Abstract
We enrolled 150 patients in a prospective multi-center study of children with acute myeloid leukemia undergoing hematopoietic stem cell transplant (HSCT) comparing detection of measurable residual (MRD) disease by a “Difference from Normal” flow cytometry (ΔN) approach with assessment of Wilms tumor 1 (WT-1) gene expression without access to the diagnostic specimen. Prospective analysis of the specimens using this approach showed that 23% of patients being screened for HSCT had detectable residual disease by ΔN (0.04–53%). Of those patients who proceeded to transplant as being in morphologic remission, 10 had detectable disease (0.04–14%) by ΔN. The disease free survival of this group was 10% (0–35%), compared to 55% (46–64; <0.001) for those without disease. The ΔN assay was validated using the postHSCT specimen by sorting the abnormal or suspicious cells to confirm recipient or donor origin by chimerism studies. All 15 of the patients who had confirmation of tumor detection relapsed, while the 2 patients with suspicious phenotype cells lacking this confirmation did not. The phenotype of the relapse specimen was then used retrospectively to assess the pre-HSCT specimen, allowing identification of additional samples with low levels of MRD involvement that were previously undetected. Quantitative assessment of Wilms tumor 1 gene expression was not predictive of relapse or other outcomes in either pre- or post-transplant specimens. Measureable residual disease detected by ΔN was highly specific, but did not identify a majority of relapsing patients. The application of the assay was limited by poor quality among onethird of the specimens and lack of a diagnostic phenotype for comparison.
Keywords: Cytogenetics and Molecular Genetics, Minimal Residual Disease, Stem Cell Transplantation, Laboratory Hematology, Acute Myeloid Leukemia
INTRODUCTION
In a prospective, multi-center study of children undergoing hematopoietic stem cell transplantation (HSCT) for AML, we compared the efficacy of “Difference from Normal” (ΔN) flow cytometry approach for residual disease detection with assessment of Wilms Tumor 1 (WT-1) gene expression in a setting where the diagnostic specimen was not available.
It is known that AML patients with measurable residual disease (MRD) following chemotherapy have poorer outcomes as they experience more relapse.1,2,3,4,5 Furthermore, data in children suggest that intensifying therapy in AML patients with persistent MRD after chemotherapy may improve outcomes.6 Walter et al. have shown that adult AML patients with MRD at time of HSCT experience more relapse post-HSCT in both MAC and reduced-intensity (RIC) settings.7,8 However, insufficient data exist regarding the impact of MRD at the time of HSCT or soon after HSCT in children.
The flow cytometric technique that has been most commonly used to detect MRD, leukemia-associated immunophenotype (LAIP), uses the phenotype identified at diagnosis to follow the response to therapy. LAIP cannot be used without a diagnostic specimen and is prone to errors when the leukemic phenotype changes following treatment as a result of clonal evolution or selection.9,10,11,12 The ΔN flow cytometry approach using a standardized panel of reagents avoids these pitfalls by focusing on the normal cells within the regenerating marrow first, then identifying cell populations that do not match these patterns in multidimensional data space.4,5,13
Molecular techniques have been tested for MRD detection using real-time quantitative reverse transcriptase polymerase chain reaction (RT-qPCR). RT-qPCR detects (or can detect) genetic alterations such as NPM1, FLT3-ITD, and RUNX1. This technique requires prior knowledge of the genetic mutation and is limited, however, as no more than 50% of patients have such genetic lesions, only half of the patients can be evaluated for MRD by PCR.14,15 High WT1 gene expression has been reported in 90100% of AML and 60–90% of acute lymphoblastic leukemia (ALL) blasts16 and can be considered a surrogate of tumor burden. This has led to its increasing use in measuring MRD in leukemia patients after initial therapy,17,18 with small studies suggesting a role for its use prior to HSCT to predict outcome. In a single-institution pediatric AML study, our group showed that after HSCT, 76% of patients with high pre-HSCT WT1 expression relapsed, in contrast to 0% of the patients with low pre-HSCT WT1 expression.19 While WT1 expression can be assessed using peripheral blood (PB) samples, the main concern remains that WT1 is not leukemia-specific, which may increase difficulty in identifying low-level MRD expression. Furthermore, WT1 and transplant outcomes has not been studied in a multi-institutional setting.
Validation of testing modalities using clinical studies is often confused with the application of the assay to predicting response to therapy. In a validation study, it is necessary to identify true positive from true negative specimens. HSCT provides such a setting since abnormal cells identified by flow cytometry post HSCT can be purified and tested by chimerism studies to prove the abnormal cells come from the recipient, not the donor. Specific genetic mutations are not required to confirm the presence of AML. Once the parameters of the assay are defined, the assay can be used determine the presence of tumor at various time points and compare the results with other detection methods.
In this manuscript we show that ΔN can specifically identify relapsing AML at the post HSCT stage with confirmation by chimerism studies. By identifying which patients had detectable tumor post HSCT the detection limits of the assay can be determined at other time points both pre- and post-HSCT and compared to the results of other assays. From such data it is possible to assess relapse risk throughout the course of treatment. Correlative studies with WT-1 assay provide a basis to compare relative predictive value of the approaches.
METHODS
Patients ages 0–21 years with AML in complete morphologic remission (CR) (<5% blasts in bone marrow (BM) with peripheral absolute neutrophil count (ANC) >500 cells/mm3) at time of HSCT were recruited from 34 centers. Patients with AML in CR1 or greater, including those with therapy-related AML, were allowed to enroll. Patients undergoing their first allogeneic HSCT were treated with a myeloablative conditioning (MAC) regimen, and received a BM, peripheral blood stem cell (PBSC), or cord blood unit(s) (CBUs) (single or double) graft from a human leukocyte antigen (HLA)-identical sibling donor, ≥7/8 HLA related or unrelated donor (PBSC and BM), or a ≥4/6 HLA unrelated donor (CBUs). Standard HSCT eligibility criteria were also required.20
BM and peripheral blood (PB) samples were collected <3 weeks prior to initiation of the preparative regimen, and at day 42 (±14 days) and day 100 (± 20 days) post-HSCT. BMAs were collected in heparin or ethylenediaminetetraacetic acid (EDTA). Treatment with hematopoietic growth factors was discontinued for a minimum of 48 hours prior to collecting the samples (see supplement for additional methods).
Flow Cytometry
Specimens were processed as routine clinical specimens as previously described using a FACS Calibur (Becton Dickinson Biosciences, San Jose CA).13 Standardize panels of monoclonal antibodies (mAbs) used to assess hematopoietic cells in BM are shown in Table 1. Aberrant leukemia cells could be identified at frequencies down to 0.02% based on identification of clusters of a minimum of 40 events at least 0.5 decades from their normal counterparts.
Table 1:
Monoclonal antibody combinations
| Tube | Monoclonal antibodies | |||
|---|---|---|---|---|
| FITC | PE | PERCP | APC | |
| 1 | HLADR | CD11b | CD45 | CD34 |
| Clone | L243 (BD) | D12 (BD) | 2D1 (BD) | 8G12 (BD) |
| 2 | CD36 | CD38 | CD45 | CD34 |
| Clone | FA6.152(BC) | HB7(BD) | ||
| 3 | CD16 | CD13 | CD45 | CD34 |
| Clone | 3G8(BD) | L138 (BD) | ||
| 4 | CD14 | CD33 | CD45 | CD34 |
| Clone | MΦ/P9(BD) | P67.6(BD) | ||
| 5 | CD7 | CD56 | CD45 | CD34 |
| Clone | 4H9(BD) | My31(BD) | ||
| 6 | CD38 | CD117 | CD45 | CD34 |
| Clone | HIT2(Invitro) | 104D2(BD) | ||
| 7 | CD36 | CD64 | CD45 | CD34 |
| Clone | FA6.152(BC) | 22(T) | ||
| 8 | CD19 | CD123 | CD45 | CD34 |
| Clone | 4G7(BD) | 9F5(BD) | ||
BD: BD Biosciences, San Jose, CA, USA
BC: Beckman Coulter, Brea,CA, USA
Invitro: Invitrogen™, Thermo Fisher Scientific, San Diego, CA, USA
T: Trillium Diagnostics, LLC, Brewer, ME, USA
The ΔN assay is biased for specificity rather than sensitivity since a positive detection of AML dramatically changes treatment decisions. In a clinical setting at least 2 abnormalities are required to define an abnormal cell population. The assay requires two analysts to independently analyze each specimen and then compare results. If there is uncertainty about the presence of an abnormal myeloid progenitor population by either analyst, the specimen was called negative or suspicious. The quality of each specimen was defined based on total cellularity, hemodilution, and viability. Collection of fewer than 100,000 cells in each of 8 tubes, significant hemodilution of >50%, lymphoid predominance of >50%, or significant cell death resulted in a specimen quality classification of suboptimal or inadequate (see supplement for additional methods)21.
Post-transplant specimens identified as suspicious or positive, were sorted using a FACS Aria (Becton Dickinson Biosciences) and DNA extracted from the purified cells. DNA from suspected leukemia cells, mature lymphocytes or neutrophils was extracted (or subjected to whole genome amplification if adequate DNA was not obtained) prior to molecular chimerism studies (Repli-g, QIAGEN Inc., Valencia, CA, USA). Discrimination of the origin of the aberrant cells (donor vs. recipient) was established by multiplex short tandem repeat (STR) evaluation (PowerPlex® 16 HS system, Promega Corporation, Madison WI, USA).
WT1 gene extraction and quantification
Total RNA was extracted from BM and PB using QIAamp® RNA Blood Mini Kit (QIAGEN Inc., Valencia, CA). WT1 analysis was performed by the Chimerism Laboratory at Lurie Children’s Hospital. The RNA concentration and purity was quantified by measuring the absorbance at 260 and 280 nM with a spectrophotometer (Pharmacia Biotech GeneQuant, Piscataway, NJ,). A two-step real-time quantitative polymerase chain reaction (RQ-PCR) was conducted in a 20 μL reaction volume containing 1 μg/μL of total RNA from each sample. Samples with >50 copies/μg were considered positive (see supplement for additional methods).
Statistical Analysis
Kaplan-Meier estimates and log-rank tests were used to estimate and compare OS and DFS. Cumulative incidence and Gray’s test were conducted to estimate and compare cumulative incidence of relapse and TRM. For sensitivity and specificity analysis at 24 months, R packages ‘survival ROC’ and ‘time ROC’ were used for OS/DFS22 and relapse/TRM,23 respectively. To examine the best cut points for WT1, six cut points based on 20%, 30%, …, 70% percentiles of WT1 and pre-specified cut points of 200 copies in the PB and 1300 copies in the BM were examined for each outcome using the univariate Cox model with the Sidak correction for multiple testing adjustment.
RESULTS
Relationship between flow cytometry and outcomes
For the prospective analysis of the specimens starting with the pre HSCT specimen, no phenotypic data were available for comparison to determine presence of residual disease. Although 150 patients were initially enrolled in the study, pre-HSCT specimens were not obtained from 6 patients. Of the 144 patients who submitted pre-HSCT specimens, abnormal myeloid progenitor cells were identified in 22 cases (0.04–53%). A review of the quality of the specimens in which no aberrant myeloid cells were identified revealed only 81 of the 122 negative samples (66%) to be adequate for MRD detection. Therefore, the frequency of detection of abnormal myeloid progenitor cells in patients who had completed therapy and were being screened for HSCT who sent adequate samples was 22 of 103 samples (21%). Twelve of these 22 residual disease positive patients (with abnormal cells from 0.5–53%) did not proceed to HSCT (due to lack of remission or other disqualifying conditions). Since clinicians did not receive the study MRD results real-time, these treatment decisions were based on local pathology and MRD. And since the patients that did not get HSCT did not continue on study, we do not have information on their course thereafter.
Among patients with pre-HSCT specimens who proceeded to HSCT (all of whom were defined as CR by morphology), 10 of 123 were identified as harboring aberrant myeloid progenitor cells at frequencies of 0.04–14% (median 3.4%). Four of these patients had >5% abnormal progenitor cells by flow cytometry with one additional patient at 4.7%. Of the 113 HSCT patients without MRD, only 76 (67%) had an adequate BM specimen submitted. Therefore, the frequency of detection of abnormal myeloid progenitor cells in patients undergoing HSCT with adequate submitted specimens was 11.6%. Two-year outcomes for the MRD positive patients identified pre HSCT were poor; DFS was 10% (0–35%), 7 relapsed and 2 died of TRM, compared with MRD-negative patients where DFS was observed in 55% (46–64%) (p<0.001)(Table 3).
Table 3:
Outcomes related to pre-HSCT MRD status by flow cytometry using ΔN approach
| Absence (N = 113) ‡ | Presence (N = 10) ‡ | ||||
|---|---|---|---|---|---|
| Outcomes | N Eval | Prob (95% CI) | N Eval | Prob (95% CI) | p-value |
| Transplant-related mortality | 113 | 10 | 0.43 | ||
| Day 100 | 6 (3–11)% | 20 (2–49)% | 0.22 | ||
| 6 months | 12 (7–19)% | 20 (2–49)% | 0.47 | ||
| 1-year | 14 (8–21)% | 20 (2–49)% | 0.56 | ||
| 2-year* | 14 (8–21)% | 20 (2–49)% | 0.56 | ||
| Relapse incidence | 113 | 10 | 0.03 | ||
| Day 100 | 14 (8–21)% | 30 (7–60)% | 0.29 | ||
| 6 months | 24 (17–32)% | 40 (13–70)% | 0.37 | ||
| 1-year | 29 (21–38)% | 50 (21–79)% | 0.25 | ||
| 2-year* | 32 (23–40)% | 70 (40–93)% | 0.01 | ||
| Disease free survival | 113 | 10 | 0.002 | ||
| Day 100 | 81 (74–88)% | 50 (21–79)% | 0.05 | ||
| 6 months | 64 (55–72)% | 40 (13–70)% | 0.14 | ||
| 1-year | 57 (47–66)% | 30 (7–60)% | 0.08 | ||
| 2-year* | 55 (46–64)% | 10 (0–35)% | <0.001 | ||
| Overall survival | 113 | 10 | 0.001 | ||
| Day 100 | 91 (85–96)% | 80 (51–98)% | 0.39 | ||
| 6 months | 77 (69–84)% | 70 (40–93)% | 0.64 | ||
| 1-year | 69 (60–77)% | 30 (7–60)% | 0.010 | ||
| 2-year* | 63 (54–72)% | 20 (2–49)% | 0.001 | ||
2 year: 2 patients lost to follow up
Excludes patients without pre-HSCT BM MRD assessment, 86/123 specimens were classified as adequate at the time of analysis.
Post-HSCT 21 patients were identified as having abnormal myeloid progenitor cells (0.03–78%), 5 observed on day 42, 13 detected on day 100, and 3 were identified on both days 42 and 100. Four of these 21 patients were detected pre-HSCT. Cell sorting to enrich for the aberrant cell population was successful for 17/21 of these positive specimens. DNA from these enriched cells was then subjected to STR analysis to confirm the genetic origin of the abnormal cells, donor or recipient. In 15 of the 17 positive specimens, the enriched tumor population demonstrated a majority of cells to be of recipient origin, confirming that the abnormal cell population was leukemia. All fifteen of the patients, confirmed to have aberrant myeloid progenitor cells of recipient origin, relapsed.
The 2 patients with suspicious cells that were not confirmed to be of recipient origin did not relapse. Retrospective analysis of these two specimens showed one specimen (identified at 0.03%) resulted from a technical false positive while the other specimen was identified as suspicious at 0.06% on both days 42 and 100 with co-expression of CD7 and CD34 as the sole phenotypic abnormality. The assay requires at least 2 quantitative phenotypic abnormalities in order to be called positive in a clinical setting. Therefore, the combination of ΔN followed by cell sorting was 100% predictive of relapse. The kinetics of relapse following detection of abnormal cells is shown in Figure 1.
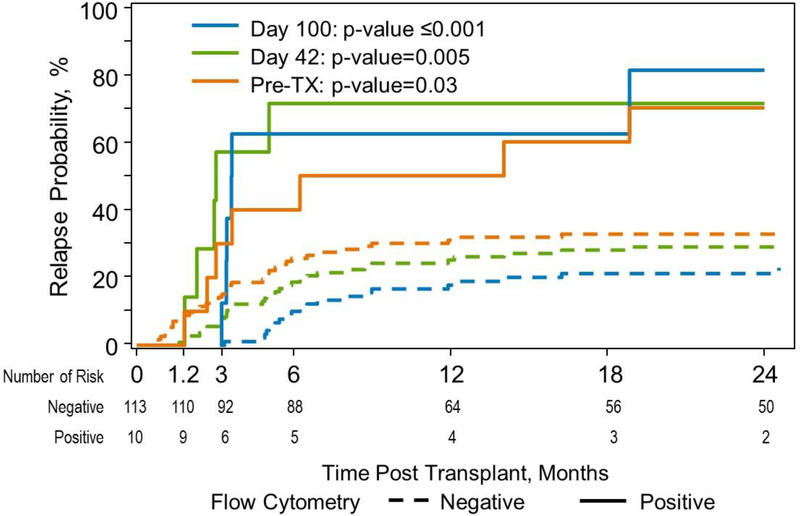
Figure 1:
Relapse Incidence at 3 timepoints for patients with abnormal myeloid progenitor cells identified by flow cytometry
The phenotypes of the cells in the 15 patients who were confirmed to harbor tumor were used as the basis for retrospective analysis of the corresponding pre HSCT specimens. Assessment of the pre HSCT specimens for quality revealed that only 9/15 identified as positive in the post-HSCT specimen had pre-HSCT specimens that were classified as adequate for assessment upon retrospective analysis. Using the relapse specimen as a guide for defining the tumor phenotype, 2 of these 9 patients were retrospectively defined as positive, both at 0.04% while 3 others were deemed suspicious at levels of 0.01–0.08%. The remaining 4 specimens had no evidence of abnormal myeloid progenitor cells with a phenotype seen in the relapse specimen, <0.02%.
Relationship between WT1 MRD and outcomes
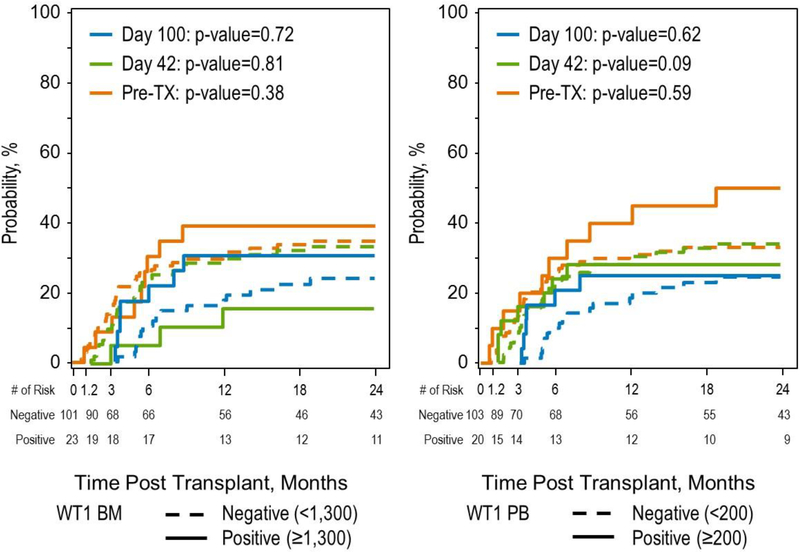
There was no statistically significant correlation of WT1 levels with outcome when measured in the pre-HSCT BM or PB specimens. The analysis was done using positive/negative as well as pre-specified cutoffs of ≥1300 copies in the BM and ≥200 copies in the PB.24 We examined whether any correlation with outcomes (TRM, RI, DFS, OS) existed between day 42 or day 100 WT1 levels, in the BM and PB, using either pre-specified cutoffs or positive/negative. The only statistically significant correlation was that patients with positive WT1 in the BM at day 42 had a 2-year OS of 58% (44–71%) versus 76% (64–86%) (p=0.05), for those with a negative WT1. There was no correlation with DFS or incidence of relapse, however. The kinetics of relapse by WT1 (BM and PB by pre-specified cutoffs) is shown in Figure 2.
Figure 2:
Relapse Incidence at 3 timepoints for patients with WT1 ≥1300 in the bone marrow and ≥200 in the peripheral blood.
DISCUSSION
In this multi-center PBMTC study, we show that a high proportion of pediatric patients who are being screened for HSCT have detectable residual disease. Although some of these patients were identified by morphology and excluded from the study, patients with up to 14% residual disease were not detected using traditional approaches. Without a diagnostic specimen the ΔN approach should be used with caution in this setting, as 10 patients were identified pre HSCT, however, when tumor burden is low, confirmation by molecular or FISH testing may be challenging. Access to the leukemia phenotype (after HSCT) allowed the detection of 2 additional patients with residual disease at levels below 0.1% while 3 additional patients were deemed suspicious pre-HSCT, again <0.1%. Four patients who relapsed post HSCT did not have detectable residual disease at levels >0.02%. These data confirm that some patients, relapsing post HSCT, have residual disease either below the assay cut off or not present in the bone marrow aspirate assessed.
This prospective, multicenter analysis shows that detection of residual AML is affected by the quality of the submitted specimen. In this study 1/3 of the specimens were not adequate for precise detection of residual disease. This characteristic of the specimen is often ignored when performing residual disease analyses, whether by flow cytometry or molecular techniques. Specimen quality was reduced by use of EDTA as an anticoagulant (reducing neutrophil and monocyte viability), use of later bone marrow draws (with subsequent hemodilution) for a test not directly related to patient stratification, or low cellularity of the specimen. The patients have received significant chemotherapy, which dramatically affects the cellularity of the bone marrow, and may have contributed to inadequate specimens. Delay in shipping was not observed to be the cause of the poor specimens. In a clinical setting where the results are used to define subsequent therapy, a small, 2ml aliquot of the first draw obtained using heparin and shipped immediately results in the highest specimen quality.
The absence of a quantitative phenotype from a diagnostic specimen in this clinical study mirrors the use of the MRD assays in a real clinical setting. The initial diagnosis and treatment is often performed at one institution while allogeneic HSCT maybe completed at a separate institution. The ΔN approach can be used even if the original phenotype is not available for comparison or if the phenotype changes. Combining residual disease detection with cell sorting for molecular confirmation increases the confidence in the results for making such an important clinical decision.
Monitoring for residual disease post HSCT using ΔN can be predictive of imminent relapse (Figure 1). The time course of relapse suggests that monthly monitoring during the first 6 months would provide an opportunity for early detection of relapse, especially if chimerism studies confirm the cell population identified is of recipient, not donor origin.
Given that prior publications have shown the WT1 assay to be predictive of relapse,15,25,26 the results of this portion of the study were unexpected. In previous studies, log reduction was measured in comparison to the diagnostic specimen run at the same time. In this study, no diagnostic specimen was available as an internal reference so only the amounts of WT1 present could be determined. The WT1 assay did not predict eventual outcomes in comparison to pre-defined thresholds or positive vs negative.
Since the WT1 assay is RNA based, degradation of RNA within the specimen is potentially problematic. Flow cytometry showed that 1/3 of the specimens obtained preHSCT were not adequate suggesting that specimen quality may play a role in interpreting the results. However, since the two assays were performed at different laboratories, the assessment of quality on one specimen may not be applicable to the specimen analyzed in a different laboratory. Following HSCT only 2 time points were analyzed and the levels of WT1 may not have been sufficient to be predictive, but this does not explain the lack of correlation pre-HSCT. If this assay is to be performed centrally for a broad number of centers, issues of specimen quality and reproducibility need to be addressed.
Table 2:
Characteristics of study population
| Variable | N (%) |
|---|---|
| Number of patients | 124 |
| Number of centers | 34 |
| Patient-related variables | |
| Age at HSCT, years | |
| Median (range) | 9 (1–21.5) |
| 0–9 | 67 (54) |
| 10–21 | 57 (46) |
| Sex | |
| Male | 71 (57) |
| Female | 53 (43) |
| Race | |
| Caucasian | 102 (82) |
| Non-Caucasian | 19 (15) |
| Declined/Unknown | 3 ( 2) |
| Performance Score | |
| ≥ 90 | 102 (82) |
| < 90 | 22 (18) |
| HCT-CI | |
| No comorbidity | 70 (56) |
| 1–2 | 31 (25) |
| ≥ 3 | 23 (18) |
| Disease-related variables | |
| Disease status | |
| CR1 | 67 (54) |
| CR2 | 46 (37) |
| Therapy-related AML at any stage | 11 ( 9) |
| Cytogenetics scoring | |
| Favorable | 10 (8) |
| Intermediate | 69 (56) |
| Poor | 41 (33) |
| Missing | 4 ( 3) |
| Transplant-related variables | |
| Graft type | |
| BM | 66 (53) |
| PBSC | 11 ( 9) |
| Single CBU | 38 (31) |
| Double CBU | 9 ( 7) |
| Donor type | |
| HLA identical sibling | 25 (20) |
| ≥ 7/8 HLA matched related | 5 ( 4) |
| ≥ 7/8 HLA matched unrelated | 94 (76) |
| Recipient CMV | |
| Negative | 55 (44) |
| Positive | 69 (56) |
| Conditioning regimen | |
| Busulfan-based | 99 (80) |
| TBI-based | 16 (13) |
| Treosulfan-based | 9 ( 7) |
| GVHD prophylaxis | |
| Post-HSCT Cyclophosphamide | 2 ( 2) |
| CNI + mycophenolate mofetil (MMF) | 55 (44) |
| CNI + methotrexate (MTX) | 59 (48) |
| CNI alone or with other | 8( 6) |
| Serotherapy | |
| Anti-thymocyte globulin (ATG) + alemtuzumab | 2 ( 2) |
| ATG alone | 56 (45) |
| alemtuzumab alone | 3 ( 2) |
| None | 63 (51) |
Highlights.
DFS was higher in pediatric AML patients that had MRD detected by the “difference-from-normal” flow cytometry (ΔN) pre-HSCT.
MRD detected by ΔN was highly specific, but did not identify a majority of relapsing patients.
Quantitative assessment of WT1 gene expression pre HSCT was not predictive of DFS.
ACKNOWLEDGEMENTS
The authors would like to thank the patients, study personnel, and care providers who participated in this study.
This work was supported by the St. Baldrick’s Foundation and the Otsuka Pharmaceutical Group.
The CIBMTR is supported primarily by Public Health Service Grant/Cooperative Agreement 5U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 4U10HL069294 from NHLBI and NCI; a contract HHSH250201200016C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014–17-1–2388 and N0014–17-1–2850 from the Office of Naval Research; and grants from *Actinium Pharmaceuticals, Inc.; *Amgen, Inc.; *Amneal Biosciences; *Angiocrine Bioscience, Inc.; Anonymous donation to the Medical College of Wisconsin; Astellas Pharma US; Atara Biotherapeutics, Inc.; Be the Match Foundation; *bluebird bio, Inc.; *Bristol Myers Squibb Oncology; *Celgene Corporation; Cerus Corporation; *Chimerix, Inc.; Fred Hutchinson Cancer Research Center; Gamida Cell Ltd.; Gilead Sciences, Inc.; HistoGenetics, Inc.; Immucor; *Incyte Corporation; Janssen Scientific Affairs, LLC; *Jazz Pharmaceuticals, Inc.; Juno Therapeutics; Karyopharm Therapeutics, Inc.; Kite Pharma, Inc.; Medac, GmbH; MedImmune; The Medical College of Wisconsin; *Mediware; *Merck & Co, Inc.; *Mesoblast; MesoScale Diagnostics, Inc.; Millennium, the Takeda Oncology Co.; *Miltenyi Biotec, Inc.; National Marrow Donor Program; *Neovii Biotech NA, Inc.; Novartis Pharmaceuticals Corporation; Otsuka Pharmaceutical Co, Ltd. – Japan; PCORI; *Pfizer, Inc; *Pharmacyclics, LLC; PIRCHE AG; *Sanofi Genzyme; *Seattle Genetics; Shire; Spectrum Pharmaceuticals, Inc.; St. Baldrick’s Foundation; *Sunesis Pharmaceuticals, Inc.; Swedish Orphan Biovitrum, Inc.; Takeda Oncology; Telomere Diagnostics, Inc.; and University of Minnesota.
*Corporate Members
Footnotes
DISCLOSURE OF CONFLICT OF INTEREST
Michael R. Loken is an employee and owner of Hematologics, Inc.
Lisa Eidenschink Brodersen is an employee of Hematologics, Inc.
Brent Logan is a grant recipient from the St. Baldrick’s Foundation
Disclaimers:
The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kern W Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood. 2004;104(10):3078–3085. doi:10.1182/blood-2004-03-1036 [DOI] [PubMed] [Google Scholar]
- 2.Laane E, Derolf AR, Björklund E, et al. The effect of allogeneic stem cell transplantation on outcome in younger acute myeloid leukemia patients with minimal residual disease detected by flow cytometry at the end of post-remission chemotherapy. Haematologica. 2006;91(6):833–836. [PubMed] [Google Scholar]
- 3.Buccisano F, Maurillo L, Gattei V, et al. The kinetics of reduction of minimal residual disease impacts on duration of response and survival of patients with acute myeloid leukemia. Leukemia. 2006;20(10):1783–1789. doi:10.1038/sj.leu.2404313 [DOI] [PubMed] [Google Scholar]
- 4.Sievers EL, Lange BJ, Buckley JD, et al. Prediction of relapse of pediatric acute myeloid leukemia by use of multidimensional flow cytometry. J Natl Cancer Inst. 1996;88(20):1483–1488. [DOI] [PubMed] [Google Scholar]
- 5.Sievers EL. Immunophenotypic evidence of leukemia after induction therapy predicts relapse: results from a prospective Children’s Cancer Group study of 252 patients with acute myeloid leukemia. Blood. 2003;101(9):3398–3406. doi:10.1182/blood-2002-10-3064 [DOI] [PubMed] [Google Scholar]
- 6.Rubnitz JE, Inaba H, Dahl G, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. 2010;11(6):543–552. doi:10.1016/S1470-2045(10)70090-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter RB, Buckley SA, Pagel JM, et al. Significance of minimal residual disease before myeloablative allogeneic hematopoietic cell transplantation for AML in first and second complete remission. Blood. 2013;122(10):1813–1821. doi:10.1182/blood-2013-06-506725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter RB, Gyurkocza B, Storer BE, et al. Comparison of minimal residual disease as outcome predictor for AML patients in first complete remission undergoing myeloablative or nonmyeloablative allogeneic hematopoietic cell transplantation. Leukemia. 2015;29(1):137–144. doi:10.1038/leu.2014.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ofran Y, Rowe JM. Introducing minimal residual disease in acute myeloid leukemia: Curr Opin Hematol. 2015;22(2):139–145. doi:10.1097/MOH.0000000000000113 [DOI] [PubMed] [Google Scholar]
- 10.Ossenkoppele G, Schuurhuis GJ. MRD in AML: does it already guide therapy decision-making? Hematology. 2016;2016(1):356–365. doi:10.1182/asheducation-2016.1.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeijlemaker W, Gratama JW, Schuurhuis GJ. Tumor heterogeneity makes AML a “moving target” for detection of residual disease: Phenotype Instability and MRD in AML. Cytometry B Clin Cytom. 2014;86(1):3–14. doi:10.1002/cyto.b.21134 [DOI] [PubMed] [Google Scholar]
- 12.Loken MR . Residual disease in AML, a target that can move in more than one direction: Letter to the Editor. Cytometry B Clin Cytom. 2014;86(1):15–17. doi:10.1002/cyto.b.21140 [DOI] [PubMed] [Google Scholar]
- 13.Loken MR, Alonzo TA, Pardo L, et al. Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: a report from Children’s Oncology Group. Blood. 2012;120(8):1581–1588. doi:10.1182/blood-2012-02-408336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimwade D, Freeman SD. Defining minimal residual disease in acute myeloid leukemia: which platforms are ready for “prime time”? Blood. 2014;124(23):3345–3355. doi:10.1182/blood-2014-05-577593 [DOI] [PubMed] [Google Scholar]
- 15.Kayser S, Walter RB, Stock W, Schlenk RF. Minimal Residual Disease in Acute Myeloid Leukemia—Current Status and Future Perspectives. Curr Hematol Malig Rep. 2015;10(2):132–144. doi:10.1007/s11899-015-0260-7 [DOI] [PubMed] [Google Scholar]
- 16.Özgen Ü, Anak S, Özbek U, et al. wt1 Gene Expression in Childhood Acute Leukemias. Acta Haematol. 2000;103(4):229–230. doi:10.1159/000041057 [DOI] [PubMed] [Google Scholar]
- 17.Kletzel M, Olzewski M, Huang W, Chou PM. Utility of WT1 as a reliable tool for the detection of minimal residual disease in children with leukemia. Pediatr Dev Pathol Off J Soc Pediatr Pathol Paediatr Pathol Soc. 2002;5(3):269–275. doi:10.1007/s10024001-0208-x [DOI] [PubMed] [Google Scholar]
- 18.Ostergaard M, Olesen LH, Hasle H, Kjeldsen E, Hokland P. WT1 gene expression: an excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukaemia patients - results from a single-centre study. Br J Haematol. 2004;125(5):590–600. doi:10.1111/j.1365-2141.2004.04952.x [DOI] [PubMed] [Google Scholar]
- 19.Jacobsohn DA, Tse WT, Chaleff S, et al. High WT1 gene expression before haematopoietic stem cell transplant in children with acute myeloid leukaemia predicts poor event-free survival. Br J Haematol. 2009;146(6):669–674. doi:10.1111/j.1365-2141.2009.07770.x [DOI] [PubMed] [Google Scholar]
- 20.Bayer L, Aplenc R, Leonard M. A Phase III Randomized Trial for Patients with de novo AML using Bortezomib and Sorafenib for Patients with High Allelic Ratio FLT3/ITD. January 2018https://www.childrensoncologygroup.org/index.php/aaml1031. Accessed March 29, 2018.
- 21.Loken MR, Chu S-C, Fritschle W, Kalnoski M, Wells DA. Normalization of bone marrow aspirates for hemodilution in flow cytometric analyses. Cytometry B Clin Cytom. 2009;76B(1):27–36. doi:10.1002/cyto.b.20429 [DOI] [PubMed] [Google Scholar]
- 22.Heagerty P, Saha-Chaudhuri P. Time-Dependent ROC Curve Estimation from Censored Survival Data.; 2013. https://CRAN.Rproject.org/package=survivalROC.
- 23.Blanche P, Dartigues J-F, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32(30):5381–5397. doi:10.1002/sim.5958 [DOI] [PubMed] [Google Scholar]
- 24.Kitamura K, Nishiyama T, Ishiyama K, et al. Clinical usefulness of WT1 mRNA expression in bone marrow detected by a new WT1 mRNA assay kit for monitoring acute myeloid leukemia: a comparison with expression of WT1 mRNA in peripheral blood. Int J Hematol. 2016;103(1):53–62. doi:10.1007/s12185-015-1882-1 [DOI] [PubMed] [Google Scholar]
- 25.Nomdedéu JF, Esquirol A, Carricondo M, et al. Bone Marrow WT1 Levels in Allogeneic Hematopoietic Stem Cell Transplantation for Acute Myelogenous Leukemia and Myelodysplasia: Clinically Relevant Time Points and 100 Copies Threshold Value. Biol Blood Marrow Transplant J Am Soc Blood Marrow Transplant. 2018;24(1):55–63. doi:10.1016/j.bbmt.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 26.Duléry R, Nibourel O, Gauthier J, et al. Impact of Wilms’ tumor 1 expression on outcome of patients undergoing allogeneic stem cell transplantation for AML. Bone Marrow Transplant. 2017;52(4):539–543. doi:10.1038/bmt.2016.318 [DOI] [PubMed] [Google Scholar]