Abstract
Background:
Prognostic biomarkers for osteosarcoma (OS) at the time of diagnosis are lacking. Necrotic response of OS to preoperative chemotherapy correlates with survival, and is determined 3–4 months after diagnosis. The purpose of this study is to identify biomarkers that will stratify patients into good or poor responders to chemotherapy at diagnosis, and to determine the role of potential biomarkers in OS pathogenesis.
Procedure:
Because OS may be caused by disruptions of osteogenic differentiation, and the Notch pathway is one regulator of bone development, we examined the link between Notch effectors, OS differentiation, and OS outcome. We probed the R2: Genomics Analysis and Visualization Platform for RNA expression levels of Notch targets in mixed high-grade OS pre-treatment biopsies. We used human OS cell lines in vitro and in mice to determine the role of the Notch target Hairy/Enhancer of Split 4 (Hes4) in OS.
Results:
We found that in OS patients, high expression of Hes4 correlated with decreased metastasis-free and overall survival. Human OS cells that overexpress Hes4 are more immature and have an increased invasive capacity in vitro. This was not universal to all Notch effectors, as Hes1 overexpression induced opposing effects. When injected into NSG mice, Hes4 overexpressing OS cells produced significantly larger, more lytic tumors and significantly more metastases than did control cells.
Conclusions:
Hes4 overexpression promotes a more aggressive tumor phenotype by preventing osteoblastic differentiation of OS cells. Hes4 expression may allow for the stratification of patients into good or poor responders to chemotherapy at diagnosis.
Keywords: Osteogenic Differentiation, Notch, Hes4, Osteosarcoma, Prognostic Factor
Introduction
Response to preoperative chemotherapy is the only validated prognostic marker that identifies osteosarcoma (OS) patients at high risk for relapse and metastasis1. Because OS patients cannot be stratified at the time of diagnosis, all pediatric and adolescent OS patients receive combination chemotherapy with cisplatin, doxorubicin, and high-dose methotrexate followed by surgical excision of the primary tumor. Neither the administration of high-dose ifosfamide with or without etoposide to patients with poor chemotherapy responses (defined as <90% tumor necrosis), nor the administration of interferon alpha-2b to patients with good chemotherapy response (>90% tumor necrosis), have improved OS patient survival2,3. It is yet to be determined, however, whether adding these agents to the preoperative chemotherapeutic regimen will improve disease response and survival for either prognosis group. A predictive biomarker assessed at the time of diagnosis is needed to meet this need. Understanding the pathways that control OS development may identify new prognostic and predictive biomarkers for OS.
Although cure rates in patients with localized OS are as high as 60–70%, patients who have metastatic relapse have 5-year overall survival rates of only 15–20%4. Currently, there is no predictive marker at initial diagnosis to identify which patients are at high risk for developing metastatic disease. The metastatic behavior of some cancers is linked to differentiation status: the more immature the cell population, the higher the likelihood for metastasis5,6. OS is a highly heterogeneous mixture of cells representing the full spectrum of differentiation, from highly proliferative mesenchymal stem cells to terminally differentiated slowly proliferating bone/osteoid-forming osteoblasts7. Normal bone development is tightly regulated by a multistep differentiation pathway in which various transcription factors control the progression of mesenchymal stem cells from an immature stem-like state through osteogenic lineage commitment to terminal differentiation8–18. Because disruption of osteogenic differentiation is thought to lead to the development and progression of OS11,19, we sought to expand our understanding of the underlying molecular mechanisms that drive tumor-cell differentiation.
The Notch signaling pathway is a mediator of cell differentiation that is critical for normal bone development. Notch signaling is activated when a membrane-bound ligand (Jag1, Jag2, Dll1, Dll3, or Dll4) on a signal-sending cell physically interacts with the extracellular domain of a membrane-bound Notch receptor (Notch1–4) on a signal-receiving cell. This interaction results in the two-step cleavage of the intracellular domain of Notch receptors. Once cleaved, the intracellular domain translocates to the nucleus to promote the expression of a number of target genes. These ‘Notch effectors’ include Hairy/Enhancer of Split 1–7 (Hes1–7), Hey1–2, and deltex. Notch receptor knockout results in severe skeletal abnormalities in mice20–22, suggesting a critical role for Notch signaling in bone development. Additionally, activation of the Notch pathway promotes the differentiation of osteoblasts from mesenchymal stem cells while inhibiting the development of osteoclasts23. Finally, the Notch downstream target Hes4 regulates the lineage commitment of normal bone marrow stromal cells to the osteogenic pathway24 and is a biomarker that identifies solid tumors likely to respond to γ-secretase inhibitor-based therapies25–27. The role of Hes4 in the development and progression of OS has yet to be defined, however.
The purpose of this study is to identify clinically relevant biomarkers that allow for the stratification of patients into good or poor responders to chemotherapy at diagnosis. We also aim to understand the biology of these markers in OS pathogenesis. We demonstrate here that high Hes4 expression promotes an immature, progenitor-like phenotype in OS cells, increasing the cell’s invasive capacity. High expression of Hes4 correlated with more aggressive and metastatic OS tumors in mice and with poor metastasis-free and overall survival in patients. Our data suggests that high Hes4 is important biologically in OS pathogenesis, and has potential as a clinically relevant prognostic biomarker in the stratification of patients into good or poor responders to chemotherapy at diagnosis.
Methods
Patient survival and metastasis probability
The R2 Genomics Analysis and Visualization platform (Academic Medical Center, Amsterdam, The Netherlands; R2: Genomics Analysis and Visualization Platform; http://r2.amc.nl) was used to generate Kaplan-Meier overall survival curves using the ‘Mixed Osteosarcoma - Kuijjer - 127 - vst - ilmnhwg6v2’ dataset28. Genome-wide gene expression analysis was performed on 84 pre-treatment high-grade osteosarcoma diagnostic biopsies, of which 29 overlapped with the 32 samples used for copy number analysis. Two different sets of control samples were used for comparison: osteoblasts (n=3) and mesenchymal stem cells (n=12, GEO accession number GSE28974). Pretreatment biopsies of high grade primary tumors from OS patient samples were analyzed on the basis of High vs Low Hes1 or Hes4. The R2 generated “scan” cut-off modus was used to determine the threshold point that most significantly separates high relative gene expression vs. low relative gene expression.
Cell culture
The human fetal osteoblastic cell line hFOB (American Type Culture Collection) was cultured at 34°C with 5% CO2 in a 1:1 mixture of phenol-free Dulbecco’s modified Eagle’s medium/Ham’s F12 medium with 2.5 mM L-glutamine (Invitrogen) supplemented with 10% fetal bovine serum and 0.3 mg/mL G418. The human OS cell lines HOS and CCHD and 293T normal kidney fibroblasts were maintained in high-glucose Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (Gemini Bio-Products). HOS, CCHD, and 293T cells were grown at 37°C in 5% CO2. HOS and 293T cells were purchased from the American Type Culture Collection. CCHD is a primary OS cell line derived from a core needle biopsy of a proximal femur lesion in an 18-year-old man who had pulmonary metastases at presentation and was a patient at the Children’s Cancer Hospital at The University of Texas MD Anderson Cancer Center.
Retroviral transduction of Hes4
Green fluorescent protein (GFP)-containing retroviral MigR1 constructs with and without Hes1 or Hes4 (MigR1-GFP, MigR1-GFP-Hes1 and MigR1-GFP-Hes4, respectively) were used to make a replication-incompetent retrovirus that was then used to infect HOS and CCHD cells as described previously29.
Proliferation of OS cells that overexpress GFP, GFP-Hes1, or Hes4
HOS and CCHD cells were transduced with MigR1-GFP, MigR1-GFP-Hes1, or MigR1-GFP-Hes4 and seeded in six-well plates in triplicate. Five days after transduction, a portion of the cells were collected and analyzed using a FACSCalibur flow cytometer (Becton Dickinson). The ratio of GFP-positive (transduced cells) to GFP-negative cells (non-transduced parental cells) was measured and normalized to 100. A portion of the cells were collected again on days 7, 9, 11, 13, 15, 17, 19, and 21 post transduction, and the ratio of GFP-positive to GFP-negative cells was calculated to determine the effect of MigR1-GFP (control), MigR1-GFP-Hes1, and MigR1-GFP-Hes4 on the rate of OS-cell growth over time.
Cellular invasion
HOS and CCHD cells transduced with MigR1-GFP, MigR1-GFP-Hes1 or MigR1-GFP-Hes4 were sorted according to GFP positivity, and their invasiveness was measured using a 24-well BioCoat Matrigel invasion chamber with an 8-μm pore size (BD Biosciences). Cells (2.5 × 104) suspended in 500 μL of serum-free Dulbecco’s modified Eagle’s medium were seeded in triplicate into the upper chamber of an assay plate. Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum was added to the bottom chamber and acted as the chemoattractant for the cells. After 48 hours of incubation at 37°C, the migrated cells were fixed, stained with Hema-3 (Thermo Fisher Scientific), and counted under a microscope at 20× magnification.
Differentiation of OS cells
Alizarin Red S staining:
HOS and CCHD cells were transduced with MigR1-GFP or MigR1-GFP-Hes4 and were sorted for GFP expression 24–48 hours post-transduction. Positive cells were seeded into 24-well plates. Once cells were 100% confluent, the cell medium was supplemented with a differentiation supplement (10 mM β-glycerophosphate and 50 μg/mL ascorbic acid), refreshed every 3 days for 21 days. On day 21, cells were gently washed with phosphate-buffered saline and fixed with 10% paraformaldehyde for 15 minutes at room temperature. Excess paraformaldehyde was removed with a phosphate-buffered saline wash. Cells were stained for calcium deposition using 40 mM Alizarin Red S (pH 4.2) for 30 minutes. Excess Alizarin Red S stain was removed via two phosphate-buffered saline washes. Water was added to the cells, and the cells were imaged using an inverted microscope (Eclipse Ti; Nikon Instruments).
Quantification of RNA in OS cells:
Reverse transcription - Quantitative polymerase chain reaction (RT-qPCR) was used to quantify changes in the expression of markers of mesenchymal stem cells (Nanog, Sox2, and Oct4), committed osteoprogenitors/preosteoblasts (RunX2 and osterix), early osteoblasts (alkaline phosphatase), and mature osteoblasts and osteocytes (osteocalcin and osteopontin). After transduction with MigR1-GFP or MigR1-GFP-Hes4, HOS and CCHD cells were sorted for GFP, and RNA was extracted using RNeasy Mini Kit (QIAGEN). cDNA was constructed with Omniscript Reverse Transcriptase Kit (QIAGEN) with oligo(dT)s (Invitrogen) according to the manufacturer’s protocol. RT-qPCR was performed using a LightCycler 480 Real-Time PCR system (Roche Life Science) with either primers (Supplemental Methods S1) and SYBR Green PCR Master Mix (Bio-Rad) or TaqMan probes (Thermo Fisher Scientific) and TaqMan Gene Expression Master Mix (Applied Biosystems).
In vivo mouse xenografts
All animal experiments were approved by the MD Anderson Institutional Animal Care and Use Committee.
Intratibial injection of OS cells:
CCHD cells (1 × 106 suspended in 15 μL of sterile phosphate-buffered saline) were injected into the right tibias of 6-week-old non-obese diabetic/severe combined immunodeficient/interleukin (IL)-2Rγ–deficient mice (The Jackson Laboratory). The mice were killed 6 weeks after inoculation, their lungs were inflated with 10% formaldehyde via transtracheal injection, and their primary tumors and lungs were fixed in formalin and embedded in paraffin. Five-micron sections of the primary tumors and metastatic lesions in the lungs were mounted on glass slides for analysis and stained with hematoxylin and eosin as well as human vimentin.
Immunohistochemical quantification of metastases:
Representative images of lung tumor burdens in the mice were obtained using a cooled charge-coupled device C5810 camera (Hamamatsu Photonics) and the Optimas imaging software program (Media Cybernetics). Lung sections obtained from mice injected with CCHD-GFP or CCHD-GFP-Hes4 cells were stained with human vimentin for easy identification. Lung sections were scanned at 10× magnification, and the positively stained lesions were counted.
Quantification of Lysis:
In order to determine the extent of lysis in the bone as a result of OS tumor burden, we used a previously developed radiographic grading scheme30. Briefly, radiographs of the tibia were taken on the day the experiment was terminated (week 6) using the In Vivo Xtreme (Bruker). A grading system using numerical values from 0 to 4 was used to quantify the extent of bone destruction, where a grade of 0 represents no lysis, a grade of 1 represents minimal bone destruction in the medullary canal, 2 is moderate bone lysis within the medullary cortex with minimal destruction to the cortex, 3 is severe bone lysis with cortical disruption, and 4 is massive destruction with soft tissue extension of the tumor.
Statistical Analysis
The significance of all experimental treatments was assessed using the Student t-test with an alpha error threshold of 0.05. All experiments were conducted at least three times unless stated otherwise.
Results
Hes4 expression correlates with poor survival and high metastasis incidence in OS patients
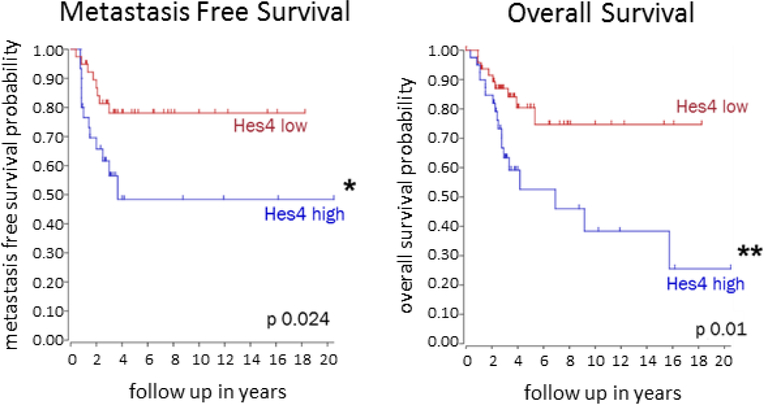
Notch signaling has been shown to regulate metastasis in several types of cancer31–36. To determine whether expression of Notch downstream targets correlates with metastasis-free and overall survival in patients with OS, we used a pre-existing RNA microarray database to examine gene expression within pretreatment high grade OS primary patient tumor biopsies. High and low expression is defined by computer generated thresholds that promote the greatest distinction between gene expression populations. High Hes4 expression, observed in 30 of 69 (43%) OS patients, correlated with significantly decreased metastasis-free and overall survival outcomes (Fig. 1). In contrast, the expression of Hes1, Hes2, Hes5, Hey1, Hey2, and DTX was not prognostically significant (Supplementary Figure S1).
Figure 1. High Hes4 expression in pretreatment high grade OS tumor biopsies correlates with decreased metastasis free and overall survival outcomes.
The R2: Genomics Analysis and Visualization Platform was used to generate Kaplan-Meier metastasis-free and overall survival curves using the Mixed Osteosarcoma - Kuijjer - 127 - vst - ilmnhwg6v2 data set. Genome-wide gene expression analysis was performed using 84 pretreatment high-grade diagnostic OS biopsy samples. Two different sets of control samples were used for comparison: osteoblasts (n = 3) and mesenchymal stem cells (n = 12; GEO accession number GSE28974). High Hes4 expression correlated with significantly lower probabilities of both overall and metastasis-free survival than low Hes4 expression (overall survival, P < 0.01; metastasis-free survival, P < 0.05).
Hes4 overexpression increases OS invasion
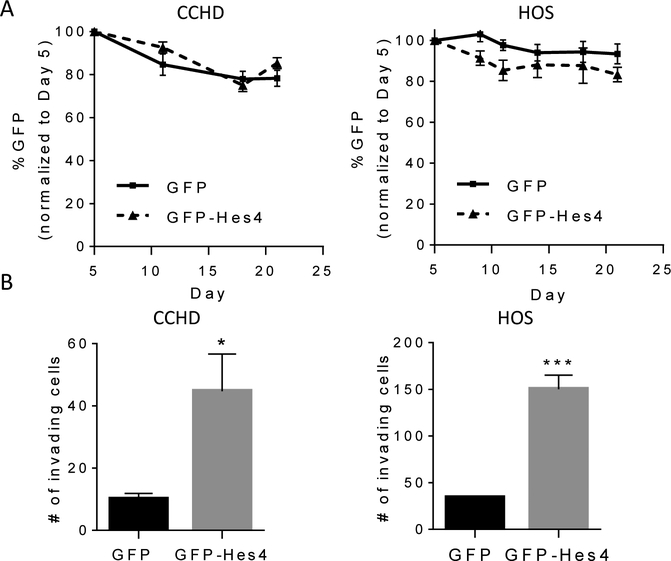
To determine whether Hes4 expression affects OS proliferative and invasive phenotypes, we transduced HOS and CCHD cells with MigR1-GFP or MigR1-GFP-Hes4 (control, CCHD/HOS-GFP; Hes4-overexpressing, CCHD/HOS-GFP-Hes4). GFP-Hes4 transduction resulted in 72- and 90-fold increases in Hes4 mRNA expression in CCHD and HOS cells, respectively (Supplementary Figure S2A–D). The proliferation of HOS and CCHD cells was not affected by overexpression of Hes4 (Fig. 2A). However, overexpression of Hes4 significantly increased the invasive capacity of HOS and CCHD cells (10.0 ± 1.9 versus 44.8 ± 11.9 [CCHD]; 13.3 ± 3.5 versus 175.3 ± 34.9 [HOS]; Fig. 2B and Supplementary Figure S2E). To determine whether this effect on invasion and proliferation was induced by other Notch effectors as well, we transduced HOS and CCHD cells with GFP-Hes1. In contrast with Hes4, overexpression of Hes1 decreased both invasion and proliferation (Supplementary Figure S3). These data indicate that the phenotypic characteristics of OS may depend on which specific Notch downstream target gene is activated. We focused on Hes4 because of the correlation between Hes4 expression and decreased OS patient survival.
Figure 2. Hes4 overexpression increases OS cell invasion but not proliferation.
(A) Hes4 overexpression does not affect proliferation. The percentages of GFP-positive CCHD and HOS cells over time after stable retroviral transduction with GFP or GFP-Hes4 (normalized to day 5 after transduction) were quantified at various time points and expressed as the mean cell number (± SEM; n = 3). (B)Hes4 overexpression increases OS in vitro invasion. CCHD-GFP, CCHD-GFP-Hes4, HOS-GFP and HOS-GFP-Hes4 cells were plated on a 24-well BioCoat Matrigel invasion chamber with an 8-μm pore size. Medium with 10% fetal bovine serum was used in the bottom well of the chamber. At 24 (HOS) or 48 (CCHD) hours, migrated cells were counted. The graph shows the mean number of migrated cells per field (±SEM; n = 3). *P ≤ 0.05; ***P ≤ 0.001.
Terminal differentiation is inhibited in Hes4-overexpressing OS cells
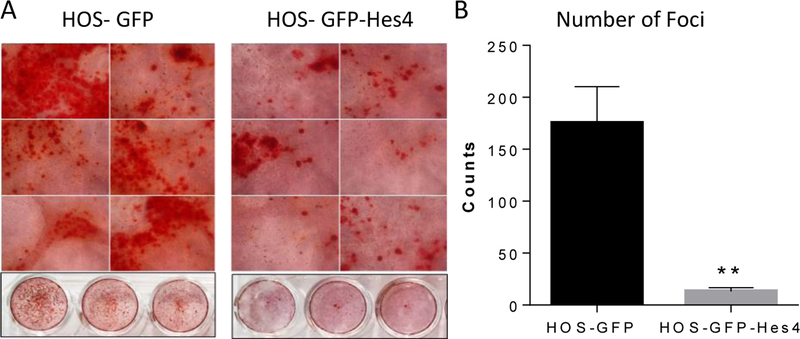
Overexpression of Hes4 correlated with an aggressive phenotype, as defined by increased invasive capacity. An aggressive phenotype has also been suggested to correlate with an immature, less differentiated state. To determine whether Hes4 affects terminal differentiation of OS cells, we used Alizarin Red S staining to measure calcium deposition, indicative of differentiation into mature osteoblasts, in HOS-GFP or HOS-GFP-Hes4 OS cells. HOS-GFP cells generated numerous calcium nodules, which appeared as red punctate points (Fig. 3). In addition to dark red focal points of thick calcium buildup, a calcium sheet appeared as a smooth bright red surface within the staining. In the presence of differentiation media, HOS-GFP-Hes4 OS cells produced fewer calcium nodules than did HOS-GFP control cells (mean ± SEM: HOS-GFP, 175.3 ± 34.9; HOS-GFP-Hes4, 13.3 ± 3.5; Fig. 3) and almost no calcium sheets. This suggested that Hes4 overexpression prevented calcium deposition, which is indicative of an immature, poorly differentiated phenotype.
Figure 3. Hes4 expression decreases osteoblastic differentiation.
Alizarin Red S staining of HOS-GFP and HOS-GFP-Hes4 cells was performed after 21 days in differentiation media. (A) Representative images of Alizarin Red S staining within 24 well plates. (B) Mean number of foci of calcium deposition per 24-well plate well (± SEM; n = 3). *P < 0.05.
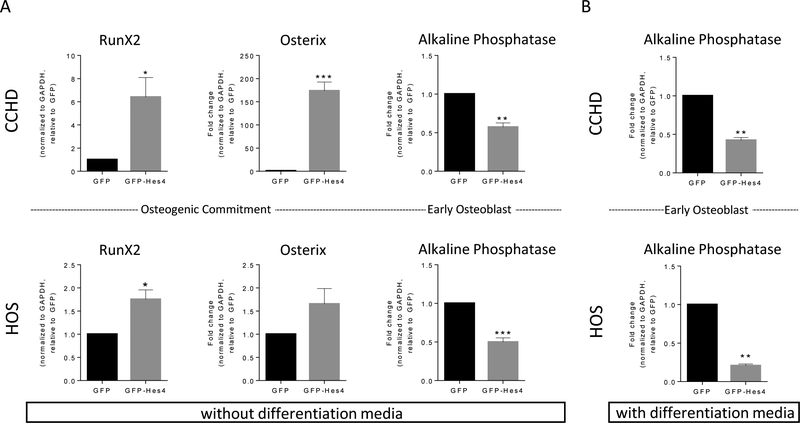
Hes4-overexpressing OS cells express increased levels of markers of committed osteoprogenitors and decreased levels of a marker of pre-osteoblasts.
The multistep progression of mesenchymal stem cells to terminally differentiated osteoblasts is delineated by the presence or absence of well-defined transcription factors (Supplementary Figure S4). To determine the stage at which Hes4 blocked OS differentiation, we used RT-qPCR to quantify markers of committed osteoprogenitors and preosteoblasts (RunX2 and osterix) and early osteoblasts (alkaline phosphatase). In both HOS and CCHD cells, overexpression of Hes4 resulted in increased expression of RunX2 and osterix, but decreased alkaline phosphatase, compared to GFP control cells. Taken together, these data suggest that there is a block in the progression of committed progenitors to early osteoblasts (Fig. 4A). We found that in the presence of differentiation media, it takes 9 days and 21 days for differentiation to occur in CCHD and HOS cell, respectively. To determine whether the expression of Hes4 blocked the maturation of osteogenic progenitors, control GFP and GFP-Hes4-overexpressing CCHD and HOS cells were incubated in differentiation media for 3 or 9 days, respectively (a time point in the middle of the differentiation process). The cells were then evaluated for alkaline phosphatase expression. Even in the differentiation media, Hes4 blocked OS-cell differentiation (Fig. 4B).
Figure 4. Overexpression of Hes4 increases the expression of RunX2 and osterix and decreases alkaline phosphatase.
(A) Hes4 overexpression increases RunX2 and osterix expression, and decreases alkaline phosphatase expression. RNA was harvested from CCHD and HOS cells 3–5 days after transduction with GFP or GFP-Hes4. RT-qPCR for RunX2, osterix, and alkaline phosphatase expression (as indicated) was normalized according to GAPDH expression relative to that in GFP-transduced control cells. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Bars, mean ± SEM (n = 3). (B) Even during forced differentiation, Hes4 prevents increased alkaline phosphatase expression. RNA harvested from CCHD and HOS cells 10–15 days after transduction with GFP or GFP-Hes4 and after 3–9 days of incubation with differentiation media. RT-qPCR was performed for alkaline phosphatase expression, normalized according to GAPDH expression relative to that in GFP-transduced control cells. **P ≤ 0.01. Bars, mean ± SEM (n = 3).
Effect of Hes4 overexpression on OS tumor growth and metastasis in vivo
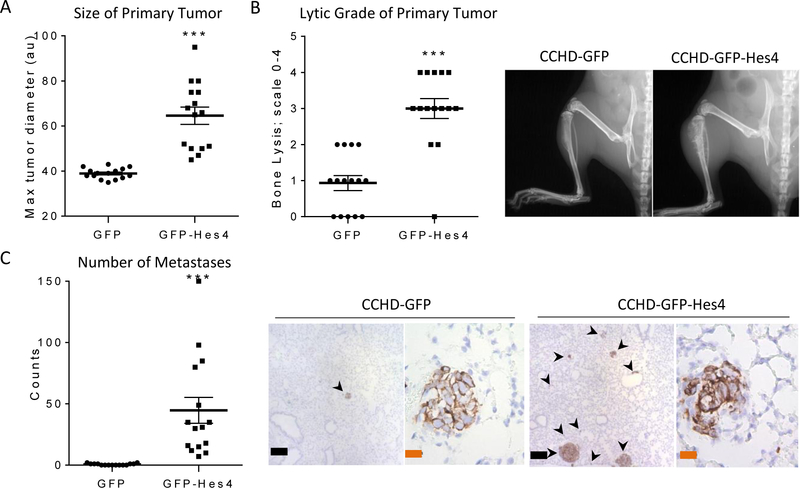
Our data indicated that Hes4-overexpressing OS cells are less differentiated and more invasive than OS cells expressing GFP alone. To determine whether overexpression of Hes4 also altered the in vivo OS tumor phenotype, we injected CCHD-GFP or CCHD-GFP-Hes4 OS cells into the tibias of NSG mice. Mice injected orthotopically with CCHD-GFP cells require euthanasia at 8 weeks due to primary and metastatic tumor burden. Mice injected with CCHD-GFP-Hes4 cells, however, required euthanasia only 4 weeks after intratibial cell injection. CCHD-GFP-Hes4 injected mice had significantly larger and more lytic primary tumors than CCHD-GFP control (mean tumor size ± SEM: 38.9 ± 0.6 arbitrary units [CCHD-GFP] versus 64.6 ± 3.9 arbitrary units [CCHD-GFP-Hes4]; P < 0.001; mean lytic grade ± SEM: 0.9 ± 0.2 [CCHD-GFP] versus 3.0 ± 0.3 [CCHD-GFP-Hes4]; P < 0.001) (Figs. 5A and5B). We also observed a 50-fold increase in the number of metastases in mice injected with CCHD-GFP-Hes4 cells over CCHD-GFP cells 4 weeks post injection (Fig. 5C).
Figure 5. Hes4 overexpression increases OS tumor aggressiveness.
Mice were injected with CCHD-GFP or CCHD-GFP-Hes4 cells into the tibia. Six-weeks post injection, all mice were sacrificed and processed for analysis. (A) CCHD-GFP-Hes4 cells formed significantly larger primary tumors than did CCHD-GFP. Each dot represents one tumor (n = 15). ***P < 0.001. (B) Mice injected with Hes4-overexpressing OS cells had significantly more lytic primary tumors than did control mice. a grading system (0–4, with 4 indicating the most lytic destruction) was used to quantify the extent of bone destruction in mice with primary CCHD-GFP or CCHD-GFP-Hes4 tumors. Each dot represents the lytic score for one tumor (mean ± SEM, n = 15). ***P < 0.001. (C) Mice injected with Hes4-overexpressing OS cells had significantly more metastases than did control mice. Metastatic lesions were quantified by counting vimentin-positive nodules on five sections per lung. Each dot represents one mouse (n = 15 per group). ***P < 0.0001. Black bars indicate 200μm, orange bars indicate 20 μm.
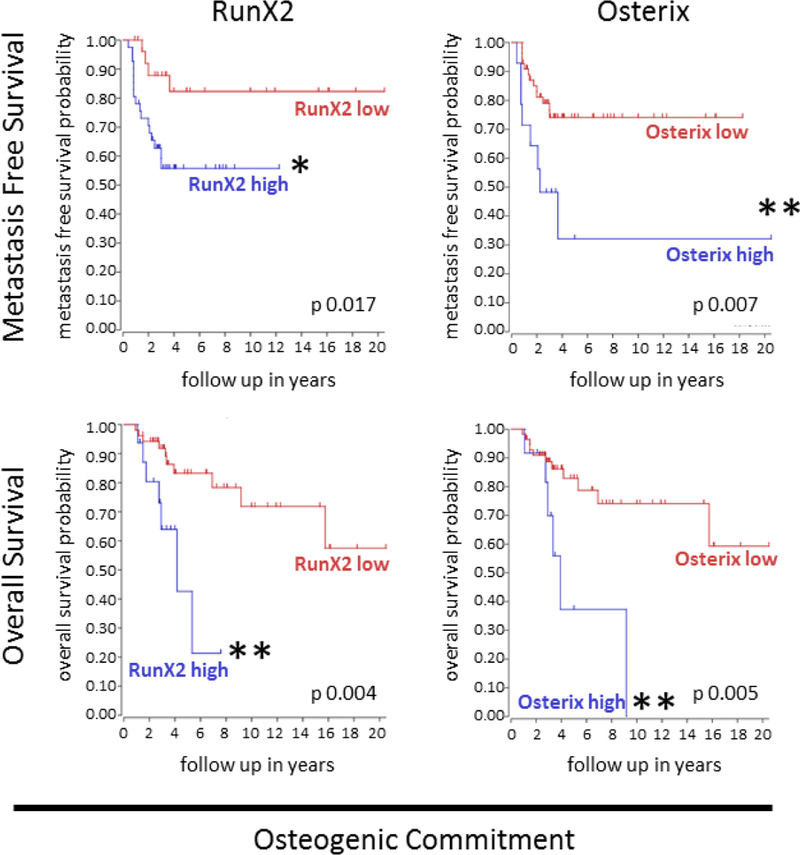
High RunX2 and osterix expression correlate with decreased metastasis-free and overall survival in OS patients
We found that overexpression of Hes4 in OS cells resulted in increased expression of RunX2 and osterix (markers of osteogenic commitment) but decreased expression of alkaline phosphatase (an early differentiation marker). This correlated with our in vitro results demonstrating decreased differentiation of OS cells following Hes4 transduction. We used the R2: Genomics Analysis and Visualization Platform to create gene-based Kaplan-Meier survival curves based on gene expression within pretreatment high grade OS biopsies. Consistent with our finding that Hes4 overexpression resulted in increased RunX2 and osterix, and a more aggressive phenotype in vivo, high gene expression of RunX2 (41/69 [59%]) or osterix (14/69 [20%]) in pretreatment high grade OS primary tumor biopsies correlated with decreased metastasis-free survival (P = 0.017 and P = 0.007, respectively) and overall survival (P = 0.0004 and P = 0.0005, respectively) than did those with low levels of RunX2 or osterix expression (Fig. 6).
Figure 6. Expression of osteogenic commitment markers in human OS samples correlates with significantly decreased overall and metastasis-free survival outcomes.
The R2: Genomics Analysis and Visualization Platform was used to generate Kaplan-Meier metastasis-free and overall survival curves using the Mixed Osteosarcoma - Kuijjer - 127 - vst - ilmnhwg6v2 data set. Genome-wide gene expression analysis was performed using 84 pretreatment high-grade diagnostic OS biopsy samples. Two different sets of control samples were used for comparison: osteoblasts (n = 3) and mesenchymal stem cells (n = 12; GEO accession number GSE28974). Patients with high RunX2 or osterix expression had significantly lower probabilities of both overall and metastasis-free survival than did those with low RunX2 or osterix expression. *P < 0.01; **P < 0.05.
Discussion
For the first time, we describe the role of Hes4 in bone differentiation in osteosarcoma. Importantly, our data suggest that expression of Hes4 may be a prognostic biomarker of patient outcome in newly diagnosed patients with high grade OS. High expression of Hes4 also correlated with decreased overall and metastasis-free patient survival. We found that overexpression of Hes4 in OS cells blocked terminal differentiation resulting in an immature preosteoblastic phenotype (Supplemental Figure S5). When injected orthotopically, Hes4-overexpressing cells produced larger tumors with increased lytic and metastatic capacity.
Defects early in osteogenic cell differentiation are hypothesized to result in more aggressive OS tumors when compared to defects at later stages37. In the present study, we confirmed this link. We showed that high Hes4 expression correlated with a less differentiated phenotype in vitro, and an increase in metastatic potential in vivo.
Overexpression of Hes4 in osteosarcoma resulted in increased expression of RunX2 and osterix, critical regulators of osteogenic differentiation. The mechanism by which Hes4 upregulates RunX2 and osterix remains unclear. There are no N-(CACNAG) or E-(CANNTG) box-binding sites (data not shown) in the promoter regions of these genes, suggesting that Hes4 may regulate these factors indirectly by binding to other Notch downstream targets. For example, Hes1, another Notch downstream target, stabilizes RunX2 within the promoter region of osterix to stimulate osteoblast differentiation38. Alternatively, Hes4 may prevent differentiation of OS via intermediate factors outside the Notch family. For example, in normal bone marrow stromal cells, Twist-1 binds to RunX2 to prevent osteogenic differentiation24. When Hes4 is overexpressed in these non-tumor cells, it binds to Twist-1 to reverse this prevention of differentiation, allowing for the osteogenic differentiation of bone marrow stromal cells. In OS cells, Hes4 may not be able to bind to Twist-1, preventing differentiation and thus resulting in a tumorigenic, immature state.
The presence of metastases at diagnosis and the response to preoperative chemotherapy are the only predictive markers of patient outcome in OS. In this study, we showed that high Hes4 expression correlated with the invasive and metastatic potential of the primary tumor, and most important, with decreased metastasis-free and overall patient survival. For OS tumors to invade and form metastases, tumor cells must first degrade bone in order to access the bloodstream. We found a significant increase in the lytic capacity of Hes4 overexpressing tumors over that of control tumors. A known contributor to lytic behavior is IL-1α, a potent cytokine secreted by OS cells39. IL-1α promotes the expression of receptor activator of nuclear factor-κB ligand (RANKL) within mature osteoblasts. When RANKL interacts with its receptor, RANK which is expressed in immature osteoclasts, RANKL facilitates the maturation of osteoclast precursors to induce osteoclast maturation39. In cells overexpressing Hes4, however, the levels of IL-1α, RANK, and RANKL RNA expression did not change (data not shown).
Our data demonstrating that Hes4 overexpression correlates with poor prognosis for OS is consistent with a previous report that inhibition of Notch signaling by γ-secretase inhibitors or the dominant-negative mutant of the Mastermind gene inhibits OS growth in mice40. However, broad inhibition of Notch signaling is likely an overly simplistic therapeutic approach in the treatment of OS. Notch signaling is known to be sensitive to context-driven cues depending on which ligands and/or receptors are expressed in the tumor cells. Therefore, hypothesizing that Notch downstream targets also work in context-dependent ways is reasonable. While Hes4 expression promotes aggressive OS, other Notch targets may have different effects on OS. For example, in a canine model of OS, Hes1 expression was elevated in most tumor samples but reduced in the most aggressive tumors41. In the present study, we showed that although Hes4 promoted increased OS invasion in vitro, Hes1 overexpression decreased both invasion and proliferation. High Hes1 expression in osteosarcoma patient samples trended toward a correlation with higher metastasis-free survival and higher overall survival. Interestingly, when Hes4 was overexpressed in osteosarcoma cells, Hes1 mRNA expression decreased (data not shown), suggesting cross-talk between Hes1 and Hes4. Taken together, these findings indicate that pan inhibition of Notch signaling in osteosarcoma may have dual effects indicating a need for a better understanding of how the downstream Notch target genes interact with each other and the different signaling pathways that each control.
Identifying ways to classify tumors and distinguish between aggressive, mostly undifferentiated tumors usually linked with poor patient outcomes and moderate, more differentiated tumors with better outcomes is important because it may allow modification of therapy at the time of diagnosis. Our data suggest that because of its significant relationship with differentiation and outcome, Hes4 is a potential prognostic factor for the early stratification and identification of osteosarcoma patients at high risk for relapsed metastasis and decreased survival.
Supplementary Material
Supplementary Figure S1. Correlation of OS patient outcome and expression of Notch downstream targets other than Hes4. The R2: Genomics Analysis and Visualization Platform was used to generate Kaplan-Meier metastasis-free and overall survival curves using the Mixed Osteosarcoma - Kuijjer - 127 - vst - ilmnhwg6v2 data set. Genome-wide gene expression analysis was performed using 84 pretreatment high-grade diagnostic OS biopsy samples. Two different sets of control samples were used for comparison: osteoblasts (n = 3) and mesenchymal stem cells (n = 12; GEO accession number GSE28974). Hes1, Hes2, Hes5, Hey1, Hey2, and DTX were classified according to high versus low expression. No significant differences were seen in any of the groups.
Supplementary Figure S2. Hes1 and Hes4 overexpression in CCHD and HOS cells. Schematic representations of Hes1 (A) and Hes4 (B) overexpression vector maps depicting the orientation of GFP and Hes1 or Hes4 within the retroviral MigR1 backbone. The expression of Hes1 and Hes4 is controlled by a constitutively active 5' LTR promoter. The presence of an IRES causes production of Hes1 or Hes4 and GFP as two separate proteins, not a fusion protein. (C) Transduction of Hes1 results in a significant increase of Hes1 RNA expression. cDNA was prepared from RNA harvested from HOS and CCHD cells after transduction with GFP or GFP-Hes1. RT-qPCR was performed to measure the levels of Hes1 expression normalized according to GAPDH expression relative to that in GFP-transduced control cells. (D) Transduction of Hes4 results in a significant increase of Hes1 or Hes4 RNA expression, respectively. cDNA was prepared from RNA harvested from HOS and CCHD cells after transduction with GFP or GFP-Hes4. RT-qPCR was performed to measure the levels of Hes4 expression normalized according to GAPDH expression relative to that in GFP-transduced control cells. *P ≤ 0.05; **P ≤ 0.01. Bars, mean ± SEM (n = 3).
Supplementary Figure S3. Hes1 overexpression in CCHD and HOS cells decreases OS invasion and proliferation. (A) Hes1 overexpression decreases invasion in CCHD and HOS cells. CCHD and HOS cells were transduced with GFP or GFP-Hes1 and sorted according to GFP positivity. Their Invasiveness was measured using a 24-well BioCoat Matrigel invasion chamber with an 8-mm pore size. A medium with 10% fetal bovine serum was used in the bottom well of the chamber as a chemoattractant. At 24 (HOS) or 48 (CCHD) hours, migrated cells were counted. The graph shows the mean number of migrated cells per field (± SEM; n = 3). *P ≤ 0.05; **P < 0.01. (B) Hes1 overexpression decreases proliferation in CCHD and HOS cells. The percentages of GFP-positive CCHD and HOS cells over time after stable retroviral transduction of GFP or GFP-Hes1 (normalized to day 5 after transduction) were quantified at various time points as described in Materials and Methods and expressed as the mean cell number ± SEM (n = 3).
Supplementary Figure S4. Schematic depicting key transcription factors involved in normal osteoblast differentiation. Differentiation stage is defined by the presence or absence of specific transcription factors and can be divided into four main stages: pluripotency, osteogenic commitment, preosteoblast/early osteoblast, and maturation
Supplemental Figure S5. Schematic depicting the highly regulated balance of osteoblasts and osteoclasts, and the role of Hes4 in the inhibition of osteogenic differentiation in OS. Bone remodeling relies on both osteoclastic and osteoblastic activity. The formation of osteoclasts and osteoblasts is highly regulated by a multistep differentiation process. Osteoclasts originate from hematopoietic stem cells while osteoblasts originate from mesenchymal stem cells. There is cross talk between osteoblasts and pre-osteoclasts (via IL-1α/RANKL/RANK signaling). Osteosarcoma is thought to arise from the disruption of osteogenic differentiation, and can occur at any point within the differentiation pathway resulting in a heterogeneous mix of OS which represents multiple maturation states. Defects at early stages within the osteogenic differentiation pathway leads to the development of more aggressive and less differentiated OS. Hes4 blocks the osteogenic differentiation pathway by preventing the maturation of pre-osteoblasts by increasing RunX2 and osterix, and decreasing alkaline phosphatase. The Hes4 mediated block of differentiation results in large primary tumors and significantly more metastases in vivo, and correlates with decreased metastasis free and overall survival in high grade OS patients.
Supplemental Methods S1. qPCR primers and Taqman probes. The primer sequences used for qPCR and unique Taqman probe identifiers are listed.
Acknowledgments
The authors would like to thank Don Norwood for his assistance as scientific editor. KS acknowledges The Center for Energy Balance and Survivorship Research, Duncan Family Institute, MD Anderson Cancer Center.
Grant Support: NIH 5R01CA141208–05 (ESK), NIH P30CA16672 (Cancer Center Support Grant)
Abbreviations
- OS
osteosarcoma
- Hes
hairy/enhancer of split
- hFOB
human fetal osteoblasts
- RANKL
receptor activator of nuclear factor-κB ligand
Footnotes
Conflict of Interest: None
References
- 1.Bielack SS, Kempf-Bielack B, Winkler K. Osteosarcoma: relationship of response to preoperative chemotherapy and type of surgery to local recurrence. J Clin Oncol. February 1996;14(2):683–684. [DOI] [PubMed] [Google Scholar]
- 2.Marina NM, Smeland S, Bielack SS, et al. Comparison of MAPIE versus MAP in patients with a poor response to preoperative chemotherapy for newly diagnosed high-grade osteosarcoma (EURAMOS-1): an open-label, international, randomised controlled trial. The Lancet Oncology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielack SS, Smeland S, Whelan JS, et al. Methotrexate, Doxorubicin, and Cisplatin (MAP) Plus Maintenance Pegylated Interferon Alfa-2b Versus MAP Alone in Patients With Resectable High-Grade Osteosarcoma and Good Histologic Response to Preoperative MAP: First Results of the EURAMOS-1 Good Response Randomized Controlled Trial. J Clin Oncol. July 10 2015;33(20):2279–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harting MT, Blakely ML, Jaffe N, et al. Long-term survival after aggressive resection of pulmonary metastases among children and adolescents with osteosarcoma. Journal of pediatric surgery. January 2006;41(1):194–199. [DOI] [PubMed] [Google Scholar]
- 5.Abarrategi A, Tornin J, Martinez-Cruzado L, et al. Osteosarcoma: Cells-of-Origin, Cancer Stem Cells, and Targeted Therapies. Stem Cells International. 2016;2016:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celià-Terrassa T, Kang Y. Distinctive properties of metastasis-initiating cells. Genes & Development. April 15, 2016. 2016;30(8):892–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorlick R, Meyers PA. Osteosarcoma necrosis following chemotherapy: innate biology versus treatment-specific. J Pediatr Hematol Oncol. November 2003;25(11):840–841. [DOI] [PubMed] [Google Scholar]
- 8.Deng Z-L, Sharff K, Tang N, et al. Regulation of osteogenic differentiation during skeletal development. Frontiers in bioscience : a journal and virtual library. 2008;13:2001–2021. [DOI] [PubMed] [Google Scholar]
- 9.Ducy P, Starbuck M, Priemel M, et al. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes & development. 1999;13(8):1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducy P, Zhang R, Geoffroy V, Ridall A, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. [DOI] [PubMed] [Google Scholar]
- 11.Haydon R, Luu H, He T-C. Osteosarcoma and osteoblastic differentiation: a new perspective on oncogenesis. Clinical orthopaedics and related research. 2007;454:237–246. [DOI] [PubMed] [Google Scholar]
- 12.Hong J-H, Hwang E, McManus M, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science (New York, N.Y.). 2005;309(5737):1074–1078. [DOI] [PubMed] [Google Scholar]
- 13.Karsenty G Role of Cbfa1 in osteoblast differentiation and function. Semin Cell Dev Biol. October 2000;11(5):343–346. [DOI] [PubMed] [Google Scholar]
- 14.Karsenty G How many factors are required to remodel bone? Nat Med. September 2000;6(9):970–971. [DOI] [PubMed] [Google Scholar]
- 15.Lian J, Stein G, Javed A, et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Reviews in endocrine & metabolic disorders. 2006;7(1–2):1–16. [DOI] [PubMed] [Google Scholar]
- 16.Nakashima K, Zhou X, Kunkel G, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. [DOI] [PubMed] [Google Scholar]
- 17.Otto F, Thornell A, Crompton T, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–771. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi A, Komori T, Suda T. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocrine reviews. 2000;21(4):393–411. [DOI] [PubMed] [Google Scholar]
- 19.Thomas D, Kansara M. Epigenetic modifications in osteogenic differentiation and transformation. Journal of cellular biochemistry. 2006;98(4):757–769. [DOI] [PubMed] [Google Scholar]
- 20.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. March 15 1994;8(6):707–719. [DOI] [PubMed] [Google Scholar]
- 21.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. May 1995;121(5):1533–1545. [DOI] [PubMed] [Google Scholar]
- 22.Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. May 16 1997;89(4):629–639. [DOI] [PubMed] [Google Scholar]
- 23.Tezuka K, Yasuda M, Watanabe N, et al. Stimulation of osteoblastic cell differentiation by Notch. J Bone Miner Res. February 2002;17(2):231–239. [DOI] [PubMed] [Google Scholar]
- 24.Cakouros D, Isenmann S, Hemming SE, et al. Novel basic Helix Loop Helix Transcription Factor Hes4 Antagonizes the Function of Twist-1 to Regulate Lineage Commitment of Bone Marrow Stromal/ Stem Cells. Stem cells and development. 2015. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Chen T, Zhang J, et al. Notch1 promotes glioma cell migration and invasion by stimulating beta-catenin and NF-kappaB signaling via AKT activation. Cancer Sci. February 2012;103(2):181–190. [DOI] [PubMed] [Google Scholar]
- 26.Stoeck A, Lejnine S, Truong A, et al. Discovery of biomarkers predictive of GSI response in triple-negative breast cancer and adenoid cystic carcinoma. Cancer discovery. 2014;4(10):1154–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messersmith WA, Shapiro GI, Cleary JM, et al. A Phase I, Dose-Finding Study in Patients with Advanced Solid Malignancies of the Oral γ-Secretase Inhibitor PF-03084014. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;21(1):60–67. [DOI] [PubMed] [Google Scholar]
- 28.Kuijjer ML, Rydbeck H, Kresse SH, et al. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes, chromosomes & cancer. July 2012;51(7):696–706. [DOI] [PubMed] [Google Scholar]
- 29.Zweidler-McKay PA, He Y, Xu L, et al. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. December 1 2005;106(12):3898–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber KL, Doucet M, Price JE, Baker C, Kim SJ, Fidler IJ. Blockade of Epidermal Growth Factor Receptor Signaling Leads to Inhibition of Renal Cell Carcinoma Growth in the Bone of Nude Mice. Cancer Research. 2003;63(11):2940–2947. [PubMed] [Google Scholar]
- 31.Yang J, Hou Y, Zhou M, et al. Twist induces epithelial-mesenchymal transition and cell motility in breast cancer via ITGB1-FAK/ILK signaling axis and its associated downstream network. Int J Biochem Cell Biol. February 2016;71:62–71. [DOI] [PubMed] [Google Scholar]
- 32.Su Q, Xin L. Notch signaling in prostate cancer: refining a therapeutic opportunity. Histology and histopathology. February 2016;31(2):149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su BH, Qu J, Song M, et al. NOTCH1 signaling contributes to cell growth, anti-apoptosis and metastasis in salivary adenoid cystic carcinoma. Oncotarget. August 30 2014;5(16):6885–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller CS. Notch signaling and malignant melanoma. Adv Exp Med Biol. 2012;727:258–264. [DOI] [PubMed] [Google Scholar]
- 35.Kranenburg O Prometastatic NOTCH Signaling in Colon Cancer. Cancer Discov. February 2015;5(2):115–117. [DOI] [PubMed] [Google Scholar]
- 36.Tang Y, Hu C, Yang H, et al. Rnd3 regulates lung cancer cell proliferation through notch signaling. PLoS One. 2014;9(11):e111897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang N, Song W-X, Luo J, Haydon R, He T-C. Osteosarcoma development and stem cell differentiation. Clinical orthopaedics and related research. 2008;466(9):2114–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh JH, Lee HW, Lee JW, Kim JB. Hes1 stimulates transcriptional activity of Runx2 by increasing protein stabilization during osteoblast differentiation. Biochem Biophys Res Commun. February 29 2008;367(1):97–102. [DOI] [PubMed] [Google Scholar]
- 39.Cao Y, Jia SF, Chakravarty G, de Crombrugghe B, Kleinerman ES. The osterix transcription factor down-regulates interleukin-1 alpha expression in mouse osteosarcoma cells. Molecular cancer research : MCR. January 2008;6(1):119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engin F, Bertin T, Ma O, et al. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet. April 15 2009;18(8):1464–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dailey DD, Anfinsen KP, Pfaff LE, et al. HES1, a target of Notch signaling, is elevated in canine osteosarcoma, but reduced in the most aggressive tumors. BMC veterinary research. 2013;9:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Correlation of OS patient outcome and expression of Notch downstream targets other than Hes4. The R2: Genomics Analysis and Visualization Platform was used to generate Kaplan-Meier metastasis-free and overall survival curves using the Mixed Osteosarcoma - Kuijjer - 127 - vst - ilmnhwg6v2 data set. Genome-wide gene expression analysis was performed using 84 pretreatment high-grade diagnostic OS biopsy samples. Two different sets of control samples were used for comparison: osteoblasts (n = 3) and mesenchymal stem cells (n = 12; GEO accession number GSE28974). Hes1, Hes2, Hes5, Hey1, Hey2, and DTX were classified according to high versus low expression. No significant differences were seen in any of the groups.
Supplementary Figure S2. Hes1 and Hes4 overexpression in CCHD and HOS cells. Schematic representations of Hes1 (A) and Hes4 (B) overexpression vector maps depicting the orientation of GFP and Hes1 or Hes4 within the retroviral MigR1 backbone. The expression of Hes1 and Hes4 is controlled by a constitutively active 5' LTR promoter. The presence of an IRES causes production of Hes1 or Hes4 and GFP as two separate proteins, not a fusion protein. (C) Transduction of Hes1 results in a significant increase of Hes1 RNA expression. cDNA was prepared from RNA harvested from HOS and CCHD cells after transduction with GFP or GFP-Hes1. RT-qPCR was performed to measure the levels of Hes1 expression normalized according to GAPDH expression relative to that in GFP-transduced control cells. (D) Transduction of Hes4 results in a significant increase of Hes1 or Hes4 RNA expression, respectively. cDNA was prepared from RNA harvested from HOS and CCHD cells after transduction with GFP or GFP-Hes4. RT-qPCR was performed to measure the levels of Hes4 expression normalized according to GAPDH expression relative to that in GFP-transduced control cells. *P ≤ 0.05; **P ≤ 0.01. Bars, mean ± SEM (n = 3).
Supplementary Figure S3. Hes1 overexpression in CCHD and HOS cells decreases OS invasion and proliferation. (A) Hes1 overexpression decreases invasion in CCHD and HOS cells. CCHD and HOS cells were transduced with GFP or GFP-Hes1 and sorted according to GFP positivity. Their Invasiveness was measured using a 24-well BioCoat Matrigel invasion chamber with an 8-mm pore size. A medium with 10% fetal bovine serum was used in the bottom well of the chamber as a chemoattractant. At 24 (HOS) or 48 (CCHD) hours, migrated cells were counted. The graph shows the mean number of migrated cells per field (± SEM; n = 3). *P ≤ 0.05; **P < 0.01. (B) Hes1 overexpression decreases proliferation in CCHD and HOS cells. The percentages of GFP-positive CCHD and HOS cells over time after stable retroviral transduction of GFP or GFP-Hes1 (normalized to day 5 after transduction) were quantified at various time points as described in Materials and Methods and expressed as the mean cell number ± SEM (n = 3).
Supplementary Figure S4. Schematic depicting key transcription factors involved in normal osteoblast differentiation. Differentiation stage is defined by the presence or absence of specific transcription factors and can be divided into four main stages: pluripotency, osteogenic commitment, preosteoblast/early osteoblast, and maturation
Supplemental Figure S5. Schematic depicting the highly regulated balance of osteoblasts and osteoclasts, and the role of Hes4 in the inhibition of osteogenic differentiation in OS. Bone remodeling relies on both osteoclastic and osteoblastic activity. The formation of osteoclasts and osteoblasts is highly regulated by a multistep differentiation process. Osteoclasts originate from hematopoietic stem cells while osteoblasts originate from mesenchymal stem cells. There is cross talk between osteoblasts and pre-osteoclasts (via IL-1α/RANKL/RANK signaling). Osteosarcoma is thought to arise from the disruption of osteogenic differentiation, and can occur at any point within the differentiation pathway resulting in a heterogeneous mix of OS which represents multiple maturation states. Defects at early stages within the osteogenic differentiation pathway leads to the development of more aggressive and less differentiated OS. Hes4 blocks the osteogenic differentiation pathway by preventing the maturation of pre-osteoblasts by increasing RunX2 and osterix, and decreasing alkaline phosphatase. The Hes4 mediated block of differentiation results in large primary tumors and significantly more metastases in vivo, and correlates with decreased metastasis free and overall survival in high grade OS patients.
Supplemental Methods S1. qPCR primers and Taqman probes. The primer sequences used for qPCR and unique Taqman probe identifiers are listed.