Abstract
Purpose
This was a study prospectively evaluating intratumoral T-cell responses to autologous somatic mutated neoepitopes expressed by human metastatic ovarian cancers.
Experimental Design
Tumor infiltrating lymphocytes (TILs) were expanded from resected ovarian cancer metastases, which were analyzed by whole exome and transcriptome sequencing to identify autologous somatic mutations. All mutated neoepitopes, independent of prediction algorithms, were expressed in autologous antigen presenting cells then co-cultured with TIL fragment cultures. Secretion of interferon-gamma or up-regulation of 41BB indicated a T-cell response.
Results
Seven women with metastatic ovarian cancer were evaluated and 5 patients had clear, dominant T-cell responses to mutated neoantigens, which were corroborated by comparison to the wild type sequence, identification of the minimal epitope, human leukocyte antigen (HLA) restriction element(s) and neoantigen-specific T-cell receptor(s). Mutated neoantigens were restricted by HLA-B, -C, -DP, -DQ and/or -DR alleles and appeared to principally arise from random, somatic mutations unique to each patient. We established that TP53 “hotspot” mutations (c.659A>G; p.Y220C and c.733G>A; p.G245S) expressed by two different patient’s tumors were immunogenic both in the context of HLA-DRB3*02:02.
Conclusions
Mutation-reactive T cells infiltrated ovarian cancer metastases at sufficient frequencies to warrant their investigation as adoptive cell therapy. Additionally, transfer of TP53 “hotspot” mutation-reactive T-cell receptors into peripheral blood T cells could be evaluated as a gene therapy for a diverse range of tumor histologies.
Introduction
Ovarian cancer is expected to account for 22,440 new cases and 14,080 deaths in the United States in 2017 (1). No curative treatments exist for patients with metastatic ovarian cancer. An increased presence of tumor infiltrating lymphocytes (TIL), composed primarily of T cells, in ovarian tumors was associated with better prognosis in patients receiving standard of care treatment suggesting T cells were actively interacting with ovarian cancer cells (2–5). Preliminary evidence suggested that adoptive cell transfer (ACT) of bulk TIL to patients with metastatic ovarian cancer was safe and well tolerated (6,7). Despite these early advances, TIL therapy for the treatment of ovarian cancer has otherwise gone untested and the specificity of ovarian cancer TIL is largely unknown.
ACT using TIL expanded from metastatic tumor lesions has led to durable, complete tumor regressions in 24% of patients with metastatic melanoma in several studies at different institutions (8–12). The infused TIL of some of the melanoma patients with complete responses recognized unique, somatic, non-synonymous mutated neoantigens, suggesting mutation-reactive T cells were critical in mediating durable tumor regressions (12–14). More recently, long-term partial regressions of metastatic colon cancer and cholangiocarcinoma were achieved following transfer of TIL recognizing autologous mutated KRASG12D and ERBB2IPE504G neoantigens, respectively (15–17). Thus, the “final common pathway” for the immunotherapy of cancer appears to be based on the targeting of somatic mutations expressed by the tumor and recognized by T cells.
Ovarian cancer falls in the middle of the continuum of numbers of expected somatic mutations stratified by cancer histology, which ranges from dozens in brain cancers to many hundreds of mutations in melanomas and lung cancers (18–20). Mutations in the TP53 tumor suppressor gene were estimated to occur in >95% of high-grade serous ovarian cancer but were less frequently observed (≤10%) in other ovarian cancer subtypes (21,22)(Catalogue of Somatic Mutations in Cancer (COSMIC) database; http://cancer.sanger.ac.uk/cosmic). Mutations occur across the TP53 gene and a single TP53 mutation is not ubiquitous to all ovarian cancer subtypes, but there are 6 codons (R175, Y220, G245, R248 and R273) which are most frequently mutated are thus called TP53 “hotspot” mutations (23–25). These TP53 “hotspot” mutations may be oncogenic because they have wild type loss-of-function and can have gain-of-function activity, although these phenomena have not been directly observed in ovarian cancer (26–29). However, there are no reports to date of intratumoral T cell responses to these TP53 “hotspot” mutations.
Intratumoral T-cell responses to human ovarian cancer mutated neoantigens have not been comprehensively evaluated. In a pilot study, one mutation T-cell response was detected amongst tumor-associated lymphocytes from ascites of 3 women with ovarian cancer (30), and a murine model of ovarian cancer neoantigen vaccination generated some murine mutation T-cell responses but was deemed an ineffective strategy due to the low mutational load of ovarian cancer (31). Recently, low frequencies (<2%) of mutated neoantigen-reactive CD8+ T cells were detected from TIL in 4 of 14 ovarian patients after screening with predicted Class-I human leukocyte antigen (HLA) neoepitopes (32), and mutation-reactive T-cells were generated from peripheral blood lymphocytes (PBL) of ovarian cancer patients vaccinated with tumor cell lysate (33). The aim of this study was to evaluate whether mutation-reactive TIL were present in resected ovarian cancer metastases independent of neoepitope prediction. All mutations identified in the cancer were introduced into both intracellular and extracellular pathways of autologous antigen presenting cells (APCs), containing all of the patient’s HLA Class-I and Class-II alleles, followed by co-culture with TIL microcultures. Mutation-reactive T cells were characterized in terms of the specificity to the mutated versus wild type variants, the minimal determinants of mutated sequences, the HLA restriction elements of the neoantigens and the identification of TCRs recognizing neoantigen:HLA complexes. Thus, a comprehensive characterization of T cell interactions with ovarian cancer-specific mutated neoantigens was performed.
Materials and Methods
Neoantigen screening with TIL
All patients were enrolled after written, informed consent was granted on NIH protocol 10-C-0166 (NCT01174121) in accordance with ethical guidelines and with approval by the NIH institutional review board. Referring physicians provided initial diagnoses and pathology, and the eligibility requirements for enrollment on the clinical trial included progressive disease at the time of surgery at the National Cancer Institute (NCI) with at least two metastases (>1 cm), one of which was amenable for harvest for TIL growth. The diagnosis date and classification (Federation of Gynecology and Obstetrics 2013), time to initial progression, histological subtype (World Health Organization 2014 Classification), therapies prior to enrollment, date of resection, metastatic sites, resection site at the NCI and key mutations (ARID1A, CCNE1, NF1, PIK3CA, PTEN, KRAS, TP53) detected in the resected metastases can be found in Table S1. Growth of TIL, exome and transcriptome sequencing and bioinformatic methods can be found in the supplementary material. After mutation calling was complete, quality control was manually performed for each called mutation with the Interactive Genomics Viewer (IGV) software (Broad Institute; Cambridge, MA; http://software.broadinstitute.org/software/igv/). Called mutations were excluded from the analysis if similar reads were observed in the normal exome or if there was an obvious mapping issue, i.e., multiple random mutations along multiple reads. A list of screened neoepitopes with patient ID, gene name, amino acid change, TMG and peptide pool is given in Table S2. Each mutated neoepitope was synthesized as a peptide or gene and cloned into a pcRNA2-SL expression vector (GenScript; Piscataway, NJ). The TMG plasmids were linearized with NotI and in vitro transcribed mRNA was made with mMESSAGE mMACHINE T7 ULTRA Transcription Kit according to manufacturer’s instructions (Thermo Fisher; Waltham, MA). Peptides were reconstituted in dimethyl sulfoxide (DMSO) then peptide pools were generated congruent to the TMG neoepitopes. Autologous immature dendritic cells (DCs) were generated by adherence method with IL-4 and GM-CSF as previously described (15). After 4–6 days, immature DCs were harvested and either transfected with TMG mRNA (80 µg/mL) using ECM 830 square electroporator (BTX; Holliston, MA) or pulsed with peptides either individually or as pools (10 µg/mL unless otherwise stated). Mutation-loaded immature DCs (0.8–1×105) were co-cultured with individual TIL microcultures (2×104) overnight in an IFNγ ELISPOT plate (Mabtech; Cincinnati, OH). The following day, the ELISPOT plate was developed according to manufacturer’s instructions after co-cultured cells were transferred to a new 96 well plate and stained for OX40, 41BB, CD3, CD4, CD8 (BD Biosciences; San Jose, CA) and/or mouse TCRβ (mTCRβ; Thermo Fisher). Stained cells were analyzed on Canto I or II or sorted by fluorescence activated cell sorting (FACS) on Aria II (BD Biosciences). FlowJo software (v.10.3) was used to analyze flow cytometry data. Spots on ELISPOT plates were enumerated with the Immunospot machine (Cellular Technology Ltd.; Shaker Heights, OH).
T-cell receptor identification and expression in PBL
Highly oligoclonal neoantigen-reactive TIL cultures were sequenced with TCRAD and TCRB surveys by Adaptive Technologies (Seattle, WA) in order to pair TCRα with TCRβ by frequency. The TCRs from bulk TIL cultures were also paired using statistical modeling on the Pairseq platform (Adaptive Biotechnologies) (34). Single-cell reverse transcriptase PCR specific for TCRα and TCRβ was performed to pair TCRs after co-culture with neoantigen (peptide or TMG) and sorting of 41BB+ T cells into single wells of 96 well plates, as was done in other studies (35,36). All TCRs were chimeras of human variable regions and mouse constant regions to enable detection with mTCRβ antibody and limit mis-pairing with the endogenous TCR. TCRs for patients 1, 4 and 6 were transduced into PBL using standard protocols with transiently produced γ-retroviral supernatants (35). TCRs for patients 5 and 7 were transposed into PBL with the Sleeping Beauty transposon/transposase system similarly to previous studies (37).
HLA restriction mapping
The HLA restriction elements were determined by transfecting COS7 tumor cell line (Green monkey; Cercopithecus aethiops) with individual Class-I alleles or both Class-II α and β chains, pulsing the mutated peptides and co-culture with T cells. Secretion of IFNγ into supernatants was evaluated by ELISA. Expression of 41BB was assayed by flow cytometry following co-cultures.
Results
Patient demographics, mutation identification and neoepitope screening
A total of seven women with metastatic ovarian cancer were enrolled on the clinical trial evaluating autologous TIL for the possible treatment of metastatic disease (NCT01174121). Each patient received standard of care (surgery and chemotherapy) and had progressive disease prior to enrollment to the trial at the NCI Surgery Branch (Table S1). At least one tumor was resected from a metastatic deposit from each patient for generation of TIL and mutation sequencing. Following whole exome and transcriptome sequencing, a total of 1,714 putative mutated neoantigens were screened and a median of 228 mutations were identified per patient (range: 64 – 332) (Table 1). Each mutation was assembled into a minigene encoding a 25 amino acid sequence by flanking the mutation with 12 amino acids of the wild type sequence as was done in previous studies (12,15). Minigenes were concatenated in tandem (median 18, range 13–21) to generate tandem minigene (TMG) constructs, which were synthesized as DNA and in vitro transcribed to mRNA. Twenty-five amino acid long peptides (LP) were also synthesized corresponding to each mutated minigene and peptide pools (PP) were assembled to increase throughput of screening. Peptides were pulsed and TMG mRNA was electroporated into autologous APCs. Thus, each mutation was screened using intracellular (TMG) and extracellular (LP) antigen presentation pathways on any possible HLA Class-I and -II molecules of the APC.
Table 1. Somatic mutated neoantigens recognized by ovarian cancer TIL.
Patient (Pt) TIL from fresh tumor (FtTu) resections were screened against putative mutated neoantigens (mut) in 25 amino acid peptides or tandem minigenes (TMGs). The gene name, genetic mutation, protein name and amino acid substitution are shown for each mutated neoantigen recognized by TIL. A minimal epitope is shown with the mutated amino acid in bold underline followed by the matching HLA restriction element. The predicted peptide:HLA complex rank and affinity (nM) were determined by NetMHCpan 3.0 for Class-I (http://www.cbs.dtu.dk/services/NetMHCpan/) and NetMHCIIpan 3.1 for Class-II (http://www.cbs.dtu.dk/services/NetMHCIIpan/).
| Pt | Fr Tu# |
# mut |
Mutated gene (location) |
Protein (AA sub.) |
Neoepitope (mutation) |
HLA | peptide:HLA Predicted |
|
|---|---|---|---|---|---|---|---|---|
| Rank | Affinity [nM] |
|||||||
| 1 | 4046 | 280 | USP9X (X:41075846 c.6026A>G) | USP9X (Y2009C) | RMQYSMECF | B*15:01:01 | 0.2 | 31.1 |
| YSMECFQFM | C*03:03:01 | 0.04 | 7.6 | |||||
| RMQYSMECFQFMKKL | DPA1*01:03:01 DPB1*04:02:01 | 0.9 | 16.4 | |||||
| 2 | 4067 | 122 | - | - | - | - | - | - |
| 3 | 4068 | 284 | - | - | - | - | - | - |
| 4 | 4097 | 317 | HIST1H1B (6:27835096 c.212C>A) | Histone H1.5 (A71D) | LADGGYDVEKNNSRI | DQA1*03:01:01 DQB1*04:02:01 | 65 | 8884.3 |
| SLAALKKALADGGYD | DRB4*01:01:01 | 19 | 387.5 | |||||
| INPP5K (17:1401379 c.526C>G) | INPP5K (L272V) | RIVWRLKRQPCAGPD | Class-II | - | - | |||
| 5 | 4098 4173 | 333 138 | RPTOR (17:78858926 c.1961A>G) | RAPTOR (D654G) | HNVAMMLAQLVSGGS | DPA1*01:03:01 DPB1*10:01:01 | 20 | 115.6 |
| DPA1*02:01:01 DPB1*10:01:01 | 17 | 113.1 | ||||||
| 6 | 4127 | 176 | TP53 (17:7577548 c.733G>A) | p53 (G245S) | HYNYMCNSSCMGSMN | DRB3*02:02:01 | 21 | 533.4 |
| 7 | 4149 | 64 | CTAGE5 (14:39796122 c.1613A>T) | CTAGE5 (E576V) | NERGVSSCDRLTDPH | DQA1*01:03:01 DQB1*06:03:01 | 95 | 4640.6 |
| HUWE1 (X:53560337 c.13058T>C) | HUWE1 (F4353S) | SEKLRHMLL | B*40:02:01 | 0.8 | 199.5 | |||
| TP53 (17:7578190 c.659A>G) | p53 (Y220C) | NTFRHSVVVPCEPPE | DRB3*02:02:01 | 17 | 433.8 | |||
Co-culture of APCs with TIL was performed with the TMG or PP to identify the recognized mutations by assaying interferon-γ (IFNγ) secretion by ELISPOT and upregulation of 41BB expression by flow cytometry. Twenty-four tumor fragments from different parts of a resected tumor were cultured in high-dose interleukin-2 (IL-2) and the resulting expanded TILs were screened against autologous APCs expressing all autologous mutations. Each TIL fragment was kept as an individual mini-culture in order to evaluate multiple, diverse, oligoclonal populations. A total of 8 neoantigens were identified from 5 of the 7 patients (Table 1). Neoantigens were not identified for patients 2 and 3. The minimal determinants of the recognized mutations (Table 1, column 6) demonstrated that the amino acid substitution could occur within the neoepitope near the N-terminus (Histone H1.5A71D, INPP5KL272V, CTAGE5E576V and HUWE1F4353S), the middle (USP9XY2009C) or the C-terminus (Histone H1.5A71D, RAPTORD654G, p53G245S, p53Y220C and USP9XY2009C). Both USP9XY2009C and Histone H1.5A71D developed more than one neoantigen minimal neoepitope. All of the mutations recognized by CD8+ T cells were predicted by an HLA binding algorithm (NetMHCpan 3.0; http://www.cbs.dtu.dk/services/NetMHCpan/) to have a rank <1 and affinity <200 nm. Five of seven HLA Class-II epitopes recognized by CD4+ T cells were predicted to have an affinity <500 nM, and all 7 had a rank >17, except for USP9XY2009C which had a rank of 0.9 (Table 1, far right column). The overall mutation burden did not predict for T-cell responsiveness as patient 3 had the third most mutations (n=284) but no corresponding mutation reactivity whereas patient 7 had three independent immunogenic neoantigens and the fewest mutated neoepitopes (n=64).
Multivalent T-cell responses to USP9XY2009C neoantigen
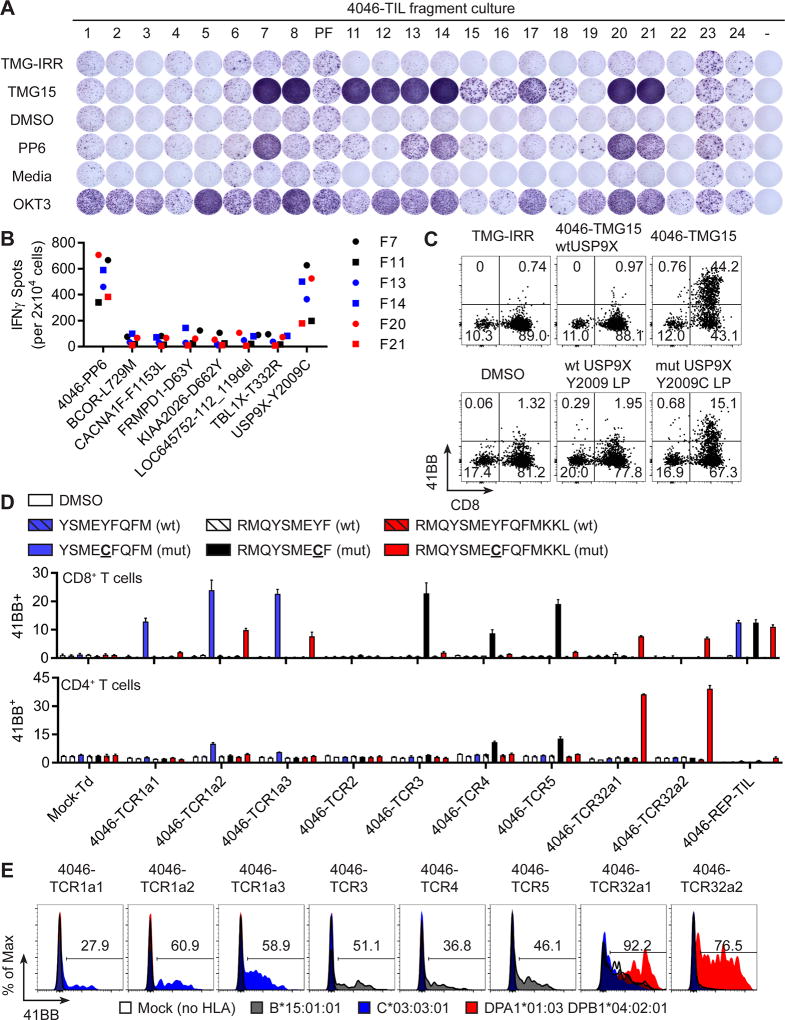
The TIL from Patient 1 demonstrated high frequency and multivalent T-cell response to USP9XY2009C, a putative driver mutation (38). She presented to the NCI Surgery Branch at 38 years-old and a metastasis was resected from an axillary lymph node for TIL growth and sequencing. The 280 identified mutations were concatenated into fifteen TMGs (4046-TMG) and the top 79 mutations were assembled into 6 peptide pools (4046-PP) after being prioritized for having high variant allele frequency in the exome and RNAseq. Fragments 7, 8, 11, 12, 13, 14, 15, 16, 17, 18, 20 and 21 displayed IFNγ secretion in response to 4046-TMG15, and fragments 7, 11, 13, 14, 20 and 21 secreted IFNγ when co-cultured with 4046-PP6 (Figure 1A). Co-culture of selected TIL fragments with individual LP congruent between 4046-TMG15 and 4046-PP6 showed USP9XY2009C as the immunogenic neoantigen (Figure 1B). In order to increase the number of T cells for further experiments, fragments 7, 11, 13, 14, 20 and 21 were grown using a rapid expansion protocol (REP) consisting of irradiated PBL feeder cells, OKT3 (agonistic pan-CD3 antibody) and IL-2. The six expanded fragments were pooled to generate the 4046-REP-TIL culture, which was specific for 4046-TMG15 and USP9XY2009C LP as evidenced by 41BB upregulation in co-cultures compared to minimal upregulation caused by irrelevant TMG (TMG-IRR) or 4046-TMG15 reverted back to wild type only at USP9X (4046-TMG15wtUSP9X), peptide vehicle (DMSO) and wild type USP9XY2009 LP (Figure 1C). Both CD4+ and CD8+ T cells in the 4046-REP-TIL expressed 41BB in response to USP9XY2009C neoantigen and suggested that a multivalent attack on USP9XY2009C occurred in Patient 1.
Figure 1. Multivalent T-cell responses to USP9XY2009C neoantigen.
(A) TIL fragments from FrTu#4046 were co-cultured with autologous APCs (i) electroporated with irrelevant TMG (TMG-IRR) or 4046-TMG15 or (ii) pulsed with DMSO (peptide vehicle) or 4046-PP6. Media (T cells only) and OKT3 were negative and positive control, respectively. Secretion of IFNγ was evaluated by ELISPOT. (B) Selected TIL fragments were co-cultured with autologous APCs pulsed with peptides and IFNγ secretion was evaluated by ELISPOT. (C) Expression of 41BB in T cells following co-culture of 4046-REP-TIL with autologous APCs electroporated with TMGs or pulsed with peptides. (D) Co-culture of TIL or TCR transduced T cells with autologous APCs pulsed with minimal neoepitope peptides and measurement of 41BB expression in gated CD8+ (top graph) or CD4+ T cells (mean ± SEM; n=3). (E) Transfection of HLA genes into COS7 cell line, pulsing of identified minimal mutated neoantigen peptides and co-culture with TCR transduced T cells. Expression of 41BB is displayed for CD8+mTCR+ (4046-TCR1a1/2/3, 4046-TCR3, 4046-TCR4 and 4046-TCR5) and CD4+mTCR+ (4046-TCR32a1/2) gates.
Putative USP9XY2009C reactive TCRs were identified by co-culture with USP9XY2009C neoantigen, isolation of 41BB+ T cells by FACS and performing single-cell RT-PCR with primers spanning the TCRα and TCRβ chains similar to previous studies (35). T-cell receptors were constructed with murine constant chains thus enabling the detection of the introduced TCR with anti-mTCRβ antibody. The most abundant clonotypes from the 4046-REP-TIL (TCRs named based on their rank) were evaluated following γ-retroviral transduction of autologous PBL. Minimal epitopes were used to evaluate TCR specificity. Peptides RMQYSMECF and YSMECFQFM (mutation in bold underline) embedded in the 25 amino acid neoantigen were predicted to bind HLA-B*15:01:01 and HLA-C*03:03:01 alleles, respectively, from patient 1 (NetMHCpan 3.0). The minimal epitope for CD4+ T cell responses to USP9XY2009C was empirically determined to be RMQYSMECFQFMKKL by co-culturing HLA Class-II restricted USP9XY2009C TIL with 15 amino acid peptides overlapping 14 amino acids (Figure S1). The top ranking TCR in the 4046-REP-TIL (23.6% by TCRB sequencing) was reactive to YSMECFQFM peptide when paired with three different TCRA chains (4046-TCR1a1, 4046-TCR1a2 and 4046-TCR1a3) (Figure 1D, blue bars). The second ranked TCR (4046-TCR2; 10.3%) was not reactive to USP9XY2009C but the 3rd (4046-TCR3; 7.0%), 4th (4046-TCR4; 6.4%) and 5th (4046-TCR5; 4.8%) ranked TCRs were each reactive to RMQYSMECF peptide (Figure 1D, black bars). T cells transduced with 4046-TCR32a1 and 4046-TCR32a2 (rank 32 and 0.2% by TCRB of 4046-REP-TIL; unique TCRα chains) reacted to RMQYSMECFQFMKKL and the majority of the response was seen by CD4+ T cells suggesting that this was a HLA Class-II restricted response (Figure 1D, red bars, bottom graph). The 4046-REP-TIL culture was reactive to all three minimal peptides whereas mock transduced T cells (open repertoire from PBL) did not respond to any of the peptides. The specificity of the responses was supported by an absence of reactivity to wild type variants of the USP9XY2009 peptides (Figure 1D, hatched bars).
The HLA restriction elements for USP9XY2009C were determined by transfecting a non-human COS7 tumor cell line with individual HLA Class-I or Class-II alleles, followed by pulsing the minimal peptides and co-culture with USP9XY2009C specific T cells. The HLA Class-I responses to RMQYSMECF and YSMECFQFM were restricted by HLA-B*15:01:01 (Figure 1E gray histograms) and HLA-C*03:03:01 (Figure 1E blue histograms), respectively, and HLA Class-II response to RMQYSMECFQFMKKL was restricted by DPA1*01:03:01 with DPB1*04:02:01 (Figure 1E red histograms). In sum, a single mutation could generate three epitopes capable of binding one HLA each and forming functional interactions with one or more TCRs.
Distinct neoepitopes within one HIST1H1BA71D mutation were recognized by CD4 T cells
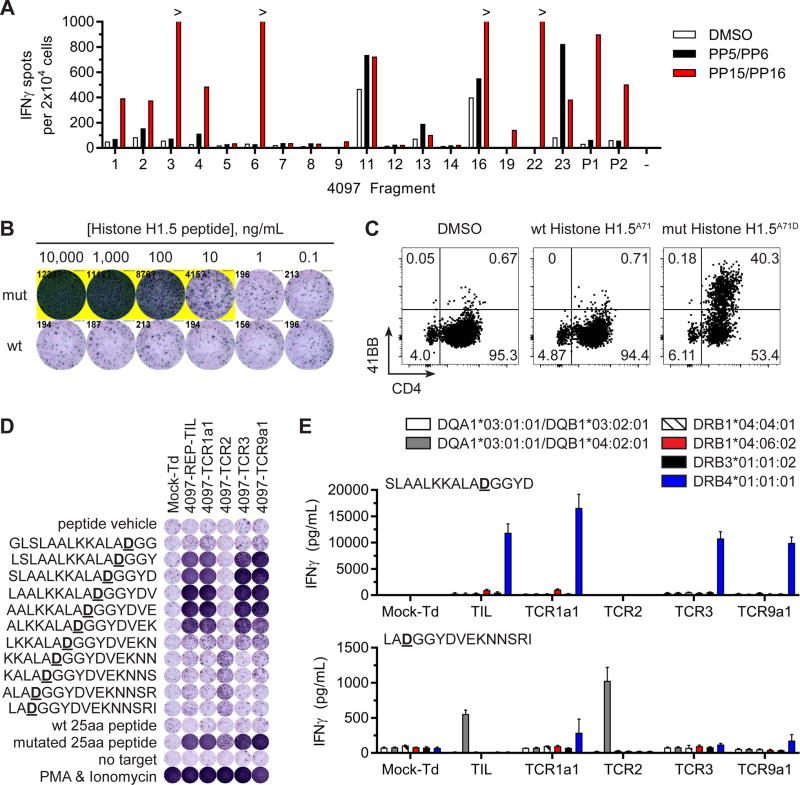
Multiple epitopes were also identified and characterized from a single Histone H1.5A71D mutation. Patient four, 59 years old, had an iliac lymph node resected for TIL growth and mutation sequencing. All 317 mutations detected from her cancer were assembled into 17 TMGs (4097-TMG) and 18 PPs (4097-PP). Because the quantity of mutations was large, two 4097-PP were pulsed separately then pooled into one co-culture with each TIL fragment micro-culture. Fragments 1, 2, 3, 4, 6, 16, 19, 22, 23 and pooled fragments P1 and P2 showed specific responses to 4097-PP15/PP16, which contained mutated Histone H1.5A71D LP (Figure 2A red bars). The screening also revealed a CD4+ T cell response in fragment 23 to 4097-PP5/PP6 (Figure 2A closed bars). Testing of the individual LP in 4097-PP6 revealed that mutated INPP5KL272V was the neoantigen, but we were unable to identify the HLA restriction element (Figure S2). To interrogate the Histone H1.5A71D response, fragments 3, 6 and 22 were expanded in REP and pooled into one 4097-REP-TIL culture, which displayed specific reactivity to mutated Histone H1.5A71D LP compared to wild type LP by IFNγ secretion (Figure 2B). Expression of 41BB by T cells co-cultured with Histone H1.5A71D LP established that this response was mediated by CD4+ T cells (Figure 2C).
Figure 2. Distinct neoepitopes within one HIST1H1BA71D mutation were recognized by CD4+ T cells.
(A) Screening of TIL fragments from FrTu#4097 against autologous APCs pulsed with DMSO, a pool of 4097-PP5 and 4097-PP6 (PP5/PP6) or a pool of 4097-PP15 and 4097-PP16 (PP15/PP16). Secretion of IFNγ was evaluated by ELISPOT. (B) 4097-REP-TIL was co-cultured with decreasing concentrations of wild type (wt) or mutated (mut) Histone H1.5A71D LP. Secretion of IFNγ was evaluated by ELISPOT. (C) Expression of 41BB in CD4+ T cells in co-cultures of 4097-REP-TIL with autologous APCs pulsed with DMSO, wt Histone H1.5A71 or mutated Histone H1.5A71D LPs. (D) Co-culture of TIL or TCR transduced T cells with autologous APCs pulsed with Histone H1.5A71D LPs (wt or mut) and 15 amino acid mutated Histone H1.5A71D peptides overlapping 14 amino acids. No target (media) and PMA & Ionomycin were negative and positive controls, respectively. Secretion of IFNγ was evaluated by ELISPOT. (E) Transfection of HLA genes into COS7 cell line, pulsing of SLAALKKALADGGYD or LADGGYDVEKNNSRI peptides and co-culture with 4097-REP-TIL or TCR transduced T cells. Secretion of IFNγ into co-culture supernatants was evaluated by ELISA (mean ± SEM; n=3).
Four TCRs in the 4097-REP-TIL culture targeting Histone H1.5A71D neoantigen were identified. A combination of TCR deep sequencing, Adaptive Biotechnologies’ Pairseq (computational TCR pairing service) and single-cell RT-PCR of TCR genes was used to reconstruct TCRs ranking 1, 2, 3, 6, 8, 9, 15, 23 and 79 in the 4097-REP-TIL. Following transduction into peripheral blood T cells it was determined that 4097-TCR1a1 (TCRβ clonotype rank and frequency in 4097-REP-TIL: #1; 36.2%), 4097-TCR2 (#2; 26.2%), 4097-TCR3 (#3, 13.2%) and 4097-TCR9a1 (#9; 0.2%) were specific for Histone H1.5A71D neoantigen (Figure S3).
The specific neoepitopes that were targeted by Histone H1.5A71D reactive TCRs were empirically characterized. Co-culture of 4097-REP-TIL and TCR transduced T cells with 15 amino acid peptides overlapping 14 amino acids demonstrated that there were two neoepitopes recognized (Figure 2D). More specifically, 4097-TCR1a1, 4097-TCR3 and 4097-TCR9a1 displayed strong responses to 5 peptides spanning LSLAALKKALADGGYDVEK whereas 4097-TCR2 responded to 4 peptides covering KKALADGGYDVEKNNSRI. All tested T cells, except for the open repertoire mock transduced culture, showed specific response to the mutated 25 amino acid Histone H1.5A71D peptide and no recognition of wild type peptide.
Given that there were 2 distinct Histone H1.5A71D neoepitopes, there was likely more than one HLA restriction element. Experiments transfecting HLA genes into COS7 cells showed that the responses by 4097-TCR1a1, 4097-TCR3 and 4097-TCR9a1 to SLAALKKALADGGYD were restricted by HLA-DRB4*01:01:01 (Figure 2E blue bars). In contrast, the 4097-TCR2 response to LADGGYDVEKNNSRI was restricted by DQA1*03:01:01 and DQB1*04:02:01 (Figure 2E gray bars). Therefore, a single Histone H1.5A71D neoantigen resulted in two distinct neoepitopes restricted by either HLA-DQ or HLA-DR.
Recognition of RAPTORD654G neoantigen by TIL from a metastasis
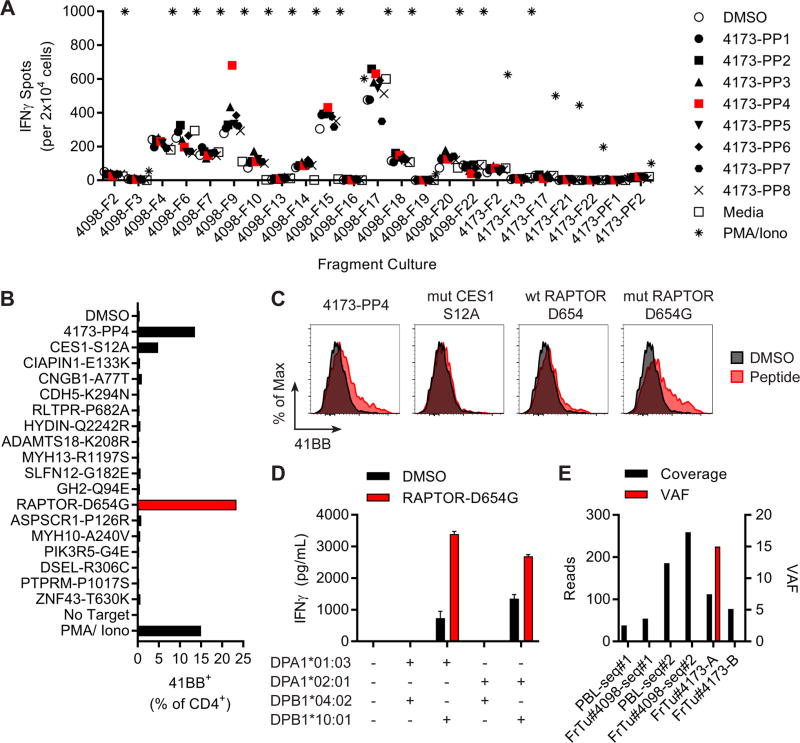
In Patient 5 we had the opportunity to evaluate the reactivity from TIL fragments from 3 spatially and temporally distinct tumors towards a common set of neoepitopes. An axillary lymph node was resected for TIL harvest (FrTu#4098), and an initial screen of TIL against 333 putative neoepitopes called from FrTu#4098 and assembled into 18 TMGs and 19 PPs resulted in no clear and/or dominant reactivities. Five months later, Patient 5 underwent another surgery to resect progressive abdominal tumor lesions (FrTu#4173-A and FrTu#4173-B). Given that there were TIL grown from fragments from 3 lesions, the TIL from fragments remaining after the initial screen (FrTu#4098) were re-screened in parallel with 7 fragments from FrTu#4173-A/B against 138 mutations present in FrTu#4173-A and/or FrTu#4173-B, which were assembled in eight TMGs (4173-TMG) and eight PPs (4173-PP).
Fragment 9 from FrTu#4098 (4098-F9) displayed IFNγ secretion in response to 4173-PP4 (Figure 3A, red squares). Parsing of the peptides present in 4173-PP4 against 4098-F9 revealed that RAPTORD654G and CES1S12A were likely the immunogenic neoantigens as measured by 41BB expression on CD4+ T cells (Figure 3B). Co-culture of 4098-F9 with highly purified peptides (>95% by HPLC) demonstrated that mutated RAPTORD654G, but not CES1S12A or wild type RAPTORD654, was responsible for the 41BB expression on CD4+ T cells (Figure 3C). Pairing of the most frequent TCRβ chain with the one of two most frequent TCRα from 4098-F9 led to the identification of 4098-F9-RAPTOR-TCRa2 (second TCRα), which was responsive to the RAPTORD654G mutation after expression in PBL with the Sleeping Beauty transposon/transposase system. T cells expressing 4098-F9-RAPTOR-TCRa2 and 4098-F9 TIL were co-cultured with overlapping 15 amino acid RAPTORD654G peptides and showed that the immunogenic neoepitope was present in the N-terminal 15 amino acids (Figure S4). COS7 cells were co-electroporated with autologous HLA Class-II alleles, pulsed with the 15 amino acid RAPTORD654G peptide (HNVAMMLAQLVSGGS) and co-cultured with 4098-F9-RAPTOR-TCRa2, which demonstrated that RAPTORD654G neoantigen was restricted by HLA-DPB1*10:01:01 in combination with either HLA-DPA1*01:03:01 or HLA-DPA1*02:01:01 (Figure 3D). Therefore, TIL from one physically and temporally distinct tumor could recognize a mutation identified in another autologous tumor.
Figure 3. Recognition of RAPTORD654G neoantigen by TIL from a metastasis.
(A) Screening of TIL fragments from FrTu#4098 and FrTu#4173 against autologous APCs pulsed with DMSO or 4173 peptide pools (PP). No target (media) and PMA & Ionomycin were negative and positive controls, respectively. Secretion of IFNγ was evaluated by ELISPOT. (B) Expression of 41BB in CD4+ T cells after co-culture of 4098-F9 with autologous APCs pulsed with individual peptides from 4173-PP4. (C) Expression of 41BB in CD4+ T cells in co-cultures of 4098-F9 with autologous APCs pulsed with DMSO, 4149-PP4, mutated CES1S12A, wild type RAPTORD654 or mutated RAPTORD654G LPs. (D) Transfection of HLA genes into COS7 cell line, pulsing of DMSO or HNVAMMLAQLVSGGS RAPTORD654G peptide and co-culture with 4098-F9-RAPTOR-TCRa2 transduced T cells. Secretion of IFNγ into co-culture supernatants was evaluated by ELISA (mean ± SEM; n=3). (E) Exome sequencing coverage (reads) and variant allele frequency (VAF) for RAPTORD654G at chromosome 17:78,858,926 (c.1961A>G).
The CD4+ T-cell response to RAPTORD654G was not observed by 4098-F9 in the original screen, because RAPTORD654G was not detected in sequencing the original FrTu#4098. Thus, FrTu#4098 was sequenced again at greater depth (FrTu#4098 seq#2), but the RAPTORD654G mutation was again only seen in FrTu#4173-A (Figure 3E). Furthermore, RAPTORD654G was also absent in another metastasis sequenced from the latter resection (FrTu#4173-B). The variant allele frequency was 15 in FrTu#4173-A, which suggested that only a fraction of the tumor cells contained the RAPTORD654G neoantigen (Figure 3E, red bars). Other studies have also shown that different ovarian cancer tumor deposits harbor distinct mutational and immunological profiles (25). This reactivity showed evidence of T-cell responsiveness to only a fraction of the patient’s tumor population, and suggested that multiple reactivities such as this one would be likely needed to effectively treat her disease.
Identification and characterization of TP53 “hotspot” mutation-reactive TCRs
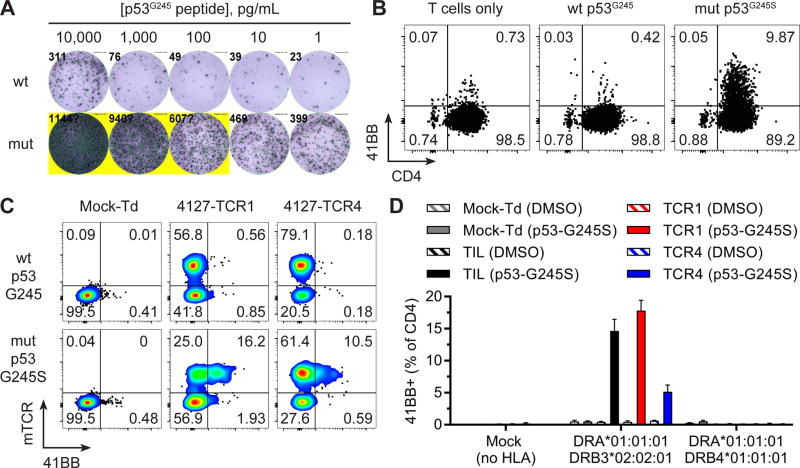
Patient 6 had a metastasis resected from the axilla from which a TP53 “hotspot” mutation (c.733G>A; p.G245S) was detected. Screening of TIL from fragments (n=24) against 176 putative neoantigens assembled in 10 TMGs and 11 PPs revealed 3% 41BB+ T cells in response to p53G245S by TIL from fragment 11, which was enriched for reactive T cells by p53G245S peptide co-culture, 41BB+ T cell sorting and REP. The resultant T cell culture displayed highly avid and specific recognition of the mutated p53G245S compared to wild type counterpart as measured by IFNγ secretion (Figure 4A). This response was mediated by CD4+ T cells as measured by 41BB expression in co-cultures (Figure 4B).
Figure 4. Identification and characterization of p53G245S neoantigen-reactive TCRs.
(A) Co-culture of 4127-F11 p53G245S-enriched TIL and autologous APCs pulsed with decreasing concentrations of p53G245S mutated (mut) or wild type (wt) peptides. Secretion of IFNγ was evaluated by ELISPOT. (B) Expression of 41BB in CD4+ T cells after co-culture with autologous APCs pulsed with p53G245S LPs. (C) Expression of 41BB in T cells expressing transduced TCRs (mTCR+) following co-culture with autologous APCs pulsed with p53G245S LPs. (D) Transfection of HLA genes into COS7 cell line, pulsing of DMSO or HYNYMCNSSCMGSMN p53G245S peptide and co-culture with 4127-F11 p53G245S-enriched TIL or mock, 4127-TCR1 or 4127-TCR4 transduced T cells. Expression of 41BB in gated CD4+ T cells is displayed (mean ± SEM; n=3).
Because TP53 (c.733G>A; p.G245S) was a “hotspot” expressed by tumors from unrelated individuals (~2.8% of cancers) (39), it was prudent to identify the TCRs for future research and clinical applications. Putative p53G245S reactive TCRs were identified from 4127-F11 by single-cell RT-PCR from 41BB+ T cells following co-culture with p53G245S peptide. T cells transduced with 4127-TCR1 and 4127-TCR4 (as measured by mTCRβ expression) demonstrated 41BB upregulation in response to p53G245S peptide (HYNYMCNSSCMGSMN) in the N-terminal 15 amino acids of the mutated p53G245S long peptide (Figure S5). Open repertoire activated T cells (mock transduced) did not express 41BB in response to p53G245S peptide supporting the specificity of the transduced TCR response, whereas 4127-TCR1 and 4127-TCR4 transduced T cells (mTCRβ+) expressed 41BB only in response to mutated p53G245S peptide (Figure 4C). The p53G245S neoepitope was restricted by HLA-DRB3*02:02:01 (Figure 4D). HLA-DRB3*02:02:01 was present in 1,367 of 3,719 patients (37%) seen at the NCI. Thus, TIL and TCR responses specific for a frequent TP53 “hotspot” mutation (c.733G>A; p.G245S) in the context of a common HLA-DR allele were observed and characterized from Patient 6.
Both unique and shared neoantigens were detected in a single patient
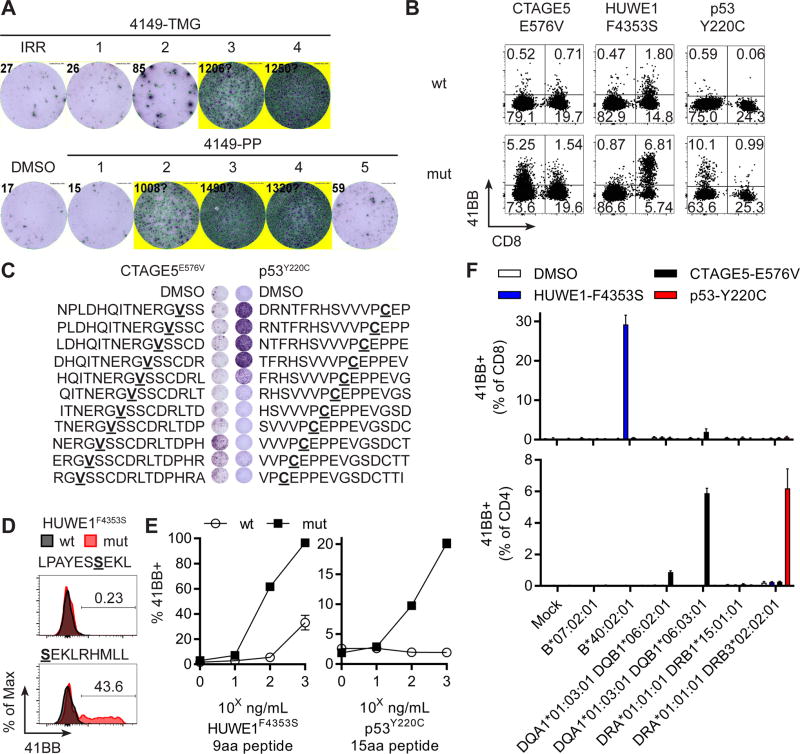
Three different immunogenic neoantigens were identified for patient 7, including a TP53 “hotspot” mutation (c.659A>G; p.Y220C; 2.4% of ovarian cancers and 1.5% of all cancers) (40,41), out of only 64 total neoepitopes identified from resected iliac lymph node metastases. An initial TIL screen with four TMGs and five PPs revealed a strong response to p53Y220C neoantigen in fragments 7, 8 and 11, and these three fragments were expanded in REP for further testing. The 4149-REP-TIL culture demonstrated a polyclonal response as evidenced by IFNγ secretion when co-cultured with three of four TMGs (4149-TMG2, 4149-TMG3 and 4149-TMG4) and three of five peptide pools (4149-PP2, 4149-PP3, and 4149-PP4) (Figure 5A). The TMGs and PPs were congruent except for 4149-PP5, which was composed of overlapping peptides from a single frameshift mutation present in TMG3. Parsing the peptides from each of the reactive TMGs or peptide pools revealed that CTAGE5E576V, p53Y220C and HUWE1F4353S were the neoantigens in 4149-TMG2/-PP2, 4149-TMG3/-PP3 and 4149-TMG4/-PP4, respectively (Figure S6). CD4+ T cells demonstrated specific recognition of CTAGE5E576V and p53Y220C whereas CD8+ T cells responded to HUWE1F4353S as evidenced by 41BB expression in mutated compared to wild type co-cultures (Figure 5B). Thus, two CD4 and one CD8 T cell populations were present in 4149-REP-TIL recognizing 3 unique mutated neoantigens.
Figure 5. Both unique and shared neoantigens were detected in a single patient.
(A) 4149-REP-TIL was screened against autologous APC (top) electroporated with TMGs or (bottom) pulsed with DMSO or peptide pools (PP). Secretion of IFNγ was evaluated by ELISPOT. (B) Expression of 41BB in T cells from 4149-REP-TIL after co-culture with mutated (mut) or wild type (wt) CTAGE5E576V, HUWE1F4353S and p53Y220C LPs. (C) Co-culture of 4149-REP-TIL with autologous APCs pulsed with 15 amino acid mutated peptides overlapping 14 amino acids from (left) CTAGE5E576V or (right) p53Y220C neoantigens. Secretion of IFNγ was evaluated by ELISPOT. (D) Co-culture of 4149-REP-TIL with autologous APCs pulsed with 9 or 10 amino acid wild type (wt) or mutated (mut) HUWE1F4353S peptides. Expression of 41BB was displayed for CD8+ T cells. (E) T cells expressing introduced TCRs (left: 4149-HUWE1-TCR1 or right: 4149-TP53-TCRa2b2) were co-cultured with autologous APCs pulsed with decreasing concentrations of minimal wild type (wt) or mutated (mut) minimal neoepitope peptides. Expression of 41BB in CD8+mTCR+ (4149-HUWE1-TCR1) or CD4+mTCR+ (4149-TP53-TCRa2b2) is displayed (mean ± SEM; n=3). (F) Transfection of HLA genes into COS7 cell line, pulsing of DMSO, NERGVSSCDRLTDPH (CTAGE5E576V), SEKLRHMLL (HUWE1F4353S) or NTFRHSVVVPCEPPE (p53Y220C) peptides and co-culture with 4149-REP-TIL. Expression of 41BB in gated (top) CD8+ or (bottom) CD4+ T cells is displayed (mean ± SEM; n=3).
Minimal epitopes were then identified for the 3 neoantigens from Patient 7. Mutations within the minimal 15 amino acid neoepitopes for CTAGE5E576V and p53Y220C were closer to the C- and N- termini, respectively (Figure 5C). Two HUWE1F4353S peptides were predicted to bind either HLA-B*07:02:01 (LPAYESSEKL) or HLA-B*40:02:01 (SEKLRHMLL), and it was the latter peptide which was specifically recognized over wild type variants by CD8+ T cells (Figure 5D). The TCRs targeting HUWE1F4353S and p53Y220C neoantigens were identified and expressed in peripheral blood T cells using the Sleeping Beauty transposon/transposase system. HUWE1F4353S and p53Y220C minimal peptides were specifically recognized by 4149-HUWE1-TCR1 and 4149-TP53-TCRa2b2, respectively, as measured by 41BB co-expression on TCR transposed (mTCRβ+) T cells with minimal response to wild type peptides (Figure 5E). Transfection of individual HLA Class-I genes or co-transfection of α and β HLA Class-II genes into COS7 cells followed by mutated peptide pulsing and co-culture with the 4149-REP-TIL demonstrated that HUWE1F4353S was restricted by HLA-B*40:02:01, CTAGE5E576V was restricted by HLA-DQA1*01:03:01 and HLA-DQB1*06:03:01 and p53Y220C was restricted by HLA-DRB3*02:02:01 (Figure 5F). In a patient with only 64 mutations, both shared (p53Y220C) and unique (HUWE1F4353S and CTAGE5E576V) neoantigens were targets of T cells.
In sum, all recognized mutations were unique to each patient. The immunogenic TP53 “hotspot” mutations, G245S in Patient 6 and Y220C in Patient 7, were presented by the same restriction element (HLA-DRB3*02:02:01), indicating that there may be commonalties for T cells to target oncogenic mutations. With the exception of the TP53 “hotspot” mutations, the neoantigens recognized by T cells arose from seemingly random, somatic, non-synonymous mutations. Therefore, the universe of potential neoantigens expressed by ovarian cancer was complex and derived from mutations unique to each patient and as well as TP53 “hotspot” mutations present in tumors of unrelated people.
Discussion
Herein we identified and characterized T cell responses to the ovarian cancer neoantigenome. The study of T cell antigens expressed by autologous epithelial cancers has been hampered by the relative inability to establish autologous tumor cell lines from resected tumors. This problem was circumvented using TMG and LP screening of all autologous cancer mutations such that the tumor cell could be, in essence, recreated in an autologous APC and used for T cell co-culture (12–17). Application of this strategy to the study of metastatic ovarian cancers established that T cells expressing TCRs capable of binding mutated neoantigens had infiltrated ovarian tumors.
The identification of T-cell responses to TP53 “hotspot” mutations from patients 6 and 7 was promising because TP53 is the most commonly mutated gene in human cancers (42) and ~95% of high-grade serous ovarian cancers will have a mutation in the TP53 gene (20,21,23,24,39). Mutated p53 is an attractive target for tumor-specific therapies because it is absent on normal tissue. However, tumor infiltrating T cell responses to TP53 “hotspot” mutations have not been characterized. The frequencies of G245S and Y220C TP53 “hotspot” mutations in all cancers are 2.8% and 1.5%, respectively, (39,40) and HLA-DRB3*02:02 was typed in 37% of patients at the NCI, which is consistent with The Allele Frequency Net Database (http://www.allelefrequencies.net). Thus, it is estimated that ~1.5% of cancers in general would have both HLA-DRB3*02:02 and either a G245S and Y220C TP53 “hotspot” mutation. In vitro sensitization of an HLA-A*02:01 donor’s PBL generated a CD8+ T cell clone capable of recognizing p53Y220C mutant epitope, suggesting that circulating T cells may bind mutated p53 epitopes (43). A library of TCRs targeting mutated p53 and other commonly altered driver genes, e.g., KRAS, could be generated to treat a wide variety of patients with T cells genetically-modified with TCRs.
All of the immunogenic mutations discovered in this study, except the p53G245S and p53Y220C neoantigens, were unique and were not reported in the COSMIC database. The Histone H1.5A71D mutation, which was in a linker H1 histone that tethers nucleosomes composed of a H2A, H2B, H3 and H4 core histones, could have impacted the chromatin structure and epigenetics (44). Linker Histone H1.3 and HUWE1, a ubiquitin E3 ligase that targets proteins for proteolysis, worked together to promote the pathogenesis of ovarian cancer in mice (45). The USP9X protein is a deubiquitinase that reverses ubiquitin-targeted protein degradation and is a central component of the centrosome (46). Transposon-mediated mutagenesis murine models of pancreatic and hepatocellular cancers were linked to USP9X and HUWE1, respectively, suggesting that these may be driver genes (38,47). CTAGE5 transports proteins from the endoplasmic reticulum and the E576V mutation could also impact the HLA Class-I pathway because this is within the proline-rich domain which is used to bind other proteins (48). Mutations in proteins involved with intracellular signaling pathways were also immunogenic. For example, RAPTOR is a critical member of the mTOR complex and the D654G mutation resides in the HEAT-3 (Huntington-EF3-PP2A-TOR1) domain that serves as vital structural support (49). The catalytic domain of was altered in the INPP5KL272V mutation and may have impacted on inositol 5-phosphate signaling and could have contributed to pathogenesis in line with the effects of other INPP5K mutations (50). A more complete understanding of the relationship between the functional consequences of a mutation and the generation of an immunogenic neoepitope may yield insights useful for T cell immunotherapy and tumor biology.
Knowledge of the identity of mutated neoantigens as described in this paper could potentially be used to improve ovarian cancer T-cell immunotherapy. The effectiveness of immunotherapy using the direct infusion of mutation-reactive TIL is likely limited by the existing differentiation status of TIL, i.e., likely senescent and/or terminally differentiated with limited proliferative potential. High frequencies of mutation-reactive T cells with improved in vivo proliferative potential could thus be infused by transducing or transposing TCRs into autologous naïve or central memory T cells. Personalized TCR gene therapy targeting a mutation expressed by the autologous tumor could be developed by transducing PBL with a TCR from transiently-produced γ-retroviral supernatant or by electroporating TCR transposons in the non-viral Sleeping Beauty transposon/transposase system (37). The landscape of somatic mutations in ovarian cancer neoantigens is large and the elucidation of their functional roles, participation in oncogenesis and metastasis, neoepitope formation, T cell responsiveness and molecular structural determinants could enable deeper understanding and the development of effective treatments for ovarian cancer.
Supplementary Material
Statement of Translational Relevance.
This study demonstrated that T cells with specificity to mutated neoantigens were present in tumor infiltrating lymphocytes in patients with metastatic ovarian cancer, sometimes at high frequencies and with multivalent reactivity, suggesting that these T cells could be used for adoptive cell therapy. Furthermore, T cells from two of the patients were specific for TP53 “hotspot” mutations (c.659A>G; p.Y220C and c.733G>A; p.G245S), which are also expressed in a broad range of tumor types in unrelated individuals. Genetic transfer of the TP53 “hotspot” mutation-specific T-cell receptors into autologous lymphocytes could be used to generate cells for use in the adoptive cell transfer immunotherapy of cancer.
Acknowledgments
We thank the TIL lab for their efforts growing the tumor infiltrating lymphocytes. We thank the FACS core members Arnold Mixon and Shawn Farid for their assistance. Thanks to Eric Tran and Sanja Stevanovic for academic and technical discussions about this study. This work was supported by intramural funding of the Center for Cancer Research, NCI.
Footnotes
Conflict of interest statement: The authors declare no competing interests.
References
- 1.American Cancer Society Facts & Figures. 2017 doi: 10.6004/jadpro.2020.11.2.1. https://wwwcancerorg/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2017html. [DOI] [PMC free article] [PubMed]
- 2.Ovarian Tumor Tissue Analysis C. Goode EL, Block MS, Kalli KR, Vierkant RA, Chen W, et al. Dose-Response Association of CD8+ Tumor-Infiltrating Lymphocytes and Survival Time in High-Grade Serous Ovarian Cancer. JAMA Oncol. 2017:e173290. doi: 10.1001/jamaoncol.2017.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol. 2012;124(2):192–8. doi: 10.1016/j.ygyno.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–13. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 5.Lo CS, Sanii S, Kroeger DR, Milne K, Talhouk A, Chiu DS, et al. Neoadjuvant Chemotherapy of Ovarian Cancer Results in Three Patterns of Tumor-Infiltrating Lymphocyte Response with Distinct Implications for Immunotherapy. Clin Cancer Res. 2017;23(4):925–34. doi: 10.1158/1078-0432.CCR-16-1433. [DOI] [PubMed] [Google Scholar]
- 6.Aoki Y, Takakuwa K, Kodama S, Tanaka K, Takahashi M, Tokunaga A, et al. Use of adoptive transfer of tumor-infiltrating lymphocytes alone or in combination with cisplatin-containing chemotherapy in patients with epithelial ovarian cancer. Cancer Res. 1991;51(7):1934–9. [PubMed] [Google Scholar]
- 7.Ikarashi H, Fujita K, Takakuwa K, Kodama S, Tokunaga A, Takahashi T, et al. Immunomodulation in patients with epithelial ovarian cancer after adoptive transfer of tumor-infiltrating lymphocytes. Cancer Res. 1994;54(1):190–6. [PubMed] [Google Scholar]
- 8.Besser MJ, Shapira-Frommer R, Itzhaki O, Treves AJ, Zippel DB, Levy D, et al. Adoptive transfer of tumor-infiltrating lymphocytes in patients with metastatic melanoma: intent-to-treat analysis and efficacy after failure to prior immunotherapies. Clin Cancer Res. 2013;19(17):4792–800. doi: 10.1158/1078-0432.CCR-13-0380. [DOI] [PubMed] [Google Scholar]
- 9.Goff SL, Dudley ME, Citrin DE, Somerville RP, Wunderlich JR, Danforth DN, et al. Randomized, Prospective Evaluation Comparing Intensity of Lymphodepletion Before Adoptive Transfer of Tumor-Infiltrating Lymphocytes for Patients With Metastatic Melanoma. J Clin Oncol. 2016;34(20):2389–97. doi: 10.1200/JCO.2016.66.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radvanyi LG, Bernatchez C, Zhang M, Fox PS, Miller P, Chacon J, et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin Cancer Res. 2012;18(24):6758–70. doi: 10.1158/1078-0432.CCR-12-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17(13):4550–7. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–52. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu YC, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20(13):3401–10. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu YC, Yao X, Li YF, El-Gamil M, Dudley ME, Yang JC, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol. 2013;190(12):6034–42. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–90. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med. 2016;375(23):2255–62. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MD, Wendl MC, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun. 2014;5:3156. doi: 10.1038/ncomms4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221(1):49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole AJ, Dwight T, Gill AJ, Dickson KA, Zhu Y, Clarkson A, et al. Assessing mutant p53 in primary high-grade serous ovarian cancer using immunohistochemistry and massively parallel sequencing. Sci Rep. 2016;6:26191. doi: 10.1038/srep26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ab Mutalib NS, Syafruddin SE, Md Zain RR, Mohd Dali AZ, Mohd Yunos RI, Saidin S, et al. Molecular characterization of serous ovarian carcinoma using a multigene next generation sequencing cancer panel approach. BMC Res Notes. 2014;7:805. doi: 10.1186/1756-0500-7-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bashashati A, Ha G, Tone A, Ding J, Prentice LM, Roth A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231(1):21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Qian J, Hu Y, Wang J, Zhou X, Chen H, et al. Heterogeneity of Li-Fraumeni syndrome links to unequal gain-of-function effects of p53 mutations. Sci Rep. 2014;4:4223. doi: 10.1038/srep04223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mello SS, Attardi LD. Not all p53 gain-of-function mutants are created equal. Cell Death Differ. 2013;20(7):855–7. doi: 10.1038/cdd.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue X, Zhao Y, Xu Y, Zheng M, Feng Z, Hu W. Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J Mol Biol. 2017;429(11):1595–606. doi: 10.1016/j.jmb.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wick DA, Webb JR, Nielsen JS, Martin SD, Kroeger DR, Milne K, et al. Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin Cancer Res. 2014;20(5):1125–34. doi: 10.1158/1078-0432.CCR-13-2147. [DOI] [PubMed] [Google Scholar]
- 31.Martin SD, Brown SD, Wick DA, Nielsen JS, Kroeger DR, Twumasi-Boateng K, et al. Low Mutation Burden in Ovarian Cancer May Limit the Utility of Neoantigen-Targeted Vaccines. PLoS One. 2016;11(5):e0155189. doi: 10.1371/journal.pone.0155189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bobisse S, Genolet R, Roberti A, Tanyi JL, Racle J, Stevenson BJ, et al. Sensitive and frequent identification of high avidity neo-epitope specific CD8 (+) T cells in immunotherapy-naive ovarian cancer. Nat Commun. 2018;9(1):1092. doi: 10.1038/s41467-018-03301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanyi JL, Bobisse S, Ophir E, Tuyaerts S, Roberti A, Genolet R, et al. Personalized cancer vaccine effectively mobilizes antitumor T cell immunity in ovarian cancer. Sci Transl Med. 2018;10(436) doi: 10.1126/scitranslmed.aao5931. [DOI] [PubMed] [Google Scholar]
- 34.Howie B, Sherwood AM, Berkebile AD, Berka J, Emerson RO, Williamson DW, et al. High-throughput pairing of T cell receptor alpha and beta sequences. Sci Transl Med. 2015;7(301):301ra131. doi: 10.1126/scitranslmed.aac5624. [DOI] [PubMed] [Google Scholar]
- 35.Pasetto A, Alena G, Robbins PF, Deniger DC, Prickett TD, Matus-Nicodemos R, et al. Tumor- and neoantigen-reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol Res. 2016 doi: 10.1158/2326-6066.CIR-16-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkhurst M, Gros A, Pasetto A, Prickett T, Crystal JS, Robbins P, et al. Isolation of T-Cell Receptors Specifically Reactive with Mutated Tumor-Associated Antigens from Tumor-Infiltrating Lymphocytes Based on CD137 Expression. Clin Cancer Res. 2017;23(10):2491–505. doi: 10.1158/1078-0432.CCR-16-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deniger DC, Pasetto A, Tran E, Parkhurst MR, Cohen CJ, Robbins PF, et al. Stable, Nonviral Expression of Mutated Tumor Neoantigen-specific T-cell Receptors Using the Sleeping Beauty Transposon/Transposase System. Mol Ther. 2016;24(6):1078–89. doi: 10.1038/mt.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486(7402):266–70. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–86. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37(9):865–76. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 41.Kobel M, Piskorz AM, Lee S, Lui S, LePage C, Marass F, et al. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J Pathol Clin Res. 2016;2(4):247–58. doi: 10.1002/cjp2.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173(2):371–85. e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito D, Visus C, Hoffmann TK, Balz V, Bier H, Appella E, et al. Immunological characterization of missense mutations occurring within cytotoxic T cell-defined p53 epitopes in HLA-A*0201+ squamous cell carcinomas of the head and neck. Int J Cancer. 2007;120(12):2618–24. doi: 10.1002/ijc.22584. [DOI] [PubMed] [Google Scholar]
- 44.Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol. 2017 doi: 10.1038/nrm.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang D, Sun B, Zhang X, Cheng D, Yu X, Yan L, et al. Huwe1 Sustains Normal Ovarian Epithelial Cell Transformation and Tumor Growth through the Histone H1.3-H19 Cascade. Cancer Res. 2017;77(18):4773–84. doi: 10.1158/0008-5472.CAN-16-2597. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Song N, Liu L, Liu X, Ding X, Song X, et al. USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat Commun. 2017;8:14866. doi: 10.1038/ncomms14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kodama T, Newberg JY, Kodama M, Rangel R, Yoshihara K, Tien JC, et al. Transposon mutagenesis identifies genes and cellular processes driving epithelial-mesenchymal transition in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2016;113(24):E3384–93. doi: 10.1073/pnas.1606876113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma W, Goldberg J. TANGO1/cTAGE5 receptor as a polyvalent template for assembly of large COPII coats. Proc Natl Acad Sci U S A. 2016;113(36):10061–6. doi: 10.1073/pnas.1605916113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou P, Zhang N, Nussinov R, Ma B. Defining the Domain Arrangement of the Mammalian Target of Rapamycin Complex Component Rictor Protein. J Comput Biol. 2015;22(9):876–86. doi: 10.1089/cmb.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiessner M, Roos A, Munn CJ, Viswanathan R, Whyte T, Cox D, et al. Mutations in INPP5K, Encoding a Phosphoinositide 5-Phosphatase, Cause Congenital Muscular Dystrophy with Cataracts and Mild Cognitive Impairment. Am J Hum Genet. 2017;100(3):523–36. doi: 10.1016/j.ajhg.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.