Abstract
Studies of chromosomal aberrations in blood or bone marrow of patients with Fanconi anemia (FA) have focused on their associations with leukemic transformation. The role of such abnormalities on outcomes after hematopoietic cell transplantation (HCT) is unclear. We used genome-wide single nucleotide polymorphism (SNP) arrays to identify chromosomal aberrations in pre-HCT blood samples from 73 patients with FA who received unrelated donor HCT for severe aplastic anemia between 1991–2007. Outcome data and blood samples were available through the Center for International Blood and Marrow Transplant Research. For survival analyses, we used the Kaplan-Meier estimator to calculate the survival probabilities and the exact log-rank test to compare the survival differences across groups. Chromosomal aberrations were detected in 16 patients (22%); most frequent were: clonal copy-loss in chromosome 7 (9.6%), clonal copy-gains in the long arm (q) of chromosome 1 (chr1q+; 8.2%), and clonal or complete copy-gains in the q arm of chromosome 3 (chr3q+; 8.2%). Seven patients (9.6%) had alterations in 3 or more chromosomes. Poor post-HCT overall survival (OS) was noted in patients with chr3q+ (p=0.04), or those with abnormalities in ≥3 chromosomes (p=0.03). The 1-yr OS=0% vs. 45% in patients with either alteration versus its absence. No statistically significant differences in OS were noted in patients carrying deletions in chr7 (1-year OS=29% vs. 42%, respectively, log-rank p=0.74). The study is limited by the small sample size. A larger, prospective study is warranted to validate our findings in light of recent improvement in transplant modalities and outcomes.
Keywords: Fanconi anemia, Chromosomal aberration, Mosaicism, Hematopoietic cell transplantation, Survival
INTRODUCTION
Fanconi anemia (FA) is a rare inherited bone marrow failure syndrome (IBMFS) caused primarily by autosomal recessive germline mutations in genes involved in DNA repair in the FA/BRCA repair pathway. Clinical manifestations of FA include congenital anomalies, aplastic anemia (AA), and increased risk of developing myelodysplasia (MDS), acute myeloid leukemia (AML) and specific solid malignancies.1,2 In a competing risk analysis study, about 70% of the patients with FA developed severe AA by age 50.2 The respective risks of MDS and AML were approximately 6,000- and 200-fold higher than in the general population.2
Cytogenetic abnormalities are frequently found in the bone marrow of patients with FA.3 Frequently reported chromosomal aberrations include gains in the long arm (“q”) of chromosome 1 (chr1q+)4,5, and chromosome 3 (chr3q+)4–6, and complete loss of chromosome 7 (monosomy 7) or partial loss of its long arm (chr7q-).4,5 Chr3q+, monosomy 7 or chr7q- have been associated with risk of progression to MDS or AML.5–7
Allogeneic hematopoietic cell transplant (HCT) remains a curative option for severe AA in patients with FA. Data are limited on the effects of pre-HCT chromosomal aberrations and outcome after HCT for FA-associated AA. A previous CIBMTR analysis of 113 patients with FA in which 73% received related donor HCT between 1985–2007 showed no statistically significant difference (log-rank p=0.2) in post-HCT survival for patients with chromosomal aberrations alone detected by clinical cytogenetics (5-year survival=58%), MDS (5-year survival=57%), or acute leukemia (5-year survival=36%).8 In the same study, analysis excluding unrelated donor subgroup and comparing those with aberrations alone to those with either MDS or acute leukemia showed a better 5-year survival in those with aberrations (67% vs. 43%, p=0.03). The study was limited by not including patients with FA who did not carry chromosomal abnormalities or enough patients with unrelated donor HCTs, where outcomes are less satisfactory.
Here, we used high-resolution SNP-arrays to identify clonal chromosomal aberrations in pre-HCT blood samples from a cohort of patients with FA who received unrelated donor HCT for severe AA, and correlated the presence of aberrations with post-HCT survival.
MATERIALS AND METHODS
Data source
The study included 73 patients with FA from the Center for International Blood and Marrow Transplant Research (CIBMTR) database and biorepository who were part of the CIBMTR-NCI Transplant Outcomes in Aplastic Anemia (TOAA) project. TOAA details are available elsewhere.9,10 The CIBMTR is a research collaboration between the National Marrow Donor Program® (NMDP)/Be The Match® and the Medical College of Wisconsin. It is one of the largest blood and marrow transplant databases, with information available on more than 450,000 transplanted patients.
Patients with FA included in this analysis received unrelated donor marrow or peripheral blood stem cell HCT for severe aplastic anemia between 1991 and 2007; none had a clinical diagnosis of AML or MDS prior to HCT. All blood samples were collected prior to HCT (median time between sample collection and HCT=7 days, range=4–65 days). The study was approved by the NMDP Institutional Review Board and the NIH office of Human Subjects Research Protections. All patients or parents provided informed consent.
Identification of chromosomal aberrations
DNA samples were genotyped using Illumina OmniExpress-24v1-0, a SNP array containing 730,525 markers with a mean distance of 4.1 kb probe spacing (Illumina, San Diego, CA). The array sample completion rate for this study was 99.3%. We used Illumina GenomeStudio software to estimate the log2 R ratio (LRR, calculates the log2 ratio of SNP observed to expected signal intensity), and B allele frequency (BAF, calculates the frequency of the B allele at a given SNP).11 LRR values > 0 indicate copy-number gain, and LRR < 0 indicates copy-number loss. A BAF value of 0.5 indicates a heterozygous biallelic genotype, and a deviation from heterozygotes band in BAF plots with LRR value of zero indicates a copy-neutral loss of heterozygosity (CN-LOH).11 To improve precision, LRR and BAF values were re-estimated after GC content correction and quantile normalization, using the method described previously.12 Re-estimated LRR and BAF data were analyzed using custom software pipelines that involved BAF Segmentation software,13 in which Circular Binary Segmentation (SBC) algorithm was used to detect chromosomal aberration events.14 To minimize false positive discovery, the analysis were restricted to chromosomal abnormalities larger than 2 Mb.12 All potential events were plotted and visualized, and false positive calls were excluded from the analysis based on manual review of each plot.
Statistical methods
We compared the demographics and clinical characteristics of patients with FA by the presence or absence of any chromosomal aberration using Chi-square or Fisher’s exact tests for small number of counts. For survival analysis, we used the Kaplan-Meier estimator to calculate the survival probabilities. Because of the small sample size, we used the exact log-rank test via the R package ‘coin’ to assess survival difference over time between groups.15 The exact test uses permutation-based technique to compute the null distribution of the test statistics without relying on asymptotic results.16 The study follow-up started at the date of HCT and ended at the date of death, last follow-up, or the end of study on August 30, 2017. Kaplan-Meier curves were plotted using R package ‘survminer’, and patient chromosomal aberrations were plotted using Partek Genomics Suite version 7.0 (Partek Incorporated, St. Louis, Missouri, http://www.partek.com/pgs). The other statistical analyses were performed using SAS 9.4 (SAS, Cary, North Carolina).
RESULTS
Patient characteristics
The median age of the 73 patients at HCT was 10.6 years (range=2.5–28.8). Approximately half were male (N=39, 53%), 41% (N=30) received 8/8 HLA matched grafts, 64% (N=47) received reduced intensity or non-myeloablative conditioning regimens, and the majority received bone marrow as their stem cell source (N=63, 86%). There were no statistically significant differences in demographic characteristics or transplant-related factors between patients with and without chromosomal aberrations (Table 1).
Table 1.
Patient characteristics by presence or absence of any chromosomal aberration
| With aberration (N=16) | Without aberration (N=57) | ||||
|---|---|---|---|---|---|
| N | % | N | % | P | |
| Patient-related | |||||
| Age, years | 0.17 | ||||
| <10 | 4 | 25.0% | 27 | 47.4% | |
| 10-<20 | 9 | 56.3% | 26 | 45.6% | |
| 20 or older | 3 | 18.8% | 4 | 7.0% | |
| Race | 0.27 | ||||
| Caucasian | 15 | 93.8% | 45 | 78.9% | |
| Other | 1 | 6.3% | 12 | 21.1% | |
| Karnofsky Performance Score1 | 0.07 | ||||
| 10–80 | 4 | 25.0% | 4 | 7.1% | |
| 90–100 | 12 | 75.0% | 52 | 92.9% | |
| Transplant-related | |||||
| Months from SAA diagnosis to HCT2 | 0.32 | ||||
| <36 | 4 | 30.8% | 24 | 42.1% | |
| 36-<72 | 3 | 23.1% | 19 | 33.3% | |
| ≥72 months | 6 | 46.2% | 14 | 24.6% | |
| Graft type, Bone marrow | 15 | 93.8% | 48 | 84.2% | 0.44 |
| Conditioning regimen | 0.44 | ||||
| Myeloablative | 7 | 43.8% | 16 | 28.1% | |
| Reduced intensity/Nonmyeloablative | 9 | 56.3% | 38 | 66.7% | |
| Other | 0 | 0% | 3 | 5.3% | |
| GvHD prophylaxis | 0.68 | ||||
| Ex vivo T-cell depletion | 10 | 62.5% | 35 | 61.4% | |
| Tacrolimus-based | 1 | 6.3% | 2 | 3.5% | |
| CSA-based | 4 | 25.0% | 18 | 31.6% | |
| Others | 1 | 6.3% | 2 | 3.5% | |
| Number of HLA matches | 0.81 | ||||
| 8/8 | 7 | 43.8% | 23 | 40.4% | |
| ≤7/8 | 9 | 56.3% | 34 | 59.6% | |
| Year of transplant | 0.76 | ||||
| 1991–1999 | 8 | 50.0% | 26 | 45.6% | |
| 2000–2007 | 8 | 50.0% | 31 | 54.4% | |
1 missing
3 missing
Detected chromosomal aberrations
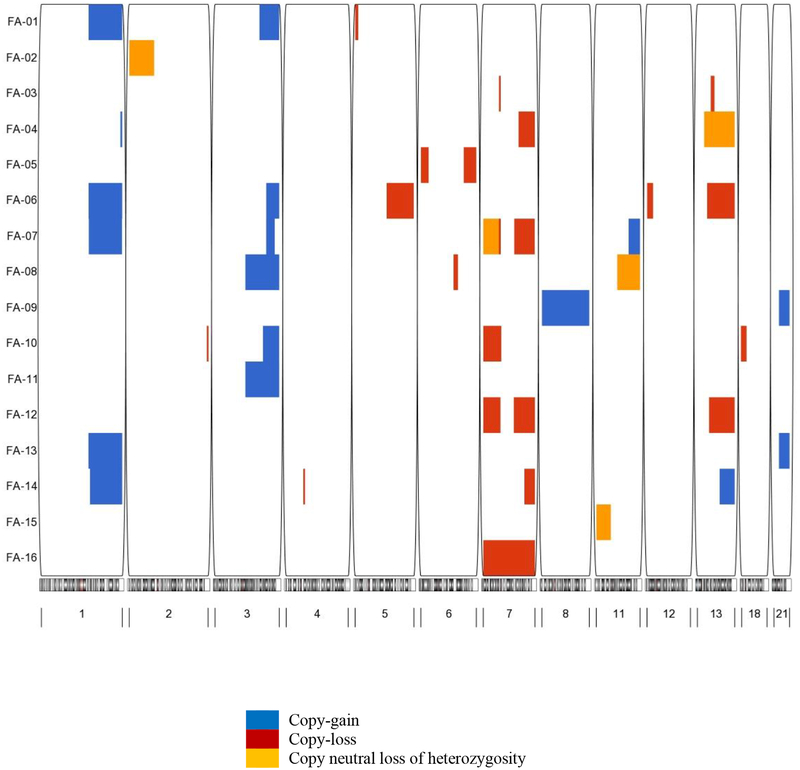
Sixteen patients (22%) had chromosomal abnormalities detected by the SNP array method, with affected cells ranging from 17% to 100%. Among those patients, 5 had cytogenetic testing (results were not available for 4 and one was negative), 2 were not tested, and data were missing for 9 patients. The most frequently detected aberrations in the SNP array analysis were clonal copy-losses in chr7 (N=7 (10%); 2 patients with chr7p- only, 2 with chr7q- only, 2 with partial deletions in both arms, and 1 with monosomy 7), followed by clonal copy gains in chr1q and/or clonal or complete copy-gains in chr3q (chr1q+, N=6 (8%); chr3q+ N=6 (8%); 3 of them carry both), and multiple aberrations affecting chr13 (N=5, 7%; 3 of these patients had chr13q-, 1 had chr13q+, and 1 had chr13q CN-LOH) (Figure 1). The proportions of cells harboring clonal chromosomal aberrations ranged from 33% to 95% for chr7 copy-losses, from 39% to 83% for chr1q+, from 37% to 100% for chr3q+, and from 34% to 73% for chr13q-. The coexistence of aberrations affecting more than 1 chromosome was common (11/16 patients); 7 patients carried aberrations in ≥3 chromosomes (Figure 1).
Figure 1. Chromosomal aberrations in patients with FA.
The plot displays the chromosomal aberration profile for each patient with FA. The rows correspond to each of the patients with chromosomal aberrations and the columns indicate the chromosome number with the location of aberrations. Blue=copy-gain, red=copy-loss, and orange=copy-neutral loss of heterozygosity.
The association between chromosome-specific aberrations and post-HCT survival
Almost all deaths (N=12, 92% of deaths) that occurred in patients with chromosomal aberrations happened within the first year post-HCT. The 1- and 3-year OS were 25% for patients with any aberration versus 46% and 37%, respectively, for patients without aberrations (E log-rank p=0.20) (Table 2). The most frequent cause of death in patients with chromosomal aberration was acute GvHD (N=3, 23.1%), followed by infection (N=2, 15.4%) and hemorrhage (N=2, 15.4%). The most common causes of death in patients without chromosomal aberration were graft failure (N=15, 34.1%), infection (N=8, 18.2%) and organ failure (N=8, 18.2%) (Table 3).
Table 2.
One-year survival probability after unrelated donor hematopoietic cell transplant by detected chromosomal aberration in patients with FA.
| With aberration | Without aberration | ||||
|---|---|---|---|---|---|
| Chromosomal aberration | N total | Probability (95% CI) | N total | Probability (95% CI) | E Log-rank p1 |
| A. Overall | |||||
| Any aberration vs. none | 16 | 0.25 (0.08–0.48) | 57 | 0.46 (0.33–0.59) | 0.20 |
| B. By chromosome (presence vs. absence) | |||||
| Chr1q+ | 6 | 0.17 (0.00–0.54) | 67 | 0.43 (0.32–0.55) | 0.10 |
| Chr3q+ | 6 | 0 | 67 | 0.45 (0.33–0.57) | 0.04 |
| Chr7 deletions2 | 7 | 0.29 (0.04–0.65) | 66 | 0.42 (0.31–0.54) | 0.74 |
| Chr13 aberration3 | 5 | 0.20 (0.00–0.62) | 68 | 0.43 (0.31–0.54) | 0.46 |
| C. Combined | |||||
| Chr1q+ or chr3q+ | 9 | 0.11 (0.00–0.38) | 64 | 0.45 (0.33–0.58) | 0.02 |
| D. By number of affected chromosomes | |||||
| 1–2 chromosomes vs. no aberration | 9 | 0.44 (0.15–0.76) | 57 | 0.46 (0.33–0.59) | 0.95 |
| ≥ 3 chromosomes vs. no aberration | 7 | 0 | 57 | 0.46 (0.33–0.59) | 0.02 |
Exact-approximation log-rank test
Includes 2 patients with chr7p- only, 2 with chr7q- only, 2 with partial deletions in both arms, and 1 with monosomy 7
Includes 3 copy-losses, 1 copy-gain and 1 copy-neutral loss of heterozygosity.
Table 3.
Causes of death by presence or absence of any chromosomal aberration
| With aberration (N of death=13) | Without aberration (N of death=44) | |||
|---|---|---|---|---|
| Cause of death | N | % | N | % |
| Graft failure | 1 | 7.7% | 15 | 34.1% |
| Acute GvHD1 | 3 | 23.1% | 1 | 2.3% |
| Chronic GvHD | 1 | 7.7% | 3 | 6.8% |
| Infection | 2 | 15.4% | 8 | 18.2% |
| IPn2/ARDS3 | 1 | 7.7% | 2 | 4.5% |
| Organ failure | 1 | 7.7% | 8 | 18.2% |
| Subsequent cancer after transplant | 1 | 7.7% | 0 | 0.0% |
| Hemorrhage | 2 | 15.4% | 4 | 9.1% |
| Other, NOS, or unknown causes | 1 | 7.7% | 3 | 6.8% |
Graft versus host disease
Interstitial pneumonitis
Adult Respiratory Distress Syndrome
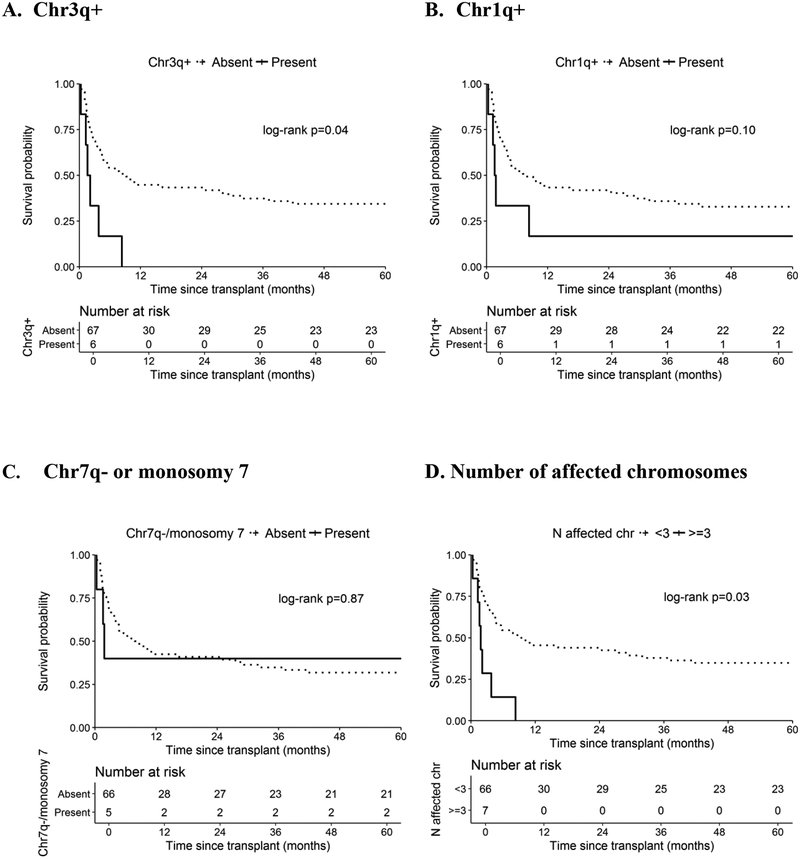
In analyses focusing on the most common aberrations (present vs. absent), the presence of chr3q+ was statistically significantly associated with worse survival (Figure 2A). Of note, no statistical significant differences in patient, disease, or HCT variables were observed between those with and without chr3q+, with the exception of patient age (median age=14.5 vs. 10.3 years, respectively, p=0.02) (Supplementary Table 1). The 1-year OS in patients with and without chr3q+ was 0% and 45%, respectively, E log-rank p=0.04). The causes of death among patients with chr3q+ were hemorrhage (N=2), interstitial pneumonitis (IPn) (N=1) and acute GVHD (N=1), and 2 patients died shortly after a 2nd HCT (1 with acute GVHD and 1 with unknown/other cause, not otherwise specified). No statistically significant differences in OS were noted with chr1q+ (1-year OS=17% vs. 43%, respectively, E log-rank p=0.10) (Figure 2B). A similar finding was noted in patients carrying deletions in chr7 (1-year OS=29% vs. 42%, respectively, E log-rank p=0.74). The survival difference was attenuated after excluding patients who also harbored chr3q+ (1-yr OS=40% in patients with copy-losses in chromosome 7 in the absence of chr3q+, E log-rank p=0.77). Similar results were noted after excluding patients with chr7p- only (1-yr OS=40% vs. 42% in those with and without chr7q- or monosomy 7, respectively, E log-rank p=0.87, Figure 2C). No statistical survival difference was noted in relation to detected aberrations in chr13 (1-year OS=20% vs. 43% in those with and without any chr13 aberration, respectively, E log-rank p=0.46) (Table 2).
Figure 2. Chromosomal aberrations and survival after unrelated donor hematopoietic cell transplant in patients with FA.
Post-HCT survival of patients with FA according to the presence and absence of chromosomal aberrations; A. Chr3q+; B. Chr1q+; C. Chr7q- or monosomy 7; D. Number of affected chromosomes. Dotted line=absence, solid line=presence of chromosomal aberrations. P-values are from exact-approximation log-rank test.
Post-HCT survival by number of affected chromosomes
The 1-year OS in patients with ≥ 3, 1–2 affected chromosomes or those without any aberration were 0%, 44% and 46%, respectively (E log-rank p=0.09). When combined, patients with ≥ 3 affected chromosomes had significantly inferior survival compared with those with <3 affected chromosomes (1-yr OS=0% vs. 45% in those with ≥ 3 and < 3 affected chromosomes, respectively, E log-rank p=0.03, Figure 2D). Of note, 71% (N=5) of patients with ≥3 affected chromosomes had chr3q+.
DISCUSSION
In this analysis of 73 patients with FA who received an unrelated donor HCT, we showed that the presence of pre-HCT chr3q+ and/or ≥ 3 affected chromosomes (71% of those had chr3q+) was associated with inferior post-transplant survival, with death primarily happening within the first year after HCT.
In agreement with previous reports of cytogenetic abnormalities in FA, we found copy-gains in chr1q, chr3q, and copy-losses in chr7 to be most frequently detected by SNP-array.4,5,17 However, the frequency of affected patients is higher in our study (22%) than in a previous report based on a SNP-array analysis in blood samples (12%).17 The higher frequency in our study may be explained, at least in part, by the over-representation of patients with severe disease, since blood samples were collected during the preparation for HCT. In the current study, the coexistence of multiple chromosomal abnormalities was not rare (9.6% of the patients had aberrations in ≥3 chromosomes, and 5/6 patients with chr3q+ had another aberration); this was also higher than observed in the previously referenced study (6/130; 4.6%).17 The cross-sectional nature of our study did not allow observation of the sequence of the occurrence of these aberrations. A previous study with follow-up molecular cytogenetic data showed that chr3q+ preceded monosomy 7 in 33% of the patients (6 out of 18); suggesting that chr3q+ may stimulate subsequent clonal aberrations in FA.6,18
Our data showed a poor post-HCT survival associated with having chr3q+. Chr3q+ in FA is a known prognostic marker, with a high risk of MDS or AML transformation, and death. FA studies of chr3q+ have shown that the affected region harbors the EVl1 oncogene.5,6,19 The detection of chr3q+ by CGH analysis in the bone marrow of patients with FA in a previous study was associated with referral to HCT (74% of the patients), and worse survival (0% without HCT and 41% with HCT for those with chr3q+ vs. 77% in those without chr3q+).18 The noticeable post-HCT survival disadvantage associated with chr3q+ in our study compared with the previous report (0% vs. 41%), could possibly be affected by transplant timing. Patients in the previous study were part of a molecular cytogenetic follow-up research study and expected to be detected and referred at an earlier stage. Our observation related to the survival disadvantage in patients with FA with multiple aberrations may also be associated with the coexistence of chr3q+ (5/7 of the patients with ≥3 affected chromosomes also carry chr3q+).
We did not observe a statistically significant difference in post-HCT survival for patients with and without copy-losses in chromosome 7 (p=0.74). Current clinical recommendations are to consider transplantation for FA patients with monosomy 7 or chr7q-.20 Our study highlights the benefit of HCT in patients with chr7q- and monosomy 7, otherwise associated with poor survival.
Our study included patients who received unrelated HCT between 1991 and 2007. Not surprisingly, post-HCT survival in the study population was poor (3-year OS in patients with and without chromosomal aberration=25% and 37%, respectively). This reflects the known historic limitations of unrelated donor HCT in patients with FA in the early years.21 Advancements in HCT practices, including the introduction of fludarabine-based reduced intensity conditioning and better supportive care resulted in significant survival improvement in the recent era.22–25 Further evaluation of recent transplant data is needed in light of current improvements in HCT modalities and outcome for patients with FA.
To our knowledge, this is the first multicenter study to evaluate the association between pre-transplant specific chromosomal aberrations in patients with FA receiving unrelated HCT and post-transplant survival. Our study took advantage of the high-quality longitudinal consolidated outcome data available through the CIBMTR. We assessed chromosomal aberration status in patient blood samples pre-HCT utilizing a high-density SNP array for precise and comprehensive detection of such events. Our study is limited by its descriptive nature and the lack of power to test for possible predictors because of the small sample size. Other limitations include the lack of more recent transplant data, and the inability to examine the impact of balanced chromosomal aberrations, as SNP-arrays do not provide information on such aberrations;26 however, a previous study showed that those are uncommon events.5
In conclusion, our results suggest that copy-gain in chromosome 3 may have a prognostic relevance in outcomes after unrelated HCT in patients with FA. In theory, it is possible that those patients may have had an unrecognized MDS that was under-treated by HCT; however, the reported causes of death in those patients do not support this hypothesis (no reported hematologic cancer deaths). Our findings highlight the importance of chromosomal analysis surveillance in patients with FA that involves chromosome 3. The study also illustrates a need to further evaluate SNP arrays as a method for detecting chromosomal aberrations in patients with FA. The study is limited by its small sample size; a larger study analyzing recent transplant data is warranted in order to better understand the role of such aberrations in the context of HCT in patients with FA.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health. The CIBMTR is supported primarily by Public Health Service Grant/Cooperative Agreement 5U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 4U10HL069294 from NHLBI and NCI; a contract HHSH250201200018C with Health Resources and Services Administration (HRSA/DHHS); and two Grants N00014-17-1-2388 and N0014-17-1-2850 from the Office of Naval Research.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The authors have no conflict of interest to declare.
REFERENCES
- 1.Alter BP, Giri N, Savage SA, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. British Journal of Haematology. 2010;150(2):179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018;103(1):30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer S, Neitzel H, Tonnies H. Chromosomal aberrations associated with clonal evolution and leukemic transformation in fanconi anemia: clinical and biological implications. Anemia. 2012;2012:349837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cioc AM, Wagner JE, MacMillan ML, DeFor T, Hirsch B. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with fanconi anemia: morphologic and cytogenetic characteristics. Am J Clin Pathol. 2010;133(1):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quentin S, Cuccuini W, Ceccaldi R, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117(15):e161–170. [DOI] [PubMed] [Google Scholar]
- 6.Tonnies H, Huber S, Kuhl JS, Gerlach A, Ebell W, Neitzel H. Clonal chromosomal aberrations in bone marrow cells of Fanconi anemia patients: gains of the chromosomal segment 3q26q29 as an adverse risk factor. Blood. 2003;101(10):3872–3874. [DOI] [PubMed] [Google Scholar]
- 7.Mehta PA, Harris RE, Davies SM, et al. Numerical chromosomal changes and risk of development of myelodysplastic syndrome--acute myeloid leukemia in patients with Fanconi anemia. Cancer Genet Cytogenet. 2010;203(2):180–186. [DOI] [PubMed] [Google Scholar]
- 8.Ayas M, Saber W, Davies SM, et al. Allogeneic hematopoietic cell transplantation for fanconi anemia in patients with pretransplantation cytogenetic abnormalities, myelodysplastic syndrome, or acute leukemia. J Clin Oncol. 2013;31(13):1669–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadalla SM, Wang T, Haagenson M, et al. Association between donor leukocyte telomere length and survival after unrelated allogeneic hematopoietic cell transplantation for severe aplastic anemia. JAMA. 2015;313(6):594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadalla SM, Wang T, Dagnall C, et al. Effect of Recipient Age and Stem Cell Source on the Association between Donor Telomere Length and Survival after Allogeneic Unrelated Hematopoietic Cell Transplantation for Severe Aplastic Anemia. Biol Blood Marrow Transplant. 2016;22(12):2276–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiffer DA, Le JM, Steemers FJ, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16(9):1136–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs KB, Yeager M, Zhou W, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44(6):651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staaf J, Lindgren D, Vallon-Christersson J, et al. Segmentation-based detection of allelic imbalance and loss-of-heterozygosity in cancer cells using whole genome SNP arrays. Genome Biol. 2008;9(9):R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. [DOI] [PubMed] [Google Scholar]
- 15.Hothorn T, Hornik K, van de Wiel MA, Zeileis A. Implementing a Class of Permutation Tests: The coin Package. J Stat Software. 2008;28(8):1–23. [Google Scholar]
- 16.Everitt BS, Hothorn T. A Handbook of Statistical Analyses Using R. https://cran.r-project.org/web/packages/HSAUR2/vignettes/Ch_survival_analysis.pdf. [Google Scholar]
- 17.Reina-Castillon J, Pujol R, Lopez-Sanchez M, et al. Detectable clonal mosaicism in blood as a biomarker of cancer risk in Fanconi anemia. Blood Adv. 2017;1(5):319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neitzel H, Kuhl JS, Gerlach A, Ebell W, Tonnies H. Clonal Chromosomal Aberrations in Bone Marrow Cells of Fanconi Anemia Patients: Results and Implications In: Fanconi anemia : a paradigmatic disease for the understanding of cancer and aging. Basel; New York: Karger; 2007, pp. 79–94. [Google Scholar]
- 19.Meyer S, Bristow C, Wappett M, et al. Fanconi anemia (FA)-associated 3q gains in leukemic transformation consistently target EVI1, but do not affect low TERC expression in FA. Blood. 2011;117(22):6047–6050. [DOI] [PubMed] [Google Scholar]
- 20.Savage SA, Dufour C. Classical inherited bone marrow failure syndromes with high risk for myelodysplastic syndrome and acute myelogenous leukemia. Semin Hematol. 2017;54(2):105–114. [DOI] [PubMed] [Google Scholar]
- 21.Wagner JE, Eapen M, MacMillan ML, et al. Unrelated donor bone marrow transplantation for the treatment of Fanconi anemia. Blood. 2007;109(5):2256–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Center for International Blood and Marrow Transplant, a contractor for the C. W. Bill Young Cell Transplantation Program operated through the U. S. Department of Health and Human Services, Health Resources and Services Administration, Healthcare Systems Bureau. U.S. Patient Survival Report; Fanconi anemia, From 2008 through 2012.
- 23.Ebens CL, DeFor TE, Tryon R, Wagner JE, MacMillan ML. Comparable Outcomes after HLA-Matched Sibling and Alternative Donor Hematopoietic Cell Transplantation for Children with Fanconi Anemia and Severe Aplastic Anemia. Biology of Blood and Marrow Transplantation. 2018;24(4):765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bierings M, Bonfim C, Latour RPD, et al. Transplant results in adults with Fanconi anaemia. British Journal of Haematology. 2018;180(1):100–109. [DOI] [PubMed] [Google Scholar]
- 25.Smetsers SE, Smiers FJ, Bresters D, Sonnevelt MC, Bierings MB. Four decades of stem cell transplantation for Fanconi anaemia in the Netherlands. British Journal of Haematology. 2016;174(6):952–961. [DOI] [PubMed] [Google Scholar]
- 26.Stevens-Kroef MJ, Olde Weghuis D, ElIdrissi-Zaynoun N, et al. Genomic array as compared to karyotyping in myelodysplastic syndromes in a prospective clinical trial. Genes Chromosomes Cancer. 2017;56(7):524–534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.