Abstract
Dietary fructose is a major contributor to the epidemic of diabetes and obesity, and it is an excellent model to study metabolic syndrome. Based on previous studies that Bgn0/− gene occupies a central position in a network of genes in the brain in response to fructose consumption, we assessed the capacity of (Bgn0/−) to modulate the action of fructose on brain and body. We exposed male biglycan knockout mice (Bgn0/−) to fructose for 7 weeks, and results showed that Bgn0/− mice compensated for a decrement in learning and memory performance when exposed to fructose. These results were consistent with an attenuation of the action of fructose on hippocampal CREB levels. Fructose also reduced the levels of CREB and BDNF in primary hippocampal neuronal cultures. Bgn siRNA treatment abolished these effects of fructose on CREB and BDNF levels, in conjunction with a reduction in a fructose-related increase in Bgn protein. In addition, fructose consumption perturbed the systemic metabolism of glucose and lipids, that were also altered in the Bgn0/ mice. Transcriptomic profiling of hypothalamus, hippocampus, and liver supported the regulatory action of Bgn on key molecular pathways involved in metabolism, immune response, and neuronal plasticity. Overall results underscore the tissue-specific role of the extracellular matrix in the regulation of metabolism and brain function, and support Bgn as a key modulator for the impact of fructose across body and brain.
Keywords: Cognition, Metabolism, Hypothalamus, Hippocampus, Liver, Transcriptome
1. Introduction
The association between metabolic disorders such as type 2 diabetes and neurological and psychiatric illnesses is becoming an alarming issue [1,2]. Dietary fructose has emerged as a major contributor to the current epidemic of metabolic disorders [3–7], based on its effects on hepatic de novo lipogenesis, abdominal obesity, cholesterol and insulin resistance [8]. More recently, fructose was found to compromise synaptic plasticity, adult neurogenesis [9], and the capacity of neural circuits to cope with the effects of brain injury [10]. Fructose appears to induce a stage of metabolic dysfunction in the brain, negatively impacting cognitive function [10]. Using systems nutrigenomics in a rodent model of high fructose consumption, we recently reported the biglycan gene (Bgn) as one of the key regulatory nodes in the hypothalamus (main brain center of metabolic control) networks affected by dietary fructose [11]. Mechanistic information about the role that Bgn plays in the metabolic action of fructose on brain is crucial to understand how diabetes and brain function are connected.
Biglycan is a member of the class I small leucine rich proteoglycans (SLRPs) present in the extracellular matrix of the CNS [12]. Biglycan has also been related with low grade inflammatory state in obesity and type 2 diabetes and development of insulin resistance [13,14]. More recently, biglycan has been associated with a broad range of neurode-generative events and disorders such as amyloid metabolism in Alzheimer’s disease pathology [15], inhibition of neurite outgrowth in sensory neurons [16], and neocortical neuronal survival [12,17,18]. Seizure-induced electrophysiological activity influences the production of biglycan [19] suggesting that biglycan can participate in activity-dependent events involved in behavior. We conducted in vivo and ex vivo studies on a Bgn knockout mouse model (Bgn0/−) to examine whether Bgn mediates the effects of fructose on the brain and peripheral metabolism. To explore the action of fructose and biglycan in the context of brain plasticity and behavior, we assessed key molecular markers of brain plasticity and metabolism. We assessed Ntkr2 which encodes the tropomyosin related kinase receptor B (TrkB) of the neurotrophin brain-derived neurotrophic factor (BDNF), and phosphorylated cAMP response element binding protein (pCREB), which together with BDNF has a key function in synaptic plasticity and memory formation [20,21]. The potential action of biglycan on metabolic and cognitive functions opens a wide range of opportunities to mitigate the deleterious effects of fructose in the brain and peripheral metabolism.
2. Materials and methods
2.1. Animals and experimental design
Male biglycan knockout mice (Bgn0/−) [22] and wild type litter-mates were received as gifts from Dr. Marian Young, National Institute Dental and Craniofacial Research and rederived at UCLA. Animals were group housed (22–24 °C and 12 h light/dark cycle). At 3 months of age an initial body composition (NMR in a Bruker minispec series mq10 machine (Bruker BioSpin, Freemont, CA) and a baseline intraperitoneal glucose tolerance test (IPGTT) were performed as previously described [11]. The male mice were divided into groups of similar mass and body composition within genotype: wild type with regular drinking water, wild type with fructose (15% w/v) in the drinking water, Bgn0/− with regular drinking water, and Bgn0/− with fructose (15% w/v) in the drinking water. Standard chow was available ad libitum throughout the study. Body composition was measured every 2 weeks for 6 weeks, and a second IPGTT was performed. Prior to sacrifice animals were tested in the Barnes Maze [23]. Animals were sacrificed either by decapitation so that brain tissue could be used for immunoblotting or by transcardial perfusion for immunohistochemistry. All experiments were performed in accordance with the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the UCLA Chancellor’s Animal Research Committee.
2.2. Genotyping
5 mm tail tip was used for DNA extraction using REDExtract-NAmp™ Tissue PCR Kit (Sigma Aldrich). Genotyping was performed by PCR (approximately 200-bp wild-type and approximately 300-bp mutant fragments) using ZymoTaq DNA Polymerase (Zymo Research) with three primers: Bgn forward 5′-CAGGAACATTGACCATG-3′, reverse 5′ GAAAGGACACATGGCACTGAA-3′ and Pgk1 reverse 5′-TGGATGTGGA ATGTGTGCGAG-3′. Touchdown PCR was performed using the following program: the first stage includes 3 min at 95 °C was followed by 2 cycles of 30 s at 95 °C, 30 s at 2 °C decrement from 68 °C to 50 °C, and 30 s at 72 °C. Then the second stage includes 20 cycles at 94 °C for 30 s, 48 °C for 30 s, and 72 °C for 30 s. The last cycle was followed by a further 3 min incubation at 72 °C. 2.5% agarose gel electrophoresis was run to check the PCR product size.
2.3. Quantitative real-time PCR (qPCR)
The disruption of full length Bgn transcript in Bgn0/− was examined with quantitative PCR using primers against exon 2 where the knockout construct was inserted. We confirmed the genotypes and the absence of full-length Bgn transcript in the Bgn0/− animals (Supplementary Fig. 1A and B). RNA was extracted from hippocampus of wild-type and Bgn0/− mice using Direct-zol RNA MiniPrep kit with Dnase I (Zymo Research). cDNA synthesis was initiated using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The quantitative real-time PCR was conducted using PowerUp SYBR Green Master Mix (ThermoFisher scientific) on Applied Biosystems QuantStudio 3, with actin as an endogenous control. Each sample was repeated with triplicates. Primers for Bgn exon 2 are: Forward 5′-TGTGGCTACTCACCTTGCTG-3′, Reverse 5′- AGGACACATGGCACTGAAGG −3′.
2.4. Primary neuronal culture, transfection of small interfering RNA (siRNA), and fructose treatment
Primary near-pure neuronal cultures were prepared from fetal wild type mice at 16 days of gestation. Briefly, dissociated hippocampal cells were plated on PLL/laminin-coated plates (8 hemispheres per 6-well plate) in plating medium (Dulbecco’s modified Eagle medium (DMEM; Gibco BRL, Rockville, MD) supplemented with 20 mM glucose, 38 mM sodium bicarbonate, 2 mM glutamine, 5% fetal bovine serum and 5% horse serum) and adding Cytosine arabinoside (Ara C, 10 μM) 3 days after plating. Dissociated cells were used 5 days (DIV 5) after seeding. For Stealth siRNA transfection, cells were transfected for 72 h using the Lipofectamine RNAiMAX reagent according to the manufacturer’s instructions (Invitrogen, Cergy-Pontosie, France). The sequence of siRNAs targeting Bgn were 5′-CCU UGC CCU UUG AGC AGA AGG GUU U −3′ (sense), 5′-AAA CCC UUC UGC UCA AAG GGC AAG G-3′ (antisense). Fructose of 1 and 5 mM concentrations were used to treat primary neurons with/without Bgn siRNAs to test whether Bgn knockdown altered the effects of fructose.
2.5. Immunofluorescence
Mice (n = 4) from each group were anesthetized with isoflurane and intracardially perfused with 0.01 M phosphate buffered saline (PBS; pH 7.4) followed by 4% paraformaldehyde in PBS. Sections were incubated with rabbit polyclonal anti-BGN (Santa Cruz Biotechnology, TX, USA), anti-NeuN, and GFAP (abcam, Cambridge, UK) primary antibody at a dilution of 0.3 μg/mL in PBS solution containing 2% BSA at 4 °C overnight. After washing, sections were incubated with rabbit secondary antibody (Cy3, Alexa 488 or 405; 1:500; Jackson Immunoresearch; West Grove, PA, USA) for 1 h at room temperature. Sections were mounted using Prolong Gold antifade reagent (Life technologies, New York, NY, USA). The staining was visualized (Zeiss Imager.Z1; Gottingen, DE) using the Axiovision software (Carl Zeiss Vision, version 4.6).
2.6. Oil red O staining
To determine hepatic lipid accumulation, frozen sections of liver (10 μm) were stained with Oil Red O for 40 min, washed, and counterstained with hematoxylin for 45 s. Quantification of tissue lipid accumulation was performed using ImageJ32 Software.
2.7. Western blot analysis
Extracted tissue proteins were homogenized in RIPA buffer containing protease inhibitors (1 μg/mL Leupeptin, 1 mM phenylmethylsulfonyl fluoride) and phosphatase inhibitors (2.5 mM sodium pyrophosphate, 1 μM Na3VO4). Protein was quantified using the BCA protein assay (BioRad). Equal amounts of protein were separated by SDS PAGE and transferred to polyvinylidene difluoride membranes. The membranes were probed with anti-BGN (1:500, Santa Cruz Biotechnology, Texas, USA), anti-actin (1:1000, Santa Cruz Biotechnology) pCREB, CREB, pTrkB (1:1000, Millipore, MA, USA), or TrkB (1:1000, Cell signaling Technology, MA, USA), as appropriate. Immunoreactive proteins were visualized using enhanced chemiluminescence reagents (Millipore, MA, USA). Band intensities were quantified using ImageJ32 Software.
2.8. Barnes maze (BM)
We used BM to assess learning and memory functions [11]. An acrylic plastic disk 1.5 cm thick and 122 cm in diameter, with 40 evenly spaced holes (diameter of 5.5 cm); the disk was brightly illuminated by four overhead halogen lamps (800 lx). Mice were trained to locate a dark escape chamber (22 × 5.5 × 8 cm) hidden underneath a hole positioned around the perimeter of the disk with two trials per day for four consecutive days. A trial was started by placing the animal in the center of the maze covered under a cylindrical start chamber; after a 10 s delay, the start chamber was raised. A training session ended after the animal had entered the escape chamber or when 5 min had elapsed, whichever came first. A trial was given four days following the last learning day to assess memory retention. The behaviors were analyzed by ANYmaze software (Stoelting CO, USA). A circle was drawn around the perimeter of each hole and number of mistakes was calculated as the number of times the animal poked his head into a hole that did not contain the escape box. Percent of time spent in the target zone was calculated by dividing the disk into 10 pie-shaped zones with the section containing the escape box labeled as the “target” zone.
2.9. Transcriptome profiling of hypothalamus, hippocampus, and liver tissues of Bgn0/− mice
Hypothalamus, hippocampus, and liver tissues were dissected from WT and Bgn0/− mice (n = 4/group). Transcriptome profiling was conducted using microarray for hypothalamus and liver and using RNA sequencing for hippocampus (different platform was used due to discontinuation of microarrays at the UCLA Neurogenomics core). RNA samples from hypothalamus and liver were assessed with Illumina MouseRef 8 v2.0 Expression BeadChip (Illumina Inc., CA, USA). Microarray gene expression data were normalized and differentially expressed genes were identified using R packages lumi [24] and maanova [25], respectively. RNA samples from hippocampus were first processed via poly-A selection and fragmentation, reverse transcribed into cDNA, and sequencing adapters ligated using the Illumina Paired-End sample prep kit. Fragments of 250–400 bp were isolated, amplified, and sequenced on an Illumina Hiseq2500 System. Paired-end RNA-Seq reads were mapped using TopHat2/Bowtie2 [26]. Differentially expressed genes (DEGs) were further identified using Cufflinks and Cuffdiff [27,28], and multiple testing correction was performed using the q-value method to control FDR. DEGs were assessed for pathway enrichment using a Fisher’s exact test and pathways curated in KEGG, Biocarta, and Reactome databases, and multiple testing was corrected using the q-value approach. The expression data was deposited to Gene Expression Ominbus (GEO) with accession number GSE59742 (hypothalamus), GSE64806 (liver) and GSE108446 (hippocampus).
2.10. Statistical analysis
The results are represented as mean ± standard error of the mean (SEM). Statistical analysis was performed by software GraphPad Prism 7.04, and two-way analysis of variance (ANOVA) (drinking solution: water vs. fructose) and (animal type: WT vs Bgn0/−) with Holm Sidak post-hoc analyses for multiple comparisons was performed for molecular and behavioral results. A level of 5% probability was considered as statistically significant.
3. Results
3.1. Action of Bgn in brain plasticity markers TrkB and CREB
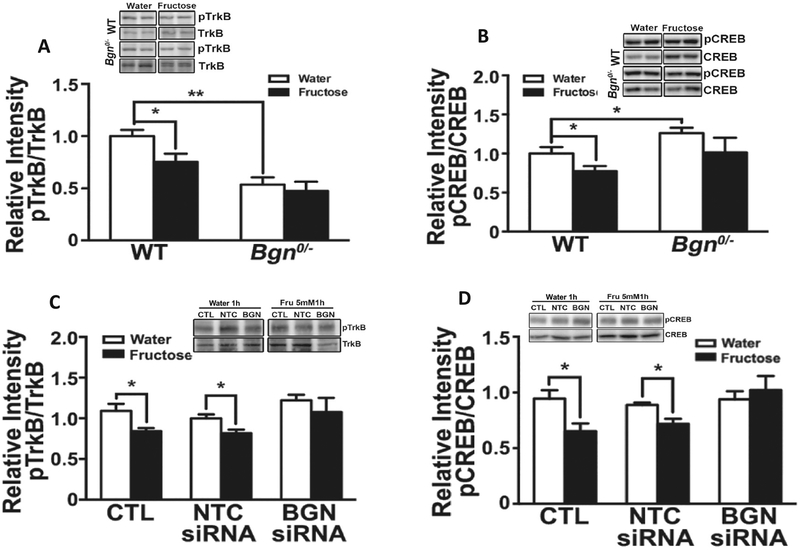
We sought to determine the role of Bgn in mediating the action of fructose on brain plasticity and function [6] based on the known roles of BDNF and CREB in brain plasticity, cell metabolism, and neurocognitive function. TrkB is the receptor of BDNF and the BDNF-TrkB signaling pathway is critical for hippocampal dependent learning and memory [29]. We assessed levels of TrkB protein and found that the ratio of phosphorylated TrkB (pTrkB) relative to total TrkB, indicative of receptor signaling, was reduced by fructose in the WT but not in the Bgn0/− animals (Fig. 1A). CREB is important for synaptic plasticity and memory formation [20,21,30] in conjunction with the activation of TrkB [10,31]. Although fructose treatment decreased the ratio pCREB/CREB in the hippocampus of WT mice (p < 0.05), it had no effects on the Bgn0/− mice (Fig. 1B), suggesting that BGN mediates the effects on fructose on CREB regulation.
Fig. 1.
Fructose treatment reduced levels of plasticity markers TrkB and CREB in hippocampus involving the action of Bgn. Fructose treatment decreased the ratio pTrkB/TrkB (A) and pCREB/CREB (B) in hippocampal neurons from WT animals but not from Bgn0/− animals. Treatment of neurons with Bgn siRNA for 72 h blocked the effects of fructose (5 mM) on pTrkB/TrkB (C) and pCREB/CREB (D). Bars denote densitometric assessments of Western blot bands. Results are presented as mean ± SEM (n = 4–6, *p < 0.05).
We assessed the action of fructose (5 mM fructose for 1 h) on hippocampal primary neuronal cells from WT mice treated with non-target control (NTC) or Bgn siRNA for 72 h. Fructose treatment reduced TrkB signaling in the control cells and those treated with NTC siRNA. In contrast, fructose did not affect TrkB signaling in cells treated with Bgn siRNA (Fig. 1C). Similarly, fructose reduced CREB levels in control (CTL) and NTC siRNA treated cells but did not affect CREB signaling in cells treated with Bgn siRNA (Fig. 1D). Taken together, both in vivo and in vitro results suggest that Bgn plays a role in regulating the expression or functionality of TrkB and downstream CREB under fructose treatment in mouse hippocampus.
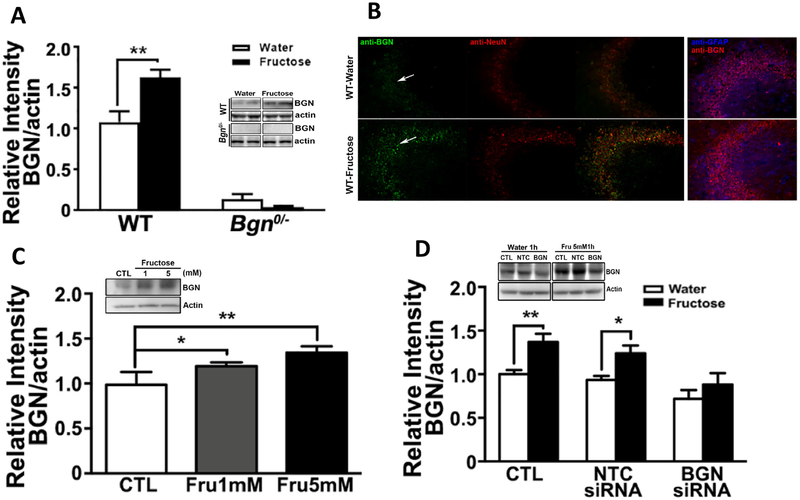
To determine whether fructose alters levels of Bgn at the protein level, we exposed wildtype (WT) mice to fructose or water and performed Western blotting in the hippocampus. Fructose consumption significantly elevated (p < 0.01) levels of Bgn in the hippocampus (Fig. 2A) whereas Bgn protein was barely detectable in Bgn0/− mice with or without fructose. Immunofluorescence showed that Bgn protein was located in neuronal elements, particularly within the CA3 region of the hippocampus. Double-labelling immunofluorescence showed that Bgn selectively co-localized within neuronal cell bodies (NeuN positive) but not in astrocytes (GFAP negative) (Fig. 2B). We tested the direct effects of fructose (1 and 5 mM, 1 h) on isolated primary hippocampal neuronal cells, and found that fructose elicited a concentration-dependent increase (p < 0.05) in Bgn levels (Fig. 2C). To confirm the cell specificity of the Bgn regulation by fructose, we treated cultures in parallel with non-target control (NTC) or Bgn siRNA for 72 h. After 72 h, in cells treated with 5 mM fructose for 1 h, agreeing with the in vivo observations, fructose increased Bgn levels in control (CTL) and NTC siRNA treated cells (p < 0.05) but had no effects in cells treated with Bgn siRNA (Fig. 2D).
Fig. 2.
Fructose elevates hippocampal BGN levels in WT mice. (A) Western blotting analysis showed an increase in BGN in WT animals treated with fructose. Bars denote the density of BGN bands, normalized to corresponding β-actin band. Results are presented as mean ± SEM (*p < 0.05, **p < 0.01, n = 4–5). (B) Immunofluorescence showed a qualitative increase in BGN intensity in neuron (NeuN) but not astrocyte (GFAP) in hippocampal tissue of mice treated with fructose. Fructose elevates BGN levels in primary hippocampal neurons, and Bgn siRNA treatment -attenuate this effect. (C) Primary hippocampal neurons were treated were exposed to 1 mM or 5 mM of fructose for 1 h, and western blotting detection showed increases in levels of BGN. Bars denote densitometric quantification of BGN bands. (D) Bgn siRNA treatment for 72 h blocked the effects of 5 mM fructose. Bars denote densitometric analysis of BGN bands. Results are presented as mean ± SEM (*p < 0.05, **p < 0.01, n = 4–5).
3.2. Role of Bgn in behavior
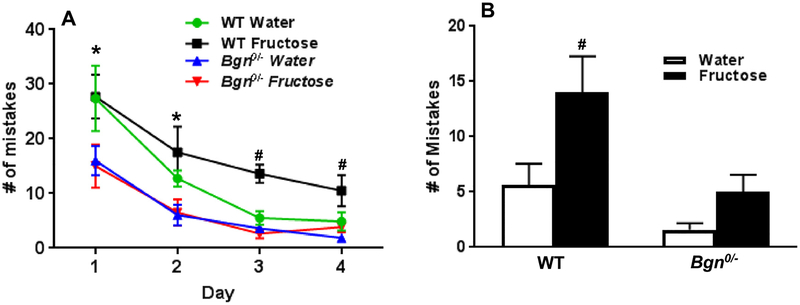
To assess the role of Bgn in the fructose-induced hippocampal-dependent learning and memory impairment, we conducted Barnes Maze (BM) in WT and Bgn0/− mice with and without fructose treatment. Bgn0/− mice have been previously reported to have impaired tendon and joints which may affect latency time [32]. Therefore, we analyzed the BM data by number of mistakes to instead of latency to eliminate the potential confound originated by differences in mobility. During the learning phase, there was a significant main effect of animal groups (p = 0.001) and day (p < 0.0001) on the number of mistakes made prior to finding the escape hole. Multiple comparison test in WT animals showed that the fructose treated group made significantly more mistakes than water treated mice in day 3 and 4 (#p < 0.05, WT Fructose vs. WT Water, Fig. 3A). Bgn0/− mice exposed to fructose did not differ from Bgn0/− mice exposed to water in terms of mistakes (Fig. 3A). Bgn0/− mice exposed to water showed significantly fewer mistakes than the WT mice fed water in day 1 and day 2 (*p < 0.05,Fig. 3A). During the memory phase, the WT fructose group made significant more mistake than WT water group (#p < 0.05, Fig. 3B). Bgn0/− mice exposed to fructose did not significantly differ from mice exposed to water in the number of mistakes (Fig. 3B).
Fig. 3.
Fructose treatment reduces learning function in the Barnes Maze and this effect was mitigated in Bgn0/− mice. (A) The number of mistakes made in four days during the learning phase. (B) The number of mistakes made during the memory phase. Results are presented as mean ± SEM (*p < 0.05, Bgn0/− Water vs. WT Water, #p < 0.05, WT Fructose vs. WT water. n = 6–12).
3.3. Molecular pathways affected by Bgn in the brain
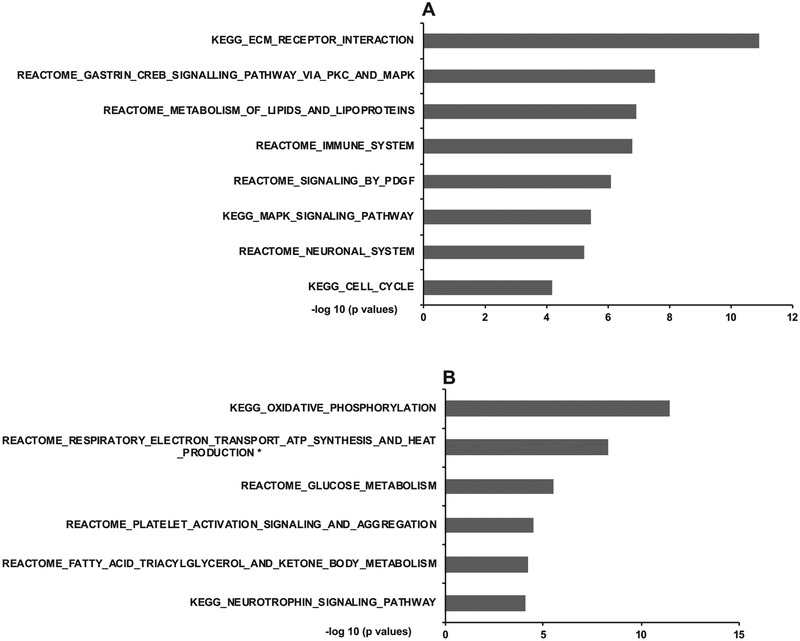
To explore the molecular processes regulated by Bgn that could contribute to its role in mediating the fructose response, we conducted transcriptome analyses of the hippocampus and hypothalamus of Bgn0/− mice. Compared to WT mice, Bgn knockdown altered 351 and 66 differentially expressed genes (DEGs) in the hippocampus and hypothalamus, respectively, at false discovery rate (FDR) < 0.05 (full DEG lists in Supplementary Table 1 for hippocampus, Supplementary Table 2 for hypothalamus (https://yanglab.ibp.ucla.edu/data-sharing/). Annotation of these DEGs affected in Bgn0/− mice revealed significant enrichment for diverse molecular pathways in each brain region. The hippocampal DEGs were enriched for pathways related to extracellular matrix (ECM), CREB, MAPK, and PDGF signaling as well as lipid metabolism, immune system, and cell cycle (Fig. 4A; full list in Supplementary Table 3, https://yanglab.ibp.ucla.edu/data-sharing/). The hypothalamic DEGs were over-represented with genes in pathways related to oxidative phosphorylation, metabolism of glucose and fatty acid triacylglycerol and ketone body, platelet function, and neurotrophin signaling (Fig. 4B; full list in Supplementary Table 4, (https://yanglab.ibp.ucla.edu/data-sharing/). These pathways significantly overlapped with those affected by fructose in the corresponding brain regions revealed in our previous study [11]. The converging pathways between fructose treatment and Bgn perturbation provide further molecular support for a mediator role of Bgn in fructose actions.
Fig. 4.
Changes in biological pathways in the hippocampus and hypothalamus of Bgn0/− mice based on transcriptome analysis. Top select significant pathways are shown for hippocampus (A) and hypothalamus (B). The −log10 (p.adj < 0.05.) of the significant pathways are show on the x-axis. Full lists of significant pathways are in Supplementary Table 3, Table 4.
3.4. Effects of Bgn0/− on body composition
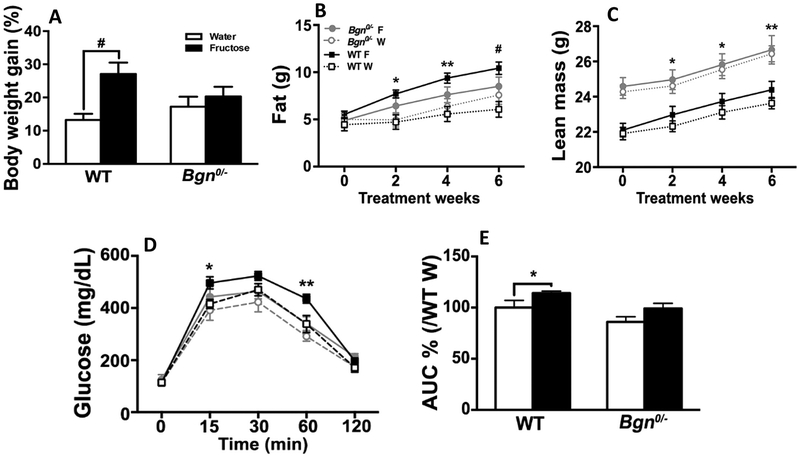
To examine the peripheral metabolic action of Bgn, we analyzed changes in body composition of Bgn0/− and WT mice over six weeks with or without fructose treatment. Fructose resulted in a significant gain in body weight in WT animals but not in Bgn0/− animals (Fig. 5A). In particular, fructose preferably increased fat mass in WT but not in Bgn0/− mice (Fig. 5B), and Bgn0/− animals had more lean mass than WT controls throughout the study. WT mice fed on fructose slightly increased lean mass as compared to WT mice that were fed on water, while Bgn0/− mice did not show changes in lean mass between fructose and water treatment (Fig. 5C).
Fig. 5.
Effects of fructose on body composition and intraperitoneal glucose tolerance test. (A) Gain of body weight of WT mice fed on fructose (F) were more significant as compared to WT mice fed on water (W) (p < 0.001), whereas Bgn0/− mice did not show much differences. (B) Fructose preferably increased fat mass in WT but not Bgn0/− mice. (C) Bgn0/− animals had more lean mass than WT controls throughout the study. WT mice fed on fructose slightly increased lean mass as compared to WT mice fed on water, while Bgn0/− mice did not show changes in lean mass between fructose and water treatment. (D) Intraperitoneal glucose tolerance test indicated WT animals in the fructose group had significantly elevated blood glucose compared with either Bgn0/− or WT animals that were fed on water at 15 and 60 min. Bgn0/− animals showed no difference in blood glucose between water and fructose treatment. (E) According to the analysis of the area under the curve (AUC), WT animals fed on fructose had a larger AUC, indicating reduced capacity of glucose clearance compared with either WT or Bgn0/− animals fed on water, and Bgn0/− animals on fructose had similar glucose clearance to the animals in the water groups. Results are presented as mean ± SEM (WT Water vs WT Fructose; *p < 0.05, **p < 0.01, #p < 0.001, n = 6–14).
3.5. Effects of Bgn0/− on intraperitoneal glucose tolerance
WT animals that were fed on fructose had a larger area under the curve (AUC) assessed using IPGTT [11], indicating reduced capacity of glucose clearance compared with either WT or Bgn0/− animals fed on water. In contrast, Bgn0/− animals on fructose had similar glucose clearance to the animals in the water groups, suggesting that Bgn0/− mice were protected against fructose-induced reductions in glucose clearance (Fig. 5D–E).
3.6. Effects of Bgn0/− on liver and peripheral metabolism
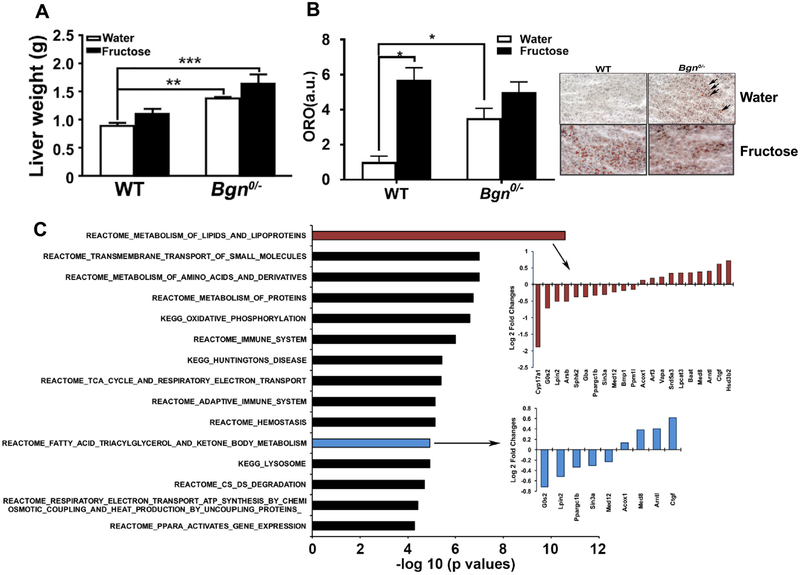
At 23 weeks, the liver of male Bgn0/− mice was heavier than that of WT mice (p < 0.001), (Fig. 6A). Although the liver weight in Bgn0/− mice treated with fructose was slightly higher than the liver in water treated WT mice, the liver weight was not significantly altered compared to Bgn0/− mice treated with water. Neutral lipids in the liver were assessed by Oil Red O (ORO) staining. A limited number of small lipid droplets was seen in the WT livers whereas much more lipid droplets were visually prominent in the Bgn0/− livers (p < 0.05) (Fig. 6B). Interestingly, fructose treatment significantly increased lipid droplets in WT (p < 0.05) but not in Bgn0/− mice (Fig. 6B). We performed transcriptome analysis with liver tissue samples from Bgn0/− and WT mice and identified 104 DEGs at FDR < 0.05 (full DEG list in Supplementary Table 5 (https://yanglab.ibp.ucla.edu/data-sharing/). Pathway analysis showed that the liver DEGs were highly enriched for pathways related to metabolism of lipids, lipoproteins, fatty acid triacylglycerol and ketone body, and amino acids, and oxidative phosphorylation, suggesting the very fundamental effects of the Bgn on metabolism in liver (Fig. 6C); full pathway list in Supplementary Table 6 (https://yanglab.ibp.ucla.edu/data-sharing/). These pathways also agree well with the results from the hypothalamus DEGs, supporting the role of Bgn in metabolic control in both the brain and periphery.
Fig. 6.
Effects of Bgn0/− on liver and peripheral metabolism. (A) The liver of Bgn0/− mice was heavier than that of WT mice and fructose consumption did not further increase liver weight in Bgn0/− mice. However, the liver weight in Bgn0/− mice treated with fructose was significantly higher than that in water treated WT mice. Neutral lipids assessed by Oil Red O (ORO) staining showed a limited number of small lipid droplets in the WT livers whereas much more lipid droplets were visually prominent in the Bgn0/− livers (B). ORO staining intensities quantified using imaging software showed statistically significant increase in Bgn0/− livers (B). Results are presented as mean ± SEM (*p < 0.05; **p < 0.01, n = 4–6). (C) Top select significant pathways affected by Bgn knockdown in Bgn0/− livers based on transcriptome analysis. The −log10 (p.adj < 0.05) of the significant pathways are show on the x-axis. Full lists of significant pathways are in Supplementary Table 6.
4. Discussion
Dietary fructose consumption is a main contributor to the epidemic of metabolic syndrome around the world [3–7]. Bgn gene was identified in our previous transcriptomic studies as the master regulator of a network of genes responsive to the effects of fructose. Through a comprehensive investigation of Bgn functions using knockout mice and cell culture experiments, here we show that Bgn plays a significant role in regulating brain plasticity and peripheral metabolism. Fructose treatment demonstrated alterations in molecular systems with a defined role in hippocampal plasticity and function, and knockout of the Bgn gene (Bgn0/−) reduced the effects of fructose. Bgn siRNA treatment also abolished the increase of Bgn protein in response to fructose treatment in primary hippocampal neuronal culture cells. Fructose consumption had several effects on the metabolism of systemic glucose and lipid metabolism, and these effects were not observed in the Bgn KO mice. In agreement with a potential regulatory action of Bgn on peripheral metabolism, our multi-tissue transcriptomic profiling supports the key role of Bgn in regulating molecular pathways involved in metabolism, immune response, and neuronal signaling pathways. The overall results emphasize the role of Bgn in connecting brain plasticity with peripheral metabolism, and the importance of Bgn in the regulation of the action of fructose.
4.1. Role of Bgn in the brain
It is known that fructose treatment impairs hippocampal-dependent learning and memory [10], which was found to be accompanied by increases in hippocampal Bgn. It is possible that Bgn could potentiate the deleterious action of fructose on the hippocampus, particularly considering that Bgn0/− mice appeared resilient to the behavioral deficits induced by fructose. Fructose treatment did not significantly increase the number of mistakes made during the learning phase or during the memory phase in Bgn0/− mice. These results suggest that the elimination of Bgn mitigated the effects of fructose on behavioral outcomes in the Barnes Maze test.
There is a paucity of information regarding the role of Bgn in cognitive processes in the literature. We assessed the potential effects of Bgn0/− and fructose on molecular systems important for synaptic plasticity and learning and memory in the hippocampus. We found that the pTrkB/TrkB ratio, that provides an indication of BDNF signaling, was reduced in the Bgn0/− mice. Fructose reduced the same ratio in the WT animals but not in the Bgn0/− animals. However, it is possible that the pTrkB/TrkB ratio had already reached floor levels in Bgn0/− mice based on the already low levels of the pTrkB/TrkB ratio in water control animals. We also assessed pCREB which plays a key function in synaptic plasticity and memory formation under regulatory control of the BDNF receptor TrkB activation [33]. Hippocampal pCREB/CREB was elevated in Bgn0/− mice. Although fructose treatment decreased pCREB/CREB in the hippocampus of WT mice, it had no effects on the Bgn0/− mice. The specificity for the effects of fructose on Bgn was also verified in primary hippocampal neurons in culture, in which we found that Bgn siRNA treatment -attenuated the effects of fructose on TrkB and CREB signaling. The overall results suggest that Bgn has the potential to influence molecular systems associated with synaptic plasticity and behavior, and that interfering the activities of Bgn in vivo and in vitro attenuates the effects of fructose on hippocampal function. It has been reported that seizure activity elevates biglycan in the mouse hippocampus [19], indicating that Bgn is susceptible to regulation by neural activity underlying cell communication and information processing. It is known that fructose treatment impairs hippocampal-dependent learning and memory which are accompanied with reductions in BDNF [10]. Given that Bgn is an extracellular matrix proteoglycan, it is likely that actions of Bgn on brain plasticity and behavior are rather indirect. The current results indicate that fructose elevates Bgn levels in hippocampal neurons in brain tissue and in culture. Although Bgn siRNA treatment did not affect basal levels of TrkB signaling in hippocampal cultured cells and protected against the effects of fructose, Bgn0/− reduced basal levels of TrkB in the brain.
To further explore the molecular mechanisms underlying the activities of Bgn in the brain, we conducted transcriptome profiling of the hippocampus of the Bgn0/− mice. We found evidence for influences on ECM receptor interaction, CREB signaling, and neuronal signaling, all of which may contribute to the modulatory role of Bgn on brain plasticity and cognitive function. Transcriptome profiling of the hypothalamus of the Bgn0/− mice found significant changes in pathways related to oxidative phosphorylation, glucose metabolism, fatty acid metabolism, and neurotrophin signaling, which could underlie the effects of Bgn on the central control of peripheral metabolism.
4.2. Effects of Bgn and fructose on peripheral metabolism
Our previous study [11] revealed lipid dysregulation in Bgn0/− mice, as shown by the elevated levels of triglycerides, total cholesterol, HDL, and unesterified cholesterol in the plasma of Bgn0/− mice. Fructose treatment elevated weight and body fat gain as well as glucose intolerance in wild type controls but not in Bgn0/−. Given that Bgn0/− animals have significantly higher lean mass than wildtype controls, the increased lean mass of Bgn0/− mice may help resist fat accumulation caused by fructose consumption, and also help to attenuate the deleterious effects of fructose on glucose dysregulation. It is perplexing that Bgn0/− animals showed lipid accumulation in liver but not in body fat mass, suggesting major differences in the activities of Bgn in different metabolic tissues and Bgn0/− may shift triglyceride storage from adipose tissue to the liver. Increased fat storage in liver is commonly associated with compromised liver insulin sensitivity, while increased lean mass may prime the Bgn0/− mice for improved insulin sensitivity. These potential tissue-specific effects of Bgn on insulin sensitivity needs to be explored in future studies. It is also interesting that considering the lipid promoting actions of Bgn0/− and fructose by separate, fructose consumption had no additional impact on liver weight in the Bgn0/− mice. It is possible that the separate actions of either Bgn0/− or fructose may be sufficient to reach a ceiling effect, thus, precluding additional effects on liver fat and weight.
Although biglycan is present in adipose tissue in the obese state, its action is poorly understood [14,34]. Pathway enrichment analysis of the differentially expressed genes in the liver tissue between Bgn0/− and WT mice indicate alterations in numerous metabolic related pathways, including metabolism of lipid and lipoprotein, metabolism of amino acids and derivatives, and fundamental cell metabolic processes. Specifically, liver Acox1 and Lpcat3 were increased in Bgn0/− animals. Acox1 is the first enzyme of the fatty acid beta-oxidation pathway [35] while Lpcat3 catalyzes the reacylation of lysophospholipids to phospholipids to modulate lysophospholipid availability and metabolism in the liver. It has been shown that LPCAT3 is an important mediator of liver X receptor effects on metabolism [36,37]. The overall results support a strong regulatory role of Bgn in peripheral metabolism.
5. Conclusions
Bgn appears as a crucial modifier of the action of fructose across brain and body metabolism, which support our previous findings revealing the central position of the Bgn gene in a network of genes responsive to fructose consumption. Bgn0/− mice appeared to recapitulate certain disruptive effects of fructose on peripheral metabolism (such as elevated lipid dysregulation) while attenuating others (such as improved glucose control). Bgn0/− mice also showed resilience to the effects of fructose on brain plasticity and function. The results of our multi-tissue transcriptomic analysis are consistent with a tissue-specific transcriptional regulatory actions of Bgn in central and peripheral control of metabolism (Fig. 7). Although most of the actions of the Bgn0/− can be considered helpful to mitigate fructose action, using Bgn as a potential therapeutic target is not straightforward since Bgn has some actions on its own such as accumulation of liver fats that can be health threatening. Therefore, tissue specificity is an important factor for any potential therapeutic targeting of Bgn.
Fig. 7.
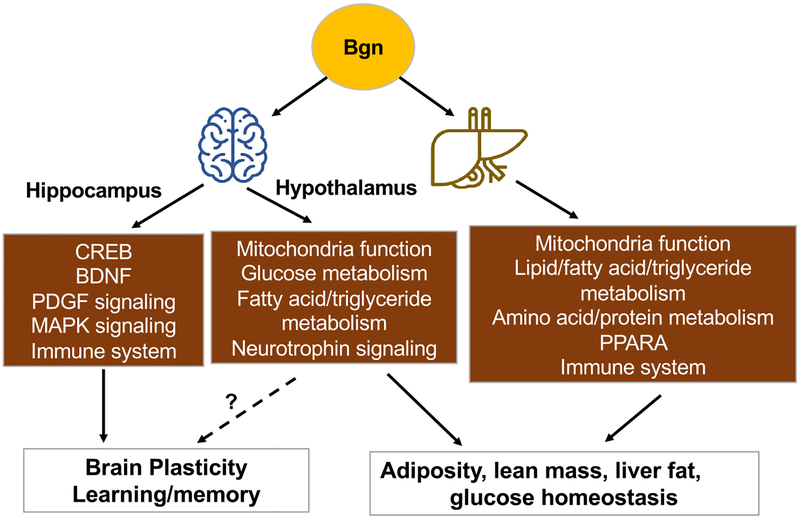
Diagram showing the effects of Bgn on the regulation of brain plasticity and peripheral metabolism. In the brain, Bgn is involved in regulation of learning and memory operation, and seems to act on molecular systems associated with brain plasticity and metabolism. In the periphery, Bgn affects the metabolism of lipids particularly acting on the liver. The transcriptomic profiling in the hypothalamus and liver, emphasizes the role of Bgn in regulating molecular pathways involved in metabolism, immune response, and neuronal signaling. The main outcomes of the actions of Bgn in the periphery seem to center on the regulations of adiposity, lean mass, and glucose metabolism. The overall results underline the role of Bgn in connecting brain plasticity with peripheral metabolism, and the importance of Bgn in the regulation of the action of fructose. PPARA: peroxisome proliferator activator receptor alpha.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbadis.2018.10.002.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health (DK104363 and NS103088 to XY. and FGP; NS50465 to FGP). We thank Dr. Oldberg Ake from Experimental Medical Sciences, University of Lund, Lund, Sweden and Dr. Marian Young from the National Institute of Dental and Craniofacial Research, National Institute of Health for kindly providing the Bgn knockout mouse model for this study.
Abbreviations:
- AUC
area under the curve
- CREB
cAMP response element binding protein
- BDNF
brain-derived neurotrophic factor
- ECM
extracellular matrix
- HDL
high-density lipoprotein
- IPGTT
intraperitoneal glucose tolerance test
- NTC
non-target control
- ORO
Oil Red O
- TrkB
tropomyosin related kinase B
- TCA
tricarboxylic acid cycle
- WT
wild type
Footnotes
Transparency document
The Transparency document associated with this article can be found, in online version.
Competing financial interests
The authors declare no competing financial interests.
References
- [1].Adeghate E, Donáth T, Adem A, Alzheimer disease and diabetes mellitus: do they have anything in common? Curr. Alzheimer Res 10 (6) (2013) 609–617. [DOI] [PubMed] [Google Scholar]
- [2].Mansur RB, Lee Y, Zhou AJ, Carmona NE, Cha DS, Rosenblat JD, Bruins R, Kakar R, Rasgon NL, Lovshin JA, Wroolie TE, Sim K, Brietzke E, Gerstein HC, Rong C, McIntyre RS, Determinants of cognitive function in individuals with type 2 diabetes mellitus: a meta-analysis, Ann. Clin. Psychiatry 30 (2018) 38–50. [PubMed] [Google Scholar]
- [3].Kelishadi R, Mansourian M, Heidari-Beni M, Association of fructose consumption and components of metabolic syndrome in human studies: a systematic review and meta-analysis, Nutrition 30 (2014) 503–510. [DOI] [PubMed] [Google Scholar]
- [4].Bray GA, Popkin BM, Calorie-sweetened beverages and fructose: what have we learned 10 years later, Pediatr. Obes 8 (2013) 242–248. [DOI] [PubMed] [Google Scholar]
- [5].Johnson RJ, Perez-Pozo SE, Sautin YY, Manitius J, Sanchez-Lozada LG, Feig DI, Shafiu M, Segal M, Glassock RJ, Shimada M, Roncal C, Nakagawa T, Hypothesis: could excessive fructose intake and uric acid cause type 2 diabetes? Endocr. Rev 30 (2009) 96–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Agrawal R, Gomez-Pinilla F, ‘Metabolic syndrome’ in the brain: deficiency in omega-3 fatty acid exacerbates dysfunctions in insulin receptor signalling and cognition, J. Physiol 590 (2012) 2485–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Malik VS, Hu FB, Fructose and cardiometabolic health: what the evidence from sugar-sweetened beverages tells us, J. Am. Coll. Cardiol 66 (2015) 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Stanhope KL, Role of fructose-containing sugars in the epidemics of obesity and metabolic syndrome, Annu. Rev. Med 63 (2012) 329–343. [DOI] [PubMed] [Google Scholar]
- [9].Cisternas P, Salazar P, Serrano FG, Montecinos-Oliva C, Arredondo SB, Varela-Nallar L, Barja S, Vio CP, Gomez-Pinilla F, Inestrosa NC, Fructose consumption reduces hippocampal synaptic plasticity underlying cognitive performance, Biochim. Biophys. Acta 1852 (2015) 2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Agrawal R, Noble E, Vergnes L, Ying Z, Reue K, Gomez-Pinilla F, Dietary fructose aggravates the pathobiology of traumatic brain injury by influencing energy homeostasis and plasticity, J. Cereb. Blood Flow Metab 36 (2016) 941–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Meng Q, Ying Z, Noble E, Zhao Y, Agrawal R, Mikhail A, Zhuang Y, Tyagi E, Zhang Q, Lee JH, Morselli M, Orozco L, Guo W, Kilts TM, Zhu J, Zhang B, Pellegrini M, Xiao X, Young MF, Gomez-Pinilla F, Yang X, Systems nutrigenomics reveals brain gene networks linking metabolic and brain disorders, EBioMedicine 7 (2016) 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Junghans U, Koops A, Westmeyer A, Kappler J, Meyer HE, Muller HW, Purification of a meningeal cell-derived chondroitin sulphate proteoglycan with neurotrophic activity for brain neurons and its identification as biglycan, Eur. J. Neurosci 7 (1995) 2341–2350. [DOI] [PubMed] [Google Scholar]
- [13].Babelova A, Moreth K, Tsalastra-Greul W, Zeng-Brouwers J, Eickelberg O, Young MF, Bruckner P, Pfeilschifter J, Schaefer RM, Grone HJ, Schaefer L, Biglycan, a danger signal that activates the NLRP3 inflammasome via toll-like and P2X receptors, J. Biol. Chem 284 (2009) 24035–24048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim J, Lee SK, Shin JM, Jeoun UW, Jang YJ, Park HS, Kim JH, Gong GY, Lee TJ, Hong JP, Lee YJ, Heo YS, Enhanced biglycan gene expression in the adipose tissues of obese women and its association with obesity-related genes and metabolic parameters, Sci. Rep 6 (2016) 30609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lam V, Takechi R, Pallebage-Gamarallage MM, Galloway S, Mamo JC, Colocalisation of plasma derived apo B lipoproteins with cerebral proteoglycans in a transgenic-amyloid model of Alzheimer’s disease, Neurosci. Lett 492 (2011) 160–164. [DOI] [PubMed] [Google Scholar]
- [16].Lemons ML, Barua S, Abanto ML, Halfter W, Condic ML, Adaptation of sensory neurons to hyalectin and decorin proteoglycans, J. Neurosci 25 (2005) 4964–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kappler J, Junghans U, Koops A, Stichel CC, Hausser HJ, Kresse H, Muller HW, Chondroitin/dermatan sulphate promotes the survival of neurons from rat embryonic neocortex, Eur. J. Neurosci 9 (1997) 306–318. [DOI] [PubMed] [Google Scholar]
- [18].Koops A, Kappler J, Junghans U, Kuhn G, Kresse H, Muller HW, Cultured astrocytes express biglycan, a chondroitin/dermatan sulfate proteoglycan supporting the survival of neocortical neurons, Brain Res. Mol. Brain Res 41 (1996) 65–73. [DOI] [PubMed] [Google Scholar]
- [19].Motti D, Le Duigou C, Eugene E, Chemaly N, Wittner L, Lazarevic D, Krmac H, Marstrand T, Valen E, Sanges R, Stupka E, Sandelin A, Cherubini E, Gustincich S, Miles R, Gene expression analysis of the emergence of epileptiform activity after focal injection of kainic acid into mouse hippocampus, Eur. J. Neurosci 32 (2010) 1364–1379. [DOI] [PubMed] [Google Scholar]
- [20].Tully T, Preat T, Boynton SC, Del Vecchio M, Genetic dissection of consolidated memory in Drosophila, Cell 79 (1994) 35–47. [DOI] [PubMed] [Google Scholar]
- [21].Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER, CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation, Cell 95 (1998) 211–223. [DOI] [PubMed] [Google Scholar]
- [22].Xu T, Bianco P, Fisher LW, Longenecker G, Smith E, Goldstein S, Bonadio J, Boskey A, Heegaard AM, Sommer B, Satomura K, Dominguez P, Zhao C, Kulkarni AB, Robey PG, Young MF, Targeted disruption of the biglycan gene leads to an osteoporosis-like phenotype in mice, Nat. Genet 20 (1998) 78–82. [DOI] [PubMed] [Google Scholar]
- [23].Barnes CA, Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat, J. Comp. Physiol. Psychol 93 (1979) 74–104. [DOI] [PubMed] [Google Scholar]
- [24].Du P, Kibbe WA, Lin SM, lumi: a pipeline for processing Illumina microarray, Bioinformatics 24 (2008) 1547–1548. [DOI] [PubMed] [Google Scholar]
- [25].Wu H, Yang H, Sheppard K, Churchill G, Kerr K, Cui X, maanova: Tools for analyzing micro array experiments, R Package version 1.28.0.
- [26].Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol. 10 (2009) R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nat. Protoc 7 (2012) 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L, Transcript assembly and quantification by RNASeq reveals unannotated transcripts and isoform switching during cell differentiation, Nat. Biotechnol 28 (2010) 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Noble EE, Billington CJ, Kotz CM, Wang C, The lighter side of BDNF, Am. J. Physiol. Regul. Integr. Comp. Physiol 300 (2011) R1053–R1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Alberini CM, Ghirardi M, Metz R, Kandel ER, C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia, Cell 76 (1994) 1099–1114. [DOI] [PubMed] [Google Scholar]
- [31].Deogracias R, Espliguero G, Iglesias T, Rodriguez-Pena A, Expression of the neurotrophin receptor trkB is regulated by the cAMP/CREB pathway in neurons, Mol. Cell. Neurosci 26 (2004) 470–480. [DOI] [PubMed] [Google Scholar]
- [32].Ameye L, Aria D, Jepsen K, Oldberg A, Xu T, Young MF, Abnormal collagen fibrils in tendons of biglycan/fibromodulin-deficient mice lead to gait impairment, ectopic ossification, and osteoarthritis, FASEB J. 16 (2002) 673–680. [DOI] [PubMed] [Google Scholar]
- [33].Finkbeiner S, CREB couples neurotrophin signals to survival messages, Neuron 25 (2000) 11–14. [DOI] [PubMed] [Google Scholar]
- [34].Bolton K, Segal D, Walder K, The small leucine-rich proteoglycan, biglycan, is highly expressed in adipose tissue of Psammomys obesus and is associated with obesity and type 2 diabetes, Biologics 6 (2012) 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fan CY, Pan J, Usuda N, Yeldandi AV, Rao MS, Reddy JK, Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. Implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism, J. Biol. Chem 273 (1998) 15639–15645. [DOI] [PubMed] [Google Scholar]
- [36].Rong X, Albert CJ, Hong C, Duerr MA, Chamberlain BT, Tarling EJ, Ito A, Gao J, Wang B, Edwards PA, Jung ME, Ford DA, Tontonoz P, LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition, Cell Metab. 18 (2013) 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cash JG, Hui DY, Liver-specific overexpression of LPCAT3 reduces postprandial hyperglycemia and improves lipoprotein metabolic profile in mice, Nutr. Diabetes 6 (2016) e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.