Abstract
The underlying physical properties of microfluidic tools have led to new biological insights through the development of microsystems that can manipulate, mimic and measure biology at a resolution that has not been possible with macroscale tools. Microsystems readily handle sub-microlitre volumes, precisely route predictable laminar fluid flows and match both perturbations and measurements to the length scales and timescales of biological systems. The advent of fabrication techniques that do not require highly specialized engineering facilities is fuelling the broad dissemination of microfluidic systems and their adaptation to specific biological questions. We describe how our understanding of molecular and cell biology is being and will continue to be advanced by precision microfluidic approaches and posit that microfluidic tools — in conjunction with advanced imaging, bioinformatics and molecular biology approaches — will transform biology into a precision science.
Over the past 60 years, biologists have measured, manipulated and even mimicked biology using microscale physics and chemistry. Microfluidic devices handle and manipulate fluids at length scales from 1 to 1,000 micrometres, with typical internal volumes of microlitres to picolitres. Biological researchers were making use of microfluidic physical phenomena long before the coining of the term ‘microfluidics’ (FIG. 1). For example, the Coulter counter and the flow cytometer revolutionized high-throughput single-cell analysis1, and DNA sequencing in small-bore capillary tubes markedly accelerated the progress of the Human Genome Project2.
Figure 1 |. A flow diagram highlighting crucial developments in microfluidics, from fabrication approaches and functional developments in the evolution of the technology platform to some of the applications discussed in this Review.
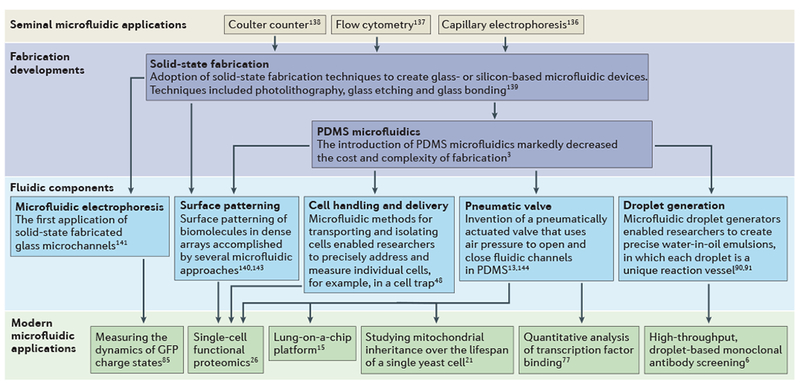
As early as the 1960s, seminal microfluidic technologies were increasing the speed and resolution of bioanalysis for a small number of powerful analytical tools136–138. Crucial fabrication developments, which began in 1990, allowed researchers to transition from using pulled glass capillaries to microfabricated glass or polymer microchannels3,139. Microfluidic tools have precisely defined features and contain highly complex channel networks and fluidic components that can execute specific functions13,48,91,140–144. In the past decade, the increasing availability of fluidic components and number of experienced microfluidic researchers have opened the door to new biological applications; these include new single-cell measurements5,12,21, high-throughput monoclonal antibody screening6, analysis of dynamic phenomena21,77,85 and organ-on-a-chip platforms15. PDMS, polydimethylsiloxane.
The advent of microfabrication techniques for the semiconductor industry (in which they are used to route electrons) inspired microscale tools for biological research (to route cells and molecules). As micro- and nano-fabrication techniques have matured, so have microfluidic tools that are specifically designed for biological investigation. For example, the development of techniques for microdevice prototyping (such as soft lithography) eliminated the need for long-duration fabrication processes in semiconductor clean room facilities, thereby making the creation of devices rapid and feasible in biological laboratories3. Such readily realizable microfabrication techniques are ‘democratizing’ microfluidic tools, thus moving tool design and creation from the engineering work bench to the biology laboratory. The dissemination of microfluidic techniques has led to the development of a wide range of new assays with unique and important advantages over conventional approaches.
The sub-microlitre volumes of microfluidic devices reduce reagent usage4 and mitigate sample loss5 (FIG. 2a). Furthermore, minimizing the level of dilution facilitates the detection of low-abundance molecules from single cells. The new tools therefore offer the opportunity to, for example, investigate more than ten protein targets in signalling pathways5 or screen the activity of secreted monoclonal antibodies6 from individual cells among the thousands of cells tested.
Figure 2 |. Physics at the microscale.
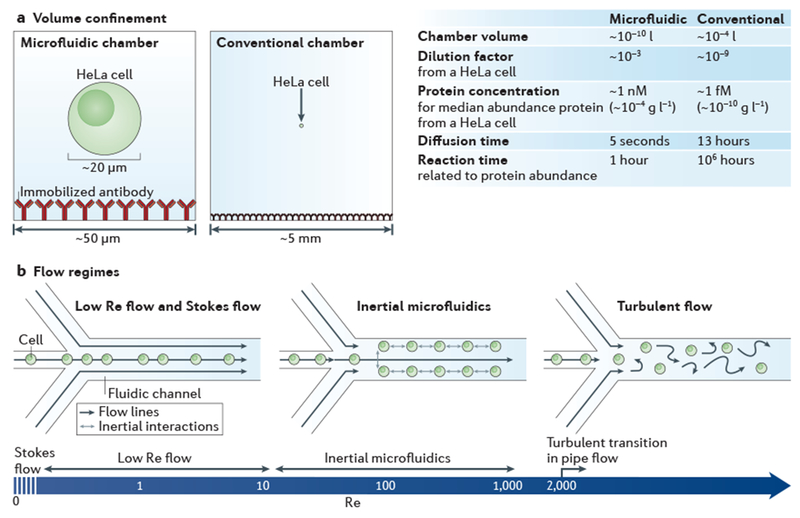
a | Bioanalysis in small, confined microfluidic volumes can enhance reaction speeds for detecting low-abundance molecules, compared with reactions conducted in a conventional chamber. In the case of detecting proteins from a single HeLa cell (median protein abundance ~170,000 molecules per cell27), a sufficient protein concentration is maintained in the microfluidic system for an antibody (assuming kon = 105 M−1 s−1 and a 60 kDa protein) to reach reaction equilibrium within ~1 hour, whereas it would take ~106 hours to reach equilibrium in the conventional system, owing to the relative concentration differences. The minimal diffusive losses that are associated with microfluidic volumes are essential for measuring proteins from single cells and other analytes that cannot be amplified, b | The predictable laminar flow that occurs in a microfluidic device is one of its most important features. The hydrodynamic flow focusing of cells is shown for different flow regimes, which vary according to the flow velocity, channel diameter or viscosity (these parameters are reflected in the Reynolds number (Re)). At near Stokes flow (Re <<1) and low Re flow (Re <10), which are the conditions in most microfluidic devices, the effects of inertial forces are minimal and for most applications can be ignored. In inertial microfluidics (typically 10 < Re <1,000), high pressures are used to create relatively high flow velocities, so that particles (such as cells) have inertial interactions with objects or flows. Inertial microfluidics can be used for several different applications145, including size-based sorting, streamline focusing and mechanical measurements12. Above a Re of 2,000 in pipe flow146, which is not practical to achieve in microfluidic devices, a transition to turbulent flow occurs.
Microfluidic devices also facilitate precise fluidic control, owing to the fact that microfluidic flows are not turbulent: they lack the eddies and vortices that churn fluids at larger scales7 (FIG. 2b). Instead, microscale fluid flows are ordered, with adjacent fluid streams moving as well-defined ‘lamina’ or layers over (relatively) long distances. This laminar flow behaviour is determined by the flow velocity, channel dimensions and fluid properties (which are grouped together in the dimensionless Reynolds number (Re)). Laminar flow has long had an important role in biological enquiry; laminar sheath flows in flow cytometry8 focus a suspension of dispersed cells into a single-file procession past the photodetector, which makes possible the interrogation of tens of thousands of single cells per second. Microfluidic laminar flow patterning directs fluid lamina of different compositions to specific regions of the device9, thus using the well-ordered behaviour of laminar flows to route each fluid without the need for confinement by micro-channel walls. More recently, microfluidic flow control formed the basis for a biophysical flow cytometer that is designed to assess single-cell mechanics for tens of thousands of cells per second10. This assay is currently being investigated as a fully automated, rapid and label-free test for certain types of malignancy11 and for stem cell pluripotency12.
Active microfluidic components (such as integrated valves or pumps with real-time control)13 can be incorporated into ‘lab-on-a-chip’ devices to deliver or extract materials from specific regions of the device with unparalleled temporal and spatial precision14. In one example, pneumatic actuators were used to recapitulate the cyclic mechanical strain of the human lung onto an in vitro alveolar–capillary interface15. Using precise microfluidic controls in ‘organ-on-a-chip’ systems, researchers have mimicked biological functions for use in drug discovery and development16. Compared with existing animal models, the microfluidic alternative may reduce costs and numbers of animals sacrificed and could offer a highly relevant humanized model system. Large-scale chips, ranging in size from square millimetre to square centimetre areas, can incorporate thousands of functional units5,17. Miniaturized integration makes feasible what would otherwise be an unwieldy system if composed of macroscale tubes and chambers.
Microfluidic arrays can be used to automate existing assays and improve assay performance, and they offer new ways to interrogate rapid biological phenomena. For example, massive multiplexing in a microfluidic array was implemented to measure the relative binding affinity of 28 yeast transcription factors for all possible 8-bp DNA sequences (65,536 sequences) to map out transcription factor specificity across the genome18. The high throughput, multiplexing and quantitative measurements of highly transient transcription factor binding that are provided by the microfluidic array would all be inaccessible by any conventional means.
Microfluidic tools are often compatible with high-resolution and real-time microscopy, owing to the planar form factor and optical transparency of microdevices. Coupled with spatial and temporal control of the chemical environment (for example, levels of soluble factors) and physical environment (for example, single-cell trapping), microfluidic live-cell imaging can offer insights into dynamic cellular processes (such as signalling dynamics19) and can be used to study long-term developments such as cell ageing20, inheritance21 and death22.
However, although microfluidic tools can reduce the time and cost of existing conventional assays and improve their reliability, microfluidic systems are by no means a replacement for all life science tools. Important drawbacks must be considered when contemplating the development of a new microfluidic assay, as discussed in BOX 1. As a general rule, the development of a microfluidic system is likely to be worthwhile only if the macroscale approach does not yield the required information or performance.
Box 1 |. Drawbacks to adopting microfluidic tools.
Microfluidic approaches can provide many advantages over conventional assays in terms of performance or capabilities (such as analysis time, throughput, reliability through automation, sensitivity or resolution); nevertheless, one approach cannot meet every need. Careful consideration of existing macroscopic approaches should be weighed against both the added capabilities of new microfluidic tools and their inherent drawbacks and risks. To assist with decision-making, we draw the reader’s attention to the common drawbacks that are associated with microfluidic approaches. Developing new microanalytical tools is a demanding and time-consuming endeavour with no guarantee of success. Therefore, novel microfluidic tools are best considered in cases in which macroscale approaches do not yield the required information or in which miniaturization of the assay can confer notable performance advantages.
Loss of sample
The high surface area to volume ratio in microsystems allows heat to dissipate effectively (from inside the channel) but can also enhance surface adsorption losses. Careful consideration of surface treatments must be made to prevent loss of sample through adsorption to channel walls. Also, microchannels can become clogged with debris or bubbles, which in many cases ruins the device.
Incompatibility with rapid processing of large sample volumes
The range of volumes and volumetric flow rates that are generally used in microfluidic devices require long durations to process large volumes of sample. For example, a channel with a 10 pm diameter would take ~1,000 minutes for a 1 ml volume to pass, compared with less than 1 minute in a 1 mm diameter channel at the same applied pressure. For high-volume applications, it may be more appropriate to consider mesofluidic approaches.
Mismatch with conventional tools
Conventional pipettes deliver microlitre volumes, whereas microdevices typically analyse nanolitre volumes, which means that the majority of the sample will not be analysed by a microfluidic device and will potentially be wasted.
Loss of historical context, standards and controls
Results from studies using new assays can be challenging or impossible to compare with published results from conventional assays. Accepted assay standards and controls may not be applicable in a new microfluidic format. Consequently, the adoption of new microfluidic assays by the research community can be slow.
Specialized requirements
Microfluidic approaches may require specialized instrumentation, expertise in engineering design and non-conventional data interpretation strategies, particularly at the earliest stages of device development.
In this Review, we investigate specific examples of how microfluidic tools have altered experimental discovery in the life sciences. We organize the Review around illustrative experimental capabilities: single-cell measurements; live-cell imaging; rapid molecular events; screening; and tailored contexts. These sections are neither exhaustive nor steeped in detailed physics. Rather, we highlight the emerging biological insight and discuss how microfluidic design — and its integration with co-evolved orthogonal techniques such as advanced imaging and bioinformatics — underpins new biological observations and fresh lines of biological enquiry.
Single-cell measurements
The precision of microfluidic devices has improved existing single-cell assays (such as immunocytochemistry23 and immunohistochemistry24,25) and has bolstered the invention of novel assays for quantitative analysis of thousands of cells with single-cell resolution26.
Protein measurements.
Confining cells in microfluidic compartments enhances the sensitivity of assays by minimizing dilution and yielding high concentrations, even with limited sample mass (FIG. 2a). Consider, for example, the enzyme-linked immunosorbent assay (ELISA). A conventional high-sensitivity ELISA detects proteins above a minimum concentration of ~1 pg ml−1 (~10 fM). In a 100 μl ELISA micro well, the 1 pg m−1 detection concentration equates to the detection of ~1 million molecules (assuming a 60 kDa target); this level of sensitivity is an order of magnitude lower than that required to detect the median protein concentration in a single mammalian cell (which has a median protein abundance of ~170,000 molecules27). However, if a single cell is lysed into a 1 nl volume microchamber (which is ~1,000 times larger than the volume of a typical mammalian cell28), a 1 pg ml−1 detection limit means that the majority of the proteome is detectable (on the basis of reported protein abundances in a single cell27). Furthermore, owing to the relative length scales of the chambers (~50 μm for the microchamber versus ~5 mm for the conventional well), it will take ~5 seconds for a uniform equilibrium concentration to be reached via diffusion within the microchamber, whereas it would take more than 10 hours to do so in the conventional chamber. These advantages conferred by small microfluidic volumes are being leveraged to provide new biological insights in single-cell analysis.
When capture reagent barcoding is combined with nanolitre volume confinement, several targets can be detected concurrently29–31. Single-cell microfluidic immunoassays have been developed to profile the cellular secretome, which is the set of all proteins secreted by a cell29,32–35. For example, the secretion activity of isolated cytotoxic T cells, which are the main effector cell type of the adaptive immune response against intracellular pathogens, was evaluated for 12 proteins in patients with metastatic melanoma and in healthy donors using capture reagent barcoding. The assay profiled the secretion of thousands of cells in parallel, with single-cell resolution, from a starting population of just 10,000 cells. Through diversity analysis, the measurements showed that there is greater cell-to-cell secretion heterogeneity in the cytotoxic T cells of patients with melanoma (45 distinct cell populations) compared with cytotoxic T cells from healthy controls (17 distinct cell populations)32. As conventional single-cell secretion assays cannot detect 12 targets at once, the microfluidic platform offers greater profiling depth for immune monitoring and clinical assessment.
In another example, profiling of the phosphorylation of 11 intracellular PI3K signalling proteins was made possible by microscale confinement of lysate from individual cells5. To understand the PI3K pathway, protein expression was evaluated in human glioma cell lines at basal levels and after drug treatment. Protein correlation maps were established for single cells to identify putative protein–protein interactions33,36. By contrast, conventional immunoassays on pooled cell populations measure protein expression as a population average. Therefore, conventional immunoassays are unable to determine whether an observed covariance between proteins is related within single cells or is a by-product of a heterogeneous pooled sample.
The performance of immunoassays depends on the availability of antibody probes with suitable specificity37. Therefore, the presence of protein isoforms (with shared antibody-binding epitopes) and nonspecific interactions are major concerns in immunoassay performance. One approach that can mitigate the limited specificity of immunoassays is western blotting. Western blotting involves an electrophoresis step (in which proteins are separated by size) followed by antibody probing (immunoassay). By reporting protein size with the subsequent immunoassay results, western blots provide improved specificity over a simple immunoassay. Single-cell resolution western blotting is made possible by microfluidic design, which achieves rapid cell lysis (within seconds) in confined volumes (microwells), followed by rapid protein electrophoresis (in seconds) and immobilization of protein in the gel (blotting). The single-cell western blot has been applied to the study of neural stem cell differentiation, from which a nestin isoform of unexpected molecular mass was reported38. When applied to the study of glioblastoma chemosensitivity and expression of proteins involved in drug resistance, the single-cell western blot detected small subpopulations of glioblastoma cells that upregulate expression of P-glycoprotein, which implicates a role for efflux pumps in a possible drug-resistance mechanism of cancer cells39.
Genetic and transcriptomic measurements.
In 2013, Nature Methods named single-cell sequencing as their ‘Method of the Year’ (REF. 40). Measurement of cell-to-cell genomic heterogeneity suggests that the genome of a multicellular organism is a composite of diverse genomes rather than one single genome41. Although single-cell sequencing does not require microfluidics, microfluidic approaches confer distinct advantages in terms of the limits of detection42, reproducibility43 and multiplexing44–46. Sequencing comprises several stages: cell isolation; cell lysis and pre-amplification; reverse transcription (for RNA detection); and secondary amplification and readout. Intriguingly, when cell lysis and pre-amplification were carried out in microfluidic nanolitre reaction volumes, fewer false positives were observed and there was less amplification bias (compared with the same assay carried out in conventional reaction volumes)42,47. False positives can arise from the indiscriminate nature of the initial amplification of nucleic acids. Microfluidic pre-amplification reduced the amplification bias of high-abundance sequences, and thus low-abundance sequences were better represented at low sequencing depths42,43,47.
Laminar flow routing of cells at a low-Re flow regime (Re <10; see FIG. 2b) to microfabricated cell traps (which are sized to house just one cell in each trap)48 has been used to position hundreds of cells within minutes for the subsequent concurrent detection of nucleic acids. Studying rapid mRNA decay49 after stimulation (for example, after trypsinization) may benefit from similarly efficient microfluidic approaches. Researchers have used microfluidics to integrate cell loading, sample preparation and fluorescent reporter readout for the measurement of several nucleic acid targets in each of 300 cells in an ~2 hour measurement period50. Precision fluid routing has been used to aliquot a 60 nl cDNA sample into discrete 25 pl reaction chambers defined by surface tension for digital PCR; carrying out cDNA dilutions in these digital PCR reaction chambers yielded a dynamic range of 0 to 10,000 molecules51.
At present, however, the multiplexing advantage of cell sorting on-chip is hampered by the bottleneck in downstream deep sequencing, for which samples are collected and analysed off-chip in serial runs. In the near future, nucleic acids from single cells will be uniquely barcoded during the pre-amplification stage, thus facilitating the sequencing of DNA from multiple cells in a single batch process. The distinct cell sequences can then be reconstructed after sequencing using the barcode identifiers52,53.
Cell mechanics.
Cell mechanics and cytoskeletal structure are crucial factors in regulating cell spreading and migration, intracellular cargo transport and cell fate54. Few analytical approaches can assess cell mechanics — and those that are available, such as micropipette aspiration55 and atomic force microscopy56, can be difficult to carry out and have a low throughput. Microfluidic flow-based approaches to mechanobiology assays are emerging as alternatives. For example, in biophysical flow cytometry57, a suspension of cells is directed through a channel that contains a micropore slightly smaller than the average cell diameter. Cell transit time through the channel is directly related to cell deformability, with a measurement throughput of ~800 cells per minute58. By combining this approach with mechanical modelling, it has been possible to measure the cortical tensions of red blood cells59. Results of another microfluidic flow cytometry study suggest that aberrant white blood cells (as defined by their size and stiffness) may be drivers of microvascular occlusion in patients with leukostasis57. In a similar approach, microbarriers that required cells to deform to be able to traverse the structures were used to sub-classify a heterogeneous breast cancer cell line on the basis of cellular deformability. Gene expression analysis of the two subpopulations of breast cancer cells showed that the more flexible cell subpopulation overexpressed several genes that are associated with cancer metastasis60. In the future, biophysical flow cytometry and sorting are likely to be combined with single-cell protein and nucleic acid measurements, perhaps establishing as-yet undiscovered biophysical and biomolecular relationships.
Contact-free measurements of single-cell deformability overcome the limitations of contact-based biophysical measurements. Instead of squeezing cells through confined spaces, contact-free tools use precise flow control to hydrodynamically stretch cells in fluid flows. The contact-free assay has fewer problems associated with channel clogging compared with contact-based assays and it can achieve throughputs of up to 65,000 cells per second10, which matches flow cytometry measurement rates. The contact-free, label-free deformability assay — operating in an inertial microfluidic flow regime with a Re of ~200 (that is, 1–2 orders of magnitude higher than in canonical microfluidic systems; see FIG. 2b) while maintaining laminar flow — has been used to accurately identify malignant pleural effusions11. Stem cell pluripotency which is currently determined by biochemical markers61, might also be ascertained by cell deformability11,55,62. High-throughput hydrodynamic cell-stretching assays have tracked changes in the deformability of human embryonic stem cells during differentiation for thousands of single cells12, approximately four orders of magnitude more measurements than are possible with existing non-microfluidic techniques. Using the large data set that was obtained, the authors created a classification strategy for identifying undifferentiated and differentiated human embryonic stem cells.
Summary.
Single-cell analysis using microfluidic devices takes advantage of minimal target dilution, automation and the integration of several assay steps and readouts, which is accomplished by either multiple concurrent steps or rapid serial processes. Further innovation is required in the interfacing of microfluidic workflows with macroscale processes. In particular, approaches are needed to integrate microfluidic systems with dispensing and collecting functions that are rapid, automated and minimize manual handling, which can otherwise perturb experimental conditions and make quantification difficult.
Live-cell imaging
Microfluidic cell culture platforms establish well-controlled cellular microenvironments that are suitable for live-cell imaging. The fine spatial and temporal control of such platforms underpins high-resolution study of dynamic phenomena and even of stable long-term cultures. On-chip culture combined with real-time microscopy and an automated microscopy stage is a practical approach for monitoring rapid phenomena and/or multiple samples in parallel.
Long-term observation.
Microfluidic cell culture arrays provide controlled microenvironments for stable long-term monitoring. Generational trends can be investigated by combining imaging with microfluidic single-cell traps63 to observe the cellular lifespan of multiple single cells. These tools have been used to investigate cellular inheritance21,64 and ageing20,22,65. For example, individual yeast cells were trapped in a commercial microfluidic device, and the mitochondrial-to-cell volume ratio was monitored for 50 hours, during which time up to 40 generations of yeast budding — for both the mother and daughter cells — were observed. The researchers noted a consistent reduction in the mitochondrial-to-cell volume ratio of mother cells with generational age. Interestingly, the mitochondrial-to-cell volume ratio of daughter cells was maintained over time. Stable inheritance despite a growing mitochondrial asymmetry between mother and daughter cells suggests that there is an active mechanism for mitochondrial sensing and feedback within budding yeast21.
Microfluidic cell culture arrays have been extended to small multicellular organisms66 to study developmental biology67,68 and as platforms for high-throughput testing in vivo69. Using a single microfluidic array, researchers grew and monitored 64 Arabidopsis thaliana seedlings expressing 12 different transgenic reporter lines. Automated confocal imaging was used to capture and reconstruct 3D time lapses of root structural development and the corresponding spatial distribution of gene expression for all 64 seedlings in parallel. In this platform, different growth media (with low pH, or iron or sulfur deficiency) were tested on genetically identical plants, and considerable heterogeneity of seedling gene expression from certain promoters was observed. Interestingly, the WUSCHEL-RELATED HOMEOBOX 5 (WOX5) promoter undergoes a burst of activity in the elongation zone of the seedling, producing a standing wave of WOX5 expression in that region, which indicates that there is a self-sustaining regulatory network for WOX5 expression67. Similarly, microfluidic systems are being developed to culture arrays of zebrafish embryos70 as a high-throughput in vivo platform for toxicology and drug screening69,71.
Dynamic response monitoring.
Even time-varying, low-Re microfluidic flows are predictable and straightforward to describe mathematically. Microscale flows typically result in Re values that are small compared with the Strouhal number, and thus both convective and unsteady parts of the describing equations do not need to be considered72. Pulsing of well-controlled flows carrying soluble signals can dynamically and precisely perturb molecular pathways (FIG. 3a). Areas of recent study include cellular feedback mechanisms, noise73, frequency-encoded responses74 and rate-limiting processes19.
Figure 3 |. Dynamic process analysis.
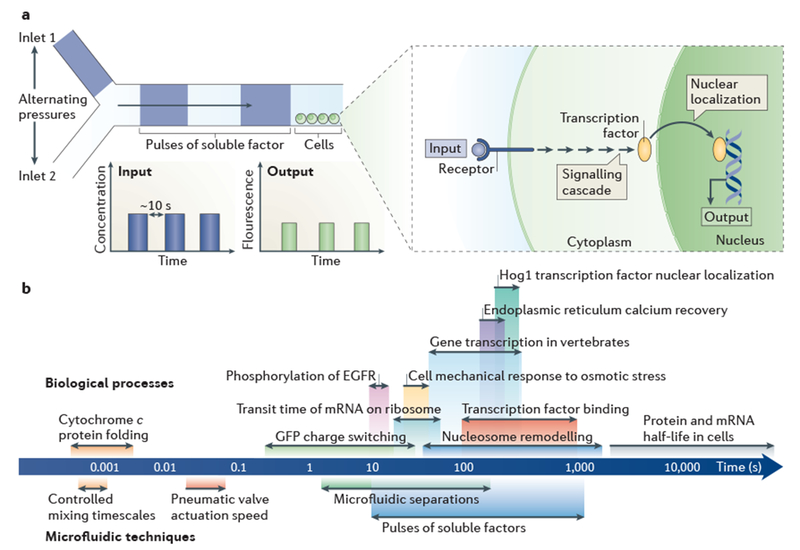
a | The schematic depicts a microfluidic device designed for the generation of pulses of a soluble factor by alternating the pressures at two separate inlets. The reproducible behaviour of fluids at the microscale enables the input to be described mathematically. Microfluidic devices easily interface with real-time microscopy (owing to their planar form factor), which enables the output of cells cultured in situ to be directly monitored as they respond to the pulses of soluble factor. Rapid pulses (< 10 seconds between pulses) have been used to study rapid biological processes such as the nuclear localization of transcription factors and their interactions with promoters75. b | The logarithmic ruler shows the timescales of dynamic biological processes and the time resolutions of the corresponding microfluidic techniques that can be used to study them (as indicated by corresponding colours). EGFR, epidermal growth factor receptor.
To study calcium signalling dynamics, two concentrations of the drug carbachol were pulsed over human embryonic kidney (HEK293) cells in a microfluidic flow chamber. Carbachol is known to regulate levels of intracellular calcium through a membrane receptor. The lower carbachol concentration resulted in an oscillating calcium signal, whereas the higher carbachol concentration resulted in an acute increase in the calcium signal that plateaued above pre-stimulus levels. A mathematical model of the pathway suggested mechanisms that might be responsible for these two behaviours: calcium replenishment and inositol trisphosphate production in response to the low carbachol concentration; and dephosphorylation of deactivated receptors in response to the high carbachol concentration19.
In another study of the high osmolarity glycerol (HOG) MAPK pathway, 1 M sorbitol was pulsed with a range of frequencies over yeast cells in a microfluidic flow chamber to study the temporal response to an osmotic shock75. The HOG MAPK pathway transmitted individual signals with periods longer than 220 seconds but integrated signals with shorter periods. At signals oscillating less than once every 220 seconds, the dynamics of Hog1–GFP nuclear localization matched those of the input signal; with higher frequency signals, the response pathway was unable to follow the input. Thus, different stimulus dynamics altered the degree of the Hog1-GFP nuclear localization response.
Rapidly switching signals in microfluidic flows have been used to probe the dynamics of transcription factor activity in yeast. Pulses of a protein kinase inhibitor controlled the nuclear localization of a fluorescently labelled transcription factor (Msn2–mCherry) for the direct study of gene expression (measured with a YFP reporter) for a range of promoters and dynamic conditions. Promoters were grouped into distinct classes on the basis of their responses, which ranged from high-threshold, slow activation to low-threshold, fast activation. The promoter activation timescale was related to nucleosome remodelling73. In another study, reductions in iron concentration led to damped oscillations of gene expression in the iron homeostasis pathway in Escherichia coli. The gene expression oscillations were independent of the cell cycle and persisted for several generations. Damping of oscillations was required to balance response speed and stability, as an underdamped oscillation was required for response speed and an over-damped response preserved only the signals that occurred with longer timescales76.
Summary.
Microfluidic approaches are uniquely well-suited for live-cell imaging. Long-term culture arrays offer a controlled environment for high-throughput biological analysis. Studying dynamic cell-signalling pathways is made possible by the negligible inertial forces and fast ‘active’ valves and switches of microfluidic devices13.
Rapid molecular events
Similar to the dynamic cellular analyses described previously, microfluidics also provides a platform for high-resolution temporal studies of molecular events. Active microfluidic functions with timescales ranging from milliseconds to seconds facilitate the direct observation of transient molecular phenomena at matching timescales (FIG. 3b) that are either not observable or not quantifiable with traditional approaches.
Transient intermolecular interactions.
Rapidly dissociating molecular interactions have a pivotal role in many biological processes. For example, transcription factor–DNA binding partners often have dissociation rates of 0.1 s− and higher. To measure rapidly dissociating interactions, a measurement must be completed in less time than is required for dissociation. If the measurement is too slow, the interacting molecules will have dissociated. One approach to overcoming this restraint is to physically trap the binding partners together. Mechanically induced trapping of molecular interactions (MITOMI)77 uses pneumatically actuated valves to rapidly squeeze the solvent out of the binding region by pressing a surface onto a spot of immobilized reagent. Removing the solvent prevents the loss of the interacting molecules into the solvent. Thus, even quickly dissociating interactions can be evaluated. Using MITOMI on a microarray of immobilized reagents, thousands of transiently associated transcription factor–DNA interactions were ‘frozen in time for subsequent quantitative evaluation77–81. MITOMI has been used in assays of dynamic molecular interactions including RNA–protein82 and protein-protein82,83 interactions.
In one MITOMI study, the dissociation constant (Kd) values of 28 yeast transcription factors were measured against all possible 8-bp DNA sequences (a total of 65,536 sequences)18. In an adaptation of the original MITOMI assay, both the on- and off-rates for binding were measured for 768 DNA–protein interactions in massively parallel microfluidic devices using periodic opening and closing of the trap17. In such studies, the minimum resolvable interaction time is determined by the valve closure time (5 ms). The quantitative transcription factor-binding kinetics that have been determined using this platform support the use of stochastic models to predict gene expression and cellular processes. The massive multiplexing, quantitative capacity and large dynamic range of MITOMI make the assay a powerful tool for the characterization and discovery of molecular interactions.
Molecular phenomena.
Microfluidic systems are suitable for interrogating rapid molecular phenomena84. Microfluidic isoelectric focusing, which separates biomolecules on the basis of charge differences, identified charge-switching in the fluorescent protein GFP upon stimulation with light85. Real-time imaging of the ~1-second microfluidic separations was crucial for observation of the charge switching and for determining switching kinetics.
Ultra-fast microfluidic mixing (that is, mixing taking place in less than 100 μs) facilitated tight control of hydrogen–deuterium exchange and thus allowed researchers to extend the dynamic range of NMR into the microsecond range84,86. Using this approach, the researchers concluded that decreases in chain dimensions during the early stages of cytochrome c folding are the result of specific α-helix interactions and do not indicate a general hydrophobic collapse of the protein.
Summary.
The temporal control afforded by microfluidics has provided new research tools for observing and quantifying rapid molecular events that are not measurable with conventional tools. We expect this to be a growth area in biologically applied microfluidics over the next decade.
Screening
Early microfluidic tools, including capillary electrophoresis87 and flow cytometry88, transformed high-throughput screening assays (FIG. 4). Recently, the emergence of droplet microfluidics89–91 has again pushed the boundaries of throughput (FIG. 4a) and is facilitating screening with new types of selection criteria that were not previously accessible. Although there are important non-droplet-based microfluidic screening platforms92–95, we focus here on the biological insights gained from droplet microfluidics because of the future potential of this platform96,97.
Figure 4 |. High-throughput microfluidics.
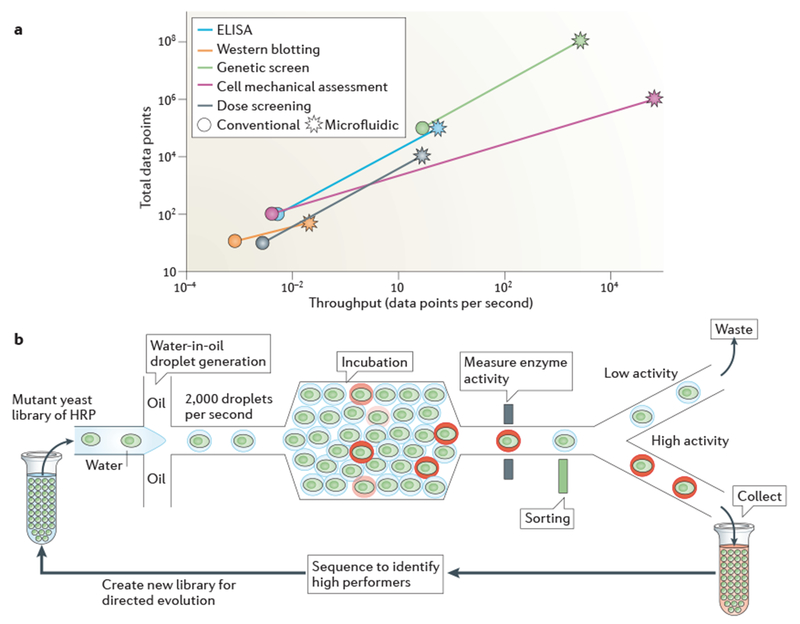
a | Integrating sample processing and analysis steps within microfluidic technologies has increased the throughput and data output of a range of analytical tools to orders of magnitude higher than those obtained using conventional approaches. For example, the mechanical properties of a cell can be measured using conventional methods (such as atomic force microscopy) at a throughput of ~10 cells per hour; by contrast, inertial microfluidics has been used to mechanically assess ~65,000 cells per second10. b | Water-in-oil droplet microfluidic technologies are at the forefront of the increase in analytical through put. Thousands of droplets are produced per second, each of which is a precisely defined experimental reaction chamber. Downstream fluidic components carryout serial high-throughput sample processing, which facilitates the analysis of hundreds of millions of samples per day. In one study4, the directed evolution of horseradish peroxidase (HRP) from a mutant yeast library was carried out using droplet microfluidics. The researchers screened 108 individual enzyme reactions in only 10 hours, which is an ~1,000-fold increase in speed and an ~10 million-fold decrease in cost compared with conventional tools. ELISA; enzyme-linked immunosorbent assay.
Functional screening of genetic libraries.
Precise control of water-in-oil emulsions facilitates the screening of hundreds of millions of samples each day96. Immiscible droplets (with volumes of 50 fl to 1 nl) reduce cross-contamination between samples and provide high local concentrations for sensitive assays. Droplet libraries are produced from suspensions of genetic elements (such as genomes of cells or viruses, or nucleic acids encoding proteins), which are diluted such that each droplet contains a single genetic element together with all other necessary reagents such as enzymes for protein production or fluorescent probes for assay readout. After droplets are selected for functional enzyme activity, the co-compartmentalized genetic elements are sequenced and used as barcode identifiers98. For example, several rounds of droplet library selection supported the directed evolution of horseradish peroxidase (depicted in FIG. 4b). This method was used to screen 100 million mutant enzymes, contained in separate droplets, in just 10 hours and to identify mutant enzymes with catalytic rates 10 times faster than those of the wild-type protein. The entire screen was 1,000-fold faster and ~10 million-fold less expensive than a comparable robotics-based screen4. Similar droplet-based screens have been applied to non-mutant libraries. For example, hundreds of thousands of single antibody-producing hybridoma cells were each encapsulated in a sub-nanolitre droplet, and the activity of secreted monoclonal antibodies was measured for each hybridoma cell6. Such functional screens may expedite and reduce the cost of high-quality monoclonal antibody production. In the future, antibody screening of non-immortalized primary B cells may also be possible, as cell proliferation is not required for this type of assay.
Droplet-based single-cell screening has been used to investigate metabolic activity in a two-stage assay to identify cells that are capable of extracellular metabolite secretion or consumption99. In the first stage, single cells were encapsulated in growth medium and cultured for several days in separate droplets. Then, in the second stage, aliquots of metabolite detection reagents were merged with droplet-encapsulated cells, facilitating the selection of cells with the desired metabolic activity.
Reaction optimization and screening.
Precisely controlled metering and mixing of reagents by microfluidics yields exquisitely controlled doses100, volumes and solvent conditions for reactions95. In particular, reactions that are sensitive to initial conditions, consume expensive reagents and/or require scrutiny over hundreds of unique conditions (for example, optimizing conditions for protein crystallization95) will benefit from microfluidic screening. Owing to time and cost limitations, conventional dose–response drug-activity screens are often carried out at approximately ten unique drug concentrations, which is a relatively low number of discrete concentration points when investigating complex relationships. To establish high-resolution (near-continuous) dose–response relationships for drugs, a bolus of drug compound with non-uniform concentration (created by diffusion-like dilution of drug concentration plugs by Taylor–Aris dispersion) was compartmentalized into 10,000 discrete water-in-oil droplets, thus encapsulating a continuum of drug concentrations, together with enzyme and substrate100. To investigate inhibition of the cancer- and diabetes-associated protein-tyrosine phosphatase 1B (also known as PTPN1), more than 700 different compounds were screened at 10,000 concentration points per compound. Novel enzyme inhibitors were discovered that are tenfold more potent under optimal conditions than are the compounds identified using conventional tools101.
Volume-sensitive reactions have also been explored using microfluidic devices. Monitoring nucleation events is possible using conventional tools; however, the precise definition of initial conditions is challenging, thus obscuring the guiding physical principles. To study fundamental mechanisms in the early formation of amyloid fibrils, individual amyloid nuclei were compartmentalized into water-in-oil immiscible droplets (with a volume of nanolitres to picolitres). Nucleation of fibrils and subsequent fibril formation and propagation were monitored by fluorescence microscopy102. A critical droplet volume of 31.8 pl was identified, above which multiple secondary nucleation sites were observed. Below this volume, primary nucleation occurred and the growth rate was limited by propagation rather than nucleation. The results suggest that cellular compartmentalization offers protection against uncontrolled protein aggregation. Furthermore, the sensitivity of fibril nucleation to initial local protein concentration underscores why secondary nucleation is not common in biological systems. Unlike for secondary nucleation, small differences in local concentration will not lead to highly variable primary nucleation.
Summary.
Precise manipulation of fluids facilitates the controlled metering of reagents and encapsulation into microfluidic droplets. The ability to image and sort droplets in real-time, multiphase microfluidic systems is leading to new high-throughput screening strategies.
Tailored contexts
Microfluidic tools can be used to reproduce customized and controlled microenvironmental contexts, which is now a mainstay of biological hypothesis testing.
Biomimetic environments.
Mimicking in vivo biological contexts in vitro is of considerable interest and includes efforts to devise organ-on-a-chip platforms. To recapitulate biological environments, researchers need precise patterning of materials to create features that are appropriate to the size of the cells and tissues (FIG. 5). Manipulating patterned regions requires the same precise control. As they are compatible with imaging microscopy, these microfluidic systems are readily monitored and measured at high resolution — something that is challenging with in vivo biological systems.
Figure 5 |. Biological length scales.
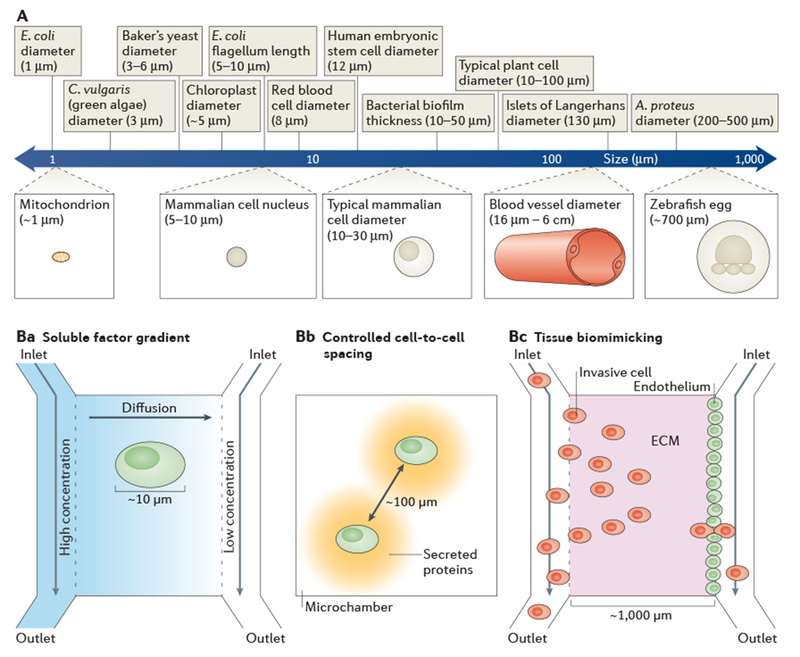
A | The logarithmic ruler shows the length scales of important biological features. The feature sizes of microfluidic devices facilitate experiments that are relevant at the micrometre scale and that are not possible with conventional techniques. Ba–c | Examples of how microfluidic length scales can match the relevant biology. Cells respond to differences in concentrations of soluble factors across their surfaces; therefore, experimental gradients of soluble factors must be generated at the same order of magnitude as the cell diameter121 (part Ba). By having reservoirs that are set at different concentrations of soluble factors at either end of a microchannel, linear concentration gradients can be formed by diffusion. Physiologically relevant cell-to-cell spacing can be realized by confining pairs of cells in microchambers132 (part Bb). Micrometre-scale tissue morphology can be replicated in microfluidic devices (part Bc). For example, events in cancer metastasis can be studied by establishing an endothelium adjacent to an extracellular matrix (ECM) in vitro and directly observing intravasation105 or extravasation106 with invasive cells. A. proteus, Amoeba proteus; C. vulgaris, Chlorella vulgaris; E. coli, Escherichia coli.
Biomimetic microfluidic systems have been developed for spheroid formation103,104, intravasation105, extravasation106 and angiogenesis107,108. For example, to investigate mechanisms by which the endothelial barrier is compromised in cancer cell invasion, researchers created a tumour-to-vascular interface using a multichannel microfluidic device. The central region of the device contained a 3D collagen matrix. Endothelial cells and fibrosarcoma cells were introduced contiguous to the collagen layer, thus concurrently forming a confluent endothelial layer and seeding cancer cells for invasion. High-resolution live-cell imaging tracked tumour cell migration into the collagen and monitored interactions with the confluent endothelial layer. Using this in vitro model, dynamic tumour cell–endothelial cell interactions were observed during intravasation, including the appearance of invasive protrusions and changes in cell shape as tumour cells exited the collagen matrix. Furthermore, intravasation was associated with endothelial barrier impairment105.
In a similar model system, breast cancer cells were introduced adjacent to the endothelium, permitting researchers to monitor the extravasation of cancer cells through an endothelial layer and into a bone-like 3D environment. Using live-cell imaging, concurrent observations were made of the extravasation process and metastatic niche formation, which was observed as micrometastases from 4 to 60 cells in size. The breast cancer cell receptor CXC-chemokine receptor 2 (CXCR2) and its ligand CXC-chemokine ligand 5 (CXCL5) were implicated in the kinetics and extent of extravasation106.
Angiogenesis is crucial to cancer progression109. To study this dynamic process, researchers created adjacent microchannels in a 3D collagen matrix107,108. One channel was endothelialized (that is, its walls were covered with a layer of endothelial cells) and adjacent channels transported angiogenic factors, thus initiating angiogenesis from the endothelialized channel into the collagen matrix. Live confocal imaging captured the dynamics of neovessel formation for various angiogenic factors. Interestingly, the requirement for vascular endothelial growth factor (VEGF) for vessel sprouting was dependent on the specific mix of angiogenic factors that were used, which may explain why blocking angiogenesis using VEGF inhibitors gives inconsistent results. Using a similar model system, a network of endothelialized channels in collagen was coupled with pressure-driven blood flow to demonstrate angiogenesis in healthy and thrombotic states107; in the future, this disease-model system could be adopted for drug testing.
Mimicking an entire organ using microfluidic design is an active area of research16,110. Organs-on-a-chip range from the biologically accurate to the abstract111, with single-organ systems including lung15, heart112, liver113, colon114, blood–brain barrier115 and muscle116. Many in vitro organ-on-a-chip systems replicate in vivo organ geometry to realize function, thus establishing a model system to test how alterations in phenotype or drugs can affect organ function. Alternatively, abstractions of the biological system in vitro, which replicate the function of the organ without duplicating biological structure, can be used to study the underlying mechanisms of biological systems111,117. Building from single-organ systems, body-on-a-chip systems consisting of multiple organs are also benefiting from microfluidic devices118,119. Although the complete elimination of animal testing may not be feasible, microfluidic organ mimics could offer an economical early-stage in vitro assessment tool for the testing of promising drug targets.
Gradients in stimuli.
Biological systems respond to average signal levels, as well as to gradients in chemical concentration120,121, material stiffness122 and pressure123. The precise flow control in microfluidic devices forms rapid, stable, linear gradients (for more than 12 hours)124. Monitoring individual mouse bone marrow-derived mast cells exposed to concentration gradients of Kit ligand revealed a previously unreported chemorepulsion that occurs during the first 90 minutes of exposure to Kit ligand concentrations below the chemoattraction activation threshold (<3 ng ml−1)120. Rapid generation of stable concentration gradients (<2 minutes) revealed the fast migration of dendritic cells125. Microfluidic gradient generators revealed the fundamental limits of chemotaxis for the eukaryote Dictyostelium discoideum. Here, the authors observed sensitivity to both gradient steepness and local concentration. Practical response limits to microfluidic gradients are likely to be controlled by the concentration and distribution of membrane receptors, a result that is supported by numerical modelling121.
Bulk microfluidic flows have been used to flush soluble cell-to-cell chemical signals away from cells, thus de-convolving a cell response to surrounding cells from the response to the chemical gradient itself. Shallow, low-concentration cAMP gradients (<5% concentration difference across a 10 μm-diameter cell) supported a chemotaxis response that was limited by external concentration fluctuations due to the sensing limits imposed by the finite number of surface receptors on each cell. For steep concentration gradients, however, the cells did not respond as predicted by a ligand-receptor binding model, suggesting that these cells have suboptimal internal concentrations (below Kd) of the signal transduction cellular machinery. Whether the chemotactic response is limited by increased fluctuations of intracellular signalling protein concentrations or by signalling pathway saturation remains under investigation121.
In another example, stimulation of individual nerve growth cones with temporally and spatially varying gradients of GABA (aminobutyric acid) showed both signal amplification behaviour (over narrow concentration ranges) and low-pass filter behaviour (cut-off frequency of 0.002 Hz) in GABAA receptors74.
Flow control.
Microfluidic systems have been designed to match both the feature sizes and flow velocities that are found in the circulatory system126. To study thrombosis, researchers embarked on a parametric analysis of thrombin and fibrinogen concentrations and flow conditions. They observed distinct fibrin deposition regimes that can be described in a state diagram: thin films of fibrin were observed with high flow rates and low concentrations of thrombin and fibrinogen, whereas 3D fibrin gels were formed with slow flow rates and increased thrombin and fibrinogen concentrations127.
Endothelialized channels were used to study the effects of flow shear on haematological diseases. Blood from patients with sickle cell disease treated with the drug hydroxyurea had increased flow velocities and reduced occlusion rates compared with blood from non-treated patients — with values approaching those observed in healthy patients128.
Microfluidic tools are also well-suited to the study of cell-surface attachment and homing in the blood stream. Such approaches show that neutrophils have decreased rolling speeds on surfaces coated with E-selectin and P-selectin when they express intercellular adhesion molecule 1 (REF. 129). Leukaemic and haematopoietic progenitor cells expressing CD44 receptors adhered to and rolled on surfaces coated with hyaluronic acid, which is a glycosaminoglycan that is found throughout connective, epithelial and neural tissues. These findings suggested a mechanism for leukocyte recruitment and homing of stem cells to the bone marrow130.
Social context.
Nearly 10,000 mouse fibroblasts were precisely arranged into 79 microchambers with various seeding densities (4,000 to 40,000 cells per square cm). Automated migration tracking of all cells over a 5 hour period enabled researchers to devise a mathematical model of cell migration. The migration model described cell migration trajectory as a function of several variables, including pseudopod direction and chemokine diffusion (from neighbouring cells). Only a small number of intercellular ‘traffic rules’ were required to replicate empirically the emergent properties of cell migration131.
The precise cell patterning that is possible with microfluidic devices is suited to the study of intracellular signalling with controlled cell-to-cell spacing. For example, glioma cells were placed at defined separation distances (of 20 to 120 μm) in microfluidic chambers (~1,000 replicates for each 20 μm interval). To assay both secreted and intracellular proteins from each of the two cells in a chamber, immunoassays were carried out on the chamber lid after cell lysis. Using thermodynamic-like considerations, the free energy minimum of epidermal growth factor receptor signalling was observed with cell spacing of 80–100 μm, indicating the most stable signalling state. The 80–100 μm spacing is consistent with the glioma cell spacing that is commonly observed in bulk cell culture132. This biological insight and platform technology could help in understanding the diffusive nature of some brain cancers.
Summary.
Microfluidics enables researchers to interrogate complex biological relationships by representing biology in tailored microenvironmental contexts that can be perturbed in a controlled manner and closely monitored using real-time, high-resolution imaging.
Perspective
Microfluidic tools, combined with advanced molecular, imaging and bioinformatics techniques, constitute a flexible ‘toolbox’ that life scientists are actively adopting and adapting. As the technology matures and becomes more robust, microfluidic tools that address open needs are progressively being applied to biological questions12,32,38,47. Although the adoption rate of microfluidic tools has historically been slow, biological research is increasingly seeing enquiry augmented (or in some cases enabled) by microfluidics. Biologists pursuing their own lines of enquiry may prefer commercially available products over early-stage prototypes, but the growing catalogue of commercially available microfluidic tools — which now includes platforms for live-cell analysis in dynamic environments (EMD Millipore) and single-cell genomics (Fluidigm) — is already having an impact on biological research21,133,134. Wider-scale adoption of microfluidic techniques is also being driven by collaboration between ‘early adopters’ in the life sciences and engineering research laboratories. Even as these technologies are being applied to questions in the life sciences, the pursuit of innovation and fundamental understanding is anticipated to continue in the engineering, materials and chemistry fields.
Despite our optimistic outlook, we do acknowledge hurdles to the widespread adoption of microfluidic assays. First, there are incompatibilities between the standards (for example, materials used) of engineers and biologists135. Second, there are gaps in expertise between microfluidic engineers and biologists; the operation and assay design of microfluidic systems might require some understanding of the underlying fundamental physics of mass transport and fluid mechanics, thus limiting access. Third, not all (or perhaps even many) biological lines of enquiry require microfluidics, and thus biologists will generally prefer to use conventional macroscale tools — even though these may be less convenient — than to learn and invest in a new technique. Given these hurdles, we emphatically acknowledge that widespread dissemination of these tools will require time, maturation and commercial availability of the underlying technologies.
For the innovators developing the next generation of microfluidic tools, we laud the efforts at collaboration (from an early stage) between physical scientists and engineers, and molecular and cell biologists. In addition to being a best practice of engineering design, such collaborations address quantitative analytical considerations (mathematical modelling, statistics and simulations), which are central to many of the studies highlighted in this Review. The continuing reframing of biological enquiry by microfluidics (and other advancing technologies) demands collaboration, the vision to propose novel questions and the increasing ‘mathematization’ of biology.
Acknowledgements
T.A.D. was supported by a US National Science Foundation Graduate Research Fellowship. A.M.T. was supported by a US Department of Homeland Security ORISE Fellowship, a Siebel Scholarship and a California Cancer Coordinating Committee Fellowship. This work was also supported by a US National Institutes of Health (NIH) New Innovator Award (DP2OD007294 to A.E.H.) and a UC Berkeley Bakar Fellowship (to A.E.H.).
Glossary
- Laminar flow
A flow regime that is Observed when the Reynolds number is less than 2,000 in pipe flow; in such cases, fluid moves in parallel stream lines or ‘lamina’
- Reynolds number
(Re), A dimensionless number that is defined as the ratio of inertial forces to viscous forces in a system. The value of Re increases with flow velocity and the characteristic length scale (Of, for example, the channel diameter) and decreases with viscosity. Microfluidic flows typically have a low Re (< 10), which results in laminar flow.
- Laminar flow patterning
Laminar patterning uses the slow mixing between multiple microfluidic flows with low Reynolds numbers for spatial patterning or to deliver soluble agents within the channel. The shape and location of patterning are controlled by modulating the relative flow rates of each fluid in the channel.
- Multiplexing
The measurement of multiple signals in parallel; that is, carrying out several assays simultaneously.
- Barcoding
The use of recognizable labels (tags) to track individual samples throughout an assay or to track the output of distinct assays from a single sample for multiplexing. Examples of such tags include DNA sequences, spectrally-encoded fluorescent beads and spatial patterning of capture reagents.
- Strouhal number
A dimensionless number that is defined as the ratio of inertial forces resulting from changes in velocity in the flow field to the inertial forces resulting from unsteady flow oscillation. The Strouhal number increases with flow velocity and decreases with the characteristic length scale of the channel and with oscillation frequency.
- Dissociation constant
(Kd). An equilibrium constant that describes the susceptibility of a complex to dissociate into its components. It is often used to describe how strongly molecules interact.
- Directed evolution
A method for the engineering of new biomolecules using the principle of natural selection. Typically, several rounds of selection are used.
- Taylor–Aris dispersion
A phenomenon that can enhance effective diffusion when there is a non-uniform flow velocity across a channel, as is typically the case in pressure-driven microfluidic flows.
- Intravasation
The invasion of cancer cells through the basement membrane into a blood or lymphatic vessel, which is a crucial step in cancer metastasis.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
References
- 1.Lanier LL Just the FACS. J. Immunol 193, 2043–2044 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Dovichi NJ & Zhang JZ How capillary electrophoresis sequenced the human genome. Angew. Chem. Int. Ed. Engl 39, 4463–4468 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Duffy DC, McDonald JC, Schueller OJ & Whitesides GM Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem 70, 4974–4984 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Agresti JJ et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl Acad. Sci. USA 107, 4004–4009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Q et al. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl Acad. Sci. USA 109, 419–424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; Multiplexed single-cell resolution immunoassays were used to directly correlate protein phosphorylation in a signalling pathway within a single cell, for thousands of cells in parallel.
- 6.El Debs B, Utharala R, Balyasnikova IV, Griffiths AD & Merten CA Functional single-cell hybridoma screening using droplet-based microfluidics. Proc. Natl Acad. Sci. USA 109, 11570–11575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; The functional screening of secreted monoclonal antibodies from hundreds of thousands of single hybridoma cells expedites the process and reduces the cost of production of high quality monoclonal antibodies.
- 7.Beebe DJ, Mensing GA & Walker GM Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng 4, 261–286 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Shapiro HM Practical Flow Cytometry (John Wiley & Sons, 2005). [Google Scholar]
- 9.Takayama S et al. Patterning cells and their environments using multiple laminar fluid flows in capillary networks. Proc. Natl Acad. Sci. USA 96, 5545–5548 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudani JS, Gossett DR, Tse HT & Di Carlo D Pinched-flow hydrodynamic stretching of single-cells. Lab Chip 13, 3728–3734 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Tse HT et al. Quantitative diagnosis of malignant pleural effusions by single-cell mechanophenotyping. Sci. Transl Med 5, 212ra163 (2013). [DOI] [PubMed] [Google Scholar]; The high-throughput cell deformability assay was developed for rapid and label-free diagnostic screening to accurately identify the malignant pleural effusions phenotype.
- 12.Gossett DR et al. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl Acad. Sci. USA 109, 7630–7635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unger MA, Chou HP, Thorsen T, Scherer A & Quake SR Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Stone HA, Stroock AD & Ajdari A Engineering flows in small devices: microfluidics toward a lab-on-a-chip. Annu. Rev. Fluid Mech 36, 381–411 (2004). [Google Scholar]
- 15.Huh D et al. Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; The biomimetic lung-on-a-chip system showed that cyclic mechanical strain accentuates toxic and inflammatory responses of the lung to silica nanoparticles — establishing the platform as a potential low-cost alternative to animal and clinical studies for drug screening and toxicology applications.
- 16.Bhatia SN & Ingber DE Microfluidic organs-on-chips. Nat. Biotechnol 32, 760–772 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Geertz M, Shore D & Maerkl SJ Massively parallel measurements of molecular interaction kinetics on a microfluidic platform. Proc. Natl Acad. Sci. USA 109, 16540–16545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fordyce PM et al. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat. Biotechnol 28, 970–975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; In a single experiment, the binding affinities of 28 yeast transcription factors were measured against all possible 8-bp DNA sequences (a total of 65,536 sequences).
- 19.Jovic A, Wade SM, Neubig RR, Linderman JJ & Takayama S Microfluidic interrogation and mathematical modeling of multi-regime calcium signaling dynamics. Integr. Biol 5, 932–939 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P et al. Robust growth of Escherichia coli. Curr. Biol 20, 1099–1103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafelski SM et al. Mitochondrial network size scaling in budding yeast. Science 338, 822–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Z et al. Molecular phenotyping of aging in single yeast cells using a novel microfluidic device. Aging Cell 11, 599–606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J et al. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res 70, 6128–6138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim MS, Kwon S, Kim T, Lee ES & Park JK Quantitative proteomic profiling of breast cancers using a multiplexed microfluidic platform for immunohistochemistry and immunocytochemistry. Biomaterials 32, 1396–1403 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Ciftlik AT, Lehr HA & Gijs MA Microfluidic processor allows rapid HER2 immunohistochemistry of breast carcinomas and significantly reduces ambiguous (2+) read-outs. Proc. Natl Acad. Sci. USA 110, 5363–5368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J et al. Microfluidics-based single-cell functional proteomics for fundamental and applied biomedical applications. Annu. Rev. Anal. Chem 7, 275–295 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Li JJ, Bickel PJ & Biggin MD System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ 2, e270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzur A, Kafri R, LeBleu VS, Lahav G & Kirschner MW Cell growth and size homeostasis in proliferating animal cells. Science 325, 167–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin L et al. Human NK cells licensed by killer Ig receptor genes have an altered cytokine program that modifies CD4+ T cell function. J. Immunol 193, 940–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerver RE et al. Programmable microfluidic synthesis of spectrally encoded microspheres. Lab Chip 12, 4716–4723 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Lee J et al. Universal process-inert encoding architecture for polymer microparticles. Nat. Mater 13, 524–529 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Ma C et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat. Med 17, 738–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin YS et al. Protein signaling networks from single cell fluctuations and information theory profiling. Biophys. J 100, 2378–2386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konry T, Dominguez-Villar M, Baecher-Allan C, Hafler DA & Yarmush ML Droplet-based microfluidic platforms for single T cell secretion analysis of IL-10 cytokine. Biosens. Bioelectron 26, 2707–2710 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hathout Y Approaches to the study of the cell secretome. Expert Rev. Proteomics 4, 239–248 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Sachs K, Perez O, Pe’er D, Lauffenburger DA & Nolan GP Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Saper CB An open letter to our readers on the use of antibodies. J. Comp. Neurol 493, 477–478 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Hughes AJ et al. Single-cell western blotting. Nat. Methods 11, 749–755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang CC, Lin JM, Xu Z, Kumar S & Herr AE Single-cell Western blotting after whole-cell imaging to assess cancer chemotherapeutic response. Anal. Chem 86, 10429–10436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.[No authors listed.] Method of the Year 2013. Nat. Methods 11, 1 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Eberwine J, Sul J-Y, Bartfai T & Kim J The promise of single-cell sequencing. Nat. Methods 11, 25–27 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Wu AR et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods 11, 41–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Streets AM et al. Microfluidic single-cell whole-transcriptome sequencing. Proc. Natl Acad. Sci. USA 111, 7048–7053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Fan HC, Behr B & Quake SR Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150, 402–412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zong C, Lu S, Chapman AR & Xie XS Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shuga J et al. Single molecule quantitation and sequencing of rare translocations using microfluidic nested digital PCR. Nucleic Acids Res 41, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcy Y et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet 3, 1702–1708 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Carlo D, Wu LY & Lee LP Dynamic single cell culture array. Lab Chip 6, 1445–1449 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Schoenberg DR & Maquat LE Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet 13, 246–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White AK et al. High-throughput microfluidic single-cell RT-qPCR. Proc. Natl Acad. Sci. USA 108, 13999–14004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White AK, Heyries KA, Doolin C, Vaninsberghe M & Hansen CL High-throughput microfluidic single-cell digital polymerase chain reaction. Anal. Chem 85, 7182–7190 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Hashimshony T, Wagner F, Sher N & Yanai I CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2, 666–673 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Islam S et al. Highly multiplexed and strand-specific single-cell RNA 5’ end sequencing. Nat. Protoc 7, 813–828 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Fletcher DA & Mullins RD Cell mechanics and the cytoskeleton. Nature 463, 485–492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ & Discher DE Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl Acad. Sci. USA 104, 15619–15624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathur AB, Collinsworth AM, Reichert WM, Kraus WE & Truskey GA Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J. Biomech 34, 1545–1553 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Rosenbluth MJ, Lam WA & Fletcher DA Analyzing cell mechanics in hematologic diseases with microfluidic biophysical flow cytometry. Lab Chip 8, 1062–1070 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adamo A et al. Microfluidics-based assessment of cell deformability. Anal. Chem 84, 6438–6443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo Q et al. Microfluidic analysis of red blood cell deformability. J. Biomech 47, 1767–1776 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Zhang W et al. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc. Natl Acad. Sci. USA 109, 18707–18712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adewumi O et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol 25, 803–816 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Chowdhury F et al. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater 9, 82–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huberts DH et al. Construction and use of a microfluidic dissection platform for long-term imaging of cellular processes in budding yeast. Nat. Protoc 8, 1019–1027 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Rowat AC, Bird JC, Agresti JJ, Rando OJ & Weitz DA Tracking lineages of single cells in lines using a microfluidic device. Proc. Natl Acad. Sci. USA 106, 18149–18154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee SS, Vizcarra IA, Huberts DH, Lee LP & Heinemann M Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proc. Natl Acad. Sci. USA 109, 4916–4920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sivagnanam V & Gijs MAM Exploring living multicellular organisms, organs, and tissues using microfluidic systems. Chem. Rev 113, 3214–3247 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Busch W et al. A microfluidic device and computational platform for high-throughput live imaging of gene expression. Nat. Methods 9, 1101–1106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choudhury D et al. Fish and Chips: a microfluidic perfusion platform for monitoring zebrafish development. Lab Chip 12, 892–900 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Akagi J et al. Integrated chip-based physiometer for automated fish embryo toxicity biotests in pharmaceutical screening and ecotoxicology. Cytometry A 85, 537–547 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Akagi J et al. Opensource lab-on-a-chip physiometer for accelerated zebrafish embryo biotests. Curr. Protoc. Cytom 67, 9.44.1–9.44.16 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Zheng C et al. Fish in chips: an automated microfluidic device to study drug dynamics in vivo using zebrafish embryos. Chem. Commun 50, 981–984 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Kirby BJ Micro-and Nanoscale Fluid Mechanics: Transport in Microfluidic Devices (Cambridge University Press, 2010). [Google Scholar]
- 73.Hansen AS & O’Shea EK Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol. Syst. Biol 9, 1–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morel M et al. Amplification and temporal filtering during gradient sensing by nerve growth cones probed with a microfluidic assay. Biophys. J 103, 1648–1656 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McClean MN, Hersen P & Ramanathan S In vivo measurement of signaling cascade dynamics. Cell Cycle 8, 373–376 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Amir A, Meshner S, Beatus T & Stavans J Damped oscillations in the adaptive response of the iron homeostasis network of E. coli. Mol. Microbiol 76, 428–436 (2010). [DOI] [PubMed] [Google Scholar]
- 77.Maerkl SJ & Quake SR A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Hens K et al. Automated protein-DNA interaction screening of Drosophila regulatory elements. Nat. Methods 8, 1065–1070 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hernday AD et al. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol. Microbiol 90, 22–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gubelmann C et al. A yeast one-hybrid and microfluidics-based pipeline to map mammalian gene regulatory networks. Mol. Syst. Biol 9, 1–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fordyce PM et al. Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc. Natl Acad. Sci. USA 109, E3084–E3093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin L et al. Systematic reconstruction of RNA functional motifs with high-throughput microfluidics. Nat. Methods 9, 1192–1194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Neveu G et al. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLos Pathog 8, e1002845 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chiang Y-Y & West J Ultrafast cell switching for recording cell surface transitions: new insights into epidermal growth factor receptor signalling. Lab Chip 13, 1031–1034 (2013). [DOI] [PubMed] [Google Scholar]
- 85.Hughes AJ, Tentori AM & Herr AE Bistable isoelectric point photoswitching in green fluorescent proteins observed by dynamic immunoprobed isoelectric focusing. J. Am. Chem. Soc 134, 17582–17591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fazelinia H, Xu M, Cheng H & Roder H Ultrafast hydrogen exchange reveals specific structural events during the initial stages of folding of cytochrome c. J. Am. Chem. Soc 136, 733–740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; Ultra-fast microfluidic mixing for hydrogen–deuterium exchange enabled researchers to determine that decreased chain dimensions during the early stages of cytochrome c folding are due to specific α-helix interactions and not a general hydrophobic collapse of the protein.
- 87.Simpson PC et al. High-throughput genetic analysis using microfabricated 96-sample capillary array electrophoresis microplates. Proc. Natl Acad. Sci. USA 95, 2256–2261 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edwards BS, Oprea T, Prossnitz ER & Sklar LA Flow cytometry for high-throughput, high-content screening. Curr. Opin. Chem. Biol 8, 392–398 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Song H, Chen DL & Ismagilov RF Reactions in droplets in microfluidic channels. Angew. Chem. Int. Ed. Engl 45, 7336–7356 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thorsen T, Roberts RW, Arnold FH & Quake SR Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett 86, 4163–4166 (2001). [DOI] [PubMed] [Google Scholar]
- 91.Anna SL, Bontoux N & Stone HA Formation of dispersions using “flow focusing” in microchannels. Appl. Phys. Lett 82, 364 (2003). [Google Scholar]
- 92.Su X et al. Microfluidic cell culture and its application in high-throughput drug screening: cardiotoxicity assay for hERG channels. J. Biomol. Screen 16, 101–111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rauch JN, Nie J, Buchholz TJ, Gestwicki JE & Kennedy RT Development of a capillary electrophoresis platform for identifying inhibitors of protein–protein interactions. Anal. Chem 85, 9824–9831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Desai B et al. Rapid discovery of a novel series of Abl kinase inhibitors by application of an integrated microfluidic synthesis and screening platform. J. Med. Chem 56, 3033–3047 (2013). [DOI] [PubMed] [Google Scholar]
- 95.Hansen CL, Skordalakes E, Berger JM & Quake SR A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. Proc. Natl Acad. Sci. USA 99, 16531–16536 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guo MT, Rotem A, Heyman JA & Weitz DA Droplet microfluidics for high-throughput biological assays. Lab Chip 12, 2146–2155 (2012). [DOI] [PubMed] [Google Scholar]
- 97.Schneider T, Kreutz J & Chiu DT The potential impact of droplet microfluidics in biology. Anal. Chem 85, 3476–3482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Granieri L, Baret JC, Griffiths AD & Merten CA High-throughput screening of enzymes by retroviral display using droplet-based microfluidics. Chem. Biol 17, 229–235 (2010). [DOI] [PubMed] [Google Scholar]
- 99.Wang BL et al. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption. Nat. Biotechnol 32, 473–478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miller OJ et al. High-resolution dose-response screening using droplet-based microfluidics. Proc. Natl Acad. Sci. USA 109, 378–383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klopfenstein SR et al. 1,2,3,4-Tetrahydroisoquinolinyl sulfamic acids as phosphatase PTP1B inhibitors. Bioorg. Med. Chem. Lett 16, 1574–1578 (2006). [DOI] [PubMed] [Google Scholar]
- 102.Knowles TP et al. Observation of spatial propagation of amyloid assembly from single nuclei. Proc. Natl Acad. Sci. USA 108, 14746–14751 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study of amyloid nucleation showed that cellular compartmentalization probably offers protection from uncontrolled fibril aggregation.
- 103.Hsiao AY et al. Microfluidic system for formation of PC-3 prostate cancer co-culture spheroids. Biomaterials 30, 3020–3027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuo CT et al. Modeling of cancer metastasis and drug resistance via biomimetic nano-cilia and microfluidics. Biomaterials 35, 1562–1571 (2014). [DOI] [PubMed] [Google Scholar]
- 105.Zervantonakis IK et al. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl Acad. Sci. USA 109, 13515–13520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; An in vitro microfluidic model of a tumour–vascular interface yielded dynamic and high-resolution images of the progression of cancer intravasation.
- 106.Bersini S et al. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 35, 2454–2461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zheng Y et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc. Natl Acad. Sci. USA 109, 9342–9347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nguyen DH et al. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl Acad. Sci. USA 110, 6712–6717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nishida N, Yano H, Nishida T, Kamura T & Kojiro M Angiogenesis in cancer. Vasc. Health Risk Manag 2, 213–219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sung JH et al. Microfabricated mammalian organ systems and their integration into models of whole animals and humans. Lab Chip 13, 1201–1212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martin JG et al. Toward an artificial Golgi: redesigning the biological activities of heparan sulfate on a digital microfluidic chip. J. Am. Chem. Soc 131, 11041–11048 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang G et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med 20, 616–623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ho CT et al. Liver-cell patterning lab chip: mimicking the morphology of liver lobule tissue. Lab Chip 13, 3578–3587 (2013). [DOI] [PubMed] [Google Scholar]
- 114.Ahmad AA et al. Optimization of 3D organotypic primary colonic cultures for organ-on-chip applications. J. Biol. Eng 8, 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Booth R & Kim H Characterization of a microfluidic in vitro model of the blood-brain barrier (muBBB). Lab Chip 12, 1784–1792 (2012). [DOI] [PubMed] [Google Scholar]
- 116.Wilson K, Das M, Wahl KJ, Colton RJ & Hickman J Measurement of contractile stress generated by cultured rat muscle on silicon cantilevers for toxin detection and muscle performance enhancement. PLoS ONE 5, e11042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Karzbrun E, Tayar AM, Noireaux V & Bar-Ziv RH Synthetic biology. Programmable on-chip DNA compartments as artificial cells. Science 345, 829–832 (2014). [DOI] [PubMed] [Google Scholar]
- 118.Esch MB, King TL & Shuler ML The role of body-on-a-chip devices in drug and toxicity studies. Annu. Rev. Biomed. Eng 13, 55–72 (2011). [DOI] [PubMed] [Google Scholar]
- 119.Sung JH et al. Using physiologically-based pharmacokinetic-guided “body-on-a-chip” systems to predict mammalian response to drug and chemical exposure. Exp. Biol. Med 239, 1225–1239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shamloo A et al. Complex chemoattractive and chemorepellent Kit signals revealed by direct imaging of murine mast cells in microfluidic gradient chambers. Integr. Biol 5, 1076–1085 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fuller D et al. External and internal constraints on eukaryotic chemotaxis. Proc. Natl Acad. Sci. USA 107, 9656–9659 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Justin RT & Engler AJ Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS ONE 6, e15978 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prentice-Mott HV et al. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. Proc. Natl Acad. Sci. USA 110, 21006–21011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu H & Heilshorn SC Microfluidic investigation of BDNF-enhanced neural stem cell chemotaxis in CXCL12 gradients. Small 9, 585–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haessler U, Pisano M, Wu M & Swartz MA Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc. Natl Acad. Sci. USA 108, 5614–5619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Giridharan GA et al. Microfluidic cardiac cell culture model (muCCCM). Anal. Chem 82, 7581–7587 (2010). [DOI] [PubMed] [Google Scholar]
- 127.Neeves KB, Illing DA & Diamond SL Thrombin flux and wall shear rate regulate fibrin fiber deposition state during polymerization under flow. Biophys. J 98, 1344–1352 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tsai M et al. In vitro modeling of the microvascular occlusion and thrombosis that occur in hematologic diseases using microfluidic technology. J. Clin. Invest 122, 408–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kuwano Y, Spelten O, Zhang H, Ley K & Zarbock A Rolling on E-or P-selectin induces the extended but not high-affinity conformation of LFA-1 in neutrophils. Blood 116, 617–624 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Christophis C et al. Shear stress regulates adhesion and rolling of CD44+ leukemic and hematopoietic progenitor cells on hyaluronan. Biophys. J 101, 585–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vedel S, Tay S, Johnston DM, Bruus H & Quake SR Migration of cells in a social context. Proc. Natl Acad. Sci. USA 110, 129–134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kravchenko-Balasha N, Wang J, Remacle F, Levine RD & Heath JR Glioblastoma cellular architectures are predicted through the characterization of two-cell interactions. Proc. Natl Acad. Sci. USA 111, 6521–6526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sen A et al. Innate immune response to homologous rotavirus infection in the small intestinal villous epithelium at single-cell resolution. Proc. Natl Acad. Sci. USA 109, 20667–20672 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Uckun FM et al. Serine phosphorylation by SYK is critical for nuclear localization and transcription factor function of Ikaros. Proc. Natl Acad. Sci. USA 109, 18072–18077 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Berthier E, Young EWK & Beebe D Engineers are from PDMS-land, biologists are from Polystyrenia. Lab Chip 12, 1224–1237 (2012). [DOI] [PubMed] [Google Scholar]
- 136.Hjerten S Free zone electrophoresis. Chromatogr. Rev 9, 122–219 (1967). [DOI] [PubMed] [Google Scholar]
- 137.Fulwyler MJ Electronic separation of biological cells by volume. Science 150, 910–911 (1965). [DOI] [PubMed] [Google Scholar]
- 138.Coulter W Means for counting particles suspended in a fluid. US patent US2656508A (1953).
- 139.Manz A, Graber N & Widmer HM Miniaturized total chemical-analysis systems: a novel concept for chemical sensing. Sens. Actuators B Chem 1, 244–248 (1990). [Google Scholar]
- 140.Kane RS, Takayama S, Ostuni E, Ingber DE & Whitesides GM Patterning proteins and cells using soft lithography. Biomaterials 20, 2363–2376 (1999). [DOI] [PubMed] [Google Scholar]
- 141.Harrison DJ, Manz A, Fan ZH, Ludi H & Widmer HM Capillary electrophoresis and sample injection systems integrated on a planar glass chip. Anal. Chem 64, 1926–1932 (1992). [Google Scholar]
- 142.Emrich CA, Tian H, Medintz IL & Mathies RA Microfabricated 384-lane capillary array electrophoresis bioanalyzer for ultrahigh-throughput genetic analysis. Anal. Chem 74, 5076–5083 (2002). [DOI] [PubMed] [Google Scholar]
- 143.Delamarche E, Bernard A, Schmid H, Michel B & Biebuyck H Patterned delivery of immunoglobulins to surfaces using microfluidic networks. Science 276, 779–781 (1997). [DOI] [PubMed] [Google Scholar]
- 144.Kazuo H & Ryutaro M A pneumatically-actuated three-way microvalve fabricated with polydimethylsiloxane using the membrane transfer technique. J. Micromech. Microeng 10, 415 (2000). [Google Scholar]
- 145.Di Carlo D Inertial microfluidics. Lab Chip 9, 3038–3046 (2009). [DOI] [PubMed] [Google Scholar]
- 146.Avila K et al. The onset of turbulence in pipe flow. Science 333, 192–196 (2011). [DOI] [PubMed] [Google Scholar]


