Abstract
In several retinal degenerative disease pathologies, such as dry age-related macular degeneration (AMD), the retinal pigment epithelium (RPE) cell monolayer becomes dysfunctional. Promising tissue engineering treatment approaches implant RPE cells on scaffolds into the subretinal space. However, these approaches are not without challenges. Two major challenges that must be addressed are RPE dedifferentiation and the inflammatory response to cell/scaffold implantation. Design and optimization of scaffold cues for the purpose of RPE transplantation remain relatively unexplored, specifically the mechanical properties of the scaffolds. Prior work from our group indicated that by varying substrate moduli significant differences could be induced in cell cytoskeleton structure, cellular activity, and expression of inflammatory markers. We hypothesized that Activin A would provide rescue effects for cells demonstrating dedifferentiated characteristics. Results demonstrated that for cells on low modulus scaffolds, the mechanical environment was the dominating factor and Activin A was unable to rescue these cells. However, Activin A did demonstrate rescue effects for cells on high modulus scaffolds. This finding indicates that when cultured on scaffolds with an appropriate modulus, exogenous factors, such as Activin A, can improve RPE cell expression, morphology, and activity, while an inappropriate scaffold modulus can have devastating effects on RPE survival regardless of chemical stimulation. These findings have broad implications for the design and optimization of scaffolds for long-term successful RPE transplantation.
Keywords: retinal pigment epithelial cells, RPE cells, Activin A, Bruch’s membrane, modulus stiffness
1. Introduction
Activin A is a member of the TGF-β super family of signaling molecules. [1] This super family of molecules participates in several biological processes including cell differentiation and proliferation, inflammatory and immune responses, and apoptosis. [2,3] Though there are different types of activins, Activin A is the most extensively studied.
The specific effects of Activin A on RPE cells has been demonstrated through several investigations. [4,5] In a study using explant cultures of chick optic vesicles, Fuhrmann et al. demonstrated that in the absence of extraocular mesenchyme signaling, Activin A promotes expression of RPE-specific genes and downregulates expression of neural markers. [5] In a separate study on the effects of Activin A on RPE cells, Sakami et al. investigated the ability of RPE cells to regenerate small defects through transition to a less mature, proliferating phenotype. [4] With the addition of Activin A, the mature RPE lost its competence to transition to the necessary regenerating phenotype. In recent years, Activin A has also proven to be a useful component in the complete media used to differentiate both embryonic and induced pluripotent stem cells into RPE cells. Idelson et al. demonstrated that the use of Activin A in differentiation media directs stem cells to form mature, functional RPE monolayers. [6] Many groups have adopted this practice in using Activin A to drive stem cells towards the mature RPE phenotype.
With dedifferentiation of cells on scaffolds being a hurdle that must be overcome in this field, the use of a signaling molecule, such as Activin A, to promote the mature, functional phenotype is desirable. In our previous work, we saw evidence of dedifferentiation on both low and high modulus scaffolds. [7] When seeded on scaffolds, RPE cells showed upregulation in SMAD3, a dedifferentiation marker, and downregulation in CRALBP, a characteristic RPE gene. These changes on both stiff and compliant substrates prompted the current investigation into whether Activin A could prevent this dedifferentiation. Though a variety of RPE scaffolds have been introduced, none have been effective at reversing age-related macular degeneration and current methods of examining these scaffolds have failed to elucidate why. Current RPE scaffolds have a variety of biological and microarchitectural cues that all influence cell behavior, thus it is impossible to tease out the effects of individual scaffold components. By probing factors singly, we can tease out which are the most important for the design of better scaffolds. Lacking in the literature is an examination of the isolated effects of Activin A; most RPE scaffolds are derived from natural sources or if synthetic, are coated with laminin or another biological protein that may influence RPE behavior, confounding any effects of Activin A. [8,9] Furthermore, no studies explore the combined effects of scaffold stiffness and presence or absence of Activin A. Most RPE scaffolds are intended to supplement or replace the diseased Bruch’s membrane, the blood-retina barrier upon which the retinal pigment epithelium is located. While the elastic moduli for the healthy Bruch’s membrane is known, the range of elastic moduli for diseased Bruch’s membranes and the scaffolds intended to replace them are poorly described. It may be that observed dedifferentiation of RPE cells on scaffolds are due to an uncharacterized divergence from the elastic moduli of the healthy Bruch’s membrane. By demonstrating what happens on different stiffness substrates, we hope to describe what may be happening with diseased native Bruch’s membranes or with poorly described RPE substrates, and whether Activin A can mitigate these effects. Therefore, this work examines the effects of Activin A on RPE cells when they are cultured on scaffolds having elastic moduli reported for healthy Bruch’s membranes (1 MPa) and also on scaffolds with elastic moduli at least one order of magnitude above (72 GPa) and below (60 kPa) that of healthy Bruch’s membranes to capture RPE behavior on a range of substrate stiffnesses. The work further examines the differential effects when Activin A is added to culture media, covalently bound to scaffolds, or physically entrapped within scaffolds.
2. Materials and Methods
All reagents were purchased from Sigma-Aldrich (Saint Louis, MO, USA) and all PEGDA was obtained from Laysan Bio, Inc (Arab, AL, USA) and used as obtained without further purification unless otherwise noted.
2.1. Acryl-PEG-RGDS Synthesis and Confirmation of Conjugation
Heterobifunctionalized Acrylate-PEG-Succinimydl Valerate (ACRL-PEG-SVA; Laysan Bio, Inc., Arab, AL, USA) was reacted with RGDS (Tocris, Bristol, UK) in a 1:1.2 molar ratio at pH 8.0 under argon. The reaction mixture was placed on a rocker on its highest tilt and speed overnight in a 4°C cold room. Following overnight reaction, the solution was then dialyzed against 4 liters of ultra-pure water in a 1000 MWCO cellulose membrane (Spectrum Labs, Rancho Dominguez, CA, USA), lyophilized, and stored at −20°C. Ninhydrin assays were performed to measure the amount of free RGDS following the conjugation reaction with ACRL-PEG-SVA. Ninhydrin reacts with free amines and produces a purple colored product. This colorimetric assay permits the measurement of unconjugated RGDS via reaction with free amines on the arginine. Briefly, prior to dialyzing the reaction solution, a 250 μL sample was lyophilized and reconstituted in 100 μL of phosphate buffered saline (PBS). This reconstituted solution was next added to sodium citrate buffer (100 μL) and 2% ninhydrin solution (200 μL) in an Eppendorf low protein binding tube. This was then placed in a boiling water bath for 15 minutes. Absorbance of the solution was read on a Beckman DTX 880 Multimode Detector at 570 nm. A standard curve was produced using known concentrations of RGDS.
2.2. Scaffold Fabrication
Scaffolds were fabricated as described previously. [7] Briefly, four molecular weights of PEGDA were evaluated to determine a low and high modulus scaffold for cell culture: 3.4 and 5 kDa at 20% or 40% (w/v) and 10 and 20 kDa at 10% or 20% (w/v). Lower PEGDA concentrations are defined as 1× and the higher concentrations as 2×. PEGDA was dissolved in HEPES-buffered saline with 10 μL/mL photoinitiator solution (2,2-dimethoxy-2-phenyl-acetophenone 300 mg/mL in N-vinylpyrrolidone) and 10 mM ACRL-PEG-RGDS. This solution was sterilized through a 0.2 μm polyethersulfone syringe filter, then pipetted into a glass-slide mold constructed of two 25 mm × 75 mm pre-cleaned glass microscope slides separated by a 500 μm thick Teflon spacer and disinfected with 70% ethanol and exposed to UV light (B-200SP UV lamp, UVP, 365 nm 10 mW2/cm2) for further sterilization for at least an hour prior to use. The solution in the mold was exposed to UV light for 3 minutes. Hydrogel elastic moduli were determined through tensile and compressive testing (described below), and high and low moduli were selected for cell culture.
For cell culture, low (60 kPa) and high (1.2 MPa) modulus scaffolds were fabricated either: 1) without Activin A; 2) with Activin A covalently bound to the hydrogel surface; or 3) with Activin A physically encapsulated within the hydrogel. These scaffolds were fabricated using a combination of 20 kDa PEGDA (10% w/v; low modulus) and 3.4 kDa PEGDA (40% w/v; high modulus). Fabrication was conducted as described above, with the addition 10 mM ACRL-PEG-RGDS to the polymer solution to permit cell adhesion. Following polymerization, rectangular-shaped hydrogel scaffolds were removed from the molds with tweezers and fully immersed in 5 mL PBS within petri dishes and allowed to swell for 24 hours in a humidified incubator.
2.2.1 Characterization of Elastic Moduli
Young’s moduli (E), or elastic moduli, of swelled scaffolds were determined by performing both tensile and compressive testing. Tensile testing was completed using a Bose Electroforce 3100 with a 1 Newton load cell as previously described. [7] Briefly, hydrogel scaffolds were removed from PBS immediately prior to testing, measured with digital calipers, then clamped at either end and mounted in the Bose Electroforce device. Scaffolds were subjected to uniaxial tensile strain applied at a rate of 6 mm/min. Compressive stress was applied following swelling using an Instron 5869 (Instron, USA) with a 50 kN load cell. A 1 mm/minute strain rate was applied to the scaffolds and they were tested to failure. The data was collected and used to calculate the elastic modulus from the slope in the linear portion of the stress-strain curve (N= 40 total; n=5 for each molecular weight concentration). The highest and lowest moduli were selected based on these measurements.
2.2.2 Covalently Bound Activin A Scaffolds
Activin A (R&D Systems, Minneapolis, MN, USA) was first conjugated to PEG-acrylate to permit covalent binding of the molecule to the surface of a PEGDA hydrogel sheet. This occurs through reaction with heterobifunctionalized Acrylate-PEG-Succinimydl Valerate in 1:35 molar ratio at pH 8.0 under argon. The reaction mixer was placed on a rocker on its highest tilt and speed overnight in 4°C cold room. Following the overnight reaction, the pH was titrated back to 7.0 and the solution was frozen at −80°C, lyophilized, and stored at −20°C.
Following hydrogel swelling, the ACRYL-PEG-Activin A was conjugated to the scaffold. First the pegylated-Activin A was resuspended at 1 mM in HEPES-buffered saline to create a stock solution. This solution was then diluted to 100 μM and 10 μL/mL of photoinitiator solution was added to it. The solution (250 μL containing 655 μg Activin A) was evenly pipetted across the entire surface of the scaffold. The surface was then exposed to UV-light for 3 minutes and placed in PBS in a humidified incubator for 24 hours. To confirm the binding of acrylated-Activin A to scaffold surfaces, an Activin A ELISA (R&D Systems, Minneapolis, MN, USA) was used to determine the amount of Activin A in the PBS of soaked scaffolds 24 hours after conjugation. This method consistently resulted in between 200 and 280 ng conjugated to hydrogel surfaces, with the excess rinsed away. High and low modulus scaffolds were designated HIGHcov and LOWcov, respectively.
2.2.3 Encapsulated Activin A Scaffolds
To encapsulate Activin A within hydrogels, the scaffold fabrication method described above was conducted with the addition of Activin A (280 ng per scaffold) to the PEGDA prepolymer solutions. The 280 ng Activin A per hydrogel was selected because this was the equivalent total mass of Activin added to the media condition. Following this addition of Activin A, scaffolds were polymerized as described. To measure Activin A release from scaffolds, Activin A loaded hydrogels were placed in PBS at 37°C and transferred to fresh solutions after 8 hours and daily after that. The amount of protein released into each solution was measured with Activin A ELISA kit (R&D Systems, Minneapolis, MN, USA) for 7 days. High and low modulus scaffolds were designated HIGHencaps and LOWencaps, respectively.
2.3. Glass Slide Functionalization
Glass slides functionalized with acrylate-RGD was used as an alternate control to tissue culture plastic to ensure observations were due to modulus differences and not cell preference for tissue culture plastic over RGD. Slide surfaces were acrylated to prepare them for covalent binding with acrylate-RGD. First, slides were incubated in a beaker of 25% nitric acid (30% solution) and 75% hydrochloric acid (30% solution) in a sonicator at 50–60°C for 5–10 minutes. After allowing the beaker to cool to room temperature, slides were removed from the acid solution and washed for 1 minute in a sonicator bath at 50 kHz (3× in ultrapure water followed by 1× in 70% ethanol). Slides were then allowed to dry completely. Next, 50 μL of 0.1% 3-(trimethoxysilyl) propyl acrylate in chloroform solution was evenly distributed onto slide surfaces. Slides were dried overnight then washed with cold ultrapure water to remove unadsorbed acrylate groups. Finally, to functionalize the surface with Acryl-PEG-RGDS, 10 mM ACRL-PEG-RGDS in HEPES-buffered saline (10 mM N-[2-hydroxyethyl] piperazine-N0-[2-ethanesulfonic acid] and NaCl in ultra-pure water), with 10 μL/mL photoinitiator solution (2,2-dimethoxy-2-phenyl-acetophenone 300 mg/mL in N-vinylpyrrolidone) was evenly distributed onto slide surfaces. Slides were then exposed to UV-light for 3 minutes and soaked in PBS overnight to remove any unbound RGD.
2.4. Cell Culture
ARPE-19 cells (ATCC, Manassas, VA, USA) were seeded on scaffolds at 10,000 cells/cm2 and maintained in 96 well plates with 200 μL complete culture media. Cells were cultured in DMEM/F12 with 2% v/v fetal bovine serum, 1% v/v antibiotic solution (10,000 Units penicillin and 10 mg streptomycin per mL). For high and low modulus scaffolds receiving Activin A in solution (HIGHfree and LOWfree, respectively), Activin A was added to culture media (100 ng/mL) such that cells would receive identical cumulative Activin A doses as cells seeded on hydrogels with bound Activin A. Scaffolds were moved to new wells after 8 hours to retain only cells attached to scaffolds and eliminate cells that had attached to well bottoms. Media was changed every day for 14 days, resulting in cells exposed to a total of 280 ng exposed to seeded cells. Cell analyses were conducted on days 1, 3, 7, and 14.
2.5. Cell Analysis
Cells were analyzed via imaging, metabolic activity assays, and measures of gene expression in cells seeded on the various substrates with and without the presence of Activin A.
2.5.1 Fluorescence Microscopy
The cytoskeleton of cells on scaffolds was visualized through phalloidin staining of actin using a cytoskeleton staining kit (EDM Millipore, Billerica, MA, USA). Cells were fixed using 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and then blocked using 1% BSA in PBS. The cells were then incubated with a 1:100 TRITC-phalloidin in PBS solution for 60 minutes. Following several washes, cells were incubated with a 1:1000 4′,6-diamidino-2-phenylindole (DAPI) in PBS solution for 5 minutes and then imaged on an Olympus IX81 Confocal Microscope.
2.5.2 Metabolic Activity Assay
A PrestoBlue mitochondrial reduction assay was performed on days 1, 7, and 14 to determine cellular activity on the scaffolds for the three different experimental groups. Control and experimental scaffolds with cells attached were immersed in assay solution and incubated for 4 hours. Controls were matched molecular weight hydrogels with no cells attached. A 100 μL sample of assay solution was aspirated from each well following the incubation period and pipetted into a fresh 96 well plate then read on a Beckman Coulter DTX 880 Multimode Detector with excitation at 560 nm and emission at 595 nm. The values read for control scaffold fluorescence were subtracted from the values read for experimental scaffold fluorescence (N=21 total; n= 3 for each scaffold).
2.5.3 qPCR
ARPE-19 RNA was isolated using Qiagen RNEasy Plus kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Briefly, cells were lysed using β-mercaptoethanol and Qiagen RLT Plus buffer and then centrifuged through a Qiashredder column to remove large debris and contaminants. Genomic DNA was removed using an eliminator column. Following this, ethanol was used to provide binding conditions for RNA to the RNeasy spin column, while other non-RNA contaminants were then washed away. The RNA was then eluted through the column and quantified using a NanoDrop™ spectrophotometer and associated software. Next, the RNA was normalized to a uniform concentration. Samples were reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biostystems). PCR was performed using SYBR Green PCR Master (Applied Biosystems) mix and PikoReal real time PCR system. The fold change relative gene expression compared to that of the control TCPS was determined using the delta-delta Ct method determining fold change compared to the housekeeping gene, GAPDH (N=21 total; n=3 for each scaffold). The expression was then normalized to day 1 expression to determine how the expression changed through the days of culture. Primer sequences are outlined in the Table.
Table.
PCR primer sequences
| Gene of Interest | Primer Sequence (5′ to 3′) |
|---|---|
| CRALBP | F: AGATCTCAGGAAGATGGTGGAC AGATCTCAGGAAGATGGTGGACAGATCTCAGGAAGATGGTGGAC |
| R: GAAGTGGATGGCTTTGAACC | |
| COL-I | F: GTCACCCACCGACCAAGAAACC |
| R: AAGTCCAGGCTGTCCAGGGATG | |
| IL-6 | F: GGCACTGGCAGAAAACAACC |
| R: GCAAGTCTCCTCATTGAATCC | |
| MCP-1 | F: GATCTCAGTCAGAGGCTCG |
| R: TGCTTGTCCAGGTGGTCCAT | |
| IL-8 | F: CTGGCCGTGGCTCTCTTG |
| R: TCCTTGGCAAAACTGCACCTT | |
| SMAD3: | F: TCCCCAGCACATAATAACTT |
| R: TGGGAGACTGGACAAAAAT | |
| αSMA | F: CTGGCATCGTGCTGGACTCT |
| R: GATCTCGGCCAGCCAGATC | |
| GAPDH | F: ACAACAGTCCATGCCATCAC |
| R: TCCACCACCCTGTTGCTGTA |
2.6. Statistical Analysis
Elastic moduli results, ELISA results, cellular metabolic activity, and gene expression were compared between groups using a Student’s t-test when comparing two groups or an analysis of variance (ANOVA) when comparing more than two groups. Following ANOVA, pairwise comparisons between groups was performed using Tukey’s post-hoc analysis. For metabolic activity and gene expression analyses, the dependent variables were control-adjusted results. p-Values less than 0.05 were considered significant and analyses were conducted in Matlab and Microsoft Excel. Statistical significance is indicated in the figures or figure legends, which are reported as mean ± standard error.
3. Results
3.1. Scaffold Physical Properties
By varying the polymer molecular weight and concentration, it was possible to vary the scaffold elastic moduli from 60 kPa to 1200 kPa. It was not possible to generate PEG scaffolds at 10 MPa, therefore PEGDA was bound to glass for the highest scaffold elastic moduli.
3.2. Activin A Binding Efficiency to Scaffolds
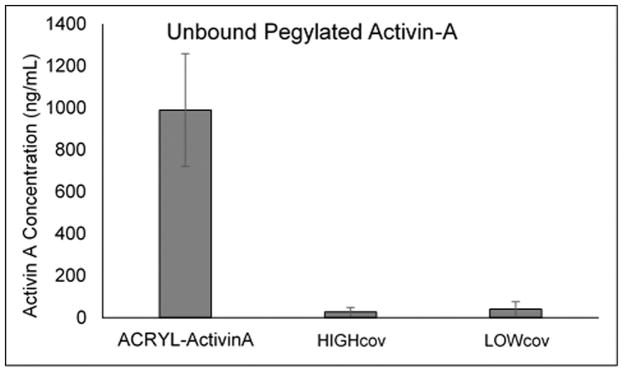
ELISA readings of PBS used to soak scaffolds with covalently bound Activin A was compared against unphotopolymerized controls (Figure 1). The concentration of Activin A in the PBS with unpolymerized acrylated-Activin A solution was calculated to be 990 ng/mL, which was equivalent to a 24% loss of Activin A during the reaction and freeze-drying process. PBS from polymerized scaffolds contained Activin A concentrations of 28 ng/mL and 41 ng/mL for high and low modulus scaffolds, respectively. From this it was determined that approximately 220 ng of Activin A was conjugated to the high modulus scaffold surface and 206 ng of Activin A conjugated to the low modulus scaffold.
Figure 1.
Concentration of Activin A in PBS containing unpolymerized acrylated-Activin A solution (Acryl-Activin A) and in PBS containing hydrogels with Activin covalently bound to the surface of high and low modulus hydrogels (HIGHcov and LOWcov, respectively). There was very little Activin A in the PBS and based on this concentration, it was determined that 220 ng and 206 ng of Activin was conjugated to the high and low modulus scaffold surface, respectively.
3.3. Entrapped Activin A Release from Scaffolds
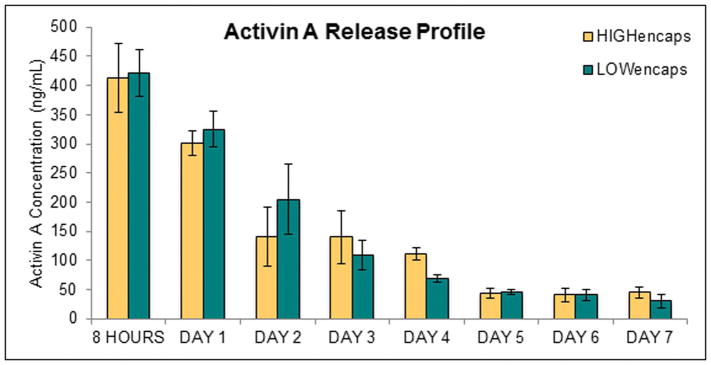
Activin A was consistently released from scaffolds for 7 days (Figure 2); by day 8, the Activin A was not detectable in sampled PBS. Calculating the cumulative mass over the 7 days showed that 88% and 91% of the Activin encapsulated was released from the high and low modulus scaffolds, respectively. During the first 8 hours, the release of Activin A was significantly higher than all other time points. There was no significant difference between the Activin A release from high and low modulus hydrogels, which both exhibited a decaying release profile.
Figure 2.
Activin A release from high and low modulus scaffolds with encapsulated Activin A (HIGHencaps and LOWencaps, respectively). There was no significant difference between the two scaffold groups. Error bars show standard deviation.
3.4. Metabolic Activity
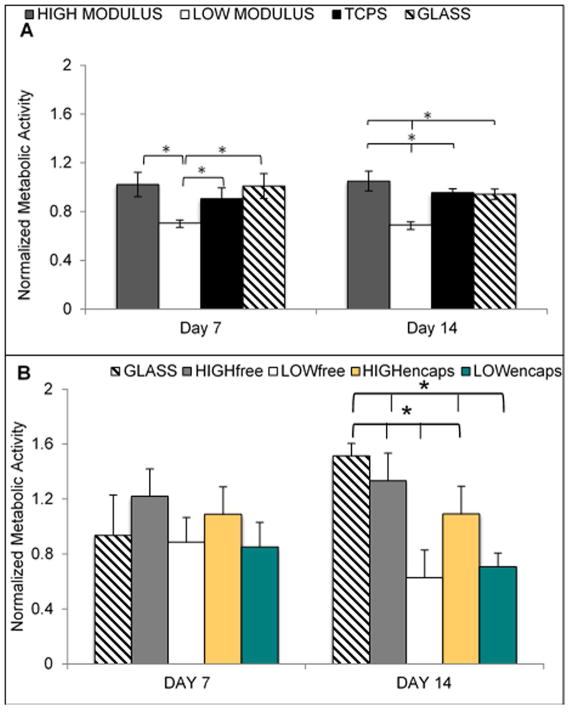
The results of metabolic activity assays were normalized to day 1 to determine how the cellular activity changed over the culture period. For cells not receiving Activin A, on both days 7 and 14, the high modulus scaffold resulted in greater cell metabolic activity than all other groups. On day 7 this increase over control was not statistically significant, but on day 14 it was significantly higher than the other groups. There was no noticeable difference in the number of cells on each substrate over the 14 days. The functionalized glass and TCPS did not show any significant differences at any time points. (Figure 3).
Figure 3.
A. ARPE-19 metabolic activity on scaffolds of varying modulus without Activin A. By day 7 of culture, the cells on the low modulus scaffold had significantly decreased activity when compared to both other conditions. The high modulus and TCPS were not different on day 7. On day 14, all three groups were significantly different from each other with the high modulus scaffold having the highest activity. B. Metabolic activity of ARPE-19 cells on varying substrates exposed to Activin A. When comparing against scaffolds with no Activin A, for all cases the presence of Activin A increased metabolic activity. When comparing between scaffolds receiving activin A, by day 14, the low modulus scaffolds had significantly lower metabolic compared to all other groups. Error bars show standard deviation. Asterisks show p < 0.05.
For cells receiving Activin A, The ARPE-19 cells cultured on high modulus scaffolds exposed to both free and encapsulated Activin A demonstrated significantly higher metabolic activity compared to cells on low modulus scaffolds exposed to Activin A. There were no significant differences between the two high modulus scaffold groups or the high modulus scaffold groups and the functionalized glass. The cells on the low modulus scaffolds had a decreased activity on day 7 and demonstrated further decrease on day 14. Although the cells in the free and encapsulated Activin A groups exhibited metabolic activity, cells in the covalently bound Activin A group had no detectable metabolic activity. Thus, since there were no cells to analyze in any covalently bound Activin A group, this condition is absent in all figures.
3.5. Cell Morphology
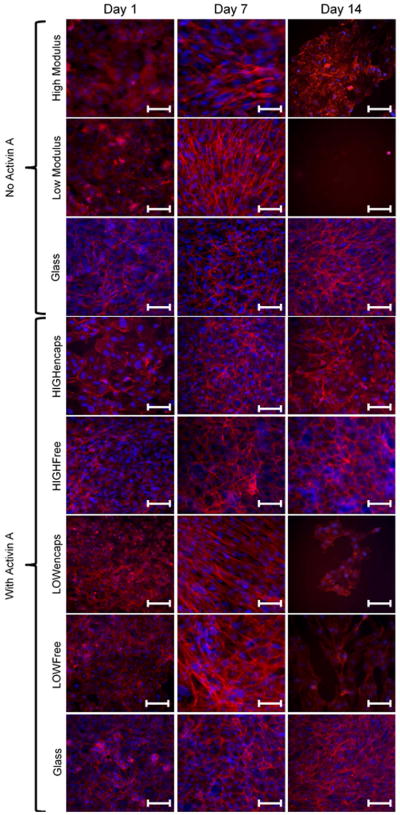
The cytoskeletons of ARPE-19 cells on different moduli scaffolds were visualized through fluorescently tagged phalloidin binding to F-actin filaments (Figure 4). In groups not receiving Activin A, cells on low modulus scaffolds exhibit elongated, parallel actin stress fibers by day 7, while cells on high modulus scaffolds show more peripheral actin fibers, less parallel stress fibers. By day 14, however, the cells on low modulus scaffolds detached and only very few cells with poor morphology could be visualized on hydrogel surfaces. On high modulus scaffolds, cells were still visible, but appeared to lose their peripheral actin filaments as more irregular actin fibers appeared. In comparison, the functionalized glass slide control exhibited strong peripheral actin fibers even on day 14. The cells on functionalized glass slides also showed the characteristic cobblestone morphology of native RPE cells.
Figure 4.
Epifluorescent images of Phalloidin (red: actin cytoskeleton) and DAPI (blue: nuclei) stains of ARPE-19 cells on various moduli substrates without Activin A or with Activin A delivered in the media or encapsulated into hydrogels. Without Activin A, the ARPE-19 cells exhibit strong parallel actin stress fibers on low modulus scaffold by day 7 and by day 14 there is little to no cell attachment. On the high modulus scaffolds, stress fibers were present, though less pronounced and there was still attachment at day 14, though cells did not display peripheral actin fibers characteristic of healthy retinal pigment epithelial cells. With Activin A, cells on low modulus scaffolds continued to display strong parallel actin stress fibers, though unlike the Activin-free case, these cells survived to day 14, albeit at fewer numbers than at day 7. Conversely, for high modulus scaffolds and functionalized glass, when Activin A was present cells seeded on these scaffolds developed peripheral actin fibers characteristic of retinal pigment epithelial cells. LOWfree, LOWencaps, HIGHfree, HIGHencaps refer to low or high modulus scaffolds with Activin A in the media or encapsulated, respectively. Scale bars are 100 μm.
In groups receiving Activin A, throughout the 14-day culture cell cytoskeletons on both high modulus scaffold groups and functionalized glass slides demonstrated a more characteristic epithelial morphology, such as increased peripheral cytoskeleton. Both low modulus scaffolds exhibited parallel actin stress fibers across the cell body. In addition, by day 14, there were little to no cells present on the low modulus scaffolds of either group. Scaffolds with Activin A covalently bound to the surface did not have any visible cells attached.
3.6. Gene Expression
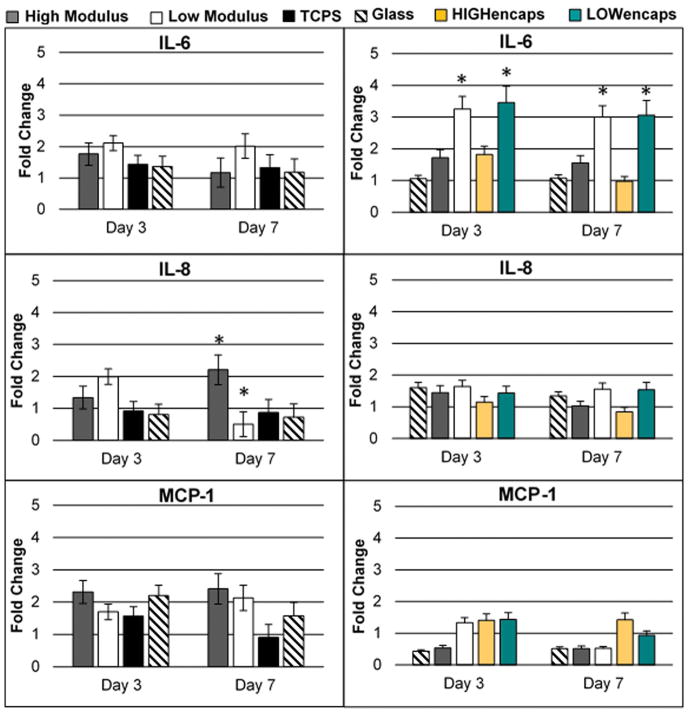
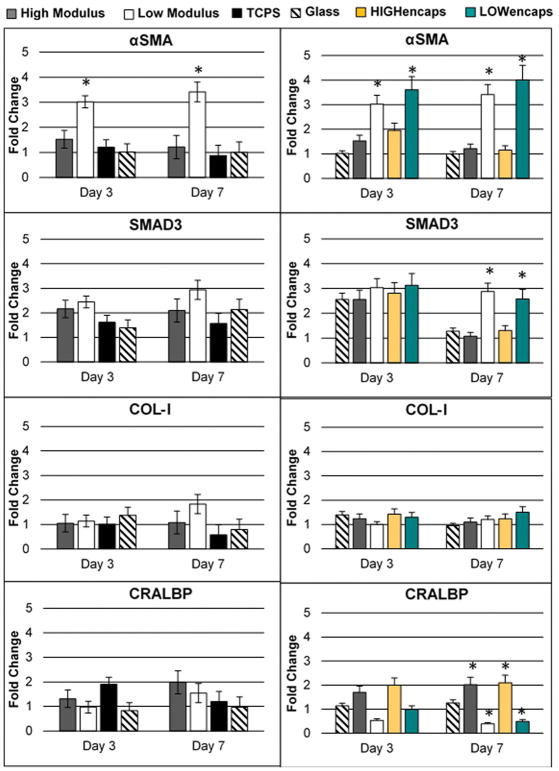
Expression of genes associated with inflammation and cell maturity was determined in ARPE-19 cells using qPCR (Figures 5 and 6). The expression of inflammatory genes, IL-6, IL-8, and MCP-1 were higher on low modulus scaffolds at all time points. The expression of dedifferentiation markers, SMAD3 and αSMA, was also significantly higher on low modulus scaffolds. While αSMA demonstrated significantly higher expression at both time points, SMAD3 was only significant at Day 7. High modulus scaffolds showed no obvious trend in genes associated with inflammation. However, both the HIGHfree and HIGHencaps groups demonstrated similar expression patterns of SMAD3 and αSMA compared to the glass slides. The expression of these dedifferentiation markers was significantly lower compared to the low modulus scaffolds.
Figure 5.
qPCR results for ARPE-19 expression of inflammatory markers IL-6, IL-8, and MCP-1 on all substrates. Left panel is without Activin A, right panel is with Activin A. On high modulus scaffolds, when Activin A was added IL-6 levels increased to significantly higher levels than all other scaffolds. Without Activin A, on day 7, IL-8 expression on high modulus scaffolds increased from day 3 levels and was significantly higher than on all other substrates, while on low modulus scaffolds, day 7 IL-8 expression decreased from day 3 levels. This was not observed when Activin A was present, and expression levels showed no differences between Day 3 and Day 7. Activin A reduced MCP-1 levels in all groups, with the greatest effect observed in substrates (glass and scaffolds) receiving Activin A in the media. Error bars show standard deviation. Asterisks show p<0.05. Legends indicating low and high modulus scaffolds refer to LOWfree and HIGHfree scaffolds for graphs with Activin A present.
Figure 6.
qPCR results for ARPE-19 expression of dedifferentiation (αSMA and SMAD3) and maturation genes (COL-1 and CRALBP) for substrates without Activin A (left panel) and substrates with Activin A (right panel). Low modulus scaffolds exhibited significantly higher levels of αSMA on both days 3 and 7 whether Activin A was present or not. The presence of Activin A reduced SMAD3 levels in day 7 higher modulus substrates, but not for low modulus substrates. Expression levels of COL-I did not show any differences between groups. Between groups with and without Activin A, CRALBP levels showed no differences in higher modulus substrates, but showed decreases in lower modulus substrates. Error bars show standard deviation. Asterisks show p<0.05. Legends indicating low and high modulus scaffolds refer to LOWfree and HIGHfree scaffolds for graphs with Activin A present.
4. Discussion
The only reported elastic modulus for a BM is that of a porcine BM, which was calculated to be approximately 1000 kPa. [10] One of the goals of this study was to present RPE cells with scaffolds approximating a normal BM Young’s modulus, and scaffolds with moduli an order of magnitude above and below this value. The low and high modulus scaffolds developed in this study were determined to have Young’s moduli an order of magnitude below and at the reported values for the BM Young’s modulus. It was not possible to achieve a Young’s modulus of 10,000 kPa with PEGDA, therefore glass was functionalized to present a stiffer substrate with surfaces presenting PEG-RGD moieties identical to those the PEGDA hydrogels. Glass has a reported modulus in the gigapascal range, thus the modulus of functionalized glass slides was on the order of 106 times higher than the high modulus hydrogels used. Tensile and compressive data of the scaffolds showed strong correlation for 2× concentration scaffolds at all molecular weights. For 1× concentration scaffolds, the Young’s modulus values were the same for 10 and 20 kDa scaffolds, but the values differed for 3.4 and 5 kDa scaffolds. It is likely these differences are due to viscoelastic effects and geometry differences between scaffolds tested in tension and compression. Tensile tests evaluated thin hydrogel sheets while compressive tests used thick hydrogel disks. This geometric difference combined with the smaller mesh size of lower molecular weight hydrogels likely increased the resistance to the expulsion of fluid while soaked hydrogels were subjected to compression.
Activin A is a signaling molecule known to promote the mature RPE phenotype and the present study examined whether chemical stimulation with Activin A could rescue the poor adhesion, morphology, and apparent dedifferentiation of ARPE-19 cells observed on varying substrate moduli. Activin A supplemented media, Activin A encapsulated in scaffolds, and Activin A covalently bound to scaffolds were examined. Activin A covalently bound to hydrogel surfaces completely inhibited cell attachment. It is likely that the larger Activin A molecule sterically hindered the much smaller PEG-RGD peptide sequence. Future studies should consider larger adhesion molecules such as laminin or fibronectin. Additionally, future studies should confirm the bioactivity of pegylated Activin A and adjust doses accordingly. When exposed to Activin A in the media or encapsulated with hydrogels, the gene expression of ARPE-19 cells on high modulus substrates (hydrogels and functionalized glass) demonstrated significantly lower inflammatory markers, IL-6 and IL-8, and dedifferentiation markers, SMAD3 and αSMA, compared to low modulus scaffolds. Our first study showed higher gene expression of IL-6 and IL-8 on high modulus scaffolds than on low modulus scaffolds at day 3, while this study showed no significance between the groups at day 3. By day 7, the current study shows that IL-6 is significantly lower on high modulus scaffolds, while IL-8 is significantly lower on low modulus scaffolds. The presence of Activin A did not significantly change the expression of cells cultured on low modulus scaffolds. When comparing high modulus scaffolds with or without the presence of Activin A, a trend emerged wherein cells exhibited a lower expression of dedifferentiation markers. In addition to this trend, the expression of CRALBP, the characteristic RPE gene, was consistently higher when the cells were exposed to Activin A.
The presence of Activin A also affected the morphology of exposed cells. Fluorescent microscopy demonstrated significant differences in the orientation of actin fiber filaments of the cytoskeleton between cells receiving Activin A and those not receiving the molecule. This is a significant observation because early studies on the effects of scaffold modulus on epithelial cells suggest that cells sense their physical environment, causing differences in focal adhesions and expression of intracellular pathways. RPE dedifferentiation has been characterized by a change in expression of cytokeratin proteins, a component of the intracellular cytoskeleton. In addition, it has been established that the actin cytoskeleton reorganizes from its characteristic hexagonal morphology to a disorganized, random morphology during dedifferentiation. [11] Therefore, it is possible that the mechanical environment experienced by RPE cells affects their adhesion and initiates or promotes dedifferentiation. This is supported by recent work demonstrating difference in phagocytic ability of RPE cells on different substrate moduli. [12]
SMAD3 and αSMA levels were significantly reduced in cells on high modulus scaffolds when exposed to Activin A, though not in cells on low modulus scaffolds. These results suggest that the right combination of molecules may promote the survival and persistence of healthy RPE cells unless they are on a scaffold that is too compliant. Thus, further study into RPE dedifferentiation mediated by scaffold modulus is warranted in the efforts to design a scaffold optimized for RPE health.
Grisanti et al. demonstrated that actin cytoskeleton reorganization occurs during dedifferentiation, whereby the dedifferentiated RPE phenotype displays elongated, linearly arranged actin fibers that span across the cell cytoplasm. [13] These linear fibers were present in ARPE-19 cells cultured on low modulus scaffolds regardless of Activin A exposure. By day 14, most of the ARPE-19 cells had detached from low modulus scaffolds; in conditions without Activin A, all cells had detached. In the high modulus groups, this cell line demonstrated a more circumferential cytoskeleton at Day 14, with cells receiving Activin A in the media displaying slightly more epithelial morphology than cells receiving encapsulated Activin A. This may be due to the reduced release of Activin A from scaffolds after Day 4 of culture, or alternately it may be due to the short in vivo half-life of Activin A (~20 min). [14]
The limitations of this investigation include the fact that the cells examined are an immortalized line (ARPE-19). Although they are an established line that exhibit many features of RPE cells, they do not completely recapitulate the morphology and electrophysiology observed in native RPE cells. In particular, ARPE-19 show a loss of pigmentation and characteristic epithelial shape. Recognizing this limitation, the goal was to perfect the system with the ARPE-19 line then to transition to a more clinically relevant RPE line. However, when primary cells harvested from chick embryos or RPE cells derived from induced pluripotent stem cells (IPSCs) were seeded on the hydrogels, they did not survive. The uniform cell death suggested that the minimalist hydrogel was insufficient to support the more finicky primary RPE cells. Since our previous study established that ARPE-19 cells were adversely affected by overly compliant scaffolds, [7] establishing whether Activin A could recover cell viability and morphology was still an important result.
Despite the limitations, the results of this study demonstrated several important findings. First, low modulus scaffolds increase inflammation and dedifferentiation marker expression and this cannot be prevented with Activin A exposure. This indicates that modulus stiffness alone can have a dominating effect over exogenous stimulation and should be carefully considered during scaffold design. The second finding is that when cultured on scaffolds with an appropriate modulus, exogenous factors, such as Activin A, can affect cell expression, morphology, and activity. In other words, scaffold modulus alone is necessary to preserve RPE fate, but not sufficient—while the wrong scaffold modulus can have devastating effects on RPE survival regardless of the presence of Activin A, the right scaffold modulus is necessary, but not sufficient to ensure RPE survival and requires additional factors such as Activin A. Our findings indicate that Activin A is capable of promoting RPE survival, and as a part of the TGF-beta family, which also plays a role in apoptotic signals, this is a finding with some support. These findings support future work in the field to determine key growth factors and signaling molecules to encapsulate in scaffolds, covalently bind to scaffolds, or deliver into the subretinal space with a cell-scaffold complex to decrease inflammation and promote mature, functional RPE cells post-transplantation.
Acknowledgments
CE White was supported by fellowships from the National Institutes of Health National Institute of General Medical Sciences (NIH NIGMS) biotechnology training program (T32 GM008339) and the US Department of Education Graduate Assistance in Areas of National Need (GAANN; award number P200A150131). B Kwok was supported by a summer fellowship by the New Jersey Space Grant Consortium (NJSGC).
References
- 1.ten Dijke P, et al. Characterization of type I receptors for transforming growth factor-beta and activin. Science. 1994;264(5155):101–4. doi: 10.1126/science.8140412. [DOI] [PubMed] [Google Scholar]
- 2.Huang SS, Huang JS. TGF-beta control of cell proliferation. J Cell Biochem. 2005;96(3):447–62. doi: 10.1002/jcb.20558. [DOI] [PubMed] [Google Scholar]
- 3.Travis MA, Sheppard D. TGF-beta activation and function in immunity. Annu Rev Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakami S, Etter P, Reh TA. Activin signaling limits the competence for retinal regeneration from the pigmented epithelium. Mech Dev. 2008;125(1–2):106–16. doi: 10.1016/j.mod.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuhrmann S, Levine EM, Reh TA. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development. 2000;127(21):4599–609. doi: 10.1242/dev.127.21.4599. [DOI] [PubMed] [Google Scholar]
- 6.Idelson M, et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5(4):396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 7.White C, DiStefano T, Olabisi R. The influence of substrate modulus on retinal pigment epithelial cells. J Biomed Mater Res A. 2017;105(5):1260–1266. doi: 10.1002/jbm.a.35992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hynes SR, Lavik EB. A tissue-engineered approach towards retinal repair: scaffolds for cell transplantation to the subretinal space. Graefes Arch Clin Exp Ophthalmol. 2010;248(6):763–78. doi: 10.1007/s00417-009-1263-7. [DOI] [PubMed] [Google Scholar]
- 9.Hotaling NA, et al. Nanofiber Scaffold-Based Tissue-Engineered Retinal Pigment Epithelium to Treat Degenerative Eye Diseases. J Ocul Pharmacol Ther. 2016;32(5):272–85. doi: 10.1089/jop.2015.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Candiello J, et al. Biomechanical properties of native basement membranes. The FEBS journal. 2007;274(11):2897–2908. doi: 10.1111/j.1742-4658.2007.05823.x. [DOI] [PubMed] [Google Scholar]
- 11.Haynes J, et al. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol Biol Cell. 2011;22(24):4750–64. doi: 10.1091/mbc.E11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boochoon KS, et al. The influence of substrate elastic modulus on retinal pigment epithelial cell phagocytosis. J Biomech. 2014;47(12):3237–40. doi: 10.1016/j.jbiomech.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grisanti S, Guidry C. Transdifferentiation of retinal pigment epithelial cells from epithelial to mesenchymal phenotype. Invest Ophthalmol Vis Sci. 1995;36(2):391–405. [PubMed] [Google Scholar]
- 14.Johnson KE, et al. Biological activity and in vivo half-life of pro-activin A in male rats. Molecular and cellular endocrinology. 2016;422:84–92. doi: 10.1016/j.mce.2015.12.007. [DOI] [PubMed] [Google Scholar]