Abstract
Objective:
Optical techniques for recording and manipulating neural activity have traditionally been constrained to superficial brain regions due to light scattering. New techniques are needed to extend optical access to large 3D volumes in deep brain areas, while retaining local connectivity.
Approach:
We have developed a method to implant bundles of hundreds or thousands of optical microfibers, each with a diameter of 8 μm. During insertion, each fiber moves independently, following a path of least resistance. The fibers achieve near total internal reflection, enabling optically interfacing with the tissue near each fiber aperture.
Main results:
At a depth of 3 mm, histology shows fibers consistently splay over 1 mm in diameter throughout the target region. Immunohistochemical staining after chronic implants reveals neurons in close proximity to the fiber tips. Models of photon fluence indicate that fibers can be used as a stimulation light source to precisely activate distinct patterns of neurons by illuminating a subset of fibers in the bundle. By recording fluorescent beads diffusing in water, we demonstrate the recording capability of the fibers.
Significance:
Our histology, modeling and fluorescent bead recordings suggest that the optical microfibers may provide a minimally invasive, stable, bidirectional interface for recording or stimulating genetic probes in deep brain regions—a hyper-localized form of fiber photometry.
1. Introduction
Optical techniques for recording and manipulating neural activity play a crucial role in advancing systems neuroscience, due to the broad field of view, the flexibility and specificity of viral and genetic targeting, and the development of novel probes offering non-overlapping spectral bandwidth and increasing temporal resolution [1, 2].
Yet these techniques are inherently limited in all but the most superficial regions of the brain, given the light scattering and absorption of tissue. New developments are helping to extend such optical techniques to deeper brain regions, but necessitate tradeoffs in terms of either more constrained experimental paradigms or increased tissue damage. Three-photon microscopy at 1300 nm has enabled recording from intact brain tissue at depths exceeding 1 mm [3, 4]. And while three-photon microscopy is not yet amenable to recording in freely behaving animals, head-mounted two-photon microscopes show promise, but are limited to depths on the order of the mean free path of near infrared photons in the brain, just a few hundred microns [5]. Greater penetration depths are now being achieved with red-shifted fluorophores [6]. At depths beyond the reach of three photon imaging, optically interfacing with deeper layers and non-cortical structures has relied on implanting miniature gradient index (GRIN) lenses or prisms [7, 8, 9, 10], or removing overlying tissue [11]. Such techniques provide optical access, but compromise or obliterate structures adjacent to the area being imaged.
Implants with a cross section greater than 50 μm cause neuronal damage or death over a zone up to 100 μm from the implant [12]; the trauma of insertion and motion of the implant after insertion trigger a range of reactions including the immune foreign-body response to non-organic material, the disruption of oxygenation due to vessel damage, the breakdown in the blood-brain barrier, and excitotoxic cell death associated with accumulation of extracellular glutamate [13, 14, 15, 16]. Due to dense local connectivity, tissue damaged by the implant and foreign body response could impact network dynamics in the imaging plane [17, 18, 19]. Yet implants with a cross section less than 10 μm have a substantially diminished tissue response [12, 20, 21, 22].
We propose a new technique to optically address deep brain regions through sub-10 μm implants. The basic idea is to cut commercially available leached fiber bundles [23], revealing the dissociated fibers, which can then be implanted. In this process, hundreds or thousands of multimode optical microfibers each with a diameter as small as 6.8 μm are implanted into the brain while the backend of the device provides optical access to the fibers. During the implant process, each fiber travels independently and finds a path of least resistance causing the implanted bundle to spread gradually. The small diameter of the fibers minimizes tissue displacement and decreases the likelihood both of evoking a tissue response and, as a result, of disrupting local network dynamics in the imaging plane.
In the approach described here, each fiber has a core and a cladding. The refractive index mismatch achieves near total internal reflection of light, enabling each fiber to interface with tissue near its aperture. Because of the splaying during insertion, the fibers will not maintain a strictly organized spatial mapping. Yet each fiber may provide a bidirectional interface with a small volume of tissue near the tip of the fiber, and potential correlations across fibers can enable reconstructing a relative spatial topography. Outside of the brain, the fibers converge to a polished imaging surface, where each fiber is arrayed in a tightly packed lattice that can interface with a traditional fluorescence microscope.
Based on histology and immunohistochemistry, we demonstrate that the bundles of optical microfibers splay during insertion into the brain, achieving a spatially distributed set of fibers throughout the target brain region. The small cross section of the individual fibers displaces less tissue than GRIN lenses (for example, a bundle of 2,000 fibers displaces half the volume of a 500 μm diameter lens), and hence may preserve more neurons and promote more natural network dynamics in the target region. Based on simulations of the optical profile of individual fibers, we assess the sensitivity of the fibers as a multi-channel, bidirectional optical interface. Finally, we show that fluorescence signals can be recorded from diffusing fluorescent beads through these small-diameter optical microfibers.
2. Methods
2.1. Fibers
The fibers we use are leached fiber bundles produced as flexible medical endoscopes. This work primarily relied on bundles of 4,500 fibers where each individual fiber has a diameter of 8 μm (Schott 1534180), although variations exist in the number of fibers (3,500–18,000) and the diameter (6.4–11.9 μm). The bundles are built for coherent imaging and constructed from three types of glass, a core (diameter: 5.1 μm, refractive index: 1.605), a cladding (thickness: 1 μm, refractive index: 1.56) and an acid soluble glass (thickness: 0.4 μm). During manufacturing, the bundles start as traditional coherent fibers, and then the acid soluble glass is dissolved for bundle flexibility [23]. The ends of the fibers come together in polished imaging surfaces held in ferrules. The dissociated fibers are covered in a flexible silicone sheathing.
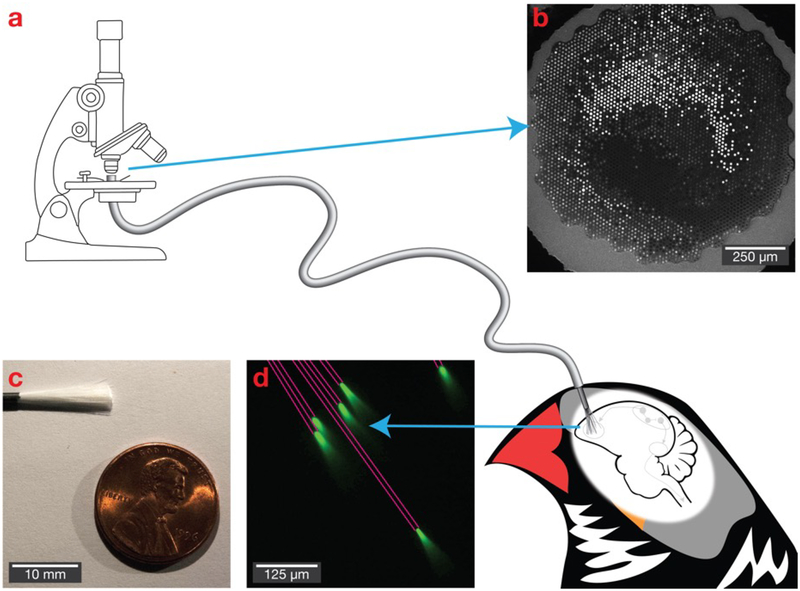
We cut the bundles in half, using a scalpel or razor, sacrificing the spatial cohesion, but providing access to the individual, dissociated fibers. The silicone sheathing was cut back to expose the fibers, and we then cut a fraction of the exposed fibers to reduce the implant size to a target number of fibers (varied over implants to assess tissue impact). The remaining fibers were secured together by forming a bead of light-cured acrylic (Flow-It ALC, Pentron Clinical) around the fibers, leaving 4–5 mm of fibers exposed (Figure 1b). The exposed fibers could be further cut using a fine scissors, shaping the bundle into a bevel. Such pre-implant shaping increases the distribution of depths of the fiber tips. The bundles used for histology were not shaped, such that all implanted fibers had a consistent length. All fiber cuts are made perpendicularly to the fiber, with the aim of producing a uniform fiber aperture; placing fibers in fluorescein (as shown in Figure 1d) reveals visually consistent fluorescence profiles, indicating that the cutting produces relatively uniform fiber tips. At this point, the dissociated fibers can be directly implanted into brain tissue; the other end of the fiber, containing the ferrule and polished surface with fibers aligned and arranged in a lattice, can be readily interfaced with a fluorescence microscope or other optical configuration (Figure 1).
Figure 1. Bundles of microfibers as a potential deep brain optical interface.
(a) The polished imaging surface is mounted in a traditional fluorescence microscope, while individual fibers with a diameter as small as 6.8 μm are implanted into the brain. (b) The polished imaging surface that connects with the microscope. (c) A bundle of 18,000 fibers. (d) Light propagates with near total internal reflection, allowing it to deliver and collect light at the tips of the fibers. Six fibers are shown in a fluorescein solution, with pink lines added to emphasize fiber path.
Optical attenuation of the fibers was measured to be 3.38 ± 0.03 dB (std. dev.) for a 840 mm long bundle (4 dB/m). Attenuation was measured using collimated light in the 446–486 nm range (relevant for exciting GFP-based indicators, such as GCaMP, or stimulating channelrhodopsin) focused on the polished imaging surface using a lens with numerical aperture matched to the fibers. In this measurement, we assumed that 19.4% of the incident light enters the cores based on the surface area of the fiber bundle, the fiber count, and the core diameter. After cutting the bundle, attenuation for the 420 mm bundle measured from the splaying fibers is 3.78 ± 0.02 dB (std. dev.), indicating that most of the light carried by the bundle is transmitted from the cut ends.
2.2. Histology
Animals.
Animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Boston University (protocols 14–028 and 14–029). Fibers were implanted in 27 adult zebra finches (> 120 days post hatch). Of the animals, eight were implanted with alternative fibers (different materials and different fiber diameters; results not shown). Of the remaining nineteen, fifteen were used for histology described in this paper with four animals being excluded due to poor slicing (tearing of the tissue when slicing through the fibers) or poor staining (during immunohistochemistry).
Fiber implant.
Anesthesia was induced with 4% isoflurane and maintained at 1–2% for the duration of the surgery. An analgesic (0.5 mg/kg meloxicam, Eloxiject) was injected intramuscularly into the breast at the start of the procedure. The animal was placed in a stereotaxic instrument and feathers were removed from the scalp. The scalp was cleaned with Betadine and ethanol. A local anesthetic (4 mg/kg bupivacaine) was injected subcutaneously into the scalp, and an incision was made along the anterior-posterior axis.
The skull over the implant point (area X) was localized based on head angle (20°) and stereotactic coordinates (5.8 mm anterior, 1.5 mm lateral), measured from the bifurcation of the mid-sagittal sinus (lambda). In order to accommodate the bundle of fibers, a 0.5–1 mm diameter craniotomy was created, with the size matched to the bundle. The craniotomy was created by first using a dental drill to remove the outer layer of bone, then by using an ophthalmic scalpel to remove the inner layer of bone [24]. The dura within the craniotomy was removed using either a dura pick constructed from sharpened tungsten or an ophthalmic scalpel.
The fiber bundle was prepared by securing the fibers together in a bead of light cured dental acrylic (Flow-It ALC, Pentron Clinical) and cut to 3–5 mm. Using a digital manipulator attached to the stereotaxic rig, the fiber bundle was positioned over the durotomy and slowly lowered into the tissue at a rate of approximately 500 μm per minute. Larger implants (more than 250 fibers) could result in a noticeable depression or “dimpling” in the tissue before the bundle passed through the surface of the brain. Such dimpling was generally observed during the first 250–350 μm of insertion; beyond that depth, the size of the depression remained consistent as we continued to lower the implant. Based on these observations, we often would slow the insertion rate for bundles with larger numbers of fibers or when dimpling was observed. In addition, we found that the visible dimpling could be alleviated by lowering the implant an additional 50 μm past the desired depth, waiting for five minutes, then returning the implant to the desired depth. We did not observe bleeding associated with the implant or the dimpling. After the fibers were lowered to a depth of 2.7–2.9 mm (measured from the point when the fibers enter the tissue), additional light-cured dental acrylic was used to secure the fiber bundle to the skull surrounding the craniotomy.
Animals received nonsteroidal anti-inflammatories (0.5 mg/kg meloxicam) both before the surgery via injection (Eloxiject) and after the surgery in their food (Metacam), as well as topical antibiotics (Pfizer Terramycin) after the surgery.
Three days post implant, animals were returned to the aviary and housed socially. Animals used to image the distribution of fibers were perfused after 21 to 331 days (mean 88 days). Animals used for immunohistochemistry staining were perfused after 77 to 395 days (mean 176 days).
Animal perfusion and fixation.
Animals were injected with 0.1 mL 10% sodium pentobarbital intramuscularly. Once anesthetized, the animals were perfused intracardially with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde in 0.1 M PBS. The skull and brain were separated from the body. Leaving the skull in place (as the fibers are anchored to the skull), small cracks were made in the bone to ensure penetration of the fixative. The skull and brain was immersed in 4% paraformaldehyde in 0.1 M PBS overnight. Next, as cryoprotection, it was immersed in 15% sucrose in 0.1 M PBS overnight, followed by 30% sucrose in 0.1 M PBS for a second night. Placing the skull upside down such that the implant trajectory was roughly perpendicular to the mounting slide, the skull was frozen (−20°C) in embedding medium (Optimal Cutting Temperature Compound, Tissue-Tek) for 30 minutes and sectioned in a cryostat (LEICA CM3050S, with Thermo Scientific MB22 microtome blades) in either 70 or 100 μm thick slices, cutting through the skull and perpendicular to the fiber bundle implant. Due to the thin, pneumatized bone of the songbird, cryosectioning through the skull was possible without any decalcifying process. For optimal cutting, blades were regularly shifted and replaced to ensure a fresh cutting surface was always in use; without such precautions, the worn blades were more likely to catch on fibers and tear surrounding tissue. Some sections were discarded because of tearing. Slices were either mounted on slides or were transferred to wells containing PBS and processed for immunohistochemical staining as described below.
Histology.
To quantify the splay of fibers, brightfield microscopy images were collected of slices mounted on slides and secured with coverslips. Images were collected from slices at various depths.
Immunohistochemistry.
In order to assess tissue health and imaging viability, a selection of slices taken at various depths were processed to label neurons via NeuN antibodies. Slices were washed in PBS, then in 0.3% Triton X-100 in PBS for 30 minutes and finally in a solution of 0.3% Triton X-100 and 5% normal donkey serum (NDS) for 45 minutes. The slices were then placed in a solution of the primary antibody (MAB377 Anti-NeuN, 1:500, EMD Millipore) made with 3% bovine serum albumin (BSA) and 0.3% Triton X-100 in PBS. The wells were placed on a rotator and allowed to incubate at 4°C overnight. Slices were washed in PBS (×3, 10 minutes each). Next, the slices were placed in a solution of the secondary antibody (715-025-150 Rhodamine [TRITC] AffiniPure Donkey Anti-Mouse IgG, 1:500, Jackson ImmunoResearch). The wells were again placed on a rotator and allowed to incubate at 4°C for one hour. Slices were washed in PBS (×3, 10 minutes each). Next, 1 mL of DAPI stain (4’,6-Diamidino-2-Phenylindole, Dihydrochloride, 300 μM solution, 1:1000, D1306, Thermo Fisher Scientific) was added to each well. After three minutes, the slices underwent a final wash (×2, 5 minutes), before being mounted on glass slides with an anti-fading mounting medium (Fluoro-Gel, EMS) and secured with a coverslip. Some immunohistochemistry samples were not usable, due to ineffective staining or fibers becoming dislodged during the washing process.
Microscopy.
Slices were imaged using an upright fluorescence microscope (Nikon Eclipse NiE, with a DS-Qi1 Monochrome camera and controlled by NIS-Elements: Advanced Research), illuminated by an LED light source (SOLA Light Engine). To assess splay, we used either a 4× (Plan Fluor, NA 0.13) or a 10 (Plan Fluor, NA 0.3) objective. To image immunohistochemistry, we used a 20× objective (Plan Apo Lambda, NA 0.75).
Qualitative and quantitative analysis.
To quantify fiber splay, brightfield images were collected from slices near the tip of the fiber. Fibers were manually annotated using a custom MATLAB program for organizing and analyzing histology. To calculate a measure indicative of the splay of the fibers, a bivariate normal distribution was fit to the position of the fibers in the slice and the area of the ellipse representing two standard deviations of the distribution (the 95% confidence interval) was calculated. The data presented are from 11 animals, reflecting the animals implanted with bundles consisting of 7–8 μm diameter fibers with at least a three week recovery period and where the tissue at the tip of the implant was cleanly sliced (see note above about sectioning).
To quantify the presence of neurons in proximity to fibers, two-channel fluorescence (with NeuN in red and DAPI in blue) and brightfield images were collected from the target implant region (area X). Control images were collected from the contralateral region (without an implant) to measure baseline neural distributions and densities. Neurons were manually annotated based on a consensus of the NeuN and DAPI signal, and fibers were manually annotated based on both the histology and brightfield images. For slices with fibers, the distance from each fiber to the nearest neuron was calculated (fibers where the edge of the image was closer than the nearest neuron were ignored), subtracting the radius of the fiber. As a control, random points were selected on the control slices without fibers, and the distance to the nearest neuron was calculated (points could be selected at or on neurons, resulting in a distance of zero).
In addition, NeuN-stained cell density was calculated for the 50 μm region surrounding each implant, normalized by densities calculated on the control slices. To account for the close proximity of neighboring fibers, the cross sectional area of neighboring fibers was subtracted from the area of the 50 μm region when calculating density surrounding implants. The data presented are based on twelve annotated slices from five animals, reflecting all animals implanted with bundles of 7–8 μm diameter fibers with at least a ten week recovery period and successful immunohistochemical staining.
2.3. Modeling
Fiber profile.
The optical profile for a single fiber was generated via a Monte Carlo simulation of 10,000,000 photon packets traveling through a 1 mm3 volume (modeled as isotropic 5 μm voxels) [25]. Photon packets enter the tissue at [500 μm, 500 μm, 200 μm] with a Gaussian distribution reflecting the NA of the fiber (0.377). Within each voxel, the photon packet can be scattered (μs = 10 mm−1 with anisotropy g = 0.9 [26]) or fractionally absorbed (μa = 0.337 mm−1 for 490 nm light, μa = 0.343 mm−1 for 512 nm light based on 3% blood volume fraction [BVf] [27], 15 g/DL hemoglobin concentration [28], an oxygenation fraction of 70% and extinction coefficients for hemoglobin [29]). The 3D path of each photon packet is averaged together, normalized and visualized as a 2D slice through the volume. The fluorescence signals received by individual fibers, given the illumination profile from the superposition of the optical profiles emitted from all of the fibers, is calculated following the procedure described in [30, 31].
Neural interface simulation.
To simulate interfacing with a neural population, a 1.2 mm3 volume of tissue was modeled. This volume is consistent with area X in the adult zebra finch [32] and is illustrative of a deep brain region. A target subpopulation of neurons of interest is modeled as uniformly distributed through the volume with a density of 780,000 neurons per mm3, based on the density of medium spiny neurons in area X in male zebra finch that are one year old [33]. All cells in the target subpopulation are assumed to express the relevant genetic probe.
Based on the histological data on splaying, the fiber bundle is assumed to have a bivariate normal distribution in xy space with standard deviation (σ) based on the number of fibers in the bundle. The fiber depth will vary based on preparation of the bundle (how the fibers are cut prior to implant) and the path of splay; this variability is modeled as a normal distribution of depths with standard deviation σ = 30 μm.
The strength of stimulation or excitation for individual neurons is calculated for each fiber by identifying the sensitivity of the voxel that corresponds with the position of the neuron relative to the tip of the fiber. The per fiber optical intensities are summed across all fibers in the bundle to calculate the total potential stimulation/excitation strength. These values are normalized as a percentage of maximum fluence in the tissue.
To evaluate the ability to uniquely address neurons through illuminating a subset of n fibers, a 20,000 iteration Monte Carlo simulation is used to select random permutations of n fibers. For each iteration, the number of neurons activated by the cumulative optical power of the selected fibers is compared with the number of neurons activated if each fiber was illuminated independently.
The round-trip fluorescence yield for pairs of fibers and neurons, a measure of expected fluorescent emission collected by the fiber from the neuron, is calculated by multiplying the total excitation strength for the neuron (as described above) by the sensitivity of the voxel that corresponds with the position of the neuron relative to the tip of the fiber (representing the time reversal of emission from the neuron reaching the fiber tip) [30, 31]. This round trip fluorescent yield is normalized based on the maximum possible yield.
2.4. Fluorescent beads
To validate the recording capability of the fiber bundles, the tips of loose fibers were immersed in a solution of water and fluorescent beads (Bangs Laboratories FSDG007, 7.32 μm diameter, 480 nm excitation, 520 nm emission). The ferrule and polished imaging surface were held below a traditional fluorescent microscope (Olympus, 20× objective) with a broadband white LED (Thorlabs SOLIS-3C) set at 60% brightness and a GFP filter cube (Semrock BrightLine GFP-4050B, 466/40 excitation, 525/50 emission, 495 dichroic). Excitation power from the objective was measured at 6.27 mW. As beads diffused in the water, changes in fluorescence were recorded by a sCMOS camera (Hamamatsu ORCA-Flash4.0 v2) with a resolution of 2048×2048 16 bit pixels and an exposure of 50 ms per frame. Saved CXD files were processed in MATLAB using a custom pipeline. Frames were motion corrected using the Scale-Invariant Feature Transform (SIFT) algorithm [34, 35, 36]. A standard deviation image created by calculating the standard deviation of pixels across frames was used to identify those fibers that were in the solution and where bead diffusion resulted in variability in the fluorescence. For the identified fibers, traces were generated by extracting and averaging all pixels that corresponded with the fiber. Traces were converted to ΔF/F0, where F0 corresponds with the 5th percentile intensity (i.e., background intensity when there is minimal fluorescence from nearby beads).
To calculate the contrast-to-noise ratio (CNR) for the bead recording, we performed a second recording to measure noise. The fibers were placed in a solution of fluorescein and water, such that the fiber brightness matched the peak brightness observed during the fluorescent bead recording. The signal was recorded, and again, traces were generated by extracting and averaging all pixels that correspond with each fiber. For the CNR, we calculate the contrast from the fluorescent bead recording by subtracting the 5th percentile from the 95th percentile intensity and averaging across fibers; we calculate the noise as the standard deviation for traces from the fluorescein recording.
3. Results
3.1. Histology
Bundles of between 50 and 5,000 microfibers were implanted into zebra finch basal ganglia (area X) at a depth of 2.9 mm. To understand the impact of the bundles, histologic samples were collected to measure the distribution of fibers in tissue and to evaluate the distance been fiber tips and the nearest NeuN-stained neurons.
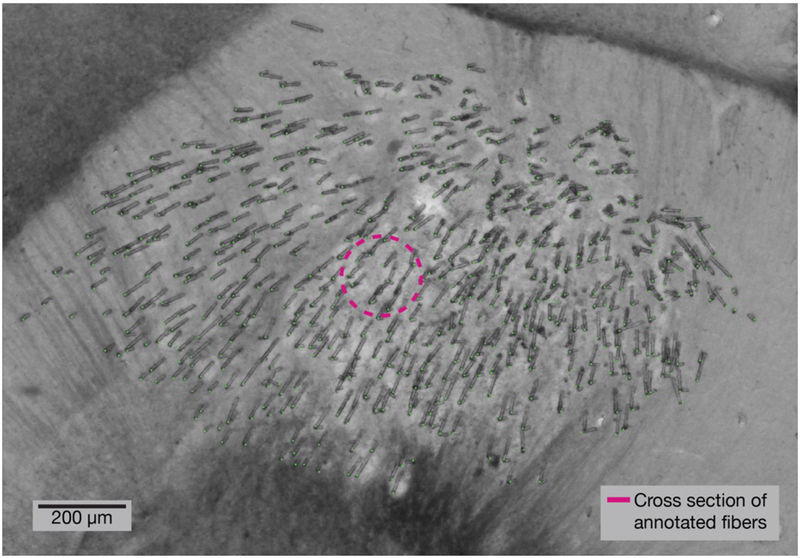
With the fibers anchored to the intact skull, the tissue was fixed and cryosectioned perpendicularly to the implant penetration angle. Sections were imaged and annotated to record the spatial distribution of microfibers at different depths. During insertion, each fiber follows a path of least resistance, splaying through the brain tissue. In these perpendicular sliced sections, the distribution of fibers resembles a bivariate normal distribution throughout the target region. In Figure 2, 530 fibers can be seen distributed spanning over 1 mm of tissue, while only displacing a cross sectional area of 26,640 μm2; a 1 mm diameter GRIN lens to access the same region would have a cross sectional area of 785,398 μm2.
Figure 2. Histology at tip of implant shows microfibers splayed throughout the target region.
A 100 μm thick brain section showing the tips of a bundle of 530 optical microfibers implanted at a depth of 2.95 mm. Before insertion, the bundle had a diameter of 570 μm. This section was collected four months after implant, and the brain sectioned perpendicularly to the insertion angle. The cross sectional area of tissue displaced by the microfibers (annotated in green) is 26,640 μm2 (pink circle).
Implant conditions account for much of the variability in the spread of the fibers. Based on anecdotal observations, the configuration of the fibers prior to implant—specifically, the spatial arrangement of fibers in the acrylic anchor point (used both to hold the fibers during the implant and the to anchor the fibers to the skull), and the spread of the fibers below this acrylic anchor point—appears to affect the final distribution of the fibers. For example, if the fibers spread in the air before coming into contact with the tissue, we tended to observe greater spread after insertion into the tissue. The configuration of the fibers in the acrylic anchor point is difficult to control, as we sought to avoid directly squeezing or stressing the fibers. But we found that we could influence the amount of spread below the anchor point by keeping the fibers dry; if the fibers get wet, there is greater adhesion during insertion and, as a result, a more narrow distribution in the tissue. As a result, we avoided wetting the fibers and minimized moisture on the surface of the tissue prior to implant (as that would get wicked into the fiber bundle and increase adhesion).
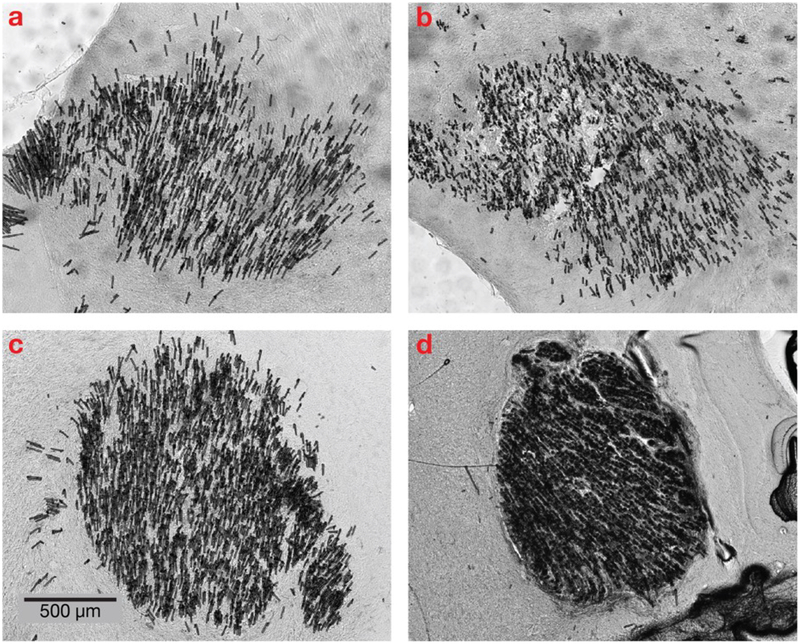
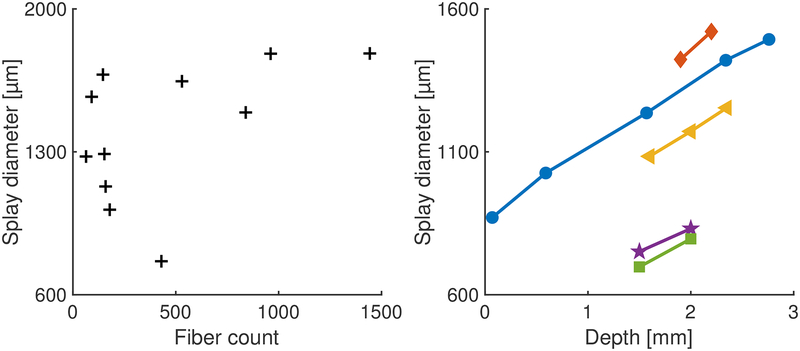
In Figure 4, the distribution of the microfibers in the tissue can be seen to increase over the four slices from different depths in the same animal; the splay area is calculated by drawing a bounding ellipse containing 95% of the fibers. For each 1 mm of implant depth, the diameter of the splay area increases by 229.1 ± 51.1 μm (std. dev., based on 9 pairs of slices from 5 animals); see Figure 3.
Figure 4. Histology at different depths as the fibers splay during insertion.
A bundle of approximately 1,125 optical microfibers implanted at a depth of 2.95 mm. Eight weeks after the implant, the animal was perfused and the brain sectioned perpendicularly to the insertion angle. At the surface, the bundle diameter was 1.03 mm. These 70 μm thick slices from depths (a) 2.76 mm, (b) 2.34 mm, (c) 1.57 mm and (d) 0.59 mm reveal a gradual spreading of the optical fibers during insertion as each fiber follows a path of least resistance.
Figure 3. Diameter of splaying increases linearly with depth.
Left: Splay of fibers at a depth of 2.9 mm, in the target region of songbird basal ganglia from 11 animals. The plot shows the diameter of the ellipse describing the splay of the fibers for various implant sizes. As the number of fibers increases, the area accessed by the fibers increases. Right: For five animals, slices were collected at multiple depths to estimate splay diameter as a function of depth. For each 1 mm of implant depth, the diameter of the splay increases by 229.1 ± 51.1 μm (std. dev.). Each color/marker pair corresponds with a distinct animal.
Tissue sections from animals with chronic implants (10+ weeks post implant) underwent NeuN staining to label neurons and DAPI staining to label nuclei. Since the red blood cells of birds contain DNA, DAPI labelled cells that are not NeuN stained include populations of glia, astrocytes, red blood cells, and any other non-NeuN stained cell nuclei. The slices show NeuN-stained neurons in close proximity to the fibers (see Figure 5). In instances where two or more fibers remain close during insertion, the proximity of the fibers may adversely affect the immediate tissue, as suggested by an increased presence of non-neural cells (DAPI stained but not NeuN stained) around such “clumps” of fibers.
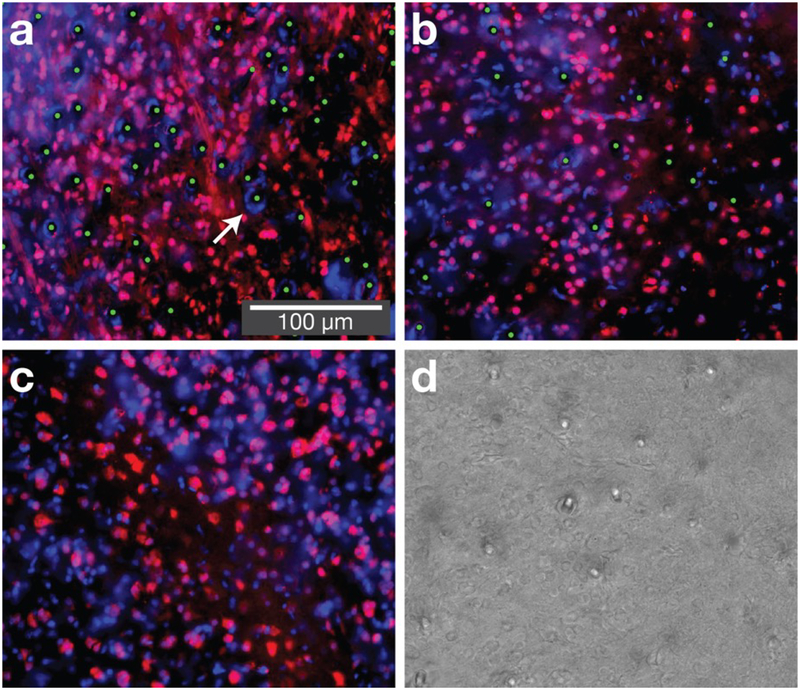
Figure 5. At chronic time points neurons are found in close proximity to fibers.
Three sections from zebra finches implanted with optical microfibers, collected at least ten weeks post-implant. Sections are near the tip of the implant, within the basal ganglia (area X, depth 2.9 mm). (a) and (b) show implants in the basal ganglia (bundle sizes of 4,500 and 1,125 fibers respectively), (c) shows unimplanted basal ganglia, and (d) shows a corresponding brightfield image used to confirm fiber locations. Red is NeuN (neurons), blue is DAPI (nuclei) and green dots are manual annotations that reveal fiber locations. The immunohistochemistry shows NeuN-stained cells in close proximity to fibers. In some cases, we observe a dense circle of DAPI stained cells in close proximity to the fibers (arrow), suggesting either bleeding (in birds, red blood cells have DNA) or a reactive tissue response (such as glia or astrocytes). This most frequently occurs at locations where multiple fibers are in close proximity (this can occur with bundles of over 4,000 fibers or when fibers are wet prior to insertion). The length scale of reactive tissue response is approximately an order of magnitude smaller than for silicon electrode shanks with a 50 μm profile [13].
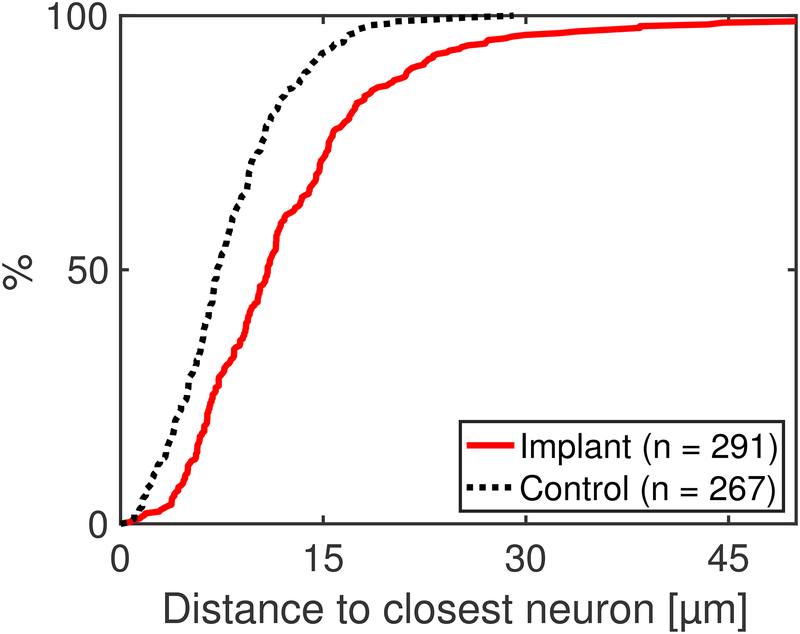
By annotating both the fibers and the neurons, the presence of NeuN-stained neurons near the fibers can be compared to control slices (same region, no implant) to evaluate tissue impact. Figure 6 compares the distance from fibers to neurons in implant slices to the distance between randomly selected points and neurons in control slices. The control measurement provides a lower bound for distance to the nearest neuron, if the implant had no impact on the tissue. For the implanted slices, the distance from a fiber to the nearest NeuN-stained neuron is on average 12.81 ± 9.22 μm (std. dev.), while on the control slice, the distance from a randomly selected point to the nearest neuron is on average 8.32 ± 4.72 μm (std. dev.).
Figure 6. Quantification of histology reveals NeuN-labeled cells near the majority of fibers.
The distribution of distances to the nearest NeuN-stained cell for implants (measuring from the edge of each fiber) and unimplanted controls (measuring from randomly selected points). The NeuN and DAPI staining shows that there are intact neurons in close proximity to the fibers; 85% of the fibers have a neuron within 20 μm. The distance from a fiber to the nearest NeuN-stained cell is 12.81 ± 9.22 μm (std. dev.); for the control, the distance from a randomly selected point to the nearest neuron is 8.32 ± 4.72 μm (std. dev.).
We also can compare the NeuN-stained cell density surrounding each fiber relative to the cell density seen in the control slices. In the 50 μm region surrounding each fiber, we observe a NeuN density of 69.8% ± 17.9 (std. dev.) the density seen in control slices (same region, no implant). Because the 50 μm surround typically includes other fibers, we subtract the cross sectional area of such neighboring fibers from the 50 μm area when calculating the density.
NeuN staining alone does not provide a comprehensive evaluation of tissue or neural health; variability in staining does not consistently indicate differences in neural populations and does not capture non-neuronal changes in tissue health [37, 38, 39, 40]. Despite having a narrow immunohistochemical tool to evaluate tissue health, our histology data are consistent with the possibility that circuits remain healthy in the vicinity of the fiber tips.
3.2. Modeling
To quantify the potential neural population accessible via the optical microfibers, we modeled the optical profile of a single fiber and a bundle of fibers throughout a volume of tissue (see section 2.3) [25].
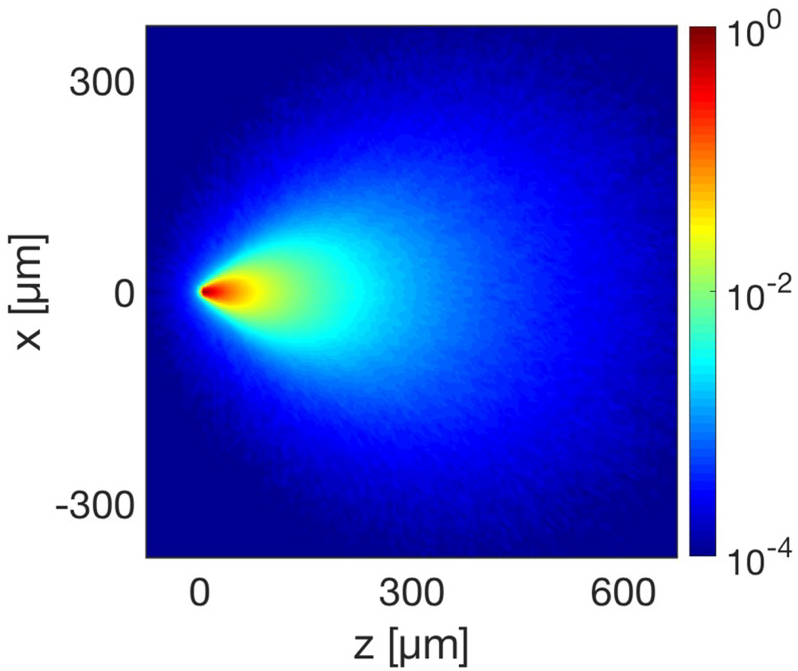
Figure 7 shows the normalized optical profile for a single fiber in tissue with spatially uniform anisotropy, scattering and absorption coefficients based on brain tissue measurements. At a distance of 40 μm from the tip of the fiber, the number of photon packets passing through an arbitrary point in the tissue drops below 10%. Although the fiber can weakly interface with a larger volume of tissue due to the scattering of light in the brain, individual fiber fluorescence will be dominated by neurons within 40 μm of the tip of the fiber. The viability of recording fluorescent signals depends on a number of additional properties that will vary based on the animal model and target region, including the indicator brightness, specificity of expression, density of the neural signal, and tissue autofluorescence.
Figure 7. A single fiber would primarily interface with neurons in close proximity to the tip, based on the tissue scattering and absorption.
The normalized log intensity emission profile of an optical microfiber with tip positioned at [0, 0]. The profile is a Monte Carlo simulation of photon packets propagating through brain tissue, with scattering and absorption properties estimated for 490 nm light. The simulated profile shows a strong interaction with tissue immediately below the tip of the fiber, enabling localized photometry or stimulation; the weak interactions with a larger volume of tissue will contribute background in recordings, or delocalized optogenetic excitation.
Similarly, stimulation through the fiber will most strongly modulate neural activity within the region immediately surrounding the tip of the fibers. Based on the coupling 2.5 μW of 470 nm light into each fiber, and accounting for attenuation measurements of the optical path and fibers, and the simulated optical profile, one fiber will provide sufficient optical power to activate channelrhodopsins in a 18,000 μm3 region surrounding the tip of the fiber given a 5 mW/mm2 activation threshold [41]. For the modeled neural subpopulation (medium spiny neurons in the zebra finch basal ganglia, with a density of 780,000 neurons per mm3 [33]), this stimulation region equates to activating approximately 14 neurons. For comparison, this stimulation region equates to activating approximately 5 neurons in mouse hippocampus CA1, based on a density of 275,000 neurons per mm3 [42, 43].
Based on the histology of splaying fibers described above, it is possible to overlay profiles for hundreds or thousands of fibers throughout a brain region to quantify the properties of the bundle as an interface.
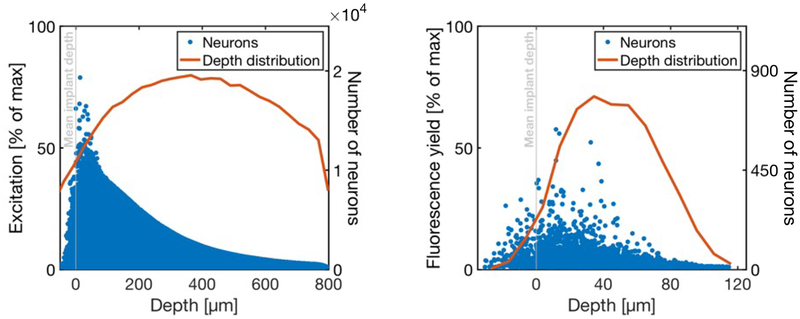
Figure 8 shows a distribution of normalized excitation/stimulation power reaching neurons for a simulated bundle of 500 fibers. Although the neurons receiving the most optical power are within the first 100 μm below the mean implant depth, the scattering properties of the tissue and the overlap in the excitation profile of fibers means that the excitation light will affect many more cells 400–600 μm below the implant depth.
Figure 8. A computational model indicates that optical microfibers would record or stimulate neurons immediately below the fiber tips.
Left: Distribution of light intensities reaching all modeled neurons, for uniform illumination of all fibers in the fiber bundle. These values are normalized by the maximum possible optical power (i.e., the power at the point in the tissue with the highest intensity). Blue dots are individual neurons, and the red line is a depth distribution of neurons that receive >1% of max excitation, indicating that for full bundle illumination, optical stimulation would activate cells far away from the fiber tips. Right: The round-trip fluorescence yield is calculated by first taking the total excitation power reaching the neuron (left) and scaling that by the strongest overlapping fiber profile (representing the collected fluorescence emission). These values are normalized by the maximum possible round-trip fluorescence yield (the maximum achievable given the excitation profile). Blue dots are individual neurons, and the red line is a depth distribution of neurons with >1% of max excitation.
In stimulation experiments, rather than illuminating all fibers, a subset of the fibers can be illuminated to produce more precisely targeted cellular modulation. Given the splay of the fibers, the vast majority of fibers can address a unique set of cells closest to the tip; yet the scattering properties of the tissue and the overlaps in the profiles mean that delivering stimulation through multiple fibers will increase activation in deeper regions and at the overlap between fiber profiles. For example, our model suggests that activating each fiber independently at non-overlapping times in a bundle of 500 fibers would serially stimulate approximately 4,600 cells; if all fibers were active simultaneously, there would be sufficient optical power to stimulate approximately 93,000 cells in the modeled neural population.
By activating small subsets of fibers, it is possible to avoid broad activation, while still exploring stimulation patterns with many degrees of freedom. By simulating overlaps in the optical profile for random sets of 10 fibers in a bundle, light delivered through the ten fibers will only activate an average of 11 more neurons (9.3%) than if the fibers were activated individually. Increasing the number of simultaneously active fibers will increase the crosstalk between the stimulation profiles. For example, sets of 50 fibers will activate an average of 506.4 more neurons (87.3%) than if the fibers were activated individually.
To evaluate the bundle as a potential recording interface, we calculate the round-trip fluorescence yield, indicative of how much fluorescent activity is collected by each fiber. Consistent with the profile for a single fiber, neurons within 40 μm of the mean implant depth have the highest fluorescence yield for recording purposes; cells up to 120 μm away will contribute to the signal, yet low fluorescence yield will likely relegate this contribution to indistinguishable background.
3.3. Fluorescent beads
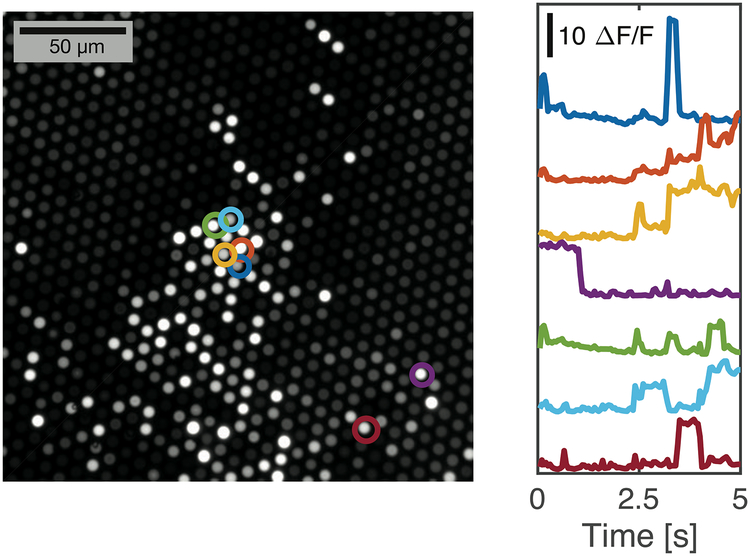
As a preliminary test of the fluorescence recording capability, we immersed dissociated fibers in a suspension of fluorescent beads in water and recorded fluorescence traces as the beads diffused through the sensitivity profile of the individual fibers. Taking a standard deviation of pixel intensities over the recording, we generated a standard deviation image of the polished imaging surface, which revealed those fibers with large fluctuations in measured fluorescence resulting from the diffusing beads (Figure 9). Extracted traces (average intensity for pixels corresponding with the fiber), shown in Figure 9, reveal minimal crosstalk between neighboring fibers and a high signal-to-noise ratio. With excitation power of 6.27 mW measured at the imaging surface of the fiber bundle, we observed fluctuations in fluorescence intensity up to 23.7× the F0 intensity.
Figure 9. Recording of diffusing fluorescent beads reveals minimal crosstalk between neighboring fibers.
The dissociated end of a bundle of fibers was immersed in a suspension of fluorescent beads in water, while the imaging surface was recorded via a traditional fluorescence microscope. The image on the left is a standard deviation image corresponding with a 1024×1024 portion of the full sCMOS sensor, where pixel brightness corresponds with variability over the 5 second recording. It accentuates those fibers immersed in water and with diffusing beads in close proximity to the fiber tips. Traces from a selection of fibers (circled) were extracted to show intensity over the recording. Fiber intensity varies as beads diffuse around the tips of the fibers, showing a high signal-to-noise ratio and minimal crosstalk between neighboring fibers.
We calculate a CNR (contrast-to-noise ratio) with contrast 25.88× the standard deviation of the noise observed during a similar recording with the fibers immersed in a uniform fluorescein solution (with fluorescent brightness matched to the peak signal in the bead recordings).
4. Discussion
Our histological results demonstrate that bundles of optical microfibers may provide an alternative to GRIN lenses to optically address 3D volumes in deep brain areas. The fibers self-splay during the implant process, achieving a distribution that resembles a bivariate normal distribution, with the diameter frequently exceeding 1 mm at an implant depth of 2.9 mm. There appears to be a relationship between the number of fibers and the diameter of the splay, but the trend does not achieve significance in the data set (r2 = 0.5, p = 0.11). Small bundles show increased variability in splay that requires further exploration. We believe that implant conditions, such as the distribution of the fibers before entering the tissue, may account for much of this variability. In addition, as visible in Figure 4, the final distribution is not always symmetric, which could reflect tissue heterogeneity, non-perpendicular implant angles or non-perpendicular sectioning of the tissue.
At the fiber tips, we find NeuN-stained cell bodies in close proximity to the fiber, suggesting the small diameter, flexible microfibers may evoke a smaller foreign body response than that seen with larger glass or electrode implants [15, 44], but more extensive histological analysis is needed in a range of species. Because our current histology is unable to identify neurons in front of (below) the fiber apertures, we instead rely on the perpendicular sections to identify stained neurons adjacent to the fibers in the section closest to the tip of the implant, providing an estimate of the presence of neural signals within range of the fiber sensitivity profile. Future experiments may be able to reconstruct the full 3D path of the fibers through the tissue, and as a result, the presence of neurons within the fiber sensitivity profile.
We are limited in our ability to compare our data on NeuN-stained cells surrounding the implant with prior literature, due to the high number and distribution of the splaying microfibers. NeuN staining is often evaluated around a single implant point, and by measuring the labeled cell density in a 50 μm region surrounding the implant; in a bundle of our splaying optical fibers, that region often contains additional optical fibers. Even with this limitation, we observe NeuN-stained cell densities in the surrounding region that exceed those described for a range of larger, less flexible probes and implants [45, 46, 47, 48, 49]. These findings suggest the splaying microfibers may provide a less invasive option for interfacing with deep brain regions.
Using a Monte Carlo simulation of photon packet propagation, we can construct a sensitivity profile for a single profile. The sensitivity profile assumes that the fibers are cut flush. Although cuts are made perpendicular to the fibers, it is likely that the cuts are not perfectly flush and result in minor imperfections in the fiber aperture, which may impact the sensitivity profile. By imaging cut fibers illuminating fluorescein solution (as shown in Figure 1d), we observe qualitatively consistent illumination profiles, suggesting that should imperfections do not result in substantial variability. Electron microscopy can be used to further investigate the geometry of the cut fiber apertures.
Our modeling results define the sensitivity profile of the fibers, indicating that fibers optically interface with a small volume of tissue near the tip of the fiber. By superimposing the per fiber sensitivity profile in a geometry consistent with our histology, our model allows evaluating the fiber bundle in terms of light delivery (relevant for stimulation) and round-trip fluorescence (relevant to recording). Although in vivo experimental performance will vary due to tissue autofluorescence, and due to indicator brightness and density, our model provides intuition for the likely interface properties. When used for stimulation, our Monte Carlo simulation suggests that patterned illumination of a handful of fibers should precisely activate distinct subsets of the neural population in the target brain region with minimal crosstalk. When used for recording, the round-trip fluorescence will be dominated by neurons in the 40 μm region surrounding the fiber aperture; collectively, a bundle should act as a high channel count fluorescence photometry interface capable of sampling fluorescent indicator activity from hundreds or thousands of points throughout the target deep brain region.
This potential for recording via the fiber bundle is prototyped by the measurements of fluorescent microbeads diffusing in solution, suggesting that it is possible to excite and measure fluorescence through the 5 μm cores. The traces from neighboring fibers show uncorrelated activity, consistent with the splaying of the fibers and minimal crosstalk between fibers. From the traces, we can calculate percentage change in fluorescence and the contrast-to-noise ratio, demonstrating performance characteristics of the fibers under the specific testbed conditions. Yet these measures are not directly comparable to in vivo performance, which will depend on indicator dynamics, tissue autofluorescence and strength of background signal. Further work is needed to demonstrate the in vivo utility of the optical microfibers for optogenetic stimulation and fluorescence imaging.
Collectively, these findings indicate that bundles of splaying optical microfibers may provide an alternative to GRIN lenses for bidirectionally interfacing with deep brain regions. Specifically, this technique may achieve a unique compromise in the set of tradeoffs associated with extending optical techniques to less superficial brain regions. The method provides a high channel count interface distributed throughout a non-superficial 3D volume with potentially reduced tissue damage relative to GRIN lenses. Additional histology in a wider range of species will be needed to compare tissue response between implanted fiber bundles and GRIN lenses.
For both recording and stimulating, the self-splaying property means that the fiber distribution will be incoherent—the position of the fibers is unknown once implanted. Because of this, the fibers are not able to elucidate absolute spatial patterns in neural activity. Despite not being able to identify the absolute position of the fibers in the tissue during usage, selectively illuminating individual fibers (e.g., by scanning a laser across the imaging surface with a galvanometer) and detecting the amount of light collected by all other fibers might serve as a proxy for measuring relative distances between fibers in the tissue [50].
When recording via the bundle, it is only possible to measure a single optical intensity value for each fiber, and each fiber will act as the optical equivalent of a local field potential. Yet, by examining correlations across fibers resulting from overlapping light fields, the recorded signals may be amenable to known source separation techniques such as independent component analysis [51] or bayesian source separation [52], achieving an optical form of “spike sorting.”
For stimulation, a digital micromirror device (DMD) can be used to project patterned light onto the polished imaging surface at the end of the fiber bundle. Given attenuation measurements and sensitivity profile modeling, 2.5 μW optical power could be coupled into a fiber to stimulate neurons localized near the fiber aperture.
The simulations presented here are limited to normalized, noise-free measurements of the optical profile for individual fibers and assume uniform and consistent optical probe expression within a target neural population modeled as point sources. Such simulations enable estimating the the region with which the fibers interface, and the relative intensity of excitation/stimulation light delivery and the relative round-trip fluorescence yield. By incorporating the expression and efficiency of the relevant fluorescent probe, the model is capable of calculating absolute power measurements and evaluating signal to noise performance.
The histology presented here suggests a new class of brain implants based on self-splaying microfibers, similar to previous work with nickel chromium aluminum microelectrode brushes used for chronic electrophysiology in primates [53], but using 10–100 times more fibers per bundle, where each fiber is approximately 6 times less stiff (based on the area moment of inertia). A large number of ultrasmall fibers can be implanted in the brain while minimizing damage near the active end of the implant. This principle can apply to optical fibers as illustrated here, or to new electrode arrays, such as the carbon fiber ultramicroelectrode array [54], or silicon carbide ultramicroelectrodes [55, 56]. As high density interconnect solutions are developed for ultramicroelectrode arrays, we anticipate seeing the principle of self-splaying microfiber interfaces extended to the electrical domain [57].
Self-splaying optical microfibers compliment a number of new techniques aiming to achieve high channel count optical interfaces, such as multi-site fiber photometry [58] and optoelectronic probes with embedded waveguides or with on-device, implantable light sources and detectors [59, 60, 61]. Technique development relevant to each of these methods will have broader repercussions, such as on-device μLED illumination and CMOS sensors to achieve fully head-mounted and wireless optical interfaces; silicon waveguides and switches for lithographic manufacture of optical implants; and optical gratings to better localize and multiplex signals [62]. The self-splaying form factor described here will benefit from these developments, with future iterations potentially moving illumination and sensing optics to a wireless, head-mounted device.
References
- [1].Emiliani Valentina, Cohen Adam E, Deisseroth Karl, and Hausser Michael. All-Optical Interrogation of Neural Circuits. Journal of Neuroscience, 35(41):13917–13926, October 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gong Yiyang, Huang Cheng, Li Jin Zhong, Grewe Benjamin F, Zhang Yanping, Eismann Stephan, and Schnitzer Mark J. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science (New York, N.Y.), 350(6266):1361–1366, November 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nicholas G Horton Ke Wang, Kobat Demirhan, Clark Catharine G, Wise Frank W, Schaffer Chris B, and Xu Chris. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nature Photonics, 7(3):205–209, March 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang Tianyu, Wang Mengran, Feng Danielle D, Horton Nicholas G, Cruz-Hern aacutendez Jean C, Cheng Yu-Ting, Reimer Jacob, Tolias Andreas S, Nishimura Nozomi, Ouzounov Dimitre G, and Xu Chris. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nature Methods, 14(4):1–5, February 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zong Weijian, Wu Runlong, Li Mingli, Hu Yanhui, Li Yijun, Li Jinghang, Rong Hao, Wu Haitao, Xu Yangyang, Lu Yang, Jia Hongbo, Fan Ming, Zhou Zhuan, Zhang Yunfeng, Wang Aimin, Chen Liangyi, and Cheng Heping. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nature Methods, 14(7):1–10, May 2017. [DOI] [PubMed] [Google Scholar]
- [6].Dana Hod, Mohar Boaz, Sun Yi, Narayan Sujatha, Gordus Andrew, Jeremy P Hasseman Getahun Tsegaye, Graham T Holt Amy Hu, Walpita Deepika, Patel Ronak, Macklin John J, Bargmann Cornelia I, Ahrens Misha B, Schreiter Eric R, Jayaraman Vivek, Looger Loren L, Svoboda Karel, and Kim Douglas S. Sensitive red protein calcium indicators for imaging neural activity. Technical report, February 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jung Juergen C, Mehta Amit D, Aksay Emre, Stepnoski Raymond, and Schnitzer Mark J. In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. Journal of Neurophysiology, 92(5):3121–3133, November 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Barretto Robert P J, Messerschmidt Bernhard, and Schnitzer Mark J. In vivo fluorescence imaging with high- resolution microlenses. Nature Publishing Group, 6(7):511–512, June 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Andermann Mark L, Gilfoy Nathan B, Goldey Glenn J, Sachdev Robert N S, Wölfel Markus, McCormick David A, Reid R Clay, and Levene Michael J. Chronic Cellular Imaging of Entire Cortical Columns in Awake Mice Using Microprisms. Neuron, 80(4):900–913, November 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cui Guohong, Jun Sang Beom, Jin Xin, Pham Michael D, Vogel Steven S, Lovinger David M, and Costa Rui M. Concurrent activation of striatal direct and indirect pathways during action initiation. 494(7436):238–242, February 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dombeck Daniel A, Harvey Christopher D, Tian Lin, Looger Loren L, and Tank David W. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nature Neuroscience, 13(11):1433–1440, November 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Seymour John P and Kipke Daryl R. Neural probe design for reduced tissue encapsulation in CNS. Biomaterials, 28(25):3594–3607, September 2007. [DOI] [PubMed] [Google Scholar]
- [13].Szarowski DH, Andersen MD, Retterer S, Spence AJ, Isaacson M, Craighead HG, Turner JN, and Shain W. Brain responses to micro-machined silicon devices. Brain Research, 983(1–2):23–35, September 2003. [DOI] [PubMed] [Google Scholar]
- [14].Polikov Vadim S, Tresco Patrick A, and Reichert William M. Response of brain tissue to chronically implanted neural electrodes. Journal of neuroscience methods, 148(1):1–18, October 2005. [DOI] [PubMed] [Google Scholar]
- [15].McConnell George C, Rees Howard D, Levey Allan I, Gutekunst Claire-Anne, Gross Robert E, and Bellamkonda Ravi V. Implanted neural electrodes cause chronic, local inflammation that is correlated with local neurodegeneration. Journal of Neural Engineering, 6(5):056003–13, August 2009. [DOI] [PubMed] [Google Scholar]
- [16].Freire Marco Aurelio M, Morya Edgard, Faber Jean, Santos Jose Ronaldo, Guimaraes Joanilson S, Lemos Nelson A M, Sameshima Koichi, Pereira Antonio, Ribeiro Sidarta, and Nicolelis Miguel A L. Comprehensive Analysis of Tissue Preservation and Recording Quality from Chronic Multielectrode Implants. PloS one, 6(11):e27554–15, November 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hayn Linda and Koch Michael. Suppression of excitotoxicity and foreign body response by memantine in chronic cannula implantation into the rat brain. Brain Research Bulletin, 117:54–68, August 2015. [DOI] [PubMed] [Google Scholar]
- [18].Hayn Linda, Deppermann Linda, and Koch Michael. Reduction of the foreign body response and neuroprotection by apyrase and minocycline in chronic cannula implantation in the rat brain. Clinical and Experimental Pharmacology and Physiology, 44(2):313–323, January 2017. [DOI] [PubMed] [Google Scholar]
- [19].Goss-Varley Monika, Dona Keith R, McMahon Justin A, Shoffstall Andrew J, Ereifej Evon S, Lindner Sydney C, and Capadona Jeffrey R. Microelectrode implantation in motor cortex causes fine motor deficit: Implications on potential considerations to Brain Computer Interfacing and Human Augmentation. Scientific Reports, 7(1):1–12, November 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Seymour John P and Kipke Daryl R. Fabrication of polymer neural probes with sub-cellular features for reduced tissue encapsulation. Conference proceedings : … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference, 1:4606–4609, 2006. [DOI] [PubMed] [Google Scholar]
- [21].Yoshida Kozai Takashi D, Langhals Nicholas B, Patel Paras R, Deng Xiaopei, Zhang Huanan, Smith Karen L, Lahann Joerg, Kotov Nicholas A, and Kipke Daryl R. Ultrasmall implantable composite microelectrodes with bioactive surfaces for chronic neural interfaces. Nature Materials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Patel Paras R, Na Kyounghwan, Zhang Huanan, Kozai Takashi D Y, Kotov Nicholas A, Yoon Euisik, and Chestek Cynthia A. Insertion of linear 8.4 μm diameter 16 channel carbon fiber electrode arrays for single unit recordings. Journal of Neural Engineering, 12(4), June 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gerstner Klaus, Plichta Armin, Schlatterbeck Dirk, Weisser Michael, Brix Peter, Sommer Martin, Rubino Robert A Jr, Bonja Jeffrey A, Strack Richard, Henze Inka, and Arsenault Paul. Leached fiber bundle and method. US Patent Office, 2004. [Google Scholar]
- [24].Long Michael A, Jin Dezhe Z, and Fee Michale S. Support for a synaptic chain model of neuronal sequence generation. Nature Publishing Group, 468(7322):394–399, November 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Boas David A, Culver J, Stott J, and Dunn Andrew. Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head. Optics express, 10(3):159–170, February 2002. [DOI] [PubMed] [Google Scholar]
- [26].Yi Ji and Backman Vadim. Imaging a full set of optical scattering properties of biological tissue by inverse spectroscopic optical coherence tomography. Optics letters, 37(21):4443–4445, November 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bouchet Audrey, Lemasson Benjamin, Le Duc Géraldine, Maisin Cécile, Bräuer-Krisch Elke, Siegbahn Erik Albert, Renaud Luc, Khalil Enam, Chantal Rémy Cathy Poillot, Bravin Alberto, Laissue Jean A, Barbier Emmanuel L, and Serduc Raphaël. Preferential effect of synchrotron microbeam radiation therapy on intracerebral 9L gliosarcoma vascular networks. International journal of radiation oncology, biology, physics, 78(5):1503–1512, December 2010. [DOI] [PubMed] [Google Scholar]
- [28].Raabe Brigitte M, Artwohl James E, Purcell Jeanette E, Lovaglio Jamie, and Fortman Jeffrey D. Effects of weekly blood collection in C57BL/6 mice. Journal of the American Association for Laboratory Animal Science : JAALAS, 50(5):680–685, September 2011. [PMC free article] [PubMed] [Google Scholar]
- [29].Kollias N and Gratzer W. Tabulated molar extinction coefficient for hemoglobin in water. Wellman Laboratories, Harvard Medical School, Boston, 1999. [Google Scholar]
- [30].Hillman Elizabeth M C, Boas David A, Dale Anders M, and Dunn Andrew K. Laminar optical tomography: demonstration of millimeter-scale depth-resolved imaging in turbid media. Optics letters, 29(14):1650–1652, July 2004. [DOI] [PubMed] [Google Scholar]
- [31].Burgess Sean A, Bouchard Matthew B, Yuan Baohong, and Hillman Elizabeth M C. Simultaneous multiwavelength laminar optical tomography. Optics letters, 33(22):2710–2712, November 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bottjer Sarah W, Glaessner Sherri L, and Arnold Arthur P. Ontogeny of brain nuclei controlling song learning and behavior in zebra finches. Journal of Neuroscience, 5(6):1556–1562, June 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kosubek-Langer Jennifer, Schulze Lydia, and Scharff Constance. Maturation, Behavioral Activation, and Connectivity of Adult-Born Medium Spiny Neurons in a Striatal Song Nucleus. Frontiers in Neuroscience, 11:101–12, June 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Vedaldi Andrea and Fulkerson Brian. VLFeat: An Open and Portable Library of Computer Vision Algorithms, 2008. [Google Scholar]
- [35].Lowe David G. Object recognition from local scale-invariant features In Computer vision, 1999. The proceedings of the seventh IEEE international conference on, pages 1150–1157. Ieee, 1999. [Google Scholar]
- [36].Lowe David G. Distinctive Image Features from Scale-Invariant Keypoints. International journal of computer vision, 60(2):91–110, 2004. [Google Scholar]
- [37].Unal-Çevik Isın, Kılınç Münire, Gursoy-Ozdemir Yasemin, Gurer Gunfer, and Dalkara Turgay. Loss of NeuN immunoreactivity after cerebral ischemia does not indicate neuronal cell loss: a cautionary note. Brain Research, 1015(1–2):169–174, July 2004. [DOI] [PubMed] [Google Scholar]
- [38].Collombet Jean-Marc, Masqueliez Catherine, Four Elise, Burckhart Marie-France, Bernabé Denis, Baubichon Dominique, and Lallement Guy. Early reduction of NeuN antigenicity induced by soman poisoning in mice can be used to predict delayed neuronal degeneration in the hippocampus. Neuroscience Letters, 398(3):337–342, May 2006. [DOI] [PubMed] [Google Scholar]
- [39].Duan Wei, Zhang Yu-Ping, Hou Zhi, Huang Chen, Zhu He, Zhang Chun-Qing, and Yin Qing. Novel Insights into NeuN: from Neuronal Marker to Splicing Regulator. Molecular Neurobiology, 53(3):1637–1647, February 2015. [DOI] [PubMed] [Google Scholar]
- [40].Michelson Nicholas J, Vazquez Alberto L, Eles James R, Salatino Joseph W, Purcell Erin K, Williams Jordan J, Cui X Tracy, and Kozai Takashi D Y. Multi-scale, multi-modal analysis uncovers complex relationship at the brain tissue-implant neural interface: new emphasis on the biological interface. Journal of Neural Engineering, 15(3):033001–24, April 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yizhar Ofer, Fenno Lief E, Davidson Thomas J, Mogri Murtaza, and Deisseroth Karl. Optogenetics in Neural Systems. Neuron, 71(1):9–34, July 2011. [DOI] [PubMed] [Google Scholar]
- [42].Kurt M Ayberk, Kafa M Ilker, Dierssen Mara, and Davies D Ceri. Deficits of neuronal density in CA1 and synaptic density in the dentate gyrus, CA3 and CA1, in a mouse model of Down syndrome. Brain Research, 1022(1–2):101–109, October 2004. [DOI] [PubMed] [Google Scholar]
- [43].Richards Kay L, Kurniawan Nyoman D, Yang Zhengyi, Kim Tae Hwan, Keller Marianne D, Low Jun, Ullmann Jeremy F P, Cole Stacey, Foong Samuel, Galloway Graham J, Reid Christopher A, Paxinos George, Reutens David C, and Petrou Steven. Hippocampal volume and cell density changes in a mouse model of human genetic epilepsy. Neurology, 80(13):1240–1246, March 2013. [DOI] [PubMed] [Google Scholar]
- [44].Lee Seon A, Holly Kevin S, Voziyanov Vladislav, Villalba Stephanie L, Tong Rudi, Grigsby Holly E, Glasscock Edward, Szele Francis G, Vlachos Ioannis, and Murray Teresa A. Gradient Index Microlens Implanted in Prefrontal Cortex of Mouse Does Not Affect Behavioral Test Performance over Time. PloS one, 11(1):e0146533–19, January 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Biran Roy, Martin David C, and Tresco Patrick A. Neuronal cell loss accompanies the brain tissue response to chronically implanted silicon microelectrode arrays. Experimental Neurology, 195(1):115–126, September 2005. [DOI] [PubMed] [Google Scholar]
- [46].Winslow Brent D and Tresco Patrick A. Quantitative analysis of the tissue response to chronically implanted microwire electrodes in rat cortex. Biomaterials, 31(7):1558–1567, March 2010. [DOI] [PubMed] [Google Scholar]
- [47].Welkenhuysen M, Andrei A, Ameye L, Eberle W, and Nuttin B. Effect of Insertion Speed on Tissue Response and Insertion Mechanics of a Chronically Implanted Silicon-Based Neural Probe. IEEE Transactions on Biomedical Engineering, 58(11):3250–3259, September 2011. [DOI] [PubMed] [Google Scholar]
- [48].Harris JP, Capadona JR, Miller RH, Healy BC, Shanmuganathan K, Rowan SJ, Weder C, and Tyler DJ. Mechanically adaptive intracortical implants improve the proximity of neuronal cell bodies. Journal of Neural Engineering, 8(6):066011–14, November 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bedell Hillary W, Hermann John K, Ravikumar Madhumitha, Lin Shushen, Rein Ashley, Li Xujia, Molinich Emily, Smith Patrick D, Selkirk Stephen M, Miller Robert H, Sidik Steven, Taylor Dawn M, and Capadona Jeffrey R. Targeting CD14 on blood derived cells improves intracortical microelectrode performance. Biomaterials, 163:163–173, May 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Heshmat Barmak, Lee Ik Hyun, and Raskar Ramesh. Optical brush: Imaging through permuted probes. Nature Publishing Group, 6:1–9, January 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hyvärinen A and Oja E. Independent component analysis: algorithms and applications. Neural networks : the official journal of the International Neural Network Society, 13(4–5):411–430, May 2000. [DOI] [PubMed] [Google Scholar]
- [52].Knuth Kevin H. A Bayesian approach to source separation. arXiv.org, 2002.
- [53].Krüger Jürgen. Seven years of recording from monkey cortex with a chronically implanted multiple microelectrode. Frontiers in Neuroengineering, pages 1–9, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Guitchounts Grigori, Markowitz Jeffrey E, Liberti William A III, and Gardner Timothy J. A carbon-fiber electrode array for long-term neural recording. Journal of Neural Engineering, 10(4):046016–14, July 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Deku Felix, Cohen Yarden, Joshi-Imre Alexandra, Kanneganti Aswini, Gardner Timothy J, and Cogan Stuart. Amorphous silicon carbide ultramicroelectrode arrays for neural stimulation and recording. Journal of Neural Engineering, pages 1–21, September 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Joseph J Pancrazio Felix Deku, Ghazavi Atefeh, Stiller Allison M, Rihani Rashed, Frewin Christopher L, Varner Victor D, Gardner Timothy J, and Cogan Stuart F. Thinking Small: Progress on Microscale Neurostimulation Technology. Neuromodulation: Technology at the Neural Interface, 84:203–8, October 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wray William. Scalable Bundle Design for Massively Parallel Neuronal Recordings In Vivo PhD thesis, December 2017. [Google Scholar]
- [58].Guo Qingchun, Zhou Jingfeng, Feng Qiru, Lin Rui, Gong Hui, Luo Qingming, Zeng Shaoqun, Luo Minmin, and Fu Ling. Multi-channel fiber photometry for population neuronal activity recording. Biomedical Optics Express, 6(10):3919–13, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Warden Melissa R, Cardin Jessica A, and Deisseroth Karl. Optical Neural Interfaces. Annual Review of Biomedical Engineering, 16(1):103–129, July 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wu Fan, Stark Eran, Ku Pei-Cheng, Wise Kensall D, Buzsáki György, and Yoon Euisik. Monolithically Integrated uLEDs on Silicon Neural Probes for High-Resolution Optogenetic Studies in Behaving Animals. Neuron, 88(6):1136–1148, December 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Segev Eran, Reimer Jacob, Moreaux Laurent C, Fowler Trevor M, Chi Derrick, Sacher Wesley D, Lo Maisie, Deisseroth Karl, Tolias Andreas S, Faraon Andrei, and Roukes Michael L. Patterned photostimulation via visible-wavelength photonic probes for deep brain optogenetics. Neurophotonics, 4(1):011002–16, January 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Segev Eran, Fowler Trevor, Faraon Andrei, and Roukes Michael L. Visible array waveguide gratings for applications of optical neural probes. In Hirschberg Henry, Madsen Steen J, Jansen E Duco, Luo Qingming, Mohanty Samarendra K, and Thakor Nitish V, editors, SPIE BiOS, pages 93052L–6. SPIE, March 2015. [Google Scholar]