Abstract
Expression of a heterologous xylose isomerase, deletion of the GRE3 aldose-reductase gene and overexpression of genes encoding xylulokinase (XKS1) and non-oxidative pentose-phosphate-pathway enzymes (RKI1, RPE1, TAL1, TKL1) enables aerobic growth of Saccharomyces cerevisiae on d-xylose. However, literature reports differ on whether anaerobic growth on d-xylose requires additional mutations. Here, CRISPR-Cas9-assisted reconstruction and physiological analysis confirmed an early report that this basic set of genetic modifications suffices to enable anaerobic growth on d-xylose in the CEN.PK genetic background. Strains that additionally carried overexpression cassettes for the transaldolase and transketolase paralogs NQM1 and TKL2 only exhibited anaerobic growth on d-xylose after a 7–10 day lag phase. This extended lag phase was eliminated by increasing inoculum concentrations from 0.02 to 0.2 g biomass L−1. Alternatively, a long lag phase could be prevented by sparging low-inoculum-density bioreactor cultures with a CO2/N2-mixture, thus mimicking initial CO2 concentrations in high-inoculum-density, nitrogen-sparged cultures, or by using l-aspartate instead of ammonium as nitrogen source. This study resolves apparent contradictions in the literature on the genetic interventions required for anaerobic growth of CEN.PK-derived strains on d-xylose. Additionally, it indicates the potential relevance of CO2 availability and anaplerotic carboxylation reactions for anaerobic growth of engineered S. cerevisiae strains on d-xylose.
Keywords: biofuels, carbon dioxide, lignocellulosic biomass, metabolic engineering, pentose metabolism
This study resolves apparent contradictions concerning the genetic interventions required for xylose fermentation by S. cerevisiae. Additionally, it indicates the potential relevance of CO2 and anaplerotic reactions for the latter.
INTRODUCTION
Over the past decades, major industrial and academic research efforts have been devoted to engineering Saccharomyces cerevisiae, a key microbial cell factory (Liu, Redden and Alper 2013; Nielsen et al.2013), for efficient conversion of lignocellulosic feedstocks into fuel ethanol, the largest-volume product of industrial biotechnology (reviewed by Alper and Stephanopoulos 2009; Young, Lee and Alper 2010; Moysés et al.2016; Jansen et al.2017). A large part of this effort was geared towards enabling S. cerevisiae strains to ferment d-xylose and l-arabinose, two pentoses that comprise a substantial fraction of the potentially fermentable sugars in lignocellulosic feedstocks (Lynd 1996; Olsson and Hahn-Hägerdal 1996; Moysés et al.2016; Jansen et al.2017). In lignocellulosic feedstocks, d-xylose is typically the most abundant sugar after glucose, often accounting for 10%–25% of the carbohydrate content (Lynd 1996). Wild-type S. cerevisiae strains can grow, albeit slowly, on the d-xylose isomer d-xylulose, whose metabolism is linked to glycolysis via the combined action of xylulokinase (Xks1) and the enzymes of the non-oxidative pentose-phosphate pathway (PPP; Wang and Schneider 1980; Hsiao et al.1982).
In naturally d-xylose-metabolizing, non-Saccharomyces yeasts such as Scheffersomyces stipitis (Toivola et al.1984), Candida shehatae (Toivola et al.1984), Pachysolen tannophilus (Smiley and Bolen 1982; Toivola et al.1984), Hansenula polymorpha (Ryabova, Chmil and Sibirny 2003) and Kluyveromyces lactis (Margaritis and Bajpai 1982), d-xylose is first converted into d-xylulose by the combined action of pyridine-nucleotide-dependent xylose reductases (XR) and xylitol dehydrogenases (XDH). Metabolic engineering strategies for enabling anaerobic fermentation of d-xylose by S. cerevisiae based on expression of heterologous XR/XDH genes, which continue to be intensively investigated, are complicated by the different redox cofactor preferences of these two oxido-reductases (Kötter and Ciriacy 1993; Hahn-Hägerdal et al.2001; Jeffries 2006).
An alternative metabolic engineering strategy is based on redox-cofactor-independent isomerisation of d-xylose to d-xylulose by a heterologously expressed xylose isomerase (XI). Implementation of this strategy was long hindered by insufficient functional expression of heterologous XI genes, under physiologically relevant conditions, in S. cerevisiae (Walfridsson et al.1996). This situation changed when Kuyper et al. (2003) demonstrated functional expression of the xylA gene of the anaerobic fungus Piromyces sp. E2 in S. cerevisiae (Kuyper et al.2003). Multi-copy overexpression of xylA, combined with overexpression of the native xylulokinase gene (XKS1) and the major paralogs encoding enzymes of the non-oxidative PPP (RKI1, RPE1, TAL1, TKL1), enabled fast aerobic growth of S. cerevisiae on d-xylose (Kuyper et al.2005). This result has since been reproduced, both with xylA and with other heterologous XI genes, and in different S. cerevisiae genetic backgrounds (Table 1). In many studies, this metabolic engineering strategy was combined with deletion of GRE3, which encodes a non-specific aldose reductase whose activity could lead to loss of carbon to xylitol (Träff et al.2001; Kuyper et al.2003; Kuyper et al.2005; Lee, Jellison and Alper 2012). Moreover, accumulation of xylitol, a known inhibitor of XI enzymes (Yamanaka 1969), might inhibit heterologously expressed XI activity.
Table 1.
Literature data on anaerobic growth of metabolically engineered, xylose-isomerase-based S. cerevisiae strains. The table summarises sets of targeted genetic modifications, aerobic-specific growth rates on d-xylose and any additional optimisation by laboratory evolution or mutagenesis required for anaerobic growth on d-xylose. NIA = no information available; * specific growth rate estimated from exponential increase of CO2 concentration in bioreactor off gas.
| Strain background | Strain | XI gene | Native genes overexpressed | Other targeted modifications | Aerobic growth rate (h−1) | Anaerobic growth | Reference |
|---|---|---|---|---|---|---|---|
| CEN.PK | RWB202 | Piromyces XylA (2 micron plasmid) | none | No | 0.005 | After extensive aerobic, oxygen-limited and anaerobic selection (μ = 0.03 h−1) | (Kuyper et al.2003) |
| CEN.PK | RWB217 | Piromyces XylA (2 micron plasmid) | RKI1, RPE1, TAL1, TKL1 (all pTPI1), XKS1 (pADH1) | gre3Δ | 0.22 | Anaerobic growth after ca. 35 h when inoculated at low cell densities (0.02 g biomass L−1, μ = 0.09 h−1). Immediate anaerobic growth when inoculated at high biomass concentration (0.2 g biomass L−1, μ = 0.09 h−1). | (Kuyper et al.2005), This study |
| CEN.PK | YEp-opt.XI-Clos-K | Clostridium phytofermentans, codon optimised | XKS1, RKI1, RPE1, TAL1, TKL1 | GAL2 overexpression | 0.057 | No | (Brat et al.2009) |
| CEN.PK | YEp-opt.XI-Piro | Piromyces XylA, codon-optimised | XKS1, RKI1, RPE1, TAL1, TKL1 | GAL2 overexpression | 0.056 | No | (Brat et al.2009) |
| BarraGrande (Industrial) | BWY10Xyl | Clostridium phytofermentans, codon optimised | NIA | NIA | 0.04 | Serial aerobic shake flask cultures (6) on d-xylose until anaerobic xylose consumption observed upon aerobic biomass production. | (Brat et al.2009) |
| CEN.PK | IMX696 | Piromyces XylA, codon-optimised | XKS1, RKI1, RPE1, TAL1, TKL1, NQM1, TKL2 | gre3Δ | 0.21 | After 12-day anaerobic adaptation phase (mutations in PMR1) | (Verhoeven et al.2017) |
| Ethanol Red (diploid) | HDY.GUF5 | Clostridium phytofermentans, codon-optimised | XKS1, RKI1, RPE1, TAL1, TKL1, TKL2, NQM1 | HXT7, S. stipitis AraA, B. licheniformis AraA, E. coli AraD, AraB | NIA | After extensive mutagenesis-, genome-shuffling and oxygen-limited selection experiments (no specific growth rates reported) | (Demeke et al.2013) |
| Natural isolate (banana) YB-210/GLBRCY0 | Y22–3 (haploid spore) | Clostridium phytofermentans XylA (ScTDH3p) | - | S. cerevisiae TAL1, S. stipitis XYL3 | NIA | No initial anaerobic growth observed. Aerobic selection in glucose–xylose media (34 transfers), anaerobic selection on same media (14 transfers). Evolved strain showed anaerobic growth in YPX medium (mutation in GRE3 obtained). | (Parreiras et al.2014) |
| BF264–15Dau (Sun et al.1989) | H131-A3 | Piromyces XylA, codon-optimised (2 micron plasmid) | RPE1, RKI1, TKL1 | P. stipitis XKS1& TAL1 | 0.031 ± 0.022 | Aerobic cultivation in anaerobic sequential batch reactors (SBRs) on SMX (2% xylose; ca. 70 transfers), transfer to microaerobic SBRs (YNBX, 60 transfers), transfer to anaerobic SBRs (YNBX, 60 transfers), transfer to anaerobic chemostat with increasing dilution rate over time for ca. 60 generation (YNBX, d = 0.02 h−1 to 0.12 h−1), 20 more generations with YNBX with 10% xylose until dilution rate of 0.148 h−1. | (Zhou et al.2012) |
| CEN.PK | TMB3361 | Piromyces XylA (2 micron plasmid) | TAL1, TKL1, RPE1, RKI1, XKS1 (all pPGK1) | E. coli XK (xylB), gre3Δ | 0.089 ± 0.002 | Anaerobic fermentations inoculated with very high cell densities (5 g L−1 CDW) resulting in partial conversion of the supplied xylose to ethanol (without an adaptation time) but without measurable growth (due to high initial cell densities). | (Parachin et al.2011) |
| CEN.PK | YRH631 (naive), YRH1114 (evolved) | Prevotella ruminicola TC2–24 XI (codon-optimised) | XKS1 | No | 0.06 (naive) 0.23 (evolved) | Six transfers in microaerobic conditions (μ unknown). | (Hector et al.2013) |
| INVSc1 (Invitrogen, USA) | INVSc1/pRS406XKS/pILSUT1/pWOXYLA (XKS, Sut1, XylA) | Orpinomyces xylA (2 μm plasmid) | XKS1 | P. stipitis SUT1 over-expression | NIA | CO2-flushed bottles inoculated with 5 g biomass L−1 showed consumption of 15.5 g L−1 xylose from a total of 50 g L−1 within 140 h. | (Madhavan et al.2009) |
| CEN.PK | TMB3066 | Piromyces XylA (2 μm plasmid) | TAL1, TKL1, RPE1, RKI1, XKS1 (all pPGK1 | gre3Δ | 0.02 | Anaerobic cultures resulting in partial conversion of the supplied xylose to ethanol (16.8 g of 50 g L−1 within 100 h, without an adaptation time) at high biomass density, no anaerobic growth reported. | (Karhumaa et al.2007) |
| CEN.PK | IMU078 | Piromyces XylA (2 μm plasmid) | TAL1, TKL1, RPE1, RKI1, NQM1, TKL2, XKS1 | gre3Δ | NIA | Anaerobic growth after ca. 7–8 d when inoculated at low biomass concentration (0.02 g biomass L−1), μ = ca. 0.09 h−1*. Immediate anaerobic growth when (i) inoculated at high biomass density (0.2 g biomass L−1, μ = 0.05 h−1*), (ii) upon supplementation with 0.1% CO2 in N2 used from sparging of bioreactors (μ = 0.05 h−1) or (iii) when l-aspartate is supplied as nitrogen source (μ = 0.05 h−1). | This study |
| CEN.PK | IMU079 | Piromyces XylA (2 μm plasmid) | TAL1, TKL1, RPE1, RKI1, XKS1 | gre3Δ | NIA | Anaerobic growth after ca. 40 h when inoculated at low cell densities (0.02 g biomass L−1, μ = 0.08 h−1). Immediate anaerobic growth when inoculated at high biomass concentration (0.2 g biomass L−1, μ = 0.07 h−1*). | This study |
| PE-2 | LVY27 LVY34.4 (evolved) LVY41.5 (evolved) | Orpinomyces sp. xylA (codon-optimised; flanked with δ LTR sequences for high copy integration) | XKS1*2, TAL1, RKI1, TKL1, RPE1 | gre3Δ | Very slow growth with 1 copy of XylA. Evolved: μ = 0.23 and 0.129 h−1 | No anaerobic growth upon integration of one copy of xylA. Selection in semi-anaerobic conditions with 5 g L−1 glucose and 40 g L−1 xylose (12 transfers). Faster growth upon selection for increased xylA copy numbers, resulting in 36 and 26 copies. | (dos Santos et al.2016) |
| BY4741 | BY4741-S2A3K | Mutated Piromyces xylA3* (2 μm plasmid) | XKS1 | gre3Δ, S. stipitis TAL1 over-expression | 0.061 h−1 | Xylose fermentation possible in high cell density, micro-aerobic conditions (no growth rates available). | (Lee et al.2012) |
| BY4741 | SXA-R2P | Mutated Piromyces xylA3* (2 copies; Lee et al.2012) | XKS1 | gre3Δ, pho13 Δ, S. stipitis TAL1 over-expression (2 copies) | 0.105 h−1 and 0.128 h−1 (evolved) | Naïve strain slowly consumed xylose in microaerobic conditions. Adaptive evolution in closed falcon tubes with media containing 20 g L−1 xylose (12 transfers). Evolved strain was capable of fast xylose consumption when inoculated at high biomass concentration in non-purged anaerobic bioreactors where initial oxygen was consumed within 12 h (no growth rates available). | (Lee et al.2014) |
| CEN.PK | BSPC095 | Piromyces xylA (2 μm plasmid) | TAL1, TKL1, RPE1, RKI1, XKS1 | gre3Δ, cox4Δ, | No initial aerobic growth | Weak aerobic growth observed in liquid xylose medium upon 10 days of aerobic incubation. Serial transfers of aerobic cultures with xylose during 1000 h resulted in aerobic growth rate of, μ = 0.11 h−1. | (Shen et al.2012) |
The literature is entirely consistent where it concerns aerobic growth on d-xylose of XI-based S. cerevisiae strains that carry the abovementioned genetic modifications. Conversely, reports on the ability of such strains to grow anaerobically on d-xylose appear to be conflicting. In their original paper, Kuyper et al. (2005) reported that S. cerevisiae RWB217, which was constructed in the CEN.PK genetic background (Entian and Kötter 2007) and harboured the complete set of genetic modifications described above as well as a gre3Δ mutation, grew anaerobically on d-xylose in synthetic media without a need for additional mutagenesis or laboratory evolution (Kuyper et al.2005). However, when other groups constructed similar S. cerevisiae strains, anaerobic growth on d-xylose was reported to require additional laboratory evolution, mutagenesis and/or genetic engineering (Table 1). Until recently, the different reported anaerobic growth performances of engineered S. cerevisiae strains might have been attributed to differences in strain background and/or XI genes (Table 1). However, the recent single-step, CRISPR-Cas9-mediated construction of a d-xylose-metabolizing strain in the CEN.PK background, based on Piromyces xylA, yielded a strain that showed instantaneous, fast aerobic growth on d-xylose but reproducibly required a 12 day adaptation period before growth on d-xylose in anaerobic bioreactor cultures set in (Verhoeven et al.2017). Whole-genome resequencing showed that this adaptation reflected selection for mutants carrying mutations in PMR1, which encodes a Golgi Mn2+/Ca2+ ATPase. These mutations increased cellular contents of Mn2+, the preferred metal cofactor of XylA (Lee et al.2017; Verhoeven et al.2017). Since Verhoeven et al. (2017) used the same S. cerevisiae genetic background and XI gene as Kuyper et al. (2005), and, moreover, also overexpressed XKS1 and PPP genes while deleting GRE3 (Table 2), their study raised questions on the genetic requirements for anaerobic d-xylose metabolism by CEN.PK-derived strains (Kuyper et al.2005; Verhoeven et al.2017).
Table 2.
Saccharomyces cerevisiae strains used in this study.
| Strain | Relevant genotype | Description | Reference |
|---|---|---|---|
| CEN.PK113-7D | MATa | Reference strain | (Entian and Kötter 2007) |
| CEN.PK113-5D | MATa ura3-52 | Uracil auxotrophic reference strain | (Entian and Kötter 2007) |
| CEN.PK102-3A | MATA ura3-52 leu2-112 | Uracil and leucine auxotrophic strain | (van Dijken et al.2000) |
| RWB217 | CEN.PK102-3A loxP-pTPI::(−266, −1)TAL1 gre3Δ::hphMX pUGpTPI-TKL1 pUGpTPI–RPE1 KanloxP–pTPI::(−40, −1)RKI1 {pAKX002, p415ADHXKS} | Metabolically engineered, non-evolved xylose consuming strain | (Kuyper et al.2005) |
| IMX975 | RWB217 can1::CAS9-tagA-loxP-natNT2-loxP | RWB217 expressing Cas9 | This study |
| IMX1366 | IMX975, sga1::pPGI1-NQM-TagB-pPYK1-TKL2 | IMX975 over-expressing NQM1 and TKL2 | This study |
| IMX581 | CEN.PK113–5D can1::CAS9-tagA-loxP-natNT2-loxP | CEN.PK113–5D expressing S. pyogenes Cas9 | (Mans et al.2015) |
| IMX696 | IMX581 gre3::pTDH3-RPE1- pPGK1-TKL1 – pTEF1-TAL1- pPGI1-NQM1 – pTPI1-RKI1 –pPYK1-TKL2-(pTPI1-xylA-tCYC1)*36)-pTEF-XKS1, pUDE335 | IMX581 over-expressing PPP genes incl. NQM1 and TKL2, expression of Piromyces xylA based on 36 integrated copies | (Verhoeven et al.2017) |
| IMX994 | IMX581 gre3::pTDH3-RPE1-tagH-pPGK1-TKL1-TagI-pTEF1-TAL1-TagA- pTPI1-RKI1-TagL- pTEF-XKS1 | IMX581 over-expressing genes from the non-oxidative pentose phosphate pathway | (Papapetridis et al.2018) |
| IMU079 | IMX994, pAKX002 (2 μm xylA) | IMX994 over-expressing xylA from a 2 μm plasmid | (Papapetridis et al.2018) |
| IMX1456 | IMX994, sga1::pPGI1-NQM-TagB-pPYK1-TKL2 | IMX994 over-expressing NQM1 and TKL2 | This study |
| IMU081 | IMX1456, pAKX002 (2 μm xylA) | IMX1456 over-expressing xylA from a 2 μm plasmid | This study |
| IMX800 | IMX581, gre3::pTDH3-RPE1-TagH-pPGK1-TKL1-TagI-pTEF1-TAL1-TagA-pPGI1-NQM1-TagB- pTPI1-RKI1-TagC-pPYK1-TKL2-TagL- pTEF-XKS1 | IMX581 over-expressing PPP genes incl. NQM1 and TKL2 | This study |
| IMU078 | IMX800, pAKX002 (2 μm xylA) | IMX800 over-expressing xylA from a 2 μm plasmid | This study |
| IMX1736 | IMU078, pck1Δ | IMU078 with a deletion in PCK1 | This study |
The goal of the present study was to critically re-examine and reproduce the genetic modifications required for anaerobic growth of S. cerevisiae on d-xylose reported by Kuyper et al. (2005) and Verhoeven et al. (2017). To this end, we constructed new engineered, d-xylose-fermenting strains to investigate the impact of subtle differences in strain construction strategies applied in the two studies. Subsequently, we compared growth of the resulting strains on d-xylose in anaerobic bioreactors, with special attention to the impact of inoculum density, initial carbon dioxide concentration and nitrogen source on anaerobic growth.
MATERIALS AND METHODS
Yeast strains and maintenance
All Saccharomycescerevisiae strains used and constructed in this study belong to the CEN.PK lineage (Entian and Kötter 2007; Nijkamp et al.2012; Salazar et al.2017). Yeast cultures were grown on synthetic medium (SM, prepared and sterilised as described previously (Verduyn et al.1992)) or YP medium (10 g L−1 Bacto yeast extract (Becton Dickinson, Sparks, MD)), 20 g L−1 Bacto peptone (Becton Dickinson); autoclaved at 121°C for 20 min. SM was supplemented with 1 mL L−1 filter-sterilised vitamin solution (Verduyn et al.1992). Concentrated sterile d-glucose or d-xylose solutions (autoclaved at 110°C for 30 min) were added to SM and YP media to a concentration of 20 g L−1, resulting in SMD or SMX and YPD or YPX, respectively. Shake-flask cultures were grown in 500-mL round-bottom flasks containing 100 mL medium and incubated in an Innova incubator (Brunswick Scientific, Edison, NJ) at 30°C and 200 rpm. Solid media contained 2% (w/v) Bacto agar (Becton Dickinson). Escherichia coli strains were grown in LB-ampicillin medium (10 g L−1 Bacto tryptone, 5 g L−1 Bacto yeast extract, 5 g L−1 NaCl, 100 mg L−1 ampicillin). For storage, sterile glycerol was added to stationary-phase cultures of yeast and E. coli to a final concentration of 30% (v/v), after which aliquots were stored at −80°C.
Molecular biology
Analytical PCR was performed with Dreamtaq polymerase (Thermo Scientific, Waltham, MA) according to the manufacturer's instructions. DNA fragments for cloning were amplified with Phusion Hot Start II High Fidelity Polymerase (Thermo Scientific, Waltham, MA) and with the desalted or PAGE-purified oligonucleotide primers (Sigma-Aldrich, St. Louis, MO) listed in Table S1, Supporting Information. DNA fragments were separated by gel electrophoresis (90 V, 35 min) on 1% (w/v) agarose gels (Thermo Scientific) buffered with 1× TAE (Thermo Scientific). When required, DNA fragments were excised from gels and purified with a Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA) or, when no unspecific products were detected, directly purified from the PCR mix with a GenElute PCR Clean-Up Kit (Sigma-Aldrich). Yeast genomic DNA was extracted with a YeaStar Genomic DNA kit (Zymo Research) and DNA for diagnostic colony PCR was extracted by boiling cells picked from colonies in 10 μL 0.2 N NaOH for 5 min. After removal of debris by centrifugation (1 min at 2000 × g), 2 μL of the supernatant was used as template in a 20 μL PCR reaction (Dreamtaq). Yeast and E. coli plasmids were extracted with a Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research, Irvine, CA) and with a Sigma GenElute Plasmid kit (Sigma-Aldrich), respectively. Yeast transformation was carried out with the lithium-acetate method (Gietz and Woods 2002). When natNT2 was used as a marker gene, the transformation procedure included an overnight incubation step in non-selective liquid media (SMG) prior to selection on solid SMG plates containing 1 g L−1 glutamic acid as the sole nitrogen source, which were supplemented with 100 mg L−1 nourseothricin (pH 6). Single-colony isolates were obtained by three consecutive re-streaks on solid selective medium and genotype confirmation by analytical PCR. Escherichiacoli DH5α was used for chemical transformation (Inoue, Nojima and Okayama 1990) or electroporation in 2 mm cuvettes (165-2086, Bio-Rad, Hercules, CA) using a Gene Pulser Xcell Electroporation system (Biorad). Isolated plasmids were routinely checked by analytical PCR and restriction analysis.
Strain and plasmid construction
Detailed genotypes of all strains and plasmids used or generated in this study are listed in Tables 2 and 3, respectively. Genomic DNA of S. cerevisiae CEN.PK113-7D was used as a template for amplification of S. cerevisiae gene-, promoter- and terminator fragments. The CEN.PK strain lineage and construction of the derived S. cerevisiae strains RWB217, IMX581, IMX994 and IMU079 are described elsewhere (Kuyper et al.2005; Entian and Kötter 2007; Mans et al.2015; Papapetridis et al.2018). Integration of overexpression cassettes for NQM1 and TKL2 (pPGI1-NQM1-TagB-pPYK1-TKL2) was achieved by targeted integration into IMX994 using 60 bp flanks homologous to SGA1. Fragments were obtained by amplification of the SGA1flank-pPGI1-NQM1-TagB-cassette from pUD344 using primers 11357 and 3276 and amplification of the TagB-pPYK1-TKL2-SGA1flank-cassette from pUD346 using primer pair 11356 and 7607. Both fragments were co-transformed with pUDR103, a plasmid expressing a gRNA targeting SGA1, into IMX994. pUDR103 was subsequently counter selected by non-selective growth in YPD medium followed by plating on solid SMG plates supplemented with uracil and 5-fluoroorotic acid (0.15 and 1 g L−1 final concentration, respectively), resulting in strain IMX1456 (IMX994 sga1::NQM1, TKL2). This strain was subsequently transformed with the xylA plasmid pAKX002, resulting in strain IMU081. Construction of strain IMX800 (IMX581 gre3::RPE1, TKL1, NQM1, RKI1, TKL2, XKS1) was similar to the construction of strain IMX994 (IMX581 gre3::RPE1, TKL1, TAL1, RKI1, XKS1; Papapetridis et al.2018) with the difference that, in the former strain, the chromosomally integrated cluster of overexpression cassettes for pentose-phosphate-pathway genes included NQM1 and TKL2. The expression cassette for TAL1 was amplified from pUD349 with primer pair 3274 and 3275, yielding the tag-flanked expression cassette TagI-pTEF1-TAL1-TagA; NQM1 was amplified from pUD344 with primers 3847 and 3276 to yield the expression cassette TagA-pPGI1-NQM1-TagB; the TagB-pTPI1-RKI1-TagC cassette was amplified from pUD345 with primers 4672 and 3277; the TagC-pPYK1-TKL2-TagL cassette was amplified from pUD346 with primers 3283 and 8285 and the TagL-pTEF-XKS1-GRE3flank cassette was amplified from pUD353 with the primer pair 7222 and 7135. In overexpression cassettes, S. cerevisiae genes retained their endogenous terminators. Transformation of strain IMX800 with the high-copy-number xylA plasmid pAKX002, which was isolated from S. cerevisiae RWB217 (Kuyper et al.2005), yielded strain IMU078. PCK1 was deleted in strain IMU078 according to the protocol described by Mans et al. (2015), resulting in strain IMX1736 (Mans et al.2015). Two plasmid fragments were transformed directly into IMU078 for in vivo assembly: a 2 μm fragment amplified from pROS13 with a double-binding primer adding a gRNA flank guiding a cut in PCK1 (primer 14234) and a backbone harbouring a kanMX marker amplified from pROS13 using the double-binding primer 6005. Plasmid fragments were transformed together with an annealed double-strand repair fragment (oligonucleotide 14235 and 14236) homologous to 60 bp up- and downstream of PCK1 to delete the ORF. Mutants were selected on solid YPD plates supplemented with 200 mg L−1 G418. Primer pair 14237 and 14238 was used to screen for positive colonies. A positive colony was grown in non-SMG, plated on solid SMG and subsequently replica-plated on solid SMG and YPD-G418 plates to check for successful plasmid loss. Cells from a colony that lost the G418-marker harbouring plasmid were double checked for a pck1Δ genotype and subsequently stocked as strain IMX1736. To enable CRISPR-Cas9-based editing of RWB217, a cas9-cassette with a can1- and a TagA- overhang amplified from p414 (DiCarlo et al.2013) with primers 2873 and 3093 and a natNT2 cassette with the same tags, amplified from pROS15 (Mans et al.2015) with primers 4653 and 5542 was integrated into the CAN1 locus by homologous recombination, resulting in strain IMX975. Integration of sga1::NQM1, TKL2 into IMX975, resulting in strain IMX1366 (IMX975 sag1::NQM1, TKL2) was done as described for strain IMU081 with the difference that the gRNA plasmid pUDR119 with an amdSYM marker was used and no pAKX002 plasmid had to be transformed due to its presence in IMX975. Selection and counter selection of the amdSYM marker cassette were performed as described previously (Solis-Escalante et al.2013). PMR1 genes, including promoters and terminators, were amplified from duplicate 200 h anaerobic bioreactor cultures of strain IMU078, with primer pair 8790 and 8791 and subsequently Sanger sequenced (BaseClear, Leiden, The Netherlands) with primers 8790, 8791 and 13459–13475.
Table 3.
Plasmids used in this study.
| Name | Relevant characteristics | Reference |
|---|---|---|
| pROS15 | 2 μm ampR natNT2 gRNA-CAN1.Y gRNA-ADE2.Y | (Mans et al.2015) |
| pROS11 | 2 μm ampR amdSYM gRNA-CAN1.Y gRNA-ADE2.Y | (Mans et al.2015) |
| pROS13 | 2 μm ampR kanMX gRNA-CAN1.Y gRNA-ADE2.Y | (Mans et al.2015) |
| pAKX002 | 2 μm, URA3, pTPI1-XylA (Piromyces spp. E2) | (Kuyper et al.2003) |
| p414 -pTEF1-Cas9-tCYC1 | CEN6/ARS4 ampR pTEF1-cas9-tCYC1 | (DiCarlo et al.2013) |
| pMEL10 | 2 μm, KlURA3, pSNR52-gRNA.CAN1.Y-tSUP4 | (Mans et al.2015) |
| pJET1.2Blunt | Multi-purpose cloning vector for storage of assembled cassettes | ThermoFisher |
| pUD344 | pJET1.2Blunt TagA-pPGI1-NQM1-tNQM1-TagB | (Verhoeven et al.2017) |
| pUD345 | pJET1.2Blunt TagB-pTPI1-RKI1-tRKI1-TagC | (Verhoeven et al.2017) |
| pUD346 | pJET1.2Blunt TagC-pPYK1-TKL2-tTKL2-TagF | (Verhoeven et al.2017) |
| pUD347 | pJET1.2Blunt TagG-pTDH3-RPE1-tRPE1-TagH | (Verhoeven et al.2017) |
| pUD348 | pJET1.2Blunt TagH-pPGK1-TKL1-tTKL1-TagI | (Verhoeven et al.2017) |
| pUD349 | pJET1.2Blunt TagI-pTEF1-TAL1-tTAL1-TagA | (Verhoeven et al.2017) |
| pUD350 | pJET1.2Blunt pTPI1-XylA-tCYC1 | (Verhoeven et al.2017) |
| pUD353 | pJET1.2Blunt pTEF1-XKS1-tXKS1 | (Verhoeven et al.2017) |
| pUDE335 | 2 μm ori, KlURA3, pSNR52-gRNA.GRE3.Y-tSUP4 | (Verhoeven et al.2017) |
| pUDR119 | 2 μm, amdSYM, pSNR52-gRNA.SGA1.Y-tSUP4 | (van Rossum et al.2016) |
| pUDR103 | 2 μm, KlURA3, pSNR52-gRNA.SGA1.Y-tSUP4 | (Papapetridis et al.2017) |
Bioreactor cultivation
Saccharomyces cerevisiae strains were physiologically characterised in anaerobic 2 L laboratory bioreactors (Applikon, Delft, The Netherlands) with a 1 L working volume. Bioreactors filled with SM were autoclaved at 121°C for 20 min. SM with l-aspartate as nitrogen source was prepared as described previously (Zelle et al.2010) and filter sterilised (Nalgene Rapid-Flow, 0.2 μm, Thermo Scientific) prior to addition to autoclaved bioreactors. Separately prepared solutions were subsequently added to final concentrations of 20 g L−1d-xylose, 1 mL L−1 vitamin solution, 0.2 g L−1 antifoam C, as well as 10 and 420 mg L−1 of the anaerobic growth factors ergosterol and Tween-80, respectively. Cultures were sparged with nitrogen (0.5 L min−1) to maintain anaerobic conditions. For physiological characterisation of strain IMU078 with CO2 supplementation, nitrogen as a sparging gas was replaced by an analytically certified (2% tolerance) mixture of 99.9% N2 and 0.1% CO2 (Linde Group, Munich, Germany). Oxygen diffusion into bioreactors was minimised by equipping them with Norprene tubing (Saint-Gobain, Courbevoie, France) and Viton O-rings (Eriks, Alkmaar, The Netherlands). Evaporation was minimised by cooling the outlet gas to 4°C. Cultures were stirred at 800 rpm, maintained at pH 5 by automatic addition of 2 M KOH and kept at 30°C. Separate inocula were prepared for each bioreactor. These inocula were obtained by growth of the respective strain, starting with a frozen stock culture, in two consecutive aerobic 100 mL shake flask cultures on SMX, at 30°C and 200 rpm. When the OD660 of the pre-cultures was between 3.5 and 5.5, they were used to inoculate the anaerobic bioreactors. The initial OD660 of bioreactor cultures was either 0.1 or 1 (0.02 or 0.2 g biomass L−1, respectively) as indicated.
Analytical techniques
Biomass dry weight measurements and optical density measurements at a wavelength of 660 nm were performed as described previously (Verhoeven et al.2017). A correlation between biomass dry weight concentration and optical density, based on at least 10 measurement points during the exponential growth phase, was used to estimate the biomass concentrations for the first few bioreactor culture samples, when the cell density was too low to allow for accurate biomass dry weight determinations (OD660 < 0.9). Specific growth rates were calculated from at least six biomass dry-weight measurements during the exponential growth phase. Concentrations of biomass, ethanol, glycerol and d-xylose, at the same sampling times, were used to calculate yields (gram product per gram xylose) from the slopes of the linear d-xylose concentration decrease versus the concentrations of the respective metabolites or biomass dry weight as described previously (Papapetridis et al.2016). CO2 and O2 concentrations in the exhaust gas of bioreactors and metabolite concentrations were measured as described previously (Verhoeven et al.2017).
RESULTS
Genetic requirements for anaerobic growth on xylose
Saccharomyces cerevisiae RWB217 was the first XI-based engineered yeast strain that was reported to grow anaerobically on d-xylose without prior laboratory evolution or mutagenesis (Kuyper et al.2005; Fig. 1A). Recently, anaerobic growth on d-xylose of a strain that was newly reconstructed in the same genetic background and contained a similar set of genetic modifications (strain IMX696 (Verhoeven et al.2017), Table 2) was reported to require mutations in PMR1. In view of this apparent discrepancy and other literature reports on additional genetic requirements for anaerobic growth of d-xylose-metabolising strains in other genetic backgrounds (Table 1), we re-investigated the requirement for anaerobic growth of CEN.PK-based S. cerevisiae strains.
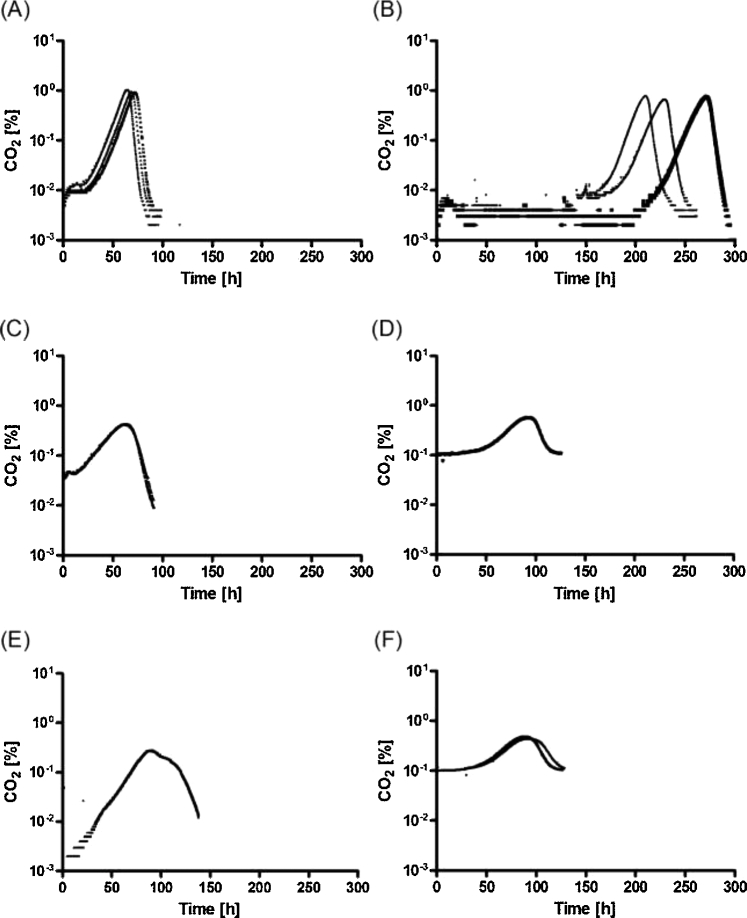
Figure 1.
Fermentation profiles, indicated as percentage of CO2 in off gas over time, of metabolically engineered, non-evolved d-xylose-metabolizing S. cerevisiae strains grown in anaerobic bioreactor batch cultures on SM supplemented with 20 g L−1d-xylose. Unless indicated otherwise, cultures were inoculated at a biomass concentration of 0.02 g L−1 and sparged with 0.5 vvm N2. (A) Strain RWB217 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, XKS1↑, xylA (pAKX002); Kuyper et al.2005). (B) Strain IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑, xylA (pAKX002)). (C) Strain IMU078 inoculated at 0.2 g biomass L−1. (D) Strain IMU078 inoculated at 0.02 g biomass L−1 and sparged with a mixture of 99.9% N2 and 0.1% CO2 at 0.5 vvm. (E) Strain IMU078 inoculated at 0.02 g biomass L−1 in media containing l-aspartate as nitrogen source instead of ammonium sulfate. Sampling of this culture for metabolite analyses (Fig. 2C) affected the CO2 profile. (F) Strain IMX1736 (IMU078 pck1Δ) inoculated at 0.02 g biomass L−1 and sparged with a mixture of 99.9% N2 and 0.1% CO2 at 0.5 vvm The panels show results from 4 (panel B, D), 3 (panel A) and 2 (panel C, E, F) independent experiments, respectively. Detailed information on strain genotypes is provided in Table 2.
While, in strain RWB217 as well as in strain IMX696, xylA was expressed from the TPI1 promoter, the xylA coding region was codon optimised in strain IMX696 (Verhoeven et al.2017) but not in strain RWB217 (Kuyper et al.2005). Moreover, in strain IMX696 the xylA expression cassettes were chromosomally integrated, resulting in ca. 36 copies per haploid genome (Verhoeven et al.2017), whereas the xylA expression cassette in strain RWB217 was expressed from a multi-copy episomal vector (Kuyper et al.2005). To investigate whether these differences affected anaerobic growth on d-xylose of these two strains, a new strain was constructed that carried the same set of genetic modifications as strain IMX696 but expressed xylA from the same episomal vector (pAKX002) as strain RWB217. Growth on d-xylose of the resulting strain, IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑; pAKX002 (2 μm xylA)), was investigated in anaerobic, nitrogen-sparged bioreactor cultures, exactly as described previously (Verhoeven et al.2017). In these cultures, strain IMU078 showed a 7–8 day adaptation phase before anaerobic growth on d-xylose set in (Fig. 1B). Under the same conditions, strain RWB217 showed anaerobic growth on d-xylose within 32 ± 6 h after inoculation (Fig. 1A), consistent with the original report by Kuyper et al. (2005). These results indicate that codon usage and/or mode of xylA overexpression were not decisive factors in causing the presence and absence of a prolonged anaerobic adaptation phase in strains IMX696 (Verhoeven et al.2017) and RWB217 (Kuyper et al.2005), respectively. To check if the integrated Cas9 expression cassette that was present in strains IMX696 and IMU078 but not in RWB217 affected anaerobic growth on d-xylose, the Cas9 cassette was integrated at the CAN1 locus of strain RWB217, yielding strain IMX975. This strain exhibited the same anaerobic growth profile as strain RWB217 (Fig. 2A).
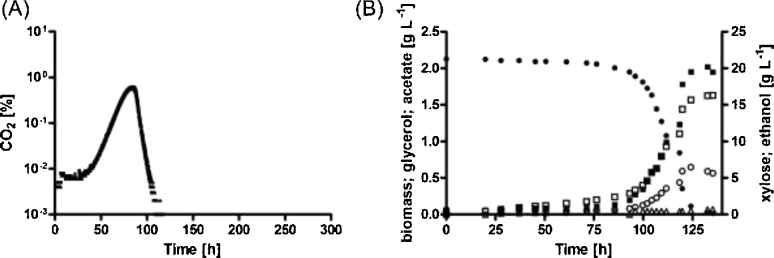
Figure 2.
Growth and product formation in anaerobic bioreactor cultures of metabolically engineered, non-evolved, d-xylose-metabolizing S. cerevisiae strains. Cultures were inoculated at a biomass concentration of 0.02 g L−1 and, unless otherwise stated, were sparged with 0.5 vvm N2. (A) Strain IMX975 (RWB217 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, XKS1↑, xylA (pAKX002), can1::Cas9)). (B) Strain IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑, xylA (pAKX002)) sparged with 0.5 vvm of a mixture of 0.1% CO2 and 99.9% N2. (C) Strain IMU078 grown in media containing l-aspartate as nitrogen source instead of ammonium sulfate. ● = d-Xylose; ▪ = Biomass; ■ = Glycerol; ○ = Ethanol; Δ = Acetate. The panels show data from single representative cultures from a set of two independent duplicate cultures for each strain. Data from replicate cultures are shown in Supplemental Fig. S1, Supporting Information.
When, after 12 days, replicate anaerobic bioreactor cultures of strain IMU078 on d-xylose had reached stationary phase, their PMR1 loci were PCR amplified and Sanger sequenced. In contrast to observations in similar experiments with strain IMX696 (Verhoeven et al.2017), no mutations were found in promoter, terminator or coding region of PMR1. This result suggested that other mutations were responsible for the eventual anaerobic growth on d-xylose of strain IMU078.
Inoculum density and resulting initial CO2 concentration affect anaerobic growth on d-xylose
In the original characterisation of strain RWB217 in anaerobic batch cultures (Kuyper et al.2005), an inoculum concentration of 0.2 g biomass L−1 was used. The experiments discussed above and the previous characterisation of strain IMX696 (Verhoeven et al.2017) were inoculated with a 10-fold lower inoculum density. Use of a 0.2 g L−1 inoculum density completely abolished the 7–8 day adaptation phase of strain IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑; pAKX002 (2 μm xylA)) in anaerobic, d-xylose-grown batch cultures (Fig. 1C). The specific growth rates of these cultures on d-xylose, estimated from CO2 concentrations in the outlet gas were, however, lower than those eventually reached by low-inoculum-density cultures after adaptation to anaerobic growth on d-xylose (0.04 ± 0.00 h−1 and 0.10 ± 0.01 h−1, respectively).
The anaerobic bioreactor cultures in this study were sparged with N2 (0.5 L min−1). As was to be expected, inoculum density positively correlated with initial CO2 concentrations in the off gas of the cultures and therefore, by inference, with concentrations of CO2 in the culture broth (Fig. 1A–F). In cultures of strain IMU078 inoculated at 0.02 g biomass L−1, the initial CO2 content in the off gas was 0.004% ± 0.001%, as compared to 0.04% ± 0.005% in cultures started at a 10-fold higher biomass concentration. To investigate whether the initial CO2 concentration influenced the onset of anaerobic growth on d-xylose, anaerobic bioreactor cultures of strain IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑; pAKX002 (2 μm xylA)), inoculated at 0.02 g biomass L−1, were sparged with a mixture of 0.1% CO2 and 99.9% N2 instead of with pure N2. This change, which led to an initial CO2 concentration in the outlet gas of 0.1%, abolished the 7–8 d lag phase observed for this strain in low-inoculum-density cultures sparged with pure N2 (Figs 1D and 2B). These results indicate that anaerobic growth of engineered, d-xylose-metabolizing S. cerevisiae can strongly depend on the concentration of CO2.
Use of l-aspartate as nitrogen source can replace CO2 supplementation in low-inoculum-density cultures of strain IMU078
As recently proposed for xylose-fermenting E. coli (Gonzalez, Long and Antoniewicz 2017), the observed CO2 requirement of engineered S. cerevisiae strains for anaerobic growth on d-xylose might reflect a pivotal role of the anaplerotic carboxylation of pyruvate or phosphoenolpyruvate (PEP) to oxaloacetate. To test this hypothesis, strain IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑; pAKX002 (2 μm xylA)) was grown on d-xylose in low-inoculum density (0.02 g biomass L−1) bioreactor cultures sparged with pure N2, in which l-aspartate instead of ammonium sulfate was used as the sole nitrogen source. In cultures grown on l-aspartate, which can be directly transaminated to oxaloacetate in a CO2-independent manner, strain IMU078 showed immediate anaerobic growth (Figs 1E and 2C).
In S. cerevisiae, oxaloacetate can be formed from pyruvate by pyruvate carboxylase (Pyc1, Pyc2) or via PEP-carboxykinase (Pck1). PCK1 expression is repressed by glucose (Gancedo and Schwerzmann 1976; Daran-Lapujade et al.2004) and elevated concentrations of CO2 are required to enable Pck1 to act as sole anaplerotic enzyme in S. cerevisiae (Zelle et al.2010). A previous transcriptome study on an evolved XI-based, xylose-fermenting strain showed an 8-fold higher transcript level of PCK1 than its non-evolved parental strain (Zhou et al.2012). However, deletion of PCK1 in strain IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑, pAKX002 (2 μm xylA)), resulting in strain IMX1736 (IMU078, pck1Δ), did not abolish the positive effect of external CO2 supplementation on anaerobic growth on d-xylose (Fig. 1F).
Omitting the over-expression of PPP paralogs enables lag-phase free anaerobic xylose fermentation at low inoculum density
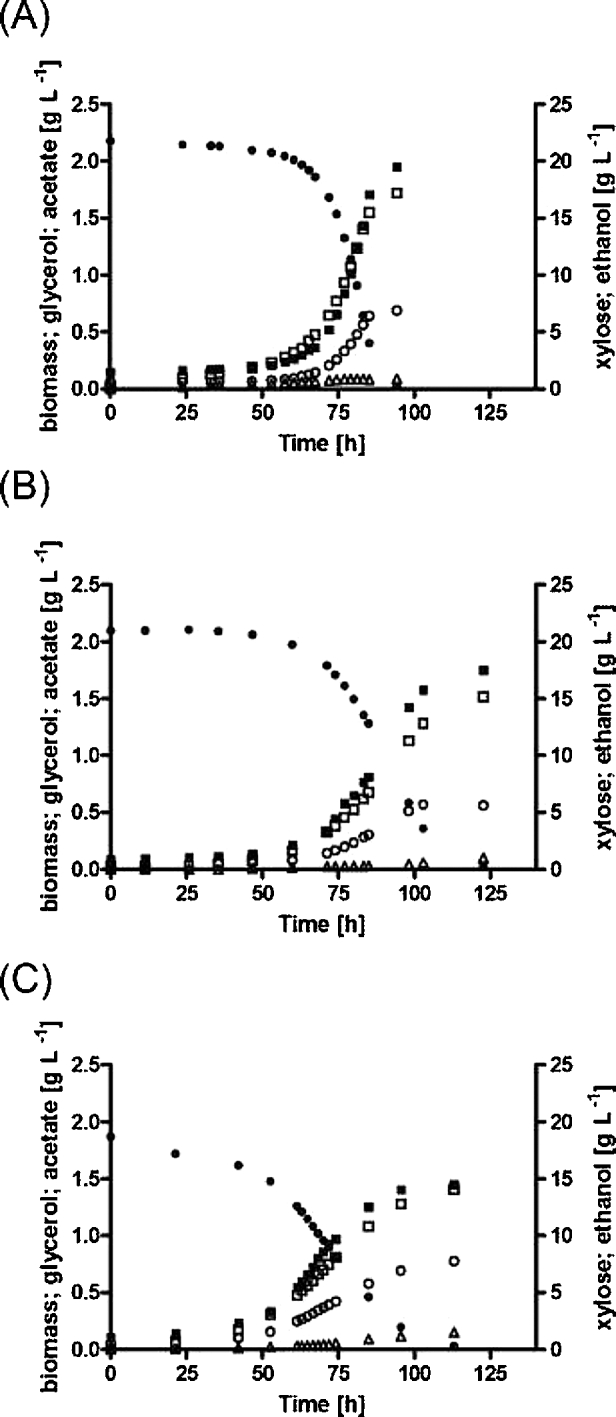
In addition to codon-optimisation and xylA expression vector, a third genetic difference exists between RWB217 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, XKS1↑; pAKX002 (2 μm xylA); Kuyper et al.2005; Table 2) and the two strains requiring a multi-day anaerobic adaptation phase prior to xylose fermentation (IMX696 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑, xylA*36 (Verhoeven et al.2017) and IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑, pAKX002 (2 μm xylA), Table 2). This difference concerns the over-expression of minor paralogs of the PPP genes TAL1 and TKL1 (NQM1 and TKL2, respectively) in the latter two strains. To evaluate the potential impact of the presence of expression cassettes for NQM1 and TKL2 in strains IMX696 and IMU078, a strain was constructed that was congenic to IMU078 except for the omission of the expression cassettes corresponding to NQM1 and TKL2 in the synthetic gene cluster harbouring the genes encoding non-oxidative PPP enzymes and XKS1 (Table 2). Although the topology of the expression cassettes was different, the relevant genotype of the resulting strain IMU079 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, XKS1↑; pAKX002 (2 μm xylA), Table 2) mimicked that of strain RWB217. Just like strain RWB217, strain IMU079 initiated anaerobic growth on d-xylose within 35 ± 5 h in low-inoculum-density, nitrogen-sparged cultures grown with ammonium sulfate as nitrogen source (Fig. 3A and B). Furthermore, specific growth rate and other physiological parameters of strain IMU079 were similar to those of the Cas9-expressing strain derived from RWB217 (IMX975, Table 4). When inoculated at a high cell density (0.2 g biomass L−1), strains IMU079 and IMU078 showed closely corresponding CO2 production profiles (Supplementary Fig. S3, Supporting Information).
Figure 3.
Fermentation profiles of the metabolically engineered, non-evolved d-xylose metabolizing S. cerevisiae strain IMU079 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, XKS1↑, xylA (pAKX002)) grown in anaerobic bioreactor batch cultures on SM supplemented with 20 g L−1d-xylose. Cultures were inoculated at a biomass concentration of 0.02 g L−1 and sparged with 0.5 vvm N2. (A) CO2 concentration in off gas. (B) Growth and product formation. ● = d-Xylose; ▪ = Biomass; ■ = Glycerol; ○ = Ethanol; Δ = Acetate. Panel (A) shows data from duplicate cultures, panel (B) shows data from a single representative culture from a set of two independent duplicate cultures. Data from replicate cultures are shown in Supplemental Fig. S2, Supporting Information.
Table 4.
Specific growth rates and yields of biomass, ethanol and glycerol of the metabolically engineered, xylose-fermenting S. cerevisiae strains IMX975 (RWB217-Cas9 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, XKS1↑, can1::CAS9, 2 μm xylA (pAKX002)), IMU079 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, XKS1↑, 2 μm xylA (pAKX002)) and IMU078 (gre3Δ, RPE1↑, RKI1↑, TAL1↑, TKL1↑, NQM1↑, TKL2↑, XKS1↑, 2 μm xylA (pAKX002)) in anaerobic bioreactor batch cultures, inoculated with an initial biomass density of 0.02 g biomass L−1 and grown on 20 g L−1 xylose. Bioreactor cultures of strains IMX975, IMU079 and IMU078 (l-Asp) were sparged with pure N2 (0.5 vvm). Cultures of strain IMU078 indicated as ‘IMU078 (+0.1% CO2)’ were sparged with a mixture of 0.1% CO2 and 99.9% N2. ‘l-Asp’ indicates that the medium contained l-aspartate instead of ammonium sulfate as sole nitrogen source. Data represent average ± SE of two independent cultures for each strain. Detailed information on strain genotypes is provided in Table 2.
| IMX975 | IMU079 | IMU078 (+ 0.1% CO2) | IMU078 (l-Asp) | |
|---|---|---|---|---|
| Specific growth rate (h−1) | 0.09 ± 0.01 | 0.08 ± 0.00 | 0.05 ± 0.01 | 0.054 ± 0.001 |
| Biomass yield (g g−1) | 0.088 ± 0.001 | 0.096 ± 0.004 | 0.093 ± 0.002 | 0.104 ± 0.002 |
| Ethanol yield (g g−1) | 0.395 ± 0.013 | 0.395 ± 0.002 | 0.382 ± 0.003 | 0.406 ± 0.002 |
| Glycerol yield (g g−1) | 0.085 ± 0.001 | 0.081 ± 0.001 | 0.074 ± 0.003 | 0.076 ± 0.000 |
To further investigate the effects of over-expression of NQM1 and/or TKL2 on anaerobic growth on d-xylose, over-expression cassettes for NQM1 and TKL2 were introduced in the SGA1 loci of strains IMX975 (RWB217, can1::Cas9) and IMU079, neither of which previously harboured these cassettes. However, characterisation of the resulting strains (IMU081 (IMU079 sga1::NQM1,TKL2), IMX1366 (IMX975 sga1::NQM1,TKL2)) in anaerobic bioreactor cultivations with xylose led to inconsistent lag-phase phenotypes (Supplementary Fig. S4, Supporting Information). In particular, introduction of the two cassettes in strains IMX975 (RWB217, can1::Cas9) and IMU079 did not yield prolonged anaerobic adaptation phases. These observations indicate that the effect of these cassettes on anaerobic growth may be related to or even caused by their immediate genetic context, e.g. by affecting expression of neighbouring genes in the pentose-pathway gene cluster in strains IMX696 and IMU078.
DISCUSSION
Repeatability of results is a key requirement for progress in all scientific research (Nature 2016). This reassessment of requirements for anaerobic growth on d-xylose of engineered, XI-based Saccharomycescerevisiae strains was prompted by results from multiple laboratories, including our own (Table 1), which appeared to contradict an early report by our group (Kuyper et al.2005). The results confirm the conclusions of Kuyper et al. (2005) by showing that the defined set of targeted genetic modifications reported in their study suffices to enable anaerobic growth of CEN.PK-based strains on d-xylose. Additionally, this study shows how anaerobic growth on d-xylose can critically depend on seemingly small differences in strain design and cultivation conditions.
Verhoeven et al. (2017) included overexpression of NQM1 and TKL2, the ‘minor’ paralogs of the PPP genes TAL1 and TKL1, respectively, in their strategy for single-step construction of a xylose-metabolising S. cerevisiae strain. Increased expression of these paralogs in evolved strains had previously been shown to contribute to improved anaerobic growth l-arabinose (Wisselink et al.2010), whose metabolism also proceeds via the non-oxidative PPP. This study shows that the mere omission of these two overexpression cassettes from the strain design of Verhoeven et al. (2017) suffices to eliminate a prolonged (12 d) adaptation phase prior to initiation of anaerobic growth on d-xylose in N2-sparged cultures grown at a low inoculum density.
A recent study showed reduced protein levels of Tkl2 and Nqm1 in xylA-based strains evolved for improved d-xylose fermentation (Sato et al.2016). However, no consistent effect on anaerobic growth performance was observed when we integrated overexpression cassettes for NQM1 and TKL2 outside rather than inside the synthetic cluster of pentose-metabolism genes that was central to the strain design used by Verhoeven et al. (2017) and in the present study. We therefore cannot exclude that the negative impact of these expression cassettes on strains IMU078 and IMX696 was related to their physical location, e.g. by transcriptional interference with other neighbouring genes in a tightly packed cluster of highly expressed genes (Kuijpers et al.2016). Transcriptome analysis would provide a logical first step in attempts to elucidate the mechanism by which integration of the NQM1 and/or TKL2 cassettes prevents anaerobic growth on xylose in strains IMU078 and IMX696. However, the requirement for controlled bioreactor batch cultivation of multiple strains, some of which exhibit long lag phases, will make this a major experimental effort. Until this issue has been resolved, the work of Sato et al. (2016) and the present study indicate that it is prudent to omit NQM1 and TKL2 overexpression from initial designs for d-xylose-fermenting strains (Sato et al.2016).
Even within the CEN.PK genetic background, literature reports differ on the ability of XI-based engineered strains to grow anaerobically on d-xylose without additional laboratory evolution (Kuyper et al.2004; Karhumaa et al.2007; Brat, Boles and Wiedemann 2009; Parachin et al.2011; Shen et al.2012; Hector et al.2013; Verhoeven et al.2017; Table 1). In this study, increasing the initial concentration of biomass or CO2 in N2-sparged, anaerobic bioreactor cultures eliminated the extended anaerobic adaptation phase of NQM1/TKL2 overexpressing strains. This observation showed that the inability of these strains to grow anaerobically on d-xylose, without first acquiring additional mutations, was conditional rather than absolute.
CO2 and bicarbonate are essential for carboxylation reactions in biosynthesis. In several bacteria, including Escherichiacoli, anaerobic growth without a long lag phase requires external supply of CO2 (Valley and Rettger 1927; Repaske, Repaske and Mayer 1974; Repaske and Clayton 1978). The critical role of biosynthetic carboxylation reactions in S. cerevisiae is illustrated by the essentiality of carbonic anhydrase (Nce103), which interconverts CO2 and HCO3−, for growth on glucose in aerated cultures at ambient atmospheric pressure (Aguilera et al.2005). Despite the key roles of inorganic carbon in microbial metabolism, sparging with CO2-free N2 is commonly applied in anaerobic laboratory bioreactor cultivation of S. cerevisiae, as sparging with CO2 complicates quantification of its production in yeast metabolism. In glucose-grown cultures of wild-type S. cerevisiae strains, endogenous CO2 production by vigorous alcoholic fermentation likely provides sufficiently high ‘sparking’ levels of CO2 to enable growth initiation even at low initial biomass concentrations. Conversely, under the same conditions, the lower fermentation rates in engineered, non-evolved xylose-fermenting strains might not be able to provide the required CO2 levels. The striking impact of endogenous CO2 generation on anaerobic performance of engineered pentose-fermenting strains indicates that, especially for strains with low endogenous CO2 production rates, supplementing anaerobic, nitrogen-sparged bioreactor batch cultures with CO2 is a complicating but necessary measure. While relevant in laboratory settings, it should be borne in mind that industrial scale bioreactors are not sparged and have increased hydrostatic pressure, which in turn increases the CO2 partial pressure. Additionally, CO2 limitation is unlikely to occur during anaerobic fermentation of lignocellulosic hydrolysates, in which pentose fermentation is typically preceded by a vigorous glucose fermentation phase (Jansen et al.2017).
A recent 13C-flux analysis study showed that anaerobic fermentation of d-xylose, but not of glucose, by E. coli required lipid turn-over by β-oxidation to provide CO2 for anaplerotic (pyruvate or PEP to oxaloacetate) carboxylation reactions (Gonzalez, Long and Antoniewicz 2017). Use of l-aspartate, whose transamination yields oxaloacetate, as the nitrogen source, completely eliminated the long anaerobic adaptation phase of NQM1/TKL2-expressing strains in N2-sparged, low-inoculum density cultures. This strong effect of bypassing a single carboxylation reaction might reflect a CO2 sparging effect that simply reduces the overall CO2 requirement for growth. Alternatively, the link between anaplerotic synthesis of oxaloacetate and d-xylose metabolism may be more specific. At physiological pH values, Mn2+, the preferred metal cofactor of Piromyces XylA (Lee et al.2017), is also a much better metal cofactor for yeast pyruvate carboxylase (Pyc1 and Pyc2) than Mg2+ (Cazzulo and Stoppani 1969). High-level expression of Piromyces xylA might therefore result in a competition between the two enzymes for Mn2+. Alternatively, lower intracellular concentrations of acetyl-CoA, a key activator of pyruvate carboxylase, in xylose-grown cultures than in glucose-grown cultures might compromise in vivo activity of Pyc1 and Pyc2 (Bergdahl et al.2012).
In comparison with other strain backgrounds, the CEN.PK lineage may have at least one specific advantage for enabling anaerobic growth on d-xylose. Using a different S. cerevisiae genetic background, Sato et al. (2016) evolved a Clostridium phytofermentans xylA-expressing strain for anaerobic xylose fermentation and analysed causal mutations (Sato et al.2016). In addition to mutations that affected GRE3, whose deletion was already in the strain design of Kuyper et al. (2005), these included mutations in IRA2 (Sato et al.2016). Loss of Ira2, an inhibitor of cAMP-PKA signalling, results in increased protein kinase A (PKA) activity (Tanaka et al.1990). The PKA/cAMP pathway is activated by glucose and regulates multiple cellular processes (Rolland, Winderickx and Thevelein 2002; Santangelo 2006). In contrast to glucose, xylose does not fully activate the cAMP/PKA pathway (Osiro et al.2018). In comparison with other S. cerevisiae genetic backgrounds, CEN.PK strains carry many sequence differences in genes involved in this signal-transduction pathway (Vanhalewyn et al.1999; Nijkamp et al.2012). Moreover, they exhibit a higher basal PKA activity than other laboratory S. cerevisiae strains (Kümmel et al.2010). The PKA/cAMP pathway regulates the glucose-dependent expression of the genes encoding several xylose-transporting members of the S. cerevisiae HXT (hexose transporter) family (i.e. HXT1, HXT2, HXT4, HXT5 and HXT7 (Hamacher et al.2002; Lee et al.2002; Kim and Johnston 2006; Saloheimo et al.2007)). A higher basal PKA activity might result in Hxt transporter landscapes that are conducive for fast xylose uptake and, thereby, enable the high rates of xylose fermentation that are required to sustain anaerobic growth.
We hope that this study will help colleagues with the design of pentose-fermenting strains and the experimental design of anaerobic yeast cultivation experiments, as well as that it will inspire further studies on the molecular basis for fast pentose fermentation in S. cerevisiae.
Supplementary Material
Acknowledgements
The authors thank Jolanda ter Horst (TUD), Ioannis Papapetridis (TUD) and Erik de Hulster (TUD) for help with fermentation set-up and sampling, Jean-Marc Daran for advice on molecular biology, Marcel van den Broek for support with bioinformatics, as well as Ioannis Papapetridis (TUD), Paul de Waal (DSM), Hans de Bruin (DSM) and Paul Klaassen (DSM) for valuable input in this project.
FUNDING
This work has been supported by the BE-Basic R&D Program, which was granted an FES subsidy from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I). The BE-Basic project ‘Omniyeast,’ within which this research was performed, received financial support from DSM.
COMPETING INTERESTS
DSM markets technology for biofuels production from lignocellulosic feedstocks, holds IP positions in this field and co-funded the research described in this publication.
AUTHORS’ CONTRIBUTIONS
J.M.B, A.J.A v. M, J.T.P together designed this study. J.M.B constructed strains IMX1736, IMX1456 and IMU081; O.A.M-R constructed strains IMX994, IMU079, IMU078 and IMX1366; M.D.V constructed strain IMX800; W.C.D constructed strain IMX975. J.M.B designed and performed most wet-lab experiments included in this publication. O.A.M-R and J.M.B performed characterisations of strains RWB217, IMU078 and IMU079. J.M.B and J.T.P wrote the manuscript. All authors read and commented a draft version of the manuscript and approved the submitted version.
Conflict of interest. None declared.
REFERENCES
- Aguilera J, Van Dijken JP, De Winde JH et al. Carbonic anhydrase (Nce103p): An essential biosynthetic enzyme for growth of Saccharomyces cerevisiae at atmospheric carbon dioxide pressure. Biochem J 2005;391:311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Stephanopoulos G. Engineering for biofuels: Exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol 2009;7:715–23. [DOI] [PubMed] [Google Scholar]
- Bergdahl B, Heer D, Sauer U et al. Dynamic metabolomics differentiates between carbon and energy starvation in recombinant Saccharomyces cerevisiae fermenting xylose. Biotechnol Biofuels 2012;5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat D, Boles E, Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microb 2009;75:2304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzulo J, Stoppani A. Effects of magnesium, manganese and adenosine triphosphate ions on pyruvate carboxylase from baker's yeast. Biochem J 1969;112:747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daran-Lapujade P, Jansen ML, Daran J-M et al. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiaeI: A chemostat culture study. J Biol Chem 2004;279:9125–38. [DOI] [PubMed] [Google Scholar]
- Demeke MM, Dietz H, Li Y et al. Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 2013;6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 2013;41:4336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos LV, Carazzolle MF, Nagamatsu ST et al. Unraveling the genetic basis of xylose consumption in engineered Saccharomyces cerevisiae strains. Sci Rep 2016;6:38676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K-D, Kötter P. 25 Yeast genetic strain and plasmid collections. Methods Microbiol 2007;36:629–66. [Google Scholar]
- Gancedo C, Schwerzmann K. Inactivation by glucose of phosphoenolpyruvate carboxykinase from Saccharomyces cerevisiae. Arch Microbiol 1976;109:221–5. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 2002;350:87–96. [DOI] [PubMed] [Google Scholar]
- Gonzalez JE, Long CP, Antoniewicz MR. Comprehensive analysis of glucose and xylose metabolism in Escherichiacoli under aerobic and anaerobic conditions by 13 C metabolic flux analysis. Metab Eng 2017;39:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn-Hägerdal B, Wahlbom CF, Gárdonyi M et al. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv Biochem Eng Biotechnol 2001;73:53–84. [DOI] [PubMed] [Google Scholar]
- Hamacher T, Becker J, Gárdonyi M et al. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 2002;148:2783–8. [DOI] [PubMed] [Google Scholar]
- Hector RE, Dien BS, Cotta MA et al. Growth and fermentation of D-xylose by Saccharomyces cerevisiae expressing a novel D-xylose isomerase originating from the bacterium Prevotella ruminicola TC2-24. Biotechnol Biofuels 2013;6:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao H-Y, Chiang L-C, Chen L-F et al. Effects of borate on isomerization and yeast fermentation of high xylulose solution and acid hydrolysate of hemicellulose. Enzyme Microb Technol 1982;4:25–31. [Google Scholar]
- Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990;96:23–8. [DOI] [PubMed] [Google Scholar]
- Jansen ML, Bracher JM, Papapetridis I et al. Saccharomyces cerevisiae strains for second-generation ethanol production: From academic exploration to industrial implementation. FEMS Yeast Res 2017;17. doi: 10.1093/femsyr/fox044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries TW. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 2006;17:320–6. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Sanchez RG, Hahn-Hägerdal B et al. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Fact 2007;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-H, Johnston M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem 2006;281:26144–9. [DOI] [PubMed] [Google Scholar]
- Kötter P, Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 1993;38:776–83. [Google Scholar]
- Kuijpers NG, Solis-Escalante D, Luttik MA et al. Pathway swapping: Toward modular engineering of essential cellular processes. Proc Natl Acad Sci 2016;113:15060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmel A, Ewald JC, Fendt S-M et al. Differential glucose repression in common yeast strains in response to HXK2 deletion. FEMS Yeast Res 2010;10:322–32. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Harhangi HR, Stave AK et al. High-level functional expression of a fungal xylose isomerase: The key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res 2003;4:69–78. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Hartog MM, Toirkens MJ et al. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 2005;5:399–409. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Winkler AA, Dijken JP et al. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: A proof of principle. FEMS Yeast Res 2004;4:655–64. [DOI] [PubMed] [Google Scholar]
- Lee M, Rozeboom HJ, de Waal PP et al. Metal dependence of the xylose isomerase from Piromyces sp. E2 axplored by activity profiling and protein crystallography. Biochemistry 2017;56:5991–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Jellison T, Alper HS. Directed Evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 2012;78:5708–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Jellison T, Alper HS. Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels 2014;7:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Kim M, Ryu Y et al. Kinetic studies on glucose and xylose transport in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 2002;60:186–91. [DOI] [PubMed] [Google Scholar]
- Liu L, Redden H, Alper HS. Frontiers of yeast metabolic engineering: Diversifying beyond ethanol and Saccharomyces. Curr Opin Biotechnol 2013;24:1023–30. [DOI] [PubMed] [Google Scholar]
- Lynd LR. Overview and evaluation of fuel ethanol from cellulosic biomass: Technology, economics, the environment, and policy. Annu Rev Energy Env 1996;21:403–65. [Google Scholar]
- Madhavan A, Tamalampudi S, Ushida K et al. Xylose isomerase from polycentric fungus Orpinomyces: Gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl Microbiol Biotechnol 2009;82:1067–78. [DOI] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman M et al. CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res 2015;15. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis A, Bajpai P. Direct fermentation of D-xylose to ethanol by Kluyveromyces marxianus strains. Appl Environ Microbiol 1982;44:1039–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moysés DN, Reis VC, de Almeida JR et al. Xylose fermentation by Saccharomyces cerevisiae: Challenges and prospects. Int J Mol Sci 2016;17:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go forth and replicate! Nature 2016;536:373. doi: 10.1038/536373a. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Larsson C, van Maris A et al. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol 2013;24:398–404. [DOI] [PubMed] [Google Scholar]
- Nijkamp JF, van den Broek M, Datema E et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN. PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 2012;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson L, Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates for ethanol production. Enzyme Microb Technol 1996;18:312–31. [Google Scholar]
- Osiro KO, Brink DP, Borgström C et al. Assessing the effect of d-xylose on the sugar signaling pathways of Saccharomyces cerevisiae in strains engineered for xylose transport and assimilation. FEMS Yeast Res 2018;18. doi: 10.1093/femsyr/fox096 [DOI] [PubMed] [Google Scholar]
- Papapetridis I, Dijk M, Dobbe AP et al. Improving ethanol yield in acetate-reducing Saccharomyces cerevisiae by cofactor engineering of 6-phosphogluconate dehydrogenase and deletion of ALD6. Microb Cell Fact 2016;15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetridis I, Dijk M, Maris AJ et al. Metabolic engineering strategies for optimizing acetate reduction, ethanol yield and osmotolerance in Saccharomyces cerevisiae. Biotechnol Biofuels 2017;10:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetridis I, Verhoeven MD, Wiersma SJ et al. Laboratory evolution for forced glucose-xylose co-consumption enables identification of mutations that improve mixed-sugar fermentation by xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res 2018;18. doi: 10.1093/femsyr/foy056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parachin NS, Bergdahl B, van Niel EW et al. Kinetic modelling reveals current limitations in the production of ethanol from xylose by recombinant Saccharomyces cerevisiae. Metab Eng 2011;13:508–17. [DOI] [PubMed] [Google Scholar]
- Parreiras LS, Breuer RJ, Narasimhan RA et al. Engineering and two-stage evolution of a lignocellulosic hydrolysate-tolerant Saccharomyces cerevisiae strain for anaerobic fermentation of xylose from AFEX pretreated corn stover. PLoS One 2014;9:e107499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repaske R, Clayton M. Control of Escherichia coli growth by CO2. J Bacteriol 1978;135:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repaske R, Repaske AC, Mayer RD. Carbon dioxide control of lag period and growth of Streptococcus sanguis. J Bacteriol 1974;117:652–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and-signalling mechanisms in yeast. FEMS Yeast Res 2002;2:183–201. [DOI] [PubMed] [Google Scholar]
- Ryabova OB, Chmil OM, Sibirny AA. Xylose and cellobiose fermentation to ethanol by the thermotolerant methylotrophic yeast Hansenula polymorpha. FEMS Yeast Res 2003;4:157–64. [DOI] [PubMed] [Google Scholar]
- Salazar AN, Gorter de Vries AR, van den Broek M et al. Nanopore sequencing enables near-complete de novo assembly of Saccharomyces cerevisiae reference strain CEN. PK113-7D. FEMS Yeast Res 2017;17. doi: 10.1093/femsyr/fox074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saloheimo A, Rauta J, Stasyk V et al. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl Microbiol Biotechnol 2007;74:1041–52. [DOI] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 2006;70:253–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Tremaine M, Parreiras LS et al. Directed evolution reveals unexpected epistatic interactions that alter metabolic regulation and enable anaerobic xylose use by Saccharomyces cerevisiae. PLoS Genet 2016;12:e1006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Chen X, Peng B et al. An efficient xylose-fermenting recombinant Saccharomycescerevisiae strain obtained through adaptive evolution and its global transcription profile. Appl Microbiol Biotechnol 2012;96:1079–91. [DOI] [PubMed] [Google Scholar]
- Smiley KL, Bolen PL. Demonstration of D-xylose reductase and D-xylitol dehydrogenase in Pachysolen tannophilus. Biotechnol Lett 1982;4:607–10. [Google Scholar]
- Solis-Escalante D, Kuijpers NG, Bongaerts N et al. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res 2013;13:126–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP et al. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 1989;338:87. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Satoh T et al. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 1990;60:803–7. [DOI] [PubMed] [Google Scholar]
- Toivola A, Yarrow D, Van Den Bosch E et al. Alcoholic fermentation of D-xylose by yeasts. Appl Environ Microbiol 1984;47:1221–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träff K, Cordero RO, Van Zyl W et al. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Environ Microbiol 2001;67:5668–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley G, Rettger LF. The influence of carbon dioxide on bacteria. J Bacteriol 1927;14:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken J, Bauer J, Brambilla L et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol 2000;26:706–14. [DOI] [PubMed] [Google Scholar]
- van Rossum HM, Kozak BU, Niemeijer MS et al. Alternative reactions at the interface of glycolysis and citric acid cycle in Saccharomyces cerevisiae. FEMS Yeast Res 2016;16:fow017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhalewyn M, Dumortier F, Debast G et al. A mutation in Saccharomyces cerevisiae adenylate cyclase, Cyr1K1876M, specifically affects glucose- and acidification-induced cAMP signalling and not the basal cAMP level. Mol Microbiol 1999;33:363–76. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA et al. Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 1992;8:501–17. [DOI] [PubMed] [Google Scholar]
- Verhoeven MD, Lee M, Kamoen L et al. Mutations in PMR1 stimulate xylose isomerase activity and anaerobic growth on xylose of engineered Saccharomyces cerevisiae by influencing manganese homeostasis. Sci Rep 2017;7:46155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walfridsson M, Bao X, Anderlund M et al. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol 1996;62:4648–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PY, Schneider H. Growth of yeasts on D-xylulose. Can J Microbiol 1980;26:1165–8. [DOI] [PubMed] [Google Scholar]
- Wisselink HW, Cipollina C, Oud B et al. Metabolome, transcriptome and metabolic flux analysis of arabinose fermentation by engineered Saccharomyces cerevisiae. Metab Eng 2010;12:537–51. [DOI] [PubMed] [Google Scholar]
- Yamanaka K. Inhibition of D-xylose isomerase by pentitols and D-lyxose. Arch Biochem Biophys 1969;131:502–6. [DOI] [PubMed] [Google Scholar]
- Young E, Lee S-M, Alper H. Optimizing pentose utilization in yeast: The need for novel tools and approaches. Biotechnol Biofuels 2010;3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelle RM, Trueheart J, Harrison JC et al. Phosphoenolpyruvate carboxykinase as the sole anaplerotic enzyme in Saccharomyces cerevisiae. Appl Environ Microbiol 2010;76:5383–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Cheng J-S, Wang BL et al. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 2012;14:611–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.