Abstract
Although Tibetans and Sherpa present several physiological adjustments evolved to cope with selective pressures imposed by the high-altitude environment, especially hypobaric hypoxia, few selective sweeps at a limited number of hypoxia related genes were confirmed by multiple genomic studies. Nevertheless, variants at these loci were found to be associated only with downregulation of the erythropoietic cascade, which represents an indirect aspect of the considered adaptive phenotype. Accordingly, the genetic basis of Tibetan/Sherpa adaptive traits remains to be fully elucidated, in part due to limitations of selection scans implemented so far and mostly relying on the hard sweep model.
In order to overcome this issue, we used whole-genome sequence data and several selection statistics as input for gene network analyses aimed at testing for the occurrence of polygenic adaptation in these high-altitude Himalayan populations. Being able to detect also subtle genomic signatures ascribable to weak positive selection at multiple genes of the same functional subnetwork, this approach allowed us to infer adaptive evolution at loci individually showing small effect sizes, but belonging to highly interconnected biological pathways overall involved in angiogenetic processes.
Therefore, these findings pinpointed a series of selective events neglected so far, which likely contributed to the augmented tissue blood perfusion observed in Tibetans and Sherpa, thus uncovering the genetic determinants of a key biological mechanism that underlies their adaptation to high altitude.
Keywords: Himalayan human populations, hypobaric hypoxia, polygenic adaptive traits
Introduction
Himalayan populations living at altitudes higher than 3,500 m above sea level (a.s.l.), such as Tibetans and Sherpa, represent one of the most iconic examples of human adaptation to a highly challenging environment. Since the first human settlement on the Tibetan Plateau, their ancestors have been subjected to strong selective pressures imposed by cold temperatures, patchy landscape, arid soil, high UV radiation, and hypobaric hypoxia. Although these populations have progressively mitigated most of such stresses thanks to technological improvements, no cultural adaptations can allow them to avoid the reduction of oxygen partial pressure (and therefore uptake) as elevation increases.
In the last decade, the genetic basis of Tibetan and Sherpa physiological adaptation to high altitude has been investigated in many population genomics and genome-wide association studies (GWAS) (Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Yi et al. 2010; Xu et al. 2011; Jeong et al. 2014; Hu et al. 2017; Yang et al. 2017). Among the genes proposed to have adaptively evolved in response to reduced oxygen availability, EPAS1 and EGLN1 resulted the most replicated candidates. In particular, their variants putatively subjected to natural selection were proved to smooth the erythropoietic cascade, which represents the ancestral physiological response observed in populations evolved at low altitude when they are exposed to hypobaric hypoxia (Beall et al. 2010; Simonson et al. 2010; Yi et al. 2010; Buroker et al. 2012; Lorenzo et al. 2014; Peng 2017; Tashi et al. 2017). Accordingly, Tibetans and Sherpa maintain lower hemoglobin concentration with respect to lowlanders who reside for long periods at high altitude (Zhuang et al. 1996). This protects them against the long-term harmful effects of polycythemia due to physiological acclimatization to high altitude (Winslow and Monge Cassinelli 1987) and reduces their susceptibility to chronic mountain sickness (Vargas and Spielvogel 2006).
Nevertheless, such findings only partially explain the complex Tibetan/Sherpa adaptive phenotype. In fact, these populations present additional biological adjustments that enable them to tolerate low inspired oxygen pressure and to live even >4,000 m a.s.l. without experiencing severe harmful consequences (reviewed in Beall 2007; Gilbert-Kawai et al. 2014). These include modifications at different levels along the oxygen transport cascade when compared with what observed in native low-altitude individuals. Such changes range from cellular adjustments (e.g., reduced number of mitochondria in muscle cells) (Hoppeler et al. 2003) to modified physiological functions (e.g., increased resting pulmonary ventilation to favor oxygen absorption) (Beall et al. 1997; Moore 2001). Furthermore, Tibetans and Sherpa show an increased concentration of exhaled nitric oxide, which acts as vasodilator in lungs (Beall et al. 2001), and augmented blood flow (Erzurum et al. 2007) especially in the brain (Jansen et al. 2007) and in uterus and placenta during pregnancy (Moore et al. 2011; Vitzthum 2013). All these characteristics, coupled with an increased capillary distribution in muscles (Kayser et al. 1991), contribute to enhance blood perfusion in their tissues. However, the genetic determinants of these key modifications remain to be elucidated, plausibly due to conceptual and methodological limitations of selection scans implemented so far and based mostly on the hard sweep model (Scheinfeldt and Tishkoff 2013). In fact, in the last few years, polygenic adaptation has been increasingly proposed to have played a more substantial role than hard selective sweeps in recent human evolution (Pritchard and Di Rienzo 2010; Pritchard et al. 2010; Hernandez et al. 2011; Schrider and Kern 2017), as well as in high-altitude adaptation of Himalayan populations (Jeong and Di Rienzo 2014; Jeong et al. 2018). This implies that in most cases positive selection may have slightly affected many genes and variants involved in the same biological pathway, each of which individually exerting a limited effect on the overall adaptive phenotype. These weak selective events produce genomic patterns around the targeted loci that are intermediate between those of neutrally evolving chromosomal regions and those due to hard selective sweeps (Pritchard and Di Rienzo 2010), so that with traditional approaches it is particularly difficult to detect them (Jeong and Di Rienzo 2014). This explains why most of the neutrality tests developed so far turned out to be inadequate to draw inferences about polygenic adaptation, suggesting that hard sweeps at EPAS1 and EGLN1 represent only a limited fraction of the evolutionary events having shaped the Tibetan and Sherpa adaptive phenotype.
To test whether this peculiar phenotype has evolved under polygenic adaptation, we combined the computation of multiple selection statistics with gene network analyses able to account for the possibility that positive selection has acted at a functional pathway or even at specific gene subnetworks involved in a given biological function rather than on single loci (Gouy et al. 2017). For this purpose, we assembled a data set composed of a newly generated whole-genome sequence (WGS) of a Sherpa individual from the Nepalese Rolwaling Himal (SRH), who works as Himalayan Guide in mountaineering expeditions to 8,000 m peaks, as well as of publicly available WGS data for high-altitude Tibetan (TBN) and Sherpa (SHP) subjects from Tibet (Lu et al. 2016). This approach enabled us to identify previously undetected signatures of positive selection having acted on genes belonging to highly interconnected functional pathways with some loci showing individually small effect sizes, but as a whole contributing to modulate angiogenetic functions. We thus provided new evidence for polygenic adaptation plausibly underlying some of the well-known Tibetan and Sherpa adaptive traits evolved in response to hypobaric hypoxia.
Materials and Methods
Samples Collection and DNA Extraction
A Sherpa individual from the Rolwaling Himal (Gaurishankar Conservation Area, GCA, Dolakha District, Nepal), who was third-generation native of the Rolwaling Sherpa community (i.e., with both parents and grandparents born in the village of Beding at 3,690 m a.s.l.), was chosen among those recruited during several sampling campaigns (Gnecchi-Ruscone et al. 2017) organized in collaboration with the ExPlora Nunaat International nonprofit organization. This subject was selected to be representative of the Rolwaling Sherpa population, presenting 100% of the “Sherpa-like” genetic component observed in previous studies (Gnecchi-Ruscone et al. 2017) and a high-altitude adapted phenotype (i.e., he is a professional mountaineer and Himalayan Guide who have climbed Mount Everest several times). DNA was extracted from blood samples by means of a Salting Out modified protocol (Miller et al. 1988) and was quantified with the Quant-iT dsDNA Broad-Range Assay Kit (Invitrogen Life Technologies, Carlsbad, CA). A DNA sample with a concentration of 48 ng/μl was then used for preparation of the molecular libraries to be submitted to whole-genome sequencing.
Whole-Genome Sequencing and Variant Calling
The NEBNextUltraDNA Library Prep protocol was used to prepare genomic DNA libraries by implementing size selection for paired-end massive parallel sequencing that was performed with the HiSeq 4000 platform (Illumina, San Diego, CA) available at the facilities of the Human Genetics Department of the University of Chicago. The experiment was designed in order to obtain 150-bp pair-end reads and an average coverage of 20X. Base calling and quality controls were performed with RTA v1.18.64.0 and CASAVA v1.8.2 (Illumina, San Diego, CA), while the generated reads were mapped onto the human reference sequence (hg19) by means of the BWA-MEM algorithm implemented in the BWA v.0.7 tool (Li and Durbin 2010). We then used SAMtools v1.3 (Li et al. 2009) to sort and index the obtained raw alignments and Picard v1.98 (http://broadinstitute.github.io/picard/; Last accessed on March 6, 2018) to mark duplicate reads. Local realignment around insertions and deletions and base quality score recalibration were performed with GATK v3.5 (DePristo et al. 2011). We finally used SAMtools to remove low-quality reads pairs showing phred-scaled mapping quality scores (-q) < 30 and the PCR/optical duplicates previously marked with Picard. Such a pipeline was then applied separately on the whole-genome 150-bp pair-end reads generated for samples representative, respectively, of previously studied SHP and TBN populations (Lu et al. 2016), to assemble a homogenous high-altitude data set. This enabled us to reliably call genotypes across all nucleotide sites of our newly generated Sherpa genome sequence and those retrieved from literature by using the GATK UnifiedGenotyper algorithm and by considering only nucleotide sites showing phred-scaled quality scores ≥ 30.
Data Curation
We performed the following quality control (QC) steps on the called genotypes with PLINK v1.07 (Purcell et al. 2007). We retained single nucleotide variants (SNVs) showing genotyping success rate >99% and no deviations from the Hardy–Weinberg Equilibrium (P > 1.5×10−9 after Bonferroni correction for multiple testing). This led to the generation of a high-quality and “high-density” data set including 6,600,121 SNVs. We then created a “low-density” data set by merging WGS data with genome-wide genotyping data for a previously described panel of Asian populations (Gnecchi-Ruscone et al. 2017) (supplementary table 1, Supplementary Material online). Accordingly, it included 1,173 samples characterized for 199,679 single nucleotide polymorphisms (SNPs) and was used to check for consistency of the assembled sequence data with genotypes already available for the populations of interest, as well as to perform fine scale clustering analyses. Both the “low-density” and the “high-density” data sets were phased to infer haplotypes with SHAPEIT2 v2.r790 (Delaneau et al. 2013) by using default parameters and HapMap phase3 recombination maps. Given the relatively low number of samples contained in the “high-density” data set, WGS data generated by the 1000 Genomes Project (The 1000 Genomes Project Consortium 2015) were used as a reference panel of phased data to ensure more robust phasing. The overlap between the 1000 Genomes Project and the assembled “high-density” data sets was of 4,077,599 SNVs, which was the final number of genetic markers used for haplotypes phasing and related downstream analyses.
Genotype-Based Population Structure Analyses
Analyses based on genotype data and aimed at dissecting patterns of population genetic structure were performed on the “low-density” data set after pruning variants in linkage disequilibrium with each other (i.e., showing r2 > 0.2). We ran the ADMIXTURE clustering algorithm (Alexander et al. 2009) on the whole “low-density” data set to test K = 2 to K = 6 putative ancestral population groups. In particular, we ran fifty replicates with different random seeds for each K to monitor for convergence and we calculated cross validation (CV) errors for each K to assess which was the most reliable number of clusters explaining the data. Only the run with the highest log-likelihood and the K showing the lowest trend of CV-errors were considered to evaluate admixture proportions for the examined samples. Principal Components Analysis (PCA) was computed on the subset of East Asian and Tibeto-Burman populations included in the “low-density” data set by using the smartpca method implemented in the EIGENSOFT package v6.0.1 (Patterson et al. 2006).
Haplotype Sharing Clustering Analyses
To assess whether the considered Tibetan and Sherpa groups form a genetically homogenous population cluster at a fine scale of analysis, we applied the haplotype-based methods implemented in the CHROMOPAINTER/fineSTRUCTURE pipeline (Lawson et al. 2012). The phased “low-density” data set was used to run CHROMOPAINTERv2 to reconstruct patterns of haplotype sharing of each individual by using all the other individuals included in the data set as potential “donors,” but excluding themselves (i.e., preventing self-copy). To account for differences in sample sizes between genotyped populations and the WGS data set, we randomly selected a subset of individuals per group to be submitted to the analysis. Moreover, we restricted the analysis to East Asian populations by removing all Tibeto-Burman groups showing signatures of generic South Asian admixture according to previous studies (Basu et al. 2016; Gnecchi-Ruscone et al. 2017). We estimated the mutation/emission and recombination/switch rates using ten steps of the Expectation Maximization algorithm on a subset of chromosomes {4, 10, 15, 22}. The mean values calculated across all the autosomes and then across individuals, weighted by the number of markers, were used to run the final CHROMOPAINTER analysis on all the chromosomes by using k = 100 to specify the number of expected chunks to define a region. We then summed the matrix of the counts of shared haplotype chunks across the 22 autosomes, which was used as input for fineSTRUCTURE version fs2.1 (Lawson et al. 2012). We ran the algorithm with 1,000,000 “burn-in” iterations of MCMC, followed by another 2,000,000 iterations and sampling the inferred clustering patterns every 10,000 runs. We finally performed 1,000,000 additional hill-climbing steps to improve posterior probability and to merge the identified clusters in a step-wise fashion.
Selection Scans on Tibetan/Sherpa Whole-Genome Sequence Data
To identify genomic signatures ascribable to the action of positive selection, we first computed two independent and complementary statistics (i.e., the segregating sites by length, nSL, and the derived intra-allelic nucleotide diversity, DIND) on the phased “high-density” data set and we then used the obtained results as input for gene network analyses (see last section of Materials and Methods). We calculated nSL, which was designed to detect both hard and soft sweeps by searching for intrapopulation patterns of extended haplotype homozygosity (Ferrer-Admetlla et al. 2014), by retrieving information on ancestral alleles from the reconstructed Homo sapiens ancestral sequence based on the Ensembl Compara EPO 6 primates whole-genome alignments. nSL scores for each SNV were then computed with the algorithms implemented in selscan v1.1.0b (Szpiech and Hernandez 2014) by setting the maximum extension parameter to 4,500 SNVs, meaning that for each variant it was considered a window of maximum 4,500 consecutive loci to calculate the nSL value. In addition, we calculated DIND, which was designed to search for the longest identical stretch of consecutive alleles by counting the number of differences between any two haplotypes belonging to the same group (Fagny et al. 2014). For this reason, it focuses on the overall differences between haplotypes carrying a derived allele and, contrarily to nSL, is particularly suited to recognize hard selective sweeps. After removing variants showing derived allele frequency <0.2, which were shown to bias DIND results (Fagny et al. 2014), we calculated DIND scores for each SNV by using self-customized Python scripts.
Selection Scans on Validation Data Sets
To validate reliability of the presented pipeline of analyses on a larger data set, and to test whether signatures of positive selection observed in Tibetan and Sherpa genomes were likely ascribable to altitude-related selective pressures, we contrasted the obtained results with those based on the computation of nSL and DIND statistics on whole-genome sequence data available for the Han Chinese (CHB) samples (N = 103) included in the 1000 Genomes Project data set (The 1000 Genomes Project Consortium 2015). In fact, CHB are known to share an ancient genetic ancestry with Tibetans and Sherpa (Hu et al. 2017), but have evolved at low altitude. Moreover, genome-wide genotyping data previously generated for Sherpa and Tamang populations from the GCA (Gnecchi-Ruscone et al. 2017) were used as additional validation data sets to confirm genomic signatures of positive selection on a larger number of samples than those included in the “high-density” data set. These data were merged with the 1000 Genomes Project data set (The 1000 Genomes Project Consortium 2015), so that the final “selection SNP-chip” data set on which we calculated the Population Brach Statistics (PBS) (Yi et al. 2010) and the Cross-Population Extended Haplotype Homozygosity (XP-EHH) (Sabeti et al. 2007) consisted of 378,174 SNPs. We used a customized Python script to calculate PBS for measuring the amount of allele frequency changes occurred after the split of two closely related populations (i.e., the Sherpa or Tamangs and CHB) with respect to an outgroup population of European origins (CEU). The two-population XP-EHH test was instead computed with the algorithms implemented in selscan v1.1.0b and was used to detect high-frequency alleles associated to long-range haplotypes in the Sherpa or Tamang genomes, but not in CHB. For this purpose, we retrieved information on the ancestral state of each of the considered variants from the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/; Last accessed on April 18, 2018) and we phased haplotypes with SHAPEIT2 v2.r790 as described in the Data curation section. Since PBS and XP-EHH provide independent evidence of putative selected alleles having risen to or near fixation in the tested population, we combined the two statistics to reduce the false positives rate by calculating the Fisher Fcs combined score (Deschamps et al. 2016) as follows: Fcs = −2 [ln(PPBS) + ln(PXP-EHH)]. Where Px were the rank P values of the two statistics defined as their rank position divided by the total number of unique values present in the related genomic distribution (i.e., one for each SNP presenting a value for both the statistics).
Gene Network Analyses
To explicitly test for occurrence of selective events under a model of polygenic adaptation, which are hardly identifiable by considering each locus independently, we used the obtained genome-wide distributions of nSL, DIND, and Fcs scores to apply the pipeline for gene network analysis implemented in the signet R package (Gouy et al. 2017). In detail, we applied it separately on the lists of nSL and DIND scores calculated on the “high-density” data set, as well as on the two lists of Fcs scores (i.e., for Sherpa and Tamangs) obtained from the analysis of the “selection SNP-chip” data set. For each variant, we retrieved information of the gene/genes located within a range of 50 kb upstream and downstream their chromosomal position. Then, for each of them, we selected the highest score (for each statistics) among those computed for all the SNVs/SNPs associated to it as the score representative of the gene of interest. This score was used as input for the signet pipeline, which takes into consideration information available for annotated biological pathways to assign genes into their network context. In particular, we selected the National Cancer Institute Nature Pathway Interaction Database (Schaefer et al. 2009) to reconstruct the functional pathways our input genes belong to. We then assessed the distribution of scores within annotated pathways in order to test whether they were significantly shifted toward extreme values. The signet pipeline was developed to rely on an iterative process that we set to 10,000 iterations, in which the first step is represented by the assignment of a score to every subnetwork of genes belonging to a wider pathway. For this purpose, scores of each gene involved were combined and normalized to allow for comparison of subnetworks of different sizes. Then, for each gene network, a simulating annealing algorithm was used to identify the highest scoring subnetwork (HSS) (Gouy et al. 2017). Finally, to calculate P values to test whether the identified HSS were larger than what expected by chance, we generated null distributions of HSS for each subnetwork of a specific size. The gene scores belonging to a network were permuted to produce gene networks with random scores and we repeated this process multiple times to obtain the final null distribution. In detail, we set this iteration to 50,000 for Fcs scores and to 20,000 for nSL and DIND (for reasons of computational complexity). We then obtained rank P values for observed HSS by comparing them with the HSS generated by the null distribution. The significant subnetworks (P < 0.05) identified for each population and according to each selection statistics were plotted with Cytoscape v3.6.0 (Shannon et al. 2003).
Results
Assessing Representativeness of Tibetan/Sherpa Whole Genomes
After application of stringent base calling and QC procedures (see Materials and Methods), we obtained a “high-density” Tibetan/Sherpa data set made up of 12 individuals characterized for 6,600,121 SNVs. To frame them into the context of the overall Asian genomic landscape, we assembled a “low-density” data set of 199,679 SNPs by merging WGS data with genome-wide genotyping data from a previously described panel of low-altitude and high-altitude Asian populations (Gnecchi-Ruscone et al. 2017) (supplementary table 1, Supplementary Material online).
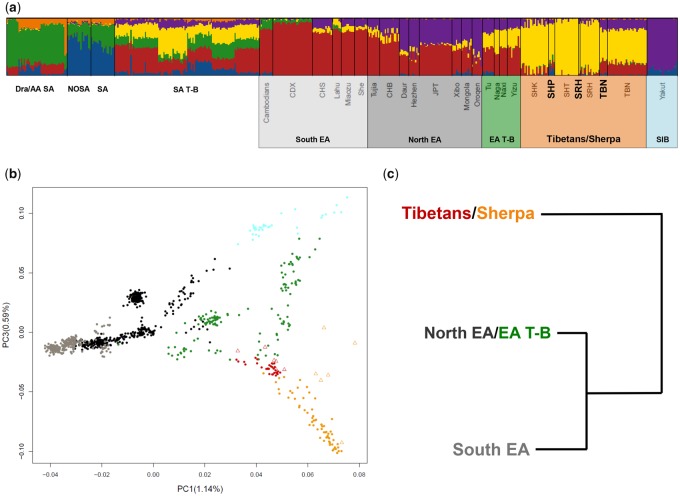
We used the “low-density” data set to perform ADMIXTURE analyses and the model showed the best predictive accuracy (i.e., the lowest CV error) when six population clusters (K = 6) were tested (supplementary fig. 1, Supplementary Material online). The observed admixture patterns pointed to a distribution of South Asian and East Asian genetic components among the considered populations that was in line with results obtained by other studies (Jeong et al. 2014, 2016; Lu et al. 2016; Gnecchi-Ruscone et al. 2017) (fig. 1A and supplementary fig. 2, Supplementary Material online). In particular, the TBN and SRH samples sequenced for the whole genome presented proportions of East Asian ancestry comparable to those inferred according to genome-wide genotyping data for their populations of origin. Instead, SHP showed higher degrees of South Asian ancestral components with respect to the other Sherpa groups, with especially two individuals presenting a cumulative percentage of Dravidian/Austro-Asiatic and Northern or Southern South Asian ancestry fractions exceeding 15% (fig. 1A).
Fig. 1.
—Population structure analyses performed on the “low-density” data set. (A) K = 6 ADMIXTURE analysis was performed on 1,173 individuals (for simplicity a subset of 520 individuals are plotted) belonging to the Asian groups included in the “low-density” data set. South Asian populations are labelled according to the following macrogroups: Dravidian and Austro-Asiatic speakers from South Asia, Dra/AA SA; Northern South Asians, NOSA; Southern South Asians, SA; South Asian Tibeto-Burmans, SA T-B. East Asian populations are reported according to their respective population label. High-altitude Tibetan/Sherpa samples sequenced for the whole genome are highlighted in bold. (B) PC1 versus PC3 performed on the East Asian and Tibeto-Burman populations included in the “low-density” data set. Samples typed with SNP-chip arrays are reported as full dots, while Tibetan/Sherpa samples sequenced for the whole genome are labelled as empty triangles. (C) fineSTRUCTRE hierarchical clustering analysis performed on a subset of East Asian populations included in the “low-density” data set. Only the upper splitting branches are reported. Shaded boxes under (A), color of points in (B), and cluster labels in (C) are color-coded according to the following macrogroups: South East Asians, light-gray; North East Asians, dark-gray/black, Siberians, light blue; Tibeto-Burmans (T-B), green; Tibetans, red; Sherpa, orange.
We then performed PCA on a subset of the East Asian and Tibeto-Burman groups included in the “low-density” data set. PC1 (accounting for 1.14% of variance) captured the main latitudinal cline of variation previously attested for East Asian populations (Li et al. 2008; HUGO Pan-Asian SNP Consortium et al. 2009) (fig. 1B). PC2 (depicting 0.66% of variance) highlighted patterns of South Asian admixture recently described for most Tibeto-Burman groups (Basu et al. 2016; Gnecchi-Ruscone et al. 2017), with the two SHP samples showing appreciable Dravidian/Austro-Asiatic and Northern or Southern South Asian ancestry components according to ADMIXTURE analyses occupying an intermediate position along this axis of variation (supplementary fig. 3, Supplementary Material online). PC3 (accounting for 0.59% of variance) instead described the genetic differentiation between Northern East Asians and high-altitude Himalayan populations (fig. 1B), thus confirming findings already pointed out by several studies (Jeong et al. 2014, 2016, 2017; Lu et al. 2016; Gnecchi-Ruscone et al. 2017; Zhang et al. 2017). WGS samples turned out to be encompassed within the range of variability shown by their respective ethnic groups, with the sole exception of the two outlier SHP individuals mentioned earlier. In detail, TBN clustered together with the other Tibetan samples from literature and the bulk of SHP subjects lay along the previously described Tibetans to Sherpa gradient of decreasing gene flow and increasing drift (Gnecchi-Ruscone et al. 2017; Jeong et al. 2017). The SRH sample was located at the end of the cline observed for the other Nepalese Sherpa from the Rolwaling Himal, Thame, and Khumjung (fig. 1B and supplementary fig. 3, Supplementary Material online).
Overall, results from ADMIXTURE and PCA suggested that the selected Tibetan and Sherpa individuals were highly representative of the gradient of genetic diversity observable for these Himalayan groups and enabled us to refine the WGS data set by filtering out two SHP subjects showing unusual Dravidian/Austro-Asiatic and Northern or Southern South Asian ancestry components.
Testing for a Homogeneous Tibetan/Sherpa Genetic Cluster
To deepen the dissection of the Tibetan/Sherpa cline of variation pointed out by ADMIXTURE and PCA, we obtained high-resolution estimation of haplotype sharing between these populations by applying the CHROMOPAINTER/fineSTRUCTURE pipeline (Lawson et al. 2012). For this purpose, we considered a representative subset of East Asian individuals included in the “low-density” data set by excluding Tibeto-Burman groups showing appreciable proportions of Dravidian/Austro-Asiatic and Northern or Southern South Asian ancestry components (see Materials and Methods). This enabled us to take into account the fact that a gradient of genetic variation could hide fine scale population structure, which is not identifiable with traditional genotype-based analyses (Leslie et al. 2015), and therefore to explicitly test whether the considered Tibetan and Sherpa groups form a genetically homogenous population cluster.
This approach led to the identification of 23 potential subclusters confirmed among the performed MCMC runs (supplementary fig. 4, Supplementary Material online) and that could be grouped into three main clades: South East Asians, North East Asians and Tibetans/Sherpa one (fig. 1C). This latter group was composed of seven subclusters, but with three of which being represented by a single individual. Moreover, the four remaining subclusters were composed of both Tibetans and Sherpa, and the WGS samples turned out to be distributed in all the Tibetan/Sherpa subclusters (supplementary fig. 5, Supplementary Material online). This further supported the representativeness of the assembled WGS data set as concerns the overall patterns of variation observable for high-altitude Himalayan groups. Moreover, although results from such a hierarchical clustering approach should not be interpreted as a strict description of relationships among populations, they suggested that clear genetic boundaries between the considered Tibetan and Sherpa groups cannot be identified based on the available data. Accordingly, we performed subsequent analyses by considering the upper fineSTRUCTURE clustering level, for which all Tibetans and Sherpa were grouped into a single clade clearly distinguishable from the rest of East Asian populations (fig. 1C).
Identifying Selective Events Mediating Tibetan/Sherpa Adaptation to High Altitude
Results from the above-mentioned population structure analyses were used to guide multiple selection scans, which were performed on the genomes of Tibetan/Sherpa samples showing low Dravidian/Austro-Asiatic and Northern or Southern South Asian ancestry components. In particular, we computed the nSL and DIND statistics on the phased “high-density” data set including 4,077,599 SNVs and we used the obtained scores as input to perform gene network analyses (see Materials and Methods).
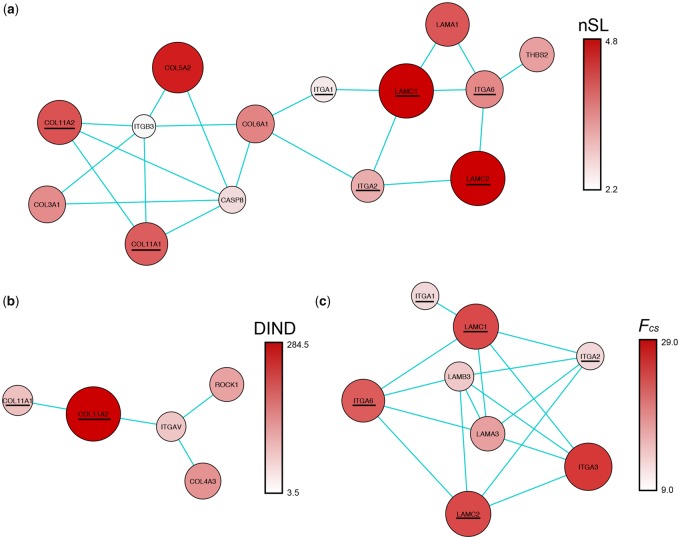
When considering results based on nSL scores (supplementary table 2 and fig. 6, Supplementary Material online), seven gene subnetworks were identified as significant, with especially three presenting P values <0.01 that belonged to nested integrin-associated pathways (i.e., Integrin β-1, Integrin α6-β4, and Integrin involved in angiogenesis), being highly overlapping and including a total of 14 genes (fig. 2A). Interestingly, ITGA6 encoding for integrin α6-β4, which was part of two of the integrin subnetworks identified, is an important receptor on platelets and is proved to play a role in angiogenesis (Avraamides et al. 2008). Other two significant subnetworks tightly linked with each other belonged to the C-MYB and C-MYC transcription factor pathways and were made up of 32 genes. These oncogene transcription factors are reported to be involved in several functions, mostly promoting the proliferation/differentiation of hematopoietic progenitor cells and sprouting angiogenesis (Bateman et al. 2017) (supplementary results, Supplementary Material online). As regards the two less significant subnetworks, one belonged to the Stabilization and expansion of the E-cadherin adherents junction pathway, being made up of six genes, and the other to the P53 pathway composed of 14 genes (supplementary results, Supplementary Material online).
Fig. 2.
—Angiogenesis-related integrin subnetworks identified as significant in Tibetans and Sherpa by the different tests performed. (A) Nested integrin subnetworks belonging to the Integrin involved in angiogenesis, β-1 integrin, and α6-β4 integrin pathways and pinpointed by nSL-based analysis performed on the “high-density” data set. (B) Nested integrin subnetworks belonging to the Integrin involved in angiogenesis and β-1 integrin pathways and pinpointed by DIND-based analysis performed on the “high-density” data set. (C) Nested integrin subnetworks belonging to the β-1 integrin and α6-β4 integrin pathways and pinpointed by replication Fcs-based analysis performed on the “selection SNP-chip” data set. The genes present in more than one of the described significant subnetworks are underlined.
Gene network analysis performed according to the computed DIND scores led to the identification of eight significant subnetworks (supplementary table 3 and fig. 7, Supplementary Material online). Among them, the Integrin β-1 and Integrin in angiogenesis pathways were represented by five genes, with COL11A1 and COL11A2 being pointed out even by nSL-based computations (fig. 2B). Furthermore, a subnetwork belonging to the CDC42 signaling cascade and made up of five genes was highlighted, including loci that encode for a GTPase complex that binds to different effectors involved in many cellular functions (Bateman et al. 2017). Another significant subnetwork was represented by three genes and belonged to the Nephrin/Neph1 signaling pathway, which plays a role in controlling the glomerular permeability (Liu et al. 2003). Three subnetworks then turned out to be composed by a few genes each (i.e., two or three), but they were all linked by ESR1 that encodes for an estrogen receptor. Interestingly, isoform 3 of the ESR1 protein is known to contribute to the activation of NOS3 and to endothelial production of nitric oxide (Bateman et al. 2017), a vasodilator showing increased concentration in the lungs of high-altitude Himalayan people (Beall et al. 2001) (supplementary results, Supplementary Material online). Finally, genes from a subnetwork belonging to the wide P73 pathway were detected as significantly enriched among the candidate targets of positive selection pinpointed by the DIND test (supplementary results, Supplementary Material online).
When applied to the low-altitude CHB population, gene network analysis based on results from the nSL test identified four significant subnetworks (supplementary results and supplementary table 4, Supplementary Material online), with especially those belonging to the Stabilization and expansion of the E-cadherin adherents junction and P73 pathways being in common with the significant subnetworks described earlier for Tibetans and Sherpa. Moreover, four macrosubnetworks were found to be significant according to gene network analysis based on DIND scores (supplementary results and supplementary table 5, Supplementary Material online), including the C-MYC transcription factor and CDC42 signaling cascade pathways previously pointed out for Tibetans and Sherpa.
To further validate results from gene network analyses, the same network enrichment approach was replicated on larger data sets consisting of results from independent neutrality statistics calculated on previously generated genome-wide genotyping data (Gnecchi-Ruscone et al. 2017). For this purpose, we used a “selection SNP-chip” data set including 378,174 SNPs (see Materials and Methods) to compute PBS and XP-EHH statistics separately on high-altitude Sherpa and medium-altitude Tamang populations from the Nepalese GCA. PBS and XP-EHH results for each group were then combined into Fcs scores before their submission to gene network analysis (see Materials and Methods).
Results for the Sherpa group (supplementary table 6 and fig. 8, Supplementary Material online) were in line with those previously obtained by several single-gene approaches (Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Yi et al. 2010; Xu et al. 2011; Hu et al. 2017; Yang et al. 2017). In fact, they pointed to the identification of significant subnetworks belonging to the HIF-1α and HIF-2α pathways and containing, respectively, the EGLN1 and EPAS1 genes among those highlighted by the highest scoring subnetwork algorithm. These two subnetworks partially overlapped and were composed of 18 genes (supplementary results, Supplementary Material online). Among the remaining significant subnetworks, three were associated to the pathway of the P53 protein family of transcription factors and tumor suppressors (i.e., P53, P63, and P73 subnetworks, for a total of 28 genes), thus confirming findings from WGS-based selection scans (supplementary results, Supplementary Material online). Other subnetworks significant for the Sherpa population were those belonging to the Integrin β-1 and Integrin α6-β4 pathways (fig. 2C). They included eight genes encoding for integrin subunits, ligands, and effectors, five of which (i.e., LAMC1, LAMC2, ITGA6, ITGA1, and ITGA2) were already pointed out by the nSL-based analyses described earlier. Finally, other Sherpa significant subnetworks included the Glucocorticoid receptor regulatory network, for a total of nine genes, and the Interleukin-1 signaling events made up of seven genes.
As regards results obtained for the Tamangs, who are not expected to have evolved adaptation to hypobaric hypoxia (Gnecchi-Ruscone et al. 2017), 11 gene subnetworks belonging to different biological pathways turned out to be significant (supplementary table 7, Supplementary Material online). In detail, three overlapping subnetworks were involved in immune activities (i.e., TCR signaling in T cells CD4+ and CD8+ and Signaling mediated by PTP1B) (Cho et al. 2013; Yang et al. 2016), while other two belonged to two P53 protein family pathways (i.e., P53 effectors and P73 network) (supplementary results, Supplementary Material online). Moreover, two additional significant subnetworks were represented by the ATF-2 transcription factor network, which is involved in the development of nervous system and the skeleton (Reimold et al. 1996), and by the Netrin-mediated signaling events that regulate axon guidance (O’Donnell et al. 2009). Finally, the RET receptor tyrosine kinase signaling events involved in several human syndromes, such as the Hirschprung’s disease, multiple endocrine neoplasia and familial thyroid carcinoma (van Weering and Bos 1998), were found to be significant in addition to the subnetwork of Vascular endothelial growth factor receptors (Abhinand et al. 2016). According to these findings, with the exclusion of those belonging to the P53 pathways (supplementary results, Supplementary Material online), the considered Sherpa and Tamang populations did not present any common subnetwork among those pointed out as significant.
Discussion
To date, traditional genome-wide selection scans and genotype–phenotype GWASs failed to replicate other genes than EPAS1 and EGLN1 as strong genetic determinants of Tibetan/Sherpa high-altitude adaptation, plausibly due to the multifaceted and polygenic nature of such an adaptive phenotype. Therefore, we aimed at explicitly testing for mechanisms of polygenic adaptation evolved by these populations in response to hypobaric hypoxia by searching for selective events simultaneously occurred at multiple loci with moderate to small effect size and involved in the same biological pathway (Pritchard and Di Rienzo 2010). In detail, we performed a series of selection scans culminating in gene network analyses that unlike previous pathway-based approaches have been designed to identify outlier subnetworks of genes involved in a wider functional pathway (Gouy et al. 2017). Accordingly, we specifically accounted for the fact that under a polygenic adaptation model positive selection likely acts on a subset of loci involved in a given biological function rather than on the entire set of genes belonging to that pathway (Gouy et al. 2017).
For this purpose, we used a panel of Tibetan/Sherpa samples sequenced for the whole genome and that genotype-based and haplotype-based population structure analyses demonstrated to be well representative of the gradient of genetic variation observable for these Himalayan groups (fig. 1). In particular, we computed nSL and DIND statistics that, with respect to traditional methods, were proved, respectively, more powerful to search for soft selective sweeps (Ferrer-Admetlla et al. 2014) and more robust to demography and variation in sequencing coverage when applied to population samples of small size (Fagny et al. 2014), as is the case in the present study. Furthermore, we validated the obtained results by taking advantage of a larger Sherpa data set (Gnecchi-Ruscone et al. 2017) (supplementary table 1, Supplementary Material online) and by performing the same gene-network analysis, but based on different selection statistics (i.e., the Fisher Fcs score calculated as combination of PBS and XP-EHH). In addition, in such a validation process, we considered as negative control groups two low-altitude populations of East Asian ancestry, such as the Han Chinese samples sequenced for the whole genome in the 1000 Genome Project and a Tamang population from the Nepalese GCA. In fact, the ancestors of Han Chinese and Tibetans are supposed to have diverged as early as 44 ka, having then maintained appreciable gene flow until ∼9 ka (Hu et al. 2017). Moreover, despite showing some genetic affinity with the Sherpa, Tamangs have been recently proposed to have originated from a low-altitude branch of the ancestral Tibeto-Burmans instead that from the Tibetan/Sherpa lineage (Gnecchi-Ruscone et al. 2017). Therefore, both Han Chinese and Tamang ethnic groups share an ancient common origin with Tibetans and Sherpa, but they have spent most of their evolutionary histories at low altitude so that they have not experienced hypoxia-related selective pressures.
According to this approach, we pinpointed some gene subnetworks belonging to highly interconnected functional pathways and that most of the performed tests (i.e., based on both WGS and validation data sets) suggested to have been pervasively subjected to positive selection only in the Tibetan and Sherpa populations. In fact, when excluding those related to the macro P-53 protein family of transcription factors (supplementary results, Supplementary Material online), the gene subnetworks plausibly involved in high-altitude adaptation were the three highly nested and belonging to different integrin pathways (fig. 2 and supplementary tables 2–4, Supplementary Material online). In particular, one of them resulted significant for Tibetans and Sherpa according to all the computed statistics and none were among the significant subnetworks observed for Han Chinese or Tamangs. These gene subnetworks are directly involved in promoting angiogenesis (i.e., the Integrin in angiogenesis pathway) or include loci encoding for integrin subunits and/or ligands, as well as proteins of the collagen family (i.e., COL genes), which contribute to angiogenetic functions. For instance, the COL6A1 candidate gene pointed out by the nSL-based test belongs to both the β-1 integrin and the Integrin in angiogenesis pathways and is known to play a role especially in muscle development, showing deleterious mutations that cause a congenital muscular dystrophy referred as the Bethlem myopathy (Jöbsis et al. 1999). The ITGA6 gene instead belongs to the β-1 integrin pathway and its subpathway Integrin α6-β4 includes an integrin subunit expressed on platelets and modulating angiogenesis (Avraamides et al. 2008). Interestingly, some ITGA6 variants were recently found to be associated with differential risk of developing polycythemia (Zhao et al. 2017) and it has been proposed that such a gene is directly regulated by hypoxia-inducible factors (Brooks et al. 2016). Subnetworks of genes belonging to these functional pathways were thus found to have experienced multiple selective events according to different tests (i.e., based on patterns of interpopulation differentiation, such as the Fcs, or of intrapopulation haplotype homozygosity, as is the case of nSL and DIND) and despite the different population samples and type of data analyzed. Therefore, this provided robust evidence supporting their adaptive evolution in the Tibetan/Sherpa lineage.
To our knowledge, and with the exclusion of ITGA6, none of the previous studies conducted on these populations have pinpointed such genes among the candidate loci contributing to Tibetan/Sherpa high-altitude adaptation. A possible explanation for this outcome is that although some of them presented quite high scores for the computed statistics, when considered independently as single genes their relation with a specific biological function (e.g., angiogenesis) is not so obvious because the proteins they encode are involved in several other basal cellular activities. Similarly, this may be the case of the significant subnetworks identified according to the DIND statistics and linked with each other by the ESR1 gene, which is known to contribute to endothelial nitric oxide production (Bateman et al. 2017). Accordingly, selective events at these subnetworks have the potential to have favored the increased blood flow observed in Tibetan/Sherpa lungs (Beall et al. 2001) (supplementary results, Supplementary Material online). Another reason for the absence of such genes from previous lists of top candidate high-altitude associated loci may relate to the fact that although at least two of the above-mentioned integrin pathways resulted significant in all tests, the genes composing these subnetworks were not always the same according to the different analyses applied and the different set of samples examined. This further corroborates the hypothesis that selective pressure imposed by hypobaric hypoxia has triggered polygenic adaptation of these Himalayan populations. Such a model of adaptation assumes that many genes were subjected to natural selection, but the intensity of the selective pressure on each of them was relaxed due to their limited individual role in shaping the adaptive phenotype (Pritchard and Di Rienzo 2010; Pritchard et al. 2010; Hernandez et al. 2011; Scheinfeldt and Tishkoff 2013; Jeong and Di Rienzo 2014; Schrider and Kern 2017). According to this evolutionary scenario, an adaptive allele could be even replaced by a different variant on another related gene without compromising the overall adaptive outcome, so that different individuals may carry different adaptive mutations on different sets of highly correlated genes (Pritchard and Di Rienzo 2010). This implies that most of these loci are expected to present moderate to low selection signatures, thus failing to result as outliers when traditional hard sweep-oriented single-gene approaches are applied (Scheinfeldt and Tishkoff 2013).
Unfortunately, with the available data and without implementing targeted functional assays it is impossible to fully elucidate the actual phenotypic trait/s modulated by variants located on the identified candidate genes. Nevertheless, it is plausible that their role in angiogenesis underlays at least some of the Tibetan/Sherpa biological adjustments observed along the oxygen transport cascade. For instance, the increased blood flow and capillary distribution already attested for Tibetans and Sherpa (Beall 2007; Gilbert-Kawai et al. 2014) may be achieved by changes in the regulation of angiogenetic factors and positive selection at gene subnetworks highlighted by the present study may have just acted in this direction by promoting increased tissue blood perfusion in response to the hypoxic stress.
In conclusion, by using whole-genome sequence data and by combining a series of complementary selection statistics with a recently developed gene network analysis, we provided new evidence for polygenic adaptation to high altitude in Tibetans and Sherpa. In particular, our results proved that such an adaptation was mediated not only by the few hard selective sweeps at genes involved in the erythropoietic cascade attested so far but also by multiple subtle selective events at loci related to functional pathways involved in regulating angiogenesis. Accordingly, the present study took a step forward into the depiction of the full spectrum of genetic determinants underlying the complex Tibetan/Sherpa adaptive phenotype and pointed to modulation of angiogenetic functions as a key evolutionary process having enabled these populations to cope with hypobaric hypoxia.
Ethics
Informed consent related to the donation of blood specimens to be processed for extraction of DNA and to be used for population genomics analyses was obtained from the subjects involved in the study within the framework of the ERC-2011-AdG 295733 project. The University of Bologna ethics committee released approval for the present study, which was designed and conducted in accordance with relevant guidelines and regulations according to the ethical principles for medical research involving human subjects stated by the WMA Declaration of Helsinki.
Supplementary Material
Acknowledgments
We would like to thank the Sherpa community of the Rolwaling Himal and the members of the Mount Everest Summiters Club who collaborated with the Explora Nunaat International team making the Extreme Malangur 2015 and Jobo Garu 2017 expeditions possible. We are also grateful to all participants of these expeditions, the Gran Sasso and Monti della Laga National Park, the Club Alpino Italiano, Massimo Izzi and Maria Giustina Palmas (“L. Galvani” Interdepartmental Centre, University of Bologna) for their valuable help in organizing the sampling of biological materials. We would like to thank Pier Massimo Zambonelli (CESIA, University of Bologna) for his IT assistance and Leone De Marco (University of Pavia) for his help in writing scripts for data analyses. Finally, a particular thank goes to Choongwon Jeong (Max Planck Institute for the Science of Human History, Jena) for his assistance in raw sequence data processing and, especially, to Anna Di Rienzo (University of Chicago), who contributed to support this work with her National Institutes of Health 1R01HL119577 grant. G.A.G.R., S.D.F., and S.S. were supported by the ERC-2011-AdG295733 to D.PET.
Literature Cited
- 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhinand CS, Raju R, Soumya SJ, Arya PS, Sudhakaran PR.. 2016. VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis. J Cell Commun Signal. 10(4):347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraamides CJ, Garmy-Susini B, Varner JA.. 2008. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. 8(8):604–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Sarkar-Roy N, Majumder PP.. 2016. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci U S A. 113(6):1594–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, et al. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM. 2007. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A. 104(Suppl 1):8655–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, et al. 1997. Ventilation and hypoxic ventilatory response of Tibetan and Aymara high altitude natives. Am J Phys Anthropol. 104(4):427–447. [DOI] [PubMed] [Google Scholar]
- Beall CM, et al. 2001. Pulmonary nitric oxide in mountain dwellers. Nature 414(6862):411–412. [DOI] [PubMed] [Google Scholar]
- Beall CM, et al. 2010. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 107(25):11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6(9):e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DLP, et al. 2016. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 15:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buroker NE, et al. 2012. EPAS1 and EGLN1 associations with high altitude sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau. Blood Cells Mol Dis. 49(2):67–73. [DOI] [PubMed] [Google Scholar]
- Cho J-H, et al. 2013. Unique features of naive CD8+ T cell activation by IL-2. J Immunol. 191(11):5559–5573. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zagury J-F, Marchini J.. 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 10(1):5–6. [DOI] [PubMed] [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps M, et al. 2016. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet. 98(1):5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzurum SC, et al. 2007. Higher blood flow and circulating NO products offset high-altitude hypoxia among Tibetans. Proc Natl Acad Sci U S A. 104(45):17593–17598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagny M, et al. 2014. Exploring the occurrence of classic selective sweeps in humans using whole-genome sequencing data sets. Mol Biol Evol. 31(7):1850–1868. [DOI] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R.. 2014. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol Biol Evol. 31(5):1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert-Kawai ET, Milledge JS, Grocott MPW, Martin DS.. 2014. King of the mountains: tibetan and Sherpa physiological adaptations for life at high altitude. Physiology (Bethesda) 29(6):388–402. [DOI] [PubMed] [Google Scholar]
- Gnecchi-Ruscone GA, et al. 2017. The genomic landscape of Nepalese Tibeto-Burmans reveals new insights into the recent peopling of Southern Himalayas. Sci Rep. 7(1):15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy A, Daub JT, Excoffier L.. 2017. Detecting gene subnetworks under selection in biological pathways. Nucleic Acids Res. 45(16):e149.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, et al. 2011. Classic selective sweeps were rare in recent human evolution. Science 331(6019):920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppeler H, Vogt M, Weibel ER, Flück M.. 2003. Response of skeletal muscle mitochondria to hypoxia. Exp Physiol. 88(1):109–119. [DOI] [PubMed] [Google Scholar]
- Hu H, et al. 2017. Evolutionary history of Tibetans inferred from whole-genome sequencing. PLoS Genet. 13(4):e1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGO Pan-Asian SNP Consortium, et al. 2009. Mapping human genetic diversity in Asia. Science 326:1541–1545. [DOI] [PubMed] [Google Scholar]
- Jansen GFA, Krins A, Basnyat B, Odoom JA, Ince C.. 2007. Role of the altitude level on cerebral autoregulation in residents at high altitude. J Appl Physiol. 103(2):518–523. [DOI] [PubMed] [Google Scholar]
- Jeong C, Di Rienzo A.. 2014. Adaptations to local environments in modern human populations. Curr Opin Genet Dev. 29:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, et al. 2014. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat Commun. 5:3281.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, et al. 2016. Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc. Proc Natl Acad Sci U S A. 113(27):7485–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, et al. 2017. A longitudinal cline characterizes the genetic structure of human populations in the Tibetan plateau. PLoS One 12(4):e0175885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, et al. 2018. Detecting past and ongoing natural selection among ethnically Tibetan women at high altitude in Nepal. PLoS Genet. 14(9):e1007650.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jöbsis GJ, Boers JM, Barth PG, de Visser M.. 1999. Bethlem myopathy: a slowly progressive congenital muscular dystrophy with contractures. Brain 122 (4):649–655. [DOI] [PubMed] [Google Scholar]
- Kayser B, Hoppeler H, Claassen H, Cerretelli P.. 1991. Muscle structure and performance capacity of Himalayan Sherpas. J Appl Physiol. 70(5):1938–1942. [DOI] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D.. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8(1):e1002453.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie S, et al. 2015. The fine-scale genetic structure of the British population. Nature 519(7543):309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319(5866):1100–1104. [DOI] [PubMed] [Google Scholar]
- Liu G, et al. 2003. Neph1 and nephrin interaction in the slit diaphragm is an important determinant of glomerular permeability. J Clin Invest. 112(2):209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo FR, et al. 2014. A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet. 46(9):951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, et al. 2016. Ancestral origins and genetic history of Tibetan highlanders. Am J Hum Genet. 99(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF.. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16(3):1215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LG. 2001. Human genetic adaptation to high altitude. High Alt Med Biol. 2(2):257–279. [DOI] [PubMed] [Google Scholar]
- Moore LG, Charles SM, Julian CG.. 2011. Humans at high altitude: hypoxia and fetal growth. Respir Physiol Neurobiol. 178(1):181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M, Chance RK, Bashaw GJ.. 2009. Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci. 32:383–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 2(12):e190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. 2017. Down-regulation of EPAS1 transcription and genetic adaptation of Tibetans to high-altitude hypoxia. Mol Biol E. 34:818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Di Rienzo A.. 2010. Adaptation – not by sweeps alone. Nat Rev Genet. 11(10):665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G.. 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 20(4):R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, et al. 1996. Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature 379(6562):262–265. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449(7164):913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, et al. 2009. PID: the Pathway Interaction Database. Nucleic Acids Res. 37(Database issue):D674–D679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Tishkoff SA.. 2013. Recent human adaptation: genomic approaches, interpretation and insights. Nat Rev Genet. 14(10):692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD.. 2017. Soft sweeps are the dominant mode of adaptation in the human genome. Mol Biol Evol. 34(8):1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 329(5987):72–75. [DOI] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD.. 2014. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 31(10):2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashi T, et al. 2017. Gain-of-function EGLN1 prolyl hydroxylase (PHD2 D4E: c 127S) in combination with EPAS1 (HIF-2α) polymorphism lowers hemoglobin concentration in Tibetan highlanders. J Mol Med. 95(6):665–670. [DOI] [PubMed] [Google Scholar]
- van Weering DH, Bos JL.. 1998. Signal transduction by the receptor tyrosine kinase Ret. Recent Results Cancer Res. 154:271–281. [DOI] [PubMed] [Google Scholar]
- Vargas E, Spielvogel H.. 2006. Chronic mountain sickness, optimal hemoglobin, and heart disease. High Alt Med Biol. 7(2):138–149. [DOI] [PubMed] [Google Scholar]
- Vitzthum VJ. 2013. Fifty fertile years: anthropologists’ studies of reproduction in high altitude natives. Am J Hum Biol. 25(2):179–189. [DOI] [PubMed] [Google Scholar]
- Winslow RM, Monge Cassinelli C.. 1987. Hypoxia, polycythemia, and chronic mountain sickness. Baltimore (MD): Johns Hopkins University Press; Available from: https://trove.nla.gov.au/version/14978659. Last accessed on April 23, 2018. [Google Scholar]
- Xu S, et al. 2011. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 28(2):1003–1011. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2017. Genetic signatures of high-altitude adaptation in Tibetans. Proc Natl Acad Sci U S A. 114(16):4189–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, et al. 2016. Protein tyrosine phosphatase 1B (PTP1B) is dispensable for IgE-mediated cutaneous reaction in vivo. Cell Immunol. 306–307:9–16. [DOI] [PubMed] [Google Scholar]
- Yi X, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, et al. 2017. Differentiated demographic histories and local adaptations between Sherpas and Tibetans. Genome Biol. 18(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. 2017. Associations of high altitude polycythemia with polymorphisms in EPAS1, ITGA6 and ERBB4 in Chinese Han and Tibetan populations. Oncotarget 8(49):86736–86746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, et al. 1996. Smaller alveolar-arterial O2 gradients in Tibetan than Han residents of Lhasa (3658 m). Respir Physiol. 103(1):75–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.