link to R-loops and the RNA helicase Sen1p
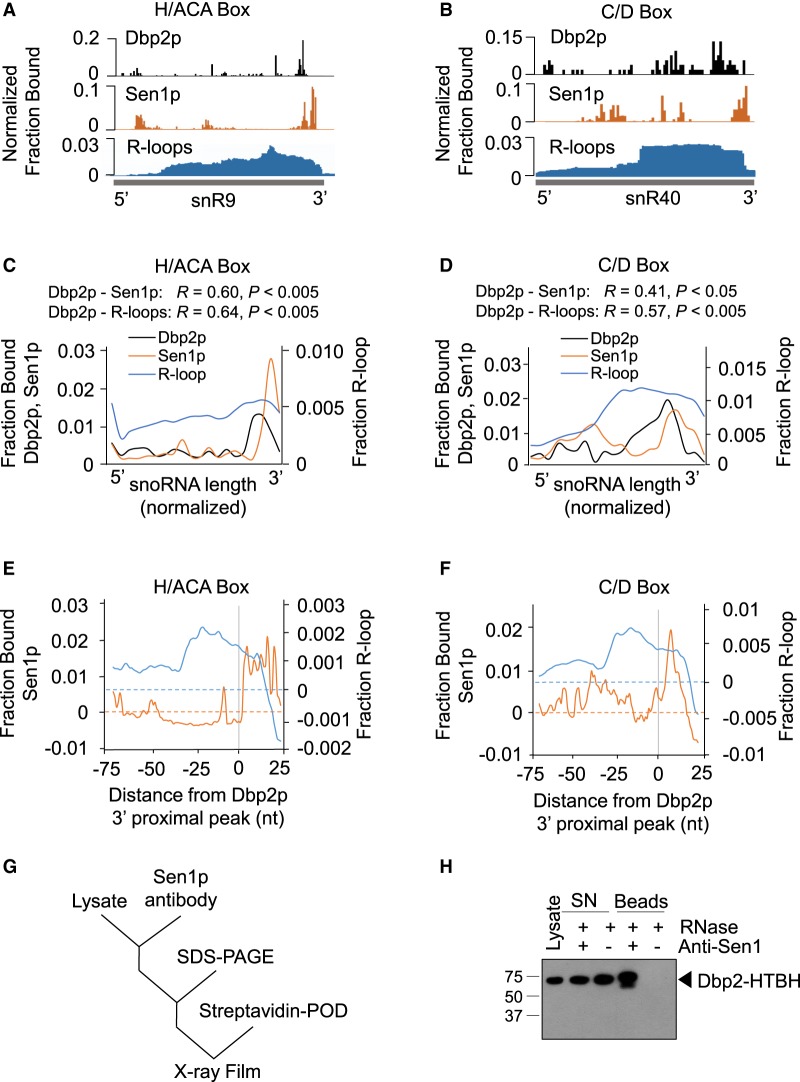
The preferential binding of Dbp2p to the 3′ ends of the snoRNAs raised the possibility of connections between Dbp2p and components of the machinery that processes snoRNAs at their 3′ termini, the NNS complex and the nuclear exosome (Kim et al. 2006; Vasiljeva et al. 2008). Dbp2p had been genetically linked to the nuclear exosome component Rrp6p (Cloutier et al. 2012). To detect links between Dbp2p and the NNS complex, we examined published, transcriptome-wide RNA binding data for NNS components (Jamonnak et al. 2011). We detected a correlation between Dbp2p binding sites on snoRNAs and the binding sites of Sen1p (Fig. 2A–F), but not with other published NNS components. The data suggested a link between Dbp2p and Sen1p.
FIGURE 2.
Correlation between Dbp2p binding sites, Sen1p binding sites, and R-loops on snoRNAs. (A) Representative traces of Dbp2p iCLIP, Sen1p PAR-CLIP (Jamonnak et al. 2011), and rnh1Δrnh201ΔS1-DRIP-Seq (R-loops) (Wahba et al. 2016) for the H/ACA-box snR9. Bars for Dbp2p represent the 5′ end of the iCLIP reads. Bars for Sen1p show the nucleotide conversion in the PAR-CLIP reads. R-loops are shown as complete reads. (B) Representative traces of Dbp2p iCLIP, Sen1p PAR-CLIP (Jamonnak et al. 2011), and rnh1Δrnh201ΔS1-DRIP-Seq (Wahba et al. 2016) for the C/D-box snR40. Data are presented as in panel A. (C) Metagene plots for Dbp2p iCLIP, Sen1p PAR-CLIP, and rnh1Δrnh201ΔS1-DRIP-Seq for H/ACA-box snoRNAs (n = 28), expressed as fractions of binding per bin (Dbp2p and Sen1p) and fraction of read density per bin (R-loops). R, Pearson correlation coefficients. P-values were calculated by a two-tailed t-test. (D) Metagene plots for Dbp2p iCLIP, Sen1p PAR-CLIP, and rnh1Δrnh201ΔS1-DRIP-Seq for C/D-box snoRNAs (n = 44). Data are presented as in panel C. (E) Proximity of Sen1p binding sites and regions of R-loop formation to Dbp2p binding sites on H/ACA-box RNAs (n = 25, three transcripts without clear 3′ peaks were excluded from the analysis). Metagene plot of normalized fraction per nucleotide of Sen1p binding and R-loop formation averaged for all H/ACA-box snoRNAs, in relation to the Dbp2p binding site, indicated by the gray line at zero. The background signal per nucleotide was subtracted by randomizing binding peaks over the RNAs three times and calculating the mean. (F) Proximity of Sen1p binding sites and regions of R-loop formation to Dbp2p binding sites on C/D-box RNAs (n = 43, one transcript without clear 3′ peak was excluded from the analysis). Data are shown as in panel E. (G) Experimental scheme for Dbp2p-Sen1p coimmunoprecipitation. Measurements were performed in the presence of RNases. (H) Representative western blot for Sen1p-Ded1p co-IP. The membrane was probed with streptavidin antibody for Dbp2p-HTBH. Experiments were performed four times for biological replicates with virtually indistinguishable results.
Sen1p is an RNA helicase of the helicase superfamily 1 (Fairman-Williams et al. 2010; Martin-Tumasz and Brow 2015). The enzyme has been implicated in transcription termination, and as part of the NNS complex, in the 3′-processing of snoRNAs and other noncoding RNAs (Ursic et al. 1997; Kim et al. 1999; Steinmetz et al. 2006; Chen et al. 2017). Deletion of Sen1p leads to a global accumulation of R-loops, and it has been proposed that Sen1p restricts R-loop formation (Mischo et al. 2011). As noted above, Dbp2p has also been shown to affect R-loop formation on the GAL gene cluster (Cloutier et al. 2016). The correlation between Dbp2p and Sen1p binding patterns on snoRNAs led us to hypothesize that a link between Dbp2p and R-loops might not be restricted to the GAL gene cluster, but might be more universal. To test this hypothesis, we compared our measured Dbp2p binding pattern on snoRNAs to recently published, transcriptome-wide maps of R-loops (Fig. 2A–F; Wahba et al. 2016). We observed a correlation between R-loop sites and Dbp2p binding sites on both, C/D- and H/ACA-box snoRNAs (Fig. 2C,D; Supplemental Fig. S1K). Metagene analyses revealed an increase in R-loop formation toward the 3′ ends of the snoRNAs. The 3′ ends of the R-loop regions coincided broadly with pronounced binding sites of Dbp2p and of Sen1p (Fig. 2C–F).
The metagene analysis also indicated that most pronounced Sen1p binding sites were immediately 3′ to Dbp2p binding sites (Fig. 2E,F). The proximity between Dbp2p and Sen1p binding sites raised the possibility of a direct interaction between both helicases. To test this notion, we immunoprecipitated Sen1p and probed for the presence of Dbp2p (Fig. 2G). We observed coimmunoprecipitation of Sen1p and Dbp2p (Fig. 2H). The experiments were performed in the presence of RNases, indicating that the Sen1p–Dbp2p interaction did not depend on RNA. While the data did not allow us to assess whether both helicases interact directly or via additional proteins, the observations established that both, Sen1p and Dbp2p are part of one multiprotein complex. This result revealed a physical link between Dbp2p and the machinery that processes the 3′ ends of snoRNAs. This physical link might play a role in the genetic connection between DBP2 and RRP6, another component of this machinery (Cloutier et al. 2012). In addition, the data raise the possibility that Dbp2p function on snoRNAs is coordinated with the NNS complex or at least with Sen1p.
Dbp2p binding to other RNA types indicates further links to R-loops
The correlation between Dbp2p binding sites with R-loops on snoRNAs indicated a link between Dbp2p and R-loops beyond the GAL cluster (Cloutier et al. 2016). R-loops are seen on transcripts by all three polymerases, and form most readily on genomic loci that are heavily transcribed, including rRNA, tRNA, snoRNAs, snRNAs, and Ty-elements (Wahba et al. 2016). This spectrum of RNA types is remarkably similar to the RNAs bound by Dbp2p (Fig. 1A), raising the possibility that links between Dbp2p and R-loops extend over the entire transcriptome.
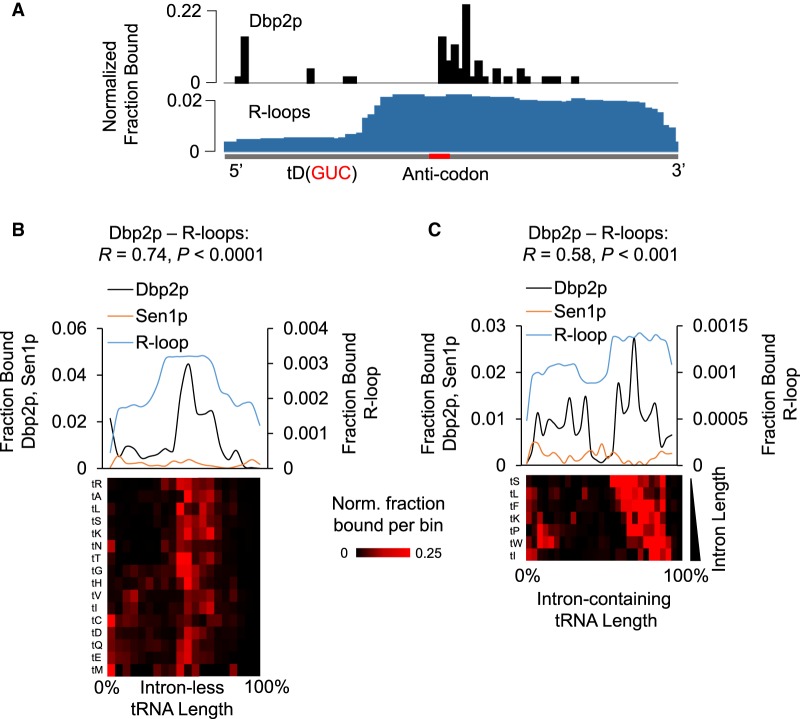
To test this possibility, we examined how Dbp2p binding to tRNAs correlated with R-loop regions (Fig. 3; Supplemental Fig. S2D). As noted, most RNA binding proteins do not extensively associate with tRNAs, unless they are involved in tRNA metabolism (Tuck and Tollervey 2013; Gerstberger et al. 2014; Mukherjee et al. 2018). Since Dbp2p had not been implicated in tRNA metabolism, tRNA binding by Dbp2p is noteworthy.
FIGURE 3.
Dbp2p binding and R-loops on tRNAs. (A) Representative traces of Dbp2p iCLIP and R-loops on the tD(GUC) tRNA. The transcript is shown in gray below, and the anti-codon is marked red. (B) Metagene plots for Dbp2p iCLIP, Sen1p PAR-CLIP, and rnh1Δrnh201ΔS1-DRIP-Seq for intronless tRNAs (n = 219), expressed as fractions of binding per bin (Dbp2p and Sen1p) and fraction of read density per bin (R-loops). Dbp2p and R-loop reads were combined for multiple tRNA isoforms for the same amino acid, because the read length did not always allow unambiguous assignment of a specific tRNA isoform. Correlation coefficients (R) and P-values were calculated as in Figure 2C,D. (Lower panel) Fraction of Dbp2p crosslinked (for all tRNA isoforms for each amino acid, averaged from biological replicates). Traces are arranged by hierarchal clustering. The amino acid for the respective tRNA isoforms is indicated on the left. (C) Metagene analysis of Dbp2p iCLIP Sen1p PAR-CLIP, and rnh1Δrnh201ΔS1-DRIP-Seq for intron-containing tRNAs (n = 64). (Upper panel) Metagene plots were generated as in Figure 1F,I, and data are presented as in panel B. Correlation coefficients and P-values were calculated as in Figure 2C,D. (Lower panel) Fraction of Dbp2p crosslinked (for all tRNA isoforms for each amino acid, averaged from biological replicates). Traces are arranged according to intron length. The amino acid for the respective tRNA isoforms is indicated on the left.
Analysis of Dbp2p binding to tRNA was performed for a composite of multiple tRNAs for each amino acid, because the limited lengths of Dbp2p iCLIP reads often precluded identification of the exact tRNA isoform. Dbp2p bound to intron-containing and intron-less tRNAs, generally in the vicinity to the anticodon, approximately 30–40 nt upstream of the 3′ ends of the tRNAs (Fig. 3A–C; Supplemental Fig. S2E). Correlation was seen between Dbp2p binding sites and R-loop regions (Fig. 3B,C). This observation supported the notion of a link between Dbp2p and R-loops beyond the GAL cluster. Of note, no significant correlation was observed between Dbp2p and Sen1p binding patterns (Fig. 3B,C), suggesting that the link between Dbp2p and Sen1p did not extend to tRNA.
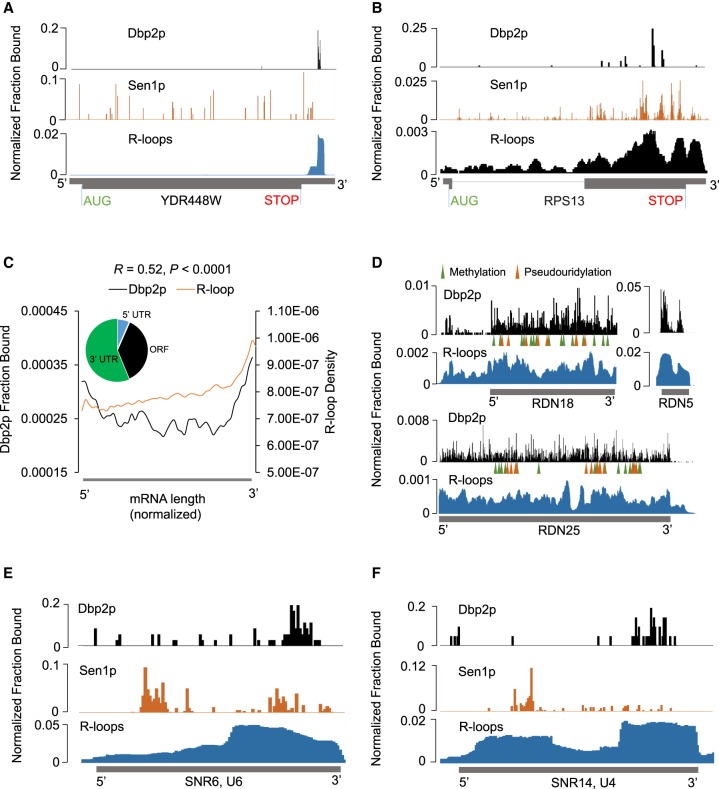
Having detected correlations between Dbp2p binding sites and regions of R-loop formation on snoRNAs and tRNAs, we next examined correlations between Dbp2p binding and R-loops for other RNA types (Fig. 4). For mRNAs, Dbp2p bound a large cross-section of transcripts (Supplemental Fig. S3A). Dbp2p crosslinking sites and R-loops accumulated toward the 3′-end consistent with a correlation (Fig. 4A–C). Dbp2p and Sen1p binding patterns on mRNAs differed significantly more than on snoRNAs (Fig. 4A,B; Supplemental Fig. S3C), suggesting that the detected interaction between Dbp2p and Sen1p might be less relevant for the function of the enzymes on mRNAs, compared to snoRNAs.
FIGURE 4.
Dbp2p binding to other RNA types indicates further links to R-loops. (A,B) Dbp2p, Sen1p binding, and rnh1Δrnh201ΔS1-DRIP-Seq on representative mRNA transcripts. Data are presented as in Figure 2A,B. (C) Metagene plot for Dbp2p iCLIP and rnh1Δrnh201ΔS1-DRIP-Seq for mRNA transcripts (n = 5055). Data were generated and are presented as in Figure 2C. Correlation coefficients and associated P-values were calculated as in Figure 2C,D. The inset shows the Dbp2p iCLIP read distribution for all mRNA transcripts in UTRs and ORF. (D) Dbp2p binding and rnh1Δrnh201ΔS1-DRIP-Seq density on rRNA transcripts. Data are presented as in Figure 2A,B. Mature rRNA transcripts are shown below the sequencing traces. Modification sites are marked according to Piekna-Przybylska et al. (2008). (E) Dbp2p, Sen1p binding, and R-loops on U6 snRNA. Data are presented as in Figure 2A,B. (F) Dbp2p, Sen1p binding, and R-loops on U4 snRNA. Data are presented as in Figure 2A,B.
For rRNA, which represented the largest fraction of Dbp2p crosslinks, Dbp2p binding sites extended at high density across the entire rRNA transcripts (Fig. 4D). We did not detect preferred binding of Dbp2p in the vicinity of modification sites, that is, at binding sites of snoRNAs (Supplemental Fig. S3D). R-loops form extensively on rRNA (Wahba et al. 2016), and also extend over the entire length of the transcripts (Fig. 4D). Extent and patterns of Dbp2p binding and R-loops on rRNA were consistent with a link between Dbp2p and R-loop formation.
In snRNAs, Dbp2p binding correlated with sites of R-loop formation (Fig. 4E,F; Supplemental Fig. S3E,F), with the exception of U2 (Supplemental Fig. S3D). Of note, the correlation was also seen for U6 (Fig. 4E), which is transcribed by RNA polymerase III (Kunkel et al. 1986). Sen1p binding to snRNAs was seen in the vicinity of Dbp2p binding sites (Fig. 4E,F; Supplemental Fig. S3E–G). However, the small number of snRNAs precludes an instructive analysis of the correlation between Dbp2p and Sen1p binding patterns on snRNAs. Finally, a correlation between sites of pronounced Dbp2p binding and sites of R-loop formation can also be seen for Ty-elements (Supplemental Fig. 3H). Collectively, the data for tRNA, rRNA, mRNA, snRNA, and Ty-elements all were consistent with Dbp2p binding to regions of preferential R-loop formation.
Conclusions
Our interrogation of the cellular RNA binding sites of Dbp2p revealed direct binding of Dbp2p to noncoding RNAs, an interaction between Dbp2p and the RNA helicase Sen1p, and a link between Dbp2p binding sites and RNA-loops for many RNA types. Direct binding of Dbp2p to noncoding RNAs, including virtually all snoRNAs and tRNAs was unexpected. Except for a reported relative reduction of C/D-box snoRNA levels in the dbp2Δ strain (Beck et al. 2014), available data provided no hint of a link of Dbp2p to these RNAs.
The direct binding of Dbp2p to snoRNAs, perhaps in conjunction with binding to rRNA, is consistent with the reported impact of Dbp2p on ribosome biogenesis (Bond et al. 2001). Diminished levels of Dbp2p lead to reductions in free 60S ribosome subunits and in mature 25S transcripts (Bond et al. 2001). It had been proposed that these effects arise from a role of Dbp2p in structural re-arrangements of rRNA during ribosome maturation (Bond et al. 2001). While this remains possible, our data suggest an additional role of Dbp2p: impact on the release of snoRNA and rRNA transcripts from the respective genomic loci.
The broad binding of Dbp2 to transcripts by all three polymerases is consistent with an expansive role of Dbp2p in the regulation of gene expression. This notion provides a rationale for the reported link of Dbp2p to energy metabolism (Beck et al. 2014; Cloutier et al. 2016). Upon glucose deprivation, RNA metabolism is significantly slowed, and Dbp2p relocates to the cytoplasm (Cloutier et al. 2016). The simultaneous loss of Dbp2p on a wide range of RNAs would be a simple way to accomplish fast and universal adaptation to the metabolic state. However, more experimentation is necessary to test this hypothesis.
In addition to the interaction between Dbp2p and a large cross-section of diverse RNAs, our data provide a physical explanation for the reported genetic link between DBP2 and RRP6, which is part of the machinery that processes 3′ ends of snoRNAs (Cloutier et al. 2012). Our data show an interaction between Dbp2p and the RNA helicase Sen1p, a component of the NNS complex, which works in conjunction with the nuclear exosome that is associated with Rrp6p (Fasken et al. 2015; Fox et al. 2015). Sen1p has also been implicated in the resolution of R-loops (Mischo et al. 2011). Our findings raise the possibility that Dbp2p might work in conjunction with Sen1p to resolve R-loops, at least on snoRNAs, where both proteins bind in close proximity.
The perhaps least anticipated finding of our study is the widespread correlation of Dbp2p binding sites with regions of R-loop formation. Dbp2p facilitates resolution of R-loops for lncRNAs on the GAL locus (Cloutier et al. 2016). Our data suggest that the connection between Dbp2p and R-loops extends beyond the GAL locus. Extrapolating from the function of Dbp2p in the removal of R-loops at the GAL locus, we speculate that Dbp2p participates in the resolution of R-loops for many transcripts. A widespread role of Dbp2p in the resolution of R-loops would provide a simple mechanistic explanation for seemingly unrelated functions of the enzyme in RNA metabolism and for the link of Dbp2p to actively transcribed chromatin (Ma et al. 2016).
MATERIALS AND METHODS
Dbp2-HTBH strain
Construction of plasmid pFA6a-HTB containing the HTB-tagged Dbp2p was described previously (Tagwerker et al. 2006). pFA6a-HTB was then used to generate the plasmid pFA6a-HTBH (BTP16). The HIS-tag was added by PCR amplification with primers HIS F and HIS R, generating pFA6a-HTBH (BTP16). This plasmid was then used as a template for PCR-mediated homologous recombination to tag DBP2 with HTBH in the yeast genome. Expression of Dbp2p-HTBH was verified by western blot using anti-RGS-His antibody (Qiagen). Growth characteristics of the DBP2-HTBH strain were examined by serial dilution experiments, compared to the wild-type strain (Supplemental Fig. S1A). Cells were grown in YP + 2%D to mid-log phase, harvested by centrifugation and re-suspended in 1× TE to an OD600 nm = 0.5. Fivefold serial dilutions were prepared, spotted on YP + 2%D plates and grown from 2 to 7 d at temperatures ranging from 16°C to 37°C (Supplemental Fig. S1A). Generation of the dbp2Δ strain has been described previously (Cloutier et al. 2012).
iCLIP
iCLIP experiments were performed with Dbp2p-HTBH on the endogenous DBP2 allele. Yeast cells containing HTBH-tagged Dbp2p were grown in rich media to OD600 nm of 0.7–0.9, harvested by brief centrifugation at 4000g, re-suspended in ice-cold YPD, transferred to a Petridish, and subjected to UV-light in a Stratalinker (600 mJ/cm2, 254 nm) on ice, mixing cells twice during crosslinking. Cells were then washed in ice-cold YPD, sedimented by centrifugation for 5 min at 5250g, frozen on dry ice, and stored at −80°C. Frozen cells were lysed in QIA-1M buffer (100 mM NaH2PO4 pH 8, 10 mM Tris, 1 M NaCl, 8 M urea, 10 mM imidazole, 0.5% (w/v) IGEPAL, 2.5 mM beta-mercaptoethanol, 1 mM PMSF, protease inhibitor cocktail [Roche]) with glass beads six times for 30 sec in a Beadbeater system (BioSpec Products). Glass beads were removed, and lysates were centrifuged at 5250g for 30 min. Cleared lysates were incubated with Ni2+-Agarose (40 μL slurry per gram dry pellet weight, pre-equilibrated in buffer QIA-1M; Qiagen) overnight at 4°C. Ni2+-beads were washed in 25 mL of wash buffer 1 (0.3 M NaCl, 10 mM Tris, 100 mM NaH2PO4, 8 M Urea, 10 mM imidazole). The sample was eluted with 10 mL elution buffer1 (0.3 M NaCl, 100 mM Tris, 50 mM NaH2PO4, 8 M Urea, 500 mM imidazole, 10% (v/v) glycerol). Eluates were incubated with 20 μL equilibrated streptavidin-conjugated magnetic Dynabeads (ThermoFisher Dynabeads-280) per gram dry pellet weight overnight at 4°C. Streptavidin beads were washed twice with 500 µL wash buffer 2 (0.3 M NaCl, 100 mM Tris, 8 M urea, 0.5 mM EDTA). Streptavidin beads were then washed twice in 500 µL 1× PNK buffer and split into two samples (80% – “L”, 20% – “H”). 200 ng RNase I was added per gram dry pellet weight to the “H”-sample (reference) and 0.2 pg RNase I to the “L”-sample, which was used for further processing.
The sample was incubated for 5 min at 37°C on a rotator. Resins were subsequently washed with 500 µL wash buffer 2 (2% [w/v] SDS), followed by 500 µL wash buffer 2 without SDS, and two washes with 500 µL 1× PNK buffer. Processing then was performed by the following methods for the two iCLIP experiments (i) using alkaline phosphatase and a 3′ RNA adaptor and (ii) using PNK and a pre-adenylated 3′ DNA adaptor. Each method involved unique reverse transcription and PCR primers.
For method (i), the supernatant was removed and beads were re-suspended in 67 μL RNase-free water, 3 μL alkaline phosphatase (NEB) and 2 μL RNasin (Roche) and incubated for 20 min at 37°C. Beads were washed twice with 500 µL 1× PNK buffer, once with 20 µL of 0.5 mM EDTA and 480 µL PNK buffer, and then twice with 500 µL of PNK buffer. 3′-ligation was performed by re-suspending the resin in 32 μL RNase-free water with 8 μL of 20 μM X12 RNA Linker (Supplemental Table S2). Reactions were performed overnight (shaking) at 4°C in a reaction volume of 40 μL (22 μL RNase-free water, 8 μL 10× T4 RNA ligase buffer, 8 μL BSA (0.2 μg/μL), 2 μL T4 RNA ligase [all NEB]).The beads were washed with 1 mL 1× wash buffer 2 (0.3 M NaCl, 100 mM Tris, 8 M urea, 0.5 mM EDTA) and twice with 1 mL 1× PNK buffer. Beads were re-suspended in 64 μL RNase-free water, 8 μL 10× PNK buffer, 4 μL γ-32P-ATP, 4 μL T4 PNK (all NEB). Reactions were performed for 60 min in a shaking thermoblock at 37°C. Beads were washed twice with 1 mL of wash buffer 2 and then twice with 1 mL of PNK.
Streptavidin beads were incubated with 200 μL proteinase K (4 mg/mL; Roche) for 20 min shaking at 37°C. The eluate was collected and the beads were incubated with another 200 µL proteinase K for 5 min at RT. The two eluates were combined and the reaction stopped by standard phenol/chloroform extraction. Released RNA was ethanol-precipitated overnight. Purified RNAs were re-suspended in 89 μL RNase-free water, 11 μL 10× DNase I Buffer, 5 μL RNasin, 5 μL RQ1 DNase (all Promega) and incubated for 20 min at 37°C. The reaction was stopped by standard phenol-chloroform extraction and RNA was ethanol-precipitated overnight.
The DNase-treated RNA was re-suspended in 1.4 μL RNase-free water, 0.2 μL 10 mM dNTPs, 1 μL 20 nM RT primer X1 (Supplemental Table S1), and incubated for 5 min at 65°C then cooled on ice. Next, 0.8 μL 5× first-strand buffer, 0.2 μL 1 M DTT, 0.2 μL RNase inhibitor, 0.2 μL Superscript III (Invitrogen) were added and incubated for 60 min at 50°C. RNA was degraded by alkaline hydrolysis (after the addition of 0.5 μL of 1 N NaOH and incubation at 98°C for 15 min). After the addition of loading buffer, cDNA was applied to 10% denaturing polyacrylamide gel electrophoresis (PAGE) and stained with SYBR Gold. Fragments of 100–125 nt were cut from the gel. The gel slices were crushed and cDNA was recovered by incubation in 500 μL diffusion buffer (20 mM Tris-HCl pH 7.5, 250 mM NaOAc, 1 mM EDTA, 0.25% (w/v) SDS) overnight at 4°C, followed by ethanol precipitation.
The cDNA was suspended in 15 μL RNase-free water and circularized with CircLigase I (Epicentre) according to the manufacturer's instructions. The circularized cDNA was utilized for amplification with Phusion polymerase (NEB) and primers X2 and X3 (Supplemental Table S1). PCR settings were: 30 sec at 98°C, followed by 24 PCR cycles (10 sec at 98°C, 30 sec at 58°C, 30 sec at 72°C). PCR products were applied to 10% nondenaturing PAGE and visualized by SYBR Gold. Products with 75–90 bp were cut, DNA was extracted from gel slices and ethanol precipitated. PCR products were amplified with Phusion polymerase and primers X4 and X5 for 7 cycles utilizing the same PCR settings as above. PCR products were then separated on a 2% Agarose gel, cut out and subjected to sequencing using primer X6.
For method (ii), after RNase digestion, Streptavidin Dynabeads were incubated with 43 µL of Tris-buffer pH 7.5, heated to 70°C for 90 sec and cooled to 37°C. Then 5 µL of 10× PNK reaction buffer, 1 µL SUPERase-IN (ThermoFisher), and 1 µL of T4 PNK (NEB) were added to the beads and incubated at 37°C for 60 min. T4 PNK was then head inactivated for 10 min at 70°C followed by two washes of the beads with 500 µL PNK buffer. The streptavidin beads were then re-suspended in 23 µL of RNase-free water containing X10r pre-adenylated linker, heated to 70°C for 90 sec and cooled to room temperature. 3′ linker ligation was then initiated by adding 4 µL 10× Rnl2 buffer, 6 µL PEG 8000 (50% w/v), 1 µL SUPERase-In, and 1 µL T4 Rnl2 (truncated) RNA ligase and incubated overnight at 16°C with rotation. The beads were washed with 500 µL of wash buffer 2 and then twice with 500 µL of PNK buffer. Proteinase K and DNase treatment was performed as described in method (i) followed by phenol/chloroform extraction and ethanol precipitation. The DNase-treated RNA was re-suspended in 15 μL RNase-free water, 2 μL 10 nM RT primer X7r (Supplemental Table S1), and incubated for 5 min at 65°C. Next, 3 μL 3.3 μM each of dGTP, dCTP, and dTTP, 6 μL 5× first-strand buffer,1 μL α-32P-dATP, 1 μL 0.1 M DTT, 1 μL RNase inhibitor, 1 μL Superscript III (all Invitrogen) were added and incubated for 30 min at 50°C. Then 1 µL of 10 mM dNTPs were added for 10min at room temperature to complete the reaction.
RNA was degraded by alkaline hydrolysis (addition of 0.5 μL of 1 N NaOH, incubation at 98°C for 15 min). After the addition of loading buffer, cDNA was applied to 10% denaturing PAGE and stained with SYBR Gold. Fragments were visualized by a PhosphorImager and cut from the gel. The gel slices were crushed and cDNA was recovered by incubation in 500 μL diffusion buffer (20 mM Tris pH 7.5, 250 mM NaOAc, 1 mM EDTA, 0.25% [w/v] SDS) overnight at 4°C, followed by ethanol precipitation.
The cDNA was suspended in 15 μL RNase-free water and circularized with CircLigase I (Epicentre) according to the manufacturer's instructions. The circularized cDNA was utilized for amplification with Phusion polymerase (NEB) and primers X8r and X9r (Supplemental Table S1). PCR settings were: 30 sec at 98°C, followed by 24 PCR cycles (10 sec at 98°C, 30 sec at 58°C, 30 sec at 72°C). PCR products were applied to 10% nondenaturing PAGE and visualized by SYBR Gold. Products were extracted from cut gel slices, ethanol precipitated, and primer X11r was used to perform Illumina sequencing (Supplemental Table S1).
Bioinformatic analysis
Raw sequencing data were analyzed in Galaxy (Goecks et al. 2010) for quality by FASTQC, followed by clipping of adaptors using Clip, collapsing of identical reads using Collapse, and trimming for low-quality bases by FastQ quality trimmer (Clip and Collapse are based on the FASTX-Toolkit [http://hannonlab.cshl.edu/fastx_toolkit/]). Reads with identical start and end positions were collapsed again by a custom workflow in Galaxy (code available upon request). Resulting reads were mapped with TopHat (BowTie2) software (Trapnell et al. 2009), allowing for two mapped read locations to account for rRNA reads and a single mismatch. Read locations were then cross-referenced as BED files with the reported gene locations of SacCer3 categorized by RNA subtype. The biological replicates contained 109,723 and 180,084 of these stringently processed, unique reads.
mRNA positional coordinates, including transcription start sites and polyadenylation sites were based on sacCer3 and reported measurements (Park et al. 2014). Genome-wide data sets were visualized using IGV software (Robinson et al. 2011). Bioinformatic analyses were performed primarily using R (R Core Team 2013) with custom scripts and RStudio Software (RStudio Team 2015). Where applicable, data sets for Dbp2p iCLIP, Sen1p PAR-CLIP, and S1-DRIP-seq were normalized per transcript by dividing read counts at each base by the number of all reads on a given transcript. Length normalization of iCLIP reads was performed by dividing read counts by transcript length in nucleotides.
Metagene profiles were calculated by binning the transcripts nucleotides to normalize for length. For snoRNAs and tRNAs, 20 bins were used. For mRNAs, 50 bins were used to normalize the length from transcription start site to polyadenylation site. Heatmaps for snoRNA were generated by hierarchical clustering of the binned genes using Cluster 3.0 (de Hoon et al. 2004) and visualized by Java Treeview (Saldanha 2004). Pearson correlation coefficients for Metagene binding patterns of Dbp2p, Sen1p, and R-loops were calculated by using corresponding bin values.
iCLIP peaks were determined by Piranha peak calling software (http://smithlabresearch.org). Parameters were optimized by visual inspection. In Figure 2E,F, Dbp2p iCLIP peaks most 3′ ends in snoRNAs were used as reference points for calculating the mean normalized binding density per nucleotide for Sen1p PAR-CLIP and S1-DRIP-Seq surrounding these reference points. Background signal was subtracted as follows: Random peak locations anywhere within snoRNA (same number as in the data set) were used as reference points. This procedure was repeated three times. The mean from these repetitions was defined as background signal. Supplemental Figure S1J was calculated by using the 3′ end of snoRNAs as a reference point and taking the mean of normalized reads for each nucleotide up and downstream of this reference point. Genome coverage was calculated using the full length reads in order to detect reads overlapping with the mature snoRNA 3′ ends. 3′ end binding plots in Supplemental Figure S2A,B were calculated as in Supplemental Figure 1E,F with individual nucleotide genome coverage.
RNA-seq
RNA-seq was performed on wild-type and dbp2Δ yeast strains (Cloutier et al. 2012) in biological duplicates. The strains were grown in YP + 2% glucose (YPD) OD600 nm = 0.4 and harvested by centrifugation. Cells were flash frozen in liquid nitrogen, and stored at −80°C. Cells were re-suspended in 150 μL of LET (25 mM Tris, pH 8.0 100 mM LiCl, 20 mM EDTA) combined with an equal volume of glass beads and vortexed for 5 min.
A total of 250 μL RNase-free water and 250 μL phenol/chloroform/LET were added and samples were vortexed for an additional 5 min before centrifugation for 2 min at 14,000 rpm. The aqueous phase was isolated and vortexed with 500 μL phenol/chloroform/LET. Centrifugation and aqueous phase extraction was repeated. The RNA was precipitated with 40 μL 3 M NaOAc/ethanol overnight at −80°C. The RNA was pelleted by centrifugation at 4°C for 25 min at 14,000 rpm and re-suspended in 50 μL of RNase-free H2O. DNA was digested with DNase I (RNase-free; Invitrogen) and RNaseOUT (Invitrogen; 10777019) in 1× DNase I reaction buffer for 1 h at 37°C. Phenol/chloroform/LET extraction, precipitation, and re-suspension was then repeated as described above.
cDNA libraries were prepared using Illumina ScriptSeq Complete Gold Kit for yeast (BGY1306) and ScriptSeq Index PCR Primers set 1 (RSBC10948) according to the manufacturer's protocol with the following adjustments. To avoid depletion of short RNAs including snoRNAs, purification of the rRNA-depleted samples was performed by ethanol precipitation instead of AMPure bead purification. cDNA was purified by the MinElute PCR Purification kit (Qiagen) in place of the Agencourt AMPure XP system. Processing of RNA-seq data was performed by mapping with Tophat followed by analysis with Cufflinks and R package edgeR.
Coimmunoprecipitation
Cultures of wild-type and Dbp2p-HTBH (50 mL) were grown at 30°C in YPD to OD600 nm = 0.5, pelleted, and resuspended with two volumes of IP Buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% [v/v] glycerol). Cells were lysed by vortexing at 4°C with glass beads for 3 min at 30 sec intervals with 2 min on ice in between vortexing. Glass beads were removed and the lysate was cleared by centrifugation at 4000 rpm for 2 min. Protein G Dynabeads (Invitrogen 10003D) were prepared according to the manufacturer's guidelines. A total of 12 μL of the Sen1p antibody was diluted up to 200 μL with PBST (10 mM Na2HPO4, 137 mM NaCl, 2 mM KH2PO4, 2.7 mM KCl, 0.05% Tween 20, pH 7.4) and incubated with protein G Dynabeads for 10 min, rotating at room temperature. Beads were immobilized with a magnet and the supernatant was discarded. Beads were then washed twice gently with 200 μL PBST and subsequently resuspended in PBST. The cell lysate was incubated with the prepared beads-antibody complex at 4°C for 3 h with gentle rocking. Beads were washed three times with wash buffer (150 mM Tris, 50 mM NaCl, 2 mM MgCl2, 0.1% NP40). Samples were eluted at 95°C for 5 min in SDS-PAGE loading buffer, run on a 10% NEXT Gel (VWR; 1B1724-500ML) according to the manufacturer's guidelines and transferred to PVDF membrane (Thermo Scientific; 88518). The PVDF membrane was blotted with Streptavidin-POD conjugate (Roche; 11089153001) and exposed to X-ray film.
DATA DEPOSITION
All sequencing data have been deposited in the GEO database under accession number GSE113489. Computer code has been deposited at GitHub (https://github.com/FrankTedeschi/Dbp2p_Rscripts).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Tran and Jankowsky groups for discussion, and Dr. Ulf-Peter Guenther, Dr. Donny Licatalosi, and Leah Zagore for advice. The α-Sen1p antibody was a kind gift from Dr. David Brow. This work was supported by the National Institutes of Health (GM118088 to E.J., GM097332 to E.J.T.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067249.118.
REFERENCES
- Barta I, Iggo R. 1995. Autoregulation of expression of the yeast Dbp2p “DEAD-box” protein is mediated by sequences in the conserved DBP2 intron. EMBO J 14: 3800–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck ZT, Cloutier SC, Schipma MJ, Petell CJ, Kit W, Ma EJ, Tran EJ. 2014. Regulation of glucose-dependent gene expression by the RNA helicase Dbp2 in Saccharomyces cerevisiae. Genetics 198: 1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Baserga SJ. 2007. The long unwinding road of RNA helicases. Mol Cell 27: 339–352. [DOI] [PubMed] [Google Scholar]
- Bond AT, Mangus DA, He F, Jacobson A. 2001. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol Cell Biol 21: 7366–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causevic M, Hislop RG, Kernohan NM, Carey FA, Kay RA, Steele RJ, Fuller-Pace FV. 2001. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene 20: 7734–7743. [DOI] [PubMed] [Google Scholar]
- Chen X, Poorey K, Carver MN, Müller U, Bekiranov S, Auble DT, Brow DA. 2017. Transcriptomes of six mutants in the Sen1 pathway reveal combinatorial control of transcription termination across the Saccharomyces cerevisiae genome. PLoS Genet 13: e1006863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, et al. 2008. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res 68: 7938–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Ma WK, Nguyen LT, Tran EJ. 2012. The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J Biol Chem 287: 26155–26166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Al Husini N, Dhoondia Z, Ansari A, Pascuzzi PE, Tran EJ. 2016. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol Cell 61: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken MB, Laribee RN, Corbett AH. 2015. Nab3 facilitates the function of the TRAMP complex in RNA processing via recruitment of Rrp6 independent of Nrd1. PLoS Genet 11: e1005044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MJ, Gao H, Smith-Kinnaman WR, Liu Y, Mosley AL. 2015. The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1-Nab3 pathway. PLoS Genet 11: e1004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace FV. 2013. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta 1829: 756–763. [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther UP, Weinberg DE, Zubradt MM, Tedeschi FA, Stawicki BN, Zagore LL, Brar GA, Licatalosi DD, Bartel DP, Weissman JS, et al. 2018. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature 559: 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. 2014. iCLIP: protein–RNA interactions at nucleotide resolution. Methods 65: 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iggo RD, Jamieson DJ, MacNeill SA, Southgate J, McPheat J, Lane DP. 1991. p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol Cell Biol 11: 1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamonnak N, Creamer TJ, Darby MM, Schaughency P, Wheelan SJ, Corden JL. 2011. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17: 2011–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Choe J, Seo YS. 1999. The sen1+ gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38: 14697–14710. [DOI] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. 2006. Distinct pathways for snoRNA and mRNA termination. Mol Cell 24: 723–734. [DOI] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Luscombe NM, Ule J. 2012. Protein–RNA interactions: new genomic technologies and perspectives. Nat Rev Genet 13: 77–83. [DOI] [PubMed] [Google Scholar]
- Kunkel GR, Maser RL, Calvet JP, Pederson T. 1986. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc Natl Acad Sci 83: 8575–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Tian L, Shen H, Gu Y, Li JL, Chen Z, Sun X, You MJ, Wu L. 2013. DDX5 is a positive regulator of oncogenic NOTCH1 signaling in T cell acute lymphoblastic leukemia. Oncogene 32: 4845–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WK, Cloutier SC, Tran EJ. 2013. The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly. J Mol Biol 425: 3824–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WK, Paudel BP, Xing Z, Sabath IG, Rueda D, Tran EJ. 2016. Recruitment, duplex unwinding and protein-mediated inhibition of the DEAD-box RNA helicase Dbp2 at actively transcribed chromatin. J Mol Biol 428: 1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tumasz S, Brow DA. 2015. Saccharomyces cerevisiae Sen1 helicase domain exhibits 5′- to 3′-helicase activity with a preference for translocation on DNA rather than RNA. J Biol Chem 290: 22880–22889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek A, Park Y, Miething C, Wilkinson JE, Gillis J, Lowe SW, Vakoc CR, Stillman B. 2014. Acquired dependence of acute myeloid leukemia on the DEAD-box RNA helicase DDX5. Cell Rep 7: 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gómez-González B, Grzechnik P, Rondón AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Wessels HM, Lebedeva S, Sajek M, Farazi T, Ghanbari M, Garzia A, Muntenau A, Spitzer J, Akat K, et al. 2018. Deciphering human ribonucleoprotein regulatory networks. bioRxiv 10.1101/295097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Morris AR, Battenhouse A, Iyer VR. 2014. Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res 42: 3736–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Decatur WA, Fournier MJ. 2008. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res 36: D178–D183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org/. [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team. 2015. RStudio: integrated development for R. RStudio Inc., Boston, MA. [Google Scholar]
- Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Shin S, Rossow KL, Grande JP, Janknecht R. 2007. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res 67: 7572–7578. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24: 735–746. [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. 2006. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol Cell Proteomics 5: 737–748. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC, Tollervey D. 2013. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. 1997. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res 25: 4778–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. 2008. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. 2016. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 30: 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortham NC, Ahamed E, Nicol SM, Thomas RS, Periyasamy M, Jiang J, Ochocka AM, Shousha S, Huson L, Bray SE, et al. 2009. The DEAD-box protein p72 regulates ERα-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERα-positive breast cancer. Oncogene 28: 4053–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.