Abstract
Phosphatase and tensin homolog (Pten) antagonizes PI3K-Akt signaling; therefore, Pten impairment causes tumorigenesis. However, the correlation between Pten deficiency and colon cancer has remained elusive due to numerous opposite observations. To study this correlation, we examined whether Pten deficiency in intestinal epithelial cells (IECs) induces tumorigenesis.
With mucosal biopsies of human colon cancer and normal colon, Pten mRNA was evaluated by quantitative PCR. Using IEC-specific Pten knockout mice (PtenΔIEC/ΔIEC), we examined the mitotic activity of IECs; and PtenΔIEC/ΔIEC; Apcmin/+ mice were generated by combining PtenΔIEC/ΔIEC with Apcmin/+ mice. Tumor-associated gene was evaluated by micro-array analysis. Fecal microbiome was analyzed through 16S rRNA gene sequencing.
We found that Pten mRNA level was reduced in human colon cancer relative to normal tissues. Augmented chromatids, increased Ki-67 and PCNA expression, and enhanced Akt activation were identified in IECs of PtenΔIEC/ΔIEC mice compared to Pten+/+ littermate. Combining PtenΔIEC/ΔIEC with Apcmin/+ condition caused rapid and aggressive intestinal tumorigenesis. However, PtenΔIEC/ΔIEC mice did not develop any tumors. While maintaining the tumor-driving potential, these data indicated that IEC-Pten deficiency alone did not induce tumorigenesis in mice. Furthermore, the expression of tumor-promoting and tumor-suppressing genes was decreased and increased, respectively, in the intestine of Pten ΔIEC/ΔIEC mice compared to controls. The abundance of Akkermansia muciniphila, capable of inducing chronic intestinal inflammation, was diminished in PtenΔIEC/ΔIEC mice compared to controls.
These findings suggested that altered tumor-associated gene expression and changed gut microbiotashape a tumor-preventive microenvironment to counteract the tumor-driving potential, leading to the tumor prevention in PtenΔIEC/ΔIEC mice.
Keywords: Microbiome, Pten, Toll-like receptor, Tumor microenvironment, Tumor prevention
1. INTRODUCTION
Phosphatase and tensin homolog (Pten) is a lipid phosphatase responsible for the conversion of phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] to PI(4,5)P2; thus, Pten activation aborts phosphatidylinositol 3-kinase (PI3K)-mediated signaling pathways [1]. Conversely, defective Pten activity perpetuates the activation of PI3K signaling, leading to enhanced cell proliferation and survival, and ultimately tumorigenesis in a variety of organs including the prostate, kidney, ovary, and skin [2–5]. Germline mutations in Pten result in hamartomas (benign growths) in several organs with an increased risk of malignant tumors [6]. Therefore, Pten is considered to be a tumor suppressor gene. On the other hand, we and other groups have demonstrated that Pten gene deletion in intestinal epithelial cells (IECs) does not cause tumorigenesis in mice [7–10], implying the possibility that Pten may play an alternative role in the intestinal epithelium, which is in constant contact with luminal milieu like microbiota and nutrients. We previously found that the level of PI(4,5)P2 generated by Pten determines the plasma membrane localization of the adapter Mal/Tirap, thereby permitting Toll-like receptor 5 (TLR5) to interact with Mal/Tirap in order to mediate gut microbe-induced immune and inflammatory responses Based on this finding, we suggested an alternative function of Pten in the gut, where it participates in the regulation of immune and inflammatory responses [7].
In accordance with the tumor suppressor function of Pten, many studies have suggested that Pten deficiency is capable of inducing tumor development in the colon. For instance, reduced Pten expression has been observed in cancer tissues from colorectal cancer patients [11]. Additionally, mutations to Pten may contribute to primary colon cancer development in humans [12, 13], and Pten gene alterations have been associated with poor prognosis in patients with rectal cancer [14]. Similarly, conditional global Pten gene deletion, accomplished by interferon α/β-responsive gene (Mx-1) promoter-driven Cre expression [15], has been suggested to induce the development of small-sized tumors in the mouse intestine [16], while global Pten knockout (KO) mice are embryonic lethal [17]. Together, these observations suggest that Pten impairment may participate in the development of tumors in the colon.
In contrast, there is considerable evidence suggesting that Pten may not be directly associated with tumorigenesis in the colon. A study examining single nucleotide polymorphisms (SNPs) in the Pten gene indicated that there may be no association between Pten and colon cancer in humans [18]. Furthermore, the Pten mRNA level is not consistent among colorectal cancer tissues and varies depending on tumor location [19], and Pten mRNA expression was seen to be preserved in human colorectal adenomas and adenocarcinomas [20]. Together, these observations indicate that the loss of Pten expression may not be directly associated with tumor development in the colon. Thus, the role of Pten deficiency in the development of colon cancer remains elusive.
Through studies using IEC-specific Villin promoter-driven Cre expression, we and other groups have demonstrated that IEC-specific Pten-KO mice are normal without tumor development [7–9]. These findings prompted us to raise two important questions regarding the role of Pten impairment in colonic tumorigenesis: 1) Whether Pten gene deletion in IECs retains the potential to promote tumorigenesis; and 2) Whether there exists a tumor-suppressing condition in the colon to counteract a tumor-promoting effect brought about by Pten deficiency. We used an array to evaluate tumor-associated gene expression and analyzed the fecal microbiome, and found that both tumor-promoting gene expression and the level of tumor-promoting microbe were substantially reduced in IEC-specific Pten-KO mice. These results indicate that the microbiota and cell signaling networks could provide tumor-suppressive intracellular and extracellular environments which inhibit the tumorigenic effect of Pten deficiency in the intestinal epithelium.
2. MATERIALS AND METHODS
2.1. Human tissues
All human tissues were collected and analyzed with the approval of the UCLA Institutional Review Board (protocol: 14–000132), Wonkwang University Sanbon Hospital Institutional Review Board (protocol: 2011–07), and Seoul Paik Hospital Institutional Review Board (protocol: IIT-233). Participants (aged 32–85) who underwent colonoscopy screening provided written informed consent to the study protocol. Patients with a personal or first-degree family history of cancer were excluded. Patients with previous chemotherapy or radiation therapy were excluded. Patients with any infectious disease or intestinal inflammatory disease such as IBD were also excluded.
Two board certified gastroenterologists performed the colonoscopy and obtained the colorectal cancer tissues and adjacent (∼10 cm away from the lesion) normal colonic mucosa tissues at two independent medical centers in South Korea (Wonkwang University Sanbon Hospital and Inje University Seoul Paik Hospital). A small piece of each specimen was sent to a pathologist to determine the pathological assessment. Remaining parts of the specimen were immediately immersed in RNAlater RNA stabilization reagent (Qiagen, Valencia, CA) and stored at 4 °C overnight and kept in liquid nitrogen until RNA isolation. Based on the pathologic determination, cancerous (adenocarcinoma) colonic tissues were selected for the experiment.
Similarly, unmatched normal colonic mucosa specimens were collected from tumor-free healthy subjects undergoing routine colonoscopy screening at Wonkwang University Sanbon Hospital. The patients examined in this study have never been included in any of the previous studies.
2.2. Animals
Pten floxed (PtenloxP/loxP) [21], Villin promoter-driven Cre expression mouse (VilCre/+) [22], and Apcmin/+mice [23] on the C57BL/6 background were obtained from the Jackson Laboratory (Bar Harbor, ME). As described previously [7–9], intestinal epithelial cell-specific Pten knockout mice (Pten ΔIEC/ΔIEC) was generated by crossing PtenloxP/loxP mice with VilCre/+ mice. Then, Pten ΔIEC/ΔIEC mice were crossed with Apcmin/+ mice to generate Pten ΔIEC/ΔIEC;Apcmin/+ mice. Mice were bred and maintained under standard SPF conditions with normal drinking water ad libitum at the animal facility of University of California Los Angeles and Oakland University under the approval of the Institutional Animal Care and Use Committees of UCLA and OU.
2.3. Quantitative real-time PCR
As described previously [8, 24], total RNA was initially isolated from human colon specimens or the full thickness of mouse intestinal tissues using RNeasy Plus Universal Midi Kit (Qiagen, Valencia, CA). Then an equal amount of RNA (4 μg in 40 μL) was transcribed into cDNA with a High Capacity Reverse Transcription Kit obtained from “Applied Biosystems” (Foster City, CA). Subsequently, quantitative real-time PCR was performed with TaqMan Universal Master Mix to measure gene expression by following the standard conditions from Applied Biosystems in 7500 Fast Real-Time PCR system. After incubating at 50 °C (2 min) and activating AmpliTaq Gold activation at 95 °C (10 min), samples were denatured at 95°C (15 sec) and annealed/extended at 60 °C (1 min) for 40 cycles. The primer pairs and FAM™ dye-labeled TaqMan® MGB (minor groove binding) probes were purchased from “Applied Biosystems”. Gapdh was included as an internal control.
Using the PCR cycle (CT) at which the probe’s fluorescent intensity passes a certain threshold value (CT) at the exponential phase, the level of expression was calculated. Through the difference in the CT values of the target genes after normalization to RNA input level, relative gene expression was determined using the CT value of control Gapdh. The delta/delta CT (2-ΔΔCT) method [25] was used to calculate the relative gene expression. Each reaction was performed in triplicate. A Mann-Whitney U-test was performed on the normalized data to check whether the relative gene expression is statistically different (P < 0.05).
2.4. Transmission Electron Microscopy
The mouse colon tissues were obtained from age- (3 months old) and gender-matched Pten ΔIEC/ΔIEC and Pten +/+ mice. As described in our previous publication [8], the dissected tissues were immersed in a solution containing 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M PBS (pH 7.4) for 2 h at room temperature then incubated at 4°C overnight. On the next day, 0.5% of tannic acid was added to the tissues and incubated for 1 h at room temperature. The tissue blocks were then washed five times in 0.1 M PBS buffer and post-fixed in a solution of 1% OsO4 in PBS (pH 7.2–7.4). The combination of tannic acid/glutaraldehyde/paraformaldehyde followed by osmification increased the staining of the membranes. The samples were washed four times in Na acetate buffer (pH 5.5), block-stained in 0.5% uranyl acetate in 0.1 M Na acetate buffer (pH 5.5) for 12 h at 4°C. The samples were dehydrated in graded ethanol (50%, 75%, 95%, 100%, 100%, 100%) 10 minutes each, passed through propylene oxide, and infiltrated in mixtures of Epon 812 and propylene oxide 1:1 and then 2:1 for 2 h each. The tissues were then infiltrated in pure Epon 812 overnight. Embedding was then performed in pure Epon 812 and curing was done in an oven at 60°C for 48 h. Sections of 60 nm thickness (gray interference color) were cut on an ultramicrotome (RMC MTX) using a diamond knife. The sections were deposited on single-hole grids coated with Formvar and carbon and double-stained in aqueous solutions of 8% uranyl acetate for 25 min at 60°C and lead citrate for 3 min at room temperature. Thin sections subsequently were examined with a 100CX JEOL transmission electron microscope.
2.5. Immunohistochemistry
Mouse colon tissues were embedded and frozen immediately. Six-micrometer sections were cut and then fixed in acetone for 15 min at 4 °C. After rehydration, sections were blocked in 2% bovine serum albumin (BSA) solution for 10 min, and incubated overnight with primary rat anti-mouse Ki-67 antibody (1:100) diluted in 2% BSA solution with 0.3% Triton X-100 at 4 °C. The negative controls received an equivalent concentration of non-immune rat IgG. After washing with PBS, sections were incubated with biotinylated anti-rat secondary antibody (Vector Laboratories, Burlingame, CA) diluted 1:200 in 2% BSA solution with 0.3% Triton X-100 for 45 min at room temperature. After inactivating endogenous peroxidase, sections were processed for peroxidase immunohistochemistry. 0.2% light green (Sigma) solution was used for counterstaining.
After the Ki-67-stained sections were visualized using the microscope, each transverse section was divided into 4 quadrants. Crypt cell nuclei with clear positive brown staining were counted. The ratio between the number of cells labeled Ki-67-positive and total cell count was expressed as a percentage, referred to as the Ki-67 proliferative cell index.
2.6. Immunofluorescence staining
To visualize the expression of phospho-AKT(Ser473) and PCNA in colon tissues, tissues were fixed with 10% neutral buffered formalin solution (Sigma-Aldrich, St. Louis, MO) at room temperature (RT). After paraffin embedment, the tissue sections (5 μm thickness) were prepared. The slides with tissue sections were dewaxed with xylene for 7 min twice and rehydrated with 100% ethanol for 2 min twice. Sections were soaked in 95% ethanol for 2 min, and then 70% ethanol for 2 min. Those were rinsed with distilled water (DW) and boiled in sodium citrate buffer at 95–99°C for 20 min to antigen retrieval. Slides were washed with TBS-T for 5 min twice and rinsed with DW. To reduce surface tension, tissues were washed with phosphate-buffered saline (pH 7.5) (PBS, Hyclone Inc., Logan, UT) containing 0.05% Trition X-100 for 5 min twice. Sections were soaked into blocking buffer (Protein Block Serum-Free, Aglient, Santa Clara, CA) containing 0.1% Trition X-100 to block non-specific binding for 2h at RT. Primary antibodies (P-AKT, #9271, Cell Signaling Technology), PCNA (sc-56, Santa Cruz Biotechnology, Inc)) were diluted in blocking buffer with 1:200 ratio. Slides containing blocked tissues were incubated in a humidified chamber, 4°C overnight. After incubation, slides were soaked in PBS-T for 5 min and repeated 4 times. Secondary antibodies; Goat anti-Rabbit IgG-heavy and light chain Antibody FITC Conjugated (A120–101F) and Goat anti-Mouse IgG-heavy and light chain Antibody FITC Conjugated (A90–116F) were from Bethyl Laboratory, Inc. (Montgomery, TX). After preparing diluted secondary antibodies with blocking buffer, the secondary antibodies were added to slides and incubated 2 h at RT. Slides were rinsed thoroughly 5 times with PBS-T, 5 min each. The slides were mounted with VECTASHIELD® Mounting Medium with 4’,6-diamidino-2-phenylindole (DAPI, Vector Laboratories, Burlingame, CA). Stained tissues were observed by FV10i (Olympus Inc, Center Valley, PA) with image analysis by FV10i FluoView software.
2.7. Blood sample collection for serum chemistry and hematologic analysis.
Blood samples were harvested by cardiac puncture from mice and collected in clot activator-containing Capillary Blood Collection Tubes, T-MG (Terumo Medical Corp. Elkton, MD). Immediately after blood collection, tubes were gently inverted several times and subsequently kept upright at 4 °C for 30 min to allow for clot formation. Then, serum was separated by centrifugation at 1,800g for 10 min at 4°C. The serum samples were subjected to serum chemistry analysis. For blood hematology procedures, mouse blood samples through cardiac puncture were collected in K2-EDTA (anticoagulant)-containing Capillary Blood Collection Tubes, T-MQK (Terumo Medical Corp. Elkton, MD). Blood serum chemistry and complete blood count were performed at DLAM Diagnostic Laboratory for blood analyses at UCLA.
2.8. Cancer-pathway focused gene analysis
To analyze the cancer-associated gene expression, the cancer pathway-focused PCR gene array analysis (Cat. No. PAMM-033Z; Qiagen, Valencia, CA) was performed in accordance with the manufacturer’s instruction. Briefly, immediately after harvesting the small intestinal tissues, total RNA was isolated using RNeasy Plus Universal Midi Kit (Qiagen, Valencia, CA). 1 μg of total RNA was treated with DNase. cDNA was synthesized using RT2 First Strand kit (Qiagen, Valencia, CA). For each analysis, cDNA samples were mixed with RT2 qPCR Master mix and distributed across the PCR array 96-well plates containing gene PCR probes and control housekeeping gene probes. The amplification cycling was performed with ABI 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA). Then, the data (fold-changes in CT values for all the genes) were analyzed with SABiosciences software that the manufacturer provided.
2.9. Fecal sample collection for microbiome analysis
Co-fostered Pten ΔIEC/ΔIEC and littermate Pten +/+ mice were obtained from a single parental breeder cage in a SPF condition with standard breeding condition. After weaning, Pten ΔIEC/ΔIEC and Pten +/+ mice were co-housed in a single cage. When the mice reached the age of 3 months old, the fecal stool pellet samples were collected from the colon, and snap-frozen in liquid nitrogen before sequencing.
2.10. DNA extraction from mouse fecal samples
Genomic DNA was purified from fecal samples with PowerSoil® DNA Isolation Kit (MoBio, Carlsbad, CA) in accordance with the manufacturer’s instructions. Approximately, 2 fecal pellets were subjected to the PowerBeads tube for cell lysis. The isolated genomic DNA was eluted from the spin filter using 50 μL of the elution buffer and stored at −20°C until PCR amplification.
2.11. PCR amplification and amplicon sequencing using next generation technology (bTEFAP®)
As described previously [26, 27], next generation technology (bTEFAP®) can be utilized to examine a broad range of microbiomes. A updated versions of bTEFAP® has adapted to non-optical sequencing technologies such as the Ion Torrent PGM as well as the Illumina MiSeq and HiSeq platforms and become one of the most widely published methods to evaluate microbiota.
V1–3 regions of Eubacterial 16S rDNA was amplified using primers the 16S universal Eubacterial primer 27F-519R set (27F AGRGTTTGATCMTGGCTCAG, and 519R GTNTTACNGCGGCKGCTG) to assess the microbial ecology of each sample on the MiSeq with methods via the bTEFAP® DNA analysis, spans [8]. A single-step 30 cycle PCR using HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA) was used under the following conditions: 94°C for 3 minutes, followed by 28 cycles of 94 °C for 30 seconds; 53 °C for 40 seconds and 72 °C for 1 minutes, final elongation 72 °C for 5 minutes. Amplicons were further purified using Agencourt Ampure beads (Agencourt Bioscience Co., Beverly, MA) and equimolar amplicons were pooled. Following PCR, all amplicon products from different samples were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Co., Beverly, MA). Sequencing was performed with the Illumina MiSeq in accordance with the manufacturer’s protocols.
2.12. Sequence analysis
The sequence data obtained from MiSeq was elaborated through a proprietary analysis pipeline (www.mrdnalab.com, MR DNA, Shallowater, TX). Barcodes and primers were trimmed, then short sequences < 200bp, sequences with ambiguous base calls, sequences with homopolymers exceeding 6bp, and chimeric sequences were removed. Sequences were then denoised. Operational taxonomic units (OTUs) were defined after removal of singleton sequences, clustering at 3% divergence (97% similarity) [26, 28, 29]. OTUs were then taxonomically classified using BLASTn against a curated GreenGenes/RDP/NCBI derived database [30] and compiled into each taxonomic level.
Normalized and denoised files were then run through QIIME (Quantitative Insights Into Microbial Ecology) to generate alpha and beta diversity data, as described previously [29, 31]. Based upon weighted unifrac analysis in the QIIME, principal coordinates analysis (PCoA) plots were generated. Additional statistical analyses were performed with NCSS2007 (NCSS, UT) and XLstat2012 (Addinsoft, NY). Significance reported for any analysis is defined as p<0.05.
2.13. Alpha and beta diversity analysis
Alpha diversity is essentially a means to evaluate how many different bacterial species are within the given sample or treatment group. Beta diversity allows comparison of the community of bacteria as a whole taking into account both how many different things are in the sample and what those things are related to phylogenetically. Alpha and beta diversity analysis were conducted as described previously[29, 31] using QIIME (www.qiime.org).
2.14. Statistical analysis:
Statistical analysis was conducted with GraphPad Prism (GraphPad Software, Inc., San Diego. CA) unless stated otherwise. Additional information regarding statistical analysis is presented in the corresponding figure legend. P values less than 0.05 were considered significant.
3. RESULTS
3.1. Pten mRNA expression was reduced in colon cancer biopsies compared to normal tissues.
Although Pten is classified as a tumor suppressor, it has remained elusive whether Pten impairment is capable of inducing tumorigenesis in the colon. To investigate the association of Pten deficiency with colonic tumor development, we evaluated the Pten mRNA expression in human colon cancer tissues. Colon cancer and adjacent normal tissues (matched) were obtained from patients with colon cancer. Normal colonic biopsies were also collected from healthy control subjects (unmatched). With quantitative real-time PCR (qPCR), we found that the Pten mRNA level was substantially lower in colon cancer tissues compared to matched normal tissues (Fig. 1A). Pten mRNA expression was also lower in colon cancer tissues when compared to the expression level in unmatched normal tissues (Fig. 1B). Meanwhile, the Pten mRNA expression level was comparable between the normal colonic biopsies obtained from colon cancer patients and control subjects (Fig. 1C).
Figure 1. Pten mRNA expression was reduced in human colon cancer tissues relative to normal tissues.
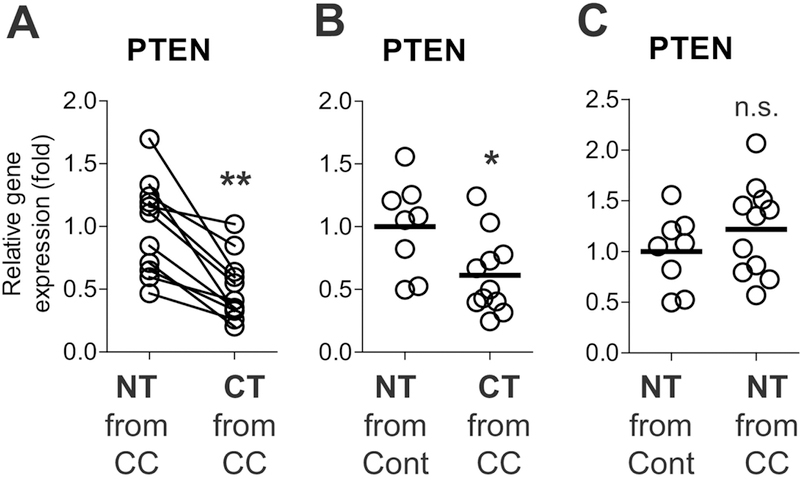
(A) Pten mRNA levels were evaluated by qPCR with matched normal tissues (NT) and colon cancer tissues (CT) obtained from colon cancer (CC) patients (11 sets). (B) The mRNA level was evaluated between unmatched normal tissues (n=8) independently obtained from healthy control subjects (Cont) and the colon cancer tissues (n=11) from colon cancer patients. (C) The mRNA expression in unmatched normal tissues (n=8) from healthy control subjects was compared with the data in the normal tissues from colon cancer patients (n=11). * P<0.05, ** P<0.01, n.s. stands for not significant (Mann-Whitney U test). Each bar indicates mean.
These data suggest that decreased Pten expression is associated with colon cancer development in humans, and are in accordance with previous research indicating that Pten deficiency may be associated with colon cancer development and metastasis in humans, and poor patient survival [13, 32].
3.2. Loss of the Pten gene in intestinal epithelial cells resulted in enhanced cell mitotic activity.
We observed that the small intestine and colon were substantially longer in mice with intestinal epithelial cell-specific Pten gene deletion (Pten ΔIEC/ΔIEC) than in littermate Pten +/+ mice (Fig. 2A and B). Similarly, the height of the villi in the colon was markedly increased in Pten ΔIEC/ΔIEC mice compared to Pten +/+ littermates (Fig. 2C). These data indicate that Pten gene deletion in IECs results in a proliferative effect within the intestine.
Figure 2. Intestinal epithelial cells of Pten ΔIEC/ΔIEC mice exhibited enhanced mitotic activity compared to those of Pten +/+ mice.
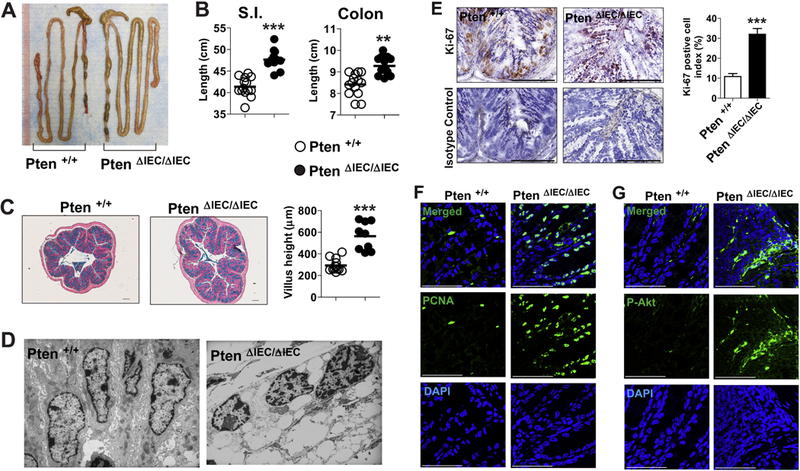
(A and B) Presented is the representative gross appearance of the intestine from Pten ΔIEC/ΔIEC and littermate Pten +/+ mice (A). Full length of the small intestine and the colon from age (1 year)- and sex- matched mice was evaluated (n=14/group) (B). (C) Paraffin-embedded sections of the mid-colon were prepared from age (3 months)-and sex-matched Pten ΔIEC/ΔIEC (n=9) and littermate Pten +/+ (n=9) mice. The sections were subjected to Alcian blue staining, followed by measurement of villus height under Axio Imager Z1 microscope (Carl Zeiss, Oberkochen, Germany). (D) Presented are electron micrographs of the colonic epithelium from Pten ΔIEC/ΔIEC and littermate Pten +/+ mice at the age of 12 month. (E) With the frozen sections of the mouse colon, expression of the cell proliferation marker Ki-67 was evaluated by immunohistochemistry with an antibody against Ki-67 and its isotype control IgG. Ki-67 positive cell index was evaluated and presented as means ± SEM. (F and G) Paraffin-embedded sections of the mid-colon were subjected to immunofluorescence staining with PCNA (F) and phospho-Akt (P-Akt) (G) antibodies. ** P<0.01, *** P<0.001 (Mann-Whitney U test). Representative images from three independent experiments were presented. Each bar in the graph indicates mean. Scale bar represents 100 μm (E) or 50 μm (F, G).
Despite the cellular function of Pten as a tumor-suppressor gene, we and other groups have already demonstrated that mice Pten ΔIEC/ΔIEC mice do not develop tumors [7–9]. Therefore, we investigated why Pten deficiency in intestinal epithelial cells is not sufficient to induce intestinal tumor development. Given the augmented Akt activation observed in Pten-deleted epithelial cell lines [7], we first examined whether the intestinal epithelial cells of Pten ΔIEC/ΔIEC mice preserve enhanced mitotic activity. Transmission electron micrographs demonstrated that the intestinal epithelial cells of Pten ΔIEC/ΔIEC mice harbor atypical nuclei with increased chromatids and chromosomal irregularity, while littermate control (Pten +/+) mice exhibited normal columnar epithelial cells with intact cell-to-cell interaction along the lateral surface (Fig. 2D). Similarly, the expression of cell proliferation marker Ki-67 and PCNA was substantially enhanced in the colonic tissues of Pten ΔIEC/ΔIEC mice compared to controls (Fig. 2E and F). We further confirmed that Akt activation was substantially increased in the colonic tissue of Pten ΔIEC/ΔIECmice compared to controls (Fig. 2G), suggesting increased proliferation of the intestinal epithelial cells. Taken together, these observations indicate that loss of the Pten gene still increases the mitotic activity of intestinal epithelial cells.
3.3. IEC-Pten gene deletion promoted intestinal tumorigenicity in an Apc min/+ mouse model of colon cancer.
Although Pten is considered a tumor suppressor gene [33], several studies have demonstrated that Pten gene deletion in IECs is not capable of inducing tumorigenesis [7–9]. This raises the question of whether defective Pten in IECs still retains the potential to promote tumorigenicity, or just participates in regulating immune and inflammatory responses without influencing tumor development [7]. To address this question, we utilized an Apc min/+ mouse model of intestinal tumorigenesis. Specifically, Pten ΔIEC/ΔIEC mice were combined with Apc min/+ mice to generate Pten ΔIEC/ΔIEC; Apc min/+ mice. We observed massive tumor development throughout the small intestine and colon of Pten ΔIEC/ΔIEC; Apc min/+ mice (Fig. 3A). The number of tumors in the intestine was dramatically higher in Pten ΔIEC/ΔIEC; Apc min/+ mice compared to littermate Pten ΔIEC/+; Apc min/+ and Pten +/+; Apc min/+ mice (Fig. 3B and C). Pten ΔIEC/ΔIEC; Apc min/+ mice also had enlarged spleens compared to age-matched littermate Pten +/+; Apc min/+ mice (Fig. 3D), which appears to be a systemic effect in response to the severe intestinal tumor development. Due to the extensive tumorigenesis in the intestine, approximately 90% of Pten ΔIEC/ΔIEC; Apc min/+ mice failed to survive, while more than 90% of littermate Pten +/+; Apc min/+ mice survived until 3 months old (Fig. 3E).
Figure 3: IEC-specific Pten deletion dramatically enhanced the intestinal tumorigenesis in the Apc min/+ background.
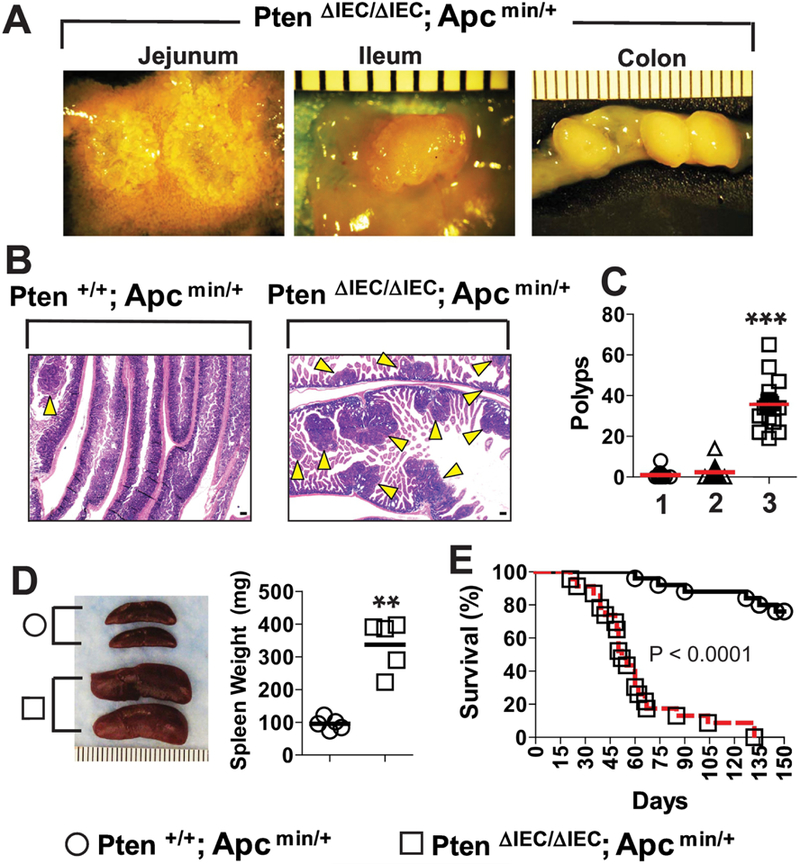
(A) Gross images of the tumors observed throughout the small intestine (jejunum and ileum) and the colon of Pten ΔIEC/ΔIEC; Apc min/+ mice. (Scale bar, 1mm). (B) The full length of the small intestine was obtained from Pten ΔIEC/ΔIEC; Apc min/+ and littermate Pten +/+; Apc min/+ mice at the age of 5–6 weeks old, and prepared in a ‘swiss-roll’ method for H&E staining. Arrowhead indicates an individual tumor mass observed under microscope. (scale bar, 100 μm) (C) Using stereoscopic microscopy, the number of visible tumors (≥ 1.0 mm in diameter) was evaluated throughout the small intestine of the age (5–6 weeks old)-matched mice: lane 1, Pten +/+; Apc min/+ (n=12); lane 2, Pten ΔIEC/+; Apc min/+ (n=12); lane 3, Pten ΔIEC/ΔIEC; Apc min/+ mice (n=19). (D) Gross images and weight of the spleen obtained from Pten +/+; Apc min/+ and Pten ΔIEC/ΔIEC; Apc min/+ mice (n=5/group). (E) The mouse mortality was evaluated between Pten +/+; Apc min/+ (n=25) and Pten ΔIEC/ΔIEC; Apc min/+ mice (n=23) over 5 months after birth. Difference in survival was shown by Kaplan-Meier plot. The log-rank (Mantel-Cox) test was used to compare significant survival difference (P< 0.0001). Data were analyzed with the results accumulated by 5 independent experiments. Presented are the representative images. ** P<0.01, *** P<0.001 (Mann-Whitney U test).
Taken together, these data suggest that the Pten gene deletion in IECs substantially promotes tumor development in an Apc min/+ background. Although tumorigenesis does not occur in mice with IEC-Pten gene deletion alone, it is of note that Pten deficiency in IECs still possesses the latent capability of promoting tumor development.
3.4. The disease development in Pten ΔIEC/ΔIEC; Apc min/+ mice was limited to the intestine.
Through hematologic analysis, we observed that red blood cells, hemoglobin, and hematocrit were markedly reduced in Pten ΔIEC/ΔIEC; Apc min/+ mice compared to littermate Pten ΔIEC/+; Apc min/+ and Pten +/+;Apc min/+ mice (Supplementary Fig. 1A). This observation is indicative of anemia resulted from severe intestinal tumor development. Pten ΔIEC/ΔIEC; Apc min/+ mice were also characterized by increased levels of RBC distribution width and a heightened percentage of monocytes, indicating malnutrition and a chronic inflammatory condition, respectively, due to the intestinal tumors. In addition, blood serum chemistry results showed reduced levels of albumin, alkaline phosphatase, and total protein in Pten ΔIEC/ΔIEC; Apc min/+ mice compared to controls (Supplementary Fig. 1B). Reduced levels of these proteins indicate protein-losing enteropathy, which is an atypical manifestation of severe intestinal tumorigenesis in Pten ΔIEC/ΔIEC; Apc min/+ mice. Alanine aminotransferase and aspartate aminotransferase, which signify liver- associated abnormality, were similar between the groups. Together, our data indicate that the tumor development observed in Pten ΔIEC/ΔIEC; Apc min/+ mice is limited to the intestine.
3.5. Tumor-promoting gene expression was reduced in the intestine of Pten ΔIEC/ΔIEC mice.
Although Pten gene deletion in IECs alone does not induce tumor development, our data demonstrated that it is capable of accelerating tumorigenesis in Apc min/+ mice. This observation prompted us to seek for an explanation for why IEC-Pten gene deletion alone does not induce intestinal tumor development at least in mice. To answer this question, we hypothesized that intracellular signaling networks and extracellular microenvironment comprised of gut microbiota would contribute to suppress the tumorigenicity driven by the Pten deficiency in the intestine.
To test this hypothesis, we first analyzed tumor-associated gene expression in the intestine of Pten ΔIEC/ΔIEC and littermate Pten +/+ mice using cancer pathway-focused PCR array analysis, followed by individual qPCR. Among various genes associated with tumorigenesis, we found that the expression of Sox10, Kdr, Mcm2, Ccnd2, and Cdc20 was downregulated in the intestine of Pten ΔIEC/ΔIEC mice compared to littermate Pten +/+ mice (Fig. 4A and B). Sox10 is a member of the SOX transcription factor family, which regulates canonical Wnt/β-catenin signaling; therefore, elevated Sox10 expression has been suggested to promote tumor progression [34]. Kdr(Vegfr2) plays a crucial role in colon cancer progression; thus, heightened Kdr expression can predict a poor prognosis in colon cancer patients [35]. Mcm2 regulates DNA replication; therefore, an enhanced Mcm complex including Mcm2 has been suggested to regulate various types of tumor development [36]. Ccnd2 promotes cell proliferation by favoring the G1/S cell cycle transition; consequently, enhanced Ccnd2 expression is linked to colon carcinogenesis [37]. Cdc20 ubiquitinates the cyclin-dependent kinase (Cdk) inhibitor p21, which is a negative regulator of the cell division cycle [38]. Thus, Cdc20 activation results in p21 degradation, leading to enhanced cell mitosis and consequent tumorigenesis [39]. The activation of Sox10, Kdr, Mcm2, Ccnd2, and Cdc20 should promote tumor development; therefore, the reduced expression of these genes observed in the intestine of Pten ΔIEC/ΔIEC mice appears to play an important role in preventing tumor development.
Figure 4: The expression of cancer-associated genes was changed in the intestine of Pten ΔIEC/ΔIEC mice.
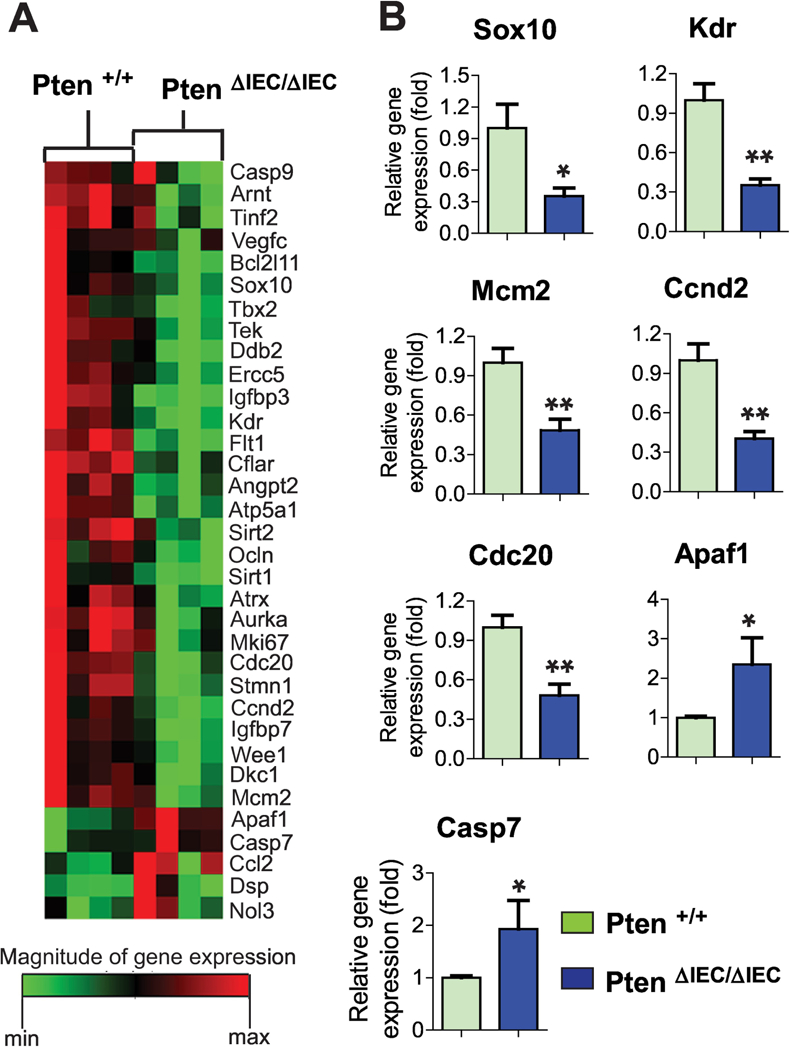
(A) An array of cancer-associated gene expression was analyzed using the mouse small intestine. Gene expression profiles with significant difference were visualized in the heat map. (n=4/group) (B) To confirm the representative gene expression exhibiting a significant difference from the array data, individual qPCR was independently performed using the mouse small intestine (n=6/group). Data are the representative of three independent experiments. Error bars indicate ± SEM. * P<0.05, ** P<0.01 (Mann-Whitney U test).
On the other hand, the expression of Apaf1 and Casp7 was up-regulated in the intestine of Pten ΔIEC/ΔIEC mice compared to controls. Apaf1 participates in the formation of apoptosomes, leading to the activation of initiator caspases, including Casp9. The executioner Casp7 is subsequently activated, thereby inducing apoptosis [40]. In light of their essential roles in apoptosis, it is likely that the elevated expression of Apaf1 and Casp7 may be capable of promoting cell apoptosis in the intestine of Pten ΔIEC/ΔIEC mice, conferring a tumor-suppressive effect.
Together, the gene expression array data suggest that Pten gene deletion in IECs diminished the expression of tumor-promoting genes, while enhancing tumor-suppressing gene expression.
3.6. Loss of the Pten gene in IECs resulted in intestinal dysbiosis.
Given the ability of certain commensal bacteria to stimulate inflammatory or anti-inflammatory responses, an emerging body of evidence indicates that the gut microflora has a sizable impact on the promotion and suppression of colon cancer in humans [41–43]. Therefore, to account for the failure of Pten deficiency in IECs to induce intestinal tumor development, we next analyzed the fecal microbiome of sex- and age (3 months old)-matched Pten ΔIEC/ΔIEC and littermate Pten +/+ mice that were co-housed since their birth. Through 16S ribosomal RNA (rRNA) gene sequencing, we evaluated the observed operational taxonomic unit (OTU) counts for Pten ΔIEC/ΔIEC and Pten +/+ mice along with the Shannon diversity index, and found no significant difference between the groups with regard to species richness (count of different species in each sample group). However, taking species richness as well as evenness (relative abundance of different species) into account revealed a significant difference between the groups (Table 1).
Table 1.
Species richness and evenness were different between Pten ΔIEC/ΔIEC and Pten +/+ mice. No significant differences between Pten ΔIEC/ΔIEC and Pten +/+ mice were observed with regards to species.However, taking species richness as well as evenness into account, there is a significant difference found between the groups. The mean rarefaction predicted OTUs data for the mouse groups were measured at a depth of 17,821 sequences. Presented are the observed species and the Shannon index showing the mean value.
| Variable n=17,821 |
Observed species | Shannon index | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean | Std. Dev. |
Mean of ranks |
Statistics | Mean | Std. Dev. |
Mean of ranks |
Statistics | |
| Pten ΔIEC/ΔIEC | 1,407.375 | 137.670 | 9.875 | P = 0.083 | 6.443 | 0.607 | 10.625 | P = 0.015 |
| Pten +/+ | 1,020.286 | 372.455 | 5.857 | 4.959 | 1.304 | 5.000 | ||
Subsequently, we analyzed the bacterial community structure using weighted UniFrac distance matrices. Principal coordinate analysis (PCoA) plots were used to visualize the data in these matrices, and analysis of similarities (ANOSIM) was utilized to determine if there was a significant difference between the bacterial communities. Pten ΔIEC/ΔIEC and littermate Pten +/+ mouse fecal samples in the weighted PCoA plot form two separate clusters (Fig. 5). With the ANOSIM R value (R=0.64, P=0.001), we determined that there is a significant difference between the sample groups. Note that an R value close to 1 indicates a high separation of microbial species composition between the samples, whereas an R value close to 0 indicates no separation.
Figure 5: The fecal microbiome of Pten ΔIEC/ΔIEC mice was distinct from that of Pten +/+ mice.
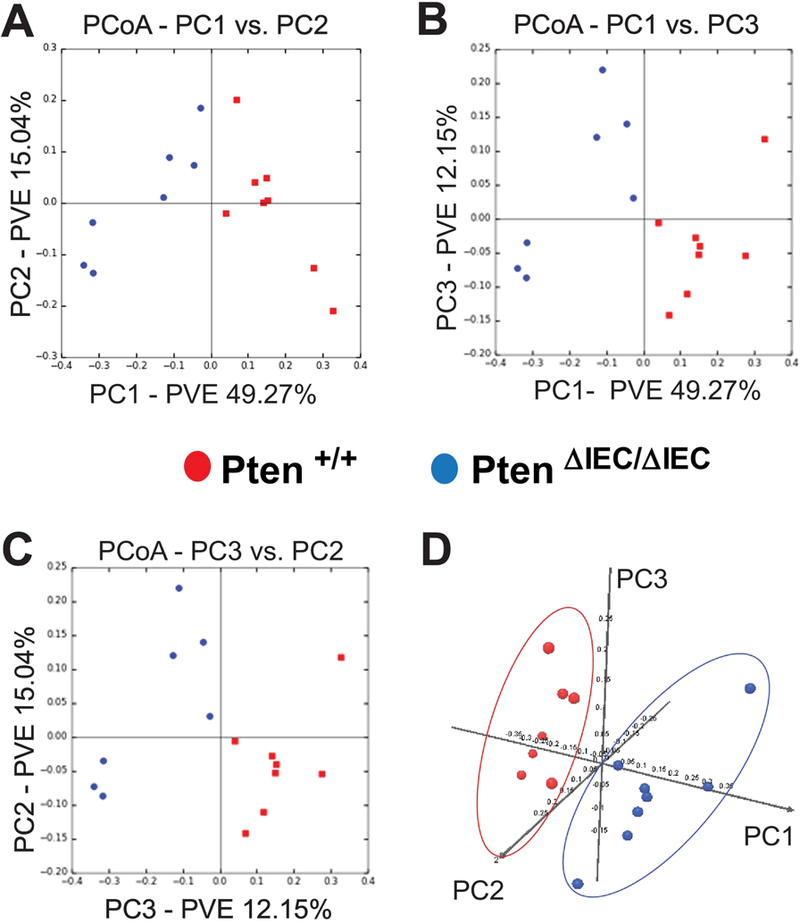
The bacterial community structure of the fecal samples from Pten +/+ and Pten ΔIEC/ΔIEC mice was analyzed using weighted UniFrac distance matrices. (A to C) Principal coordinate analysis plots represent the three highest discriminating axes. The primary vector PC1 explains 49.27% of the variation between the groups, while PC2 and PC3 represent 15.04% and 12.15%, respectively. PVE, percent variance explained. (D) The first three vectors together exhibit 76.4% of the variation among the groups. Each dot represents individual microbiota samples from the mouse. (n=8/each group).
While the species diversity itself was comparable between the fecal samples of Pten ΔIEC/ΔIEC and Pten +/+ mice, our data taken together demonstrate that Pten gene deficiency in IECs did alter the species composition of the fecal microbiome.
3.7. The fecal microbiota communities of Pten ΔIEC/ΔIEC mice were different from those of Pten +/+ mice.
We found that 100% of analyzed sequences from all Pten ΔIEC/ΔIEC and Pten +/+ mouse samples belonged to the bacteria kingdom, with no detectable archaeal or eukaryotic contamination. The majority (>98.7%) of analyzed sequences from the mice were assigned to 4 major phyla: Bacteroidetes (Pten ΔIEC/ΔIEC vs. Pten +/+, 45.07% vs. 36.34%), Firmicutes (47.47% vs. 31.30%), Verrucomicrobia (2.88% vs. 28.61%), and Proteobacteria (3.46% vs. 2.55%) (Fig. 6A). Among these major phyla constituting the fecal microbiota, it is worth noting that the phylum of Verrucomicrobia was substantially reduced in Pten ΔIEC/ΔIEC mice compared to Pten +/+ mice.
Figure 6: Relative abundance of taxonomic groups observed in the fecal samples were different between Pten ΔIEC/ΔIEC and Pten +/+ mice.
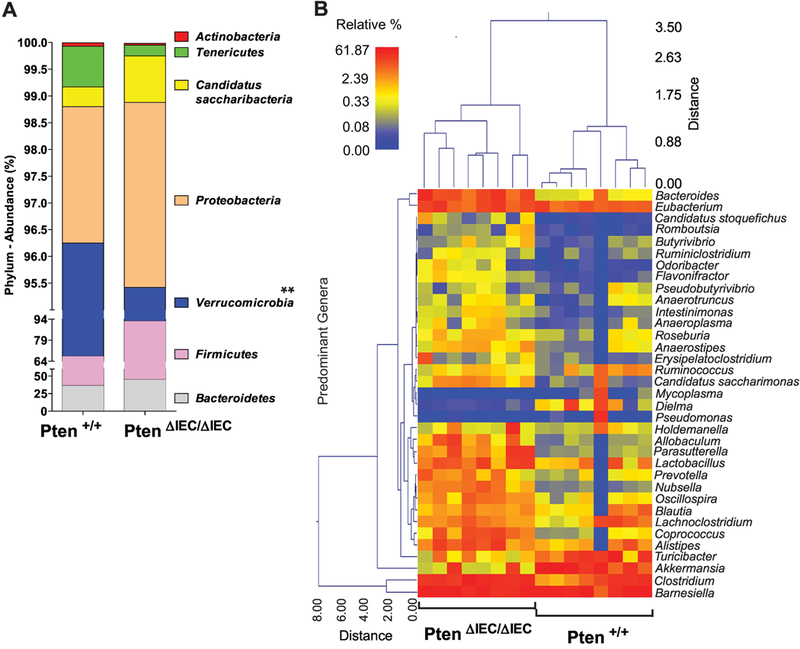
(A) The abundance (% of sequences) of the phyla detected in the fecal samples of Pten ΔIEC/ΔIEC and Pten +/+ mice. Bacteroidetes, Firmicutes, Verrucomicrobia, and Proteobacteria are the 4 major phyla encompassing the majority of sequences (> 98.7 %). A statistically significant difference between the groups was identified in Verrucomicrobia. ** P<0.01 (Mann-Whitney U test). (B) A dual hierarchal dendrogram was generated based on the predominant genera using Ward’s minimum variance clustering and Manhattan distances. Samples with more similar microbial populations are mathematically clustered closer together. The samples with a more similar consortium of genera cluster closer together with the length of connecting lines (top of heatmap) related to the similarity; shorter lines between two samples indicate closely matched microbial consortia. The heatmap represents the relative percentages of each genus. The legend for the heatmap is provided in the upper left corner. The predominant genera are represented along the right Y-axis.
Next, we examined whether any specific bacterial genera were significantly different between the samples. We identified a wide range of genera whose abundances were significantly different between the groups. To provide a visual overview combined with analysis, we generated a dual hierarchical dendrogram to display the data for the predominant genera with clustering related to the different groups. Based on the distinct clustering (Fig. 6B), we observed a significant difference in the predominant genera detected between Pten ΔIEC/ΔIEC and Pten +/+ mice.
Taken together with the PCoA plots, these data suggest that the fecal microbial communities from Pten ΔIEC/ΔIEC mice are easily distinguished and indeed different from Pten +/+ mice, further demonstrating that Pten gene deletion in IECs changed the fecal microbiome.
3.8. The colon cancer-associated bacteria Akkermansia muciniphila was dramatically decreased in the feces of Pten ΔIEC/ΔIEC mice compared to Pten +/+ mice.
Subsequently, we examined bacterial genera whose abundances were significantly changed between Pten ΔIEC/ΔIEC and littermate Pten +/+ mice. Although there were a wide range of genera that were found to be significantly different between the mouse groups, the two most notable differences were found in the relative abundances of Akkermansia (28.61% in Pten +/+ → 2.88% in Pten ΔIEC/ΔIEC) and Clostridium(7.89% → 20.27%)(Fig. 7A). At the level of bacterial species, we observed that the abundance of Akkermansia muciniphila (28.61% in Pten +/+ → 2.88% in Pten ΔIEC/ΔIEC) was greatly reduced in the fecal samples of Pten ΔIEC/ΔIEC mice compared to Pten +/+ mice, while the abundance of Clostridium sp. was increased (6.97% in Pten +/+ → 19.23% in Pten ΔIEC/ΔIEC) (Fig. 7B).
Figure 7: The abundance of the colitogenic bacteria Akkermansia muciniphila was dramatically reduced in Pten ΔIEC/ΔIEC mice compared to Pten +/+ mice.
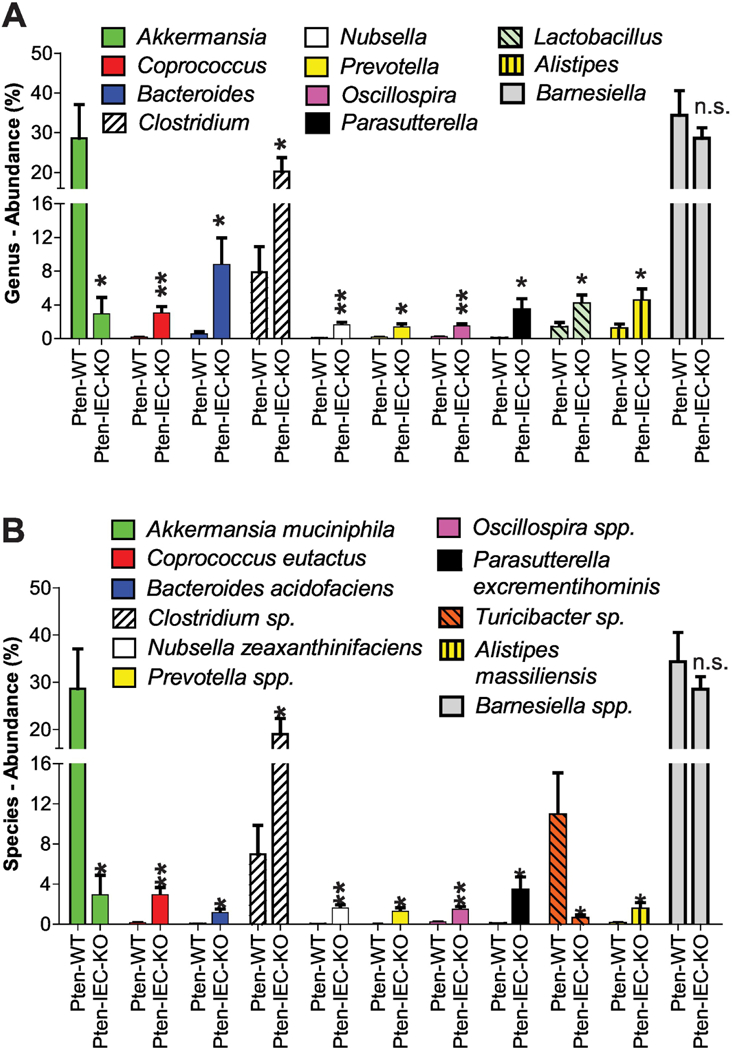
(A and B) Major bacteria identified in the fecal samples of Pten +/+ (Pten-WT) and Pten ΔIEC/ΔIEC (Pten-IEC-KO) mice (n=8/group) were analyzed to compare the abundance at the genus (A) and species (B) level. Results are means ± SD, * P<0.05, **P<0.01 (Mann-Whitney U test). n.s., not significant.
Akkermansia muciniphila, the representative of the Verrucomicrobia phylum, is a mucin-degrading bacterium [44], and thus has great potential to induce colonic inflammation [45]. It has been suggested that Akkermansia muciniphila colonization can markedly increase tumor development in an Apc mouse model of colon cancer [46] and may promote tumorigenesis in human colon cancer [47]. In light of this notion, it is reasonable to believe that the greatly decreased abundance of Akkermansia muciniphila would participate in providing a tumor-suppressing microenvironment to prevent tumor development in Pten ΔIEC/ΔIEC mice.
4. DISCUSSION
Tumorigenesis in the colon is influenced by a variety of factors, including chronic inflammatory conditions, genetic aberrations, compromised tumor-immune responses, and carcinogenic environmental agents. Amid these factors, an emerging body of evidence indicates that the complex interplay between the gut microbiome and the host confers a significant impact on tumor prevention and progression. For instance, Fusobacterium nucleatum is enriched in colorectal cancer tissues where it promotes the malignancy [48]. Bacteroides fragilis has been observed at an increased abundance in stool samples from colorectal cancer (CRC) patients compared to controls [49], and is known to promote colon tumorigenesis by enhancing Th17 inflammatory responses [50]. Monocolonization with the commensal microbe Escherichia coli NC101 has been suggested to promote chronic inflammation in the colon, leading to enhanced tumorigenesis in azoxymethane-treated IL-10-KO mice [51]. Enterococcus faecalis is capable of causing a chromosomal aberration (aneuploidy) in colonic epithelial cells in IL-10-KO mice [52, 53]. The abundance of Akkermansia muciniphila has been measured to be 4 times higher in the stool samples of CRC patients than healthy subjects[47]; moreover, Akkermansia muciniphila colonization exacerbates tumor development in the intestine of Apc min/+ mice [46]. In contrast, probiotic bacteria, including Lactobacillus and Bifidobacterium species, have been suggested to confer a tumor-suppressing effect in the colon [54].
The mechanisms by which gut microbes mediate tumor-promoting or tumor-suppressing effects are not clear yet. Given the fact that long-term inflammatory conditions cause DNA damage and chromosomal instability, early genetic events in a pathway toward colon cancer, it appears that the bacterial capability of inducing a chronic inflammatory condition is a major culprit behind the promotion of tumor development [54]. In agreement with this notion, it has been demonstrated that Lactobacillus and Bifidobacterium species, which have a potent anti-inflammatory effect in the intestine, are capable of suppressing tumorigenesis [55, 56].
Akkermansia muciniphila is a relatively abundant bacteria found in the colon of healthy humans, where it comprises approximately 1–4% of the colonic microbiota [46, 57, 58] and has strong clinical relevance to a variety of diseases, including inflammatory bowel diseases (IBD) and metabolic disorders [44]. Akkermansia muciniphila has the clinically significant function of degrading high molecular weight glycoproteins released as mucins from epithelial cells in the intestinal epithelial layer, which allows the bacteria to modulate the selectively permeable, physical barrier system that separates the host from luminal milieu like gut microbes, toxins, and dietary antigens [44]. Due to its mucin-degrading capability, enriched Akkermansia muciniphila in the colon can act as a pathobiont to develop inflammatory conditions[45, 59], which substantially increase the risk of tumor development[8, 60]. Notably, our own fecal microbiome analysis data identified a dramatically reduced abundance of Akkermansia muciniphila in Pten ΔIEC/ΔIEC (2.88%) mice compared to Pten +/+ (28.61%) littermates. Given this bacterial species’ inflammatory impact, it is reasonable to believe that the decreased abundance of Akkermansia muciniphila has a role in preventing intestinal tumorigenesis in Pten ΔIEC/ΔIEC mice. This notion is supported by the differing rates of intestinal tumor development that have been observed in Pten ΔIEC/ΔIEC mice. Although several studies have demonstrated that Pten ΔIEC/ΔIEC mice do not develop tumors in the intestine during the first 12 months of life[7–9], Byun et al. have suggested that approximately 19% of Pten ΔIEC/ΔIEC mice develop tumors in the duodenum and jejunum of the intestine by the age of 12 months[61] The sizable discrepancy between these data strongly indicates that gut microbial differences between housing facilities may have a significant impact on tumorigenesis in mice with the same genetic predisposition.
A genetic predisposition can mediate the activation of tumor-promoting signaling pathways, leading to enhanced cell proliferation and consequent tumor development. Conversely, tumor-suppressing signaling pathways can be activated to induce apoptotic cell death, leading to tumor prevention or suppression. Therefore, a dynamic balance between tumor-promoting and tumor-suppressing pathways has a crucial role in inhibiting the initial cellular event of tumor development [62]. Likewise, changed expression of tumor-promoting or tumor-suppressing genes has an important role in signaling networks contributing to tumorigenesis. Numerous studies have demonstrated that the activation of Sox10, Kdr, Mcm2, Ccnd2, and Cdc20 has an important function in signaling networks promoting tumor development [34–39]. In contrast, the activation of Apaf1 and Casp7 mediates cell apoptosis, resulting in a tumor-suppressing effect [40]. We identified reduced expression of Sox10, Kdr, Mcm2, Ccnd2 and Cdc20 genes, and elevated expression of Apaf1 and Casp7 genes in the intestine of Pten ΔIEC/ΔIEC mice compared to Pten +/+ mice. Therefore, along with the reduced abundance of the inflammation-inducing bacteria Akkermansia muciniphila in the colon of Pten ΔIEC/ΔIEC mice, decreased levels of tumor-promoting genes and elevated levels of tumor-suppressing genes may explain why intestinal tumors did not develop in Pten ΔIEC/ΔIEC mice.
Although it is unable to induce intestinal tumorigenesis in mice, Pten deficiency in humans has a well-characterized deleterious impact in patients with CRC. In fact, Pten gene alteration can be indicative of relapse [12], metastasis [63], and poor prognosis [14] in CRC patients. Although the underlying mechanisms remain elusive, impaired TGF-β signaling pathways [10, 63], promoted PI3K-Akt signaling pathways [12], or altered cell migration and interaction with the extracellular matrix [64] may contribute to the deleterious impacts of Pten deficiency in CRC patients.
5. CONCLUSION
In this study, we identified reduced mRNA expression of the tumor suppressor gene Pten in human colon cancer tissues relative to normal tissues, implying the involvement of Pten impairment in the development of colon cancer. However, Pten deficiency in IECs did not induce tumorigenesis in mice.We found that both the intestinal microflora and tumor-associated gene expression had been shifted favorably toward a tumor-suppressing condition in Pten ΔIEC/ΔIEC mice. Therefore, together with the changed expression of tumor-associated gene expression, our findings suggest that the gut microbiome plays an important part in shaping the tumor microenvironment to suppress tumorigenesis in the colon.
Supplementary Material
4) Highlights.
Pten gene expression is reduced in human colon cancer biopsies.
However, IEC-specific Pten-KO does not induce tumorigenesis in mice.
IEC-Pten-KO mice differentially express tumor-associated genes in the gut.
IEC-Pten-KO mice harbor low level of colitogenic bacteria, Akkermansia muciniphila.
Altered gut microbiome and gene expression shape a tumor-preventive environment.
ACKNOWLEDGEMENTS
This research was supported by a grant from Oakland University and the National Institutes of Health (DK079015, S.H.R).
Abbreviations:
- Apaf1
apoptotic peptidase activating factor 1
- Casp7
caspase-7
- Ccnd2
Cyclin D2
- Cdc20
cell division cycle 20 homologue
- IEC
Intestinal epithelial cells
- Kdr
kinase insert domain receptor
- Pten
Phosphatase and tensin homolog
- Mcm2
minichromosome maintenance protein 2
- Mal
MyD88 adaptor-like
- PCNA
proliferating cell nuclear antigen
- Sox10
SRY-box 10
- Tirap
TIR domain–containing adaptor protein
- TLR5
Toll-like receptor 5
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- [1].Hsu F, Mao Y, The structure of phosphoinositide phosphatases: Insights into substrate specificity and catalysis, Biochimica et biophysica acta, 1851 (2015) 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vecchio L, Seke Etet PF, Kipanyula MJ, Krampera M, Nwabo Kamdje AH, Importance of epigenetic changes in cancer etiology, pathogenesis, clinical profiling, and treatment: what can be learned from hematologic malignancies?, Biochimica et biophysica acta, 1836 (2013) 90–104. [DOI] [PubMed] [Google Scholar]
- [3].Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, Lotan TL, Clinical implications of PTEN loss in prostate cancer, Nat Rev Urol, 15 (2018) 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haddadi N, Lin Y, Travis G, Simpson AM, Nassif NT, McGowan EM, PTEN/PTENP1: ‘Regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy, Mol Cancer, 17 (2018) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Que WC, Qiu HQ, Cheng Y, Liu MB, Wu CY, PTEN in kidney cancer: A review and meta-analysis, Clin Chim Acta, 480 (2018) 92–98. [DOI] [PubMed] [Google Scholar]
- [6].Hobert JA, Eng C, PTEN hamartoma tumor syndrome: an overview, Genet Med, 11 (2009) 687–694. [DOI] [PubMed] [Google Scholar]
- [7].Choi YJ, Jung J, Chung HK, Im E, Rhee SH, PTEN regulates TLR5-induced intestinal inflammation by controlling Mal/TIRAP recruitment, FASEB J, 27 (2013) 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Im E, Jung J, Pothoulakis C, Rhee SH, Disruption of pten speeds onset and increases severity of spontaneous colitis in il10(−/−) mice, Gastroenterology, 147 (2014) 667–679 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Langlois MJ, Roy SA, Auclair BA, Jones C, Boudreau F, Carrier JC, Rivard N, Perreault N, Epithelial phosphatase and tensin homolog regulates intestinal architecture and secretory cell commitment and acts as a modifier gene in neoplasia, FASEB J, 23 (2009) 1835–1844. [DOI] [PubMed] [Google Scholar]
- [10].Yu M, Trobridge P, Wang Y, Kanngurn S, Morris SM, Knoblaugh S, Grady WM, Inactivation of TGF-beta signaling and loss of PTEN cooperate to induce colon cancer in vivo, Oncogene, 33 (2014) 1538–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lin PC, Lin JK, Lin HH, Lan YT, Lin CC, Yang SH, Chen WS, Liang WY, Jiang JK, Chang SC, A comprehensive analysis of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) loss in colorectal cancer, World J Surg Oncol, 13 (2015) 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Colakoglu T, Yildirim S, Kayaselcuk F, Nursal TZ, Ezer A, Noyan T, Karakayali H, Haberal M, Clinicopathological significance of PTEN loss and the phosphoinositide 3-kinase/Akt pathway in sporadic colorectal neoplasms: is PTEN loss predictor of local recurrence?, Am J Surg, 195 (2008) 719–725. [DOI] [PubMed] [Google Scholar]
- [13].Zhou XP, Loukola A, Salovaara R, Nystrom-Lahti M, Peltomaki P, de la Chapelle A, Aaltonen LA, Eng C, PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers, The American journal of pathology, 161 (2002) 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bohn BA, Mina S, Krohn A, Simon R, Kluth M, Harasimowicz S, Quaas A, Bockhorn M, Izbicki JR, Sauter G, Marx A, Stahl PR, Altered PTEN function caused by deletion or gene disruption is associated with poor prognosis in rectal but not in colon cancer, Human pathology, 44 (2013) 1524–1533. [DOI] [PubMed] [Google Scholar]
- [15].Asano A, Jin HK, Watanabe T, Mouse Mx2 gene: organization, mRNA expression and the role of the interferon-response promoter in its regulation, Gene, 306 (2003) 105–113. [DOI] [PubMed] [Google Scholar]
- [16].He XC, Yin T, Grindley JC, Tian Q, Sato T, Tao WA, Dirisina R, Porter-Westpfahl KS, Hembree M, Johnson T, Wiedemann LM, Barrett TA, Hood L, Wu H, Li L, PTEN-deficient intestinal stem cells initiate intestinal polyposis, Nat Genet, 39 (2007) 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP, Pten is essential for embryonic development and tumour suppression, Nat Genet, 19 (1998) 348–355. [DOI] [PubMed] [Google Scholar]
- [18].Phillips LS, Thompson CL, Merkulova A, Plummer SJ, Tucker TC, Casey G, Li L, No association between phosphatase and tensin homolog genetic polymorphisms and colon cancer, World J Gastroenterol, 15 (2009) 3771–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kuramochi H, Nakamura A, Nakajima G, Kaneko Y, Araida T, Yamamoto M, Hayashi K, PTEN mRNA expression is less pronounced in left- than right-sided colon cancer: a retrospective observational study, BMC cancer, 16 (2016) 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Taniyama K, Goodison S, Ito R, Bookstein R, Miyoshi N, Tahara E, Tarin D, Urquidi V, PTEN expression is maintained in sporadic colorectal tumours, J Pathol, 194 (2001) 341–348. [DOI] [PubMed] [Google Scholar]
- [21].Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H, Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo, Science, 294 (2001) 2186–2189. [DOI] [PubMed] [Google Scholar]
- [22].Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL, Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine, J Biol Chem, 277 (2002) 33275–33283. [DOI] [PubMed] [Google Scholar]
- [23].Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W, Interleukin-10-deficient mice develop chronic enterocolitis, Cell, 75 (1993) 263–274. [DOI] [PubMed] [Google Scholar]
- [24].Choi YJ, Im E, Pothoulakis C, Rhee SH, TRIF modulates TLR5-dependent responses by inducing proteolytic degradation of TLR5, J Biol Chem, 285 (2010) 21382–21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods, 25 (2001) 402–408. [DOI] [PubMed] [Google Scholar]
- [26].Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS, Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP), BMC microbiology, 8 (2008) 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA, Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs, Foodborne pathogens and disease, 5 (2008) 459–472. [DOI] [PubMed] [Google Scholar]
- [28].Edgar RC, Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 26 (2010) 2460–2461. [DOI] [PubMed] [Google Scholar]
- [29].Swanson KS, Dowd SE, Suchodolski JS, Middelbos IS, Vester BM, Barry KA, Nelson KE, Torralba M, Henrissat B, Coutinho PM, Cann IK, White BA, Fahey GC Jr., Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice, The ISME journal, 5 (2011) 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL, Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB, Applied and environmental microbiology, 72 (2006) 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Callaway TR, Dowd SE, Edrington TS, Anderson RC, Krueger N, Bauer N, Kononoff PJ, Nisbet DJ, Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing, J Anim Sci, 88 (2010) 3977–3983. [DOI] [PubMed] [Google Scholar]
- [32].Wang ZJ, Taylor F, Churchman M, Norbury G, Tomlinson I, Genetic pathways of colorectal carcinogenesis rarely involve the PTEN and LKB1 genes outside the inherited hamartoma syndromes, The American journal of pathology, 153 (1998) 363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Carracedo A, Pandolfi PP, The PTEN-PI3K pathway: of feedbacks and cross-talks, Oncogene, 27 (2008) 5527–5541. [DOI] [PubMed] [Google Scholar]
- [34].Cronin JC, Loftus SK, Baxter LL, Swatkoski S, Gucek M, Pavan WJ, Identification and functional analysis of SOX10 phosphorylation sites in melanoma, PLoS One, 13 (2018) e0190834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang SD, McCrudden CM, Meng C, Lin Y, Kwok HF, The significance of combining VEGFA, FLT1, and KDR expressions in colon cancer patient prognosis and predicting response to bevacizumab, Onco Targets Ther, 8 (2015) 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu Y, He G, Wang Y, Guan X, Pang X, Zhang B, MCM-2 is a therapeutic target of Trichostatin A in colon cancer cells, Toxicol Lett, 221 (2013) 23–30. [DOI] [PubMed] [Google Scholar]
- [37].Mermelshtein A, Gerson A, Walfisch S, Delgado B, Shechter-Maor G, Delgado J, Fich A, Gheber L, Expression of D-type cyclins in colon cancer and in cell lines from colon carcinomas, British journal of cancer, 93 (2005) 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pei XH, Xiong Y, Biochemical and cellular mechanisms of mammalian CDK inhibitors: a few unresolved issues, Oncogene, 24 (2005) 2787–2795. [DOI] [PubMed] [Google Scholar]
- [39].Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M, APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase, Molecular cell, 27 (2007) 462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Galluzzi L, Lopez-Soto A, Kumar S, Kroemer G, Caspases Connect Cell-Death Signaling to Organismal Homeostasis, Immunity, 44 (2016) 221–231. [DOI] [PubMed] [Google Scholar]
- [41].Buchta Rosean CM, Rutkowski MR, The influence of the commensal microbiota on distal tumor-promoting inflammation, Semin Immunol, 32 (2017) 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Van Raay T, Allen-Vercoe E, Microbial Interactions and Interventions in Colorectal Cancer, Microbiol Spectr, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen J, Pitmon E, Wang K, Microbiome, inflammation and colorectal cancer, Semin Immunol, 32 (2017) 43–53. [DOI] [PubMed] [Google Scholar]
- [44].Derrien M, Belzer C, de Vos WM, Akkermansia muciniphila and its role in regulating host functions, Microb Pathog, (2016). [DOI] [PubMed]
- [45].Ganesh BP, Klopfleisch R, Loh G, Blaut M, Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice, PLoS One, 8 (2013) e74963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dingemanse C, Belzer C, van Hijum SA, Gunthel M, Salvatori D, den Dunnen JT, Kuijper EJ, Devilee P, de Vos WM, van Ommen GB, Robanus-Maandag EC, Akkermansia muciniphila and Helicobacter typhlonius modulate intestinal tumor development in mice, Carcinogenesis, 36 (2015) 1388–1396. [DOI] [PubMed] [Google Scholar]
- [47].Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP, Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults, PLoS One, 8 (2013) e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M, Genomic analysis identifies association of Fusobacterium with colorectal carcinoma, Genome Res, 22 (2012) 292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, Soyletir G, A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer, Clin Microbiol Infect, 12 (2006) 782–786. [DOI] [PubMed] [Google Scholar]
- [50].Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL, A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses, Nature medicine, 15 (2009) 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C, Intestinal inflammation targets cancer-inducing activity of the microbiota, Science, 338 (2012) 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Balish E, Warner T, Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice, The American journal of pathology, 160 (2002) 2253–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB, Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria, Gastroenterology, 128 (2005) 891–906. [DOI] [PubMed] [Google Scholar]
- [54].Cipe G, Idiz UO, Firat D, Bektasoglu H, Relationship between intestinal microbiota and colorectal cancer, World J Gastrointest Oncol, 7 (2015) 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ringel-Kulka T, Palsson OS, Maier D, Carroll I, Galanko JA, Leyer G, Ringel Y, Probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium lactis Bi-07 versus placebo for the symptoms of bloating in patients with functional bowel disorders: a double-blind study, J Clin Gastroenterol, 45 (2011) 518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lightfoot YL, Yang T, Sahay B, Mohamadzadeh M, Targeting aberrant colon cancer-specific DNA methylation with lipoteichoic acid-deficient Lactobacillus acidophilus, Gut microbes, 4 (2013) 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM, The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract, Applied and environmental microbiology, 74 (2008) 1646–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S, Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly, Applied and environmental microbiology, 73 (2007) 7767–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, Baxter NT, Zhao L, Schloss PD, Martens EC, Eaton KA, Chen GY, NLRP6 Protects Il10(−/−) Mice from Colitis by Limiting Colonization of Akkermansia muciniphila, Cell Rep, 19 (2017) 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Long MD, Sands BE, When Do You Start and When Do You Stop Screening for Colon Cancer in Inflammatory Bowel Disease?, Clin Gastroenterol Hepatol, (2018). [DOI] [PubMed]
- [61].Byun DS, Ahmed N, Nasser S, Shin J, Al-Obaidi S, Goel S, Corner GA, Wilson AJ, Flanagan DJ, Williams DS, Augenlicht LH, Vincan E, Mariadason JM, Intestinal epithelial-specific PTEN inactivation results in tumor formation, Am J Physiol Gastrointest Liver Physiol, 301 (2011) G856–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ucker DS, Levine JS, Exploitation of Apoptotic Regulation in Cancer, Frontiers in immunology, 9 (2018) 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dicuonzo G, Angeletti S, Garcia-Foncillas J, Brugarolas A, Okrouzhnov Y, Santini D, Tonini G, Lorino G, De Cesaris M, Baldi A, Colorectal carcinomas and PTEN/MMAC1 gene mutations, Clinical cancer research : an official journal of the American Association for Cancer Research, 7 (2001) 4049–4053. [PubMed] [Google Scholar]
- [64].Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM, Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN, Science, 280 (1998) 1614–1617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


