Misfolded proteins in amyloid plaques in transgenic Alzheimer’s disease mouse brains are visualized directly without labeling.
Abstract
One of the key pathological features of Alzheimer’s disease (AD) is the existence of extracellular deposition of amyloid plaques formed with misfolded amyloid-β (Aβ). The conformational change of proteins leads to enriched contents of β sheets, resulting in remarkable changes of vibrational spectra, especially the spectral shifts of the amide I mode. Here, we applied stimulated Raman scattering (SRS) microscopy to image amyloid plaques in the brain tissue of an AD mouse model. We have demonstrated the capability of SRS microscopy as a rapid, label-free imaging modality to differentiate misfolded from normal proteins based on the blue shift (~10 cm−1) of amide I SRS spectra. Furthermore, SRS imaging of Aβ plaques was verified by antibody staining of frozen thin sections and fluorescence imaging of fresh tissues. Our method may provide a new approach for studies of AD pathology, as well as other neurodegenerative diseases associated with protein misfolding.
INTRODUCTION
The two major pathological hallmarks of Alzheimer’s disease (AD) are the deposition of amyloid-β (Aβ) plaques and the formation of neurofibrillary tangles (1). The initial event of the amyloid cascade hypothesis is the misfolding of Aβ peptides, followed by their aggregation to form insoluble plaques, leading to further neuronal loss and, ultimately, AD dementia (2–4). The misfolded polypeptides are usually rich in β sheet conformation and aggregate to form oligomers, fibrils, and senile plaques (SPs). Such a protein aggregation mechanism has been increasingly recognized to exist in a range of neural degenerative diseases, including AD, Huntington’s disease, Parkinson’s disease, prion disease, etc (5–8). However, the amyloid hypothesis has been long debated, indicating the lack of thorough understanding of the fate of Aβ peptides and their relationship with other parameters of the disease, such as the tau pathology (9–14). Novel imaging techniques that are sensitive to protein misfolding may benefit AD research and shed new light on the biology and pathology of protein conformational diseases.
Several imaging methods have been developed to detect Aβ in various conditions. Positron emission tomography imaging has been used to probe brain-wide Aβ contents with the aid of radiotracers (15, 16). Different staining methods have been readily established for histological studies of Aβ plaques. Congo red is a commonly used dye for staining misfolded proteins because of its strong binding affinity to β sheet conformation (17). Congo red is also commonly combined with polarization microscopy to study the linear optical birefringence of the plaques (18). A variety of antibodies have been developed for the immunohistochemical detection of Aβ in AD (19). Moreover, several dyes, including thioflavin S and methoxy XO4, have been used for two-photon imaging and longitudinal in vivo observations of the progressive aggregation of SPs (20–22). Despite these successes, the potential interference of the labeling molecules with protein misfolding and aggregation remains a concern and introduces complexities in the experimental design and data interpretation (17, 20). Label-free imaging methods would effectively avoid the issues induced by exogenous molecules and may provide new tools for AD imaging.
Until now, vibrational spectroscopies have been the most promising label-free methods to probe the misfolding of proteins (23–26). Infrared (IR) and Raman spectroscopy are capable of revealing the conformational changes of proteins via the spectral changes of the amide bands (24, 27–29). In particular, the amide I vibration of the polypeptide backbone has been found to be most sensitive to the secondary structural changes of proteins (23, 30–32). While IR spectra show a red shift of the amide I band upon fibril formation (24), Raman spectra demonstrate a blue shift of the same vibrational mode (29). Although IR absorption is advantageous in strong signal intensity due to optical resonance, it suffers from overwhelming water absorption in biological tissues and requires thin sectioning or the use of deuterated water. On the other hand, Raman spectroscopy is much more compatible with biological specimens, but its weak signal intensity has resulted in slow imaging speed and limited the direct applications in biomedical imaging. Various methods have been developed to enhance the Raman process, such as surface-enhanced Raman scattering (33), tip-enhanced Raman scattering (34), resonance Raman scattering (35), and coherent Raman scattering (CRS) spectroscopy and microscopy (36, 37). CRS microscopy takes advantage of the coherent nature of the nonlinear optical process with Raman signal enhanced by several orders of magnitude and hence is capable of performing rapid, label-free chemical imaging for various biomedical applications (38–40). Although there exist two major types of CRS microscopes, coherent anti-Stokes Raman scattering (CARS) microscopy and stimulated Raman scattering (SRS) microscopy, SRS is generally considered to be superior in maintaining undistorted Raman spectra and more convenient to conduct quantitative analysis (36, 41, 42).
In this study, we investigated the capability of SRS microscopy to rapidly image Aβ plaques in brain tissue from the APPswe:PS1dE9 (APP:PS1) mouse model of AD. We found that the amide I band of Aβ protein is blue shifted by ~10 cm−1 when forming fibrils and SPs, which could be effectively applied to distinguish native and misfolded proteins in AD brain tissues via multicolor SRS imaging.
RESULTS
SRS spectral characterization of Aβ
The optical setup and principle of SRS microscopy are illustrated in Fig. 1A and explained in detail in Materials and Methods. SRS is capable of selectively imaging different biomolecules based on their intrinsic Raman spectra, such as lipids, proteins, and nucleic acids (39, 42). Most previous studies were using Raman-active vibrational modes from various chemical bonds and groups, including the CH bonds of lipids and proteins, amide group of proteins, O–P–O group of nucleic acids, C=C bonds of unsaturated lipids, C≡C bonds of alkyne-tagged molecules, etc (43–45). Among these vibrational modes, amide I band is known to be highly sensitive to the secondary structural changes of proteins, that is, α helix to β sheet transitions. Therefore, SRS microscopy targeting the amide I region has the potential to image protein misfolding. We have synthesized Aβ fibrils with their morphologies verified by atomic force microscopy (AFM), as shown in Fig. 1B. Spontaneous Raman spectra of lipids, Aβ peptides, and misfolded fibrils are shown in Fig. 1C. As can be seen, the formation of β sheet–rich fibrils results in a narrowed and blue-shifted peak in the amide I band, with the peak position changed from 1658 to 1670 cm−1, which agrees with previous studies (27, 28, 31). In contrast, the CH stretch modes are insensitive to the secondary structural changes of proteins, showing almost identical spectra in the region (2800 to 3200 cm−1; fig. S1). We measured SRS spectra of the above three components with a spectral resolution of ~8 cm−1, and the spectral shapes were well preserved (Fig. 1D), which is one of the most important advantages of SRS compared with CARS microscopy. Because the spectral shift of amide I band is around 10 cm−1, it requires an SRS microscope with sufficient spectral resolution to differentiate the two Raman bands. At this point, picosecond narrow band laser–based SRS has the best spectral resolutions, while femtosecond laser–based methods usually have worse than resolutions of 20 cm−1 and hence may not be suited for these studies. These SRS spectra of different components will serve as the basis for quantitative numerical decomposition for the later spectral imaging data analysis based on linear algebra.
Fig. 1. Schematics of the experimental design.
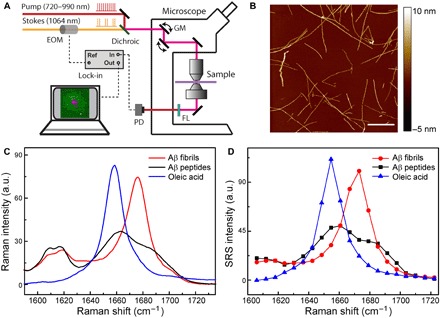
(A) Optical setup of the SRS microscope. (B) AFM image of incubated Aβ fibrils. (C) Spontaneous Raman spectra of lipid, Aβ in the forms of soluble peptides, and fibrils. (D) SRS spectra of the same species in (C). Scale bar, 1 μm. EOM, electro-optical modulator; PD, photo diode; FL, optical filter; GM, galvo mirror; a.u., arbitrary units.
Imaging amyloid plaques in thin frozen sections via SRS microscopy
We first evaluated the capability of SRS microscopy to image Aβ plaques in transgenic mouse brain tissues. Ten-micrometer-thick, snap-frozen sections were imaged with a hyperspectral SRS microscope that ranged from 1602 to 1725 cm−1. We selected three SRS images at the Raman frequencies of 1640, 1658, and 1670 cm−1 to extract the three components of lipids, normal proteins, and Aβ plaques. In images taken at 1640 cm−1, the general tissue architectures can be seen but appear relatively homogeneous, with minimum contrast of the amyloid plaque (Fig. 2A). In images taken at 1658 cm−1, the overall signal strength of normal tissues has reached the maximum, and the shape of the plaques starts to appear (Fig. 2B). In addition, in images taken at 1670 cm−1, while the intensities of normal tissues slightly decrease, the plaques reach their maximum brightness and show the highest contrast (Fig. 2C). This Raman frequency representing β sheet conformation agrees very well with previous spontaneous Raman measurements (26, 28, 31). To extract the contents of lipids, normal proteins, and amyloid plaques, we applied the linear decomposition algorithm that has been commonly used previously (39, 42, 46). The algorithm requires preknowledge of the SRS spectral intensities of each target chemical component, which has been measured on pure lipids, bovine serum albumin, and amyloid fibrils. A representative three-color image showing the distributions of the aforementioned components is shown in Fig. 2D, where the amyloid plaques could be visualized (cyan). In addition, protein-rich cellular structures (blue) and lipid-rich white matters (green) are also revealed, providing microscopic structural and chemical contrast of APP:PS1 transgenic mouse brain tissues. SRS spectra at marked locations are shown in Fig. 2E, verifying the spectral signatures of cells, white matters, and amyloid plaques in the fingerprint region.
Fig. 2. Multicolor SRS images acquired on frozen AD mouse brain section.
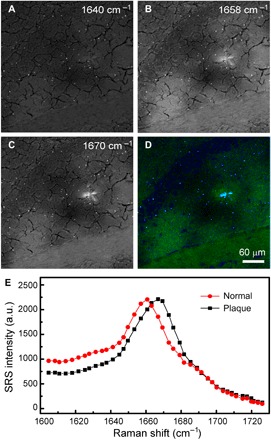
Individual SRS images at (A) 1640 cm−1, (B) 1658 cm−1, and (C) 1670 cm−1 and (D) the composite three-color image showing the distribution of lipid (green), normal protein (blue), and amyloid plaque (cyan). (E) SRS spectra of the plaque and normal tissue. Image size, 512 by 512 pixels (5 s per frame).
To further confirm that the plaques detected by SRS spectral imaging are amyloid plaques, we performed both antibody staining and SRS on the same frozen tissue sections for comparison. Since SRS microscopy is noninvasive, tissue sections were first imaged with SRS and then followed by staining with anti-Aβ antibody (ab2539, Abcam) to selectively label Aβ. Stitched large-area images of coronal brain sections from an APP:PS1 mouse are shown in Fig. 3A. Normal brain architectures are easily visualized in both images, including the cortex, white matters, hippocampus, and dentate gyrus. In regions where amyloid plaques are detected immunohistochemically, three-color SRS could also successfully image them with one-to-one correlation (Fig. 3B). This indicates that, by extracting fine spectral information, SRS microscopy could robustly probe misfolded proteins in amyloid plaques in frozen brain tissues. However, the amide I spectral signature is so sensitive to protein conformational change that extra care needs to be taken if the tissue processing will induce additional structural changes. For instance, we observed that tissue fixation might have disrupted protein conformations, resulting in the loss of SRS spectral signature of the amyloid plaques, although their macroscopic morphologies remain visible (fig. S2).
Fig. 3. Comparison between SRS and antibody staining results on the same tissue section.
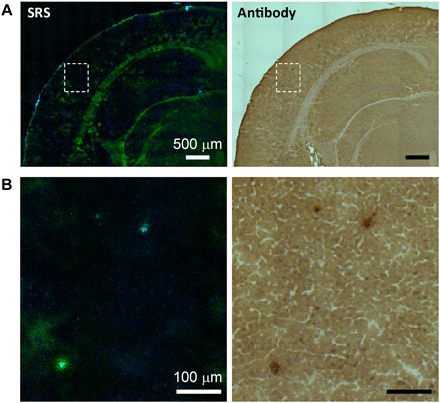
(A) Large-scale images and (B) zoom-in images showing three plaques.
SRS microscopy of amyloid plaques in fresh brain tissues
We next imaged fresh, unprocessed brain tissues from the same transgenic mouse model. Compared with frozen tissues, fresh specimens have the advantages of better-preserved tissue architectures, free of the artifacts caused by tissue processing such as freezing and sectioning artifacts that result in cracks and unevenness in SRS images (Figs. 2 and 3). Three-color SRS images of 1-mm-thick fresh brain sections are demonstrated in Fig. 4, showing the raw images taken at 1658, 1670, and 1680 cm−1 (Fig. 4, A to C). Similar to the results in frozen tissues, images at 1658 cm−1 have the maximum intensity of normal tissues; images at 1670 cm−1 show the maximum brightness of the plaques; and images taken at 1680 cm−1 demonstrate increased contrast of the surrounding cells, although the overall intensities have dropped to about 50%. The same numerical decomposition method was applied to decompose the raw images into the distributions of lipids (green), normal proteins (blue), and amyloid plaques (magenta). Major tissue histological features are visualized, including the normal tissues, cells, and dense SPs (Fig. 4D), with their SRS spectra plotted in Fig. 4E to prove their distinct spectral identities. One interesting phenomenon revealed by the three-color SRS is that each plaque is surrounded by a lipid-rich halo structure (fig. S3), which might originate from the degenerated neurites and myelin sheaths. However, the relationship between this halo structure and the amyloid plaque is not yet clear, nor is the role this structure plays in the development of disease; resolution of these questions awaits further investigation. More quantitative chemical analysis of the plaque, halo, and normal tissues can be found in fig. S4.
Fig. 4. Multicolor SRS images acquired on fresh AD mouse brain.
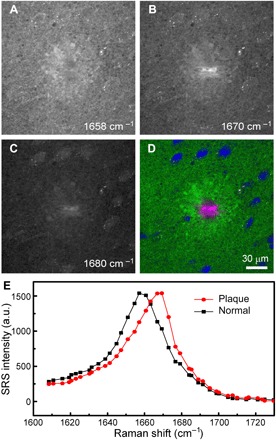
Individual SRS images of a 1-mm-thick fresh mouse brain section at (A) 1658 cm−1, (B) 1670 cm−1, and (C) 1680 cm−1 and (D) the composite three-color image showing the distribution of lipid (green), normal protein (blue), and amyloid plaque (magenta). (E) SRS spectra of the plaque and normal tissue. Image size, 512 by 512 pixels (5 s per frame).
We further compared the results of our label-free imaging technique with that of staining methods. Since antibody staining is difficult to perform on thick tissues, we applied fluorescence dye labeling with thioflavin S, a molecule that binds to β-pleated sheet conformation. After we imaged the fresh thick tissues with SRS, they were fixed and labeled with thioflavin S and then imaged with two-photon–excited fluorescence (TPEF) microscopy. Here, we mainly use a two-color SRS at 1658 and 1670 cm−1 to extract the content of amyloid plaques, while normal proteins with similar Raman peak frequency to lipids are included in the lipid channel. Both SRS and TPEF images of the same plaques are shown in Fig. 5, with near to perfect correlation between the two imaging modalities. Minor differences might be due to the changes induced by tissue processing and slightly different imaging depths. These results strongly support that SRS microscopy could indeed image misfolded amyloid proteins in fresh brain tissues without any exogenous labels.
Fig. 5. Comparison between the SRS and thioflavin S–labeled two-photon imaging results on the same tissue.
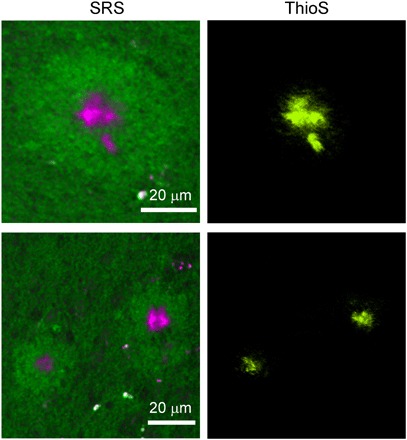
ThioS, thioflavin S.
Most SRS and CARS studies were conducted in the high-frequency CH stretch region (2800 to 3100 cm−1) because of the highest signal intensity acquired in this window. We also performed hyperspectral SRS microscopy in the high-frequency region on amyloid plaques in fresh APP:PS1 mouse brain tissues. Figure 6 shows the results of the same plaque that has been imaged in Fig. 4 in the fingerprint region, replicating major tissue structures. In the image taken at 2850 cm−1, lipid-rich normal brain and the halo structures are seen, while the dense plaque and cell nucleus appear dark (Fig. 6A). In contrast, the image taken at 2930 cm−1 shows a strong signal of both proteins and lipids; thus, both the plaque and the cells, as well as the overall tissue, appear bright (Fig. 6C). An additional image taken at 2880 cm−1 is also shown in Fig. 6B, where the dense plaque has slightly higher relative peak intensity at this frequency due to higher protein concentration. SRS spectra of the plaque, cell nucleus, and the halo are shown in Fig. 6E. Unlike the spectra in the fingerprint region, we observed no frequency shift between normal proteins and dense plaques in the high-frequency region, echoing with the spontaneous Raman measurements where normal and misfolded proteins appear roughly the same (fig. S1). Therefore, it is difficult to spectrally differentiate amyloid plaques and normal intracellular proteins in the CH window with simple linear decomposition algorithm (39, 42), even when using the three-channel images taken at the above Raman frequencies. Only two-color SRS images could be reconstructed with lipid (green) and protein (magenta) contrast, as shown in Fig. 6D, where the proteins in the plaques and cell nucleus are not well distinguished. Therefore, although CH stretch region gives higher signal intensity, it is not preferred to image protein conformational changes. Moreover, from the lipid/protein analysis (fig. S4), we could readily see that the surrounding halo structures are rich in lipids, which is likely due to the deposition of degenerated myelin after neuronal death.
Fig. 6. Two-color SRS images acquired on fresh AD mouse brain in the CH region.
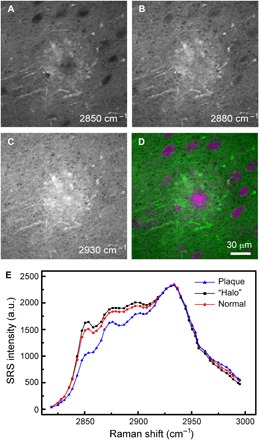
Individual SRS images at (A) 2850 cm−1, (B) 2880 cm−1, and (C) 2930 cm−1 and (D) the composite two-color image showing the distribution of lipids (green) and total protein (magenta). (E) SRS spectra of the plaque, surrounding halo and normal tissue. Image size, 512 by 512 pixels (1 s per frame).
Careful comparison between the images taken in the fingerprint and CH stretch regions reveals several differences between them. First, myelinated axons are visualized more clearly in the CH than in the fingerprint region. The Raman signal (2850 cm−1) is mainly contributed from the CH2 stretch modes of lipids, whereas the Raman band (1660 cm−1) is mainly from C=C bonds of unsaturated lipids and the amide I band of proteins. Since myelin lipids are mostly composed of cholesterol and cerebroside (47), the much higher number of CH bonds than C=C bonds leads to better contrast of myelin sheaths in the channel (2850 cm−1). Second, the morphologies of the amyloid plaques are not identical in the two spectral windows. This might be due to the mixed contents of normal and misfolded proteins detected in the CH stretch region, in contrast to the more β sheet–specific imaging of amyloid proteins in the fingerprint region.
DISCUSSIONS
Among all the specimens we measured, fresh tissues have the best SRS spectral and imaging qualities. A comparison between Figs. 2E and 4E indicates that the signal-to-background ratio I1670/I1610 increases from ~4 in frozen tissues to ~6 in fresh tissues. Relatively higher background of frozen tissues might come from higher cross-phase modulation of dry tissue sections without water immersion (see Materials and Methods). These spectral differences, although insignificant, have resulted in slightly different choices of spectral channels to optimize three-color images (Figs. 2 and 4). Our current work does not present in vivo imaging results, which is mainly because of the limited imaging speed (~5 s per frame) in the weak amide I region that would cause severe motion artifacts. Nevertheless, with improved laser parameters (such as the repetition rate and pulse duration) and SRS signal intensities, it is possible to achieve live animal imaging just as we have demonstrated in the CH stretch region (38–40).
Compared with labeling methods, SRS microscopy detects more histological changes induced by diseases, some of which may not be revealed by specific labeling. For instance, thioflavin S staining could efficiently label SPs, but it does not detect the halo structures surrounding the plaque cores. In contrast, SRS microscopy not only detects the Aβ plaques but also reveals the morphological changes of the nearby tissues (Fig. 5). Hence, SRS has the advantage of visualizing histological features, containing biomolecules that are insensitive to various staining methods, and may be able to discover new phenomena at the tissue level.
Spontaneous Raman spectroscopy is advantageous in acquiring spectra with much wider range and higher spectral resolution and hence provides better chemical resolution and specificity when combined with proper spectral analysis techniques (26–28). In contrast, current SRS microscopy is superior in rapid, high-quality imaging to resolve only a few chemical components based on more obvious spectral differences within a narrower range (39, 40). Although recent spontaneous Raman microscopy has shown promise in imaging amyloid plaques and neurofibrillary tangles, it took ~50 ms per pixel to acquire decent images (31). Whereas in our work, only 10 μs per pixel was needed to achieve better image contrast.
It is much more important but challenging to probe trace amount of Aβ in AD tissues because Aβ oligomers are believed to be the more toxic species and to play a key role in disease development. The limited sensitivity, spectroscopy, and large tissue backgrounds in current SRS systems have prevented the detection of small aggregates of Aβ fibrils. Therefore, improving the spectral quality of SRS microscopy by pushing forward the hyperspectral imaging capability might ultimately enable the detection of oligomers and diffuse plaques (46, 48–50), even holding promise for imaging of the hyperphosphorylated tau in neurofibrillary tangles based on their Raman signatures (31, 51).
CONCLUSIONS
In summary, we have applied multicolor SRS microscopy to detect amyloid plaques in brain tissue from a transgenic mouse model of AD. The spectral shift of the amide I band of β sheets allows us to differentiate misfolded Aβ from normal proteins and lipids. Thus, the three-color SRS in the fingerprint region could successfully image the histo-architectures of SPs, cells, and normal brain tissues in both frozen sections and fresh brain tissues. In contrast, Raman spectra in the high-frequency CH stretch region are insensitive to protein conformational changes and thus could not be easily applied for imaging plaques. Our work provides a new method to image amyloid plaques in a label-free manner and may open up new opportunities for studying AD and other protein misfolding diseases.
MATERIALS AND METHODS
SRS microscope
A detailed description of the SRS microscope can be found in previous works (36, 39, 43) and is illustrated in Fig. 1A. We used the Stokes beam from the fundamental output at 1064 nm and a tunable pump beam (650 to 1000 nm) from an optical parametric oscillator (picoEmerald, APE GmbH, Germany). The two pulse trains were spatially and temporally overlapped, delivered into a laser scanning microscope (FV300, Olympus), and focused onto the sample. The pump beam was modulated at a high frequency (10 MHz) using an electro-optical modulator. The small fraction of SRS signal over the large pump intensity (ΔI/I < 10−4) was detected by a home-built lock-in amplifier with a time constant of ~1 μs (38). A pump power of ~60 mW and a Stokes power of ~100 mW were used at the sample, after the microscope objective (numerical aperture, 1.2; UPLSAPO 60XWIR, Olympus). SRS images at the amide I region were taken at ~5 s per frame, with an image size of 512 by 512 pixels and a dwell time of 10 μs per pixel; a spectral imaging of 40 images took ~4 min. Same-sized images in the high-frequency CH stretch region were taken at ~1 s per frame with a dwell time of 2 μs per pixel. The lateral image resolution is ~400 nm, and the axial resolution is ~2 μm. The spectral resolution of our SRS system is ~8 cm−1.
Synthesis of Aβ fibrils
Aβ peptides were purchased from Chinese Peptide Ltd. (Hangzhou, China). Aβ peptides were first dissolved in phosphate-buffered saline, followed by incubation at 37°C for ~10 days to form mature fibrils.
Spontaneous Raman measurements
A home-built Raman spectrometer, including a monochromator (iHR320, Horiba), a charge-coupled device camera (Symphony, Horiba), and a microscope (IX71, Olympus), was used to measure all the spontaneous Raman scattering spectra. Aβ fibrils were condensed by evaporating the water in vacuum, and the residual thin film was used for spontaneous Raman measurements. To reduce autofluorescence, quartz slides and cover slips were used instead of normal glass.
Transgenic mouse model of AD
APP:PS1 double transgenic mice were purchased from the Jackson Laboratories. These mice overexpressed mutant human amyloid precursor protein gene containing the Swedish mutation K594N/M595L and the delta E9 mutant human presenilin 1 gene under the control of the prion promoter. Their AD neuropathology is well characterized, with plaque deposition starting at 5 months of age (52, 53).
Fresh tissue preparations
Fresh APP:PS1 mouse brains were taken from sacrificed mice, put in a brain slicer, and sliced to 1-mm-thick tissue sections with razor blades. Each fresh tissue section was placed in a half-inch hole in a glass slide and sealed in between two cover slips sandwiching the soft specimens, making homogenous contact between the tissue and the coverslips to ensure optimum imaging planes for SRS imaging.
Frozen sectioning and antibody staining
APP:PS1 mouse brains were snap frozen in liquid nitrogen after they were freshly taken from sacrificed mice. Thin frozen sections (~10 um) were prepared with a cryotome, dried in vaccum, covered with a coverslip, and imaged directly under an SRS microscope. Anti-Aβ antibody ab2539 was purchased from Abcam (Cambridge, MA) and used to stain the frozen sections after they were imaged with an SRS microscopy.
Dye labeling of thick tissues
Fresh sections from APP:PS1 mouse brain were fixed in 4% paraformaldehyde for 15 min. Amyloid plaques were stained with 0.025% thioflavin S in ethanol for 8 min. Sections were washed with water for 3 min and then mounted onto cover slides.
Two-photon imaging of labeled plaques
Two-photon fluorescence images were obtained using a Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss MicroImaging, Jena, Germany). Thioflavin S was excited at 750 nm with a mode-locked Ti-sapphire laser (Chameleon, Coherent Inc., Santa Clara, CA), and fluorescence emission was collected in the range of 535 to 590 nm.
Supplementary Material
Acknowledgments
We thank B. T. Hyman for stimulating discussions. We also thank D. Fu, F.-k. Lu, and W. Yang for technical supports. Funding: Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering (R01EB017254) to X.S.X.; NIH grants EB000768 and S10 RR025645 to B.J.B.; and the National Key R&D Program of China (2016YFC0102100), National Natural Science Foundation of China (81671725), and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01) to M.J. Author contributions: M.J., B.J.B., and X.S.X. conceived the project. M.J., L.Z., and M.A. performed the measurements and the data analysis. M.J. wrote the manuscript with inputs from B.J.B. and X.S.X. All authors read and approved the final manuscript. Competing Interests: X.S.X. and C.W.F. are shareholders of Invenio Imaging Inc., a company developing SRS microscopy systems. All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaat7715/DC1
Fig. S1. Spontaneous Raman spectra of Aβ in the CH stretching region.
Fig. S2. SRS images of a fixed AD brain section.
Fig. S3. More SRS images of SPs in fresh brain tissues.
Fig. S4. Lipid/protein analysis of the plaque and surrounding tissue.
REFERENCES AND NOTES
- 1.Jarvik L., Greenson H., About a peculiar disease of the cerebral cortex. By Alois Alzheimer, 1907 (Translated by L. Jarvik and H. Greenson). Alzheimer Dis. Assoc. Disord. 1, 3–8 (1987). [PubMed] [Google Scholar]
- 2.Selkoe D. J., Amyloid β protein precursor and the pathogenesis of Alzheimer’s disease. Cell 58, 611–612 (1989). [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D. J., Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 81, 741–766 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Eisenberg D., Jucker M., The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiti F., Dobson C. M., Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Prusiner S. B., Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 (1982). [DOI] [PubMed] [Google Scholar]
- 7.Ross C. A., Poirier M. A., Protein aggregation and neurodegenerative disease. Nat. Med. 10 Suppl, S10–S17 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Gregersen N., Bross P., Vang S., Christensen J. H., Protein misfolding and human disease. Annu. Rev. Genomics Hum. Genet. 7, 103–124 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Hardy J., Alzheimer’s disease: The amyloid cascade hypothesis: An update and reappraisal. J. Alzheimers Dis. 9, 151–153 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Reitz C., Alzheimer’s disease and the amyloid cascade hypothesis: A critical review. Int. J. Alzheimers Dis. 2012, 369808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Checler F., Turner A. J., Journal of Neurochemistry special issue on Alzheimer’s disease: ’Amyloid cascade hypothesis—20 years on’. J. Neurochem. 120 Suppl 1, iii–iv (2012). [DOI] [PubMed] [Google Scholar]
- 12.Tayeb H. O., Murray E. D., Price B. H., Tarazi F. I., Bapineuzumab and solanezumab for Alzheimer’s disease: Is the ’amyloid cascade hypothesis’ still alive? Expert. Opin. Biol. Ther. 13, 1075–1084 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Hardy J., Selkoe D. J., The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 297, 353–356 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Karran E., Mercken M., De Strooper B., The amyloid cascade hypothesis for Alzheimer’s disease: An appraisal for the development of therapeutics. Nat. Rev. Drug Discov. 10, 698–712 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Herholz K., Ebmeier K., Clinical amyloid imaging in Alzheimer’s disease. Lancet Neurol. 10, 667–670 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Villemagne V. L., Klunk W. E., Mathis C. A., Rowe C. C., Brooks D. J., Hyman B. T., Ikonomovic M. D., Ishii K., Jack C. R., Jagust W. J., Johnson K. A., Koeppe R. A., Lowe V. J., Masters C. L., Montine T. J., Morris J. C., Nordberg A., Petersen R. C., Reiman E. M., Selkoe D. J., Sperling R. A., Van Laere K., Weiner M. W., Drzezga A., Aβ imaging: Feasible, pertinent, and vital to progress in Alzheimer’s disease. Eur. J. Nucl. Med. Mol. Imaging 39, 209–219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frid P., Anisimov S. V., Popovic N., Congo red and protein aggregation in neurodegenerative diseases. Brain Res. Rev. 53, 135–160 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Jin L.-W., Claborn K. A., Kurimoto M., Geday M. A., Maezawa I., Sohraby F., Estrada M., Kaminksy W., Kahr B., Imaging linear birefringence and dichroism in cerebral amyloid pathologies. Proc. Natl. Acad. Sci. U.S.A. 100, 15294–15298 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barelli H., Lebeau A., Vizzavona J., Delaere P., Chevallier N., Drouot C., Marambaud P., Ancolio K., Buxbaum J. D., Khorkova O., Heroux J., Sahasrabudhe S., Martinez J., Warter J. M., Mohr M., Checler F., Characterization of new polyclonal antibodies specific for 40 and 42 amino acid-long amyloid β peptides: Their use to examine the cell biology of presenilins and the immunohistochemistry of sporadic Alzheimer’s disease and cerebral amyloid angiopathy cases. Mol. Med. 3, 695–707 (1997). [PMC free article] [PubMed] [Google Scholar]
- 20.Liebscher S., Meyer-Luehmann M., A peephole into the brain: Neuropathological features of Alzheimer’s disease revealed by in vivo two-photon imaging. Front. Psychiatry 3, 26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer-Luehmann M., Spires-Jones T. L., Prada C., Garcia-Alloza M., de Calignon A., Rozkalne A., Koenigsknecht-Talboo J., Holtzman D. M., Bacskai B. J., Hyman B. T., Rapid appearance and local toxicity of amyloid-β plaques in a mouse model of Alzheimer’s disease. Nature 451, 720–724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spires-Jones T. L., de Calignon A., Meyer-Luehmann M., Bacskai B. J., Hyman B. T., Monitoring protein aggregation and toxicity in Alzheimer’s disease mouse models using in vivo imaging. Methods 53, 201–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barth A., Zscherp C., What vibrations tell us about proteins. Q. Rev. Biophys. 35, 369–430 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Bouchard M., Zurdo J., Nettleton E. J., Dobson C. M., Robinson C. V., Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 9, 1960–1967 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams R. W., Estimation of protein secondary structure from the laser Raman amide I spectrum. J. Mol. Biol. 166, 581–603 (1983). [DOI] [PubMed] [Google Scholar]
- 26.Rivas-Arancibia S., Rodríguez-Martínez E., Badillo-Ramírez I., López-González U., Saniger J. M., Structural changes of amyloid beta in hippocampus of rats exposed to ozone: A Raman spectroscopy study. Front. Mol. Neurosci. 10, 137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong J., Atwood C. S., Anderson V. E., Siedlak S. L., Smith M. A., Perry G., Carey P. R., Metal binding and oxidation of amyloid-β within isolated senile plaque cores: Raman microscopic evidence. Biochemistry 42, 2768–2773 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Chen P., Shen A., Zhao W., Baek S.-J., Yuan H., Hu J., Raman signature from brain hippocampus could aid Alzheimer’s disease diagnosis. Appl. Opt. 48, 4743–4748 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Ortiz C., Zhang D., Ribbe A. E., Xie Y., Ben-Amotz D., Analysis of insulin amyloid fibrils by Raman spectroscopy. Biophys. Chem. 128, 150–155 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Maiti N. C., Apetri M. M., Zagorski M. G., Carey P. R., Anderson V. E., Raman spectroscopic characterization of secondary structure in natively unfolded proteins: α-Synuclein. J. Am. Chem. Soc. 126, 2399–2408 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Michael R., Lenferink A., Vrensen G. F. J. M., Gelpi E., Barraquer R. I., Otto C., Hyperspectral Raman imaging of neuritic plaques and neurofibrillary tangles in brain tissue from Alzheimer’s disease patients. Sci. Rep. 7, 15603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kast R. E., Auner G. W., Rosenblum M. L., Mikkelsen T., Yurgelevic S. M., Raghunathan A., Poisson L. M., Kalkanis S. N., Raman molecular imaging of brain frozen tissue sections. J. Neurooncol. 120, 55–62 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Beier H. T., Cowan C. B., Chou I.-H., Pallikal J., Henry J. E., Benford M. E., Jackson J. B., Good T. A., Coté G. L., Application of surface-enhanced Raman spectroscopy for detection of beta amyloid using nanoshells. Plasmonics 2, 55–64 (2007). [Google Scholar]
- 34.Deckert-Gaudig T., Kämmer E., Deckert V., Tracking of nanoscale structural variations on a single amyloid fibril with tip-enhanced Raman scattering. J. Biophotonics 5, 215–219 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Xu M., Shashilov V., Lednev I. K., Probing the cross-β core structure of amyloid fibrils by hydrogen–deuterium exchange deep ultraviolet resonance Raman spectroscopy. J. Am. Chem. Soc. 129, 11002–11003 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Freudiger C. W., Min W., Saar B. G., Lu S., Holtom G. R., He C., Tsai J. C., Kang J. X., Xie X. S., Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zumbusch A., Holtom G. R., Xie X. S., Three-dimensional vibrational imaging by coherent anti-Stokes Raman scattering. Phys. Rev. Lett. 82, 4142–4145 (1999). [Google Scholar]
- 38.Saar B. G., Freudiger C. W., Reichman J., Michael Stanley C., Holtom G. R., Xie X. S., Video-rate molecular imaging in vivo with stimulated Raman scattering. Science 330, 1368–1370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji M., Orringer D. A., Freudiger C. W., Ramkissoon S., Liu X., Lau D., Golby A. J., Norton I., Hayashi M., Agar N. Y. R., Young G. S., Spino C., Santagata S., Camelo-Piragua S., Ligon K. L., Sagher O., Xie X. S., Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci. Transl. Med. 5, 201ra119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He R., Xu Y., Zhang L., Ma S., Wang X., Ye D., Ji M., Dual-phase stimulated Raman scattering microscopy for real-time two-color imaging. Optica 4, 44–47 (2017). [Google Scholar]
- 41.Fu D., Lu F.-K., Zhang X., Freudiger C., Pernik D. R., Holtom G., Xie X. S., Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy. J. Am. Chem. Soc. 134, 3623–3626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu F.-K., Basu S., Igras V., Hoang M. P., Ji M., Fu D., Holtom G. R., Neel V. A., Freudiger C. W., Fisher D. E., Xie X. S., Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. U.S.A. 112, 11624–11629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji M., Lewis S., Camelo-Piragua S., Ramkissoon S. H., Snuderl M., Venneti S., Fisher-Hubbard A., Garrard M., Fu D., Wang A. C., Heth J. A., Maher C. O., Sanai N., Johnson T. D., Freudiger C. W., Sagher O., Xie X. S., Orringer D. A., Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci. Transl. Med. 7, 309ra163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei L., Hu F., Shen Y., Chen Z., Yu Y., Lin C.-C., Wang M. C., Min W., Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 11, 410–412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X., Roeffaers M. B., Basu S., Daniele J. R., Fu D., Freudiger C. W., Holtom G. R., Xie X. S., Label-free live-cell imaging of nucleic acids using stimulated Raman scattering microscopy. ChemPhysChem 13, 1054–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu F.-K., Ji M., Fu D., Ni X., Freudiger C. W., Holtom G., Xie X. S., Multicolor stimulated Raman scattering (SRS) microscopy. Mol. Phys. 110, 1927–1932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.G. J. Siegel, B. W. Agranoff, R. W. Albers, Basic Neurochemistry: Molecular, Cellular, and Medical Aspects (Lippincott-Raven, 1999). [Google Scholar]
- 48.He R., Liu Z., Xu Y., Huang W., Ma H., Ji M., Stimulated Raman scattering microscopy and spectroscopy with a rapid scanning optical delay line. Opt. Lett. 42, 659–662 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Liao C.-S., Slipchenko M. N., Wang P., Li J., Lee S.-Y., Oglesbee R. A., Cheng J.-X., Microsecond scale vibrational spectroscopic imaging by multiplex stimulated Raman scattering microscopy. Light Sci. Appl. 4, e265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozeki Y., Umemura W., Otsuka Y., Satoh S., Hashimoto H., Sumimura K., Nishizawa N., Fukui K., Itoh K., High-speed molecular spectral imaging of tissue with stimulated Raman scattering. Nat. Photonics 6, 845–851 (2012). [Google Scholar]
- 51.Ramachandran G., Milán-Garcés E. A., Udgaonkar J. B., Puranik M., Resonance Raman spectroscopic measurements delineate the structural changes that occur during tau fibril formation. Biochemistry 53, 6550–6565 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Alloza M., Robbins E. M., Zhang-Nunes S. X., Purcell S. M., Betensky R. A., Raju S., Prada C., Greenberg S. M., Bacskai B. J., Frosch M. P., Characterization of amyloid deposition in the APPswe/PS1dE9 mouse model of Alzheimer disease. Neurobiol. Dis. 24, 516–524 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Jankowsky J. L., Slunt H. H., Ratovitski T., Jenkins N. A., Copeland N. G., Borchelt D. R., Co-expression of multiple transgenes in mouse CNS: A comparison of strategies. Biomol. Eng. 17, 157–165 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/11/eaat7715/DC1
Fig. S1. Spontaneous Raman spectra of Aβ in the CH stretching region.
Fig. S2. SRS images of a fixed AD brain section.
Fig. S3. More SRS images of SPs in fresh brain tissues.
Fig. S4. Lipid/protein analysis of the plaque and surrounding tissue.


