Abstract
Precision medicine holds great promise to harness genetic and epigenetic cues for targeted treatment of a variety of diseases, ranging from many types of cancers, neurodegenerative diseases, to cardiovascular diseases. The proteomic profiles resulting from the unique genetic and epigenetic signatures represent a class of relatively well accessible molecular targets for both interrogation (e.g., diagnosis, prognosis) and intervention (e.g., targeted therapy) of these diseases. Aptamers are promising for such applications by specific binding with cognate disease biomarkers. Nucleic acid aptamers are a class of DNA or RNA with unique three-dimensional conformations that allow them to tightly bind with target molecules. Aptamers can be relatively easily screened, synthesized, programmably designed, and chemically modified for various biomedical applications, including targeted therapy. Aptamers can be chemically modified to resist enzymatic degradation or optimize their pharmacological behaviors, which ensured their chemical integrity and bioavailability under physiological conditions. In this review, we will focus on recent progress and discuss the challenges and opportunities in the research areas of aptamer-based targeted therapy in the forms of aptamer therapeutics or aptamer-drug conjugates (ApDCs).
Keywords: Aptamer, nucleic acid therapeutics, aptamer-drug conjugate, drug delivery, targeted immunotherapy
1. Introduction
Aptamers are single-stranded DNA or RNA (ssDNA or ssRNA) with unique tertiary structures that enable them to specifically bind with cognate molecular targets. Aptamers are typically screened via Systematic Evolution of Ligands by Exponential Enrichment (SELEX) [1, 2], using targets ranging from small molecules [1, 3, 4] to biomacromolecules [2, 5–10], infected cells [11], stem cells [12], and cancer cells [13–20]. Such aptamers, especially if modified with hydrophobic groups to improve aptamer binding and targetability in the case of slow off-rate modified aptamers (SOMAmers), can have decent binding affinities with dissociation constants (Kds) typically in the pico to nanomolar range [21]. Resolution of the atomic structures of aptamer/target complexes suggested that such strong aptamertarget binding can be attributed to a combination of noncovalent interactions such as van der Waals force, hydrogen bonding, and stacking interactions [4, 22–25].
1.1. Why aptamers?
Advancements in nucleic acid chemistry have resulted in the development of versatile tools for chemical modification and bioconjugation. Such tools have further empowered aptamers as candidates of molecular theranostic agents via, for example, expanding the library diversity for aptamer screening, dramatically enhancing the biostability of nucleic acid aptamers, lowering the dissociation rate of aptamers from target molecules and thus increasing their binding affinities, tagging aptamers with fluorogenic or radioisotope reporters, and conjugating aptamers with therapeutic agents for targeted drug delivery. Importantly, many of these chemical modifications or bioconjugations can be automated on a DNA/RNA synthesizer or can be conducted under biofriendly conditions [26, 27]. These tools pave the way for aptamers to be developed for versatile biomedical applications such as bioimaging and targeted therapy.
As a class of molecular ligands, aptamers have some characteristic features compared with other classes of ligands such as antibodies or peptides [11]. Aptamers can be screened for a wide array of molecular targets, including toxins or poorly immunogenic targets that are difficult to generate antibodies against. Aptamer screening is efficient (ideally can be within a week) and cost-effective. Current technologies of automated DNA/RNA synthesis also enable easy, cost-effective, large-scale manufacture and automated chemical modification of aptamers with low batch-to-batch variations. The relatively simple chemical structures of aptamers entail full conformational recovery even after thermal or chemical denaturation. Aptamers typically have long shelf life. Further, the nature of nucleic acid aptamers permits straightforward antidote development [28, 29]. These features make aptamers intriguing for targeted therapy. In addition, aptamers have been widely utilized in many other biomedical scenarios, including in vitro bioanalysis and in vitro diagnostics (IVD), bioimaging, biomarker discovery, and aptamer-aided drug discovery. In this article, we will primarily discuss the applications of aptamers in targeted therapy.
1.2. Aptamer development
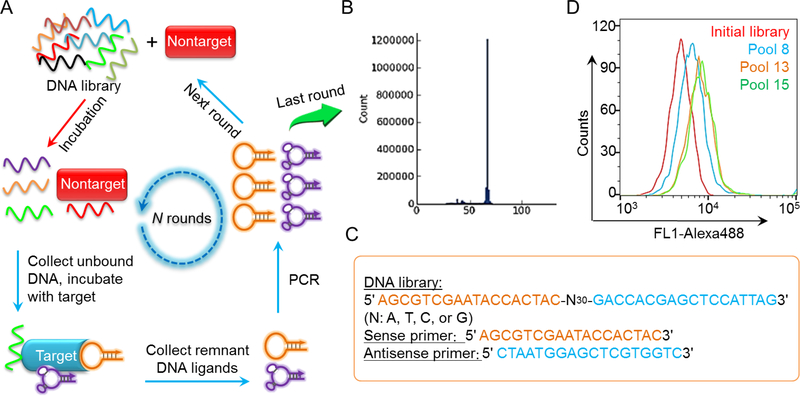
Aptamers are often developed via SELEX [1, 2] for a variety of molecular or cellular targets (Figure 1). The past few decades have witnessed the advancement of genetics and proteomics that have mapped out the profile of upregulated biomarkers in many diseases, including many types of cancers. Aptamers have been screened for a wide variety of these disease-related biomarkers (Table 1) such as epidermal growth factor receptor (EGFR) [30–32], human epidermal growth factor receptor 2 (HER2) [33], prostate-specific membrane antigen (PSMA) [34], protein tyrosine kinase (PTK7) [35], and vascular endothelial growth factor (VEGF) [36], all of which are important biomarkers for diseases including cancer. Aptamer AS1411 that binds to nucleolin [37] was discovered serendipitously, rather than by screening. Aptamer screening has substantially benefited from technology advancement in chemical or enzymatic nucleic acid synthesis and amplification (e.g., PCR), sequencing, as well as bioinformatic analysis. For example, by taking advantage of technology platforms such as capillary electrophoresis [38], microfluidics [39], and flow cytometry [40, 41], the isolation of aptamer candidate probes during selection can be further facilitated. SELEX may even be automated so that aptamers can be identified ideally within only a few days [42].
Figure 1.
Workflow of SELEX. (A) Schematic illustration of in vitro SELEX for aptamer screening. (B) The oligonucleotide pool from the last round of SELEX was subject to sequencing and bioinformatic analysis of homology and frequency. (C) Exemplary sequences of library and primers used for SELEX. (D) An example of flow cytometry results that demonstrate the progressive enrichment of aptamer candidates to target cells.
Table 1.
Examples of aptamers selected with potential for targeted therapy.
| Molecular targets | Names (if not self-explaining) of aptamer examples | Disease indication | References |
|---|---|---|---|
| Nucleolin | AS1411 | Cancer | [37] |
| VEGF | Pegaptanib (PEGylated), VEap121 | AMD, cancer | [36] |
| PTK7 | Sgc8 | Cancer | [35] |
| IGHM | TD05 | Cancer | [51] |
| PSMA | A9, A10 | Cancer | [34] |
| Tenascin C | Cancer | [6] | |
| Mucin 1 | AptA, AptB | Cancer | [52] |
| αvβ3 integrin | Apt-αvβ3 | Cancer | [53] |
| NF-κB | Y1, Y3 | Cancer | [54] |
| E2F3 transcription factor | aptamer 8–2 | Cancer | [55, 56] |
| HER2 HER3 |
Herceptamers A30 |
Cancer Cancer |
[33, 57–60] [10] |
| EGFR EpCAM CD30 |
E07 SYL3C, Ep1 C2, NGS6.0 |
Cancer Cancer Cancer |
[30–32] [61, 62] [63] |
| HIV gp120 | B40, UCLA1 | Viral infection | [64] |
| IgE | D17.4 | Allergy | [65] |
| Osteoblasts | CH6 | Metabolic skeletal disorders | [66] |
| TrR | GS24, DW4, C2.min | Lysosomal storage diseases | [67] |
| 4–1BB | Immune diseases, including cancer | [68] | |
| CTLA-4 | CTLA4apt, aptCTLA-4 | Immune diseases, including cancer | [69, 70] |
| OX40 | 9C7, 11F11, 9D9 | Immune diseases, including cancer | [71, 72] |
| PD-1 | MP7 | Immune diseases, including cancer | [73] |
| PD-L1 | aptPD-L1 | Immune diseases, including cancer | [74] |
| Tim-3 | TIM3Apt | Immune diseases, including cancer | [75] |
| LAG3 | Apt1, Apt2, Apt4, Apt5 | Immune diseases, including cancer | [76] |
| CD28 | AptCD28 | Immune diseases, including cancer | [77] |
| DEC205 | Immune diseases, including cancer | [78] | |
| IL-4Ra | cl.42 | Immune diseases, including cancer | [79] |
| IL-6R | AIR-3 | Immune diseases, including cancer | [43] |
| FIXa | Ch-9.3t | Thrombosis | [80–83] |
| Plasminogen activator inhibitor 1 | SM20, WT15 | Thrombosis | [84] |
| α-thrombin | TBA (thrombin-binding aptamers) | Thrombosis | [8] |
EpCAM: epithelial cell adhesion molecule. PTK7: protein tyrosine kinase 7. IGHM: immunoglobulin μ heavy chains. HER: human epidermal growth factor receptor. IL-6R: interleukin 6 receptor. PSMA: prostate-specific membrane antigen. VEGF: vascular endothelial growth factor. AMD: age-related macular degeneration. IgE: Immunoglobulin E. TrR: transferrin receptor. PD-1: programmed death receptor I. PD-L1: programmed death ligand I. CTLA4: cytotoxic T-lymphocyte associated protein 4. TIM-3: T cell immunoglobulin-3. LAG3: lymphocyteactivation gene 3. FIXa: Factor IXa.
Aptamer screening can be conducted using known molecular targets, such as proteins or peptides or small molecules. Using this strategy, aptamers have been identified for various disease-related biomarkers, such as interleukin 6 receptor (IL-6R) [43], PSMA [34], and VEGF [36]. An obvious advantage of directly using molecular targets for aptamer screening is that the resulting aptamers are certainly able to bind to the corresponding targets. To ensure binding selectivity of aptamers, a negative screening using non-target molecules can be performed to remove any unwanted ligands that binds to these non-target molecules as well. To screen aptamers against molecular targets that are exposed on cell surfaces or in tumor tissues, SELEX has also been conducted using cell fragments, live cells [44–46], or even tumor tissues [47]. For instance, cell-SELEX was developed to use whole live cells to screen aptamers for specific target cells [40], including diseased cells [13–20, 45, 48, 49]. One prominent characteristic of Cell-SELEX is its ability to generate aptamers for any cells of interest without reliance on the prior knowledge of their molecular signatures. Further, aptamers can be identified for cell surface functional molecules which require cofactors or post-translational modifications to configure functional conformations on cell surfaces. Moreover, cell-SELEX can in turn be used for the discovery of biomarkers that are previously unknown or whose roles in pathogenesis have not been recognized yet [46, 50]. Interestingly, aptamers can not only be used as targeting ligands, but also as therapeutics. A good example is pegaptanib (i.e., Macugen®), an VEGF165inhibiting aptamer that was approved by FDA to treat age-related macular degeneration (AMD) [36].
1.3. Versatile biomedical applications of aptamers
Aptamers have been explored for a wide range of biomedical applications. The high specificity of aptamer-mediated molecular recognition entails their application to distinguish subtle molecular differences, which may be fundamentally critical under situations such as IVD and diagnostic imaging for patient stratification to select patient responders to a certain targeted therapy. A proof of such concept is using aptamers to distinguish three different but closely-related and morphologically similar types of acute myeloid leukemia (AML) cells [16]. In this study, an aptamer was identified to selectively bind to target AML cells spiked in bone marrow aspirates, but not to the two subtypes of nontarget AML cells. Such ability to precisely distinguish the molecular signatures could empower the elucidation of molecular basis underlying biological development and pathogenesis.
IVD might be one of the most extensively explored areas for aptamer-based biomedical applications [85–87]. This is in a large part motivated by the unique advantages of aptamers over antibodies or peptide ligand counterparts, in terms of, for example, probe screening, synthesis, modification, and stability including recoverability after denaturation. For instance, aptamers can be developed into aptasensors that sensitively and selectively report the presence of cognate targets [88]. Such concept can be expanded and combined with nanobiotechnology, microfluidics, as well as synthetic biology [89]. For instance, aptamer-coated microfluidics have been studied to detect, enrich, and isolate target cells, such as circulating tumor cells (CTCs) [90–92].
Aside from in vitro detection, aptamers have also been explored for in vivo bioimaging [93]. Bioimaging is critical for disease diagnosis, prognosis, patient stratification, as well as the evaluation of therapy responses. Compared with antibodies, the ease of bioconjugation, fast tissue penetration, and rapid body clearance represent potential advantages of aptamers for applications in bioimaging. By modifying aptamers with fluorescent dyes, radioisotopes, or magnetic nanomaterial reporters, aptamers have been explored for molecular imaging by optical bioimaging [94, 95], positron emission tomography (PET) or single-photon emission computed tomography (SPECT) [33, 93, 96–98], as well as MRI [99, 100].
Aptamers have also been developed as therapeutics. Examples are aptamers for the treatment of AMD (e.g., pegaptanib), treatment of cancer (e.g., AS1411), and as anticoagulants (e.g., pegnivacogin). Aptamers can also aid the screening of small molecule compounds against drug targets, especially those conventionally regarded as undruggable. The elucidation of the tertiary structures of aptamer/target complexes (e.g., thrombin [22, 23], HIV TAR [24], and NF-κB [25]) has facilitated the understanding of aptamer-target interactions at the molecular and atomic levels and assisted structurebased drug discovery. For instance, using aptamers as either templates or substitutes, the compounds that can compete with aptamers for binding to target molecules can then be identified as drug candidates in high-throughput drug screening [101–110].
Aptamers have facilitated the studies of nucleic acid molecular biology, which in the long term could facilitate aptamers’ contributions to biomedical applications. For example, RNA aptamers were screened to specifically bind with a fluorogenic dye derived from green fluorescence protein (GFP), as such this dye derivative can fluoresce upon binding with these RNA aptamers [111, 112]. By tuning the chromatographic spectra of dye derivatives, aptamers have been developed to emit fluorescence signals of a broad spectrum of wavelengths. This group of aptamers essentially established a toolbox of the RNA mimics of fluorescent proteins, and have interesting indications for studying RNA biology [89, 113, 114].
Another interesting application of aptamers is the discovery of biomarkers, largely because aptamers can be selected by cell-SELEX without prior knowledge of the molecular identities. For example, a biotinylated aptamer, sgc8 [13], was utilized to identify PTK7 in many types of cancer cells. The biology of PTK7, a pseudokinase without tyrosine kinase activity, was previously rarely studied [35, 115]. The discovery, enabled by aptamers, of the differentiation of PTK7 expression in cancer cells and many healthy cells implied potential correlation of upregulated PTK7 expression in cancer development and the potential value of PTK7 as a diagnosis or therapy targets.
2. Aptamers as therapeutics
Aptamers have also been developed as therapeutics. Such therapeutic aptamers can readily modulate the biological pathways for the intervention of many types of diseases such as cancer, infectious diseases, and cardiovascular diseases. Pegaptanib (Macugen®) is one example, which was approved by the U.S. FDA to treat AMD. Pegaptanib is an anti-VEGF antagonist RNA aptamer of 28 nucleotides in length. VEGF binds and activates its receptors that are primarily on vascular endothelial cells, induces angiogenesis, and increases vascular permeability and inflammation, leading to neovascular (wet) form of AMD. Pegaptanib sodium solution is intravitreally injected to treat AMD by blocking VEGF. Note that, to increase its in vivo half-life, pegaptanib is PEGylated by conjugating two 20-kilodalton (20 KDa) monomethoxy polyethylene glycol (PEG) units at the end of the aptamer molecule; and to increase the biostability of aptamer, pegaptanib is synthesized with 2’-deoxy-2’-fluoro cytidine and uridine, several 2’-deoxy-2’-O-Methyl adenosine and guanosine, and an inverted dT to cap the 3’-end of the aptamer. Indeed, this aptamer was originally selected with 2’-fluoro purines and pyrimidines and further modified with 2’-O-methyl, and during the latter step, some RNA purines were found unable to be exchanged with 2-O-methyl.
For the development of aptamers as anticancer therapeutics, one example is AS1411 that is under clinical testing for the treatment of cancers including AML [116]. Unlike many other aptamers, AS1411 was discovered not by SELEX, but serendipitously by screening antisense oligonucleotides for antiproliferation effect [117]. This 26-nucleotide AS1411 has only guanines and thymines, and forms guanine-mediated quadraplex structures in solution. This aptamer was thought to bind with nucleolin [118] and can be internalized into many types of cancer cells [119]. The underlying mechanisms of AS1411-mediated antiproliferation effect have not been fully understood, but have been shown to involve the inhibition of cell proliferation via multiple signaling pathways involving BCL-2 mRNA destabilization and NF-κB inhibition [119]. In addition to developing aptamers for direct interaction with cancer cells for cancer therapy, aptamers can also be developed to modulate the immune system and indirectly inhibit cancer cell growth for cancer therapy. Example of this class of aptamers include multimeric aptamers that binds to 4–1BB (CD137), a costimulatory factor that can be expressed on activated T cells and can enhance T cell proliferation, IL-2 secretion, survival and cytolytic activity of T cells [120].
Aptamers therapeutics have also been developed for the intervention of infectious diseases by targeting proteins related to inflammation and immunity. For instance, HIV-1 viral infection was inhibited using a trans-activation response (TAR) nucleic acid decoy that is essentially an aptamer [121], or by an aptamer selected against the TAR element [122]. In addition, aptamers have been developed for reverse transcriptase, HIV-1 Gag protein, nucleocapsid protein, integrase, Tat protein and gp120 [35,36,49], all of which are closely related to viral infection. For example, gp120, a viral surface protein that mediates virus binding to target cells via CD4 receptor and a co-receptor such as CCR5 or CXCR4, has been studied as a molecular target and selected anti-gp120 aptamers that displayed viral inhibition abilities. Such aptamers with dual ability of binding and biological inhibition have been further explored for targeted delivery of viral inhibiting siRNA [158], which will be discussed later.
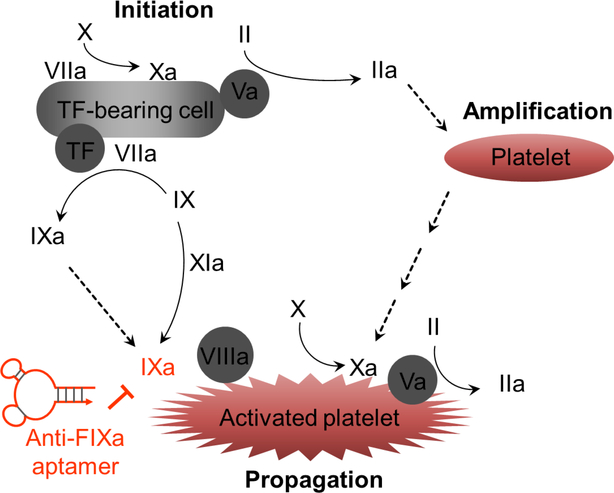
In the broad area of cardiovascular diseases, aptamers have also been extensively sought after. For instance, aptamers have been developed for a panel of molecules involved in coagulation. For example, aptamers have been developed as anticoagulants by binding to and blocking Factor IXa (FIXa) (Figure 2) [80–83, 123]. Current clinical practice of anticoagulation uses heparin, which however has drawbacks such as the lack of safe and well-controllable antidotes. Indeed, 3–5% patients receiving the standard-of-care heparin treatment have haparin-induced thrombocytopenia. Factor IX is an inactive precursor. In the contact pathway, the signal peptide of Factor IX is removed, and then Factor IX is glycosylated and cleaved by factor XIa, thus producing the active form of Factor IX; alternatively, in the tissue factor pathway, after signal peptide cleavage, glycosylated Factor IX is cleaved by factor VIIa to produce the active form. Activated FIXa processes factor X to factor Xa. IIIVa/IXa complexes convert Factor X to factor Xa, which is pivotal to assemble prothrombinase and generate thrombin on the surfaces of tissue factor-bearing cells and activated platelets. Under these physiological scenarios, antiFIXa therapy may thus prevent the intravascular thrombosis that requires the contribution of intrinsic (contact) pathway. Towards this end, anti-FIXa aptamers have been developed. As an example, an anti-FIXa aptamer called pegnivacogin was developed to inhibit the function of FIXa to catalyze the conversion of FX to FXa [80–83, 123], which enables pegnivacogin to inhibit the intrinsic pathway activation and reduce the propagation of thrombin generation. Pegnivacogin is a 31-nucleotide RNA aptamer that is additionally PEGylated to increase blood circulation half-life, and modified with 2’-fluoro on pyrimidine (C, U) and an inverted dT on the 3’-end of the aptamer to enhance the biostability of the aptamer. Interestingly, the nucleic acid nature of this aptamer allowed straightforward development of an antidote against this anticoagulant aptamer, by simply using a strand of oligonucleotides that is complementary to part of the aptamer to disrupt the tertiary aptamer structure [81, 123, 124]. Pegnivacogin has been clinically investigated for applications such as prevention from arterial thrombosis or venous thrombosis. The phase I and II studies of pegnivacogin showed promising results, but the phase III trial had to be terminated due to serious side effects likely caused by PEG-induced allergy [83]. Nevertheless, if appropriately modified to have reasonable circulation half-life and safety, this aptamer should still be a valuable candidate of anticoagulant drugs.
Figure 2.
Mechanism of the development of aptamers as anticoagulants by targeting FIXa. Shown are a brief description of blood coagulation cascades, in which IXa can be inhibited by anti-FIXa aptamers, thereby inhibiting blood coagulation.
3. Aptamer-drug conjugates (ApDCs) for targeted drug delivery
In addition to serving as therapeutics by themselves, aptamers have also been extensively studied as targeting ligands for drug delivery in the form of ApDCs. Compared with counterparts of antibody-drug conjugates (ADCs) [125, 126], a few of which have been already approved by US FDA for cancer treatment, ApDCs present multiple potential advantages. For example, aptamers can be chemically or enzymatically modified with versatile functional groups for bioconjugation with therapeutics or for optimization of biostability; aptamers can be recovered from some extreme thermal or chemical conditions for possible efficient bioconjugation; and the relatively small molecular weights of aptamers and ApDCs make them promising for faster and deeper tissue penetration than ADCs. Typical ApDCs are composed of three parts: aptamer ligands, drug moieties, and linkers between aptamers and the drug moieties. ApDCs have been studied for multiple modalities of therapy, such as chemotherapy, immunotherapy, radiotherapy, and phototherapy [127–136]. Depending on disease-related biomarkers, ApDCs can be applied in a wide range of diseases such as cancer and acquired immune deficiency syndrome (AIDS). In addition, the introduction of nanotechnology to ApDCs can further advance the development of ApDCs by increasing the drug loading capacity and passive drug delivery efficiency.
3.1. ApDCs for chemotherapy
Chemotherapy is one of the mainstream treatment modalities for cancer, and has dramatically improved the outcome of cancer treatment. A common limitation with conventional chemotherapy is their toxicity in healthy tissues and the consequent adverse side effects that compromise the overall therapeutic efficacy. Reducing the exposure of healthy tissues to these drugs is thus expected to reduce these side effects and improve therapeutic efficacy. Towards this goal, ApDC-mediated targeted drug delivery has been explored by specifically delivering drugs to diseased tissues or cells, but not to healthy tissues.
The programmability and ease of synthesis and modification of aptamers permit versatile designs of ApDCs. For instance, since a well-defined aptamer/drug ratio is critical to achieve consistently high efficiency of drug delivery, ApDCs can be constructed with a 1:1 ratio of aptamer: drug by site-specific chemical conjugation or by physical complexation of drugs with aptamers. As an example, by exploring the characteristic aptamer conformation, chemotherapeutic agent doxorubicin (Dox) have been physically complexed with a PSMA-targeting aptamer at a 1:1 ratio via intercalation of Dox into an intrinsic double-stranded 5’-(GC)-3’ sites of this aptamer. ApDCs can also be developed by covalently conjugating aptamers with drugs. In an example of Dox, an ApDC was synthesized by conjugating Dox with a DNA aptamer sgc8 that specifically binds to biomarker PTK7 [137, 138]. Interestingly, during the conjugation of aptamers with drugs, the linkers can be designed to conditionally release the drug cargo at the desired tissue or subcellular organelles. For instance, an acid-labile hydrazone linker was utilized as the linker of sgc8-Dox ApDCs, such that Dox can be conditionally released in acidic tumor environment or in the acidic endosome or lysosome. It is also worth noting that, owing to the ability of aptamer sgc8 to be internalized into cancer cells, the corresponding ApDCs were also able to be internalized into cancer cells and thus ensured efficient intracellular delivery of drugs.
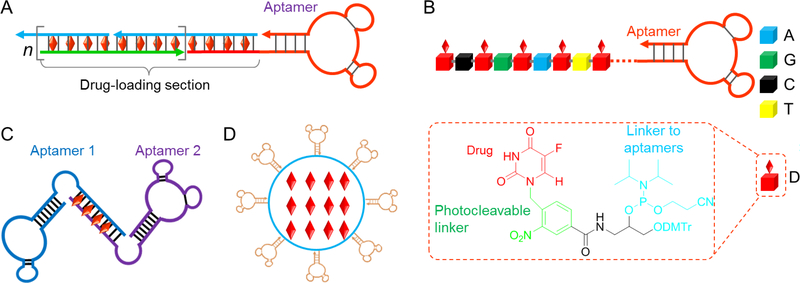
To maximize drug delivery efficiency, ApDCs can also be developed at a 1: n ratio of aptamer: drug, so that one aptamer can carry multiple copies of drugs. During conventional screening of chemotherapeutic drugs, highly toxic compounds are likely excluded due to severe toxicity to healthy tissues. A resulting trade-off is that the therapeutic potency might be moderate, and thus a relatively high dose of such drugs is required for optimal therapeutic efficacy. The ability of one ApDC construct to deliver multiple copies of drugs can partially overcome this limitation and increase therapeutic efficacy. For instance, taking advantage of physical drug complexation with the tandem 5’-(GC)-3’ or 3’-(GC)-5’ sites, a variety of ApDC derivatives have been developed by either employing the multiple intrinsic drug-intercalating sites in aptamers or by intentionally engineering a defined number of such drug-intercalating sites on aptamers. In one example, via a hybridization chain reaction (HCR), an ApDC, also called nanotrain in this case, was developed in which one aptamer was tethered with a long double-stranded DNA (dsDNA) that are composed with nearly 100% of drug-intercalating sites [133] (Figure 3A). Such design maximized the drug loading capacity and thus increase the drug delivery efficiency. These nanotrains proved efficient and specific for specific delivery of Dox into target cancer cells in a xenograft tumor model in mice. ApDCs can also be developed by chemically conjugating multiple drug copies with one aptamer molecule. In an example of ApDCs, multiple drug copies were site-specifically conjugated onto one aptamer molecule (Figure 3B) via programmed automated nucleic acid synthesis using a prodrug-incorporated phosphoramidite [139]. Specifically, to develop an ApDC to deliver 5-fluorouracil (5-FU) for colon cancer therapy, a phosphoramidite prodrug for 5-FU was synthesized. The resulting phosphoramidite prodrug can then be synthesized at any predesigned positions in ApDCs on an automated DNA synthesizer using standard solid-phase DNA synthesis chemistry. As a result, ApDCs can be synthesized with multiple copies of drugs on one ApDC molecule. Note that, a photocleavable linker was also synthesized between 5-FU moiety and the backbone of phosphoramidite prodrug, thus allowing controllable drug release [139]. The resulting ApDC has proven to be selective and efficient in delivering 5-FU prodrug into target HCT116 colon cancer cells. As another example of constructing ApDCs with multiple drug copies on each aptamer, aptamer-drug adducts were synthesized using natural aptamer without any additional chemical modification [140]. Particularly, just like natural drug-genome adduct formation during drug-induced apoptosis, ApDC adducts can be synthesized using aptamers and anthracycline drugs (e.g., Dox) and cisplatin, in a biofriendly reaction condition and in the presence of formaldehyde. The formaldehyde contributed a conditionally cleavable crosslinker between the Dox and the nucleotides on aptamers. It was further found that these anthracycline drugs were mainly conjugated on the guanosine, thus revealing an approach to the programmable and predictable synthesis of ApDCs. Note that, since the drug conjugation sites on aptamer structures are critical to ensure the functionalities of ApDCs and the specificity of drug delivery, the ability of chemically-defined ApDCs to be programmably synthesized represents another characteristic feature that make ApDCs promising for large-scale manufacture as well as clinical translation.
Figure 3.
ApDCs for chemotherapy. (A) Via noncovalent complexation of aptamer-tethered nanotrains and drugs, an ApDC was developed to deliver a large payload of DNA-intercalating drugs into target tumor cells. (B) An example of ApDCs that was programmably synthesized using a phosphoramidite that carried a 5-FU prodrug via a photocleavable linker (inset: molecular structure of a prodrugincorporating phosphoramidite). (C) As a mimic of bispecific antibody, a bi-specific ApDC was developed by simply linking two independent aptamers via a dsDNA linker, which was additionally harnessed for drug loading in bi-specific drug delivery. (D) A schematic representation of aptamerfunctionalized nanocarriers for targeted drug delivery.
The ability to easily develop bispecific ApDCs is another example that testifies the potential of ApDCs. Indeed, bispecific antibody therapeutics have been sought after in the past decade [141, 142]. Bispecific antibody therapeutics [143] can be developed to target two different druggable sites, or deliver a therapeutic antibody via a targeting antibody to target cells, or engage cytotoxic T cell lymphocytes with target cancer cells via a bispecific T-cell engager (BiTE) [144]. To demonstrate the principle of bispecific ApDCs, a bivalent aptamer was constructed by simply conjugating two aptamers that target two different biomarkers via a dsDNA linker that can additionally load drugs for delivery (Figure 3C) [145]. The resulting bispecific ApDCs demonstrated the ability of bispecific cancer cell detection and anticancer drug delivery. Bivalent ApDCs may provide a well programmable, and simple and cost-effective alternative to bispecific antibodies.
As a class of relatively small targeting ligands, aptamers work by active targeting of ApDCs to target sites. The drug delivery efficiency can be further enhanced by combining such ability of aptamers with the capabilities of nanocarriers for high-capacity drug loading and efficient passive targeted drug delivery by exploiting the characteristic enhanced permeation and retention (EPR) effect in many tumors as well as tissues that are undergoing inflammation. Towards this end, aptamer-nanocarrier conjugates have been extensively explored for targeted drug delivery, based on nanoplatforms such as liposomes, DNA/RNA nanostructures, and inorganic gold or silicon nanomaterials [146–148] (Figure 3D). For instance, biocompatible and biodegradable poly(ethylene glycol) - poly(lactic-co-glycolic-acid) (PEG-PLGA) nanoparticles have been functionalized with aptamers for targeted drug delivery [128, 136, 149–151] [149, 150]. The resulting nanoparticulate ApDCs proved the ability of targeted cisplatin delivery into PSMA-positive cancer cells [150, 152, 153].
3.2. ApDCs for gene therapy
Gene therapy is an integral part of precision medicine. The past decades have witnessed historical breakthrough in mapping out the genetic foundations of many human diseases. Gene therapy is therefore a promising approach to treat diseases by correcting the corresponding genetic errors. The approaches to gene therapy can be at the genome level or transcriptome level. At the genome level, plasmids can be delivered into nuclei to allow genes of interest to be integrated within genomic DNA, or alternatively, the genome can be edited by technologies such as transcription activator-like effector nucleases (TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems. In transcriptome approaches, mRNA functions can be inhibited by technologies such as antisense oligonucleotide technology, RNA interference (RNAi), as well as most recently CRISPR interference (CRISPRi). Like many other therapeutic agents, gene therapy agents by themselves lack specificity towards diseased cells, which make it critical to deliver the delivery of gene therapy agents specifically to diseased cells. To this end, aptamers may selectively deliver gene therapeutics into diseased cells.
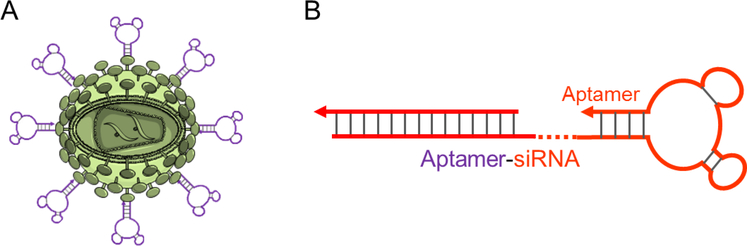
Viral gene therapy has been investigated for a long time. Despite the concerns over its safety and some setback in some clinical tests, recent progress has again proved that it is promising to treat many genetic diseases. This has been further evidenced by the recent US FDA’s approval of chimeric antigen receptor T (CAR-T) cell therapy which uses ex vivo expanded effector T cells that are developed by viral transduction of CAR genes into CD8+ T cells. Unlike ex vivo viral transduction in the production of CAR-T cells, gene therapy of many other human diseases likely requires in vivo administration of gene therapeutic viruses. ApDC approaches may been utilized to reduce the potential “off-target” effect in viral gene delivery. In this line of research, adeno-associated virus 2 (AAV2) vectors have been conjugated with multiple sgc8 aptamers, and the resulting sgc8-AAV2 construct significantly enhanced the delivery efficiency of AAV2, as demonstrated using a GFP gene as a model [154] (Figure 4A). In another example, aptamers were covalently modified on viral capsid surfaces for multivalent drug delivery [155]. Though no gene therapeutics have been used in these studies, they established the methodology to construct aptamer-viral vehicle conjugates that are potential for applications in targeted delivery of viral gene therapeutics.
Figure 4.
ApDCs for gene therapy. (A) Schematic representation of an aptamer-modified viral carrier for potential application in targeted gene delivery. (B) A schematic illustration of a chimeric aptamersiRNA as ApDC for targeted siRNA delivery.
Perhaps ApDC-mediated gene therapy is more often studied in transcriptome approaches recently. By molecular engineering of nucleic acids, DNA/RNA aptamers can be programmably conjugated with nucleic acid gene therapeutics, ranging from small interfering RNA (siRNA), small hairpin RNA (shRNA), and microRNA (miRNA). The resulting gene therapeutic ApDCs have been studied for RNAi-based gene therapy in cancer as well as AIDS [58, 156–159]. In an early example, aptamer-siRNA conjugates were constructed for specific delivery of siRNA into target cancer cells (Figure 4B) [158]. In this study, by conjugating a PSMA-targeting aptamer with siRNAs that silence polo-like kinase 1 (Plk1) and B-cell lymphoma 2 (Bcl2), respectively, the resulting ApDCs specifically delivered siRNA into PSMA-positive LNCaP cells without using any transfection agents, leading to target genes silencing and cell apoptosis. By contrast, these ApDCs siRNA constructs did not induce detectable apoptosis in PSMA-negative PC-3 cells. Remarkably, when intratumorally or intravenously administered to LNCaP tumor-bearing mice, these ApDCs inhibited tumor growth and provided a strong preclinical evidence of ApDC-based gene therapy. Note that, for systemic administration, these aptamer-siRNA conjugates were systematically optimized by truncation of the aptamer sequence to reduce manufacture cost, by 2’-fluoropyrimidine modification to improve biostability, by PEGylation to increase circulation half-life, and by engineering the aptamer-siRNA conjugate structures to optimize the intracellular processing by the RNA-induced silencing complex. Aptamer-siRNA conjugates have further been explored for cancer therapy in combination with chemotherapy or radiotherapy. In an independent study, a similar PSMA-targeting aptamer-siRNABcl2 conjugate was studied to knockdown Bcl2 and sensitize cancer cells to cisplatin [58]. As another example, an ApDC was developed using PSMA-targeting aptamer and a shRNA, which is another form of RNA interfering gene therapeutics that can also be processed by RISC machinery [160, 161]. Particularly, the shRNA was designed to silence a DNA-activated protein kinase gene, which served as a target for radiosensitization, such that target prostate cancer tissues were selectively sensitized to radiation (Figure 4B) [160]. As a result, combination of these ApDCs with radiotherapy enhanced the therapeutic efficacy selectively in PSMA-positive tumor and reduced nontarget toxicity in healthy tissues.
siRNA/shRNA-based ApDCs have also been studied to treat other types of diseases such as bone diseases and infectious diseases. Again, the implementation of nanocarriers in ApDC-mediated gene therapy may increase the loading capacity of gene therapeutics and hence enhance the therapeutic efficacy. Liposome is one of the most successful drug nanocarriers. The protective hollow compartments in liposomes are especially valuable to ensure the in vivo integrity and half-life of nucleic acid therapeutics, which, if not protected, can be rapidly degraded by nucleases and cleared from the body [66, 162, 163]. In one such study, as a potential bone anabolic strategy, liposome was modified with aptamers that specifically bind to osteoblast and loaded with an siRNA to silence osteogenic pleckstrin homology domain-containing family O member 1 (Plekho1) as a potential bone anabolic strategy (Figure 4B) [66]. This nanoconjugates specifically delivered siRNA into target osteoblasts, silenced target Plekho1, and consequently improved bone formation and microarchitecture, and increased bone mass. In another example, siRNA/shRNA-based ApDCs were studied for the treatment of AIDS [145, 161, 164, 165]. Specifically, an aptamer-siRNA conjugate was delivered to T cells with an aptamer that inhibited glycoprotein 120 (gp120) and an siRNA that silenced HIV-1 tat/rev common exon sequence, thereby inhibiting HIV infection [164].
3.3. Aptamer conjugates for immunotherapy
Immunotherapy has made historical breakthrough in the past few years, especially for the treatment of cancer [166–169]. While current approaches of immunotherapy can modulate the functionalities of the immune system, most of these approaches are also accompanied with significant immune toxicities that can be even escalated in the scenarios of combination immunotherapy. To address this challenge, one strategy is to specifically deliver immunotherapeutics to the target tumor or immune cells. Towards this end, ApDCs are a promising platform. ApDCs have been studied to deliver immunomodulatory agents to confine immune costimulation to the tumor region, induce neoantigens in tumor, block exhaustioninducing immune checkpoints to reinvigorate functional immune cells, and prolong antitumor immunity. Note that, in addition to serving as targeting ligands, aptamers themselves can also work as immunotherapeutics and directly participate in the modulation of immune functions. The elucidation of critical molecular targets, such as immune checkpoints or immunostimulatory targets, opens the opportunities to develop immunotherapy by targeting these molecular sites. A variety of aptamers have been developed for immune checkpoints that are involved in immune suppression in cancer patients, such as programmed death receptor I (PD-1) or its ligand programmed death ligand I (PD-L1), cytotoxic T-lymphocyte associated protein 4 (CTLA4), and T cell immunoglobulin-3 (TIM-3) [68, 69, 71–76, 170]. By molecular engineering of these aptamers with immunotherapeutics, ApDCs can be developed for specific delivery of immunotherapeutics. The principle of immunotherapy to treat diseases by modulating the immune system can be used for various diseases, including many types of cancer, infectious diseases, autoimmune diseases. In this article, we will focus on recent progress of ApDCs for cancer immunotherapy.
As mentioned above, aptamers can also serve as immunotherapeutics themselves (Figure 5A). In the context of cancer immunotherapy, therapeutic aptamers have been reported for immune checkpoints such as CTLA-4, PD-1, PD-L1, and Tim-3 for tumor immunotherapy [68, 69, 72–76, 170]. Given the unique features of aptamer therapeutics in terms of screening, manufacture, stability, as well as fast tissue penetration, aptamer inhibitors of immune checkpoints provide unique opportunities to develop cancer immunotherapy as an alternative to the current mainstream of antibody therapeutics. For instance, to develop aptamer antagonist to block CTLA-4 in cancer immunotherapy, a DNA aptamer was developed by cell-SELEX with a strong binding affinity (Kd = 11.84 nM) [69]. This aptamer was also able to be internalized into lymphocytes, and inhibited tumor cell growth both in cultured cells and in tumor-bearing mice models. In another study, DNA aptamers were developed for PD-1 to block the immunoinhibitory PD-1/PD-L1 pathway, as a potential alternative approach to using antibody blockers for reversing immune evasion and provoking antitumor immunity [73]. The reported aptamer specifically binds to the extracellular domain of murine PD-1 and prevent PD-1 from complexation with PD-L1. PEGylated aptamer significantly inhibited the growth of colon carcinoma. Similarly, aptamers have been developed against PD-L1 to block the binding between human PD-1 and PD-L1 [74]. In mouse models, these aptamers promoted lymphocyte proliferation and suppressed the tumor growth. Further immune profiling revealed that these aptamers skewed the tumor immune microenvironment and potentiated the antitumor immunity, as evidenced by elevated levels of tumor infiltrating T lymphocytes, as well as intratumoral antitumor cytokines and chemokines such as tumor necrosis factor-α, interferon-γ (IFN-γ), interleukin-2 (IL-2), chemokine (C-X-C motif) ligand 9 (CXCL9), and CXCL10. TIM-3, an immune checkpoint that work by negatively regulation of effector CD4+ T cells and CD8+ CTLs, can also be blocked by aptamers [170]. An engineered trimeric anti-TIM-3 aptamer was found to block the functional interaction of TIM-3 with Galectin-9, leading to reduced cell death and enhanced survival of T cells and improved therapeutic efficacy in tumor-bearing mice. Beside immune checkpoints, costimulatory factors are also a class of critical molecular targets involved in antitumor immunostimulation. For instance, aptamers have been developed against costimulatory molecules OX40 and 4–1BB [68, 171, 172]. The activation of immunity that can eliminate malignant cells or virusinfected cells demands the induction of antigen-specific effector T cells. Launching potent and durable antitumor immunity requires at least two signals: first, a specificity signal from the complexes of antigen epitopes and major histocompatibility complexes (MHC), and second, the engagement of costimulatory molecules on activated T cells. For instance, the association of OX40 with its ligand is critical for optimal immunoactivation and immunotherapy efficacy [71]. Therefore, agonistic agents, including aptamers, have been developed to specifically stimulate OX40 on T cells, enhance cell proliferation, and promote the production of cytotoxic IFN-γ.
Figure 5.
Aptamers for immunotherapy. (A) Aptamers as therapeutics that bind to and modulate the biological functions of immunomodulatory molecules. (B) ApDCs developed for targeted delivery of immunotherapeutic drugs.
Using aptamers as targeting and internalizing ligands, ApDCs have also been studied for intracellular delivery of immunotherapeutics (Figure 5B). Cancer development has been accompanied with the formation of multi-tier immunosuppression network that involved not only immune checkpoints on the cell surfaces, but perhaps even more inside cells. Targeting these intracellular immunomodulatory molecules is thus attractive for cancer immunotherapy, but delivering non-membrane-penetrable immunomodulatory drugs into cells can be challenged by multiple cell surface or subcellular membrane barriers. Aptamers can be internalized into target cells together with their internalizing receptors, which makes the corresponding ApDCs promising for intracellular delivery of immunotherapeutics. Towards this goal, ApDCs have been developed using an anti-CTLA4 aptamer and an siRNA for signal transducer and activator of transcription 3 (STAT3) [70, 173]. CTLA4 was overexpressed in tumor infiltrating CD8+ T cells in both mice and human patients, and thus the anti-CLTA4 aptamer was able to deliver siRNA cargo into these T cells and get internalized into these cells. Internalized siRNA then mediated the silencing of STAT3, an intracellular mediator during tumor immunosuppression in intratumoral exhausted CD8+ T cells and Tregs, and CTLA4-expressing malignant T cells. In preclinical animal studies, optimized ApDCs can be administered locally or systemically, resulting in dramatically reduced repertoire of tumor-associated regulatory T cells (Tregs). Such immunomodulation eventually contributed to potent inhibition of tumor growth and metastasis in multiple tumor models in mice. Remarkably, in immunodeficient mice bearing human T cell lymphomas, where CD8+ T cells overexpressed CTLA4, treatment with these ApDCs promoted tumor cell apoptosis and inhibited tumor growth. In another study, DNA CpG oligonucleotide [174], which recognizes and stimulates the Toll-like receptor 9 (TLR9) pathway, was explored as an aptamer-like targeting ligand to construct CpGsiRNASTAT3. STAT3 is immunosuppressive also in many antigen presenting cells (APCs) by multiple mechanisms such as inducing antigen-specific T cell tolerance [175–177]. Moreover, STAT3 can also suppresses CpG-mediated immunostimulation [178]. To simultaneously activate TLR9 pathway and inhibit intracellular STAT3 pathway, CpG-siRNASTAT3 conjugate was developed to activate TLR9 by CpG, mediate intracellular delivery by CpG as well, and silence STAT3 upon internalization of CpGsiRNASTAT3. Consequently, the dual functional CpG-siRNASTAT3 conjugate synergistically activated TLR9 pathway and disarmed STAT3 pathway in APCs [179–181].
Another interesting application of siRNA-based ApDC immunotherapeutics is enhancing the antitumor immune memory and hence increasing the persistence of immunotherapy. Antitumor memory T cells are critical for the eradication of cancer cells and the prevention of cancer recurrence. Such memory cells have been proved beneficial for many approaches of immunotherapy ranging from adoptive cell transfer to tumor therapeutic vaccines. In addition to isolating natural memory T cells or inducing such cells by immunostimulatory agents, drugs have been developed to promote the differentiation of activated CD8+ T cells into memory cells. Such compounds, however, often lack of specificity and may cause adverse side effects if distributed to healthy tissues, and even undesirable immunosuppressive effect for drugs like mTOR inhibitor rapamycin. Targeted delivery of these drugs holds the potential to overcome these drawbacks. In one example, an aptamer-siRNA conjugate was developed using an aptamer that targets 4–1BB and an siRNA that silences mTOR complex 1 (mTORC1) [182]. 4–1BB is a costimulatory factor on CD8+ T cells, and can thus guide the specific drug delivery into these T cells. Inhibition of mTORC1, but not its closely-related mTOR complex 2 (mTORC2), has been shown to promote CD8+ T cell memory. It is critical that the siRNA selectively silence mTORC1, but not mTORC2. As a result, the ApDCs were delivered into 4–1BB-positive CD8+ T cells, promoted the CD8+ T cell memory, and eventually enhanced vaccine-mediated antitumor immunity. By contrast, nonselective mTOR inhibitor, rapamycin, led to the inhibition of both mTORC1 and mTORC2 in CD8+ T cells, compromised the cytotoxicity of these CD8+ CTLs. It is worth noting that the ability of such ApDCs to delivery nucleic acid therapeutics such as siRNA into T cells, which are typically extremely difficult to transfect without additional transfection agents, make ApDCs particularly attractive for targeted immunomodulation and immunotherapy. In a similar study, ApDCs were developed for intracellular delivery of siRNA to silence IL-2 receptor (IL-2Ra), in order to attenuate IL-2 signaling in CD8+ T cells and then enhance the persistence of their antitumor efficacy [183].
Bi-specific aptamer conjugates are another class of ApDCs that hold unique potentials for cancer immunotherapy, due to the ease of bi-specific aptamer engineering and the versatile functionalities of aptamers. For instance, bi-specific aptamers have been developed for tumor-specific delivery of immunotherapeutic aptamers, so that the immunomodulation can be largely confined in the tumor lesion and the off-target side effects can be attenuated. This is increasingly being appreciated in clinic management of cancer immunotherapy. Current cancer immunotherapeutics, such as immune checkpoint inhibitors, high-dose IL-2, or adoptive cell transfer, can all efficiently potentiate immunity, which promotes the antitumor immunity but at the same time can also activate self-reactive T cell responses (autoimmunity). Such drawback not only precludes dose escalation that may be necessary to exhausts the therapeutic potential, but may also lead to severe autoimmune diseases, including but not limited to type I diabetes resulting from autoimmunity against one’s own beta cells. To address this limitation, a bi-specific aptamer was constructed using one aptamer that target VEGF, which is broadly expressed in tumor stroma, and another aptamer that can modulate costimulatory molecules 4–1BB [172]. The resulting construct can specifically deliver immunomodulatory agonistic 4–1BB aptamers into tumor lesions for confined immunomodulation. Such construct showed potent and wide applicable tumor-confined costimulation, and elicited potent and relatively confined antitumor immunity in multiple preclinical models of subcutaneous tumors, postsurgical metastasis, drug-induced fibrosarcoma, and oncogene-induced autochthonous glioma. A similar approach was also studied using PSMA-targeting aptamers for tumor-targeted delivery of 4–1BB-modulating aptamers, and showed synergistic antitumor immunomodulation with anticancer vaccines [184]. This study suggests that potentiating naturally occurring antitumor immunity via tumor-targeted costimulation could be an effective approach to elicit protective immunity to control tumor progression in cancer patients. These studies highlight the unique potential of bi-specific aptamer-based unique tumor immunotherapeutics.
Aside from nucleic acid therapeutics, ApDCs have the potential for targeted delivery of other types of therapeutics, such as peptides. For example, aptamers have been used for targeted delivery of peptide antigens. Efficient and specific delivery of peptide antigens to antigen presenting cells (APCs), especially dendritic cells (DCs) that are considered as “professional” APCs, is critical for effective antigen presentation and vaccine-mediated immunomodulation [185]. This is especially necessary for subunit vaccines, in which the subunit peptide antigens themselves can rarely be delivered to the desirable APCs. Targeted antigen delivery to DCs has been studied to improve antigen delivery. For example, an ApDC was developed by conjugating an aptamer with SIINFEKL, an MHC-I-restricted antigen epitope of ovalbumin (OVA) to target CD8+ dendritic cells (Figure 5B) [78]. The aptamer was selected to specifically bind to murine DEC205, a C-type lectin that is predominantly expressed on CD8α+ DCs, a subpopulation of DCs that are efficient at antigen cross-presentation. The targeted antigen delivery led to efficient T cell proliferation and production of IFN-γ and IL-2, and was able to inhibit the growth of an OVA-expressing B16F10 murine melanoma in mice.
Tregs are relatively less often studied as the targets for anticancer immunomodulation and cancer immunotherapy. Part of the reason might be the difficulty to ensure the specificity of immunomodulation as there has thus far no a single biomarker that is exclusively on Tregs. One study exploited Foxp3, a marker that is mostly expressed in Tregs. Yet still, Foxp3 is an intracellular biomolecule that precludes ligand-mediated cell-specific drug delivery. An aptamer that target CD28 was studied in conjugation with a Foxp3 inhibitor called P60. Proof-of-the-principle studies have shown that such CD28-targeting ApDCs were able to deliver P60 to CD28-positive cells, including Tregs, and improve the therapeutic efficacy of P60 in combination of immunostimulatory anticancer vaccines [77].
Like some other therapy modalities, nanotechnology has the potential to tremendously benefit ApDCbased cancer immunotherapy [186–189]. One such benefit is to improve the drug delivery efficiency to tissues and cells because of the features of nanomedicines such as typically high drug loading capacity, efficient passive targeting, and rapid intracellular delivery. In one example, liposome was modified with an aptamer that targets IL-4Ra, which is overexpressed in tumor, for targeted delivery of immunostimulatory CpG to overcome the tumor immunosuppressive microenvironments [79]. The resulting nanoscale ApDCs were efficiently delivered into tumor tissues, and inhibited myeloid-derived suppressor cells (MDSCs) and Tregs [190] in tumor. Consequently, the skewed tumor immune microenvironment contributed to the improvement of tumor immunotherapy in syngeneic mouse models.
3.4. ApDCs for radiotherapy
Radiotherapy is one of the main modalities for cancer treatment in clinic. Radiotherapy typically uses high-energy radiation to kill cancer cells, modulate the tumor immune microenvironment, and shrink tumors. The sources of high-energy radiation may be X-rays, gamma rays, and charged particles. Radiotherapy can be given in the form of external-beam radiotherapy, internal radiotherapy, as well as systemic radiotherapy. While external-beam radiotherapy, internal radiotherapy can be localized to specific regions of tumor lesions, systemic radiotherapy, which typically involves the injection of radioisotopes to allow the delivery of radioisotopes into diseased lesions, is often less specific (with exceptions such as the use of radioactive iodine (131I) for systemic radiotherapy of thyroid cancer, as thyroid cells naturally take up 131I.) Such less specific delivery of radioisotopes would harm healthy tissues and cells and cause adverse side effects, making targeted delivery of radioisotopes significant to reduce such side effects and improve the therapeutic efficacy of systemic radiotherapy. Targeting ligands, such as antibodies and aptamers, are promising for targeted delivery of therapeutic radioisotopes. One such example is ibritumomab tiuxetan (Zevalin®), which is a conjugate of ibritumomab that target CD20 on B lymphocytes and a tiuxetan that allow the chelation of radioisotopes, has been approved by the FDA to treat certain types of B-cell non-Hodgkin lymphoma (NHL) [191]. As “chemical antibodies”, aptamers have also been investigated for targeted delivery of radiopharmaceuticals for bioimaging and radiotherapy. For radiotherapy, one study conjugated an anti-PSMA aptamer to PEGylated liposomes, which were then loaded with α-particle generating actinium-225 (225Ac, half-life: 10 days), for specific delivery of 225Ac to PSMA-positive cancer [192]. In vitro studies demonstrated that these conjugates specifically delivered 225Ac and killed PSMA-positive prostate cancer cells as well as human umbilical vein endothelial cells that were induced to express PSMA. While this study seems promising, vigorous in vivo studies are demanded to evaluate and optimize ApDC-based targeted radioisotopes. Some key challenges of systemic nucleic acid therapeutics, such as fast clearance from the body and typically high accumulation in liver and gall bladder, ought to be addressed for effective in vivo targeted radiotherapy. With the advancement of nucleic acid chemistry and the development of versatile nucleic acid modifications that enable resistance to nuclease degradation and prolong circulation, it is anticipated that ApDC-based radiotherapy could be substantially advanced.
3.5. ApDCs for phototherapy
Phototherapy, such as photodynamic therapy (PDT) and photothermal therapy (PTT), holds great potential for the treatment of a variety of diseases. In PDT, photosensitizers are activated by photons to generate reactive oxygen species (ROS) [193, 194] that can subsequently attack their surrounding diseased cells. PTT appears to work by directly generating heat from photon energy, thereby killing cells. Due to the potential off-target toxicities of photosensitizers and PTT agents, there remains a need to specifically deliver these agents into diseased tissues or cells [43, 195–202]. Therefore, targeted delivery of phototherapeutics is potential to minimize drug toxicity to healthy tissues both by targetspecific drug delivery and by precisely controlling phototherapy-initiating external light source. For example, an ApDC of photosensitizer chlorin e6 (Ce6) was synthesized using an aptamer for interleukin 6 receptor (IL-6R) [43]. The resulting ApDC was specifically delivered to and internalized by target cells. Light irradiation induced apoptosis specifically in targeted cells treated with these ApDCs underwent, but cytotoxicity was undetectable in target cells treated with free Ce6 or in nontarget cells treated with the ApDCs. Since IL-6 and IL-6R play a pivotal role in the immune responses involved in many types of cancers and inflammatory diseases, such ApDCs might be promising to selectively kill target diseased cells. The application of ApDCs in PTT has been studied mainly by exploiting the ability of some nanomaterials to convert light energy into heat, resulting in hyperthermia-mediated toxicity. Nanomaterials studied for PTT include many types of gold nanomaterials [203–205], polydopamine nanomaterials [206], as well as carbon nanomaterials [207] that can be conjugated with aptamers for ligand-mediated active targeted delivery and EPR effect-mediated passive targeted delivery. In one example, cancer cell-targeting aptamers were conjugated with photothermal gold nanorods (AuNRs). The resulting aptamer-AuNR conjugates were thus coupled with both delivery specificity and high hyperthermia efficacy [208], which consequently contributed to selective cytotoxicity upon irradiation with near infrared light. While using light irradiation can further increase the selectivity of phototherapy and reduce the toxicity in healthy tissue, one intrinsic drawback of phototherapy is that its application is often restrained to light accessible diseased tissues such as skin diseases or esophagus diseases.
4. Conclusions
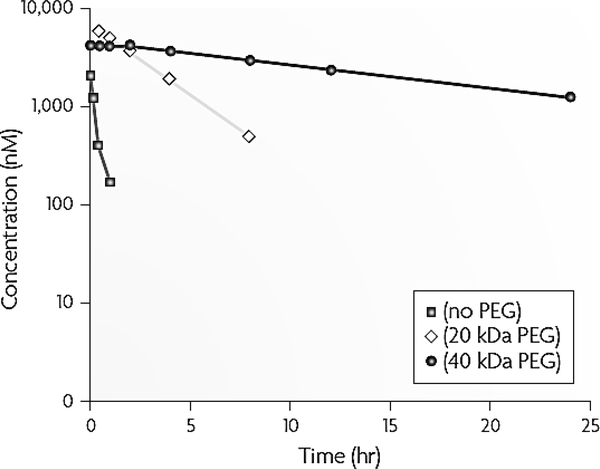
It has been over two decades since aptamers were first described. Multiple unique features of aptamers arouse extensive interest in the development of aptamers as nucleic acids-based alternatives to antibodies and peptide ligands for versatile biomedical applications ranging from in vitro diagnosis (IVD), molecular imaging, drug delivery, to biomarker discovery and drug screening. Specifically, IVD could be benefited by fast aptamer probe generation, rapid and high-throughput large-scale manufacture in solution or on solid phase, straightforward molecular engineering and chemical modification, and robust chemical integrity and ability of conformational reconfiguration upon denaturation; bioimaging could be benefited by the fast aptamer screening and labeling, as well as rapid systemic clearance from the body; and targeted therapy (including targeted drug delivery) may be benefited by the versatility of molecular targets of therapy interest, easy development of versatile ApDCs or aptamer therapeutics, tunable in vivo retention half-lives, as well as rapid and deep tissue penetration. While tremendous achievements have been achieved, multiple challenges have been revealed, such as limited biostability of natural nucleic acids, short in vivo half-lives in the context of aptamer-mediated therapy, as well as potential unwanted immunogenicity triggered via the recognition of nucleic acid by the immune system. Advancement of nucleic acid chemistry and biotechnology has made significant breakthrough to stabilize nucleic acids under physiological conditions, prolong in vivo retention, and mitigate any unwanted immunogenicity. For example, the biostability of nucleic acids can be dramatically enhanced by 2’-O-methyl and 2’-fluoro modifications on nucleotides [11, 209–211], phosphorothioate backbone, as well as L-nucleotides. Moreover, the pharmacokinetics and biodistribution of nucleic acid therapeutics can be improved by modification with chemical moieties or nanomaterials. For example, aptamers modified with PEG (40 kDa) resulted in prolonged half-lives to up to 1 day in rodents and 10 days in human [11, 210–212] (Figure 6). These breakthroughs have set the stage for aptamers to be implemented in a wide variety of biomedical fields, including bioimaging and targeted therapy. Note that despite the FDA approval of multiple PEG-modified biomedicines, PEG may still elicit adverse immune responses. One aforementioned example is pegnivacogin anticoagulant, the phase III clinical trial of which was terminated partially because of PEG-induced immune responses [83]. This scenario calls for alternative technologies to improve the pharmacological behaviors while avoiding immune responses. Endogenous biomacromolecule-based drug delivery systems may have the potential to be tolerated by the immune system while still improving the in vivo pharmacological properties. Examples of such include bioconjugates of drugs with constant fragment (Fc) of antibodies and albumin. In the past few years, we have demonstrated a class of molecular albumin binders, namely Evans blue derivatives, were capable to dramatically prolong the in vivo half-lives of theranostic agents, such as chemotherapeutics [213], immunotherapeutics [214], radiotherapeutics [215, 216], and imaging agents [217–219], and consequently improve the therapeutic efficacy or imaging contrast. Our ongoing studies have demonstrated that such albumin-binding technology also dramatically enhanced the circulation half-life and the tumor accumulation of a cancer-specific nucleic acid aptamer (unpublished).
Figure 6.
Improve the pharmacokinetics of aptamers by PEGylation. A 39-mer aptamer composing of 2’-deoxy purine and 2’-O-methyl pyrimidine was conjugated with PEG (20 kDa and 40 kDa, respectively). The resulting conjugates were administered intravenously to mice at 10 mg/kg. Pharmacokinetics profiles showed that PEGylation dramatically enhanced the half-lives of the aptamers [11].
Acknowledgement
This work was supported by Intramural Research Programs of National Institutes of Health and start-up fund from the School of Pharmacy at Virginia Commonwealth University.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ellington AD, Szostak JW, In vitro selection of RNA molecules that bind specific ligands, Nature, 346 (1990) 818–822. [DOI] [PubMed] [Google Scholar]
- [2].Tuerk C, Gold L, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase, Science, 249 (1990) 505–510. [DOI] [PubMed] [Google Scholar]
- [3].Huizenga DE, Szostak JW, A DNA Aptamer That Binds Adenosine and ATP, Biochemistry, 34 (1995) 656–665. [DOI] [PubMed] [Google Scholar]
- [4].Hermann T, Patel DJ, Adaptive Recognition by Nucleic Acid Aptamers, Science, 287 (2000) 820–825. [DOI] [PubMed] [Google Scholar]
- [5].Mallikaratchy P, Stahelin RV, Cao Z, Cho W, Tan W, Selection of DNA ligands for protein kinase C-delta, Chem. Commun, (2006) 3229–3231. [DOI] [PubMed] [Google Scholar]
- [6].Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L, A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment, Proc. Natl. Acad. Sci. U.S.A, 100 (2003) 15416–15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Parekh P, Tang Z, Turner PC, Moyer RW, Tan W, Aptamers Recognizing Glycosylated Hemagglutinin Expressed on the Surface of Vaccinia Virus-Infected Cells, Anal. Chem, 82 (2010) 8642–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ, Selection of single-stranded DNA molecules that bind and inhibit human thrombin, Nature, 355 (1992) 564–566. [DOI] [PubMed] [Google Scholar]
- [9].Hu J, Wu J, Li C, Zhu L, Zhang WY, Kong G, Lu Z, Yang CJ, A G-Quadruplex Aptamer Inhibits the Phosphatase Activity of Oncogenic Protein Shp2 in vitro, Chembiochem, 12 (2011) 424–430. [DOI] [PubMed] [Google Scholar]
- [10].Chen C.-h.B., Chernis GA, Hoang VQ, Landgraf R, Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3, Proc. Natl. Acad. Sci. U.S.A, 100 (2003) 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Keefe AD, Pai S, Ellington A, Aptamers as therapeutics, Nature Review Drug Discovery, 9 (2010) 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Guo K-T, SchÄfer R, Paul A, Gerber A, Ziemer G, Wendel HP, A New Technique for the Isolation and Surface Immobilization of Mesenchymal Stem Cells from Whole Bone Marrow Using High-Specific DNA Aptamers, Stem Cells, 24 (2006) 2220–2231. [DOI] [PubMed] [Google Scholar]
- [13].Shangguan D, Li Y, Tang Z, Cao Z, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W, Aptamers evolved from live cells as effective molecular probes for cancer study, Proc. Natl. Acad. Sci. U.S.A, 103 (2006) 11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikaratchy P, Chen HW, Li Y, Tan W, Selection of aptamers for molecular recognition and characterization of cancer cells, Anal. Chem, 79 (2007) 4900–4907. [DOI] [PubMed] [Google Scholar]
- [15].Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y, Cardona D, Witek RP, Liu C, Tan W, Identification of liver cancer-specific aptamers using whole live cells, Anal. Chem, 80 (2008) 721–728. [DOI] [PubMed] [Google Scholar]
- [16].Sefah K, Tang Z, Shangguan D, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips JA, Kim Y, Tan W, Molecular recognition of acute myeloid leukemia using aptamers, Leukemia, 23 (2009) 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, Smith JE, Tan W, Molecular recognition of small-cell lung cancer cells using aptamers, Chemmedchem, 3 (2008) 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA, Tan W, In Vitro Selection of DNA Aptamers to Glioblastoma Multiforme, ACS Chem. Neurosci, 2 (2011) 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Simaeys DV, López-Colón D, Sefah K, Sutphen R, Jimenez E, Tan W, Study of the Molecular Recognition of Aptamers Selected through Ovarian Cancer Cell-SELEX, Plos One, 5 (2010) e13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D, Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells, Analyst, 134 (2009) 1808–1814. [DOI] [PubMed] [Google Scholar]
- [21].Dunn M, Jimenez R, Chaput J, Analysis of aptamer discovery and technology, Nat. Rev. Chem, 1 (2017) 0076. [Google Scholar]
- [22].Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A, The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer, J. Biol. Chem, 268 (1993) 17651–17654. [DOI] [PubMed] [Google Scholar]
- [23].Long SB, Long MB, White RR, Sullenger BA, Crystal structure of an RNA aptamer bound to thrombin, RNA, 14 (2008) 2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lebars I, Legrand P, Aime A, Pinaud N, Fribourg S, Di Primo C, Exploring TAR-RNA aptamer loop-loop interaction by X-ray crystallography, UV spectroscopy and surface plasmon resonance, Nucleic Acids Res, 36 (2008) 7146–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, Ghosh G, Crystal structure of NF-kappa B (p50)(2) complexed to a high-affinity RNA aptamer, Proc. Natl. Acad. Sci. U.S.A, 100 (2003) 9268–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dellafiore MA, Montserrat JM, Iribarren AM, Modified Nucleoside Triphosphates for In-vitro Selection Techniques, Frontiers Chem., 4 (2016) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Singh Y, Murat P, Defrancq E, Recent developments in oligonucleotide conjugation, Chem. Soc. Rev, 39 (2010) 2054–2070. [DOI] [PubMed] [Google Scholar]
- [28].Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, Scardino E, Fay WP, Sullenger BA, Antidote-mediated control of an anticoagulant aptamer in vivo, Nat. Biotechnol, 22 (2004) 1423–1428. [DOI] [PubMed] [Google Scholar]
- [29].Oney S, Lam RTS, Bompiani KM, Blake CM, Quick G, Heidel JD, Liu JY-C, Mack BC, Davis ME, Leong KW, Sullenger BA, Development of universal antidotes to control aptamer activity, Nat. Med, 15 (2009) 1224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang D-LL, Song Y-LL, Zhu Z, Li X-LL, Zou Y, Yang H-TT, Wang J-JJ, Yao P-SS, Pan R-JJ, Yang CJ, Kang D-ZZ, Selection of DNA aptamers against epidermal growth factor receptor with high affinity and specificity, Biochem. Biophy. Res. Commun, 453 (2014) 681–685. [DOI] [PubMed] [Google Scholar]
- [31].Esposito CL, Passaro D, Longobardo I, Condorelli G, Marotta P, Affuso A, de Franciscis V, Cerchia L, A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death, PLoS ONE, 6 (2010) e24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li N, Nguyen HH, Byrom M, Ellington AD. Inhibition of Cell Proliferation by an Anti-EGFR Aptamer, Plos One, 6 (2011) e20299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhu G, Zhang H, Jacobson O, Wang Z, Chen H, Yang X, Niu G, Chen X, Combinatorial Screening of DNA Aptamers for Molecular Imaging of HER2 in Cancer, Bioconjug. Chem, 28 (2017) 1068–1075. [DOI] [PubMed] [Google Scholar]
- [34].Lupold SE, Hicke BJ, Lin Y, Coffey DS, Identification and Characterization of Nuclease-stabilized RNA Molecules That Bind Human Prostate Cancer Cells via the Prostate-specific Membrane Antigen, Cancer Res, 62 (2002) 4029–4033. [PubMed] [Google Scholar]
- [35].Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W, Cell-specific aptamer probes for membrane protein elucidation in cancer cells,J. Proteome Res, 7 (2008) 2133–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gragoudas ES, Adamis AP, Cunningham ET, Feinsod M, Guyer DR, Pegaptanib for Neovascular AgeRelated Macular Degeneration, N. Engl. J. Med, 351 (2004) 2805–2816. [DOI] [PubMed] [Google Scholar]
- [37].Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO, Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer, Exp. Mol. Pathol, 86 (2009) 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mosing RK, Mendonsa SD, Bowser MT, Capillary Electrophoresis-SELEX Selection of Aptamers with Affinity for HIV-1 Reverse Transcriptase, Anal. Chem, 77 (2005) 6107–6112. [DOI] [PubMed] [Google Scholar]
- [39].Mosing RK, Bowser MT, Microfluidic selection and applications of aptamers, J. Separation Sci, 30 (2007) 1420–1426. [DOI] [PubMed] [Google Scholar]
- [40].Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W, Development of DNA aptamers using CellSELEX, Nat. Protoc, 5 (2009) 1169–1185. [DOI] [PubMed] [Google Scholar]
- [41].Mayer G, Ahmed M-SL, Dolf A, Endl E, Knolle PA, Famulok M, Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures, Nat. Protoc, 5 (2010) 1993–2004. [DOI] [PubMed] [Google Scholar]
- [42].Cox JC, Rudolph P, Ellington AD, Automated RNA Selection, Biotechnol. Prog, 14 (1998) 845–850. [DOI] [PubMed] [Google Scholar]
- [43].Kruspe S, Meyer C, Hahn U, Chlorin e6 conjugated interleukin-6 receptor aptamers selectively kill target cells upon irradiation, Mol. Ther. Nucleic Acids, 3 (2014) e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shamah SM, Healy JM, Cload ST, Complex Target SELEX, Acc. Chem. Res, 41 (2008) 130–138. [DOI] [PubMed] [Google Scholar]
- [45].Fang X, Tan W, Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach, Acc. Chem. Res, 43 (2009) 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Blank M, Weinschenk T, Priemer M, Schluesener H, Systematic Evolution of a DNA Aptamer Binding to Rat Brain Tumor Microvessels, J. Biol. Chem, 276 (2001) 16464–16468. [DOI] [PubMed] [Google Scholar]
- [47].Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM, In vivo selection of tumor-targeting RNA motifs, Nat. Chem. Biol, 6 (2010) 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tang Z, Parekh P, Turner P, Moyer RW, Tan W, Generating Aptamers for Recognition of Virus-Infected Cells, Clin. Chem, 55 (2009) 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shangguan D, Cao Z, Li Y, Tan W, Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples, Clin. Chem, 53 (2007) 1153–1155. [DOI] [PubMed] [Google Scholar]
- [50].Berezovski MV, Lechmann M, Musheev MU, Mak TW, Krylov SN, Aptamer-Facilitated Biomarker Discovery (AptaBiD), J. Am. Chem. Soc, 130 (2008) 9137–9143. [DOI] [PubMed] [Google Scholar]
- [51].Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W, Aptamer Directly Evolved from Live Cells Recognizes Membrane Bound Immunoglobin Heavy Mu Chain in Burkitt’s Lymphoma Cells, Mol. Cell. Proteomics, 6 (2007) 2230–2238. [DOI] [PubMed] [Google Scholar]
- [52].Ferreira CSM, Matthews CS, Missailidis S, DNA aptamers that bind to MUC1 tumour marker: Design and characterization of MUC1-binding single-stranded DNA aptamers, Tumor Biol., 27 (2006) 289–301. [DOI] [PubMed] [Google Scholar]
- [53].Mi J, Zhang X, Giangrande PH, McNamara Ii JO, Nimjee SM, Sarraf-Yazdi S, Sullenger BA, Clary BM, Targeted inhibition of αvβ3 integrin with an RNA aptamer impairs endothelial cell growth and survival, Biochem. Biophys. Res. Commun, 338 (2005) 956–963. [DOI] [PubMed] [Google Scholar]
- [54].Lebruska LL, Maher LJ, Selection and Characterization of an RNA Decoy for Transcription Factor NF-κB†, Biochem., 38 (1999) 3168–3174. [DOI] [PubMed] [Google Scholar]
- [55].Martell RE, Nevins JR, Sullenger BA, Optimizing Aptamer Activity for Gene Therapy Applications Using Expression Cassette SELEX, Mol. Ther, 6 (2002) 30–34. [DOI] [PubMed] [Google Scholar]
- [56].Ishizaki J, Nevins JR, Sullenger BA, Inhibition of cell proliferation by an RNA ligand that selectively blocks E2F function, Nat. Med, 2 (1996) 1386–1389. [DOI] [PubMed] [Google Scholar]
- [57].Chi-hong BC, George AC, Van QH, Ralf L, Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3, Proc. Natl. Acad. Sci. U.S.A, 100 (2003) 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, Giangrande PH, Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers, Nucleic Acids Res, 40 (2012) 6319–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Varmira K, Hosseinimehr SJ, Noaparast Z, Abedi SM, A HER2-targeted RNA aptamer molecule labeled with 99mTc for single-photon imaging in malignant tumors, Nucl. Med. Biol, 40 (2013) 980–986. [DOI] [PubMed] [Google Scholar]
- [60].Varmira K, Hosseinimehr SJ, Noaparast Z, Abedi SM, An improved radiolabelled RNA aptamer molecule for HER2 imaging in cancers, J. Drug Targeting, 22 (2014) 116–122. [DOI] [PubMed] [Google Scholar]
- [61].Song Y, Zhu Z, An Y, Zhang W, Zhang H, Liu D, Yu C, Duan W, Yang CJ, Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture, Anal. Chem, 85 (2013) 4141–4149. [DOI] [PubMed] [Google Scholar]
- [62].Xiang D, Shigdar S, Qiao G, Wang T, Kouzani AZ, Zhou S-FF, Kong L, Li Y, Pu C, Duan W, Nucleic acid aptamer-guided cancer therapeutics and diagnostics: the next generation of cancer medicine, Theranostics, 5 (2014) 23–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang P, Zhao N, Zeng Z, Feng Y, Tung C-HH, Chang C-CC, Zu Y, Using an RNA aptamer probe for flow cytometry detection of CD30-expressing lymphoma cells, Lab. Invest, 89 (2009) 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Khati M, Schüman M, Ibrahim J, Sattentau Q, Gordon S, James W, Neutralization of Infectivity of Diverse R5 Clinical Isolates of Human Immunodeficiency Virus Type 1 by gp120-Binding 2’ F-RNA Aptamers, J. Virol, 77 (2003) 12692–12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wiegand T, Williams P, Dreskin S, Jouvin M, Kinet J, Tasset D, High-affinity oligonucleotide ligands to human IgE inhibit binding to Fc epsilon receptor I, J. Immunol, 157 (1996) 221–230. [PubMed] [Google Scholar]
- [66].Liang C, Guo B, Wu H, Shao N, Li D, Liu J, Dang L, Wang C, Li H, Li S, Lau WK, Cao Y, Yang Z, Lu C, He X, Au DW, Pan X, Zhang B-TT, Lu C, Zhang H, Yue K, Qian A, Shang P, Xu J, Xiao L, Bian Z, Tan W, Liang Z, He F, Zhang L, Lu A, Zhang G, Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy, Nat. Med, 21 (2015) 288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen C.-h.B.H., Dellamaggiore KR, Ouellette CP, Sedano CD, Lizadjohry M, Chernis GA, Gonzales M, Baltasar FE, Fan AL, Myerowitz R, Neufeld EF, Aptamer-based endocytosis of a lysosomal enzyme, Proc. Natl. Acad. Sci. U.S.A, 105 (2008) 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Benaduce A, Brenneman R, Schrand B, Pollack A, Gilboa E, Ishkanian A, 4–1BB Aptamer-Based Immunomodulation Enhances the Therapeutic Index of Radiation Therapy in Murine Tumor Models, Int. J. Radiat. Oncol. Biol. Phys, 96 (2016) 458–461. [DOI] [PubMed] [Google Scholar]
- [69].Huang B-T, Lai W-Y, Chang Y-C, Wang J-W, Yeh S-D, Lin E, Yang P-C, A CTLA-4 Antagonizing DNA Aptamer with Antitumor Effect, Mol. Ther, 8 (2017) 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Herrmann A, Priceman SJ, Swiderski P, Kujawski M, Xin H, Cherryholmes GA, Zhang W, Zhang C, Lahtz C, Kowolik C, Forman SJ, Kortylewski M, Yu H, CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells, J. Clin. Invest, 124 (2014) 2977–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Soldevilla M, Villanueva H, Bendandi M, Inoges S, de Cerio A, Pastor F, 2-fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia, Biomaterials, 67 (2015) 274–285. [DOI] [PubMed] [Google Scholar]
- [72].Pratico ED, Sullenger BA, Nair SK, Identification and Characterization of an Agonistic Aptamer Against the T Cell Costimulatory Receptor, OX40, Nucleic Acid Ther., 23 (2013) 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Prodeus A, Abdul-Wahid A, Fischer NW, Huang EH, Cydzik M, Gariépy J, Targeting the PD-1/PD-L1 Immune Evasion Axis With DNA Aptamers as a Novel Therapeutic Strategy for the Treatment of Disseminated Cancers, Mol. Ther, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lai W-Y, Huang B-T, Wang J-W, Lin P-Y, Yang P-C, A Novel PD-L1-targeting Antagonistic DNA Aptamer With Antitumor Effects, Mol. Ther, 4 (2015) e237. [DOI] [PubMed] [Google Scholar]
- [75].Gefen T, Castro I, Muharemagic D, Puplampu-Dove Y, Patel S, Gilboa E, A TIM-3 Oligonucleotide Aptamer Enhances T Cell Functions and Potentiates Tumor Immunity in Mice, Mol. Ther, 25 (2017) 2280–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Soldevilla M, Hervas S, Villanueva H, Lozano T, Rabal O, Oyarzabal J, Lasarte J, Bendandi M, Inoges S, de Cerio A, Pastor F, Identification of LAG3 high affinity aptamers by HT-SELEX and Conserved Motif Accumulation (CMA), PLOS ONE, 12 (2017) e0185169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lozano T, Soldevilla M, Casares N, Villanueva H, Bendandi M, Lasarte J, Pastor F, Targeting inhibition of Foxp3 by a CD28 2′-Fluro oligonucleotide aptamer conjugated to P60-peptide enhances active cancer immunotherapy, Biomaterials, 91 (2016) 73–80. [DOI] [PubMed] [Google Scholar]
- [78].Wengerter BC, Katakowski JA, Rosenberg JM, Park CG, Almo SC, Palliser D, Levy M, Aptamertargeted antigen delivery, Mol. Ther, 22 (2014) 1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Liu Y-J, Dou X-Q, Wang F, Zhang J, Wang X-L, Xu G-L, Xiang S-S, Gao X, Fu J, Song H-F, IL-4Rα aptamer-liposome-CpG oligodeoxynucleotides suppress tumour growth by targeting the tumour microenvironment, J. Drug Targeting, (2016) 1–26. [DOI] [PubMed] [Google Scholar]
- [80].Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA, RNA aptamers as reversible antagonists of coagulation factor IXa, Nature, 419 (2002) 90–94. [DOI] [PubMed] [Google Scholar]
- [81].Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, Scardino E, Fay WP, Sullenger BA, Antidote-mediated control of an anticoagulant aptamer in vivo, Nat. Biotechnol, 22 (2004) 1423–1428. [DOI] [PubMed] [Google Scholar]
- [82].Blake CM, Wang H, Laskowitz DT, Sullenger BA, A reversible aptamer improves outcome and safety in murine models of stroke and hemorrhage, Oligonucleotides, 21 (2010) 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lincoff MA, Mehran R, Povsic TJ, Zelenkofske SL, Huang Z, Armstrong PW, Steg GP, Bode C, Cohen MG, Buller C, Laanmets P, Valgimigli M, Marandi T, Fridrich V, Cantor WJ, Merkely B, Lopez-Sendon J, Cornel JH, Kasprzak JD, Aschermann M, Guetta V, Morais J, Sinnaeve PR, Huber K, Stables R, Sellers M, Borgman M, Glenn L, Levinson AI, Lopes RD, Hasselblad V, Becker RC, Alexander JH, Investigators R-P, Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): a randomised clinical trial, Lancet, 387 (2016) 349–356. [DOI] [PubMed] [Google Scholar]
- [84].Blake CM, Sullenger BA, Lawrence DA, Fortenberry YM, Antimetastatic Potential of PAI-1-Specific RNA Aptamers, Oligonucleotides, 19 (2009) 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Tan W, Donovan MJ, Jiang J, Aptamers from Cell-Based Selection for Bioanalytical Applications, Chem. Rev, 113 (2013) 2842–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, Kim N, Saccomano NA, Wilcox SK, Zichi D, Sanders GM, From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay, PloS one, 6 (2011) e26332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gold L, Walker JJ, Wilcox SK, Williams S, Advances in human proteomics at high scale with the SOMAscan proteomics platform, N. Biotechnol, 29 (2012) 543–549. [DOI] [PubMed] [Google Scholar]
- [88].Huang J, Zhu Z, Bamrungsap S, Zhu G, You M, He X, Wang K, Competition-Mediated Pyrene-Switching Aptasensor: Probing Lysozyme in Human Serum with a Monomer-Excimer Fluorescence Switch, Anal. Chem, 82 (2010) 10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kellenberger C, Chen C, Whiteley A, Portnoy D, Hammond M, RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic di-AMP, J. Am. Chem. Soc, 137 (2015) 6432–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Phillips JA, Xu Y, Xia Z, Fan ZH, Tan W, Enrichment of cancer cells using aptamers immobilized on a microfluidic channel, Anal. Chem, 81 (2009) 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sheng W, Chen T, Kamath R, Xiong X, Tan W, Fan ZH, Aptamer-enabled efficient isolation of cancer cells from whole blood using a microfluidic device, Anal. Chem, 84 (2012) 4199–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W, Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells, Anal. Chem, 81 (2009) 7436–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wang AZ, Farokhzad OC, Current progress of aptamer-based molecular imaging, J. Nucl. Med, 55 (2014) 353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Shi H, He X, Wang K, Wu X, Ye X, Guo Q, Tan W, Qing Z, Yang X, Zhou B, Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration, Proc. Natl. Acad. Sci. U.S.A, 108 (2011) 3900–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhang J, Smaga LP, Satyavolu NSRSR, Chan J, Lu Y, DNA Aptamer-Based Activatable Probes for Photoacoustic Imaging in Living Mice, J. Am. Chem. Soc, 139 (2017) 17225–17228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hicke BJ, Stephens AW, Gould T, Chang Y-F, Lynott CK, Heil J, Borkowski S, Hilger C-S, Cook G, Warren S, Schmidt PG, Tumor Targeting by an Aptamer, J. Nucl. Med, 47 (2006) 668–678. [PubMed] [Google Scholar]
- [97].Jacobson O, Yan X, Niu G, Weiss ID, Ma Y, Szajek LP, Shen B, Kiesewetter DO, Chen X, PET imaging of tenascin-C with a radiolabeled single-stranded DNA aptamer, J. Nucl. Med, 56 (2015) 616–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Jacobson O, Weiss I, Wang L, Wang Z, Yang X, Dewhurst A, Ma Y, Zhu G, Niu G, Kiesewetter DO, Vasdev N, Liang S, Chen X, 18F-labeled Single-Stranded DNA Aptamer for PET Imaging of Protein Tyrosine Kinase7 Expression, J. Nucl. Med, 56 (2015) 1780–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Roy K, Kanwar RK, Kanwar JR, LNA aptamer based multi-modal, Fe3O4-saturated lactoferrin (Fe3O4-bLf) nanocarriers for triple positive (EpCAM, CD133, CD44) colon tumor targeting and NIR, MRI and CT imaging, Biomaterials, 71 (2015) 84–99. [DOI] [PubMed] [Google Scholar]
- [100].Wang AZ, Bagalkot V, Vasilliou CC, Gu F, Alexis F, Zhang L, Shaikh M, Yuet K, Cima MJ, Langer R, Kantoff PW, Bander NH, Jon S, Farokhzad OC, Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for combined prostate cancer imaging and therapy, ChemMedChem, 3 (2008) 1311–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Green LS, Bell C, Janjic N, Aptamers as reagents for high-throughput screening, Biotechniques, 30 (2001) 1094–1096. [DOI] [PubMed] [Google Scholar]
- [102].Srivatsan SG, Famulok M, Functional nucleic acids in high throughput screening and drug discovery, Comb. Chem. High Throughput Screen, 10 (2007) 698–705. [DOI] [PubMed] [Google Scholar]
- [103].Yamazaki S, Tan L, Mayer G, Hartig JS, Song J-N, Reuter S, Restle T, Laufer SD, Grohmann D, Kraeusslich H-G, Bajorath J, Famulok M, Alternative small-molecule target sites aptamer displacement identifies that escape viral resistance, Chem. Biol, 14 (2007) 804–812. [DOI] [PubMed] [Google Scholar]
- [104].Hafner M, Vianini E, Albertoni B, Marchetti L, Gruene I, Gloeckner C, Famulok M, Displacement of protein-bound aptamers with small molecules screened by fluorescence polarization, Nat. Protoc, 3 (2008) 579–587. [DOI] [PubMed] [Google Scholar]
- [105].Famulok M, Exploring Chemical Space with Aptamers, J. Med. Chem, 52 (2009) 6951–6957. [DOI] [PubMed] [Google Scholar]
- [106].Mayer G, Faulhammer D, Graettinger M, Fessele S, Blind M, A RNA-Based Approach towards SmallMolecule Inhibitors, ChemBioChem, 10 (2009) 1993–1996. [DOI] [PubMed] [Google Scholar]
- [107].Yamazaki S, Famulok M, Screening of Novel Inhibitors of HIV-1 Reverse Transcriptase with a Reporter Ribozyme Assay, in: Mayer G (Ed.) Methods Mol. Biol 2009, pp. 187–199. [DOI] [PubMed] [Google Scholar]
- [108].Niebel B, Lentz C, Pofahl M, Mayer G, Hoerauf A, Pfarr KM, Famulok M, ADLOC: An Aptamer-Displacement Assay Based on Luminescent Oxygen Channeling, Chem. Eur. J, 16 (2010) 11100–11107. [DOI] [PubMed] [Google Scholar]
- [109].Auslaender D, Wieland M, Auslaender S, Tigges M, Fussenegger M, Rational design of a small moleculeresponsive intramer controlling transgene expression in mammalian cells, Nucleic Acids Res, 39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Mueller M, Ackermann D, Famulok M, Nucleic acid based tools for pharmacology and nano-engineering, C. R. Chim, 14 (2011) 819–825. [Google Scholar]
- [111].Paige JS, Wu KY, Jaffrey SR, RNA mimics of green fluorescent protein, Science, 333 (2011). 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Filonov GS, Moon JD, Svensen N, Jaffrey SR, Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution, J. Am. Chem. Soc, 136 (2014) 16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Paige JS, Nguyen-Duc T, Song W, Jaffrey SR, Fluorescence imaging of cellular metabolites with RNA, Science, 335 (2012) 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Song W, Strack RL, Jaffrey SR, Imaging bacterial protein expression using genetically encoded RNA sensors, Nat. Meth, 10 (2013) 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Shin W-S, Maeng Y-S, Jung J-W, Min J-K, Kwon Y-G, Lee S-T, Soluble PTK7 inhibits tube formation, migration, and invasion of endothelial cells and angiogenesis, Biochem. Biophys. Res. Commun, 371 (2008) 793–798. [DOI] [PubMed] [Google Scholar]
- [116].Ireson CR, Kelland LR, Discovery and development of anticancer aptamers, Mol. Cancer Ther, 5 (2006) 2957–2962. [DOI] [PubMed] [Google Scholar]
- [117].Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM, Antiproliferative activity of G-rich oligonucleotides correlates with protein binding, J. Biol. Chem, 274 (1999) 26369–26377. [DOI] [PubMed] [Google Scholar]
- [118].Soundararajan S, Wang L, Sridharan V, Chen W, Courtenay-Luck N, Jones D, Spicer EK, Fernandes DJ, Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4–11 leukemia cells, Mol. Pharmacol, 76 (2009) 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ, The Nucleolin Targeting Aptamer AS1411 Destabilizes Bcl-2 Messenger RNA in Human Breast Cancer Cells, Cancer Res, 68 (2008) 2358–2365. [DOI] [PubMed] [Google Scholar]
- [120].McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E, Multivalent 4–1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice, J. Clin. Invest, 118 (2008) 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Sullenger BA, Gallardo HF, Ungers GE, Gilboa E, Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication, Cell, 63 (1990) 601–608. [DOI] [PubMed] [Google Scholar]
- [122].Kolb G, Reigadas S, Castanotto D, Faure A, Ventura M, Rossi JJ, Toulme JJ, Endogenous Expression of an Anti-TAR Aptamer Reduces HIV-1 Replication, RNA Biol, 3 (2006) 150–156. [DOI] [PubMed] [Google Scholar]
- [123].Dyke CK, Steinhubl SR, Kleiman NS, Cannon RO, Aberle LG, Lin M, Myles SK, Melloni C, Harrington RA, Alexander JH, Becker RC, Rusconi CP, First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of Factor IXa activity, Circulation, 114 (2006) 2490–2497. [DOI] [PubMed] [Google Scholar]
- [124].Nimjee SM, Lohrmann JD, Wang H, Snyder DJ, Cummings TJ, Becker RC, Oney S, Sullenger BA, Rapidly regulating platelet activity in vivo with an antidote controlled platelet inhibitor, Mol. Ther, 20 (2011) 391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Hughes B, Antibody-drug conjugates for cancer: poised to deliver?, Nat. Rev. Drug Discov, 9 (2010) 665–667. [DOI] [PubMed] [Google Scholar]
- [126].Webb S, Pharma interest surges in antibody drug conjugates, Nat. Biotechnol, 29 (2011) 297–298. [DOI] [PubMed] [Google Scholar]
- [127].Hu R, Zhang X, Zhao Z, Zhu G, Chen T, Fu T, Tan W, DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery, Angew. Chem. Int. Ed, 53 (2014) 5821–5826. [DOI] [PubMed] [Google Scholar]
- [128].Huang F, You M, Chen T, Zhu G, Liang H, Tan W, Self-assembled hybrid nanoparticles for targeted codelivery of two drugs into cancer cells, Chem. Commun, 50 (2014) 3103–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wang J, Zhu G, You M, Song E, Shukoor MI, Zhang K, Altman MB, Chen Y, Zhu Z, Huang CZ, Tan W, Assembly of aptamer switch probes and photosensitizer on gold nanorods for targeted photothermal and photodynamic cancer therapy, ACS Nano, 6 (2012) 5070–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Zheng J, Zhu G, Li Y, Li C, You M, Chen T, Song E, Yang R, Tan W, A spherical nucleic acid platform based on self-assembled DNA biopolymer for high-performance cancer therapy, ACS Nano, 7 (2013) 6545–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zhou C, Chen T, Wu C, Zhu G, Qiu L, Cui C, Hou W, Tan W, Aptamer CaCO3 nanostructures: a facile, pH-responsive, specific platform for targeted anticancer theranostics, Chem. Asian J, 10 (2014) 166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Zhu G, Hu R, Zhao Z, Chen Z, Zhang X, Tan W, Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications, J. Am. Chem. Soc, 135 (2013) 16438–16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Zhu G, Zheng J, Song E, Donovan M, Zhang K, Liu C, Tan W, Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics, Proc. Natl. Acad. Sci. U.S.A, 110 (2013) 7998–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Ryou S-MM, Yeom J-HH, Kang HJ, Won M, Kim J-SS, Lee B, Seong M-JJ, Ha N-CC, Bae J, Lee K, Gold nanoparticle-DNA aptamer composites as a universal carrier for in vivo delivery of biologically functional proteins, J Control. Release, 196 (2014) 287–294. [DOI] [PubMed] [Google Scholar]
- [135].Shieh Y-A, Yang S-J, Wei M-F, Shieh M-J, Aptamer-based tumor-targeted drug delivery for photodynamic therapy, ACS Nano, 4 (2010) 1433–1442. [DOI] [PubMed] [Google Scholar]
- [136].Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC, Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers, Proc. Natl. Acad. Sci. U.S.A, 105 (2008) 2586–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Huang Y-F, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y, Tan W, Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells, ChemBioChem, 10 (2009) 862–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Meng L, Sefah K, O’Donoghue MB, Zhu G, Shangguan D, Noorali A, Chen Y, Zhou L, Tan W, Silencing of PTK7 in colon cancer cells: caspase-10-dependent apoptosis via mitochondrial pathway, Plos One, 5 (2010) e14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M, Qing F-L, Tan W, Automated modular synthesis of aptamerdrug conjugates for targeted drug delivery, J. Am. Chem. Soc, 136 (2014) 2731–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Zhu G, Cansiz S, You M, Qiu L, Han D, Zhang L, Mei L, Fu T, Chen Z, Tan W, Nuclease-resistant synthetic drug-DNA adducts: programmable drug-DNA conjugation for targeted anticancer drug delivery, NPG Asia Mater, 7 (2015) e169 [Google Scholar]
- [141].Wolf E, Hofmeister R, Kufer P, Schlereth B, Baeuerle PA, BiTEs: bispecific antibody constructs with unique anti-tumor activity, Drug Discov. Today, 10 (2005) 1237–1244. [DOI] [PubMed] [Google Scholar]
- [142].Beck A, Wurch T, Bailly C, Corvaia N, Strategies and challenges for the next generation of therapeutic antibodies, Nat. Rev. Immunol, 10 (2010) 345–352. [DOI] [PubMed] [Google Scholar]
- [143].Segal DM, Weiner GJ, opinion in immunology W-LM, Bispecific antibodies in cancer therapy, Curr. Opin. Immunol, 11 (1999) 558–562.. [DOI] [PubMed] [Google Scholar]
- [144].Baeuerle PA, Reinhardt C, Bispecific T-cell engaging antibodies for cancer therapy, Cancer Res, 69 (2009) 4941–4944. [DOI] [PubMed] [Google Scholar]
- [145].Nguyen DN, Mahon KP, Chikh G, Kim P, Chung H, Vicari AP, Love KT, Goldberg M, Chen S, Krieg AM, Chen J, Langer R, Anderson DG, Lipid-derived nanoparticles for immunostimulatory RNA adjuvant delivery, Proc. Natl. Acad. Sci. U.S.A, 109 (2012) E797–E803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Cao Z, Tong R, Mishra A, Xu W, Wong GC, Cheng J, Lu Y, Reversible cell-specific drug delivery with aptamer-functionalized liposomes, Angew. Chem. Int. Ed, 48 (2008) 6494–6498. [DOI] [PubMed] [Google Scholar]
- [147].Kang H, O’Donoghue MB, Liu H, Tan W, A liposome-based nanostructure for aptamer directed delivery, Chem. Commun, 46 (2010) 249–251. [DOI] [PubMed] [Google Scholar]
- [148].Alshaer W, Hillaireau H, Vergnaud J, Ismail S, Fattal E, Functionalizing Liposomes with anti-CD44 Aptamer for Selective Targeting of Cancer Cells, Bioconjug. Chem, 26 (2014) 1307–1313. [DOI] [PubMed] [Google Scholar]
- [149].Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ, Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles, Proc. Natl. Acad. Sci. U.S.A, 105 (2008) 17356–17361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC, Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo, Proc. Natl. Acad. Sci. U.S.A, 108 (2011) 1850–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R, Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo, Proc. Natl. Acad. Sci. U.S.A, 103 (2006) 6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Farokhzad OC, Karp JM, Langer R, Nanoparticle-aptamer bioconjugates for cancer targeting, Expert Opin. Drug Deliv, 3 (2006) 311–324. [DOI] [PubMed] [Google Scholar]
- [153].Farokhzad OC, Jon S, Khademhosseini A, Tran T-NTN, Lavan DA, Langer R, Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells, Cancer Res, 64 (2004) 7668–7672. [DOI] [PubMed] [Google Scholar]
- [154].Wu Y, Zhang L, Cui C, Cansiz S, Liang H, Wu C, Teng IT, Chen W, Liu Y, Hou W, Enhanced Targeted Gene Transduction: AAV2 Vectors Conjugated to Multiple Aptamers via Reducible Disulfide Linkages, J. Am. Chem. Soc, 140 (2017) 2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Tong GJ, Hsiao SC, Carrico ZM, Francis MB, Viral Capsid DNA Aptamer Conjugates as Multivalent Cell-Targeting Vehicles, J. Am. Chem. Soc, 131 (2009) 11174–11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Li X, Zhao Q, Qiu L, Smart ligand: aptamer-mediated targeted delivery of chemotherapeutic drugs and siRNA for cancer therapy, J Control. Release, 171 (2013) 152–162. [DOI] [PubMed] [Google Scholar]
- [157].Hong Yan L, Xiaohu G, A Universal Protein Tag for Delivery of SiRNA-Aptamer Chimeras, Sci. Rep, 3 (2013) 3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH, Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras, Nat. Biotechnol, 24 (2006) 1005–1015. [DOI] [PubMed] [Google Scholar]
- [159].Dassie JP, Liu X-YY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, Giangrande PH, Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors, Nat. Biotechnol, 27 (2009) 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Povsic TJ, Cohen MG, Mehran R, Buller CE, Bode C, Cornel JH, Kasprzak JD, Montalescot G, Joseph D, Wargin WA, Rusconi CP, Zelenkofske SL, Becker RC, Alexander JH, A randomized, partially blinded, multicenter, active-controlled, dose-ranging study assessing the safety, efficacy, and pharmacodynamics of the REG1 anticoagulation system in patients with acute coronary syndromes: Design and rationale of the RADAR Phase IIb trial, Am. Heart J, 161 (2011) 261–26800. [DOI] [PubMed] [Google Scholar]
- [161].Song P, Chou YK, Zhang X, Meza-Romero R, Yomogida K, Benedek G, Chu C-QQ, CD4 aptamer-RORγt shRNA chimera inhibits IL-17 synthesis by human CD4(+) T cells, Biochem. Biophys. Res. Commun, 452 (2014) 1040–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Subramanian N, Kanwar JR, Athalya P, Janakiraman N, Khetan V, Kanwar RK, Eluchuri S, Krishnakumar S, EpCAM aptamer mediated cancer cell specific delivery of EpCAM siRNA using polymeric nanocomplex, J. Biomed. Sci, 22 (2015) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Shu D, Shu Y, Haque F, Abdelmawla S, Guo P, Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nat. Nanotechnol, 6 (2011) 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Zhou J, Li H, Li S, Zaia J, Rossi JJ, Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy, Mol. Ther, 16 (2008) 1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zhou J, Rossi JJ, Aptamer-targeted RNAi for HIV-1 therapy, Meth. Mol. Biol, 721 (2010) 355–371. [DOI] [PubMed] [Google Scholar]
- [166].Schumacher TN, Schreiber RD, Neoantigens in cancer immunotherapy, Science, 348 (2015) 69–74. [DOI] [PubMed] [Google Scholar]
- [167].Sharma P, Allison JP, The future of immune checkpoint therapy, Science, 348 (2015) 56–61. [DOI] [PubMed] [Google Scholar]
- [168].Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon R-AA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M, Nivolumab plus ipilimumab in advanced melanoma, N. Engl. J. Med, 369 (2013) 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Rosenberg SA, Raising the bar: the curative potential of human cancer immunotherapy, Sci. Transl. Med, 4 (2012) 127–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Hervas-Stubbs S, Soldevilla MM, Villanueva H, Mancheño U, Bendandi M, Pastor F, Identification of TIM3 2’-fluoro oligonucleotide aptamer by HT-SELEX for cancer immunotherapy, Oncotarget, 7 (2015) 4522–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Schrand B, Berezhnoy A, Brenneman R, Williams A, Levay A, Gilboa E, Reducing toxicity of 4–1BB costimulation: targeting 4–1BB ligands to the tumor stroma with bi-specific aptamer conjugates, OncoImmunol., 4 (2015) e970918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Schrand B, Berezhnoy A, Brenneman R, Williams A, Levay A, Kong L-Y, Rao G, Zhou S, Heimberger AB, Gilboa E, Targeting 4–1BB Costimulation to the Tumor Stroma with Bispecific Aptamer Conjugates Enhances the Therapeutic Index of Tumor Immunotherapy, Cancer Immunol, 2 (2014) 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Hua Y, Drew P, Richard J, STATs in cancer inflammation and immunity: a leading role for STAT3, Nat. Rev. Cancer, 9 (2009) 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Pashenkov M, Goëss G, Wagner C, Hörmann M, Jandl T, Moser A, Britten CM, Smolle J, Koller S, Mauch C, Tantcheva-Poor I, Grabbe S, Loquai C, Esser S, Franckson T, Schneeberger A, Haarmann C, Krieg AM, Stingl G, Wagner SN, Phase II trial of a toll-like receptor 9-activating oligonucleotide in patients with metastatic melanoma, J. Clin. Oncol, 24 (2006) 5716–5724. [DOI] [PubMed] [Google Scholar]
- [175].Alshamsan A, Hamdy S, Samuel J, El-Kadi AO, Lavasanifar A, Uludağ H, The induction of tumor apoptosis in B16 melanoma following STAT3 siRNA delivery with a lipid-substituted polyethylenimine, Biomaterials, 31 (2010) 1420–1428. [DOI] [PubMed] [Google Scholar]
- [176].Hua Y, Heehyoung L, Andreas H, Ralf B, Richard J, Revisiting STAT3 signalling in cancer: new and unexpected biological functions, Nat. Rev. Cancer, 14 (2014) 736–746. [DOI] [PubMed] [Google Scholar]
- [177].Cheng F, Wang H-WW, Cuenca A, Huang M, Ghansah T, Brayer J, Kerr WG, Takeda K, Akira S, Schoenberger SP, Yu H, Jove R, Sotomayor EM, A critical role for Stat3 signaling in immune tolerance, Immunity, 19 (2003) 425–436. [DOI] [PubMed] [Google Scholar]
- [178].Zhang L, Alizadeh D, Van Handel M, Kortylewski M, Yu H, Badie B, Stat3 inhibition activates tumor macrophages and abrogates glioma growth in mice, Glia, 57 (2009) 1458–1467. [DOI] [PubMed] [Google Scholar]
- [179].Forman S, Rossi JJ, Pardoll DM, Jove R, Yu H, In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses, Nat. Biotechnol, 27 (2009). 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Kim JH, Noh Y-W, Heo MB, Cho MY, Lim YT, Multifunctional Hybrid Nanoconjugates for Efficient In Vivo Delivery of Immunomodulating Oligonucleotides and Enhanced Antitumor Immunity, Angew. Chem. Int. Ed, 51 (2012) 9670–9673. [DOI] [PubMed] [Google Scholar]
- [181].Kortylewski M, Kujawski M, Herrmann A, Yang C, Wang L, Liu Y, Salcedo R, Yu H, Toll-like receptor 9 activation of signal transducer and activator of transcription 3 constrains its agonist-based immunotherapy, Cancer Res, 69 (2009) 2497–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Berezhnoy A, Castro I, Levay A, Malek TR, Gilboa E, Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity, J. Clin. Invest, 124 (2014) 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Rajagopalan A, Berezhnoy A, Schrand B, Puplampu-Dove Y, Gilboa E, Aptamer-Targeted Attenuation of IL-2 Signaling in CD8+ T Cells Enhances Antitumor Immunity, Mol. Ther, 25 (2017) 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Gilboa E, Berezhnoy A, Schrand B, Reducing Toxicity of Immune Therapy Using Aptamer-Targeted Drug Delivery, Cancer Immunol. Res, 3 (2015) 1195–1200. [DOI] [PubMed] [Google Scholar]
- [185].Keler T, He L, Ramakrishna V, Champion B, Antibody-targeted vaccines, Oncogene, 26 (2007) 3758–3767. [DOI] [PubMed] [Google Scholar]
- [186].Irvine DJ, Hanson MC, Rakhra K, Tokatlian T, Synthetic Nanoparticles for Vaccines and Immunotherapy, Chem. Rev, 115 (2015) 11109–11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Zhu G, Zhang F, Ni Q, Niu G, Chen X, Efficient Nanovaccine Delivery in Cancer Immunotherapy, ACS Nano, 11 (2017) 2387–2392. [DOI] [PubMed] [Google Scholar]
- [188].Zhu G, Mei L, Vishwasrao HD, Jacobson O, Wang Z, Liu Y, Yung BC, Fu X, Jin A, Niu G, Wang Q, Zhang F, Shroff H, Chen X, Intertwining DNA-RNA nanocapsules loaded with tumor neoantigens as synergistic nanovaccines for cancer immunotherapy, Nat. Commun, 8 (2017) 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Chen H, Zhang W, Zhu G, Xie J, Chen X, Rethinking cancer nanotheranostics, Nat. Rev. Mater, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Zhu G, Liu Y, Yang X, Kim Y-HH, Zhang H, Jia R, Liao H-SS, Jin A, Lin J, Aronova M, Leapman R, Nie Z, Niu G, Chen X, DNA-inorganic hybrid nanovaccine for cancer immunotherapy, Nanoscale, 8 (2016) 6684–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Alcindor T, Witzig TE, Radioimmunotherapy with yttrium-90 ibritumomab tiuxetan for patients with relapsed CD20+ B-cell non-Hodgkin’s lymphoma, Curr. Treat. Options Oncol, 3 (2002) 275–282. [DOI] [PubMed] [Google Scholar]
- [192].Bandekar A, Zhu C, Anti-prostate-specific membrane antigen liposomes loaded with 225Ac for potential targeted antivascular α-particle therapy of cancer, J. Nucl. Med, 55 (2014) 107–114. [DOI] [PubMed] [Google Scholar]
- [193].Mallikaratchy P, Tang Z, Tan W, Cell specific aptamer-photosensitizer conjugates as a molecular tool in photodynamic therapy, ChemMedChem, 3 (2008) 425–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Niagara Muhammad I, Muthu Kumara G, Jing Z, Paul CH, Ratha M, Yong Z, In vivo photodynamic therapy using upconversion nanoparticles as remote-controlled nanotransducers, Nat. Med, 18 (2012) 1580–1585. [DOI] [PubMed] [Google Scholar]
- [195].Cohen BA, Bergkvist M, Targeted in vitro photodynamic therapy via aptamer-labeled, porphyrin-loaded virus capsids, J. Photochem. Photobiol. B, 121 (2013) 67–74. [DOI] [PubMed] [Google Scholar]
- [196].Ferreira CSM, Cheung MC, Missailidis S, Phototoxic aptamers selectively enter and kill epithelial cancer cells, Nucleic Acid Res, 37 (2009) 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Han D, Zhu G, Wu C, Zhu Z, Chen T, Zhang X, Tan W, Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy, ACS Nano, 7 (2013) 2312–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Stephanopoulos N, Tong GJ, Hsiao SC, Francis MB, Dual-surface modified virus capsids for targeted delivery of photodynamic agents to cancer cells, ACS Nano, 4 (2010) 6014–6020. [DOI] [PubMed] [Google Scholar]
- [199].Wang K, You M, Chen Y, Han D, Zhu Z, Huang J, Williams K, Yang CJ, Tan W. Self-assembly of a bifunctional DNA carrier for drug delivery, Angew. Chem. Int. Ed, 50 (2011) 6098–6101. [DOI] [PubMed] [Google Scholar]
- [200].Yuan Q, Wu Y, Wang J, Lu D, Zhao Z, Liu T, Zhang X, Tan W. Targeted bioimaging and photodynamic therapy nanoplatform using an aptamer-guided G-quadruplex DNA carrier and near-infrared light, Angew. Chem. Int. Ed, 52 (2013) 13965–13969. [DOI] [PubMed] [Google Scholar]
- [201].Han D, Zhu G, Wu C, Zhu Z, Chen T, Zhang X, Tan W, Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy, ACS Nano, 7 (2013) 2312–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].You M, Zhu G, Chen T, Donovan MJ, Tan W, Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy, J. Am. Chem. Soc, 137 (2015) 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Huang P, Lin J, Li W, Rong P, Wang Z, Wang S, Wang X, Sun X, Aronova M, Niu G, Leapman RD, Nie Z, Chen X, Biodegradable gold nanovesicles with an ultrastrong plasmonic coupling effect for photoacoustic imaging and photothermal therapy, Angew. Chem. Int. Ed, 52 (2013) 13958–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Huang X, El-Sayed IH, Qian W, El-Sayed MA, Cancer cell imaging and photothermal therapy in the nearinfrared region by using gold nanorods, J. Am. Chem. Soc, 128 (2006) 2115–2120. [DOI] [PubMed] [Google Scholar]
- [205].Yang L, Tseng Y-TT, Suo G, Chen L, Yu J, Chiu W-JJ, Huang C-CC, Lin C-HH, Photothermal Therapeutic Response of Cancer Cells to Aptamer-Gold Nanoparticle-Hybridized Graphene Oxide under NIR Illumination, ACS Appl. Mater. Interfaces, 7 (2015) 5097–5106. [DOI] [PubMed] [Google Scholar]
- [206].Lin L-SS, Cong Z-XX, Cao J-BB, Ke K-MM, Peng Q-LL, Gao J, Yang H-HH, Liu G, Chen X, Multifunctional Fe₃O₄@polydopamine core-shell nanocomposites for intracellular mRNA detection and imagingguided photothermal therapy, ACS Nano, 8 (2014) 3876–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Rong P, Yang K, Srivastan A, Kiesewetter DO, Yue X, Wang F, Nie L, Bhirde A, Wang Z, Liu Z, Niu G, Wang W, Chen X, Photosensitizer loaded nano-graphene for multimodality imaging guided tumor photodynamic therapy, Theranostics, 4 (2013) 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Huang Y-F, Sefah K, Bamrungsap S, Chang H-T, Tan W, Selective Photothermal Therapy for Mixed Cancer Cells Using Aptamer-Conjugated Nanorods, Langmuir, 24 (2008) 11860–11865. [DOI] [PubMed] [Google Scholar]
- [209].Wilson C, Keefe AD, Building oligonucleotide therapeutics using non-natural chemistries, Curr. Opinion Chem. Biol, 10 (2006) 607–614. [DOI] [PubMed] [Google Scholar]
- [210].Da Pieve C, Blackshaw E, Missailidis S, Perkins AC, PEGylation and biodistribution of an anti-MUC1 aptamer in MCF-7 tumor-bearing mice, Bioconjug. Chem, 23 (2012) 1377–1381. [DOI] [PubMed] [Google Scholar]
- [211].McCauley TG, Kurz JC, Merlino PG, Lewis SD, Gilbert M, Epstein DM, Marsh HN, Pharmacologic and pharmacokinetic assessment of anti-TGFbeta2 aptamers in rabbit plasma and aqueous humor, Pharm. Res, 23 (2006) 303–311. [DOI] [PubMed] [Google Scholar]
- [212].Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD, Direct in vitro selection of a 2’-O-methyl aptamer to VEGF, Chem. Biol, 12 (2004) 25–33. [DOI] [PubMed] [Google Scholar]
- [213].Zhang F, Zhu G, Jacobson O, Liu Y, Chen K, Yu G, Ni Q, Fan J, Yang Z, Xu F, Fu X, Wang Z, Ma Y, Niu G, Zhao X, Chen X, Transformative nanomedicine of an amphiphilic camptothecin prodrug for long circulation and high tumor uptake in cancer therapy, ACS Nano, 11 (2017) 8838–8848. [DOI] [PubMed] [Google Scholar]
- [214].Zhu G, Lynn GM, Jacobson O, Chen K, Liu Y, Zhang H, Ma Y, Zhang F, Tian R, Ni Q, Cheng S, Wang Z, Lu N, Yung BC, Wang Z, Lang L, Fu X, Jin A, Weiss ID, Vishwasrao H, Niu G, Shroff H, Klinman DM, Seder RA, Chen X, Albumin/vaccine nanocomplexes that assemble in vivo for combination cancer immunotherapy, Nat. Commun, 8 (2017) 1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Tian R, Jacobson O, Niu G, Kiesewetter DO, Wang Z, Zhu G, Ma Y, Liu G, Chen X, Evans Blue Attachment Enhances Somatostatin Receptor Subtype-2 Imaging and Radiotherapy, Theranostics, 8 (2018) 735745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Chen H, Jacobson O, Niu G, Weiss I, Kiesewetter DO, Liu Y, Ma Y, Wu H, Chen X, Novel molecular “add-on” based on Evans Blue confers superior pharmacokinetics and transforms drugs to theranostic agents, J. Nucl. Med, 58 (2016) 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Zhang J, Lang L, Zhu Z, Li F, Niu G, Chen X, Clinical Translation of an Albumin-Binding PET Radiotracer 68Ga-NEB, J. Nucl. Med, 56 (2015) 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Wang Y, Lang L, Huang P, Wang Z, Jacobson O, DO K, IU A, G T, G N, X C, In vivo albumin labeling and lymphatic imaging, Proc. Natl. Acad. Sci. U.S.A, 112 (2015) 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Niu G, Lang L, Kiesewetter DO, Ma Y, Sun Z, Guo N, Guo J, Wu C, Chen X, In Vivo Labeling of Serum Albumin for PET, J. Nucl. Med, 55 (2014) 1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]