ABSTRACT
Filopodia are actin-dependent finger-like structures that protrude from the plasma membrane. Actin filament barbed-end-binding proteins localized to filopodial tips are key to filopodial assembly. Two classes of barbed-end-binding proteins are formins and Ena/VASP proteins, and both classes have been localized to filopodial tips in specific cellular contexts. Here, we examine the filopodial roles of the FMNL formins and Ena/VASP proteins in U2OS cells. FMNL3 suppression reduces filopodial assembly by 90%, and FMNL3 is enriched at >95% of filopodial tips. Suppression of VASP or Mena (also known as ENAH) reduces filopodial assembly by >75%. However, VASP and Mena do not display consistent filopodial tip localization, but are enriched in focal adhesions (FAs). Interestingly, >85% of FMNL3-containing filopodia are associated with FAs. Two situations increase Ena/VASP filopodial localization: (1) expression of myosin-X, and (2) actively spreading cells. In spreading cells, filopodia often mark sites of nascent adhesions. Interestingly, VASP suppression in spreading cells causes a significant increase in adhesion assembly at filopodial tips. This work demonstrates that, in U2OS cells, Ena/VASP proteins play roles in filopodia beyond those at filopodial tips.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: MYO10, Myosin-10, FMNL1, FMNL2, ENAH, Adhesion
Summary: Both formins and ENA/VASP proteins mediate filopodial assembly. In U2OS cells, the formin FMNL3 functions at filopodial tips, whereas ENA/VASP proteins may influence filopodia at focal adhesions.
INTRODUCTION
Filopodia are finger-like protrusions from the plasma membrane, with diameters generally less than 200 nm. These structures serve multiple purposes, including cell migration, cell–substratum adhesion and cell–cell adhesion (Blanchoin et al., 2014; Chhabra and Higgs, 2007; Faix et al., 2009; Yang and Svitkina, 2011). In addition, filopodia are often the initial sites of microbial contact during phagocytosis or viral infection (Chang et al., 2016; Horsthemke et al., 2017). Both the length and the dynamics of filopodia can vary widely depending on cell type and context.
An essential component of filopodia is a bundle of actin filaments that runs the length of the filopodium. Filaments in this bundle are parallel, with barbed ends at the tip (Blanchoin et al., 2014; Chhabra and Higgs, 2007; Faix et al., 2009; Yang and Svitkina, 2011). Actin filament elongation occurs through actin monomer addition to these barbed ends, and is necessary both for initial filopodial assembly and to maintain filopodial length (Mallavarapu and Mitchison, 1999).
A number of mechanisms have been demonstrated or proposed for actin filament assembly and elongation in filopodia (Faix et al., 2009; Faix and Rottner, 2006; Yang and Svitkina, 2011). The convergent elongation model postulates that filopodial actin filaments assemble from an Arp2/3 complex-nucleated branched actin network (Svitkina et al., 2003), whereby a subset of filaments in the actin network are protected from capping, allowing sustained filament elongation. The tip nucleation model proposes that both nucleation and elongation are mediated by formin proteins, making this model independent of Arp2/3 complex (Yang and Svitkina, 2011). In addition to these two models, myosin-X (symbol MYO10) has been shown to be involved in filopodial assembly in multiple cell lines. Overexpression of myosin-X leads to increased filopodial assembly (Berg and Cheney, 2002; Bohil et al., 2006; Dent et al., 2007; Tokuo et al., 2007), whereas myosin-X suppression or deletion leads to a decrease in filopodia number (Heimsath et al., 2017; Horsthemke et al., 2017; Tokuo et al., 2007). The mechanism by which myosin-X promotes filopodial assembly is unclear. Both endogenous and overexpressed myosin-X are enriched at filopodial tips (Berg and Cheney, 2002; Tokuo and Ikebe, 2004), suggesting that the role of myosin-X might include enhancing barbed-end growth. Additionally, myosin-X has a number of filopodia-associated cargo including integrins and VASP (Tokuo and Ikebe, 2004; Zhang et al., 2004).
All filopodial assembly requires an anti-capping mechanism, which allows sustained filopodial elongation in the presence of barbed-end-capping proteins (Faix et al., 2009; Yang and Svitkina, 2011). Formins and Ena/VASP proteins are prime candidates for this role, being capable of anti-capping biochemically and in cells (Edwards et al., 2014). There is evidence for both formins and Ena/VASP proteins being involved in filopodial assembly. Of the 15 mammalian formins, six have been shown to promote filopodial assembly: mDia1 (also known as DIAPH1), mDia2 (also known as DIAPH3), DAAM1, FMN2, FMNL2 and FMNL3 (Block et al., 2012, 2008; Gauvin et al., 2015; Goh et al., 2012; Harris et al., 2010; Hoffmann et al., 2014; Jaiswal et al., 2013; Matusek et al., 2008; Naj et al., 2013; Pellegrin and Mellor, 2005; Sahasrabudhe et al., 2016; Sarmiento et al., 2008; Yang et al., 2007; Young et al., 2015). In addition, a link between formins and filopodia has been shown in zebrafish (Wakayama et al., 2015), Drosophila (Barzik et al., 2014; Bilancia et al., 2014; Homem and Peifer, 2009; Nowotarski et al., 2014) and Dictyostelium (Schirenbeck et al., 2005).
Of the three mammalian Ena/VASP proteins [VASP, Mena (also known as ENAH) and EVL], all have been shown to contribute to mammalian filopodial assembly (Applewhite et al., 2007; Dent et al., 2007; Kwiatkowski et al., 2007; Lebrand et al., 2004), with additional evidence for filopodial roles for the Ena/VASP family members in Drosophila (Homem and Peifer, 2009) and Dictyostelium (Han et al., 2002; Schirenbeck et al., 2006).
It is unclear, however, whether formins and Ena/VASP proteins play complementary or redundant roles in filopodia assembly. In Drosophila, Enabled and Diaphanous proteins appear to regulate different filopodial populations (Nowotarski et al., 2014), and can act antagonistically in some cases (Bilancia et al., 2014). Additionally, these proteins are not universally enriched in filopodial tips, where the actin filament barbed ends reside. Ena/VASP proteins display clear filopodial tip enrichment in some situations (Applewhite et al., 2007; Bear et al., 2002; Disanza et al., 2013; Lanier et al., 1999; Reinhard et al., 1992; Rottner et al., 1999; Schäfer et al., 2009; Schirenbeck et al., 2006), but are absent from >90% of filopodia generated upon the overexpression of the constitutively active mDia2 (Barzik et al., 2014). VASP also enriches in focal adhesions (FAs), where it plays roles in dorsal stress fiber assembly (Gateva et al., 2014; Tojkander et al., 2015) and focal adhesion dynamics (Young and Higgs, 2018). For the formin family, mDia2, FMNL2 and FMNL3 have been shown to enrich at filopodial tips (Block et al., 2012, 2008; Gauvin et al., 2015; Pellegrin and Mellor, 2005; Yang et al., 2007; Young et al., 2015).
Our previous work has shown a link between FMNL3 and filopodia in mammalian cells. Suppression of FMNL3 in U2OS cells eliminates the majority of filopodia, whereas expression of constitutively active FMNL3 induces filopodia in multiple cell types (Gauvin et al., 2015; Harris et al., 2010; Young et al., 2015). The mechanism for filopodial assembly by constitutively active FMNL3 is dependent on the cell type. Arp2/3 complex is required for optimal filopodial assembly in both suspension-growing (Jurkat T cells) and adherent cells (U2OS and 3T3 fibroblasts), yet the formin mDia1 is only required for filopodial assembly in adherent cells (Young et al., 2015). Overexpression of VASP inhibits FMNL3-induced filopodial assembly in suspension-growing cells but stimulates FMNL3-induced filopodial assembly in adherent cells. Despite this stimulation, VASP localizes predominantly to FAs in adherent cells, and only rarely to filopodial tips that are induced by constitutively active FMNL3. Finally, the majority of filopodia assembled upon expression of constitutively active FMNL3 originate at FAs (Young et al., 2015). These results suggest that FAs could play a role in filopodial assembly in U2OS cells.
In this study, we continue to use U2OS cells as a model system for filopodial assembly, focusing on the roles of the FMNL formins and Ena/VASP proteins. As determined by siRNA-mediated knockdown, FMNL3 is the major FMNL family member contributing to filopodial assembly, with FMNL1 and FMNL2 knockdown having little effect. FMNL3 is present at the tips of almost all filopodia, and the vast majority of filopodia are associated with FAs. Knockdown of either VASP or Mena eliminates the majority of filopodia. However, neither VASP nor Mena enrich consistently in the filopodium itself and are even less frequently found at filopodial tips. In contrast, expression of myosin-X causes VASP and Mena to colocalize with FMNL3 in virtually all filopodial tips. Additionally, when cells are actively spreading, a higher percentage of filopodial tips contain VASP. Interestingly, in spreading cells VASP knockdown significantly raises the percentage of filopodia containing FA proteins at filopodia tips. These results suggest that the roles of Ena/VASP proteins in filopodia are not exclusively at the filopodial tip, and that their FA-associated functions might also be significant for filopodia in certain cell types.
RESULTS
FMNL3-enriched filopodia preferentially associate with focal adhesions
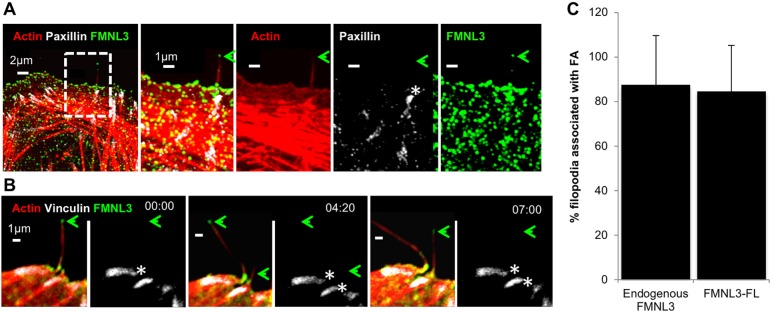
We previously showed that a GFP fusion of the constitutively active FMNL3-FF (FH1-FH2) construct causes assembly of filopodia that are disproportionally associated with FAs (Young et al., 2015). To examine whether filopodia containing endogenous FMNL3 also display a preferential association with FAs, we examined U2OS cells that were fixed after 18 h of plating on fibronectin (FN) and stained for FMNL3, paxillin and actin filaments. Here, the majority of filopodia trace back to FA sites (Fig. 1A,C, 87.5+22.5%, mean±s.d.). We also examined FMNL3-associated filopodia by live-cell microscopy, transfecting U2OS cells with full-length FMNL3–GFP (called FMNL3–GFP throughout), mApple–F-tractin and BFP–vinculin. Similar to what was observed with endogenous FMNL3, a significant proportion of filopodia arise from FAs (Fig. 1B,C, 84.4+20.4%; Movie 1). Together, these findings show that the majority of FMNL3-enriched filopodia associate with FAs and suggest that FAs could be sites for filopodial assembly.
Fig. 1.
FMNL3-containing filopodia preferentially associate with FAs. (A) Endogenous FMNL3 localization. U2OS cells were plated on FN-treated coverslips for 18 h before fixation and staining with TRITC–phalloidin (red), paxillin (white) and FMNL3 (green). The green arrow indicates a FMNL3-enriched filopodium. The white asterisk indicates an FA at the base of the filopodium. Micrographs are maximum intensity projections (MIPs) of 13 0.18 μm z-slices. (B) FMNL3–GFP localization. Timelapse confocal montage of U2OS cell expressing mApple–F-tractin (red), BFP–vinculin (white) and FMNL3–GFP (green), plated on an FN-treated coverslip at 18 h prior to imaging. The green arrow shows filopodia tips enriched with FMNL3. The white asterisks indicate a FA at the base of the filopodium. Times are in min:sec. These images are stills from Movie 1. (C) Quantification of the percentage FMNL3-containing filopodia that associate with focal adhesions. Endogenous FMNL3, n=58 filopodia, 14 cells, four experiments. FMNL3–GFP: n=126 filopodia, 12 cells, three experiments. Error bars show the s.d.
VASP does not localize to most filopodial tips in spread U2OS cells
We have found that FMNL3 enriches in filopodial tips in several contexts (Fig. 1A,B; Gauvin et al., 2015; Harris et al., 2010; Young et al., 2015). ENA/VASP proteins are also associated with filopodial tips (Applewhite et al., 2007; Barzik et al., 2014; Bear et al., 2002; Disanza et al., 2013; Lanier et al., 1999; Reinhard et al., 1992; Rottner et al., 1999; Schäfer et al., 2009; Schirenbeck et al., 2006), potentially due to their barbed-end-binding properties. Additionally, VASP is a well-known component of FAs (Brindle et al., 1996; Kanchanawong et al., 2010; Reinhard et al., 1992).
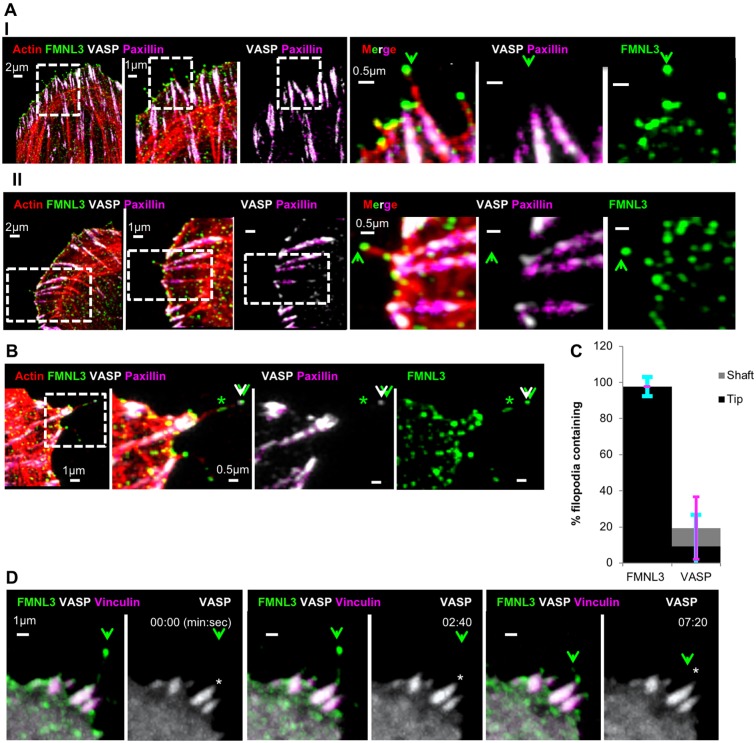
For these reasons, we examined FMNL3 and VASP localization in filopodia and the associated FA in U2OS cells. As determined by fixed-cell immunofluorescence, the majority of filopodia contain FMNL3 at their tips (97.7±5.2%, Fig. 2A–C), and FMNL3 occasionally is present along filopodial shafts (Fig. 2B). VASP displays significant localization at FAs (Fig. 2A,B), but is infrequently found in filopodia, with tip localization in only 9.3±17.4% and shaft localization in 9.6±17.3% of filopodial examined (Fig. 2B,C; Fig. S1A). This shaft localization is most often near the base, and somewhat contiguous with the FAs (Fig. S1A).
Fig. 2.
FMNL3 localizes to filopodia tips, whereas VASP localizes to associated focal adhesions. (A) Endogenous proteins. U2OS cells were plated on FN-treated coverslips for 18 h before fixation and staining for TRITC–phalloidin (red), VASP (white), FMNL3 (green) and paxillin (magenta). Two representative image sets are provided (I and II). Each set contains both an expanded view (left) and a magnified view (right). The green arrowhead indicates filopodia with FMNL3 enriched at the tip. Micrographs are maximum intensity projections (MIPs) of 0.18 μm z-slices (I, 8; II, 11 slices). (B) Endogenous protein example (similar to in A) showing both FMNL3 and VASP enriched at a filopodial tip (white/green arrowheads), as well as FMNL3 shaft enrichment (green asterisk). Expanded view on left, and magnified images on right. Micrographs are MIPs of 0.18 μm z-slices, 7 slices. (C) Quantification of filopodia containing endogenous FMNL3 or VASP within their shafts (gray) or tips (black), from fixed-cell images. n=73 filopodia, 16 cells, three experiments. These results are P<0.0001 for comparative tip localization for FMNL3 and VASP, and not significant, for comparative shaft localization for FMNL3 and VASP, as calculated by two sample t-test. Error bars show the s.d.: cyan indicates tip, magenta indicates shaft. (D) Fluorescent fusion proteins. Time-lapse confocal montage of U2OS cell expressing FMNL3–GFP (green), mCherry–VASP (white) and BFP–vinculin (magenta), plated on FN-treated coverslips at 18 h prior to imaging. The green arrowhead indicates FMNL3-enriched filopodium. White asterisks indicate VASP localization in focal adhesion at base of filopodium. Times are in min:sec. These images are stills from Movie 2.
One possibility for our low detection of VASP in filopodia is that VASP concentration is lower at filopodial tips versus FAs, which might mask the tip signal. However, over-saturating the FA-VASP signal does not reveal any filopodial tip VASP signal above background levels (Fig. S1B). Similar patterns of FMNL3 and VASP localization occur when cells are plated on collagen (Fig. S1C,D).
We also used live-cell microscopy to examine the localization of a fluorescently tagged VASP construct in relation to fluorescently tagged FMNL3 and vinculin. FMNL3–GFP enriches primarily as bright puncta at the filopodial tip. In contrast, mCherry–VASP is not detectable at the filopodial tip but displays significant accumulation at the associated FAs (Fig. 2D; Movie 2).
Our low level of VASP localization in filopodia is similar to that obtained upon transfection of a constitutively active mDia2 construct into Ena/VASP-null cells re-expressing fluorescently tagged VASP, in which VASP displays tip localization in <10% of filopodia (Barzik et al., 2014). We obtain similar results in U2OS cells when using the constitutively active GFP–mDia2-FFC construct (Harris et al., 2010), as determined by both immunofluorescence of endogenous VASP (Fig. S1E) and when using transfected mCherry–VASP (Fig. S1F). Taken together, these results suggest that VASP is not consistently present at filopodial tip protein in spread U2OS cells.
Suppression of FMNL3, VASP or Mena reduces filopodial number
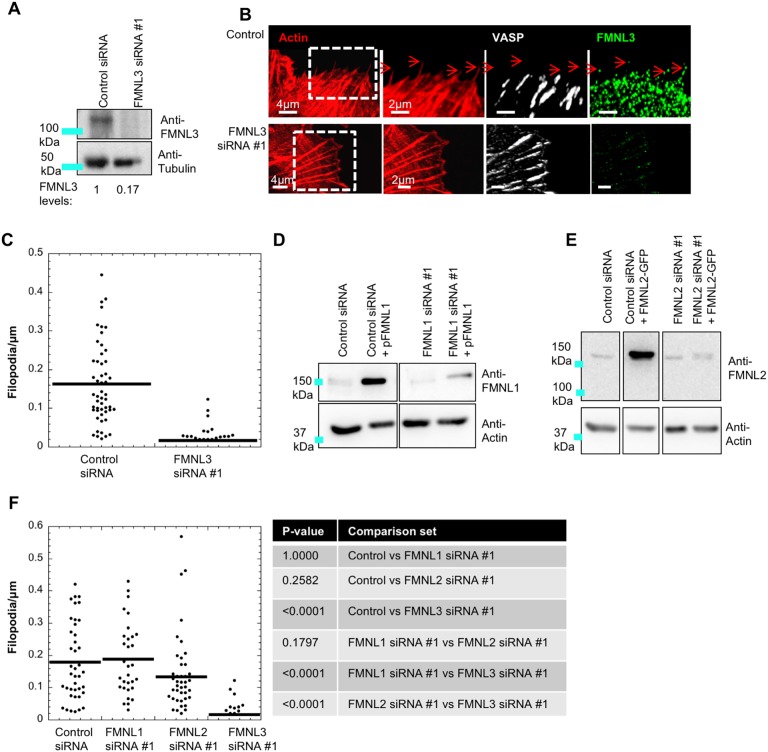
Given that FMNL and Ena/VASP proteins have been shown to contribute to filopodial assembly in multiple contexts, we examined their importance for filopodia assembly in spread U2OS cells via siRNA-mediated knockdown. We first examined the dependence of U2OS filopodia on the FMNL family of formins. Similar to what we found in our previous work (Gauvin et al., 2015), FMNL3 suppression results in a 90% reduction in filopodia, as quantified by filopodial density at the cell edge (Fig. 3A–C). In contrast to FMNL3, FMNL1 and FMNL2 are less readily detectable by western blot analysis of U2OS extracts (Fig. 3D,E). We tested siRNA effectiveness using transfected FMNL1 and FMNL2 constructs and established efficient knockdown of the exogenously expressed proteins (Fig. 3D,E). For endogenous FMNL1, siRNA reduces a faint band at the expected size (150 kDa), suggesting low endogenous FMNL1 levels (Fig. 3D). For endogenous FMNL2, siRNA does not reduce band intensity in the expected size range (100–150 kDa, Fig. 3E), suggesting that endogenous FMNL2 levels are below the level of detection. Since the siRNAs did reduce expression of exogenous FMNL1 and FMNL2, we compared their effectiveness against that of FMNL3 knockdown. Neither siRNA treatment for FMNL1 nor FMNL2 causes a significant change to filopodia density compared to control cells, which is significantly different to the FMNL3 effect (as calculated by one-way ANOVA analysis followed by post hoc Dunn's multiple comparison test; Fig. 3F). A second siRNA for FMNL1, FMNL2 and FMNL3 produces similar effects (Fig. S2A–D). These results suggest that FMNL3 is the predominant FMNL formin required for filopodial assembly in U2OS cells. Interestingly, one FMNL2 siRNA causes a significant increase in FMNL3 protein levels (Fig. S2B).
Fig. 3.
Suppression of FMNL3 causes significant reduction in filopodia density in U2OS cells. (A) Western blot of U2OS cells treated with control siRNA or FMNL3 siRNA #1, probed for FMNL3 or tubulin (loading control). Numbers beneath lanes indicate the relative amount of FMNL3 remaining. (B) U2OS cells treated with control siRNA or FMNL3 siRNA #1 cells were fixed and stained for TRITC–phalloidin (red), FMNL3 (green) and VASP (white). Micrographs are maximum intensity projections (MIPs) of 0.18 μm z-slices (6–12 slices); red arrows indicate filopodia. (C) Dot plot of filopodial density for cells treated with control (mean 0.162±0.101) and FMNL3 siRNA #1 (mean 0.016±0.012). n=52 (control siRNA) and 44 (FMNL3 siRNA #1) cells, four experiments. These results are P<0.001, as calculated by a Mann–Whitney U-test. Errors are given as s.d. (D) Western blot for FMNL1 in U2OS cells transfected with either control siRNA or FMNL1-directed siRNA #1. Two samples also transfected with plasmid expressing human FMNL1 are shown (pFMNL1). Western blots are probed for FMNL1 (top) or actin (loading control). All lanes are from one gel, with intervening lanes removed. Image processing of all lanes is identical. (E) Western blot for FMNL2 in U2OS cells, transfected with either control siRNA or FMNL2-directed siRNA #1. Two samples also transfected with plasmid expressing human FMNL2 (FMNL2–GFP) are shown. Western blots are probed for FMNL2 (top) or actin (loading control). All lanes are from one gel, with intervening lanes cut out. Image processing of all lanes is identical. (F) Dot plot of filopodia density following treatment with control (mean 0.180±0.118), FMNL1 #1 (mean 0.187±0.111), FMNL2 #1 (mean 0.133±0.120), and FMNL3 #1 (mean 0.017±0.032) siRNAs. n=41 (control), 31 (FMNL1 #1), 44 (FMNL2 #1) and 32 (FMNL3 #1) cells, three experiments. Errors are given as s.d. The table shows P-values calculated by one-way ANOVA analysis followed by post hoc Dunn's multiple comparison test.
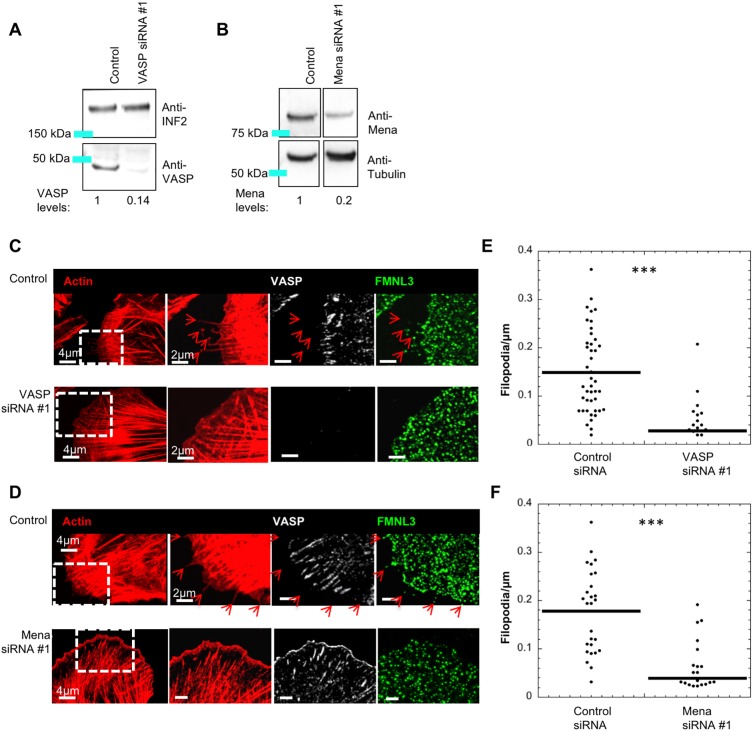
We next examined the role of Ena/VASP proteins via siRNA-mediated knockdown. Both VASP and Mena are readily detectable in U2OS cells, and are depleted by siRNA treatment (Fig. 4A,B). In contrast, we could not definitively identify a western blot band for EVL that was reduced by siRNA treatment (not shown), so we did not analyze the role of EVL in filopodial assembly further. We evaluated the effect of VASP or Mena suppression on filopodial density in U2OS cells. Suppression of either protein results in a significant decrease in filopodial density, with depletion of both proteins in combination not causing a significantly further decrease in filopodia density (Fig. 4C–F; Fig. S2F). A second siRNA for VASP and Mena produces similar effects (Fig. S2A,E,F). These results suggest that both VASP and Mena play roles in filopodial assembly in spread U2OS cells, and that their specific roles cannot be fully compensated for by the other family member.
Fig. 4.
Suppression of VASP or Mena causes filopodial reduction in U2OS cells. (A) Western blot of U2OS cells treated with control siRNA or VASP siRNA#1, then probed for VASP or INF2 (loading control). Numbers beneath lanes indicate the relative amount of VASP remaining. (B) Western blot of U2OS cells treated with control siRNA or Mena-siRNA #1, then probed for Mena or tubulin (loading control). Numbers beneath western lanes indicate the relative amount of of Mena remaining. All lanes are from one gel, with intervening lanes removed. Image processing of all lanes is identical. (C,D) U2OS cells treated with control siRNA, VASP siRNA #1 or Mena siRNA #1 were fixed and stained with TRITC–phalloidin (red), and anti-FMNL3 and anti-VASP antibodies. Micrographs are maximum intensity projections (MIPs) of 0.18 μm z-slices (4–8 slices); red arrows indicate filopodia. (E,F) Dot plots of filopodial density for VASP and Mena knockdown, respectively. (E) Control (mean 0.152±0.086) and VASP siRNA #1 (mean 0.028±0.043) n=30 cells, five experiments. (F) Control (mean 0.171±0.079) and Mena siRNA #1 (mean 0.039±0.051) n=32 cells, three experiments. ***P-value <0.0001 as calculated by Mann–Whitney U-test. Errors are given as s.d.
Having identified that Mena also plays a role in filopodial abundance in U2OS cells, we examined Mena localization in spread U2OS cells, focusing on filopodia and associated FAs. Both endogenous Mena (Fig. S3A) and plasmid-expressed GFP–Mena (Fig. S3B) display no apparent enrichment in filopodia but are abundant in FAs. Plasmid-expressed GFP–EVL similarly displays FA enrichment but is not detectable in filopodia (Fig. S3C). Taken together, our results suggest that Ena/VASP proteins are not constitutively present at filopodial tips in U2OS cells, yet still significantly contribute toward filopodial assembly.
Ena/VASP localization to filopodial tips upon myosin-X expression
The low percentage of filopodial tips displaying Ena/VASP enrichment in U2OS cells was surprising. One possibility is that U2OS cells have low levels of myosin-X, which is known to promote tip localization of VASP in HeLa cells (Tokuo and Ikebe, 2004). Supporting this possibility, western blotting shows that U2OS whole-cell extract contains less myosin-X than HeLa whole-cell extract (Fig. S4A).
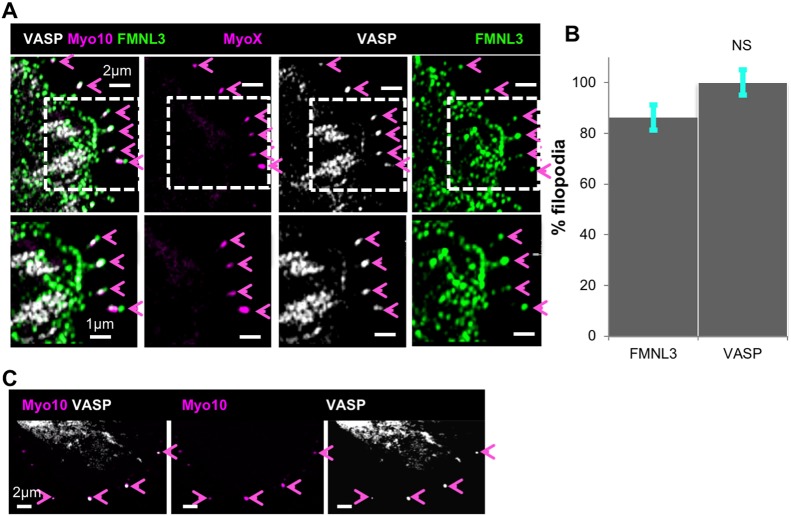
We next asked whether filopodia induced by myosin-X overexpression in U2OS cells displayed greater VASP filopodial enrichment than those in untransfected cells. U2OS cells were transfected with GFP–myosin-X, then plated on FN for 18 h before fixation and staining for VASP and FMNL3 (Fig. 5A), or VASP and actin filaments (Fig. S4B). As reported previously (Berg et al., 2000; Bohil et al., 2006), transfection with myosin-X induces filopodia assembly (Fig. S4C), with myosin-X enriching at filopodial tips (Fig. S4B). In 100% of myosin-X-containing filopodia, VASP is also found at filopodia tips, where it is often observed as a bright puncta (Fig. 5A,B). In comparison, FMNL3 localizes to 86±18% (mean±s.d.) of myosin-X-containing filopodia tips (Fig. 5A,B). Similar results are observed on collagen-plated cells (Fig. S4D,E). In addition, live-cell imaging reveals that mCherry–VASP and GFP–myosin-X co-localize in dynamic filopodial tips (Fig. 5C; Movie 3).
Fig. 5.
VASP enriches at filopodial tips upon myosin-X expression. (A) U2OS cells were transfected with GFP–myosin-X (magenta) and plated on FN-treated coverslips for 18 h before fixation and staining with anti-VASP (white) and anti-FMNL3 (green). Magenta arrowheads indicate myosin-X enriched filopodia tips, which in all cases contain VASP and FMNL3. Bottom panels are magnified views of the areas indicated in the top panels. Micrographs are maximum intensity projections (MIPs) of 0.18 μm z-slices, 5 slices. (B) Quantification of FMNL3 and VASP at filopodial tips in GFP–myosin-X-transfected cells. n=130 filopodia, 17 cells, three experiments. NS, not significant, as calculated by a two-sample t-test. Error bars show s.d. (C) Single time point from time-lapse montage of a U2OS cell expressing GFP–myosin-X (magenta) and mCherry–VASP (white), plated on an FN-treated coverslip for 18 h before imaging. Magenta arrowheads indicate VASP and myosin-X-enriched filopodia. Images were acquired through Airyscan microscopy. Time is 9:00 (min:sec), taken from Movie 3.
We also examined GFP–Mena and GFP–EVL localization in mCherry–myosin-X co-transfected cells, and observe that they too localize to filopodial tips under these conditions (Fig. S4F,G). These results suggest that our inability to detect Ena/VASP proteins at filopodial tips is not due to insufficient probe sensitivity, since these proteins are readily detected at filopodial tips upon myosin-X expression.
VASP depletion alters the filopodia–adhesion relationship during spreading
We next examined whether VASP and FMNL3 show similar filopodial localization patterns in actively spreading cells, using a collagen-based system we recently developed (Young and Higgs, 2018). Here, cells spread rapidly and uniformly in the first hour after plating, and collagen-plated cells typically display higher filopodial density during spreading (Fig. S5A–C).
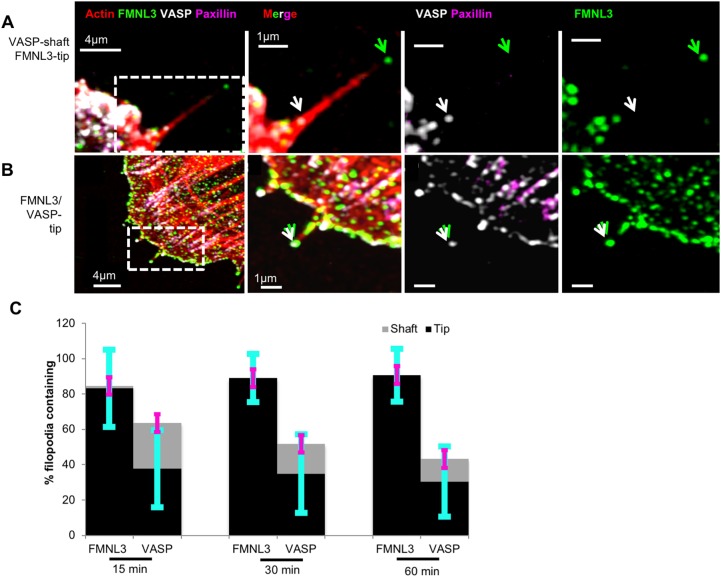
U2OS cells were fixed at various times up to 1 h post-seeding, then co-stained for FMNL3, VASP, paxillin and actin filaments. As with 18 h plated cells, FMNL3 is enriched in the majority of filopodia, where it faithfully localizes to filopodia tips (15 min: 83%, 30 min: 89%, 1 h: 90%, Fig. 6A–C). Interestingly, at these time-points a significantly higher number of filopodia contain VASP compared to at 18 h post-seeding (15 min, 64%; 30 min, 52%; 1 h, 33%; Fig. 6C), with a higher percentage of these occurrences being at filopodial tips (15 min, 38%; 30 min, 35%; 1 h, 20%; Fig. 6A–C). As with collagen-plated cells, there is an increase in VASP localization to filopodia in spreading FN-plated cells (Fig. S5D).
Fig. 6.
Percentage of filopodia containing VASP increases during cell spreading. U2OS cells were plated on collagen-treated coverslips for various times (15 min, 30 min and 1 h) before fixation and staining with TRITC–phalloidin (red), anti-VASP (white), anti-FMNL3 (green) and anti-paxillin (magenta) antibodies. The green arrowhead indicates FMNL3 at filopodial tips. (A) Example of VASP in the shaft (white arrowhead). (B) Example of VASP at the tip (white arrowhead). (C) Percentage of filopodia at leading edge of spreading cells containing FMNL3 or VASP within their tip or shafts. 15 min, n=212 filopodia, 22 cells; 30 min: n=168 filopodia, 22 cells; 1 h: n=197 filopodia, 24 cells, three experiments. All results are P<0.0001 (comparative tip and shaft localization for FMNL3 and VASP), as calculated with a two-sample t-test. Error bars are s.d.: cyan indicates tip, magenta indicates shaft.
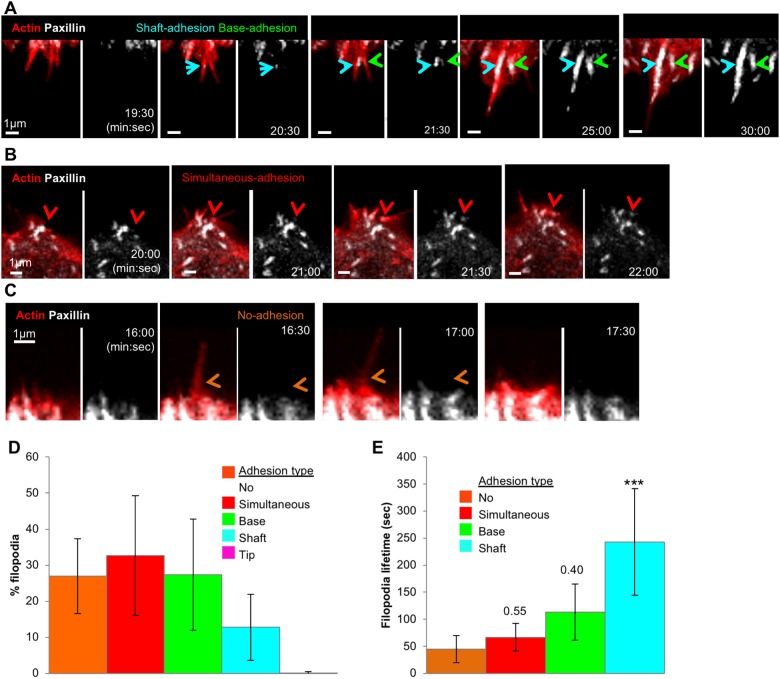
Given the relationship between filopodia and FAs in spread U2OS cells (Young et al., 2015 and Fig. 1 of this work), we analyzed the relationship between filopodia and adhesions during cell spreading. By performing live-cell imaging of cells co-transfected with mApple–F-tractin and GFP–paxillin, we determined that the majority of filopodia (73%) are associated with the assembly of nascent adhesions, which we divide into three main classes: adhesions that initially appear at the base of pre-formed filopodia (‘base adhesion’, 27.4±15.4%, Fig. 7A; Movie 4), adhesions that initially appear within pre-formed filopodial shafts (‘shaft adhesion’, 12.8±9.1%, Fig. 7A; Movie 4), and adhesions that assemble simultaneously with filopodia (‘simultaneous adhesion’, 32.7±16.8%, Fig. 7B; Movie 5). Another population of filopodia is not accompanied by clear adhesion assembly (‘no adhesion’, 26.9±10.3%, Fig. 7C; Movie 6). An extremely infrequent event is the assembly of new adhesions at filopodial tips (‘tip adhesion’) with only two such instances out of 542 total filopodia (Fig. 7D). Interestingly, filopodial lifetime changes with the position of the adhesion, with filopodia containing shaft adhesions being appreciably more stable than the other filopodia–adhesion classes (243±98 s for shaft adhesions, versus 113±51, 67±52 and 45±26 s for base adhesions, simultaneous adhesions and no adhesions, respectively, mean±s.d.; Fig. 7E).
Fig. 7.
Most filopodial assembly precedes adhesion assembly during early spreading. Time-lapse Airyscan microscopy examination of U2OS cells, expressing GFP–paxillin (white) and mApple–F-tractin (red), and spreading on collagen. All imagining conducted using Airyscan microscopy. Time is in min:sec. (A) Example of both a shaft adhesion (cyan arrow) and base adhesion (green arrow). These images are stills from Movie 4. (B) Example of a simultaneous adhesion (red arrow). These images are stills from Movie 5. (C) Example of filopodium assembling with no accompanying adhesion (orange arrows). These images are stills from Movie 6. (D) Quantification of five categories of filopodium–adhesion relationship during cell spreading, including: no adhesion, simultaneous adhesion, base adhesion, shaft adhesion and tip adhesion. n=542 filopodia, 15 ROIs, eight cells, five experiments. Error bars are s.d. (E) Filopodial lifetimes by category during cell spreading. n=358 filopodia, 12 ROIs, five cells, three experiments. ***P<0.0001 compared to no adhesion filopodia (one-way ANOVA analysis followed by post hoc Dunnett multiple comparison test). Error bars are s.d.
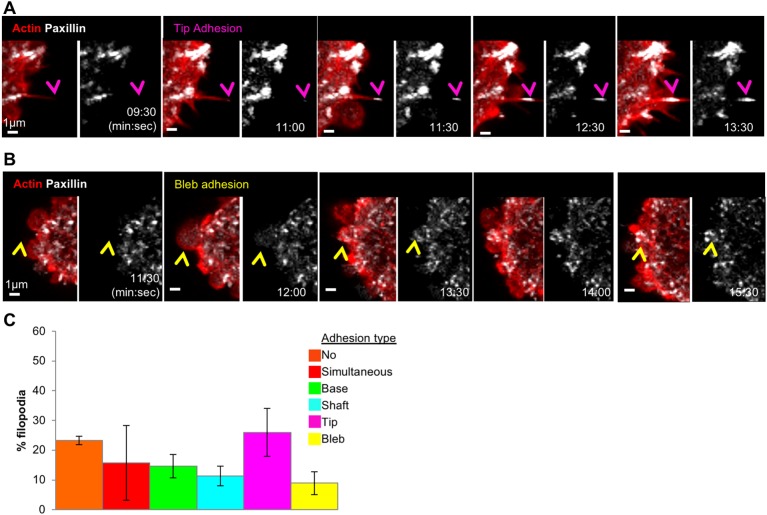
We next examined adhesion assembly during cell spreading in VASP-depleted cells. VASP depletion leads to a significant increase in the percentage of tip adhesions (26.0±8.1% for VASP-depleted versus 0.13±0.34% for control cells, mean±s.d.; P=0.002, Fig. 8A,C; Movie 7). Similar results were obtained for a second siRNA (Fig. S6A). In VASP-depleted cells, the filopodial lifetime for filopodia with tip adhesion assembly is 141.1±94 s (n=45 filopodia). Another subpopulation of VASP-depleted cells produce blebs that lead to new adhesion assembly (Fig. 8B,C). VASP depletion does not significantly reduce the number of filopodia assembled during spreading (control 28.3±14.4 filopodia/cell; VASP-depleted 22.8±14.0; Fig. S6A) or the spreading rate (Fig. S6B). These results suggest that VASP may play a different role in filopodial assembly during early U2OS cell spreading, which is related to its function in adhesions.
Fig. 8.
VASP depletion causes increased tip adhesion assembly during cell spreading. Time-lapse Airyscan microscopy examination of VASP-depleted (siRNA #1-treated) U2OS cells expressing GFP–paxillin (white) and mApple–F-tractin (red), and spreading on collagen. (A) Example of tip adhesion (magenta arrowhead). These images are stills from Movie 7. Images were acquired through Airyscan microscopy. Time is in min:sec. (B) Example of bleb adhesion (yellow arrowhead). Images were acquired through Airyscan microscopy. Time is in min:sec. (C) Quantification of filopodial–adhesion relationship categories in VASP-depleted U2OS cells spreading on collagen. n=182 filopodia, five cells, three experiments. Error bars are s.d.
DISCUSSION
In this work, we focus on U2OS cells specifically, in order to understand filopodial assembly mechanisms in one cell type. Other cell types might share some mechanistic features. However, as filopodia are quite diverse in both morphology and assembly mechanisms (Blanchoin et al., 2014; Chhabra and Higgs, 2007; Faix et al., 2009; Yang and Svitkina, 2011; Young et al., 2015), a number of details are likely to be cell type specific. For example, the fact that U2OS cells are derived from bone may make their mechanical requirements different from other cell types commonly used.
We examine all three FMNL family members for filopodial roles here. We find FMNL3 is an important filopodial-associated formin in spread U2OS cells, being required for optimal filopodial assembly and localizing to the tips of most filopodia. These results are consistent with our previous studies (Gauvin et al., 2015; Harris et al., 2010; Young et al., 2015) and with studies on zebrafish endothelial cells (Wakayama et al., 2015). FMNL2 has also been associated with filopodia in other cell types (Block et al., 2012), while there has been no previous evidence for FMNL1 in filopodial function. In U2OS cells, FMNL3 is readily detectable, whereas western blot analysis shows low levels of FMNL1 and no clear band for FMNL2. By performing siRNA treatment, we find that only FMNL3 knockdown has a significant effect on filopodial density. We conclude that FMNL1 and FMNL2 do not play major roles in the assembly of filopodia in U2OS cells under these conditions, which could be due to their low expression levels. Alternatively, the increase in FMNL3 levels upon FMNL2 siRNA treatment suggests the expression of a low level of FMNL2 that might play a defined role in filopodial assembly, which is counteracted by increased FMNL3.
Both VASP and Mena play roles in filopodia as well, since their suppression causes >75% reduction in filopodial density. However, we detect neither VASP nor Mena in the tips of most filopodia examined in U2OS cells, either when probing for endogenous protein by immunofluorescence or when tracking fluorescently tagged protein by live-cell microscopy. These results surprised us, given the tip localization of ENA/VASP in other studies (Applewhite et al., 2007; Barzik et al., 2014; Bear et al., 2002; Disanza et al., 2013; Lanier et al., 1999; Reinhard et al., 1992; Rottner et al., 1999; Schäfer et al., 2009; Schirenbeck et al., 2006). One possibility is that there are low levels of ENA/VASP at filopodial tips, which might be masked by high levels at the FAs. However, we could not detect any signal above background level at filopodial tips even with extensive adjustments of fluorescence levels. This situation is in contrast to that for FMNL3 or mDia2, in which endogenous or GFP fusion proteins display intense tip staining.
Overexpression of myosin-X results in VASP and Mena localization to all filopodial tips, confirming our ability to detect ENA/VASP at filopodial tips. This result agrees with previous studies showing that VASP binds the C-terminal region of myosin-X, and that myosin-X is required for efficient transport of VASP to filopodial tips (Bohil et al., 2006; Tokuo et al., 2018; Tokuo and Ikebe, 2004). Furthermore, we find comparatively low levels of myosin-X in U2OS cells compared with HeLa cells, which have previously been shown to contain endogenous myosin-X and VASP colocalized at filopodial tips (Tokuo and Ikebe, 2004). These results suggest that myosin-X is required for efficient Ena/VASP tip localization, at least in U2OS cells. Other proteins have been shown to bind VASP and mediate its localization to filopodial tips, such as IRSp53 (Disanza et al., 2013) and dDia2 in Dictyostelium (Schirenbeck et al., 2006). Interestingly, mDia2 has also been shown to interact with VASP (Barzik et al., 2014), but that study also reported that VASP was largely absent from filopodial tips induced by expression of constitutively active mDia2. Additionally, lamellipodin can bind VASP and localize it to the lamellipodial leading edge (Krause et al., 2004).
We note, the mechanism by which myosin-X stimulates filopodial assembly is still unclear at present, with the interaction with Ena/VASP being one possibility. However, this interaction is not essential for myosin-X-mediated filopodial assembly, since myosin-X constructs lacking the C-terminus (missing the VASP-binding domain) still induce filopodia (Bohil et al., 2006; Tokuo et al., 2007) and myosin-X expression can induce filopodia in neurons lacking all Ena/VASP proteins (Dent et al., 2007). Nonetheless, Ena/VASP proteins may stimulate filopodial elongation in myosin-X-transfected cells, since expression of myosin-X constructs lacking its C-terminus produce shorter filopodia (Tokuo et al., 2007).
An interesting question is whether two classes of barbed-end elongation factors are needed for the 10–30 barbed ends that are present at the filopodial tip. Two studies have suggested that formins and VASP might control distinct filopodial pools with differing properties (Barzik et al., 2014; Nowotarski et al., 2014). Formins are, in general, more processive barbed-end elongation factors than ENA/VASP proteins (Breitsprecher et al., 2008; Hansen and Mullins, 2010; Kovar et al., 2006). However, the processivity of VASP can be enhanced by a number of mechanisms, including clustering (Breitsprecher et al., 2008; Disanza et al., 2013), fascin-mediated filament crosslinking (Winkelman et al., 2014) and tethering to filaments by lamellipodin (Hansen and Mullins, 2015). Concerning filopodia specifically, the actions of myosin-X (Bohil et al., 2006; Tokuo and Ikebe, 2004) and IRSp53 (Disanza et al., 2013) might increase the ability of Ena/VASP proteins to serve as barbed-end elongation factors at filopodial tips, through clustering. Interestingly, we find that FMNL3 also enriches at the majority of filopodial tips in myosin-X-expressing cells, showing that their simultaneous presence is possible. The coordinated regulation of formins and Ena/VASP proteins might dictate the balance of their activities at the filopodial tip.
Despite Ena/VASP proteins not being localized to a high percentage of filopodia in U2OS cells, it is clear that both VASP and Mena play roles in filopodial assembly or maintenance in this cell type. One possibility is that VASP and/or Mena contribute to filopodial assembly through their functions at the associated FAs. We find that most FMNL3-containing filopodia are associated with FAs at the base, in agreement with other studies linking filopodia and FAs (He et al., 2017; Kanchanawong et al., 2010; Reinhard et al., 1995; Steketee et al., 2001; Steketee and Tosney, 2002; Young et al., 2015). These results could suggest that FAs provide components for filopodial assembly. Furthermore, a recent publication shows myosin-X accumulation at nascent adhesions prior to filopodia assembly, but following initial actin accumulation (He et al., 2017). FAs contain populations of short actin filaments (Cramer et al., 1997; Patla et al., 2010) and are the site of dorsal stress fiber assembly (Hotulainen and Lappalainen, 2006). Interestingly, two proteins have been shown to play roles in both filopodial and dorsal stress fiber assembly, mDia1 (Hotulainen and Lappalainen, 2006; Young et al., 2015) and VASP (Gateva et al., 2014 and this work). It is possible that short filaments assembled at the FAs could be used for both filopodia and/or dorsal stress fiber assembly.
It is interesting that knockdown of either VASP or Mena results in loss of the majority of filopodia. This result suggests that the two proteins play non-redundant roles in filopodial assembly. One possibility is that VASP and Mena might bind barbed-ends at different times during filopodial assembly, or that one of the proteins is indirectly linked to filopodia. For example, we recently showed that VASP plays a role in the FA ‘splitting’ process, whereby FAs longitudinally divide into 300 nm width units (Young and Higgs, 2018). Alternately, VASP and Mena might work together in the same process, with specific functions for each. Interestingly, VASP and Mena are capable of assembling into mixed oligomers, with unclear functional consequences (Riquelme et al., 2015).
Finally, we find an interrelationship between filopodia and FAs in spreading cells, in which filopodia are frequently the sites of new adhesion assembly, and filopodia that are associated with adhesion assembly have longer lifetimes. These results agree with previous studies (Galbraith et al., 2007; Johnson et al., 2015; Schäfer et al., 2009; Steketee and Tosney, 2002). An interesting finding is that VASP depletion significantly increases the percentage of ‘tip adhesions’; adhesions that first appear at the filopodial tip. Tip adhesions have been observed in other cells (Steketee and Tosney, 2002), but are rarely detected in wild-type U2OS cells. The mechanism regulating the positioning of new adhesion assembly in relation to filopodia is unclear at present. Interestingly, another formin, FMN2, has recently been reported to contribute to tip adhesions in growth cones (Sahasrabudhe et al., 2016).
In summary, our results suggest a relationship between filopodia and FAs in which actin polymerization factors are utilized in ways that are distinct from their canonical roles.
MATERIALS AND METHODS
Cell culture
Human osteosarcoma U2OS cells and human cervical cancer HeLa cells (American Type Culture Collection HTB96 and CCL2, respectively) were grown in DMEM (Invitrogen) supplemented with 10% newborn calf serum (Hyclone) and at 37°C and 5% CO2. Cell lines used for a maximum of 25 passages, and were routinely tested for contamination.
DNA transfections
DNA constructs were introduced using JetPrime transfection reagent (PolyPlus); a total of 0.3 μg of each plasmid DNA was used for all transfections. Cells were transfected for 24 h before fixation or live-cell imaging.
The following expression constructs were used: FL-FMNL3-GFPint (called FMNL3–-GFP throughout, described previously in Gauvin et al., 2015); mouse mCherry-VASP (Addgene #55151; deposited by Michael Davidson); chicken paxillin-GFP (Addgene #15233; Laukaitis et al., 2001); F-tractin-mApple (gift from Clare Waterman, NIH, Bethesda, MD); human BFP-vinculin (Addgene #55252; deposited by Michael Davidson); mouse GFP-mDia2-FFC described previously in Harris et al. (2010); human GFP-MyosinX (Addgene #47608, Bennett et al., 2007); bovine Myosin10-mCherry (gift from Johanna Ivaska, University of Turku, Finland; Plantard et al., 2010); mouse GFP-Mena and GFP-EVL (gifts from Stephanie Gupton, University of North Carolina-Chapel Hill, NC; Loureiro et al., 2002); human FMNL1 in pcDNA-3.1 vector (gift from Angela Krackhardt, Technische Universitat Munchen, Germany; Han et al., 2009); and human FMNL2-GFP (gift from Robert Grosse, Phillips Universitat Marburg, Germany; Wang et al., 2015).
Before fixation or live-cell imaging, cells were plated onto glass bottom 35 mm dishes (MatTek P35G-1.5-14-C) pre-coated for 1 h (4°C) with 50 μg/ml collagen (PureCol, Advanced BioMatrix, #5005) or 10 μg/ml fibronectin (Sigma-Aldrich, FL1141). For ‘spread’ cells, ∼120,000 cells were plated at least 18 h prior to fixation or live-cell imaging. For ‘spreading’ cells, ∼120,000 cells were plated and cells were fixed at 15 min, 30 min or 1 h after plating, or live-cell imaging was started 8 min after plating.
siRNA treatment
50 nM of each siRNA oligonucleotide was introduced using Lipofectamine® RNAiMAX following the manufacturer's instructions. Cells were analyzed 72 h post transfection. Oligonucleotides for siRNA included: human FMNL1 (synthesized by IDT, hs.Ri.FMNL1.13.5, siRNA #1 5′-GUGGUACAUUCGGUGGAGAACAUGA-3′ and hs.Ri.FMNL1.13.4, siRNA #2 5′-GGCAACUACAUGAACAGUGCUUGCUACUGUUCAUGU-3′); human FMNL2 (synthesized by IDT, hs.Ri.FMNL2.13.1, siRNA #1 5′-CAUGAUGCAGUUUAGUAA-3′, and hs.Ri.FMNL2.13.6, siRNA #2 5′-AAAAAAAGAUUCAUGAGC-3′); human FMNL3 (synthesized by Ambion, s40551, siRNA #1 5′-GCAUCAAGGAGACAUAUGA, and s40549, siRNA #2 5′-CATTCGTTCTTACAAGGAA-3′); human VASP (synthesized by Dharmacon, siRNA #1 J-019763-09, 5′-GAGUGAAUCUGUGCGGAGA-3′, and synthesized by IDT hs.Ri.VASP.13.1, siRNA #2 5′-CCUCUACUUGACUUGGAAUUGGC-3′); human Mena (synthesized by IDT, siRNA #1 hs.Ri.ENAH.13.3, 5′-CCUAAAGGGUUGAAGUACAAUCAAG-3′, and hs.Ri.ENAH.13.1, siRNA #2 5′-CCAGUGUUGAAUCAUGCUAUAUU-3′); and negative control (synthesized by IDT, #51-01-14-04 5′-CGUUAAUCGCGUAUAAUACGCGUAU-3′).
Antibodies
Mouse anti-Mena [Santa Cruz Biotechnology; sc-135988, 1:150 for western blotting (WB), 1:50 for immunofluorescence (IF)]; rabbit mouse anti-VASP 9A2 (Cell Signaling Technology, #3132, 1:1000 for WB, 1:50 for IF); guinea-pig anti-FMNL3 [as described in Harris et al. (2010), 1:1000 for WB, 1:500 for IF); mouse anti-FMNL2 (Santa Cruz Biotechnology, sc-390208, 1:700 for WB); rabbit anti-FMNL1, raised against the FH1-FH2-DAD-C fragment (amino acids 449–1094) of mouse FMNL1 (Harris et al., 2004), used at 0.4 μg/ml for WB; rabbit anti-INF2 [as described in Ramabhadran et al., (2011), 1/1000 for WB]; mouse anti-tubulin DM1-α (Sigma-Aldrich MABT205, 1:10,000 for WB); mouse anti-paxillin (BD Biosciences, #610569, 1:50 for IF); rabbit anti-myosin-X (Novus Biologicals, #22430002, 1:800 for WB); mouse anti-actin (Millipore MAB1501, 1:5000 for WB). Secondary antibodies were: Alexa Fluor 405-conjugated anti-mouse IgG (Invitrogen, #A-31553, 1:500), Alexa Fluor 488-conjugated anti-guinea-pig IgG (Invitrogen, #A-11073, 1:500), Alexa Fluor 647-conjugated anti-rabbit IgG (Invitrogen, #A32728, 1/500), goat anti-mouse IgG horseradish peroxidase (HRP) conjugate (Bio-Rad, #1705047, 1:2000), goat anti-rabbit IgG HRP conjugate (Bio-Rad, #170-6515, 1:5000) and peroxidase-conjugated anti-guinea-pig IgG (Jackson Laboratories, #106-035-003, 1:2000).
Western blot analysis
For detection of siRNA-mediated depletion, U2OS cells were extracted 72 h post transfection. Cells from a six-well culture plate were trypsinized, washed with PBS and resuspended 200 μl 1× DB (denaturing buffer; 50 mM Tris-HCl, pH 6.8, 2 mM EDTA, 20% glycerol, 0.8% SDS, 0.02% Bromophenol Blue, 1000 mM NaCl and 4 M urea). Approximately 50,000 cells were loaded. Proteins were separated by 7.5% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA). The membrane was blocked with TBS-T (20 mM Tris-HCl, pH 7.6, 136 mM NaCl and 0.1% Tween-20) containing 3% bovine serum albumin (Research Organics) for 1 h, then incubated with primary antibody at 4°C overnight. After washing with TBS-T, the membrane was incubated with HRP-conjugated secondary antibody (Bio-Rad, Hercules, CA) for 2 h at room temperature. Protein signal was detected by chemiluminescence and western band intensity was quantified using ImageJ. Each sample density was calculated by normalizing to their relative loading control bands.
Immunofluorescence microscopy
Cells were fixed on collagen or FN-coated glass bottom 35 mm dishes (MatTek, P35G-1.5-14-C) by removal of medium and addition of 4% paraformaldehyde (Electron Microscopy Sciences, #15710) in PBS for 20 min. Cells were washed 3× PBS before permeabilization with 0.25% Triton X-100 in PBS for 15 min. Cells were washed three times with PBS and incubated in blocking buffer (10% calf serum, 0.1% saponin and 0.02% sodium azide in PBS) for 30 min. Cells were incubated with primary treatment for 1.5 h, followed by three PBS washes, and incubation in secondary antibody for 1.5 h with 2.5 μM TRITC–phalloidin (Sigma-Aldrich). Cells were washed three times with PBS and imaged in PBS. All steps conducted at 23°C.
Imaging
For Airyscan imaging, images were acquired using a Zeiss LSM 880 Airyscan microscope using a 100×1.4 NA objective and collecting through a 32-channel GaAsP detector configured as 0.2 Airy Units per channel. Cells were imaged with the 405 nm laser and BP 420-480/BP 495-550 filter for BFP, 488 nm laser and BP 420-480/BP 495-620 filter for GFP, 561 nm laser and BP 495-550/LP 570 filter for RFP, and 633 nm laser and BP 570-620/LP 645 filter for far-red signals. All fixed-cell imaging was undertaken by Airyscan microscopy. For fixed-cell imaging, multiple z-slices were acquired of 0.18 μm step size (number of slices specified in figure legends). For live-cell microscopy, imaging was conducted at 37°C and 5% CO2, with single ventral slices acquired. Images subsequently processed using Zen2 software. Raw data were processed using Airyscan processing in ‘auto strength’ mode (mean strength±s.d.=5.5±1.3) with Zen Black software version 2.3.
For live-cell confocal imaging, images were acquired using a Wave FX spinning disc confocal microscope (Quorum Technologies, on a Nikon Eclipse microscope), equipped with Hamamatsu ImageM EM CCD cameras and Bionomic Controller (20/20 Technology, Inc.) temperature-controlled stage set to 37°C. After equilibrating to temperature for 10 min, cells were imaged through the 60×1.4 NA Plan Apo objective (Nikonov et al., 2007) using the 403 nm laser and 435-485 filter for BFP, the 491 nm laser and 525/20 filter for GFP, and the 561 nm laser and 593/40 filter for mRFP/mApple. Single ventral slices were acquired.
For live-cell imaging of filopodia–FA dynamics (confocal), cells were co-transfected with GFP–FMNL3, mApple–F-tractin and BFP-vinculin and plated on FN for 18 h before imaging. Cells were imaged in live-cell medium [DMEM minus phenol red (Sigma-Aldrich #10-012-CV), 25 mM HEPES (Sigma-Aldrich), 10% NCS (HyClone)] for 10 min with 20 s interval. Single ventral slices were acquired.
For live-cell spreading (Airyscan), ∼100,000 cells were trypsinized in 0.05% trypsin/EDTA (Sigma-Aldrich) for 5 min at 24 h post-transfection and plated directly onto collagen-coated glass bottom 35 mm MatTek dishes in live-cell medium. Cells were allowed to adhere for varying times (indicated for individual experiment) before imaging by Airyscan microscopy. Single ventral slices were acquired.
Quantification methods
Unless otherwise stated all image analysis was performed on Imaris Software (8.1.4, Bitplane Inc.)
Filopodia–FA association
For live-cell analysis, cells were co-transfected with GFP–FMNL3, mApple–F-tractin and BFP–vinculin, and were tracked using the FMNL3 and F-tractin signal to follow filopodia, and the vinculin signal to follow FAs. At each timeframe (20 s intervals), filopodia in the field of view were examined for association with FAs (<0.3 μm separation). No filopodia were counted twice (n=126 filopodia, 12 cells, three independent experiments). For fixed-cell analysis, cells were fixed after 18 h of plating on FN-treated coverslips, co-stained for FMNL3, paxillin and TRITC-phalloidin. Filopodia that were within 0.3 μm from a FAs at the filopodia base were classed as FA-associated. Maximum intensity projections of Airyscan-acquired images were used to examine filopodia/FA association. N=58 filopodia, 14 cells, four independent experiments.
VASP and FMNL3 localization in filopodia
U2OS cells were plated on collagen or FN-coated glass bottom 35 mm dishes (MatTek) for 15 min, 30 min, 1 h or 18 h before fixation and staining for FMNL3, VASP, paxillin and actin filaments (TRITC–phalloidin). Filopodia were identified as actin protrusions that were >1 μm from the cell edge. The protrusion had to be of a uniform diameter from base to tip, nominally <300 nm diameter, to be considered a filopodium. Classification of ‘tip’ localization required the staining to be indistinguishable from the most distal actin staining following background subtraction.
Filopodia density
Filopodia density was calculated from fixed-cell images following 18 h of plating on FN-treated coverslips and staining with TRITC–phalloidin (actin filaments). Cell edge length was measured by using TRITC–phalloidin as an outline for the cell boundary (the line was drawn using the ‘Polygon’ function). Cell edges chosen as edges free from cell–cell contact and not containing stress fibers running parallel to the cell edge. The filopodia number in this region was counted, identified as actin protrusions that were >1 μm from the cell edge. The protrusion had to be of a uniform diameter from base to tip, nominally <300 nm diameter, to be considered a filopodium. Filopodial density was calculated by dividing the number of filopodia by length of cell edge.
Filopodia–adhesion assembly in spreading cells
Filopodia–adhesion association was examined from analysis of live-cell videos of GFP–paxillin- and RFP–F-tractin-expressing cells. Imaging began 8 min (for U2OS cells plated on collagen) after seeding and was for a duration of 47 min at 30 s intervals. Randomly chosen regions of interest (ROIs) of spreading cells were analyzed. Filopodia assembled in these ROIs were examined for new adhesion assembly. Only new adhesions that persisted for longer than 60 s were counted. Adhesion classes include: shaft adhesion, whereby filopodia precede the appearance of detectable paxillin within the filopodial shaft; base adhesion, whereby filopodia precede the appearance of detectable paxillin at the filopodial base; simultaneous adhesion, whereby filopodia and a new adhesion appear at the same time; tip adhesion, whereby filopodia precede the appearance of detectable paxillin at the filopodial tip; and no adhesion, whereby filopodia assembly does not correlate to new adhesion assembly. In addition, a subset of VASP-depleted cells resulted in bleb adhesion, whereby a cell bleb leads to new adhesion assembly. Filopodial lifetimes were measured from the frame of initial appearance to frame of disappearance, with no allowable frame gaps. Filopodia number was measured from 8–18 min and 18–28 min post-seeding, with the total number of new filopodia counted over the entire cell edge. Cell spreading area was calculated from frames starting at 8 min (for U2OS cells plated on collagen) post seeding for a duration of 47 min at 30 s intervals. At each time-point, the total cell area was measured by using the ‘Cell detection’ function, with RFP as the source channel. The following analysis settings were used: 0.1 μm smooth width, auto-threshold, voxel number filter=5. Results were from n=5 cells and three independent experiments.
Supplementary Material
Acknowledgements
We thank past and present members of the Higgs lab, Phil Aldridge and Ann Lavanway for microscopy help, Stephanie Gupton for Mena/EVL plasmids, and Eva Naps for her valuable tips. Purchase of the Airyscan microscope was possible through generous support from NIGMS (Supplement to GM109965), the Provost and Dean of Sciences at Dartmouth College, and the Norris Cotton Cancer Center.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: L.E.Y., H.N.H.; Methodology: L.E.Y., H.N.H.; Validation: L.E.Y.; Formal analysis: L.E.Y.; Investigation: L.E.Y., C.J.L.; Writing - original draft: L.E.Y., H.N.H.; Writing - review & editing: L.E.Y., H.N.H.; Visualization: L.E.Y.; Supervision: H.N.H.; Project administration: H.N.H.; Funding acquisition: H.N.H.
Funding
This work was supported by National Institutes of Health (NIH R01 GM109965 and NIH R35 GM122545 to H.N.H., as well as NIH P20 GM113132. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.220814.supplemental
References
- Applewhite D. A., Barzik M., Kojima S.-I., Svitkina T. M., Gertler F. B. and Borisy G. G. (2007). Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol. Biol. Cell 18, 2579-2591. 10.1091/mbc.e06-11-0990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M., McClain L. M., Gupton S. L., Gertler F. B. and Ginsberg M. H. (2014). Ena/VASP regulates mDia2-initiated filopodial length, dynamics, and function. Mol. Biol. Cell 25, 2604-2619. 10.1091/mbc.e14-02-0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J. E., Svitkina T. M., Krause M., Schäfer D. A., Loureiro J. J., Strasser G. A., Maly I. V., Chaga O. Y., Cooper J. A., Borisy G. G. et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509-521. 10.1016/S0092-8674(02)00731-6 [DOI] [PubMed] [Google Scholar]
- Bennett R. D., Mauer A. S. and Strehler E. E. (2007). Calmodulin-like Protein Increases Filopodia-dependent Cell Motility via Up-regulation of Myosin-10. J. Biol. Chem. 282, 3205-3212. 10.1074/jbc.M607174200 [DOI] [PubMed] [Google Scholar]
- Berg J. S. and Cheney R. E. (2002). Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 4, 246-250. 10.1038/ncb762 [DOI] [PubMed] [Google Scholar]
- Berg J. S., Derfler B. H., Pennisi C. M., Corey D. P. and Cheney R. E. (2000). Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J. Cell Sci. 113, 3439-3451. [DOI] [PubMed] [Google Scholar]
- Bilancia C. G., Winkelman J. D., Tsygankov D., Nowotarski S. H., Sees J. A., Comber K., Evans I., Lakhani V., Wood W., Elston T. C. et al. (2014). Enabled negatively regulates diaphanous-driven actin dynamics in vitro and in vivo. Dev. Cell 28, 394-408. 10.1016/j.devcel.2014.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Sykes C. and Plastino J. (2014). Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235-263. 10.1152/physrev.00018.2013 [DOI] [PubMed] [Google Scholar]
- Block J., Stradal T. E., Hänisch J., Geffers R., Köstler S. A., Urban E., Small J. V., Rottner K. and Faix J. (2008). Filopodia formation induced by active mDia2/Drf3. J. Microsc. 231, 506-517. 10.1111/j.1365-2818.2008.02063.x [DOI] [PubMed] [Google Scholar]
- Block J., Breitsprecher D., Kuhn S., Winterhoff M., Kage F., Geffers R., Duwe P., Rohn J. L., Baum B., Brakebusch C. et al. (2012). FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 22, 1005-1012. 10.1016/j.cub.2012.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohil A. B., Robertson B. W. and Cheney R. E. (2006). Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl. Acad. Sci. USA 103, 12411-12416. 10.1073/pnas.0602443103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D., Kiesewetter A. K., Linkner J., Urbanke C., Resch G. P., Small J. V. and Faix J. (2008). Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 27, 2943-2954. 10.1038/emboj.2008.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle N. P., Holt M. R., Davies J. E., Price C. J. and Critchley D. R. (1996). The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 318, 753-757. 10.1042/bj3180753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K., Baginski J., Hassan S. F., Volin M., Shukla D. and Tiwari V. (2016). Filopodia and viruses: an analysis of membrane processes in entry mechanisms. Front. Microbiol. 7, 300 10.3389/fmicb.2016.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra E. S. and Higgs H. N. (2007). The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9, 1110-1121. 10.1038/ncb1007-1110 [DOI] [PubMed] [Google Scholar]
- Cramer L. P., Siebert M. and Mitchison T. J. (1997). Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 136, 1287-1305. 10.1083/jcb.136.6.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E. W., Kwiatkowski A. V., Mebane L. M., Philippar U., Barzik M., Rubinson D. A., Gupton S., Van Veen J. E., Furman C., Zhang J. et al. (2007). Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 9, 1347 10.1038/ncb1654 [DOI] [PubMed] [Google Scholar]
- Disanza A., Bisi S., Winterhoff M., Milanesi F., Ushakov D. S., Kast D., Marighetti P., Romet-Lemonne G., Müller H.-M., Nickel W. et al. (2013). CDC42 switches IRSp53 from inhibition of actin growth to elongation by clustering of VASP. EMBO J. 32, 2735-2750. 10.1038/emboj.2013.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M., Zwolak A., Schäfer D. A., Sept D., Dominguez R. and Cooper J. A. (2014). Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 15, 677 10.1038/nrm3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J. and Rottner K. (2006). The making of filopodia. Curr. Opin. Cell Biol. 18, 18-25. 10.1016/j.ceb.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Faix J., Breitsprecher D., Stradal T. E. and Rottner K. (2009). Filopodia: complex models for simple rods. Int. J. Biochem. Cell Biol. 41, 1656-1664. 10.1016/j.biocel.2009.02.012 [DOI] [PubMed] [Google Scholar]
- Galbraith C. G., Yamada K. M. and Galbraith J. A. (2007). Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science 315, 992-995. 10.1126/science.1137904 [DOI] [PubMed] [Google Scholar]
- Gateva G., Tojkander S., Koho S., Carpen O. and Lappalainen P. (2014). Palladin promotes assembly of non-contractile dorsal stress fibers through VASP recruitment. J. Cell Sci. 127, 1887 10.1242/jcs.135780 [DOI] [PubMed] [Google Scholar]
- Gauvin T. J., Young L. E. and Higgs H. N. (2015). The formin FMNL3 assembles plasma membrane protrusions that participate in cell–cell adhesion. Mol. Biol. Cell 26, 467-477. 10.1091/mbc.e14-07-1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh W. I., Lim K. B., Sudhaharan T., Sem K. P., Bu W., Chou A. M. and Ahmed S. (2012). mDia1 and WAVE2 Proteins Interact Directly with IRSp53 in Filopodia and Are Involved in Filopodium Formation. J. Biol. Chem. 287, 4702-4714. 10.1074/jbc.M111.305102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-H., Chung C. Y., Wessels D., Stephens S., Titus M. A., Soll D. R. and Firtel R. A. (2002). Requirement of a Vasodilator-stimulated Phosphoprotein Family Member for Cell Adhesion, the Formation of Filopodia, and Chemotaxis in Dictyostelium. J. Biol. Chem. 277, 49877-49887. 10.1074/jbc.M209107200 [DOI] [PubMed] [Google Scholar]
- Han Y., Eppinger E., Schuster I. G., Weigand L. U., Liang X., Kremmer E., Peschel C. and Krackhardt A. M. (2009). Formin-like 1 (FMNL1) is regulated by N-terminal myristoylation and induces polarized membrane blebbing. J. Biol. Chem. 284, 33409-33417. 10.1074/jbc.M109.060699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. D. and Mullins R. D. (2010). VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191, 571-584. 10.1083/jcb.201003014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. D. and Mullins R. D. (2015). Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments. eLife 4, e06585 10.7554/eLife.06585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. S., Li F. and Higgs H. N. (2004). The mouse formin, FRLa, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem 279, 20076-20087. 10.1074/jbc.M312718200 [DOI] [PubMed] [Google Scholar]
- Harris E. S., Gauvin T. J., Heimsath E. G. and Higgs H. N. (2010). Assembly of filopodia by the formin FRL2 (FMNL3). Cytoskeleton 67, 755-772. 10.1002/cm.20485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Sakai T., Tsukasaki Y., Watanabe T. M. and Ikebe M. (2017). Myosin X is recruited to nascent focal adhesions at the leading edge and induces multi-cycle filopodial elongation. Sci. Rep. 7, 13685 10.1038/s41598-017-06147-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimsath E. G., Yim Y.-I., Mustapha M., Hammer J. A. and Cheney R. E. (2017). Myosin-X knockout is semi-lethal and demonstrates that myosin-X functions in neural tube closure, pigmentation, hyaloid vasculature regression, and filopodia formation. Sci. Rep. 7, 17354 10.1038/s41598-017-17638-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A.-K., Naj X. and Linder S. (2014). Daam1 is a regulator of filopodia formation and phagocytic uptake of Borrelia burgdorferi by primary human macrophages. FASEB J. 28, 3075-3089. 10.1096/fj.13-247049 [DOI] [PubMed] [Google Scholar]
- Homem C. C. and Peifer M. (2009). Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol. Biol. Cell 20, 5138-5155. 10.1091/mbc.e09-02-0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke M., Bachg A. C., Groll K., Moyzio S., Müther B., Hemkemeyer S. A., Wedlich-Söldner R., Sixt M., Tacke S., Bähler M. et al. (2017). Multiple roles of filopodial dynamics in particle capture and phagocytosis, and phenotypes of Cdc42 and Myo10 deletion. J. Biol. Chem. 292, 7258-7273. 10.1074/jbc.M116.766923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P. and Lappalainen P. (2006). Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 173, 383-394. 10.1083/jcb.200511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal R., Breitsprecher D., Collins A., Corrêa I. R., Xu M.-Q. and Goode B. L. (2013). The formin daam1 and fascin directly collaborate to promote filopodia formation. Curr. Biol. 23, 1373-1379. 10.1016/j.cub.2013.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. E., King S. J., Asokan S. B., Rotty J. D., Bear J. E. and Haugh J. M. (2015). F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. J. Cell Biol. 208, 443 10.1083/jcb.201406102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A. M., Ramko E. B., Davidson M. W., Hess H. F. and Waterman C. M. (2010). Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580-584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N. and Pollard T. D. (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423-435. 10.1016/j.cell.2005.11.038 [DOI] [PubMed] [Google Scholar]
- Krause M., Leslie J. D., Stewart M., Lafuente E. M., Valderrama F., Jagannathan R., Strasser G. A., Rubinson D. A., Liu H., Way M. et al. (2004). Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell 7, 571-583. 10.1016/j.devcel.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski A. V., Rubinson D. A., Dent E. W., Edward van Veen J., Leslie J. D., Zhang J., Mebane L. M., Philippar U., Pinheiro E. M., Burds A. A. et al. (2007). Ena/VASP Is Required for neuritogenesis in the developing cortex. Neuron 56, 441-455. 10.1016/j.neuron.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Lanier L. M., Gates M. A., Witke W., Menzies A. S., Wehman A. M., Macklis J. D., Kwiatkowski D., Soriano P. and Gertler F. B. (1999). Mena is required for neurulation and commissure formation. Neuron 22, 313-325. 10.1016/S0896-6273(00)81092-2 [DOI] [PubMed] [Google Scholar]
- Laukaitis C. M., Webb D. J., Donais K. and Horwitz A. F. (2001). Differential dynamics of α5 integrin, paxillin, and α-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153, 1427 10.1083/jcb.153.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrand C., Dent E. W., Strasser G. A., Lanier L. M., Krause M., Svitkina T. M., Borisy G. G. and Gertler F. B. (2004). Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron 42, 37-49. 10.1016/S0896-6273(04)00108-4 [DOI] [PubMed] [Google Scholar]
- Loureiro J. J., Rubinson D. A., Bear J. E., Baltus G. A., Kwiatkowski A. V. and Gertler F. B. (2002). Critical roles of phosphorylation and actin binding motifs, but not the central proline-rich region, for Ena/Vasodilator-stimulated phosphoprotein (VASP) function during cell migration. Mol. Biol. Cell 13, 2533-2546. 10.1091/mbc.e01-10-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A. and Mitchison T. (1999). Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 146, 1097-1106. 10.1083/jcb.146.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusek T., Gombos R., Szécsényi A., Sánchez-Soriano N., Czibula Á., Pataki C., Gedai A., Prokop A., Raskó I. and Mihály J. (2008). Formin proteins of the DAAM subfamily play a role during axon growth. J. Neurosci. 28, 13310-13319. 10.1523/JNEUROSCI.2727-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj X., Hoffmann A.-K., Himmel M. and Linder S. (2013). The formins FMNL1 and mDia1 regulate coiling phagocytosis of borrelia burgdorferi by primary human macrophages. Infect. Immun. 81, 1683-1695. 10.1128/IAI.01411-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonov A. V., Hauri H.-P., Lauring B. and Kreibich G. (2007). Climp-63-mediated binding of microtubules to the ER affects the lateral mobility of translocon complexes. J. Cell Sci. 120, 2248-2258. 10.1242/jcs.008979 [DOI] [PubMed] [Google Scholar]
- Nowotarski S. H., McKeon N., Moser R. J. and Peifer M. (2014). The actin regulators Enabled and Diaphanous direct distinct protrusive behaviors in different tissues during Drosophila development. Mol. Biol. Cell 25, 3147-3165. 10.1091/mbc.e14-05-0951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patla I., Volberg T., Elad N., Hirschfeld-Warneken V., Grashoff C., Fässler R., Spatz J. P., Geiger B. and Medalia O. (2010). Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat. Cell Biol. 12, 909 10.1038/ncb2095 [DOI] [PubMed] [Google Scholar]
- Pellegrin S. and Mellor H. (2005). The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol. 15, 129-133. 10.1016/j.cub.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Plantard L., Arjonen A., Lock J. G., Nurani G., Ivaska J. and Strömblad S. (2010). PtdIns(3,4,5)P3 is a regulator of myosin-X localization and filopodia formation. J. Cell Sci. 123, 3525 10.1242/jcs.069609 [DOI] [PubMed] [Google Scholar]
- Ramabhadran V., Korobova F., Rahme G. J. and Higgs H. N. (2011). Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell 22, 4822-4833. 10.1091/mbc.e11-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M., Halbrügge M., Scheer U., Wiegand C., Jockusch B. M. and Walter U. (1992). The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 11, 2063-2070. 10.1002/j.1460-2075.1992.tb05264.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M., Giehl K., Abel K., Haffner C., Jarchau T., Hoppe V., Jockusch B. M. and Walter U. (1995). The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 14, 1583-1589. 10.1002/j.1460-2075.1995.tb07146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme D. N., Meyer A. S., Barzik M., Keating A. and Gertler F. B. (2015). Selectivity in subunit composition of Ena/VASP tetramers. Biosci. Rep. 35 10.1042/BSR20150149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K., Behrendt B., Small J. V. and Wehland J. (1999). VASP dynamics during lamellipodia protrusion. Nat. Cell Biol. 1, 321-322. 10.1038/13040 [DOI] [PubMed] [Google Scholar]
- Sahasrabudhe A., Ghate K., Mutalik S., Jacob A. and Ghose A. (2016). Formin 2 regulates the stabilization of filopodial tip adhesions in growth cones and affects neuronal outgrowth and pathfinding in vivo. Development 143, 449 10.1242/dev.130104 [DOI] [PubMed] [Google Scholar]
- Sarmiento C., Wang W., Dovas A., Yamaguchi H., Sidani M., El-Sibai M., Desmarais V., Holman H. A., Kitchen S., Backer J. M. et al. (2008). WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J. Cell Biol. 180, 1245-1260. 10.1083/jcb.200708123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer C., Borm B., Born S., Mohl C., Eibl E. M. and Hoffmann B. (2009). One step ahead: role of filopodia in adhesion formation during cell migration of keratinocytes. Exp. Cell Res. 315, 1212-1224. 10.1016/j.yexcr.2008.11.008 [DOI] [PubMed] [Google Scholar]
- Schirenbeck A., Bretschneider T., Arasada R., Schleicher M. and Faix J. (2005). The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 7, 619-625. 10.1038/ncb1266 [DOI] [PubMed] [Google Scholar]
- Schirenbeck A., Arasada R., Bretschneider T., Stradal T. E. B., Schleicher M. and Faix J. (2006). The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc. Natl Acad. Sci. USA 103, 7694-7699. 10.1073/pnas.0511243103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steketee M. B. and Tosney K. W. (2002). Three functionally distinct adhesions in filopodia: shaft adhesions control lamellar extension. J. Neurosci. 22, 8071 10.1523/JNEUROSCI.22-18-08071.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steketee M., Balazovich K. and Tosney K. W. (2001). Filopodial initiation and a novel filament-organizing center, the focal ring. Mol. Biol. Cell 12, 2378-2395. 10.1091/mbc.12.8.2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M. and Borisy G. G. (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409-421. 10.1083/jcb.200210174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S., Gateva G., Husain A., Krishnan R. and Lappalainen P. (2015). Generation of contractile actomyosin bundles depends on mechanosensitive actin filament assembly and disassembly. eLife 4, e06126 10.7554/eLife.06126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuo H. and Ikebe M. (2004). Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 319, 214-220. 10.1016/j.bbrc.2004.04.167 [DOI] [PubMed] [Google Scholar]
- Tokuo H., Mabuchi K. and Ikebe M. (2007). The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J. Cell Biol. 179, 229-238. 10.1083/jcb.200703178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuo H., Bhawan J. and Coluccio L. M. (2018). Myosin X is required for efficient melanoblast migration and melanoma initiation and metastasis. Sci. Rep. 8, 10449 10.1038/s41598-018-28717-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama Y., Fukuhara S., Ando K., Matsuda M. and Mochizuki N. (2015). Cdc42 mediates Bmp-induced sprouting angiogenesis through Fmnl3-driven assembly of endothelial filopodia in zebrafish. Dev. Cell 32, 109-122. 10.1016/j.devcel.2014.11.024 [DOI] [PubMed] [Google Scholar]
- Wang Y., Arjonen A., Pouwels J., Ta H., Pausch P., Bange G., Engel U., Pan X., Fackler O. T., Ivaska J.. et al. (2015). Formin-like 2 promotes β1-integrin trafficking and invasive motility downstream of PKCα. Dev. Cell 34, 475-483. 10.1016/j.devcel.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Winkelman J. D., Bilancia C. G., Peifer M. and Kovar D. R. (2014). Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc. Natl. Acad. Sci. USA 111, 4121-4126. 10.1073/pnas.1322093111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. and Svitkina T. (2011). Filopodia initiation: focus on the Arp2/3 complex and formins. Cell Adh. Migr. 5, 402-408. 10.4161/cam.5.5.16971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Czech L., Gerboth S., Kojima S., Scita G. and Svitkina T. (2007). Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5, 2624-2645. 10.1371/journal.pbio.0050317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. E. and Higgs H. N. (2018). Focal adhesions undergo longitudinal splitting into fixed-width units. Curr. Biol. 10.1016/j.cub.2018.04.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. E., Heimsath E. G. and Higgs H. N. (2015). Cell type–dependent mechanisms for formin-mediated assembly of filopodia. Mol. Biol. Cell 26, 4646-4659. 10.1091/mbc.e15-09-0626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Berg J. S., Li Z., Wang Y., Lång P., Sousa A. D., Bhaskar A., Cheney R. E. and Strömblad S. (2004). Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 6, 523 10.1038/ncb1136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.