Abstract
Amyloids may be regarded as native nanomaterials that form in the presence of complex protein mixtures. By drawing an analogy with the physicochemical properties of nanoparticles in biological fluids, we hypothesized that amyloids should form a protein corona in vivo that would imbue the underlying amyloid with a modified biological identity. To explore this hypothesis we characterized the protein corona of human islet amyloid polypeptide (IAPP) fibrils in FBS using two complementary methodologies developed herein; quartz crystal microbalance and ‘centrifugal capture’, coupled with nano-liquid chromatography tandem mass spectroscopy. Clear evidence for a significant protein corona was obtained. No trends were identified for amyloid corona proteins based on their physicochemical properties, while strong binding with IAPP fibrils occurred for linear proteins or multi-domain proteins with structural plasticity. Proteomic analysis identified amyloid-enriched proteins that are known to play significant roles in mediating cellular machinery and processing, potentially leading to pathological outcomes and therapeutic targets.
Keywords: amyloid, protein corona, liquid chromatography, mass spectrometry, amyloidogenesis
The dynamic and nonspecific adsorption of proteins on a nanoscale substrate renders a ‘protein corona’, which consequently defines the biological identity of the nanostructure.1 The physicochemical and biological properties of the protein corona have been extensively examined in the last decade,2 and characterizations of these aspects have become a necessity for establishing any engineered nanomaterial designed for biological applications.
Native biomaterials resemble engineered nanoparticles in mechanical and physicochemical attributes, but acquire these traits through endogenous processes that include the formation of ‘native’ protein coronae. The capacity for biomaterials to interact with surrounding biological media supports the hypothesis that, similar to nanomaterials, the surface properties and biological behavior of biomaterial-corona complexes diverges from that observed for isolated materials prepared in vitro.3,4 This is exemplified by extracellular matrix component fibronectin, which demonstrated the ability to foul upon mechanically-induced unwinding.5 Viral particles also displayed the capacity to form protein coronae in biological fluids, and such coronae appeared to direct viral pathogenesis and catalyze amyloid aggregation.6 Whereas protein absorption to synthetic nanomaterials can result in rapid clearance from the circulation,7 protein corona formation on native nanomaterials as they are produced and secreted into intra- or extracellular environment may play a role in their endogenous function, and in maintaining their biological stability.
Amyloid fibrils represent a major class of native nanomaterial. They manifest as long, semi-flexible, hydrophobic nanofibers formed through aggregation of amyloid proteins that are ubiquitously present in the body.8 While amyloids can be involved in functional processes, such as in melanin production,9 amyloidogenesis is a deleterious process implicated in metabolic and neurodegenerative pathologies that present a major burden on global health.8
Here, we consider human islet amyloid polypeptide (IAPP): a 37-residue amyloidogenic peptide that is co-secreted with insulin from pancreatic β-cells in vivo, and stabilized within intracellular β-cell granules prior to secretion by a combination of low pH, insulin and complexation with C-peptide and zinc ions at a specific ratio.10 IAPP plays a functional role in glycemic control,11 and a pathological role in the development and morbidity of type 2 diabetes (T2D)12 through their amyloid aggregation within the extracellular space, a process toxic to the β-cells.13 Although IAPP amyloid deposition is largely associated with cell death and dysfunction in the pancreas,13 extra-pancreatic plaques and associated cytotoxicity have also been observed in the heart,14 kidney15 and brain.16,17 Given that plasma amylin concentration is either unchanged,18 or even decreased,19 in patients with T2D when compared to non-diabetics, it is likely that amyloid nucleation components from pancreatic deposits, including fibrils, are capable of translocating into the circulation where they subsequently develop into distal plaques that contribute to extra-pancreatic T2D pathologies. Given the long biological lifetime of amyloid deposits,20,21 particularly when compared to synthetic nanomaterials,22,23 the formation and implications therein of an ‘amyloid-protein corona’ may elicit extensive and wide-ranging effects on biological processes, thus belying a more complex role in amyloid-associated disease pathologies.
IAPP amyloids are capable of associating with numerous proteins and co-factors in vivo, including serum amyloid P component,24 apolipoprotein E,25 and glycosaminoglycans (GAGs), in particular heparan sulfate proteoglycans.26 GAGs have demonstrated the capacity to enhance fibrillization of both functional27,28 and incompletely processed IAPP,31 and are subsequently implicated in IAPP-associated cytotoxicity.29,30 We have recently established the capacity for IAPP amyloids to form a protein corona in vitro to impact the amyloid morphology and cytotoxicity.32 As an extension, herein we present a comprehensive qualitative annotation of the complex protein corona formed on the major pathological amyloid IAPP. Fetal bovine serum (FBS) was utilized as a representative medium, mimicking the in vivo milieu and providing context for amyloid studies in vitro. Time point-based analyses of corona deposition by transmission electron microscopy (TEM) and quartz crystal microbalance (QCM) revealed rapid formation of a viscoelastic, non-uniform protein corona on IAPP amyloids within seconds of exposure to FBS.33 Helium ion microscopy (HIM) imaging highlighted the capacity for the amyloid-protein corona to disrupt cell morphology within minutes of exposure. Modified QCM and ‘centrifugal capture’ (CC) techniques provided independent work flows for ‘one-pot’ amyloid-protein corona isolation, elution and tryptic digestion for subsequent analysis by nano-liquid chromatography tandem mass spectroscopy (nLC-MS/MS). Replicate QCM and CC experiments revealed that the IAPP amyloid corona is composed of multiple protein types with varied biological functions, including adhesion and signal transduction. This study provides insights into the influence of proteins on the properties of IAPP amyloids, including their toxicity. In addition, methods in this study offer a prototype pipeline applicable to other amyloid proteins, and thus have implications for the rational design of effective strategies against amyloid pathologies in vivo.
Results and Discussion
Amyloids rapidly formed a protein corona in complex solution
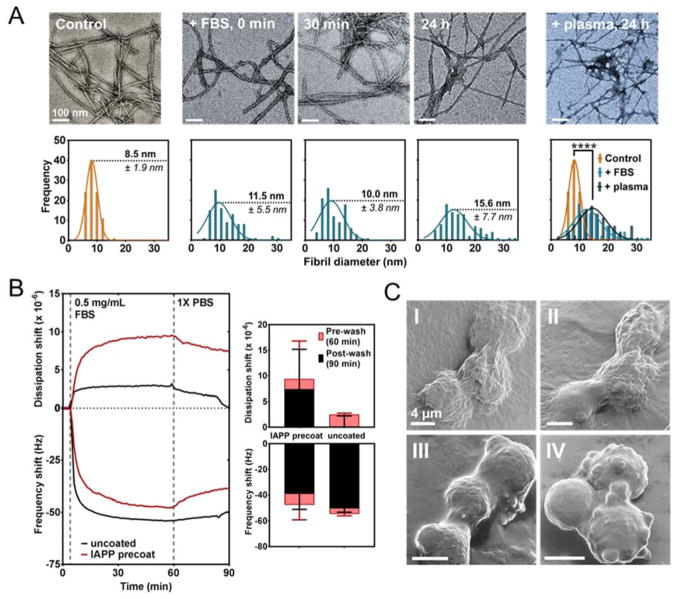
TEM imaging revealed serum proteins associating with mature IAPP amyloids within seconds of mixing (Fig. 1A). After 24 h an amyloid-protein corona was established that visibly changed amyloid morphology (Fig. 1A, upper panels). IAPP amyloids formed in water were long, semi-flexible nanofibers with a narrow width distribution, presenting an average diameter of 8.5 ± 1.9 nm (Fig. 1A, lower panel); in contrast, introduction of FBS immediately (T = 0 min) increased the average fibril width by 3 nm. By 24 h, the average recorded fibril width nearly doubled that of the control amyloids. This significant shift in fibril diameter upon formation of the IAPP amyloid-FBS protein corona showed high concordance with an independently conducted assay (Fig. 1A, far right panels), wherein IAPP amyloids were incubated with human plasma proteins at an analogous mass ratio and measured after 24 h (average fibril diameter: 16 ± 5.53 nm).
Figure 1.
Formation and characterization of IAPP amyloid-protein corona in FBS. A: TEM imaging (upper panel; scale: 100 nm) and associated analysis of fibril diameter (lower panel; n = 100 points sampled) of amyloid exposed to FBS over 24 h, with a comparative measurement after 24 h exposure to human plasma proteins. **** p < 0.0001. B: QCM characterization of amyloid corona formation on an IAPP amyloid functionalized surface (IAPP pre-coat) or gold (uncoated) under flow over 1 h (left panel) with comparison of the soft (pre-wash) and hard (post-wash) coronae (right panel). Error: SEM (n = 4). C: HIM imaging of βTC6 cells exposed for 30 min in vitro to either amyloid (I), amyloid + FBS, pre-incubated for 2 h at RT (II), FBS alone (III), or no treatment applied (IV). Scale: 4 μm. Concentration of IAPP amyloid in all experiments: 0.1 mg/mL; FBS: 0.5 mg/mL.
The rapid formation and large structural variance of the IAPP amyloid-protein corona observed with TEM was corroborated by QCM measurements (Fig. 1B). Gold-coated QCM sensors were functionalized with IAPP amyloids in a two-step process to ensure coverage of the full sensor surface: an initial incubation period under static conditions over 16 h, followed by a second passivation step under flow for 1 h (Fig. S1, Supporting Information). IAPP amyloid displayed cumulative deposition as per ‘classical’ behavior of pathological amyloids in vivo.34 Passivation of FBS onto the IAPP amyloid-functionalized QCM sensor demonstrated an exponential protein deposition onto amyloid within the first 15 min of exposure at a flow rate of 100 μL/min. Saturation under these conditions was observed to occur at circa 1 h. Surprisingly, these kinetics were very comparable to protein deposition onto the non-functionalized gold surface, to which proteins had a strong binding affinity due to the ubiquity of thiol-rich cysteine residues, suggesting amyloids may initiate similar protein interactions in vivo. Upon removal of transiently bound proteins through a wash step, protein deposition onto non-functionalized control sensors correlated to a dissipation shift of close to zero, indicating formation of a rigid, compressed protein layer, with little variation observed over individual runs. In contrast, the final mean dissipation shift when serum proteins bound to IAPP amyloids was 7.4×10−6 with extensive variations between independent runs, indicating that a viscoelastic, elongated surface coating of proteins was able to form on IAPP amyloid plaques and that this surface was non-uniform in character, thus concordant with TEM observations. Furthermore, after removal of unbound FBS, little change in the frequency shift was observed, providing strong evidence that protein association to amyloid was not transient, but most likely involved at minimum the formation of a ‘hard’ corona within an hour of protein exposure. The facile association of proteins with the IAPP amyloid surface observed here can be attributed to the physicochemical properties of the amyloid exposed moieties. Namely, basic lysine (Lys1; pKa=10.54) and arginine (Arg11; pKa=12.48) are protonated at physiological conditions and can form H-bonds; concordantly, polar tryptophan (Thr4, Thr6 and Thr9) and aromatic phenylalanine (Phe15) are known to form hydrophobic and π-π stacking interactions.35,36
IAPP amyloid with associated proteins affected pancreatic β-cell morphology
IAPP amyloids with no pre-exposure to protein solution (Fig. 1C, panel I), amyloids pre-incubated with FBS for 2 h at room temperature (II), and FBS in absence of amyloid (III) were incubated with insulin-producing pancreatic β-cell line βTC6 in suspension for 30 min. Cells were then fixed and imaged by helium ion microscopy (HIM). βTC6 cells exposed to IAPP amyloids, with or without preformed coronae, displayed a shift in morphology from the rounded, smoother cell surface – visualized for those where amyloid was absent (III) and for untreated controls (IV) – to a flattened morphology with a highly folded cell membrane (III&IV). Pathological amyloids have been shown to affect cell membrane fluidity, through means of intercalation and lipid-stripping;37–39 as such, these data suggest that rapid amyloid-protein corona formation in cell culture media over the 30 min treatment period was no less protective against amyloid-membrane interactions than the introduction of amyloids with a preformed corona over 2 h.
Amyloid-protein corona was effectively isolated using two independent methodologies
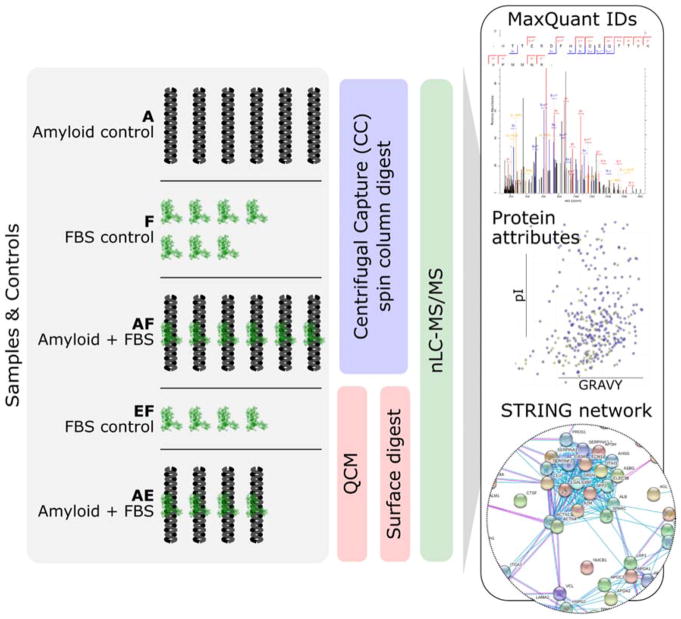
Two distinct methods were optimized to isolate a complex protein-protein corona for composition analysis by nLC-MS/MS. The sample preparation goal was to adopt orthogonal approaches for achieving ‘one-pot’ – and therefore streamlined – isolation, digestion and elution of corona proteins from captured amyloid. The first of these methods, CC, utilized high molecular weight cut-off (MWCO) spin columns (1 MDa) to separate amyloid and amyloid-protein complexes from free, unbound proteins via low-impact benchtop centrifugation, following in-solution formation of coronae over 2 h. CC demonstrated up to 90% amyloid retention and less than 10% nonspecific serum protein retention on the filter after four wash cycles (Fig. S2). In parallel, a second method incorporating quartz crystal microbalance (QCM) allowed the study of protein corona formation under flow; wherein IAPP amyloid captured on the sensor surface as large contiguous ‘plaques’ was able to collect serum proteins over time, thus providing further insights on amyloid-protein interactions combined with physical exposure conditions that more closely mimic those in vivo.40 For both CC and QCM, amyloid-protein corona isolation was followed by an in situ (on-filter for CC; on-sensor for QCM) protein elution and ‘soft’ digestion protocol which maximized collection of corona proteins while minimizing the digestion of amyloid. This limited amyloid contamination for the subsequent nLC-MS/MS measurements, the results of which were submitted for protein database queries using the Andromeda search engine (MaxQuant)41,42 to determine which proteins could be identified in the amyloid-protein corona complex isolates. Together, CC and QCM provided different perspectives on the protein corona of a proteinaceous amyloid system – a complex analytical challenge – resulting in evidence that directly implicates these coronae in key protein pathways. The qualitative nature of shotgun nLC-MS/MS made this a post-hoc exploratory analysis of amyloid coronae components with the aim of generating testable hypotheses regarding the implications of coronal enrichment. Figure 2 highlights the workflow used to assess each experiment type: for CC these were amyloid only (A), FBS only (F) and amyloid-corona complexes (AF), while for QCM these were FBS only (EF) and amyloid-corona complexes (AE). In brief, further analysis included consideration of identification overlaps between experiments; namely, pooled searches for each experiment type to maximize IDs, and independent searches for technical replicates of experiments F and AF to establish the robustness of identification across replicates (see Supporting Information). In addition, trends were sought in the relationships between Grand Average of Hydropathy (GRAVY), isoelectric point (pI) and molecular weight (MW), the proportion contribution of specific amino acid classes to corona protein sequences, as well as the existence of enriched gene ontologies (GO), pathways and protein domains in the protein networks (STRING analysis) formed by the individual proteins or peptides identified as being part of the amyloid protein corona and found lacking in the background controls, hereafter referred to as ‘unique’ proteins (see Supporting Information for details of processing and presentation).
Figure 2.
Sample nomenclature, replicates, methods and analytical pipeline implemented for centrifugal capture (CC) and quartz crystal micro-balance (QCM) amyloid-corona characterization by nano-liquid chromatography MS/MS (nLC-MS/MS), including MaxQuant Andromeda searches, protein attribute comparisons and protein network analysis (STRING). Experimental nomenclature is provided, including amyloid control (A), fetal bovine serum (FBS) only control (F) and amyloid plus FBS (AF) for CC preparations, as well as FBS control (EF) and amyloid plus FBS (AE) for QCM preparations.
Isolation method impacts protein identification rates and similarity of composition
A primary reason for using both CC and QCM approaches was the potential for producing vastly different protein coronae: through corona formation in different environmental conditions. Given that little is known regarding the composition of amyloid coronae, the nLC-MS/MS identifications for these orthogonal approaches were contrasted in the context of both experimental design and the existing knowledgebase regarding synthetic nanomaterial coronae.
The CC method – inspired by separation approaches applied to synthetic nanomaterials43 – was hypothesized to drive the formation of relatively ‘simple’ amyloid-protein complexes in-solution, which were both formed and washed under relatively turbulent conditions (i.e. rinsing/centrifugation) leaving only the ‘hard’ corona intact.43 This was considered in concert with the concept of a relatively large MWCO filter being able to effectively retain fibril complexes,44 where the remainder of the proteins in-solution would bypass the filter. In sharp contrast, the gentle QCM micro-flow conditions – for amyloid deposition, FBS application and PBS rinsing – not only generated complex surface associated plaque structures, but created conditions more conducive to preserving a triad of binders: high-affinity ‘hard’ corona proteins, any sterically captured proteins as well as secondary/tertiary protein-protein ‘soft’ corona binders.40
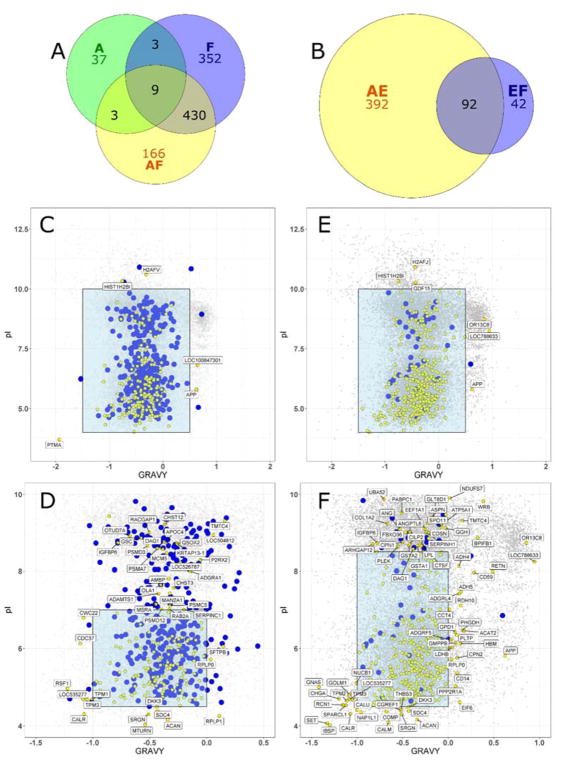
CC controls were considered first. A total of 50 proteins were identified in the amyloid fibrils only (A) sample, which may have been derived from column carryover or present as contaminants in solution. Not unexpectedly, using a Human *.fasta file, sample A was also found to also contain multiple Keratins (see Supporting Information). Overall, the CC (F/AF) and QCM (EF/AE) experiments resulted in 961 (Fig. 3A) and 526 (Fig. 3B) protein identifications, respectively. The overall difference can be attributed to either inter-experimental variation, or to the greater number of F/AF nLC-MS/MS analyses: 13, as compared to 8 for EF/AE. The overlap between the F and AF conditions accounted for 45.6% of the total identifications made with the CC method, while only 17.5% were shared between the FBS only (EF, N = 134 total and 42 unique proteins) and FBS in the presence of amyloid plaques (AE, N = 484 total and 392 unique proteins). This difference was likely caused by a combination of non-specific protein binding to the MWCO filter – see the retention of protein mass in Fig. S2 – in addition to trapping of protein aggregates in the spin filter. Here, given the robust wash steps being used for the CC method, the unique identifications may represent the “hard” protein corona formed around the fibrils, whereas the overlap proteins are part of the aforementioned “non-specific” noise in the preparation. As such, the unique AF proteins were marked as potentially interesting, given their absence across the numerous F sample control measurements. However, it could not be ruled out that these proteins were present in the F samples, but below the concentration required for detection by the applied nLC-MS/MS method. Next-stage application of label free quantitation (LFQ) by nLC-MS/MS may highlight the contribution of the F/AF overlap proteins to the corona, as relative quantity can provide enrichment information.
Figure 3.
Overlap of protein identifications made using nLC-MS/MS analysis of centrifugal capture (CC, A) and quartz crystal microbalance (QCM, B) amyloid corona experiments. The GRAVY/pI relationships for unique CC (AF) and QCM (AE) amyloid serum coronae are shown in yellow in panels D–F. Blue markers in C–D are all identified AF experiment proteins and in E–F are unique serum only control (EF) proteins. All plots are overlaid onto the Bos taurus proteome background (gray). Labels are specific to designated regions of the plots (panels C/E: labels outside blue box, GRAVY ≤ −1.5 and ≥ 0.5, pI ≤ 4 and pI ≥ 10 | panel D: labels outside ranges GRAVY ≤ −1 and ≥ 0, pI ≤ 4.5 and ≥ 7, panel F: labels outside ranges GRAVY ≤ −1 and ≥ 0, pI ≤ 4.5 and ≥ 8.5).
The sizeable difference in overlap profiles for the QCM experiments (EF/AE), as compared to the CC methods (F/AF), indicated that QCM micro-flow conditions may prevent the buildup and capture of large non-specific protein aggregates, but also preserve a different kind of binding complex: one combining high-affinity corona binders, proteins that become sterically trapped in the plaque-protein complex, and finally any secondary and tertiary protein-protein interactions that comprise a ‘soft’ amyloid corona.45 An alternative hypothesis, given that coronae are implicated in conformational change46 is that the amyloids catalyze the formation of protein aggregates that subsequently form a cloud around the fibril. A natural extension of this is that the use of alternative wash fluids could remove the lower affinity binders to reveal the ‘hard’ corona. These are exciting possibilities, as they suggest the data herein may provide a glimpse of a hard and soft coronae dichotomy for amyloid fibrils.
Sequence-dependent characteristics do not indicate significant trends in properties for amyloid-corona proteins
Two independent analytical approaches were used to qualitatively evaluate the unique protein populations for the CC (AF) and QCM (AE) prepared coronae. The first of these was to consider the physicochemical properties of the protein sequences identified. Figure 3C shows the GRAVY – pI relationships for unique AF proteins (yellow) overlaid onto unique F proteins (blue) and the Bos taurus proteome, with a zoom view in Fig. 3D. Although proteins with outlier values were observed for both GRAVY (PTMA, APP) and pI (PTMA, HIST1H2B1, H2AFV, Fig. 3C), no significant general trend was observed. The identification of Amyloid beta A4 protein (APP; see Fig. S4) here is interesting given the potential for assembly of heterogeneous amyloid structures comprised of both IAPP and APP.47
There was also no clearly discernible pattern for an overlay of unique AE (yellow) and unique EF (blue) proteins onto the Bos taurus proteome (Fig. 3E, zoom in 3F). The AF/F and AE/EF GRAVY – MW comparisons (Fig. S5A/S5B) also demonstrated no significant outliers, with the exception of TTN identification for AF: a likely false positive identification. Furthermore, the amino acid proportions in the identified protein sequences did not show any noteworthy trends related to residue charge or hydrophobicity (see Supporting Information). An overall nonspecific loss of serum proteins from a Blue-Native gel upon IAPP amyloid-corona formation was additionally observed in an electrophoresis-based pilot study (Fig. S6).
Protein networks for amyloid-corona complexes
The second qualitative analysis undertaken used the STRING resource (string-db.org)48,49 to identify molecular actions and enriched functional associations across the corona protein network. Multiple network annotations were enriched for the unique AF amyloid corona proteins (CC method, see Fig. S7). GO biological process Ubiquitin-dependent catabolic process (red) was enriched in the core network, in addition to the GO molecular function Carbohydrate derivative binding (blue) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for ECM-receptor interaction (green), focal adhesion (yellow), proteasome (pink), PI3K/Akt (dark green) and the INTERPRO protein domain Concanavalin A-like lectin/glucanase domain (light blue). The proteins bound uniquely to amyloid fibrils may therefore interact with key elements of the cellular machinery including the proteasomal system, membrane adhesion factors and signal transduction pathways. These observations support the hypothesis that, similar to synthetic nanomaterial coronae, the amyloid-corona plays a role in changing biological identity to impact cellular interactions.45,50 By extension, if this interaction causes dysregulation, then this may provide a mechanistic foundation for pathology. As suggested earlier, this might include conformational changes to key proteins in these annotated pathways, the artificial concentration of proteins at amyloid plaque locations, or the ‘activation’ of plaques to act as scaffolds that support damaging molecular functions. The specific protein components driving such mechanisms are a key stage-gate going forward for this line of research.
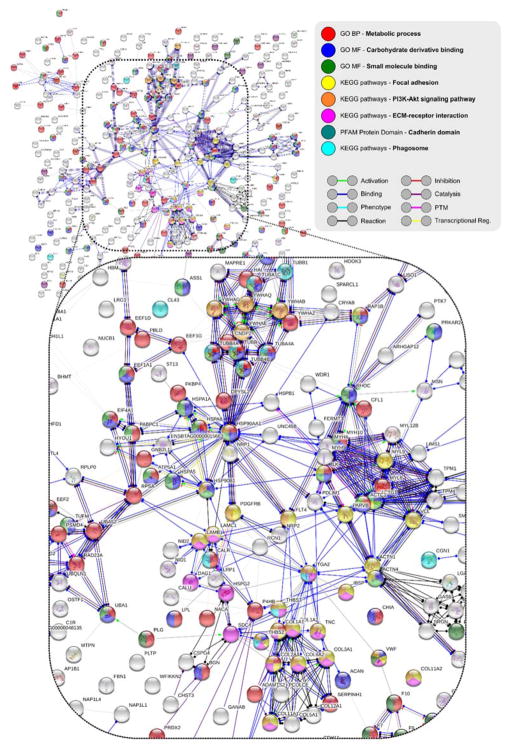
These observations were mirrored for the unique STRING protein network generated from the unique QCM protein list (experiment AE), albeit with many more functional enrichments (Figure S8A/S8B). Figure 4 shows the enrichment of GO annotations for biological process Metabolic process (red) as well as molecular functions that include Small molecule binding (green) and Carbohydrate derivative binding (blue). The KEGG pathways, Focal adhesion (yellow), PI3K/Akt signaling pathway (orange), ECM-receptor interaction (pink), Phagosome (light blue) and PFAM protein domain Cadherin domain (dark green) were also enriched. The core network of AE interactors thus contained proteins that may influence cellular metabolic networks. The AE network also mirrored elements enriched in the AF amyloid coronae, including Focal adhesion and PI3K/Akt signaling (Figure S7, Figure S8B). In addition to the functions annotated in Figure 4, it was not surprising to find elements of the coagulation cascade (see Figure S8C). Finally, in a departure from the AF network in Figure S7, catalytic and hydrolase activity were both annotated for the AE network (Figure S8B/S8C, suggesting a possible role for amyloid fibrils in the concentration, promotion or depletion of enzymatic activity through co-localization and potential protein conformational changes in the amyloid-corona.46
Figure 4.
Quartz crystal microbalance (QCM) method amyloid and serum corona (AE) STRING48,49 (string-db.org) protein network (molecular action) produced using database and experimental interactors, with a minimum interaction score of 0.400 and no additional interactors, against a whole Bos taurus genome background. Enrichment analysis and molecular action legends are included, in addition to predicted action effects – positive (arrowhead), negative (endpoint line), unspecified (endpoint circle).
The occurrence of overlapping GO/KEGG annotations across the unique AF and AE protein networks, in tandem with those annotations present in AE but missing in AF, gives additional credence to the hypothesis that the unique AF binders may in fact be hard corona proteins. The additional AE proteins may thus be part of either an extended hard corona network preserved by the less rigorous isolation conditions of the QCM method, or the postulated “soft” corona.
The sum of these data provide evidence to suggest that the amyloid corona, and potentially its soft component, interacts not only with the coagulation cascades – as synthetic nanomaterials are known to51,52 – but the coronae may cause amyloid to interfere in metabolic processes as well as adhesion and signal transduction pathways. It was promising to observe the latter results across two very different analytical approaches, including some shared proteins contributing to these network annotations: one example was 14-3-3 protein beta/alpha for the PI3K annotation (YWHAB). These observations cement the need to quantitatively evaluate the composition of these coronae in future.
Structural analysis reveals key characteristics of the proteins enriched on amyloid
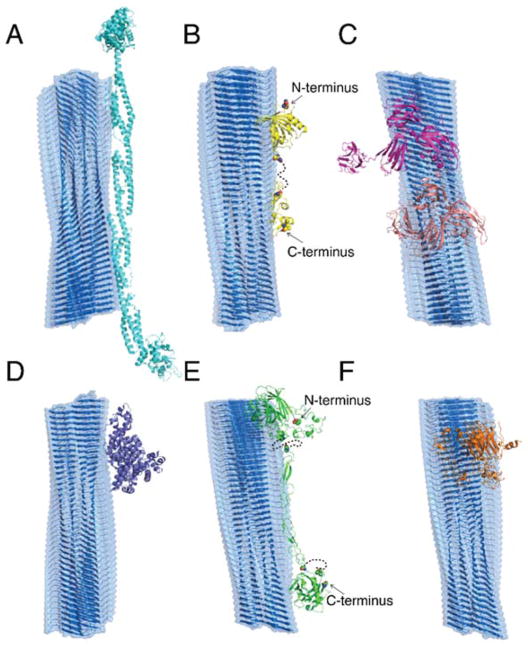
For the top ten serum proteins (by unique peptide count) in amyloid coronae identified by either CC (AF) or QCM (AE) methods, structural information available in the order of whole sequences, the sequence homologs, or sub-sequences was obtained by searching the protein databank (PDB) (Tables S1&S2). Corresponding three-dimensional (3D) structures of the AF and AE proteins were shown in Figs. S9&S10, respectively. For comparison, available structures of top FBS proteins (Table S3) were also shown in Fig. S11. Structural examination of the AF proteins (Fig. S9) suggests two salient features, having either fibrillar morphology (e.g., Figs. S9A,E,F) or multi-domains (e.g., Figs. S9B,C,D,F) with conformational flexibilities (e.g., the “open” and “close” conformations of neuropilin in Figs. S9C&D), in contrast to mostly globular structures of top FBS proteins (Fig. S11). A protein with either of the observed structural properties, such as the linear alpha-actinin-4 with coiled coils (Fig. 5A) or the multi-domain neuropilin with inter-domain plasticity (Fig. 5C), was able to make a large number of contacts with the fibrillar amyloid (Fig. 5), rendering a strong binding corona. On the other hand, available structures of the AE proteins (Fig. S10) included not only the strong amyloid-binding features observed in the AF proteins (Fig. S10B) but also globular proteins (e.g., the highly abundant serum albumin in Fig. S10A), the latter of which made less contacts and thus comparably weaker binding with amyloid fibrils (Figs. 5D,F) but still demonstrated a dynamic shift in secondary structure when its capacity for binding to IAPP fibrils was individually assessed (Fig. S12). The observed differential amyloid-binding features of proteins by different experimental methodologies are consistent with their corresponding approaches to prepare the amyloid coronae - i.e., the relatively turbulent condition under CC left the ‘hard’ corona intact, while the less rigorous isolation conditions of QCM resulted in both ‘soft’ and ‘hard’ coronae. Additionally, it is notable that almost all of the top AF and AE proteins were negatively charged (Tables S1&S2), while the net charges of IAPP fibrils are positive. Hence, as proposed in our previous study with model proteins,32 electrostatic attractions likely play an important role in concordance with structural morphology and conformational plasticity in the formation of amyloid coronae.
Figure 5. Schematic binding between IAPP amyloid fibril and AF/AE proteins with maximal contact interface.
The amyloid fibril is formed by β-sheets of composite peptides (grey). The binding of AF proteins – (A) alpha-actinin-4 (cyan), (B) protein AMBP (yellow) and (C) neuropilin in open (magenta) and closed (light pink) conformations – and AE proteins – (D) serum albumin (purple), (E) thrombospondin-1 (blue) and (F) cartilage oligomeric matrix protein (orange) – with the amyloid fibril was estimated by aligning them with maximum contact surface areas. Serum proteins are shown in cartoon format with dashed lines representing missing sequences without available structural information.
Conclusion
Until recently, the idea that endogenous nanomaterials form their own protein corona in vivo, independent of specific biological interactions that would define their inherent functionality, has been largely overlooked. This is particularly important for amyloids, as they are ubiquitous components of not only human biology but even single-celled bacteria.53 Collectively, the data of this study described the rapid generation of a largely heterogeneous coating of protein on the surface of pathogenic IAPP amyloid fibrils and plaques within a model environment, and demonstrated a multivariate array of amyloid-associative proteins pertaining to metabolic and cellular pathway functions. In addition, we have also introduced two optimized methodologies for streamlined isolation and identification of the amyloid-protein corona that produced different compositions, albeit with concordant aspects that lend weight not only to the results reported, but to the importance of considering multiple approaches for every research question addressed.
The association of a single protein with a given material has the potential to significantly impact its biological properties in vivo. For example, the human immunodeficiency virus (HIV) envelope protein Tat, secreted by virus-infected cells, has been shown to associate with mature Aβ amyloid fibrils and induce increased neurotoxicity comparative to either the amyloid or protein alone,54 demonstrating a secondary source to amyloid-associated pathologies. Native protein-amyloid interactions can be analogously deleterious: clusterin, for example, a protein with increased expression in Alzheimer’s disease, has been shown to co-localize with Aβ plaques in vivo, impacting both their localization and pathology.55
An amyloid-protein corona may have the capacity to become biologically dynamic; the β-sheet rich surface of amyloid structures has recently been shown to bind zinc ions and act as a potential catalyst for biochemical modification of interacting ligands, e.g. as an esterase.56,57 If modulation of known binding sites, and exposure of cryptic sequences, were to occur through the unfolding of protein bound to the amyloid surface, this may impact the targeting capacity of anti-amyloid agents, and further introduce immunoreactivity if surface-exposed moieties are recognized as foreign. Furthermore, an amyloid-protein corona may undergo protein exchange during translocation, which could potentially modulate amyloid-associated biological interactions through the association of protective or cytotoxic proteins enriched in different milieu. For example, highly biocompatible synthetic nanomaterials, e.g. graphene, have been found capable of cytotoxic activity under certain conditions,58,59 of which their capacity to add cytotoxic proteins, such as amyloid species,60 to a preformed corona upon exposure to a new environment may play a contributory role. The innate structural polymorphism of amyloid species36,61 could also contribute to the diversity of the formed amyloid-protein corona, and how it may then affect biological processes in the surrounding milieu.
The recent failure of Eli Lilly’s anti-amyloid antibody for treatment of AD during Phase III clinical trials62 underscores the need to pursue fundamental questions regarding amyloid-cell interactions, including the role of the protein corona on amyloid species in vivo. As such, the methodologies we have introduced can be expanded, in conjunction with single-molecule techniques such as liquid-phase atomic force microscopy,63,64 to assess corona formation (kinetics, topology and enriched proteome) of other pathological amyloid in human plasma and sera from different environments – and, furthermore, the mechanisms by which coronal proteins and other factors potentially found associated with amyloids (e.g. metal ions) specifically impact key pathways at the cellular interface – thus advancing our understanding of amyloid pathologies in major diseases and supporting the development of anti-amyloid strategies that have so far eluded success.
Methods
Materials
Lyophilized human islet amyloid polypeptide (IAPP) monomers (37-residue: KCNTATCATQRLANFLVHSSNNFGAILSSTNVGSNTY, disulfide bridge: 2–7; MW: 3,906; sourced from either AnaSpec or Abcam) were allowed to fibrillate in Milli-Q water (pH 6.5) for > 1 week at room temperature to allow complete conversion of monomers and lower order amyloid pathway species to mature amyloid fibrils. Stock concentrations of IAPP amyloids were either 200 μM (~ 0.8 mg/mL) or 256 μM (~ 1 mg/mL). Fetal bovine serum (FBS; charcoal stripped), sourced from Sigma Aldrich, was passed through a 0.45 μm filter (PFTE membrane) to remove large aggregates and stored as 1 mL aliquots at -20 °C. Concentrations of both IAPP amyloid and FBS stocks utilized during individual experiments were established utilizing a NanoPhotometer (NanoDrop) with built-in Implemen software, and then diluted accordingly to their respective experimental concentrations in 1× phosphate buffered saline (PBS; pH 7). DL-1,4-Dithiothreitol (DTT) was purchased from Acros Organics. Trifluoroacetic acid (TFA) was acquired through Alfa Aesar. Iodoacetamide (IAA), porcine trypsin and Dulbecco’s Modified Eagle’s Media (DMEM) were purchased from Sigma Aldrich. Human plasma was kindly donated by Joshua Glass.
Transmission electron microscopy
Carbon-coated formvar copper grids (400 mesh) were glow-discharged to promote hydrophilicity. Mature IAPP amyloids (final concentration: 0.1 mg/mL) were mixed with FBS at a 1:5 ratio by protein mass, and 10 μL aliquots were taken at 0 min (immediately), 5 min, 30 min, 2 h and 24 h, and allowed to absorb to the grid for 60 s before excess liquid was drawn off using filter paper. Grids washed in 10 μL Milli-Q water and liquid drawn off as previously. Samples were then double-stained with 1% aqueous uranyl acetate by touching the grids to one 5 μL droplet and then drawing liquid off immediately, before placing the grid on a second droplet for 15 s in the dark and subsequently removing excess stain. TEM images were taken on a Tecnai F20 transmission electron microscope (FEI) utilizing an UltraScan 1000 (2k × 2k) CCD camera (Gatan). IAPP amyloid fibril diameters were subsequently analyzed using Gatan Microscopy Suite software, with 100 points taken for each sample set assessed. Gaussian modeling of fibril diameter distribution (least squares fit) was applied using Prism (GraphPad).
Helium ion microscopy
Sample preparation: a suspension of 20,000 βTC6 cells in 150 μL DMEM (supplemented with 15% FBS) was incubated for 30 min at 37 oC with either water, IAPP amyloids (final applied concentration: 40 μM; ~ 0.16 mg/mL), 0.45 μm-filtered FBS (final applied concentration: 0.8 mg/mL) or IAPP amyloids pre-incubated with FBS (1:5 ratio by mass) for 2 h at room temperature. PFA was then added to a final concentration of 2.5% and cells fixed at 4 °C overnight. Cells were then pelleted at 1.0 g for 3 min, supernatant removed, pellet resuspended through gentle pipetting in 20% ethanol, and incubated for 1 h at room temperature. This sequence was then repeated, increasing ethanol concentration stepwise by 20% until 80% was reached, with one further step at 90% ethanol then performed. Samples were then applied to carbon tape atop a specimen stub, and allowed to dry completely before imaging. HIM imaging was undertaken on a Zeiss Orion NanoFab instrument.
Quartz crystal microbalance
QSense QSX 301 Gold quartz crystal sensors (Biolin) were incubated with either 0.1 mg/mL mature IAPP amyloids in PBS buffer (n = 4) or PBS buffer alone controls (n = 4) for 17 h at room temperature. Excess liquid was then removed and sensors placed into a temperature-controlled measurement chamber (Q-Sense Standard Flow module; 40 μL crystal surface interface volume) set at 25 °C. Experiments were run on a QSense E4 microbalance, utilizing QSoft 401 software to monitor the frequency shifts and resonance harmonics (recorded simultaneously at seven harmonics, two data points per second). Flow was controlled by an ISMATEC IPC high precision multichannel dispenser pump.
The conditions applied to sensors were as follows: sensors were flushed with 1× PBS buffer for 10 min at 300 μL/min prior to the start of the experiment to remove any unbound amyloid, before lowering flow rate to 100 μL/min for one hour to establish a stable baseline. Subsequently, sensors functionalized with IAPP amyloids were further passivated with IAPP amyloid solution (0.1 mg/mL) for 10 min at 100 μL/min and a further hour at 10 μL/min, in order to ensure amyloid coverage of the entire sensor surface. Controls were run with PBS during this time. Flow rate was returned to 100 μL/min for 5 min to equilibrate the flow chamber. Sensors were then washed with PBS at 100 μL/min for 30 min. FBS (0.5 mg/mL, in PBS) was passivated across both sample sets at 100 μL/min for 55 min, and then sets were again flushed with PBS for 50 min. Sensors were then removed from the flow chambers for on-chip tryptic digestion of amyloid- or sensor-bound protein.
For the presentation of frequency shift and dissipation shift data, the third harmonic was utilized from each sample set and one data point per minute displayed on the plot. For data presentation, experiments were normalized to zero as the starting value for each plot to provide a common baseline. Error is represented as standard error of mean (SEM; n = 4).
Protein corona isolation through centrifugal capture
VivaSpin 2 1,000 kDa spin columns were equilibrated with 1× PBS and centrifuged for 10 min at 6461 g on a SciQuip 2–16 centrifuge (Sigma). A 400 μL aliquot of IAPP amyloid (0.5 mg/mL), FBS (2.5 mg/mL; pre-filtered through 0.45 μm filter) or IAPP amyloids pre-incubated with FBS at a mass ratio of 1:5 for 2 h at room temperature was added to the spin column, mixed well, and then samples were centrifuged at 6461 g for 10 min. The filtrate was recovered from the bottom section of the apparatus, and the volume in the upper section was made up to 400 μL with 1× PBS, mixed well, and then centrifuged as previously. This process was repeated three additional times. The lower section of the spin column was then washed out with HPLC water to remove any remaining free protein prior to on-filter tryptic digestion of captured amyloid-bound protein.
‘Soft’ tryptic digestion of amyloid protein corona
To separate and digest corona proteins from captured amyloid, tryptic digestion was performed under mild conditions, hereby referred to as a ‘soft’ digestion. For amyloid-protein corona samples collected by the centrifugal capture method, all steps were carried out at the spin column filter interface; concordantly, for the QCM method, reagents were added sequentially to a liquid meniscus atop the sensor chip.
Captured amyloid on spin column filters and quartz crystal sensors was incubated with 0.1% RapiGest SF surfactant (Waters) in 50 mM NH4HCO3 at room temperature for 1 h. Disulfide reduction was carried out at room temperature for 1 h through addition of DTT (in 50 mM NH4HCO3) to a final concentration of 10 mM. Proteins were subsequently alkylated using IAA (in 50 mM NH4HCO3) over 15 min in the dark at room temperature, with the mixture gently agitated every 5 min. Reduced and alkylated corona proteins were washed from quartz crystal sensors using RapiGest and collected in Eppendorf tubes prior to tryptic digestion. Porcine trypsin (in 50 mM NH4HCO3) was added to an approximate final concentration ratio of 1:100 (trypsin : corona proteins) and proteins were digested for 13–17 h at 37 °C. Digested proteins from centrifugal capture samples were eluted from the spin column filter through centrifugation (5 min, 6461 g), and all digests were then quenched with TFA. Samples were then concentrated using a Savant SpeedVac Plus (model SC110AR) vacuum centrifuge and reconstituted in 2% acetonitrile (ACN), 0.1% TFA, for analysis using MS.
Nano liquid crystal electrospray tandem mass spectroscopy (nLC-ESI-MS-MS)
All samples were first desalted on C18 StageTips as described elsewhere,65 and approximately 10 μg of tryptic peptides solubilized in 50 μL 2% ACN/0.1% TFA were collected from all controls for proteomic analysis. Tryptic peptides were separated by reversed phase chromatography prior to mass spectrometry analysis, utilizing two columns: an Acclaim PepMap μ-precolumn cartridge 300 μm i.d. × 5 mm, 5 μm, 100 Å, and an Acclaim PepMap RSLC 75 μm × 50 cm, 2 μm, 100 Å (Thermo Scientific) installed on an Ultimate 3000 RSLCnano system (Dionex). Mobile phase buffer A was composed of 0.1% aqueous formic acid (FA) and mobile phase B was composed of 80% ACN containing 0.1% FA. Samples were loaded onto the μ-precolumn and equilibrated in 2% aqueous ACN containing 0.1% TFA for 8 min at 10 μL min−1, and peptides were subsequently eluted onto the analytical column at 250 nL min−1 by increasing the mobile phase B concentration from 8% B to 35% over 53 min then to 80% B over 2 min, followed by a 15 min re-equilibration at 4% B.
Eluting peptides were converted to gas-phase ions by means of electrospray ionization (ESI) and analyzed on a Thermo Orbitrap Fusion (Q-OT-qIT, Thermo Scientific). Survey scans of peptide precursors from 375 to 1500 m/z were performed at 120K resolution (at 200 m/z) with a 5 × 105 ion count target. Tandem MS was performed by isolation at 1.6 Th using the quadrupole, HCD fragmentation with normalized collision energy of 33, and rapid scan MS analysis in the ion trap. The MS-MS ion count target was set to 5 × 103 and the max injection time was 150 ms. Precursors with charge state 2–6 were selected and sampled for MS-MS. The dynamic exclusion duration was set to 35 s with a 10 ppm tolerance around the selected precursor and its isotopes. Monoisotopic precursor selection was turned on. The instrument was run in top speed mode with 2 s cycles.
nLC-MS/MS queries, analysis and informatics
Base peak and average ion chromatograms were created for each nLC-MS/MS analysis following conversion of *.RAW files to *.mzML (MSconvert, part of ProteoWizard v3.0.9248) using the R package xcms66 (see Supporting Information for script and chromatograms).
*.RAW files for each experiment were searched together using the inbuilt Andromeda engine in MaxQuant (v1.6.0.16).41 Contaminants considered: Yes, Enzyme: Trypsin/P, Fixed modifications: Carbamidomethylation, Variable modifications: N-acetylation, Methionine oxidation. Additional MaxQuant parameters are reported in the Supporting Information. The Bos taurus reference proteome dated 21/12/2017 (UP000009136_9913.fasta) was used for searches – except the repeat queries for experiment A (amyloid only CC), which used a Homo sapiens reference proteome dated 21/12/2017 (UP000005640_9606.fasta).
Reference proteome(s) source: ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/reference_proteomes/.
Summary.txt and proteinGroup.txt files from each MaxQuant search were combined using a custom R67 script, which is formatted as an Rmarkdown68 *.html report (available as Supporting Information, along with details of analysis and custom R scripts).
Unique protein ID lists for experimental subsets AE and AF were matched to STRING identifiers using the uniprot ID mapping tool (http://www.uniprot.org/uploadlists/) and these identifiers were submitted to the online STRING resource for protein network analysis.48,49 Experimental and database interactions were considered for a minimum interaction score of 0.400, with no additional interactors (only submitted list), against a whole genome background. Gene ontology (GO), pathway and protein domain enrichments were annotated using the online interface and downloaded as high resolution annotated protein network images for inclusion in this manuscript. Molecular action and evidence (functional association) networks were considered as indicated. Figures (Venn, pI/GRAVY/MW, networks) were formatted, annotated and re-exported using InkScape (v0.91, inkscape.org).
Structural analysis
For a given serum protein, the 3D structure information was extracted from the protein databank (PDB) by searching in the order of the whole sequence, the close homologs (e.g., human), or the sub-sequences. PDB IDs for top AE, AF and FBS proteins were listed in Tables S1, S2 & S3, respectively. The net charge of a given protein in Tables S1–3 was estimated by counting the number of basic and acidic amino acids under physiological conditions, i.e. Arg and Lys residues were assigned +1, Asp and Glu were assigned −1, while His was neutral. The corresponding 3D structures of AE and AF proteins were shown in Figs. S9&S10.
The IAPP fibril model was constructed with 80 IAPPs forming a two-layered fibril using the solid-state NMR-derived constraints,36 which was assigned a ~1.5 °C left-handed twist between consecutive IAPP beta-sheets according the experimentally identified fibril morphologies.69 The binding structure between a protein and the amyloid fibril in Fig. 5 was approximated by aligning the two molecules with maximum contacts.
Statistics
Where applicable, a one-way analysis of variance (ANOVA) test with Tukey’s correction for multiple comparisons was performed; for amyloid fibril diameter comparisons, n = 100, degrees of freedom = 699 (treatment = 693, residual = 6), F = 36.05. Data were considered significant when p < 0.05.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Table 1A.
Top 12 unique amyloid serum coronae (AF) proteins from the CC method.
| # | Uniprot ID | Protein name | Unique peptides |
|---|---|---|---|
| 1 | A5D7D1 | Alpha-actinin-4 | 22 |
| 2 | P41361 | Antithrombin-III | 15 |
| 3 | P00978 | Protein AMBP | 14 |
| 4 | E1BMX5 | Neuropilin | 13 |
| 5 | E1BEL6 | Neuropilin | 12 |
| 6 | F1MMT2 | Laminin subunit alpha 2 | 8 |
| 7 | P13213 | SPARC | 7 |
| 8 | F1MSA6 | Uncharacterized protein | 7 |
| 9 | F1MC48 | IQ motif containing GTPase activating protein 1 | 7 |
| 10 | F1MKG2 | Collagen type VI alpha 2 chain | 6 |
| 11 | F1MHP5 | Fms related tyrosine kinase 4 | 6 |
| 12 | F1MD77 | Laminin subunit gamma 1 | 6 |
Ranked by the number of unique peptides matched to sequences using an Andromeda database query.
Table 1B.
Top 12 unique amyloid serum coronae (AE) proteins from the QCM method.
| # | Uniprot ID | Protein name | Unique peptides |
|---|---|---|---|
| 1 | A0A140T897 | Serum albumin | 72 |
| 2 | F1N3A1 | Thrombospondin-1 | 40 |
| 3 | E1BGJ0 | LDL receptor related protein 1 | 36 |
| 4 | P07224 | Vitamin K-dependent protein S | 33 |
| 5 | F1MJK3 | Uncharacterized protein | 29 |
| 6 | F1MMP5 | Inter-alpha-trypsin inhibitor heavy chain H1 | 28 |
| 7 | P01267 | Thyroglobulin | 27 |
| 8 | E1BKL9 | Uncharacterized protein | 24 |
| 9 | Q0P569 | Nucleobindin-1 | 23 |
| 10 | P35445 | Cartilage oligomeric matrix protein | 23 |
| 11 | Q3SWX5 | Cadherin-6 | 19 |
| 12 | G3MYZ3 | Afamin | 19 |
Ranked by the number of unique peptides matched to sequences using an Andromeda database query.
Acknowledgments
This work was supported by ARC Project CE140100036 (Davis), NSF CAREER CBET 1553945 (Ding) and NIH MIRA R35GM119691 (Ding). Davis is thankful for an ARC Australian Laureate Fellowship. Pilkington acknowledges the support of an Australian Government RTP scholarship. TEM imaging was undertaken at the Bio21 Advanced Microscopy Facility. HIM imaging was performed at the UniMelb MCFP platform by B. Nasr. Mass spectrometry was undertaken at the Warwick Proteomics Research Technology Platform (UK).
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
TPD, PCK and PW conceived the project. EHP, OJRG, YX and PCK wrote the manuscript. EHP performed TEM/QCM/CC measurements and analyses; JHF and CZ performed nLC-MS/MS measurements; OJRG performed data analyses (nLC-MS/MS and protein network); YX and FD performed structural analysis; AK performed CD measurement and Blue-Native PAGE; AF performed cell culture.
Additional experimental data and methods, additional structural analysis, in-depth proteomic analyses and associated methods, MaxQuant data parameters, script for generation of full Rmarkdown html report, additional references. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cedervall T, Lynch I, Lindman S, Berggård T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc Natl Acad Sci USA. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ke PC, Lin S, Parak WJ, Davis TP, Caruso F. A Decade of the Protein Corona. ACS Nano. 2017;11:11773–11776. doi: 10.1021/acsnano.7b08008. [DOI] [PubMed] [Google Scholar]
- 3.Du Z, Gao N, Guan Y, Ding C, Sun Y, Ren J, Qu X. Rational Design of a “Sense and Treat” System to Target Amyloid Aggregates Related to Alzheimer’s Disease. Nano Res. 2018;11:1987–1997. [Google Scholar]
- 4.Zhou L, Chen Z, Dong K, Yin M, Ren J, Qu X. DNA-Mediated Construction of Hollow Upconversion Nanoparticles for Protein Harvesting and Near-Infrared Light Triggered Release. Adv Mater. 2014;26:2424–2430. doi: 10.1002/adma.201304437. [DOI] [PubMed] [Google Scholar]
- 5.Little WC, Schwartlander R, Smith ML, Gourdon D, Vogel V. Stretched Extracellular Matrix Proteins Turn Fouling and Are Functionally Rescued by the Chaperones Albumin and Casein. Nano Lett. 2009;9:4158–4167. doi: 10.1021/nl902365z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezzat K, Pernemalm M, Palsson S, Roberts TC, Jarver P, Dondalska A, Bestas B, Sobkowiak MJ, Levanen B, Skold M, Thompson EA, Wood MJA, Power UF, Masich S, Linden A, Sandberg JK, Lehtio J, Spetz A-L, Andaloussi SEL. [accessed Feb 6, 2018];The Viral Protein Corona Directs Viral Pathogenesis and Amyloid Aggregation. 2018 doi: 10.1038/s41467-019-10192-2. bioRxiv: 246785. bioRxiv.org e-Print archive. https://www.biorxiv.org/content/early/2018/01/16/246785. [DOI] [PMC free article] [PubMed]
- 7.Moghimi SM, Hunter AC, Murray JC. Long-Circulating and Target-Specific Nanoparticles: Theory to Practice. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 8.Ke PC, Sani MA, Ding F, Käkinen A, Javed I, Separovic F, Davis TP, Mezzenga R. Implications of Peptide Assemblies in Amyloid Diseases. Chem Soc Rev. 2017;46:6492–6531. doi: 10.1039/c7cs00372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional Amyloid Formation Within Mammalian Tissue. PLoS Biol. 2006;4:0100–0107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge X, Käkinen A, Gurzov EN, Pang L, Pilkington EH, Separovic F, Davis TP, Ke PC, Ding F. Zinc-Coordination and C-Peptide Complexation: A Potential Mechanism for the Endogenous Inhibition of IAPP Aggregation. Chem Commun. 2017;53:9394–9397. doi: 10.1039/c7cc04291d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz O, Brock B, Rungby J. Amylin Agonists: A Novel Approach in the Treatment of Diabetes. Diabetes. 2004;53:S233–S238. doi: 10.2337/diabetes.53.suppl_3.s233. [DOI] [PubMed] [Google Scholar]
- 12.Raleigh D, Zhang X, Hastoy B, Clark A. The β-Cell Assassin: IAPP Cytotoxicity. J Mol Endocrinol. 2017;59:R121–R140. doi: 10.1530/JME-17-0105. [DOI] [PubMed] [Google Scholar]
- 13.Kahn SE, Andrikopoulos S, Verchere CB. Islet Amyloid: A Long-Recognized But Underappreciated Pathological Feature of Type 2 Diabetes. Diabetes. 1999;48:241–253. doi: 10.2337/diabetes.48.2.241. [DOI] [PubMed] [Google Scholar]
- 14.Despa S, Margulies KB, Chen L, Knowlton AA, Havel PJ, Taegtmeyer H, Bers DM, Despa F. Hyperamylinemia Contributes to Cardiac Dysfunction in Obesity and Diabetes: A Study in Humans and Rats. Circ Res. 2012;110:598–608. doi: 10.1161/CIRCRESAHA.111.258285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong W, Liu ZH, Zeng CH, Peng A, Chen HP, Zhou H, Li LS. Amylin Deposition in the Kidney of Patients With Diabetic Nephropathy. Kidney Int. 2007;72:213–218. doi: 10.1038/sj.ki.5002305. [DOI] [PubMed] [Google Scholar]
- 16.Jackson K, Barisone GA, Diaz E, Jin LW, DeCarli C, Despa F. Amylin Deposition in the Brain: A Second Amyloid in Alzheimer Disease? Ann Neurol. 2013;74:517–526. doi: 10.1002/ana.23956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ly H, Despa F. Hyperamylinemia as a Risk Factor for Accelerated Cognitive Decline in Diabetes. Expert Rev Proteomics. 2015;12:575–577. doi: 10.1586/14789450.2015.1104251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartter E, Svoboda T, Ludvik B, Schuller M, Lell B, Kuenburg E, Brunnbauer M, Woloszczuk W, Prager R. Basal and Stimulated Plasma Levels of Pancreatic Amylin Indicate Its Co-Secretion With Insulin in Humans. Diabetologia. 1991;34:52–54. doi: 10.1007/BF00404025. [DOI] [PubMed] [Google Scholar]
- 19.Sanke T, Hanabusa T, Nakano Y, Oki C, Okai K, Nishimura S, Kondo M, Nanjo K. Plasma Islet Amyloid Polypeptide (Amylin) Levels and Their Responses to Oral Glucose in Type 2 (Non-Insulin-Dependent) Diabetic Patients. Diabetologia. 1991;34:129–132. doi: 10.1007/BF00500385. [DOI] [PubMed] [Google Scholar]
- 20.Jansen WJ, Ossenkoppele R, Knol DL, Tijms BM, Scheltens P, Verhey FRJ, Visser PJ The Amyloid Biomarker Study Group. Prevalence of Cerebral Amyloid Pathology in Persons Without Dementia: A Meta-Analysis. J Am Med Assoc. 2015;313:1924–1938. doi: 10.1001/jama.2015.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan P, Bero AW, Cirrito JR, Xiao Q, Hu X, Wang Y, Gonzales E, Holtzman DM, Lee JM. Characterizing the Appearance and Growth of Amyloid Plaques in APP/PS1 Mice. J Neurosci. 2009;29:10706–10714. doi: 10.1523/JNEUROSCI.2637-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertrand N, Grenier P, Mahmoudi M, Lima EM, Appel EA, Dormont F, Lim J-M, Karnik R, Langer R, Farokhzad OC. Mechanistic Understanding of In Vivo Protein Corona Formation on Polymeric Nanoparticles and Impact on Pharmacokinetics. Nat Commun. 2017;8(777) doi: 10.1038/s41467-017-00600-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors Affecting the Clearance and Biodistribution of Polymeric Nanoparticles. Mol Pharmaceutics. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westermark P, Skinner M, Cohen AS. The P-Component of Amyloid of Human Islets of Langerhans. Scand J Immunol. 1975;4:95–97. doi: 10.1111/j.1365-3083.1975.tb02604.x. [DOI] [PubMed] [Google Scholar]
- 25.Chargé SBP, Esiri MM, Bethune CA, Hansen BC, Clark A. Apolipoprotein E is Associated With Islet Amyloid and Other Amyloidoses: Implications for Alzheimer’s Disease. J Pathol. 1996;179:443–447. doi: 10.1002/(SICI)1096-9896(199608)179:4<443::AID-PATH612>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 26.Young ID, Ailles L, Narindrasorasak S, Tan R, Kisilevsky R. Localization of the Basement Membrane Heparan Sulfate Proteoglycan in Islet Amyloid Deposits in Type II Diabetes Mellitus. Arch Path Lab Med. 1992;116:951–954. [PubMed] [Google Scholar]
- 27.Jha S, Patil SM, Gibson J, Nelson CE, Alder NN, Alexandrescu AT. Mechanism of Amylin Fibrillization Enhancement by Heparin. J Biol Chem. 2011;286:22894–22904. doi: 10.1074/jbc.M110.215814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Wang L, Lu T, Wei Y, Li F. The Effects of Chondroitin Sulfate and Serum Albumin on the Fibrillation of Human Islet Amyloid Polypeptide at the Phospholipid Membranes. Phys Chem Chem Phys. 2016;18:12000–12008. doi: 10.1039/c5cp07642k. [DOI] [PubMed] [Google Scholar]
- 29.Oskarsson ME, Singh K, Wang J, Vlodavsky I, Li JP, Westermark GT. Heparan Sulfate Proteoglycans are important for Islet Amyloid Formation and Islet Amyloid Polypeptide-Induced Apoptosis. J Biol Chem. 2015;290:15121–15132. doi: 10.1074/jbc.M114.631697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vidal J, Verchere CB, Andrikopoulos S, Wang F, Hull RL, Cnop M, Olin KL, LeBoeuf RC, O’Brien KD, Chait A, Kahn SE. The Effect of Apolipoprotein E Deficiency on Islet Amyloid Deposition in Human Islet Amyloid Polypeptide Transgenic Mice. Diabetologia. 2003;46:71–79. doi: 10.1007/s00125-002-0984-5. [DOI] [PubMed] [Google Scholar]
- 31.Meng F, Abedini A, Song B, Raleigh DP. Amyloid Formation by Pro-Islet Amyloid Polypeptide Processing Intermediates: Examination of the Role of Protein Heparan Sulfate Interactions and Implications for Islet Amyloid Formation in Type 2 Diabetes. Biochemistry. 2007;46:12091–12099. doi: 10.1021/bi7004834. [DOI] [PubMed] [Google Scholar]
- 32.Pilkington EH, Xing Y, Wang B, Käkinen A, Wang M, Davis TP, Ding F, Ke PC. Effects of Protein Corona on IAPP Amyloid Aggregation, Fibril Remodelling, and Cytotoxicity. Sci Rep. 2017;7(2455) doi: 10.1038/s41598-017-02597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pisani C, Gaillard JC, Odorico M, Nyalosaso JL, Charnay C, Guari Y, Chopineau J, Devoisselle JM, Armengaud J, Prat O. The Timeline of Corona Formation Around Silica Nanocarriers Highlights the Role of the Protein Interactome. Nanoscale. 2017;9:1840–1851. doi: 10.1039/c6nr04765c. [DOI] [PubMed] [Google Scholar]
- 34.Jurgens CA, Toukatly MN, Fligner CL, Udayasankar J, Subramanian SL, Zraika S, Aston-Mourney K, Carr DB, Westermark P, Westermark GT. β-Cell Loss and β-Cell Apoptosis in Human Type 2 Diabetes Are Related to Islet Amyloid Deposition. Am J Pathol. 2011;178:2632–2640. doi: 10.1016/j.ajpath.2011.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandrescu AT. Amide Proton Solvent Protection in Amylin Fibrils Probed by Quenched Hydrogen Exchange NMR. PLoS ONE. 2013;8(e56467) doi: 10.1371/journal.pone.0056467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luca S, Yau WM, Leapman R, Tycko R. Peptide Conformation and Supramolecular Organization in Amylin Fibrils: Constraints From Solid-State NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sparr E, Engel MFM, Sakharov DV, Sprong M, Jacobs J, de Kruijff B, Höppener JWM, Killian JA. Islet Amyloid Polypeptide-Induced Membrane Leakage Involves Uptake of Lipids by Forming Amyloid Fibres. FEBS Lett. 2004;577:117–120. doi: 10.1016/j.febslet.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 38.Engel MFM, Khemtémourian L, Kleijer CC, Meeldijk HJD, Jacobs J, Verkleij AJ, de Kruijff B, Killian JA, Höppener JWM. Membrane Damage by Human Islet Amyloid Polypeptide Through Fibril Growth at the Membrane. Proc Natl Acad Sci USA. 2008;105:6033–6038. doi: 10.1073/pnas.0708354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilkington EH, Gurzov EN, Käkinen A, Litwak SA, Stanley WJ, Davis TP, Ke PC. Pancreatic β-Cell Membrane Fluidity and Toxicity Induced by Human Islet Amyloid Polypeptide Species. Sci Rep. 2016;6(21274) doi: 10.1038/srep21274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gianneli M, Yan Y, Polo E, Peiris D, Aastrup T, Dawson KA. Novel QCM-Based Method to Predict In Vivo behaviour of nanoparticles. Proc Technol. 2017;27:197–200. [Google Scholar]
- 41.Cox J, Mann M. MaxQuant Enables High Peptide Identification Rates, Individualized P.P.B.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 42.Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A Peptide Search Engine Integrated Into the MaxQuant Environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 43.Hadjidemetriou M, Al-Ahmady Z, Mazza M, Collins RF, Dawson K, Kostarelos K. In Vivo Biomolecule Corona Around Blood-Circulating, Clinically Used and Antibody-Targeted Lipid Bilayer Nanoscale Vesicles. ACS Nano. 2015;9:8142–8156. doi: 10.1021/acsnano.5b03300. [DOI] [PubMed] [Google Scholar]
- 44.Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, Wan W, Stubbs G, Schwieters CD, Lee VMY, George JM, Rienstra CM. Solid-State NMR Structure of a Pathogenic Fibril of Full-Length Human α-Synuclein. Nat Struc Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Docter D, Westmeier D, Markiewicz M, Stolte S, Knauer SK, Stauber RH. The Nanoparticle Biomolecule Corona: Lessons Learned – Challenge Accepted? Chem Soc Rev. 2015;44:6094–6121. doi: 10.1039/c5cs00217f. [DOI] [PubMed] [Google Scholar]
- 46.Saptarshi SR, Duschi A, Lopata AL. Interaction of nanoparticles with proteins: relation to bio-reactivity of the nanoparticle. J Nanobiotechnol. 2013;11(26) doi: 10.1186/1477-3155-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young LM, Mahood RA, Saunders JC, Tu LH, Raleigh DP, Radford SE, Ashcroft AE. Insights Into the Consequences of Co-Polymerisation in the Early Stages of IAPP and Aβ Peptide Assembly From Mass Spectrometry. Analyst. 2015;140:6990–6999. doi: 10.1039/c5an00865d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: Protein-Protein Interaction Networks, Integrated Over the Tree of Life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING Database in 2017: Quality-Controlled Protein–Protein Association Networks, Made Broadly Accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walkey CD, Chan CW. Understanding and Controlling the Interaction of Nanomaterials With Proteins in a Physiological Environment. Chem Soc Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 51.Oslakovic C, Cedervall T, Linse S, Dahlbäck B. Polystyrene Nanoparticles Affecting Blood Coagulation. Nanomedicine. 2012;8:981–986. doi: 10.1016/j.nano.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona With Possible Implications for Biological Impacts. Proc Natl Acad Sci USA. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Diversity, Biogenesis and Function of Microbial Amyloids. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hategan A, Bianchet MA, Steiner J, Karnaukhova E, Masliah E, Fields A, Lee MH, Dickens AM, Haughey N, Dimitriadis EK, Nath A. HIV Tat Protein and Amyloid-β Peptide Form Multifibrillar Structures That Cause Neurotoxicity. Nat Struc Mol Biol. 2017;24:379–386. doi: 10.1038/nsmb.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wojtas AM, Kang SS, Olley BM, Gatherer M, Shinohara M, Loranzo PA, Liu CC, Kurti A, Baker KE, Dickson DW, Yue M, Petrucelli L, Bu G, Carare RO, Fryer JD. Loss of Clusterin Shifts Amyloid Deposition to the Cerebrovasculature via Disruption of Perivascular Drainage Pathways. Proc Natl Acad Sci USA. 2017;114:E6962–E6971. doi: 10.1073/pnas.1701137114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Garawi ZS, McIntosh BA, Neill-Hall D, Hatimy AA, Sweet SM, Bagley MC, Serpell LC. The Amyloid Architecture Provides a Scaffold for Enzyme-Like Catalysts. Nanoscale. 2017;9:10773–10783. doi: 10.1039/c7nr02675g. [DOI] [PubMed] [Google Scholar]
- 57.Lee M, Wang T, Makhlynets OV, Wu Y, Polizzi NF, Wu H, Gosavi PM, Stöhr J, Korendovych IV, DeGrado WF, Hong M. Zinc-Binding Structure of a Catalytic Amyloid From Solid-State NMR. Proc Natl Acad Sci USA. 2017;114:6191–6196. doi: 10.1073/pnas.1706179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akhavan O, Ghaderi E, Akhavan A. Size-Dependent Genotoxicity of Graphene Nanoplatelets in Human Stem Cells. Biomaterials. 2012;33:8017–8025. doi: 10.1016/j.biomaterials.2012.07.040. [DOI] [PubMed] [Google Scholar]
- 59.Akhavan O, Ghaderi E, Hashemi E, Akbari E. Dose-Dependent Effects of Nanoscale Graphene Oxide on Reproduction Capability of Mammals. Carbon. 2015;95:309–317. [Google Scholar]
- 60.Mahmoudi M, Akhavan O, Ghavami M, Rezaee F, Ghiasi SMA. Graphene Oxide Strongly Inhibits Amyloid Beta Fibrillation. Nanoscale. 2012;4:7322–7325. doi: 10.1039/c2nr31657a. [DOI] [PubMed] [Google Scholar]
- 61.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR Provides Evidence for Mobile Water Molecules in β-Amyloid Fibrils. Proc Natl Acad Sci USA. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abbott A, Dolgin E. Leading Alzheimer’s Theory Survives Drug Failure. Nature. 2016;540:15–16. doi: 10.1038/nature.2016.21045. [DOI] [PubMed] [Google Scholar]
- 63.Yu YP, Zhang S, Liu Q, Li YM, Wang C, Besenbacher F, Dong M. 2D Amyloid Aggregation of Human Islet Amyloid Polypeptide at the Solid-Liquid Interface. Soft Matter. 2012;8:1616–1622. [Google Scholar]
- 64.Liu P, Zhang S, Chen MS, Liu Q, Wang C, Wang C, Li YM, Besenbacher F, Dong M. Co-Assembly of Human Islet Amyloid Polypeptide (hIAPP)/Insulin. Chem Commun. 2012;48:191–193. doi: 10.1039/c1cc14285b. [DOI] [PubMed] [Google Scholar]
- 65.Rappsilber J, Mann M, Ishihama Y. Protocol for Micro-Purification, Enrichment, Pre-Fractionation and Storage of Peptides for Proteomics Using StageTips. Nat Protoc. 2007;8:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 66.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 67.R Core Team. [accessed Feb 19: 2018];R: A Language and Environment for Statistical Computing. https://www.R-project.org/
- 68.Allaire JJ, Xie Y, McPherson J, Luraschi J, Ushey K, Atkins A, Wickham H, Cheng J, Chang W. [accessed Feb 19: 2018];Rmarkdown: Dynamic Documents for R. https://CRAN.R-project.org/package=rmarkdown.
- 69.Käkinen A, Adamcik J, Wang B, Ge X, Mezzenga R, Davis TP, Ding F, Ke PC. Nanoscale Inhibition of Polymorphic and Ambidextrous IAPP Amyloid Aggregation With Small Molecules. [accessed March 25, 2018];Nano Res. 2017 doi: 10.1007/s12274-017-1930-7. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.