Summary
Objectives: Electronic health records (EHRs) have increasingly emerged as a powerful source of clinical data that can be leveraged for reuse in research and in modular health apps that integrate into diverse health information technologies. A key challenge to these use cases is representing the knowledge contained within data from different EHR systems in a uniform fashion.
Method: We reviewed several recent studies covering the knowledge representation in the common data models for the Observational Medical Outcomes Partnership (OMOP) and its Observational Health Data Sciences and Informatics program, and the United States Patient Centered Outcomes Research Network (PCORNet). We also reviewed the Health Level 7 Fast Healthcare Interoperability Resource standard supporting app-like programs that can be used across multiple EHR and research systems.
Results: There has been a recent growth in high-impact efforts to support quality-assured and standardized clinical data sharing across different institutions and EHR systems. We focused on three major efforts as part of a larger landscape moving towards shareable, transportable, and computable clinical data.
Conclusion: The growth in approaches to developing common data models to support interoperable knowledge representation portends an increasing availability of high-quality clinical data in support of research. Building on these efforts will allow a future whereby significant portions of the populations in the world may be able to share their data for research.
Keywords: Knowledge representation, Common Data Model, interoperability, Electronic Health Records
Introduction
Electronic Health Records (EHRs) have emerged as a powerful tool to facilitate discoveries that can improve health. Challenges in data representation, quality control, and the derivation of clinical knowledge and phenotypes have been addressed in many successful studies. While initial efforts in this space were largely based within single institutions, more recent efforts in the United States (US) and internationally have focused on linking and federating data across multiple sites. Endeavors such as the Electronic Medical Records and Genomics (eMERGE) network and linked Informatics for Integrating Biology and the Bedside (i2b2) sites via the Shared Health Research Information Network (SHRINE) have shown the power of combining clinical data across institutions to make numerous discoveries that would not have been adequately powered at single sites [ 1 – 3 ]. A key challenge these efforts have is to represent the knowledge in EHR-based data in a uniform fashion. The eMERGE Network has addressed this challenge through queries of local bespoke data models with result sets mapped to standardized data dictionaries. The establishment of larger networks, such as the US Patient Centered Outcomes Research Network (PCORNet) [ 4 , 5 ] and the Health Care Systems Research Network [ 6 ], has advanced alternate models of early data harmonization.
Over the past two years, work in the domain of clinical knowledge representation has crystallized around a number of modern standards established to maximize the consistency and utility of clinical data. These efforts have largely focused on supporting data models that can facilitate exchange for clinical use and research, and interoperability standards that allow novel modular health apps to integrate into diverse health information technologies. Results of these efforts and early research evaluating the use of these novel models are beginning to appear in the biomedical literature. In this focused review, we will discuss some key studies covering high-impact examples of these topics. First, we will summarize several recent studies evaluating the latest versions of the international Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), initially designed to support drug surveillance and now adopted across numerous other research use cases. Second, we will summarize several studies describing the new PCORNet network and CDM; while these are primarily US-based, they still can inform international efforts. Together these data models have begun to support large federated research networks that allow consistent phenotype characterization across sites. Third, we will review and relate the emerging Health Level Seven, International (HL7) Fast Healthcare Interoperability Resource (FHIR) standard, which can leverage the standards used in these data models when performing specific tasks in dedicated app-like programs functioning across multiple EHRs and research systems. The relationship between CDMs and interoperability standards are shown in the Figure 1 .
Fig. 1 Representation of the PCORnet and OMOP Common Data Model (CDM) domains alongside the FHIR interoperability standard. CDMs are filled with clinical data derived from EHR systems via an extract, transform, and load (ETL) procedure. Clinical data domains are stored as tables within each of the CDMs, noted by the interior boxes. In the figure, the horizontal boxes presented in gray shading connect overlapping domain groupings between the models. Both CDMs also include data domains not represented here, such as cohort definitions. The FHIR standard allows transmission among EHRs, CDMs, Electronic Data Warehouses (EDWs), and data lakes.
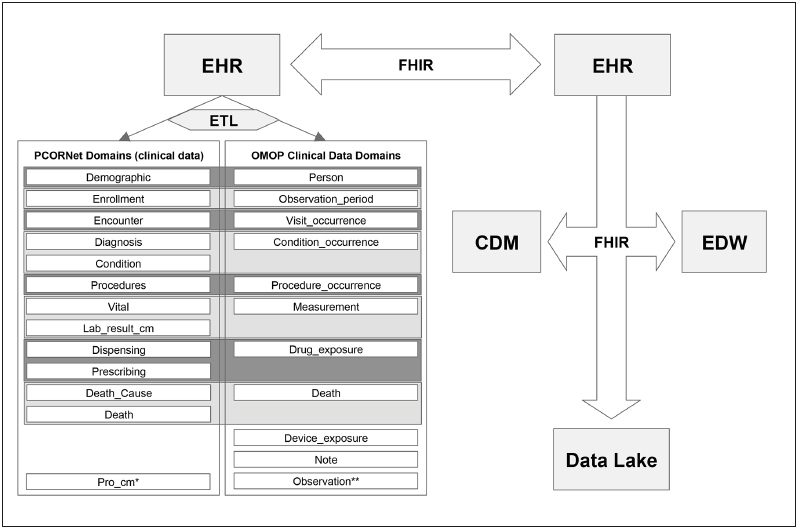
. * Patient-Reported Outcome Common Measures** The Observation domain allows storage of data not represented elsewhere in the CDM.
Scope of the Review
The intent of this review is to highlight emerging data models that we believe represent the future of representing clinical knowledge in raw EHR data. To identify references relevant for inclusion in this focused review, 1) we generally limited the search to references published within the past two years (i.e., early 2015 through early 2017); 2) we selected references identified in keyword searches using “Knowledge Representation”, “Data Model”, “Data Standard”, “FHIR”, “OMOP”, “OHDSI”, “CDM”, and “PCORNet” in PubMed and Google Scholar; and, 3) we added additional references known to the authors to be relevant. Because of the focus on newer versions of common data models, this report also limits discussion about those that are well established and discussed at length elsewhere, such as i2b2 [ 7 ]. This review does not recommend one data model to be generally adopted for all use cases. There are two primary reasons for this. First, a CDM’s success is intrinsically tied to the use case for which it is designed. As a result, its success in one knowledge domain does not necessarily indicate that it will be useful in another. Second, the choice of a CDM for a particular use case often involves considerations beyond pure knowledge representation, including the culture, existing technical infrastructural investments, and history of the organization adopting the data model. Until such a time that empiric evidence indicates that a particular CDM can be used generally across disparate use cases, any recommendation for one would be premature and would have the potential to disrupt other cultural considerations without a clear benefit in doing so.
Among the research informing this review, a recent 2016 study by Garza and colleagues supports our focus on the OMOP and PCORNet data models. In this study, investigators evaluated four CDMs for their content coverage, integrity, flexibility, ease of querying, standards compatibility, and ease and extent of implementation. The study evaluated how well these CDMs performed across these metrics when used to represent data stored in a registry of over 12,000 participants’ self-reported medical data, and corresponding and linked EHR data from numerous regional healthcare facilities. Among the CDMs studied, the OMOP model performed the best across all measures, and the PCORNet model performed reasonably in terms of coverage, and well for measures of flexibility, ease of querying, and implementation. These results are consistent with a 2013 study by Ogunyemi and colleagues, which found that OMOP performed best among CDMs evaluated for their coverage of comparative effectiveness research data types [ 8 ], and that FHIR, which has a flexible internal data model, represented the interoperability standard most likely to integrate seamlessly with clinical data repositories and operational clinical systems.
Data Model Architectures and Design Considerations
The informatics community is increasingly pursuing data management strategies intended to balance costs and accessibility. A recent approach, termed “data lakes”, creates large, un-transformed, electronic data storage within data warehouses [ 9 ]. Such data lakes allow inexpensive storage and ready access to broad data resources [ 10 ]. However, the non-standardized nature of data lakes means that repeated re-use of the data they contain has the potential to become infeasible if extensive data transformation has to recur frequently. Addressing this limitation, common data models can represent abstracts of the commonly used data in a structured format optimized for common use cases. In this way, common data models sit atop—and abstract knowledge from—data lakes, thereby providing standard structured formats. CDMs targeted in this review, including the PCORNet and OMOP CDMs, are designed as a hybrid entity-relationship (ER) [ 11 ] and entity-attribute-value (EAV) [ 12 ] database models, and are considered relational database models. These data models are highly flexible, and the use of the EAV model allows for efficient use of data storage for sparse data.
We note that the implementation of these models within traditional relational databases can incur processing limitations as the volume of data in each table grows into the multi-terabyte range or beyond. Addressing this, there is a growing use of other data representation solutions that scale into petabytes; these representations were originally developed for semantic data for the web, such as NoSQL and the Resource Description Framework (RDF). While these solutions were originally used when data are not highly relational, such as unstructured text data and graph databases, there are emerging solutions that create a relational database layer that sits on top of the NoSQL architecture that allow the common benefits of data scalability and processing speed along with the more mature SQL query language, transactional operations, and relational integrity. For this reason, we do not consider the implementation and use of these data models to be limited to only traditional relational database architectures. We also note that there is a need to efficiently move data between database and storage systems in a flexible but rigorous format that accounts for the richness of health care data. A popular recent example of this is FHIR, which is discussed below.
In addition, because of the large effort and cost of building the first CDM representation, there have been an increasing number of crosswalks and translation procedures across CDMs designed to allow data partners to participate in queries developed across multiple CDMs. For example, PCORNet community members have developed an i2b2 information model designed to map with PCORNet, allowing i2b2 users to have both models simultaneously operational [ 7 ]. Other efforts include cross-walks between the OMOP Version 4 and Version 5 CDM’s to both PCORNet CDM Version 2.0 and Version 3.x by members of the Clinical Data Research Network (CDRN) community that are also using the OMOP CDM.
Observational Medical Outcomes Partnership Data Model
The first common data model we review is the one developed for OMOP. Officially launched in 2008, OMOP started as a five-year public/private partnership seeking to create a framework for collaborative study in the growing set of EHR, federal, and commercial databases [ 13 , 14 ]. A primary goal for OMOP was to integrate data from multiple resources to improve surveillance for adverse events related to drugs by first overcoming major barriers related to the disparate data sources. Perhaps the most important product of this partnership was the development of the OMOP CDM. The CDM was designed to enable collaboration across multiple sites by unifying data structures and mapping data to common standardized vocabularies when possible. This is achieved by six gatherings of database tables: clinical data, health system data, health economics, derived elements, meta-data, and standardized vocabularies. The majority of tables are person-centric, with connections to the health system and vocabulary tables to provide further information. While CDMs generally lose some information when mapping from source representations, OMOP strives to mitigate this by preserving the original data representation as “source” values. For example, the standard vocabulary used for “conditions” is SNOMED-CT; to record historical ICD-9-CM billing code data related to conditions, codes would be translated to SNOMED-CT terms using the available map, but the original ICD-9-CM values would be retained as well. An important early study demonstrated that the CDM could successfully serve the purpose of drug safety surveillance by integrating data from across EHR datasets, commercial research databases, and datasets containing insurance and pharmacy administrative claims [ 13 ].
Building from the successful foundation of OMOP, a new collaborative was formed, with a first meeting in 2014: the Observational Health Data Sciences and Informatics (OHDSI) program [ 15 ]. The OHDSI program updated the OMOP CDM from version 4 (originally released in 2012) to version 5 (released in 2014), adding new tables for relationships among existing tables (e.g., family relationships for persons and relationships between observations), tables for full text data (e.g., clinical notes) and laboratory test results, expanded cost tables, and more. Additionally, it continued and expanded the work from OMOP including updating terminology mappings, supporting groups interested in research in observational health data, creating new techniques and tools to assist in the analysis of such data, and working together to study areas of interest. OHDSI provided a suite of open source analytic tools designed to operate on the OMOP CDM, with live demonstrations available at http://www.ohdsi.org/analytic-tools/ . The first released tool, the Automated Characterization of Health Information at Large-scale Longitudinal Evidence Systems (ACHILLES), presents users with descriptive statistics about the data stored in the CDM, both in R and as web-based HTML5 reports [ 16 ]. ACHILLES is commonly leveraged to study and compare data quality to help identify issues with consistency of data availability and representation among sites or databases.
In the past few years, a number of large studies have leveraged OHDSI to characterize and evaluate clinical cohorts and to study novel hypotheses. One of the most notable studies was conducted by Hripcsak and colleagues in 2016. They created an international distributed data network that complied with the OMOP CDM. The data network consisted of 11 data sources from four countries (Japan, South Korea, the United Kingdom, and the US), including EHR and administrative claims data on 250 million patients. This study characterized treatment pathways for three diseases, hypertension, type 2 diabetes, and depression, and identified differences in treatment patterns among and within countries (some of which were dramatic) [ 17 ]. This study would have been much more challenging, if not impossible, without a standardized data model and a set of nomenclatures, especially given the increased difficulty in sharing data across international borders. The group from Ajou University School of Medicine that participated in the above study has also described their work in converting from their local EHR system to the CDM [ 18 ]. One of the major obstacles encountered was the lack of an existing map from the Korean Standard Classification of Diseases version 5 (KCD-5), a modified version of ICD-10, to the CDM standard for conditions of SNOMED-CT. In addition, there was no perfect mapping of local drug codes, and no procedure codes, though overall coverage was strong. This example demonstrates a common challenge and strength of the OMOP CDM: while there was an initial barrier to implementation due to local data differences, overcoming this barrier facilitated the participation in a large scale international collaboration. Any such collaboration would have required similar mapping; the CDM provided the additional benefit of enabling other groups and data sets to map their data and participate as an OHDSI collaborator as well.
In another study, Boland and colleagues systematically explored in an OMOP-formatted database the relationship between season at birth and lifetime disease risk for 1,688 clinical conditions [ 19 ]. In this “Season-Wide Association Study” (SeaWAS), the investigators identified 55 diseases with incidence was closely associated with birth month, many of which being chronic diseases of adults. Among these, 16 had not been previously described in the biomedical literature, including atrial fibrillation, hypertension, and congestive heart failure. For example, the study found that adults with atrial fibrillation were more likely to have been born in the months January through July, and adults with chronic myocardial ischemia were more likely to have been born in March through June.
The OHDSI standard for observational health data also makes the integration of additional non-person level data sources more feasible by the utilization of existing terminologies. Any external database that provides information for concepts represented by standardized terminologies can be used freely, and mappings from these terminologies to other databases can be shared providing great benefit to the community. One example is an OHDSI project to integrate a drug knowledge base related to drug safety, described by Boyce and colleagues in a 2016 article [ 20 ]. This project targeted RxNorm, which is a standard vocabulary used for representing medication exposures [ 21 ]. The OMOP CDM includes mappings between RxNorm and a number of other medication data sources, such as First Databank, Anatomical Therapeutic Chemical (ATC) Classification System, and National Drug File - Reference Terminology (NDF-RT). By creating an additional mapping between RxNorm and DrugBank, this project made it possible to link drugs to chemicals, protein targets, genes, and disease associations. This integration provided a connection back to the observational health data in the CDM.
Another example is in the space of drug safety, where groups have integrated adverse drug reaction (ADR) data with observational data. In a 2015 study by Li and colleagues, the research team leveraged the adverse drug reports available in the FDA’s Adverse Event Reporting System (FAERS) to determine whether it was feasible to combine standardized information from EHR systems and other sources to identify true ADRs [ 22 ].The study observed that EHR data standardized to the OMOP CDM, when combined with other data sources, led to a significant improvement in the accuracy of detecting four clinically serious ADRs. In an other 2016 project by Voss and colleagues, a multinational team integrated a number of sources and adverse drug event databases for study [ 23 ]. These resources include United States and European product labels, the US FAERS database, and a data set created by processing the scientific literature. This project demonstrated that the research team could use existing resources and reference sets to predict a series of associations between drugs and health outcomes of interest. In this project, more data sources were associated with an improvement in prediction quality. This provided value as a companion resource that may improve the detection of adverse drug events by identifying negative and positive controls for use in large-scale studies.
From the start, a primary goal of OHDSI was to expand groups’ abilities to share tools and collaborate on projects. The initial OHDSI collaboration involved a number of public and privately funded organizations, including academic medical centers, government groups, insurance organizations, and pharmaceutical companies. One recent extension is the inclusion of data from skilled nursing facilities, where the minimum data set required for the US Centers Medicare and Medicaid data is a valuable commonly available target [ 24 ]. Other health data are becoming more available electronically as well. The number of groups implementing the OMOP CDM and OHDSI toolkit has been growing. This expansion makes it a more attractive option for projects in the observational health data space. Notably, the US Precision Medicine Initiative All of US 1 Research Program is adopting the OMOP CDM as a part of its data repositories. With the increasing number of opportunities for access to more open and available data sources, the impact of newly developed methods and tools that function with the OMOP CDM can be much greater over those developed with less standardized approaches.
The OMOP CDM’s primary limitations are related to the structure and content of the model. The defined structure has a set of data domains that receive particular attention, e.g., conditions, procedures, and medications. Currently, these domains generally cover common EHR domains, and there are planned expansions to continue to improve the model. The content limitation requires mapping to one of the defined standard vocabularies for each domain to enable the most features and interconnectivity, requiring either the overhead to map to these concepts or accepting the loss. The CDM preserves the native representations, which can mitigate the loss of information through these mappings. These limitations are generally necessary in multi-site collaborations, however, and the OMOP CDM permits flexibility where possible while still enabling consistency across data sets. Implementations of the CDM can include additional tables and fields as necessary to supplement with locally available information not represented. A study by Garza and colleagues found that the OMOP CDM best fits the criteria they established for longitudinal EHR-based registry studies [ 25 ]. These criteria were based on prior work and included a number of categories critical to CDM use: content coverage, integrity, flexibility, simplicity, integration, and implementability.
The Patient Centered Outcomes Research Network Data Model
The second common data model we review is the one developed to support a federated network of research networks across the US and launched by the US Patient-Centered Outcomes Research Institute (PCORI). Established by the US Congress through the Patient Protection and Affordable Care Act of 2010 [ 26 ], PCORI’s mandate is to conduct patient-centered comparative clinical effectiveness research, to provide evidence for patients and their families to make informed medical decisions, and to engage patients and families directly in the research enterprise. To fulfill this mandate, PCORI developed and funded PCORNet (The National Patient-Centered Clinical Research Network) in 2013 to conduct faster, easier, and less costly clinical effectiveness research in both observational cohorts and clinical trial frameworks. The PCORNet initiative funded two types of clinical research networks: Clinical Data Research Networks (CDRNs) and Patient-Powered Research Networks (PPRNs). While both types of research networks are intended to be patient-centered, CDRNs are focused around leveraging health care systems and EHR data, while PPRNs are focused around recruiting groups of engaged patient cohorts for sustained research [ 27 ]. In a subsequent phase of the funding awarded in 2015, PCORNet expanded to include additional CDRNs and PPRNs, as well as two health insurers. There are currently a total of 13 CDRNs and 20 PPRNs in operation across the US, and this model of a modular network can be extrapolated to other countries and international regions.
The core informatics infrastructure for PCORNet is made up of a CDM and an open-source workflow engine built on Pop-MedNet ( http://www.popmednet.org ), which allows federated query distribution and result retrieval. Among the 11 CDRNs funded in Phase 1, seven built their infrastructure on top of i2b2-based data repositories, and four on top of OMOP-based repositories. To harmonize data infrastructures and representation, the PCORNet Coordinating Center made the strategic decision to develop a new CDM that allows more control and adaptability for specific clinical trial and patient reported outcomes. Further, the PCORNet CDM provided a conceptual anchor for PCORNet. Because the initial PCORNet Coordinating Center was closely aligned with the US Mini-Sentinel program, it established the PCORNet CDM as an extension of the Mini-Sentinel CDM Version 4 [ 28 , 29 ]. The Mini-Sentinel is a program sponsored by the US Food and Drug Administration that uses standard EHR data to monitor the safety of medications and medical devices across the country. The PCORNet CDM has diverged from Mini-Sentinel based on data collection requirements within the PCORNet user community and the needs for PCORNet-approved research studies. In particular, the PCORNet CDM design is optimized for patient-centered comparative effectiveness research. For this reason, it contains some data domains that are relatively more highly specialized to target specific use cases when compared to other common CDMs. For example, the PCORNet CDM includes tobacco use and patient-reported outcomes among its data domains in support of specific research studies it is anticipated to cover.
The PCORNet model centers its schema on the patient entity, and enforces data mapping to controlled vocabularies, such as Current Procedural Terminology (CPT), SNOMED CT, Healthcare Common Procedure Coding System (HCPCS), the ICD versions 9 and 10, Logical Observation Identifiers Names and Codes (LOINC), and RxNorm. This enforced mapping is the core distributed data network innovation that allows efficient query and analysis execution across different instances of the data model. The PCORNet CDM has gone through a series of releases as it has evolved for broader use cases [ 30 , 31 ]. Versions 1.0 and 2.0 were released in 2014, and included data domains for patient demographics, coverage enrollment, outpatient medication dispensing, vital signs, conditions, procedures, inpatient and outpatient encounters, diagnoses, laboratory tests, and patient-recorded outcomes. In Version 3.0, released in 2015, the primary keys were standardized for a number of tables, and a number of additional data domains were added. These include timing of death, cause of death, medication prescribing (orders), trial meta-data, and data provenance meta-data domains. In the latest incremental update, the V3.1 in 2016, clarifications of ETL (Extract, Transform, Load) conventions were included for enrollment, death, and encounters, as well as added granularity to sexual and gender orientation based on the Institute of Medicine and PCORNet community feedback, the first CDM to do so.
Data quality and consistency across instantiations of the data model are critical issues for any community using a CDM. In 2013, PCORI commissioned a data inventory survey of initial CDRNs and PPRNs to get a sense of data variation, representations, and challenges likely to be faced. This survey applied the lessons learned from prior efforts with Mini-Sentinel, the Agency for Healthcare Research and Quality, and other networks, and the data inventory findings. To perform the survey, the PCORNet Coordinating Center and the PCORNet Data Committee developed and released a series of Data Quality Characterization (DQC) queries. The queries included increasingly sophisticated data summarization, tabulation, standardized vocabulary mapping assessment, and temporal trend analysis. Aggregated results from the sites were returned to the Coordinating Center to be reviewed and used to make recommendations for data model instance fixes and for providing basic preparatory to research data about patient demographics, enrollment, and administrative coding.
The first such large DQC query submitted data back to the coordinating center without embedded error reporting, and the results were reviewed by the Coordinating Center afterwards. However, this one-time review did not allow for iterative quality improvement at the site prior to submission, and based on community feedback, potential errors and problem reporting was built into the second phase queries to allow data partners to iteratively fix potential problems prior to result submission, improving efficiency. Another issue that was addressed was to determine the minimum dataset bin size for what constitutes de-identified aggregate data. For this, the PCORNet Data Committee reviewed institutional policies from 1) the US Department of Veterans Affairs, and 2) the US Center for Medicare and Medicaid Services and member networks, and made a recommendation to the PCORnet Coordinating Center and Governance board for a final minimum bin size of 10 for aggregate counts of patient characteristics for network approved studies, which was approved.
PCORNet has a number of high profile ongoing embedded research studies and a few completed efforts. Perhaps the most well-known is the ADAPTABLE trial, a pragmatic clinical trial of aspirin dosing optimal for the secondary prevention of cardiovascular events [ 32 ]. This trial is notable in addressing via a comparative effectiveness study an important patient-centered question where clear clinical guidelines do not exist and which would not reasonably ever be fundable as a randomized controlled trial. The ADAPTABLE trial should serve as a model for future research evaluating the comparative effectiveness of other highly used medications that are generic and lowcost. The trial is leveraging the PCORNet CDM data model to identify patients for recruitment and to ascertain outcomes. Two other large ongoing patient-centered PCORNet comparative effectiveness studies are focusing on healthy body weight. One is a large observational multi-CDRN study evaluating three methods for bariatric surgery to promote weight loss in morbidly obese patients [ 33 ]. The other is an observational study evaluating the relationship between antibiotic use and weight gain in later childhood [ 34 ].
There are a number of limitations to the PCORNet CDM. First, as is true of any common data model, the general set of use cases drives the data representation and thus optimizes its use for some activities but makes others more challenges. For this reason, no single CDM can serve all needs. PCORNet was developed to serve comparative effectiveness analyses, to support the collection and use of patient-reported outcomes and data, and to support pragmatic clinical trial data collection and use. The emphasis in the CDM is still on comparative effectiveness, with a focus on medications, laboratory data, and administrative codes, so other areas, such as medical devices, microbiology, and pathology data, are not currently represented. In addition, registry data, such as cancer registries, cardiac catheterization registries, and others, are not easily represented outside of the existing tables. Last, the data model lacks a general fact table that can be used for unusual data or irregular data, which is both a strength and a weakness. It prevents fragmentation and heterogeneity in the data being placed in the CDM, but also prevents users of the model to have a flexible place to put data they require for their use case that does not fit into the existing data model. In addition, the PCORNet CDM has not been used outside the US, so the degree to which it has an international utility is unclear. We also note that PCORI and PCORNet are transitioning management of this research network to the newly established People-Centered Research Foundation (PCRF, http://www.pcrfoundation.org ). At the time of this publication, the specific impact this transition will have on the PCORNet CDM is unknown.
The Health Level 7: Fast Healthcare Interoperability Resource Standard
Standards to support the interoperable exchange of clinical data have existed for some time, but have not been widely adopted. Among these, the Health Level Seven International (HL7) Reference Information Model (RIM) was developed iteratively from 1996 onwards to support HL7 V3 standards, including document-level and message-based exchange. A subset of the most widely used constraint on the RIM, the Consolidated Clinical Document Architecture (C-CDA), was cited in the US Meaningful Use Stage 2 and Stage 3 regulations, which set forth criteria for demonstrating that EHR technology can be used in ways that enhance patient health and engagement [ 35 ].The ability to produce C-CDAs containing elements from a core set of clinical data elements termed the “Common Clinical Data Set” [ 36 ] is now required for EHRs to be certified under the 2015 Edition of Certified Electronic Health Record Technology (CEHRT), which is a set of companion regulations to ensure that EHRs can meet Meaningful Use Stage 3 requirements [ 35 ]. Even so, it is widely accepted that these elements are necessary but not sufficient to represent all relevant clinical knowledge. Further, a 2014 study raised concerns over the accuracy and reproducibility of C-CDAs in identifying errors and permissible heterogeneity in C-CDA documents that can limit semantic interoperability [ 37 ].
Prior to and in anticipation of these concerns, HL7 authorized a “Fresh Look” Task Force in 2011. This Task Force’s work ultimately led to the Fast Healthcare Interoperability Resource (FHIR) standard [ 38 ]. A key characteristic of FHIR is that it was designed to support many of the popular components of prior HL7 standards, such as messaging and document-level exchange capabilities, but it also introduced new means of exchanging data based on RESTful web services (Representational State Transfer) and Application Programming Interfaces (APIs). This is important because RESTful web services are familiar to most software application developers, allowing for a better alignment of healthcare standards with modern technology and interfaces. FHIR has developed more rapidly than previous generations of HL7 standards, partly through an ambitious effort called the Argonaut Project [ 39 ]. The Argonaut Project, which evolved from recommendations put forth by the Joint HIT Standards and Policy Committee’s JASON Task Force Report, was chartered with the goal of rapidly developing a first-generation FHIR-based API “based on Internet standards and architectural patterns and styles” [ 39 ].
One of the most important recent developments regarding FHIR was the adoption of a FHIR-based API by the Substitutable Medical Apps Reusable Technologies (SMART) project. The SMART team had initially developed a custom API solution, which was changed to a FHIR-based API. This change, along with the implementation of modern authorization (OAuth2) and authentication (OpenID Connect) protocols, led to the creation of SMART on FHIR [ 40 ]. SMART on FHIR has become increasingly recognized as the preferred solution to enable app-based health information tools [ 41 , 42 ]; the SMART app gallery now has 46 apps, the majority of which are conformant to SMART on FHIR specifications ( https://gallery.smarthealthit.org/ ). Over the past two years, a number of studies have demonstrated SMART on FHIR being implemented in a variety of high-impact settings. For example, it has been implemented within the widely used i2b2 framework [ 43 , 44 ]. Further, Bloomfield and colleagues recently described implementing SMART on FHIR within Duke University’s production EHR [ 45 ]. In addition, recent studies presented efforts to create a SMART on FHIR API that integrates with the OpenMRS EHR [ 46 , 47 ]. Several of the largest EHR vendors have committed to the implementation of a patient-facing SMART on FHIR app called “Sync for Science” (S4S), which is intended to facilitate transmission of clinical EHR data to the All of Us Research Program [ 48 ]. With FHIR more widely supported within EHRs, one logical next step is to enable “on-demand” clinical decision support (CDS) from within a clinician’s workflow. An effort called CDS Hooks™ has created excitement within the SMART on FHIR community and has been the most popular track at FHIR Connectathons, which take place every 4 months, over the past year ( http://cds-hooks.org ).
The FHIR standard was conceived as a set of core resources that would encompass approximately 80% of clinical data elements (i.e., the “80/20 rule”) along with a system of extensions to easily and predictably capture the remaining 20%. As such, the initial iterations of FHIR focused on demographic, clinical, and transactional (e.g., financial and administrative) data. In 2015, Alterovitz and colleagues described extending the FHIR model to include the representation of genomic data [ 49 ]. Partially as a result of this work, the latest build of FHIR (STU3) has now a dedicated sequence resource for describing DNA and protein sequences, as well as enhanced specifications for genomic test reports and interpretations. Subsequently, Warner and colleagues described a SMART on FHIR app called SMART Precision Cancer Medicine that enables population-level queries for somatic mutation data within the context of individual patients [ 50 ]. The importance of this app was recognized in the 2016 US President’s Cancer Panel report [ 51 ].
There have been several additional noteworthy efforts involving the integration of FHIR into existing knowledge management or representation frameworks over the past two years. For example, existing knowledge representation paradigms used for clinical decision support, such as the Arden Syntax, can be conveyed as FHIR objects [ 52 , 53 ]. In addition, Khalilia and colleagues demonstrated that the Multipa-rameter Intelligent Monitoring in Intensive Care II (MIMIC-II) Intensive Care Unit database [ 54 ] can be transformed into an OMOP CDM and subsequently used in clinical predictive models using FHIR web services [ 55 ]. Several research teams have described different standards-based semantic metadata repositories that integrate FHIR data elements [ 55 – 57 ]. The DeepPhe cancer phenotype information model is based on a subset of FHIR resources that have been translated into OWL 2 Description Logic (DL) representation language [ 58 ]. This enables integration with extensive existing knowledge resource ontologies, such as the NCI Thesaurus.
The main challenge of adopting the FHIR standard is that it continues to evolve rapidly. Recognizing the usefulness of the C-CDA standard, efforts to enable the continued use of CDA and C-CDA within FHIR-based services are ongoing [ 59 ]. However, unlike other HL7 standards (e.g., V2 and V3), backwards compatibility in FHIR is not guaranteed. To address the natural tendency to hesitate to implement such a rapidly changing standard, HL7 recently introduced the FHIR Maturity Model (FMM) based on the Capability Maturity Model (CMM SM , Carnegie Mellon University) [ 60 ]. Note that once a Resource or Profile achieves FMM4, non-backwards compatible changes are strongly discouraged. As of FHIR DSTU2, no individual Resource has achieved FMM4, but several are at FMM3 (e.g., Observation, DiagnosticReport, Patient, ValueSet). It is anticipated that FHIR will continue to mature and to be widely adopted as a solution for knowledge representation tasks over the next few years. In particular, FHIR exists as a ready means to transport data to and from CDM repositories, and can act as a translation layer to bring data into a CDM from any of a variety of new sources.
Conclusion
Clinical data is increasingly being used to advance the goals of precision medicine. As we describe in this paper, there has been a recent growth in high-impact efforts to support quality-assured and standardized clinical data sharing across different institutions and EHR systems. This growth portends an increasing availability of high-quality clinical data in support of research. Here, we have focused on three major efforts as part of a larger landscape moving towards shareable, transportable, and computable clinical data. In addition to supporting drug surveillance and patient-centered comparative effectiveness research, such efforts will be critical for large programs such as the All of Us Research Program to be a success. Building on these efforts will allow a future whereby significant portions of populations in the world may be able to share their data for research.
Acknowledgements
VA HSR&D VINCI
PCORI CDRN-1501-26498
PCORI CDRN-1306-04819
NIH BCHI R01-HL-130828
Footnotes
Precision Medicine Initiative, PMI, All of Us, the All of Us logo, and The Future of Health Begins With You are service marks of the U.S. Department of Health and Human Services.
References
- 1.Weber GM, Murphy SN, McMurry AJ, Macfadden D, Nigrin DJ, Churchill S et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc. 2009;16:624–30. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;03:79re1.. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21:576–7. doi: 10.1136/amiajnl-2014-002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21:578–82. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogt TM, Lafata JE, Tolsma DD, Greene SM. The Role of Research in Integrated Health Care Systems: The HMO Research Network. Perm J. 2004;08:10–7. doi: 10.7812/tpp/04.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klann JG, Abend A, Raghavan VA, Mandl KD, Murphy SN. Data interchange using i2b2. J Am Med Inform Assoc. 2016;23:909–15. doi: 10.1093/jamia/ocv188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogunyemi OI, Meeker D, Kim H-E, Ashish N, Farzaneh S, Boxwala A. Identifying appropriate reference data models for comparative effectiveness research (CER) studies based on data from clinical information systems. Med Care. 2013;51:S45–52. doi: 10.1097/MLR.0b013e31829b1e0b. [DOI] [PubMed] [Google Scholar]
- 9.Jiang G, Evans J, Endle CM, Solbrig HR, Chute CG. Using Semantic Web technologies for the generation of domain-specific templates to support clinical study metadata standards. J Biomed Semant. 2016;07:10. doi: 10.1186/s13326-016-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Why Hasn’t Big Data Come to the Rescue in Clinical Data Unification? [Internet][cited 2017 Apr 7]. Available from:http://www.clinicalinformaticsnews.com/cln/comment/why-big-data-rescue-clinical-data.html
- 11.Chen PP-S. The Entity-relationship Model—Toward a Unified View of Data. ACM Trans Database Syst. 1976;01:9–36. [Google Scholar]
- 12.Nadkarni PM, Marenco L, Chen R, Skoufos E, Shepherd G, Miller P. Organization of Heterogeneous Scientific Data Using the EAV/CR Representation. J Am Med Inform Assoc. 1999;06:478–93. doi: 10.1136/jamia.1999.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19:54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FitzHenry F, Resnic FS, Robbins SL, Denton J, Nookala L, Meeker D et al. Creating a Common Data Model for Comparative Effectiveness with the Observational Medical Outcomes Partnership. Appl Clin Inform. 2015;06:536–47. doi: 10.4338/ACI-2014-12-CR-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 16.ACHILLES for data characterization [Internet] Obs. Health Data Sci. Inform 2017. Available from:http://www.ohdsi.org/analytic-tools/achilles-for-data-characterization/
- 17.Hripcsak G, Ryan PB, Duke JD, Shah NH, Park RW, Huser V et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci. 2016;113:7329–36. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon D, Ahn EK, Park MY, Cho SY, Ryan P, Schuemie MJ et al. Conversion and Data Quality Assessment of Electronic Health Record Data at a Korean Tertiary Teaching Hospital to a Common Data Model for Distributed Network Research. Healthc Inform Res. 2016;22:54–8. doi: 10.4258/hir.2016.22.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boland MR, Shahn Z, Madigan D, Hripcsak G, Tatonetti NP. Birth month affects lifetime disease risk: a phenome-wide method. J Am Med Inform Assoc. 2015;22:1042–53. doi: 10.1093/jamia/ocv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyce RD, Ryan PB, Norén GN, Schuemie MJ, Reich C, Duke J et al. Bridging Islands of Information to Establish an Integrated Knowledge Base of Drugs and Health Outcomes of Interest. Drug Saf. 2014;37:557–67. doi: 10.1007/s40264-014-0189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18:441–8. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Ryan PB, Wei Y, Friedman C. A Method to Combine Signals from Spontaneous Reporting Systems and Observational Healthcare Data to Detect Adverse Drug Reactions. Drug Saf. 2015;38:895–908. doi: 10.1007/s40264-015-0314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voss EA, Boyce RD, Ryan PB, van der Lei J, Rijnbeek PR, Schuemie MJ.Accuracy of an Automated Knowledge Base for Identifying Drug Adverse Reactions. J Biomed Inform [Internet][cited 2017 Jan 3]; Available from:http://www.sciencedirect.com/science/article/pii/S1532046416301794 [DOI] [PMC free article] [PubMed]
- 24.Boyce RD, Handler SM, Karp JF, Perera S, Reynolds CF.Preparing Nursing Home Data from Multiple Sites for Clinical Research – A Case Study Using Observational Health Data Sciences and Informatics. eGEMs [Internet] 2016[cited 2017 Jan 3];4. Available from:http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5108634/ [DOI] [PMC free article] [PubMed]
- 25.Garza M, Del Fiol G, Tenenbaum J, Walden A, Zozus MN. Evaluating common data models for use with a longitudinal community registry. J Biomed Inform. 2016;64:333–41. doi: 10.1016/j.jbi.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Compilation of Patient Protection and Affordable Care Act Extracted sections concerning Patient-Centered Outcomes Research and the Authorization of the Patient-Centered Outcomes Research Institute (PCORI) [Internet]. pcori.org 2017. Available from:http://www.pcori.org/sites/default/files/PCORI_Authorizing_Legislation.pdf
- 27.Selby JV, Beal AC, Frank L. The Patient-Centered Outcomes Research Institute (PCORI) national priorities for research and initial research agenda. JAMA. 2012;307:1583–4. doi: 10.1001/jama.2012.500. [DOI] [PubMed] [Google Scholar]
- 28.Curtis LH, Weiner MG, Boudreau DM, Cooper WO, Daniel GW, Nair VP et al. Design considerations, architecture, and use of the Mini-Sentinel distributed data system. Pharmacoepidemiol. Drug Saf. 2012;21 01:23–31. doi: 10.1002/pds.2336. [DOI] [PubMed] [Google Scholar]
- 29.FDA’s Sentinel Initiative [Internet] [cited 2017 Apr 7]. Available from:https://www.fda.gov/safety/fdassentinelinitiative/ucm2007250.htm
- 30.PCORnet Common Data Model 3.0 User Guide [Internet] 2017. Available from:http://pcornet.org/wp-content/uploads/2014/07/PCORnet_CDM_3_Lay_Guide_FINAL.pdf
- 31.PCORnet Community Rallies to Launch New Version of Common Data Model. pcori.org. 2017.
- 32.Johnston A, Jones WS, Hernandez AF. The ADAPTABLE Trial and Aspirin Dosing in Secondary Prevention for Patients with Coronary Artery Disease. Curr Cardiol Rep. 2016;18:81. doi: 10.1007/s11886-016-0749-2. [DOI] [PubMed] [Google Scholar]
- 33.PCORnet Bariatric Study (PBS) [Internet] ClinicalTrials.gov 2017. Available from:https://clinicaltrials.gov/ct2/show/NCT02741674
- 34.Short- and Long-Term Effects of Antibiotics on Childhood Growth (ABX) [Internet] ClinicalTrials.gov 2017. Available from:https://clinicaltrials.gov/ct2/show/NCT02744846
- 35.Blumenthal D, Tavenner M. The “Meaningful Use” Regulation for Electronic Health Records. N Engl J Med. 2010;363:501–4. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 36.Common Clinical Data Set [Internet] Off Natl Coord Health Inf Technol 2017. Available from:https://www.healthit.gov/sites/default/files/commonclinicaldataset_ml_11-4-15.pdf
- 37.D’Amore JD, Mandel JC, Kreda DA, Swain A, Koromia GA, Sundareswaran S et al. Are Meaningful Use Stage 2 certified EHRs ready for interoperability? Findings from the SMART C-CDA Collaborative. J Am Med Inform Assoc. 2014;21:1060–8. doi: 10.1136/amiajnl-2014-002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bender D, Sartipi K. HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. IEEE 26th International Symposium on Computer-Based Medical Systems (CBMS) 2013:326–31. [Google Scholar]
- 39.HL7 launches Argonaut Project to advance FHIR interoperability standard. Health Manag Technol. 2015;36:26. [PubMed] [Google Scholar]
- 40.Mandel JC, Kreda DA, Mandl KD, Kohane IS, Ramoni RB. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc. 2016;23:899–908. doi: 10.1093/jamia/ocv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandl KD, Mandel JC, Kohane IS. Driving Innovation in Health Systems through an Apps-Based Information Economy. Cell Syst. 2015;01:8–13. doi: 10.1016/j.cels.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandl KD, Kohane IS. Time for a Patient-Driven Health Information Economy? N Engl J Med. 2016;374:205–8. doi: 10.1056/NEJMp1512142. [DOI] [PubMed] [Google Scholar]
- 43.Pfiffner PB, Pinyol I, Natter MD, Mandl KD. C3-PRO: Connecting ResearchKit to the Health System Using i2b2 and FHIR. PloS One. 2016;11:e0152722.. doi: 10.1371/journal.pone.0152722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagholikar KB, Mandel JC, Klann JG, Wattanasin N, Mendis M, Chute CG SMART-on-FHIR implemented over i2b2. J Am Med Inform Assoc. 2016. [DOI] [PMC free article] [PubMed]
- 45.Bloomfield RA, Polo-Wood F, Mandel JC, Mandl KD. Opening the Duke electronic health record to apps: Implementing SMART on FHIR. Int J Med Inform. 2017;99:1–10. doi: 10.1016/j.ijmedinf.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Kasthurirathne SN, Mamlin B, Grieve G, Biondich P. Towards Standardized Patient Data Exchange: Integrating a FHIR Based API for the Open Medical Record System. Stud Health Technol Inform. 2015;216:932. [PubMed] [Google Scholar]
- 47.Kasthurirathne SN, Mamlin B, Kumara H, Grieve G, Biondich P. Enabling Better Interoperability for HealthCare: Lessons in Developing a Standards Based Application Programing Interface for Electronic Medical Record Systems. J Med Syst. 2015;39:182. doi: 10.1007/s10916-015-0356-6. [DOI] [PubMed] [Google Scholar]
- 48.PMI Cohort Program announces new name: the All of Us Research Program [Internet] 2016. Available from:https://www.nih.gov/allofus-research-program/pmi-cohort-program-announces-new-name-all-us-research-program
- 49.Alterovitz G, Warner J, Zhang P, Chen Y, Ullman-Cullere M, Kreda D et al. SMART on FHIR Genomics: facilitating standardized clinico-genomic apps. J Am Med Inform Assoc. 2015;22:1173–8. doi: 10.1093/jamia/ocv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warner JL, Rioth MJ, Mandl KD, Mandel JC, Kreda DA, Kohane IS et al. SMART precision cancer medicine: a FHIR-based app to provide genomic information at the point of care. J Am Med Inform Assoc. 2016;23:701–10. doi: 10.1093/jamia/ocw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rimer BK.Improving Cancer-Related Outcomes with Connected Health [Internet] 2016. Nov. Available from:https://deainfo.nci.nih.gov/Advisory/pcp/annualReports/2016/ConnHealth_FullRpt.pdf
- 52.Kimura E, Ishihara K. Internal domain-specific language based on Arden Syntax and FHIR. Stud Health Technol Inform. 2015;216:955. [PubMed] [Google Scholar]
- 53.Gaebel J, Cypko MA, Lemke HU. Accessing Patient Information for Probabilistic Patient Models Using Existing Standards. Stud Health Technol Inform. 2016;223:107–12. [PubMed] [Google Scholar]
- 54.Lee J, Scott DJ, Villarroel M, Clifford GD, Saeed M, Mark RG. Open-access MIMIC-II database for intensive care research. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:8315–8. doi: 10.1109/IEMBS.2011.6092050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khalilia M, Choi M, Henderson A, Iyengar S, Braunstein M, Sun J. Clinical Predictive Modeling Development and Deployment through FHIR Web Services. AMIA Annu Symp Proc. 2015;2015:717–26. [PMC free article] [PubMed] [Google Scholar]
- 56.Doods J, Neuhaus P, Dugas M. Converting ODM Metadata to FHIR Questionnaire Resources. Stud Health Technol Inform. 2016;228:456–60. [PubMed] [Google Scholar]
- 57.Ulrich H, Kock A-K, Duhm-Harbeck P, Habermann JK, Ingenerf J. Metadata Repository for Improved Data Sharing and Reuse Based on HL7 FHIR. Stud Health Technol Inform. 2016;228:162–6. [PubMed] [Google Scholar]
- 58.Hochheiser H, Castine M, Harris D, Savova G, Jacobson RS. An information model for computable cancer phenotypes. BMC Med Inform Decis Mak. 2016;16:121. doi: 10.1186/s12911-016-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rinner C, Duftschmid G. Bridging the Gap between HL7 CDA and HL7 FHIR: A JSON Based Mapping. Stud Health Technol Inform. 2016;223:100–6. [PubMed] [Google Scholar]
- 60.FHIR Maturity Model [Internet] HL7 FHIR 2017. Available from:http://hl7.org/fhir/resource.html#maturity


