Abstract
The Cancer Genome Atlas (TCGA) represents one of several international consortia dedicated to performing comprehensive genomic and epigenomic analyses of selected tumour types to advance our understanding of disease and provide an open-access resource for worldwide cancer research. Thirty-three tumour types (selected by histology or tissue of origin, to include both common and rare diseases), comprising >11 000 specimens, were subjected to DNA sequencing, copy number and methylation analysis, and transcriptomic, proteomic and histological evaluation. Each cancer type was analysed individually to identify tissue-specific alterations, and make correlations across different molecular platforms. The final dataset was then normalized and combined for the PanCancer Initiative, which seeks to identify commonalities across different cancer types or cells of origin/lineage, or within anatomically or morphologically related groups. An important resource generated along with the rich molecular studies is an extensive digital pathology slide archive, composed of frozen section tissue directly related to the tissues analysed as part of TCGA, and representative formalin-fixed paraffin-embedded, haematoxylin and eosin (H&E)-stained diagnostic slides. These H&E image resources have primarily been used to verify diagnoses and histological subtypes with some limited extraction of standard pathological variables such as mitotic activity, grade, and lymphocytic infiltrates. Largely overlooked is the richness of these scanned images for more sophisticated feature extraction approaches coupled with machine learning, and ultimately correlation with molecular features and clinical endpoints. Here, we document initial attempts to exploit TCGA imaging archives, and describe some of the tools, and the rapidly evolving image analysis/feature extraction landscape. Our hope is to inform, and ultimately inspire and challenge, the pathology and cancer research communities to exploit these imaging resources so that the full potential of this integral platform of TCGA can be used to complement and enhance the insightful integrated analyses from the genomic and epigenomic platforms.
Keywords: The Cancer Genome Atlas, TCGA, genomics, digital pathology, image analysis, computational histology, PanCancer
Introduction
Contemporary anatomical or surgical pathology practice encompasses not only the traditional morphological approach to diagnosis, but also, increasingly, molecular and genomic approaches for diagnostic, prognostic and theragnostic purposes. Digital pathology is also making advances in clinical practice and research, allowing an increased utilization of telepathology, and adding new analytical tools to the pathology toolkit. The past few years have seen the completion of The Cancer Genome Atlas (TCGA) Project, which has generated a wealth of raw molecular data and digital image data. Much insightful integration across the genomic and epigenomic platforms has occurred, and is published in numerous articles – both through primary TCGA publications and by extensive use of TCGA datasets to enhance the work of groups worldwide. The molecular resources of TCGA have contributed greatly to our understanding of the tumour types included, and represent a vast resource for advancing the field of pathology research and clinical practice. Other similar large-scale genomic analyses include the International Cancer Genome Consortium (ICGC), which was established with the goal of generating whole genome sequencing data, whole exome sequencing data, transcriptomic data and DNA methylation data on >25 000 tumour samples. Integrated projects such as the TCGA PanCancer Initiative and the ICGC PanCancer Analysis of Whole Genomes are further expanding the initial tumour type-specific analysis to discover commonalities across different lines of differentiation and tissues of origin. Although TCGA has generated digital slide images for the majority of tumours, the digital imaging resource has been underrepresented among this impressive productivity and distinctly underutilized. This review will briefly introduce TCGA and the PanCancer initiatives, and their application to the field of pathology, but will focus primarily on the largely untapped resource of digital histology analysis and the promise that it brings to improving our understanding of cancer biology and ultimately patient care.
Overview of TCGA and the PanCancer Initiative
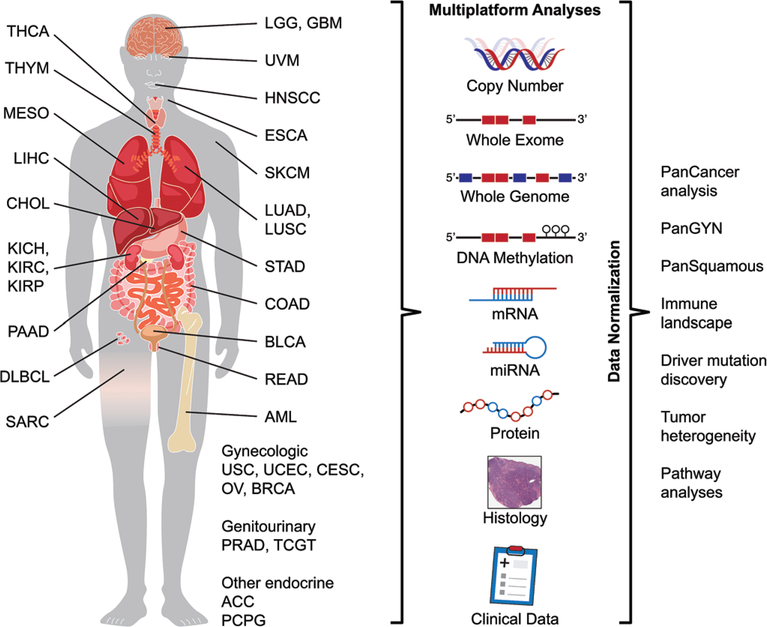
TCGA was begun as a collaboration between the National Cancer Institute and the National Human Genome Research Institute. The goal of this massive project was to comprehensively characterize multiple molecular aspects of 33 selected cancer types (Table 1; Figure 1), including DNA sequence (all exomes and low-pass genomes for a subset), copy number and methylation, mRNA and microRNA (miRNA) expression, and the expression of selected proteins. Fresh frozen tumour and non-neoplastic tissues were submitted to the Biospecimen Core Resource (BCR) from contributing sites, along with pathology reports and clinical data, including treatment and patient outcomes. From these frozen tumour specimens, top and bottom frozen sections were obtained and scanned as part of the specimen curation protocol. In most cases, selected representative haematoxylin and eosin (H&E)-stained slides were also submitted for review, and these were digitally scanned. Molecular analyses were undertaken at designated Genome Characterization Centers and Genome Sequencing Centers. Raw data were integrated and analysed at Genome Data Analysis Centers. Disease Working Groups initially defined the key problems to be addressed, and the type and format of clinical data to be gathered for each tumour type from contributing sites. Finally, Analysis Working Groups composed of clinical and scientific experts reviewed the multiple platform analyses in order to better understand the findings, and developed resource publications to introduce the findings and encourage further work in the field. All datasets are publically available for further analysis through the Genomic Data Commons Data Portal (https://gdc.cancer.gov), and have been widely utilized. In addition, a number of exceedingly useful data visualization and analysis tools were developed at the Genome Data Analysis Centers to allow data access and exploration through online interactive tools (examples: http://firebrowse.org; http://cbioportal.org; http://explorer.cancerregulome.org; http://bioinformatics.mdanderson.org/tcgambatch). Other multinational genomic efforts such as the ICGC followed similar workflows and resulted in equally valuable data resources, although an in-depth discussion is beyond the scope of this review. The ICGC datasets are publically available through the ICGC data portal (https://dcc.icgc.org/).
Table 1.
TCGA cancer types*
| Tumour type | TCGA code | Number of cases with-Omic data† | Number of cases with digital pathology images |
|---|---|---|---|
| Haematopoietic | |||
| Acute myeloid leukaemia | AML | 191 | NA |
| Diffuse large B-cell lymphoma | DLBC | 48 | 48 |
| Thymoma | THYM | 124 | 124 |
| Gynaecological | |||
| Cervical squamous cell carcinoma Endocervical adenocarcinoma | CESC | 307 | 308 |
| Uterine carcinosarcoma | UCS | 57 | 57 |
| Uterine corpus endometrial carcinoma | UCEC | 559 | 560 |
| Ovarian serous cystadenocarcinoma | OV | 608 | 590 |
| Urinary tract | |||
| Bladder urothelial carcinoma | BLCA | 412 | 412 |
| Renal chromophobe carcinoma | KICH | 66 | 113 |
| Clear cell renal cell carcinoma | KIRC | 535 | 537 |
| Papillary renal cell carcinoma | KIRP | 291 | 291 |
| Prostate/testis | |||
| Prostate adenocarcinoma | PRAD | 498 | 500 |
| Testicular germ cell tumours | TGCT | 150 | 150 |
| Endocrine | |||
| Thyroid carcinoma | THCA | 507 | 507 |
| Adrenocortical carcinoma | ACC | 92 | 92 |
| Phaeochromocytoma Paraganglioma | PCPG | 179 | 179 |
| Breast | |||
| Breast invasive carcinoma | BRCA | 1098 | 1103 |
| Gastrointestinal tract | |||
| Oesophageal carcinoma | ESCA | 185 | 185 |
| Stomach adenocarcinoma | STAD | 443 | 478 |
| Colonic adenocarcinoma | COAD | 460 | 462 |
| Rectal adenocarcinoma | READ | 171 | 283 |
| Liver, pancreaticobiliary | |||
| Cholangiocarcinoma | CHOL | 36 | 39 |
| Hepatocellular carcinoma | LIHC | 377 | 377 |
| Pancreatic adenocarcinoma | PAAD | 185 | 191 |
| Pulmonary | |||
| Lung adenocarcinoma | LUAD | 585 | 523 |
| Lung squamous cell carcinoma | LUSC | 504 | 512 |
| Mesothelioma | MESO | 87 | 87 |
| Head and neck | |||
| Head and neck squamous cell carcinoma | HNSC | 528 | 523 |
| Melanocytic malignancies | |||
| Cutaneous melanoma | SKCM | 470 | 471 |
| Uveal melanoma | UVM | 80 | 80 |
| Brain | |||
| Lower-grade glioma | LGG | 516 | 521 |
| Glioblastoma multiforme | GBM | 617 | 613 |
| Mesenchymal | |||
| Adult soft tissue sarcoma | SARC | 261 | 261 |
From https://portal.gdc.cancer.gov (accessed 27 October 2017).
Numbers of cases reflect the total with data, including cases excluded from TCGA analyses for quality control or pathology review. -Omic data for these studies includes genomic, transcriptomic, epigenomic, and/or proteomic data. Not all cases with data were analysable across all platforms.
Figure 1.
Overview of TCGA. Schematic representation of the 33 cancers analysed by the TCGA/PanCancer Initiative organized by tissue of origin, and the data types acquired. Examples of the PanCancer analyses undertaken are listed on the right. TCGA tumour type abbreviation codes are as follows: ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; ESCA, oesophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, chromophobe renal cell carcinoma; KIRC, clear cell renal clear cell carcinoma; KIRP, papillary renal cell carcinoma; LAML, acute myeloid leukaemia; LGG, lower-grade glioma; LIHC, hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous adenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, phaeochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectal adenocarcinoma; SARC, adult soft tissue sarcoma; SKCM, cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumour; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
TCGA platforms
The core dataset generated by TCGA for each tumour sample included pathology data, digitized histology images, patient clinical and outcome data, whole exome sequencing on all cases, whole genome sequencing on a selected subset of cases, DNA copy number generated by the use of single-nucleotide polymorphism arrays, mRNA and miRNA expression data generated through microarrays in early analyses, or, subsequently, mRNA sequencing, miRNA sequencing and DNA methylation data generated via methylation arrays. Expression of selected proteins and phosphorylated epitopes was assessed with reverse-phase protein arrays. Data were assessed and correlated across the analytical platforms to create an integrated genomic and epigenomic view of cancer. Extensive high-order analysis has been performed for all platforms except the scanned pathology images, which have primarily been used for analog analysis by specialist pathologists to confirm the diagnosis and extract specific pathological characteristics.
The PanCancer analysis project
After examination of genomic, epigenomic, transcriptomic and proteomic alterations within individual tumour types segregated by tissue of origin, the second phase of the TCGA Research Network examined similarities and differences between tumour types and tissue sites under the umbrella of the PanCancer analysis project (PanCancer Initiative). The initial PanCancer12 effort published in 2013 incorporated data from 12 of the initial tumour types analysed in the TCGA marker articles [1]. The ongoing PanCancer33 initiative is characterizing all 11 000+ cases across 33 cancer types. It includes histology-specific analyses, such as the pan-squamous cell analysis incorporating data on squamous cell carcinomas of all available sites (head and neck, oesophagus, lung, cervix, and bladder urothelial carcinomas with squamous elements), as well as broad analyses across all cancers such as the mutational driver effort harnessing the power of the large datasets to identify rare driver mutations, or the panimmune initiative seeking to characterize the patterns and significance of immune infiltration across malignancies. These analyses, uniting datasets generated on different platforms at different institutions, and harnessing technological advances made over a period of multiple years, are made possible by broad normalization efforts to mitigate batch effects across all TCGA samples, such as those applied to the exome sequencing data for mutational calling.
Major findings from the PanCancer analysis project
Contemporary pathology must provide not only diagnostic information, but also prognostic and theragnostic features where possible. TCGA and PanCancer efforts incorporate the strengths of both traditional cancer studies by performing tissue-specific analyses (TCGA), and contemporary ‘basket’ approaches to biomarker investigation by investigating alterations affecting a small percentage of tumours with widely disparate origins to identify commonalities than can inform our understanding of tumour biology or clinical management (PanCancer analyses). Another advantage of this broad-based multiplatform approach is the ability to more comprehensively profile disparate alterations across cancers that affect the same cell signalling pathways; such convergence might be targetable by similar classes of agents.
Among the many insights gleaned from the initial PanCancer12 initiative was the demonstration that, for some cancer histologies, such as clear cell renal cell carcinoma, molecular alterations are very distinct from those of all other carcinomas, and that the tumours form a relatively homogeneous group. In contrast, other carcinomas, such as breast invasive ductal carcinoma, show considerable molecular divergence within a relatively homogeneous morphological group, despite the common site of origin, supporting different clinical approaches to therapy [2,3]. Moreover, carcinomas with similar histological features arising at different anatomical sites (e.g. squamous cell carcinoma of any site, or colonic and rectal adenocarcinoma) are more similar than they are different [4]. To some extent, these findings reflect an intuitive observation concerning the histological features of these morphologically similar groups of tumours, and provide a rationale for interpreting histological findings in the context of revealed molecular signatures. It is of note that gene expression and methylation signatures do seem to reflect tissue of origin to a greater extent than genomic alterations in most cases, supporting the importance of cellular context in determining tumour cell phenotypes [4].
Molecular convergence across tissue types
At the genomic level, malignancies appear to be divided into two broad categories reflecting divergent oncogenic processes: copy number-driven and mutation-driven [5]. Malignancies in the PanCancer12 analysis with high levels of somatic copy number alterations were associated with early TP53 mutations, reflecting the importance of TP53 in regulating genomic stability, and included ovarian carcinoma, squamous cell carcinoma, breast invasive ductal carcinoma, uterine serous carcinoma, uterine carcinosarcoma, and pleomorphic adult sarcomas [6,7]. Mutation-driven malignancies included clear cell renal cell carcinoma, glioblastoma, acute myeloid leukaemia, colorectal adenocarcinoma, and uterine non-serous carcinomas. This analysis appears to hold whether one looks solely at defined driver mutations or considers the overall genomic structure and features of a cancer type [7].
Common somatic copy number alterations across malignancies include amplification of regions containing oncogenes [CCND1, CCNE1, MYC, epidermal growth factor receptor (EGFR), ERBB2, MCL1, and MDM2], or genes involved in telomere maintenance, histone modification, or chromatin remodelling (TERC, RMRP, WHSC1L1, BRD4, KAT6A, KAT6B, NSD1, and PHF1), emphasizing the importance of epigenetic factors in tumourigenesis [8]. Likewise, recurrent hotspot mutations in the chromatin modifier genes ARID1 and CTCF are frequent across multiple cancer lineages [5,9].
Squamous cell-like molecular classification
One of the major findings of the PanCancer12 initiative was the way in which carcinomas from different anatomical sites converged into molecularly similar types. For instance, in the multiplatform analysis, squamous cell carcinomas from the lung and head and neck clustered together with a subset of bladder carcinomas into a molecular subtype characterized by TP53 alterations, amplification of TP63, and high expression of immune-related and proliferation genes [4]. To pathologists, this finding was not surprising, given that urothelial carcinoma commonly shows squamous differentiation, and up to 50% divergent squamous differentiation was allowed in the specimens subjected to expert pathology review for the bladder urothelial carcinoma tissue-specific analysis [10]. Pathologists are commonly asked in their daily practice to identify the site of origin for metastatic squamous cell carcinoma, and the molecular findings from this initial report reflect the challenges of deciphering tissue of origin for squamous cell carcinomas, which essentially comprise two relatively homogeneous molecular phenotypes [human papilloma virus (HPV)-associated (typically affecting squamous cell carcinomas of the oropharynx and female gynecologic squamous mucosal sites), and non-HPV associated]. These common alitiesmay reflect the essential final common pathways driving squamous differentiation in the malignant setting. The PanCancer33 analysis promises to expand the PanSquamous analysis to include cervical squamous cell carcinoma, oesophageal squamous cell carcinoma, and the minor subset of bladder cancer showing squamous differentiation, in an attempt to identify additional commonalities across all anatomical sites, new molecular subtypes within squamous cell carcinoma, and tissue of origin-specific alterations.
Colorectal adenocarcinoma
Less surprisingly, colonic adenocarcinoma and rectal adenocarcinoma were noted to be nearly indistinguishable at the molecular level, reflecting the continuity of the lower gastrointestinal tract and known histological similarities. The main distinction within colorectal carcinoma is the increased frequency of hypermethylated and hypermutated DNA mismatch repair-deficient (resulting in microsatellite instability) and DNA polymerase-ε-deficient carcinomas in the right colon; otherwise, non-hypermutated carcinomas showed identical genomic, epigenomic and transcriptomic alterations, independently of rectal or colon origin [11]. Interestingly, microsatellite-unstable colon cancers do show different histological features from microsatellite-stable cases [12]. This genotype– morphology association was recently demonstrated again by the use of scanned images of colorectal carcinoma contained in TCGA image archives [13].
BRAF mutations
One of the hopes of TCGA and the PanCancer Initiative is to identify alterations present across multiple cancer types that can be used to guide therapy. Indeed, the effort has identified numerous mutations that are present across disparate tumour types. Some of these alterations represent potentially attractive therapeutic targets, such as mutant BRAF, EGFR, or ARID1A. BRAF mutations have been found in multiple cancer types, including melanoma, thyroid carcinoma, and colorectal adenocarcinoma, as well as in a subset of pancreatic and lung adenocarcinomas [14–17]. The dramatic response of melanoma positive for BRAF p.V600E mutations to BRAF inhibitors such as vemurafenib or dabrafenib [18–21] suggested that other malignancies harbouring the identical alteration might also respond. Unfortunately, reality has been somewhat more nuanced. Whereas BRAF inhibitors show promise in BRAF-mutated papillary thyroid carcinoma [22], patients with colorectal carcinomas with BRAF mutations (representing 10% of all colorectal adenocarcinoma) have shorter survival, and are less responsive to conventional chemotherapy than patients with other colorectal carcinomas [23]. In contrast to melanoma, which shows initial response rates of up to 80% prior to the development of resistance, colorectal adenocarcinomas with the identical BRAF p.V600E mutation respond to vemurafenib as a sole agent in <5% of cases [24]. Subsequent studies have shown that this resistance is most frequently due to an extracellular signal-related kinase-mediated feedback resulting in increased epidermal growth factor receptor activation – a process that does not occur in melanoma [23,25–28]. Others have reported that resistance in colorectal adenocarcinoma may also be due to phosphoinositide 3-kinase–AKT pathway activation, or alterations in other mitogen-activated protein kinase pathway genes [23,26]. Studies are ongoing to identify combinatorial therapies to overcome these mechanisms of resistance. Thus, although disparate tumours may harbour several identical mutations or genomic alterations, creating the temptation to treat them in a similar fashion, these alterations must still be considered in the context of other pathways that are active in the individual tumour type. Although this contextual information can be gleaned from genomic and epigenomic characterization, which sometimes also provides explanatory power, on a practical level the fastest, and most economical, way to assess tumour context is by pathological H&E evaluation, whereby a colorectal carcinoma can readily be distinguished from melanoma or papillary thyroid carcinoma with a rapid glance.
Obstacles such as the different responses of genetically similar but histologically different malignancies to targeted therapy illustrate the challenges of personalized medicine. We must learn from these experiences, and determine how to better predict how disparate tumour types with similar underlying molecular features will respond to targeted agents, and understand the mechanisms of any differential response. TCGA, the ICGC, and the PanCancer and PanCancer Analysis of Whole Genomes Initiatives, among other cancer type-specific initiatives, are producing molecular tools and cancer models that, coupled to histological evaluation, can probe these mysteries and build hypotheses that can then be tested with directed prospective or mechanistic studies.
Limitations of TCGA and PanCancer analysis projects
With the sole exception of cutaneous melanoma, all of the malignancies included in TGCA were required to be primary, untreated tumours. In addition, specimens were garnered from available frozen materials present at contributing tissue source sites. Therefore, the specimens included in TCGA may reflect bias in institutional biorepository collections, resulting from institutional research interests, operative patterns, or patient populations. Moreover, tumours routinely subjected to neoadjuvant therapy may not have been able to be included in TCGA, because of limited availability of untreated specimens. Because of the non-inclusion of metastatic disease or aggressive primary tumours subjected to neoadjuvant therapy, the mutational frequencies or prevalences may not translate to modern clinical oncology practices, in which, in many instances, genomic data and targeted therapy are driven more by metastatic, high-stage cases than by primary disease.
Finally, although TCGA and the PanCancer Initiative, as well as groups involved in the ICGC, have performed in-depth analysis and multidimensional correlations between genomic, transcriptomic, epigenomic and proteomic data, the rich dataset provided by the digital pathology images collected for quality assurance remains underutilized, although some of the current PanCancer projects have initiated efforts to extract features and data from this resource. Digital images of frozen sections, obtained from tissues immediately adjacent to materials submitted for genomic analysis, were evaluated by pathologists to broadly confirm the diagnosis, and to estimate the extent of necrosis and proportion of tumour cells to stroma and immune infiltrates, to ensure that minimum purity standards required for genomic analysis are being met (Figure 2). In many TCGA analyses, submitted diagnostic H&E pathology images were used by expert pathology committees to confirm the diagnosis of submitted tissues, or to identify specific pathological features, such as grade, mitotic activity, degree of lymphocytic infiltration, and the presence and type of heterologous elements (e.g. in uterine carcinosarcoma) or variant morphology (e.g. squamous or neuroendocrine differentiation in urothelial bladder carcinoma) [6,7,10,29,30]. These facets are of undoubted importance, both for quality control of the analysed data, and to provide morphological correlates to molecular findings that might assist in translating molecular findings to clinical diagnostics. However, they barely scratch the surface of the potential of digital pathology images using computational histological approaches, which will be described in depth below.
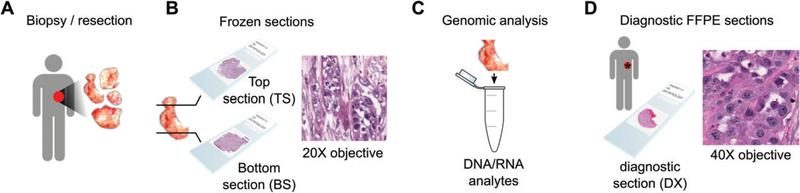
Figure 2.
Tissue procurement in TCGA. (A) A Tissue Source Site (TSS) obtains samples from surgical resection. (B) A portion of this tissue is selected for submission to TCGA, and the BCR produces ‘top-section’ (TS) and ‘bottom-section’ (BS) slides for review to determine that the percentage necrosis and abundance and proportion of tumour cells are adequate for genomic analysis. (C) The middle portion of this tissue is used to extract RNA and DNA analytes for genomic analysis. (D) One or more ‘diagnostic’ formalin-fixed paraffin-embedded (FFPE) slides are submitted to the BCR by the TSS for confirmation of histological diagnosis. These diagnostic slides originate from the same tumour, but their relationship to the material submitted for genomic analysis is unknown. The frozen sections provide the best representation of the tissue contents reflected in genomic signatures. However, the freezing artefacts in these slides can confound routine pathological examination or image analysis algorithms. The FFPE sections reveal cytological details, and have sufficient quality to confirm diagnosis, but the relationship or molecular similarity of these sections to the tissues submitted for genomic analysis is not as precise, as larger tumours may have considerable heterogeneity, and it is not always clear where the frozen tissue was sampled from relative to these H&E sections. The tradeoff between image quality and adjacency to genomic materials is an important consideration in designing an image analysis study of TCGA, and should be weighed on the basis of intratumoural heterogeneity and sensitivity of the image analysis algorithms to artefacts
Historical and contemporary clinical applications of digital pathology
To better understand the potential applications of the TCGA digital slide archive dataset, it is first helpful to review some historical highlights of digital pathology and contemporary applications in clinical diagnostics and research settings. One of the first applications of computer-assisted diagnosis was the invention of automated screening machines for Pap smear diagnosis; although devices first began trials in the 1950s, it was not until the 1990s that devices became commercially available. These devices rely on liquid cytology preparations for optimal visualization, and perform image segmentation to identify nuclei and extract features that can then be used to classify the cells as normal or atypical [31]. These automated screening machines are widely used today, but cases flagged as atypical must undergo rescreening by a cytotechologist or pathologist, and, for the most part, they are not intended to be used as a stand-alone replacement for human diagnosis.
Early clinical applications of digital pathology included semiquantitative analysis of immunohistochemical markers [e.g. oestrogen receptor (ER), progesterone receptor (PR), HER2, and Ki67] [32], although these have been somewhat slow to achieve widespread use. More recently, digital pathology applications have gained US Food and Drug Administration approval, including the Ventana Image Analysis System for HER2 [33] and programmed death-ligand 1 [34] scoring, and the Aperio eIHC IVD system for HER2 and ER/PR scoring [35], as well as the use of whole slide imaging for general review and interpretation of surgical pathology slides (https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm552742.htm). Digital pathology is mainly used in the USA for remote diagnosis of frozen sections during surgery, and to provide remote consultation services. In Europe, some practices have transitioned to fully digital workflows [36,37], establishing the feasibility of large-scale digital practice, and have reported improved ergonomics and efficiency. In addition to diagnostic uses, digital imaging has also created large archives that can be utilized for teaching purposes, such that multiple trainees can view the same slide simultaneously, and without risk of damage to the original patient materials. In many academic practices, the practice of scanning consultation cases for retention in the archives for future comparison with subsequent resections or recurrences has increased the ease of confirming diagnoses or assessing response to therapy.
Advances in slide-scanning microscopes, computing and image analysis algorithms over the past decade have greatly increased interest in digital pathology. Slide-scanning microscopes can now digitize an entire histological section at ×40 objective magnification, producing a ‘whole slide image’ in under a minute, and are common fixtures in academic histology laboratories (Figure 3A). Images produced by slide-scanning microscopes typically contain billions of pixels and, as a result, are several gigabytes in size, even following image compression. As with genomic sequencing, increased storage density and processor performance have made it possible to process multi-terabyte digital pathology datasets with intensive algorithms. This has produced remarkably advanced capabilities for extracting quantitative descriptions of histology from images produced by slide-scanningmicroscopes. Contemporary algorithms can identify and describe the shape, staining and texture of a million or more cell nuclei in a single image, or accurately delineate stromal regions or multicellular structures such as blood vessels.
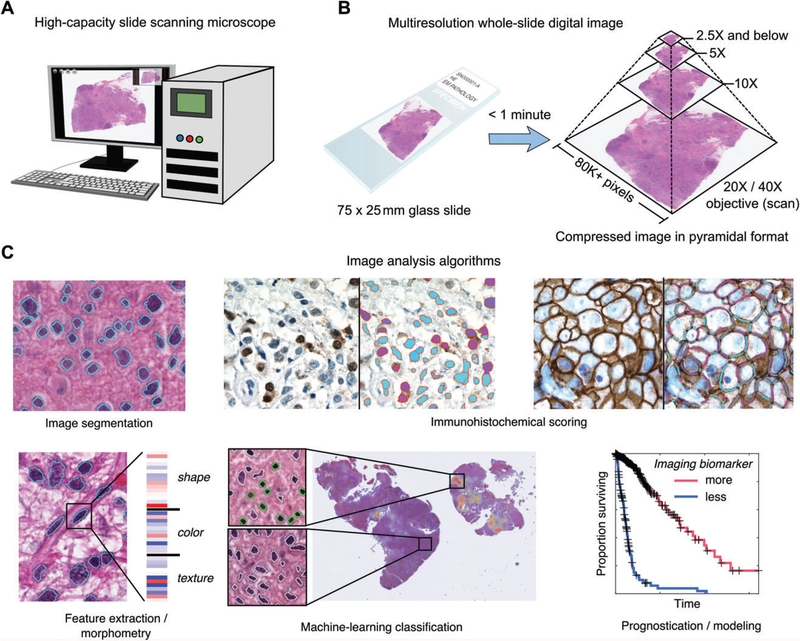
Figure 3.
Whole slide imaging and image analysis. (A) Slide-scanning microscopes can rapidly digitize an entire glass slide, producing a ‘whole slide’ digital image. These devices can scan large batches of slides, producing >1000 scans in a single day. (B) Slides are digitized with a × 20 or ×40 objective, and this base magnification is used by the scanner software to produce a multiresolution image pyramid containing downsampled magnifications. This pyramidal format enables smooth zooming and interaction with the image, and provides additional resolutions for image analysis. (C) A large number of image analysis algorithms exist for analysing whole slide images (from left to right, top to bottom): image segmentation algorithms are used to automatically delineate the boundaries of structures such as cell nuclei; immunohistochemical scoring algorithms can be used to measure the subcellular localization and intensity of antigens; feature extraction can be used to calculate quantitative features describing the shape, colour and texture of tissue elements; machine-learning algorithms can be used with imaging features to classify objects – here, a classifier was trained to identify mononuclear cells (green) in a glioma, and a heatmap indicating the concentration of positively classified cells in the slide is shown; measurements made by image analysis can be used to build prognostic models that can objectively discriminate patient outcomes.
An overview, albeit not exhaustive, of algorithmic capabilities is shown in Figure 3B. Image segmentation algorithms aim to explicitly delineate the boundaries of histological structures or image regions by using salient properties such as shape, colour, or texture, and constitute the first step in many computational histology analyses. Segmentation of nuclei or membrane segments can be used for immune-histochemical scoring. Feature extraction refers to the process of calculating quantitative descriptions of objects or image regions that capture shape, staining intensity or textural patterns with precise numerical values. Extracted features can be used to train machine-learning algorithms to classify different types of cells or tissues, or to make patient-level predictions about prognosis, grade, or histological subtype.
Examples of computer-aided diagnosis utilizing such algorithms and machine learning include: a neuroblastoma grading system for analysing stromal development and degree of differentiation [38], a system for identifying and counting centroblasts in follicular lymphoma [39], multiple systems for grading prostate adenocarcinoma by using nuclear morphology and glandular structure [40,41], and a grading system for breast cancer that characterizes epithelium, stroma, and their interfaces [42].
Computational pathology in TCGA
TCGA is unmatched in scale as a digital pathology dataset, containing >32 000 H&E-stained images (20 099 frozen; 11 916 diagnostic) from >11 000 specimens, with >16 000 gigabytes of pathology imaging data in total. These images are publically available through the Cancer Digital Slide Archive (http://cancer.digitalslidearchive.net/), a web-based resource for hosting digital pathology images [43]. The Cancer Digital Slide Archive enables users to search for slides by project and sample ID, and to review these multi-gigapixel images within a web browser interface that provides fluid multiresolution zooming and panning. Combined with newly emerging image analysis capabilities, the TCGA digital pathology data present unique opportunities to investigate relationships between histological patterns and clinical outcomes, and the molecular basis of cancer phenomena such as immune infiltration, angiogenesis, and tumour–stromal interactions.
Several studies have been performed with TCGA digital pathology data to investigate genotype–phenotype correlations or histological biomarkers of patient outcomes. Although it is not possible to provide a complete review of these studies, we highlight several illustrative examples below with a focus on investigations using image analysis algorithms.
The molecular correlates of nuclear pleomorphism in sarcomas
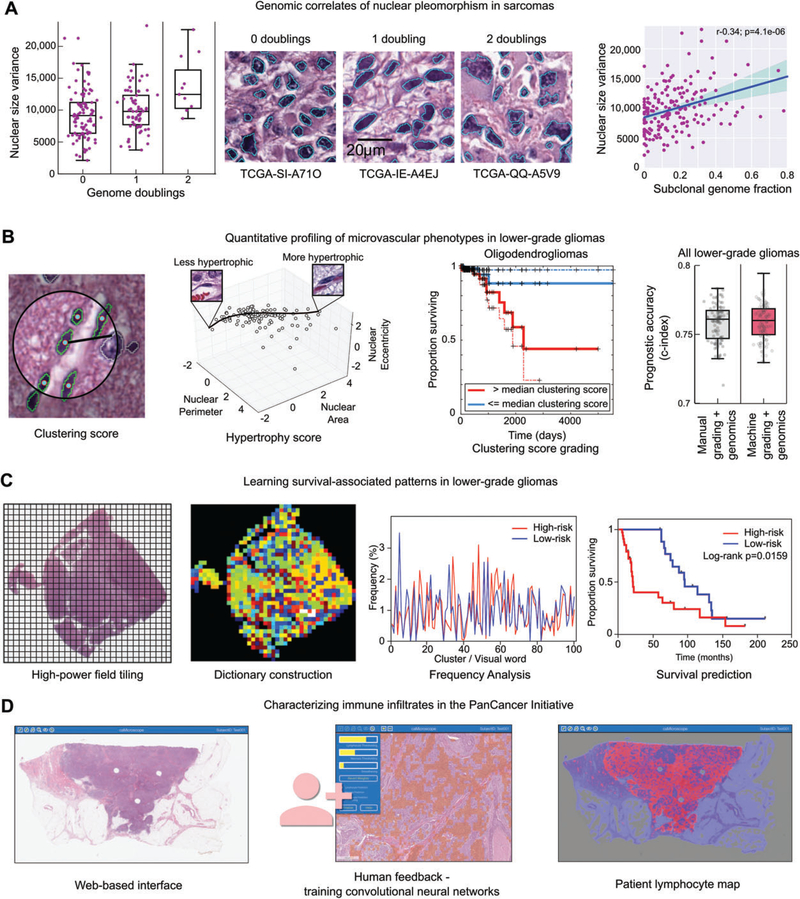
Nuclear pleomorphism has long been appreciated as a prognostic indicator in many cancers, and is routinely evaluated as part of the grading of sarcoma, breast carcinoma, and other malignancies. Although studies in a variety of carcinomas showed that nuclear pleomorphism correlated with increased DNA content (ploidy) [44–48], the more precise molecular correlates of pleomorphism are not well understood. The Sarcoma TCGA Analysis Working Group used image analysis methods to assess the relationships between nuclear pleomorphism, clonality, and genomic instability (Figure 4A). A nuclear morphology approach was used to segment >500 million nuclei in diagnostic images of sarcoma samples, and to calculate morphological features for each nucleus. A histogram of nuclear size was calculated for each patient, and used to generate variance, skew and kurtosis statistics that describe the variations in nuclear size within each tumour. These statistics were compared with estimates of subclonal genome fraction, genome doublings and unbalanced copy number segments obtained from copy number and DNA sequencing data to evaluate associations between nuclear pleomorphism and genomic complexity. This analysis revealed that pleomorphism in sarcomas is significantly correlated with both ploidy and genomic complexity, and that increasing pleomorphism is associated most strongly with increases in subclonality and increases in the number of unbalanced copy number segments [7].
Figure 4.
Image analysis studies of TCGA. (A) Nuclear morphometry was used to study the genomic correlates of nuclear pleomorphism in sarcomas. Image segmentation was used to delineate >500 million nuclei in diagnostic sarcoma images, and the area of each nucleus was calculated. The variance of nuclear area was calculated for 235 sarcomas, and compared with measurements of genome doublings and subclonality obtained from sequencing and copy number data. Increased pleomorphism was significantly associated with measures of genomic complexity, including genome doublings, subclonality, and aneuploidy. (B) Machine learning was used to investigate microvascular phenotypes in lower-grade gliomas. A classifier was developed to identify vascular endothelial cells in gliomas (green). These classifications were used to measure to the clustering of endothelial cells and to model the morphological spectrum of endothelial nuclei in order to describe the extent of endothelial hyperplasia and hypertrophy in TCGA samples. These measurements were used as a biomarker to stratify overall survival, and were as effective at predicting outcomes as manual histological grading when combined with diagnostic genetic biomarkers. (C) Unsupervised machine learning was used to identify survival-associated patterns in lower-grade gliomas using TCGA data. Features describing the texture of haematoxylin were analysed in tiled high-power fields. These features were used to cluster the fields to define a dictionary of ‘visual words’ that captures the frequent patterns in the tissue. The frequency of these words in each slide were used to predict patient survival and to identify molecular correlates of histological patterns. (D) Convolutional networks were used to map the spatial distribution of TILs in 13 cancer types as part of the recent PanCancer immune working group. A web-based interface was used to train convolutional neural networks to identify patches containing TILs. These algorithms were then used to map the presence of TILs in >6000 whole slide images.
Prognostic morphological and molecular correlates of triple-negative breast carcinoma
Digital pathology analysis was also used to identify survival-associated morphological features in frozen section images of triple-negative breast cancer [49]. Images were first segmented into ‘superpixels’ – small irregular image patches whose boundaries adapt to follow sharp gradients such as tumour–stroma interfaces. In dense superpixels that correspond to tumour regions, nuclei are segmented, and morphometric features are calculated to describe nuclear shape, staining, and texture. Stromal superpixels are identified according to their homogeneous texture, and the shape and texture of these stromal compartments are also measured. A total of 44 TCGA breast invasive ductal carcinoma samples were used as a discovery set to identify survival-associated features, and the prognostic accuracy of these features was validated with an institutional set of 143 breast invasive ductal carcinoma tissue microarrays. To further validate their findings, gene expression data from the TCGA were used to learn surrogate gene expression signatures of survival-associated morphological features. These surrogate signatures were validated in two additional independent datasets containing gene expression and outcomes. A similar analysis was performed with non-TCGA data [42].
Characterizing immune infiltrates in the PanCancer Initiative
As part of the PanCancer33 analysis effort, an immune working group was formed to describe and analyse the correlates of immune infiltration across TCGA projects. In addition to performing gene expression deconvolution analyses to measure the presence of immune infiltrates in TCGA, the group developed image analysis-based approaches to identify tumour-infiltrating lymphocytes (TILs) in H&E images, and to describe their spatial distributions. A convolutional neural network was developed to classify 50-μm-square image patches for TIL content, and was trained and evaluated on 176 whole slide images of lung adenocarcinoma sections by use of a web-based interface [50,51]. This convolutional neural network was then adapted to 13 other tissue types to map the presence of TILs in >6000 whole slide images. Spatial statistics of these TIL maps were used to compare TIL distributions in different cancer types, and with genomic profiles and patient outcomes. These analyses will be featured in the next PanCancer publication.
Necrosis and hypoxia in glioblastoma
In glioblastoma, the extent of necrosis and angiogenesis was measured in frozen section images by use of a combination of manual annotations and image analysis algorithms [52]. These measurements were correlated with gene expression profiles derived from adjacent tissues to determine the genes and molecular pathways that are correlated with hypoxia and the development of necrosis and angiogenesis. This study found that the mesenchymal gene expression-based subtype was highly correlated with the extent of necrosis in a tissue sample, and that master transcriptional regulators of the mesenchymal subtype, including CEBPB and STAT3, were among the genes most strongly correlated with necrosis. Immunohistochemical studies revealed that CEBPB was highly expressed in hypoxic perinecrotic cells, suggesting that the tumour microenvironment can significantly impact on gene expression-based classifications of glioma.
Microvascular phenotypes predict survival in lower-grade gliomas
Gliomas are highly vascular solid tumours, and the appearance of microvascular structures reflects the response of endothelial cells to aberrant signalling from neoplastic cells. Microvascular hypertrophy, visible as nuclear and cytoplasmic enlargement of endothelial cells, is an early transformation and indicator of increased transcriptional activation. Microvascular hyperplasia follows, as endothelial cells proliferate and become more clustered, producing multilayered microvascular structures. These changes are known to accompany disease progression, and precision computational histology approaches can be used to describe these non-tumour elements as prognostic biomarkers.
The authors developed a cytological classifier to identify vascular endothelial cell nuclei in diagnostic images of lower-grade gliomas, and generated morphological and spatial statistics of endothelial nuclei to describe the extent of microvascular hypertrophy and microvascular hyperplasia in each tumour (Figure 4B) [53]. An interactive classification system that enables pathologists to interact with large-scale nuclear morphometry data was used to generate training data, and to evaluate the accuracy of the vascular endothelial nuclei classifier. This system uses a framework called active learning, which directs users to label examples of cell nuclei. After classification of 360 million cell nuclei in 464 tumours from the TCGA lower-grade glioma project, the morphometric features and locations of positively classified cells were used to generate hypertrophy and hyperplasia scores for each tumour. Hypertrophy was scored by learning a non-linear model based on nuclear area, eccentricity and shape that scores individual nuclei in terms of hypertrophy. Hyperplasia was scored by measuring the spatial clustering of vascular endothelial nuclei by use of the Ripley’s K-function statistic. Prognostic models based on these scores were able to predict overall survival as accurately as manual histological grading.
Learning survival-associated patterns in lower-grade gliomas
Powell et al developed an approach to learn visual patterns associated with overall survival by using the lower-grade glioma data from TCGA, and a combination of image analysis and unsupervised machine learning (Figure 4C) [54]. Both cell of origin and grade are important predictors of outcome in lower-grade glioma, and the morphologies of neoplastic nuclei and their spatial distribution are important prognostic indicators. A colour deconvolution algorithm was used to digitally separate the H&E stains, generating a separate intensity image for each. Fields measuring 256×256μm were sampled at ×10 objective magnification. The haematoxylin component of each patch was used to derive textural features without explicitly segmenting the cell nuclei, and the eosin component was used to remove fields corresponding to artefacts (i.e. tissue folds) or fields containing primarily glass. The features generated from each field were used with a clustering algorithm to generate a ‘visual dictionary’ of clusters, each cluster representing a visual ‘word’ that describes some histological pattern observed in gliomas. This dictionary was used to describe the frequencies of visual words in each whole slide image. This study found that many of these words were associated with overall survival, and correlation between word frequency and gene expression identified signalling cascades associated with the visual words.
Conclusions
TGCA and PanCancer Initiatives have generated a rich and deep resource to enable future cancer studies. The publications and the associated publically available datasets are not intended to be the ‘last word’ in understanding of these diseases, but are meant to provide a stepping-stone to future analyses. Data types available include DNA sequence, copy number, and methylation data, transcriptomic data, including mRNA, miRNA and, in a subset of cases, long non-coding RNA, and proteomic data from reverse-phase protein array analysis. A major untapped resource of TCGA comprises the digital pathology images amassed from all 33 cancer types; emerging computational histological techniques to extract high-level data from these images combined with the detailed associated molecular data promise the ability to conduct detailed morphological–molecular studies to provide a deeper understanding of the mechanisms informing both histological appearance and tumour behaviour. In this review, we have presented a few tangible examples of how imaging data from TCGA have been used to gain unique insights into cancer biology, and it is our sincere hope that use of this data will grow in volume and strength over the coming months and years. The exploitation of this data will help to achieve the full potential of TCGA analysis and, ultimately, along with other work in the digital pathology space, influence clinical pathology practice by providing new tools and approaches for decision support. With appropriate clinical validation, these approaches can help to provide the information necessary for patient management in contemporary oncological pathology practice. Although these new methods are certainly not going to soon replace routine H&E diagnosis, pathologists must remain open to testing, adopting and utilizing these new methods as appropriate in their practices, to provide optimal care for our patients.
Acknowledgements
The authors would like to thank Kim Anh Vu for her assistance with figure preparation. This work was supported by the National Cancer Institute Informatics Technology for Cancer Research (ITCR) grant U24CA194362.
Footnotes
No conflicts of interest were declared.
References
- 1.Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013; 45: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015; 163: 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014; 158: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciriello G, Miller ML, Aksoy BA, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 2013; 45: 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherniack AD, Shen H, Walter V, et al. Integrated molecular characterization of uterine carcinosarcoma. Cancer Cell 2017; 31: 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Cancer Genome Atlas Network. Comprehensive and integratedgenomiccharacterizationofadultsofttissuesarcoma.Cell 2017;171: 950–965.29100075 [Google Scholar]
- 8.Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013; 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev 2017; 42: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Cancer Genome Atlas Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014; 507: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander J, Watanabe T, Wu TT, et al. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol 2001; 158: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shia J, Schultz N, Kuk D, et al. Morphological characterization of colorectal cancers in The Cancer Genome Atlas reveals distinct morphology–molecular associations: clinical and biological implications. Mod Pathol 2017; 30: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002; 417: 949–954. [DOI] [PubMed] [Google Scholar]
- 15.Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 2002; 62: 6451–6455. [PubMed] [Google Scholar]
- 16.Kimura ET, Nikiforova MN, Zhu Z, et al. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res 2003; 63: 1454–1457. [PubMed] [Google Scholar]
- 17.The Cancer Genome Atlas Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017; 32: 185–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman PB, Robert C, Larkin J, et al. Vemurafenib in patients with BRAFV600 mutation-positive metastatic melanoma: final overall survival results of the randomized BRIM-3 study. Ann Oncol 2017; 28: 2581–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364: 2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012; 367: 1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long GV, Eroglu Z, Infante J, et al. Long-term outcomes in patients with BRAF V600-mutant metastatic melanoma who received dabrafenib combined with trametinib. J Clin Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brose MS, Cabanillas ME, Cohen EE, et al. Vemurafenib in patients with BRAF(V600E)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: a non-randomised, multicentre, open-label, phase 2 trial. Lancet Oncol 2016; 17: 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke CN, Kopetz ES. BRAF mutant colorectal cancer as a distinct subset of colorectal cancer: clinical characteristics, clinical behavior, and response to targeted therapies. J Gastrointest Oncol 2015; 6: 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopetz S, Desai J, Chan E, et al. LX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. ASCO Meeting Abstracts 2010; 28: 3534. [Google Scholar]
- 25.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012; 483: 100–103. [DOI] [PubMed] [Google Scholar]
- 26.Ahronian LG, Sennott EM, Van Allen EM, et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov 2015; 5: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012; 2: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das Thakur M, Stuart DD. The evolution of melanoma resistance reveals therapeutic opportunities. Cancer Res 2013; 73: 6106–6110. [DOI] [PubMed] [Google Scholar]
- 29.The Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015; 161: 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 2017; 32: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bengtsson E, Malm P. Screening for cervical cancer using automated analysis of PAP-smears. Comput Math Methods Med 2014; 2014: 842037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farris AB, Cohen C, Rogers TE, et al. Whole slide imaging for analytical anatomic pathology and telepathology: practical applications today, promises, and perils. Arch Pathol Lab Med 2017; 141: 542–550. [DOI] [PubMed] [Google Scholar]
- 33.Ventana. Ventana Medical Systems, Inc. receives FDA approval for the first fully automated diagnostic assay for HER2 gene status determination in breast cancer patients. [Accessed 31 October 2017]. Available from: http://www.ventana.com/wp-content/uploads/pr_2011-06-14_INFORM-HER2.pdf
- 34.Ventana. Roche receives FDA approval for complementary PD-L1 (SP263) biomarker test in urothelial carcinoma. 2017. [Accessed 31 October 2017]. Available from: http://www.ventana.com/rochereceives-fda-approval-complementary-pd-l1-sp263-biomarkertest-urothelial-carcinoma/
- 35.Administration UFaD. Susbstantial Equivalence Determination 510K K071128. [Accessed 31 October 2017]. Available from: https://www.accessdata.fda.gov/cdrh_docs/reviews/K071128.pdf
- 36.Thorstenson S, Molin J, Lundstrom C. Implementation of large-scaleroutine diagnostics using whole slide imaging in Sweden: digital pathology experiences 2006. –2013. J Pathol Inform 2014; 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stathonikos N, Veta M, Huisman A, et al. Going fully digital: perspective of a Dutch academic pathology lab. J Pathol Inform 2013; 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurcan MN, Kong J, Sertel O, et al. Computerized pathological image analysis for neuroblastoma prognosis. AMIA Annu Symp Proc 2007; 304–308. [PMC free article] [PubMed] [Google Scholar]
- 39.Fauzi MF, Pennell M, Sahiner B, et al. Classification of follicular lymphoma: the effect of computer aid on pathologists grading. BMC Med Inform Decis Mak 2015; 15: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparks R, Madabhushi A. Statistical shape model for manifold regularization: Gleason grading of prostate histology. Comput Vis Image Underst 2013; 117: 1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali S, Veltri R, Epstein JI, et al. Adaptive energy selective active contour with shape priors for nuclear segmentation and Gleason grading of prostate cancer. Med Image Comput Comput Assist Interv 2011; 14: 661–669. [DOI] [PubMed] [Google Scholar]
- 42.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 2011; 3: 108–113. [DOI] [PubMed] [Google Scholar]
- 43.Gutman DA, Cobb J, Somanna D, et al. Cancer Digital Slide Archive: an informatics resource to support integrated in silico analysis of TCGA pathology data. J Am Med Inform Assoc 2013; 20: 1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen I, Kotb WF, Friedrich KH, et al. Core classification of lung cancer: correlating nuclear size and mitoses with ploidy and clinicopathological parameters. Lung Cancer 2009; 65: 312–318. [DOI] [PubMed] [Google Scholar]
- 45.Diwakar N, Sperandio M, Sherriff M, et al. Heterogeneity, histological features and DNA ploidy in oral carcinoma by image-based analysis. Oral Oncol 2005; 41: 416–422. [DOI] [PubMed] [Google Scholar]
- 46.Helliwell TR, Atkinson MW, Cooke TG, et al. Morphometric analysis, ploidy and response to chemotherapy in squamous carcinomas of the head and neck. Pathol Res Pract 1989; 185: 755–759. [DOI] [PubMed] [Google Scholar]
- 47.Gustafsson U, Einarsson C, Eriksson LC, et al. DNA ploidy and S-phase fraction in carcinoma of the gallbladder related to histopathology, number of gallstones and survival. Anal Cell Pathol 2001; 23: 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grignon DJ, Ayala AG, Ro JY, et al. Primary sarcomas of the kidney. A clinicopathologic and DNA flow cytometric study of 17 cases. Cancer 1990; 65: 1611–1618. [DOI] [PubMed] [Google Scholar]
- 49.Wang C, Pecot T, Zynger DL, et al. Identifying survival associated morphological features of triple negative breast cancer using multiple datasets. J Am Med Inform Assoc 2013; 20: 680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saltz J, Almeida J, Gao Y, et al. Towards generation, management, and exploration of combined radiomics and pathomics datasets for cancer research. AMIA Jt Summits Transl Sci Proc 2017; 2017: 85–94. [PMC free article] [PubMed] [Google Scholar]
- 51.Saltz J, Sharma A, Iyer G, et al. A containerized software system for generation, management, and exploration of features from whole slide tissue images. Cancer Res 2017; 77: e79–e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cooper LA, Gutman DA, Chisolm C, et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol 2012; 180: 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nalisnik M, Amgad M, Lee S, et al. Interactive phenotyping of large-scale histology imaging data with HistomicsML. Sci Rep 2017; 7: 14588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Powell RT, Olar A, Narang S, et al. Identification of histological correlates of overall survival in lower grade gliomas using a bag-of-words paradigm: a preliminary analysis based on hematoxylin & eosin stained slides from the lower grade glioma cohort of The Cancer Genome Atlas. J Pathol Inform 2017; 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]