Abstract
Chronic obstructive pulmonary disease (COPD) is a leading cause of death worldwide, and long-term oxygen therapy has been shown to reduce mortality in COPD patients with severe hypoxemia. However, the Long-term Oxygen Treatment Trial (LOTT), a large randomized trial, found no benefit of oxygen therapy in COPD patients with moderate hypoxemia. We hypothesized there may be differences in response to oxygen which depend on genotype or gene expression. In a genome-wide time-to-event analysis of the primary outcome of death or hospitalization in 331 subjects, 97 single nucleotide polymorphisms (SNPs) showed evidence of interaction with oxygen therapy at p<1e-5, including 7 SNPs near Arylsulfatase B (ARSB; p=6e-6). In microarray expression profiling on 51 whole blood samples from 37 individuals, at screening and/or at 12-month follow-up, ARSB expression was associated with the primary outcome depending on oxygen treatment. The significant SNPs were conditional expression quantitative trait loci for ARSB expression. In a network analysis of genes affected by long-term oxygen, two observed clusters including 26 co-expressed genes were enriched in mitochondrial function. Using data from the observational COPDGene Study, we validated the expression of 25 of these 26 genes, plus ARSB. The effect of long-term oxygen therapy in COPD varied based on ARSB expression and genotype. ARSB has previously been shown to be associated with hypoxemia in human bronchial and colonic epithelial cells and in a mouse model. In peripheral blood, long-term oxygen treatment affected expression of mitochondrial-related genes, a biologically relevant pathway in COPD.
Keywords: Arylsulfatase B, pharmacogenomics, expression quantitative trait loci, genome-wide association study, microarray, oxygen
Background
Chronic obstructive pulmonary disease (COPD) is a leading cause of death worldwide, and mortality from COPD continues to rise. Since the early 1980s, when the Nocturnal Oxygen Treatment Trial and the Medical Research Council Study both found that long-term oxygen therapy reduced mortality in COPD patients with severe hypoxemia, supplemental oxygen became one of the only treatments for COPD with a proven survival benefit [1, 2]. In the 1990s, two trials were performed to investigate whether domiciliary or nocturnal oxygen had a similar survival benefits for COPD patients with moderate hypoxemia [3, 4]. Though no overall survival benefit was seen in these two studies, the results in moderately hypoxemic patients suggested heterogeneity in response to oxygen, based on clinical characteristics. In the oxygen-treated group in the Gorecka et al. trial, survivors had better lung function and higher body mass index than non-survivors [3]. Data from the National Emphysema Treatment Trial further demonstrate heterogeneity in response to oxygen, finding that supplemental oxygen use by non-hypoxemic patients was associated with reduced survival [5].
As reported in 2016, the Long-term Oxygen Treatment Trial (LOTT) enrolled 738 COPD patients with moderate hypoxemia into a randomized clinical trial with two arms [6]. Subjects with hypoxemia at rest were randomized to supplemental oxygen or no supplemental oxygen 24 hours daily, while those with hypoxemia upon exertion were randomized to oxygen with exertion and with sleep. This largest randomized trial to date concluded long-term oxygen therapy did not prolong the time to death or first hospitalization among COPD patients with moderate hypoxemia. However, subgroup analysis in LOTT showed benefits of supplemental oxygen in subjects age 71 or older, subjects who experienced a COPD exacerbation in the 1-3 months prior to enrollment and subjects with lower quality of life.
Considering the history of studies of long-term supplemental oxygen in COPD, there is clear heterogeneity in response to oxygen based on resting oxygen saturation, and there may also be heterogeneity based on other patient characteristics. There could be differential responses to oxygen therapy in COPD patients based on genetics, particularly variation in oxygen-responsive genes. Our group has previously identified genetic variants associated with resting oxygen saturation [7].
However, pharmacogenomic studies have not yet been conducted to investigate the heterogeneity of oxygen effects in COPD patients. In other diseases, genetic influences on oxygen toxicity and oxygen benefit have been demonstrated in both human and animal studies [8-12]. Based on this background, we hypothesize that a set of oxygen-responsive genes will be differentially expressed in COPD patients treated with long-term supplemental oxygen therapy, and genetic variation and gene expression could underlie inter-individual variation in response to oxygen. Herein, we examined genome-wide genetic variation and microarray gene expression in a subset of the LOTT study population. We used multi-omics data from the observational COPDGene study [13, 14] to validate some of the findings from LOTT. Some of the results from this study have been previously reported as an abstract [15].
Materials and Methods
For detailed descriptions of the methods, see the supplementary material. Figure 1 shows an overview of the study design.
Figure 1. Overview of the study design.
NHW: Non-Hispanic White. AA: African American.
LOTT Genomics Study
Ancillary study subjects were enrolled from LOTT [6]. In LOTT, 738 COPD patients with moderate resting hypoxemia or exercise-induced desaturation were randomized to long-term supplemental oxygen or no supplemental oxygen therapy, either 24 hours daily or with exercise and sleep, respectively. A total of 354 subjects provided DNA samples and 37 subjects provided 68 whole blood RNA samples for expression studies. Subjects gave written informed consent, and the genetics study was approved by the institutional review board at each participating center.
DNA samples were genotyped on the Infinium Multi-Ethnic Global-8 chip (Illumina, San Diego, CA). At four centers, whole blood samples were collected in PaxGene RNA tubes. Genome-wide gene expression levels were measured using the Illumina HumanHT-12 v4 BeadChip. The raw genotype data was preprocessed by GenomeStudio for quality control (See supplemental methods for detailed quality control criteria). Genotype imputation was performed using Minimac3 based on the 1000 Genome Project Phase3 panel. Raw microarray data were preprocessed using the lumi R package (See supplemental methods). To measure relative gene expression level, variance stabilization and quantile normalization was performed. LOTT microarray data are available in the Gene Expression Omnibus database (accession GSE111790).
Statistical analysis for time-to-event variable
The genetics study used the same primary outcome as LOTT: time to death or first hospitalization. With the SNP data, genome-wide time-to-event analysis was performed using Cox proportional hazards models with an interaction term between additive genotype and an indicator variable for oxygen therapy. Covariates included age, sex, and principal components derived from genetic data along with main effect terms for oxygen therapy and each SNP. Analysis was performed separately in the two racial groups (White and Black). Using matched coordinates of SNPs and transcripts on the hg19 reference genome, we identified transcripts located ±50kb of the SNPs (based on the probe coordinates), after removing uncharacterized transcripts, poorly mapped probes and SNPs violating the assumption of proportional hazards in time-to-event analysis. Transcriptome analysis examined the statistical interaction between gene expression and oxygen treatment (See supplemental methods).
Conditional expression quantitative trait loci (eQTL) analysis in COPDGene
In the COPDGene study, 6,752 White subjects were genotyped [13] and gene expression levels were measured in 510 individuals using RNA-sequencing [14]. COPDGene RNA-sequencing data are available in GEO (GSE97531). Using the data from 376 White subjects, two types of eQTL analyses were performed. In the conventional eQTL analysis, we performed linear association tests to assess whether the SNP genotype is associated with gene expression. The conditional eQTL model included a genotype-by-oxygen interaction term.
Gene expression affected by long-term oxygen therapy in LOTT
In LOTT, a total of 51 gene expression samples were available for the longitudinal analysis after quality control. The gene expression data consisted of 37 COPD patients treated with oxygen vs. no treatment, either at screening or 12-month follow-up, or at both time points. Due to the unbalanced paired data, we used a mixed-effects model, with individual as the random effect to incorporate the longitudinal design. Oxygen therapy and study time points were the fixed effects. Sex, age, and smoking pack-years were considered as covariates. Functional enrichment analysis was performed using DAVID [16] and network-based clustering analysis was conducted to characterize genes affected by oxygen therapy. We also tested for association between gene expression baseline resting oxygen saturation (SpO2) and 6 minute walk distance, using robust regression based on Huber’s M-estimator [17].
RESULTS
Study subjects
After quality control, genome-wide genotype data were available for 331 LOTT subjects (Supplementary Tables S1 and S2) and microarray gene expression data were available for 51 whole blood samples from 37 individuals, either at screening, 1 year follow-up, or both time points (Tables 1, Supplementary Table S3). Of the 331 genotyped subjects, 168 and 163 COPD patients were randomly assigned to the oxygen and no-oxygen groups, respectively. Transcriptome data were available for 14 oxygen and 16 no-oxygen randomized subjects from the screening time-point, and 11 and 10 subjects at the 12-month follow-up. Most subjects had moderate to severe COPD (GOLD spirometry grades 2 and 3) [18].
Table 1.
LOTT Pharmacogenomics Study subjects
| Characteristics | Genetic data (n=331) |
Expression data (n=51) |
|
|---|---|---|---|
| Measured time points | At screening | At screening | 12-month Follow-up |
| Randomized to oxygen vs. control | 168/163 | 14/16 | 11/10 |
| Desaturation*
Resting only Exercise only Resting and exercise |
48 164 119 |
6 17 7 |
4 11 6 |
| Age, years | 70.1±6.82 | 71.8±8.05 | 71.1±5.75 |
| Sex | Male: 194 Female: 137 |
Male: 16 Female: 14 |
Male: 14 Female: 7 |
| BMI, kg/m^2 | 28.2±6.14 | 27.7±5.77 | 30.3±6.03 |
| Race | White: 290 Black: 41 |
White: 20 Black: 10 |
White: 13 Black: 7 Other: 1 |
| GOLD Spirometry Grade | 0: 5 1: 18 2: 115 3: 147 4: 45 NA: 1 |
0: 1 1: 3 2: 8 3: 16 4: 2 |
1: 3 2: 5 3: 9 4: 3 |
| Pack-years of smoking | 59.3±32.3 | 56.9±46.51 | 68.8±55.6 |
| Resting oxygen saturation (SpO2) | 93.7±2.08 | 94.3±2.07 | 93.6±l.94 |
| 6 minute walk distance, feet | 1060.7±326.96 | 951.4±271.66 | 904.1±3 16.44 |
| Post Bronchodilator FEV1 percent predicted | 48.2±l7.38 | 53.6±23.13 | 56.4±26.29 |
Plus-minus values represent means ± SD and categorical values represent number of subjects.
BMI=body mass index; GOLD= Global Initiative for Obstructive Lung Disease; FEV1= forced expiratory volume in 1 second
Moderate resting desaturation defined as SpO2 89-93%. Moderate exercise-induced desaturation defined as SpO2 ≥80% for >5 minutes and <90% for ≥10 seconds during 6-minute walk test [6].
Genome-wide time-to-event analysis for the LOTT primary outcome
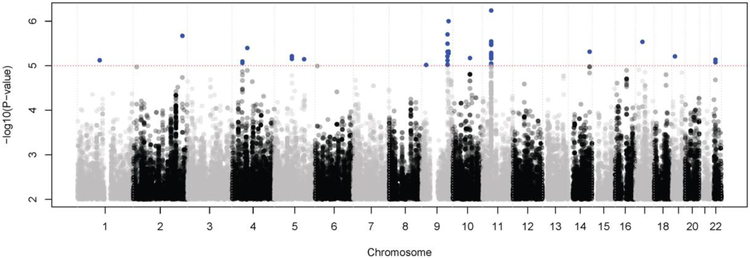
Time-to-event analysis for the LOTT primary outcome was performed with an interaction term between genotype and oxygen therapy. Examination of the quantile-quantile plots of –logl0(p-values) showed no evidence of inflation among White subjects, but there appeared to be inflation in the Black subjects, likely due to the smaller sample size (Supplementary Figure S1); therefore, we focused only on the white subjects. At p <le-5, 97 SNPs showed evidence of an interaction between genotype and oxygen therapy, which represented 15 different genomic regions (Figure 2 and Supplementary Table S4). In these regions, we searched for annotated transcripts located ±50 kb from the chromosomal location of each SNP, resulting in 12 SNPs and five transcripts falling in 5 linkage disequilibrium (LD) blocks: CDK5R2, ARSB, MLLT3, SLC31A2, and TLR4 (Table 2, Supplementary Figure S2). No adjacent transcripts were found for the remaining 85 SNPs. Time-to-event analysis of these 5 transcripts showed that only the ARSB (arylsulfatase B) gene is significantly associated with the primary outcome according to oxygen treatment at Bonferroni adjusted P < 0.05.
Figure 2. Manhattan plot of genome-wide time-to-event analysis for LOTT primary outcome.
Blue dots indicate SNPs with P < 1e-5.
Table 2.
Result of time-to-event analysis with five candidate transcripts resulting from the genetic analysis
| Gene symbol | Chromosome | Strand | Probe coordinate | P-value* | P-adjusted† |
|---|---|---|---|---|---|
| CDK5R2 | 2 | + | 219826469-219826518 | 0.059 | 0.298 |
| ARSB | 5 | − | 78073232-78073281 | 0.001 | 0.005 |
| MLLT3 | 9 | − | 20345151-20345200 | 0.462 | 1 |
| SLC31A2 | 9 | + | 115926028-115926077 | 0.075 | 0.379 |
| TLR4 | 9 | + | 120477004-120477053 | 0.344 | 1.000 |
P-value derived for interaction term between gene expression levels and indicator of oxygen use in Cox proportional hazards model for the LOTT primary outcome.
Bonfcrroni adjusted P-value.
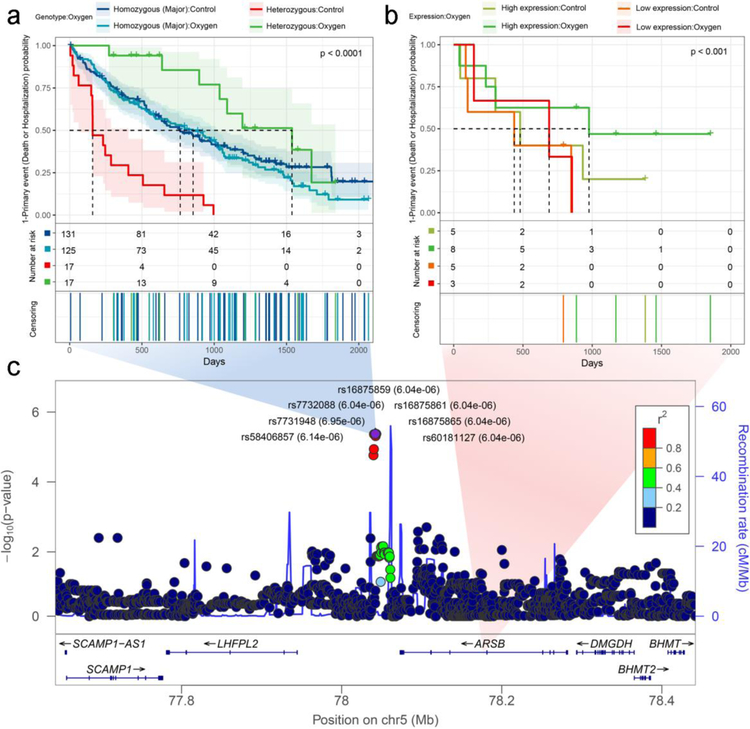
In the genetic analysis, there were 7 SNPs near the ARSB gene which were in nearly complete LD (r2= 0.97 to 1), with average minor allele frequency of 0.155. Therefore, we considered these 7 SNPs as one LD block and selected one of five perfectly correlated SNPs (rs7732088) as a representative in the subsequent analyses. There was a difference in the effects of oxygen therapy in heterozygous subjects (Bonferroni adjusted P= 1.06e-04), with improved outcomes in oxygen-treated subjects (Figure 3a and Supplementary Table S5). There was no difference in oxygen effect in the homozygotes for the major allele. There was a nominally significant difference in outcomes between oxygen and no oxygen treatment groups for subjects with high or low ARSB expression (Figure 3, Supplementary Table S5). We identified the same LD block in the 41 African American LOTT subjects with genotype data (average r2=0.89). In a parallel SNP analysis to Table S5, we confirmed similar directions for the hazard ratios, though none of the results were statistically significant (data not shown).
Figure 3. Time-to-event analyses of the ARSB gene and adjacent 7 SNPs in LOTT.
(a) Kaplan-Meier plot of ARSB SNPs in genetic data analysis. These SNPs are located at 5q 14.1 and exhibit strong linkage disequilibrium; therefore, rs7732088 was used as representative SNP. (b) Kaplan-Meier plot of ARSB gene expression. (c) LocusZoom plot for genome-wide time-to-event analysis for SNPs identified near ARSB.
eQTL analysis in the COPDGene study
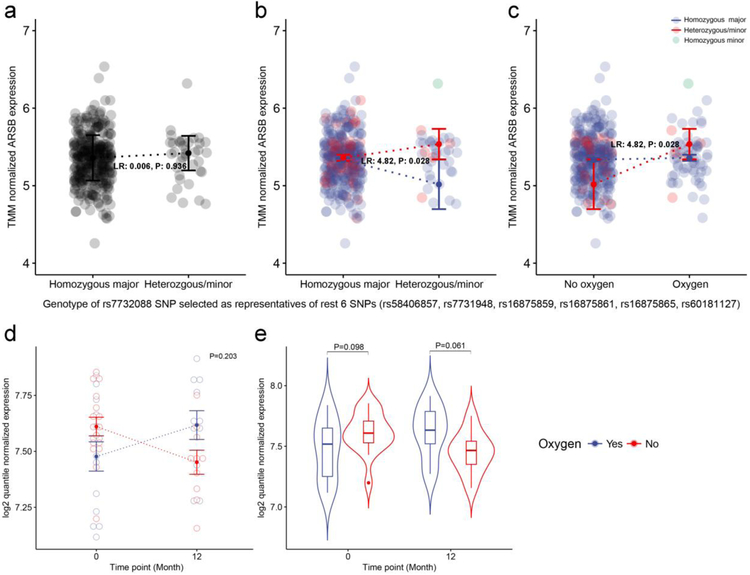
We hypothesized that there will be a cis-regulatory mechanism between ARSB and identified SNPs, which we tested using the larger sample size of GWAS and RNA-sequencing data from the COPDGene study (Supplementary Table S6). There was no evidence of eQTL associations between ARSB and nearby SNPs in the conventional eQTL analysis (Figure 4a). However, the conditional eQTL analysis based on current oxygen therapy showed significant cis-regulatory relationships (LR 4.82, P=0.028) (Figure 4bc). This result is consistent with the directionality of interaction effects in the LOTT time-to-event analyses. These results suggest COPD patients carrying the minor allele at the 7 SNPs near ARSB may have a longer or shorter time to death or hospitalization based on up- or down-regulation of expression levels of the ARSB, depending on oxygen therapy.
Figure 4. Conventional and conditional eQTL analysis of ARSB gene in the COPDGene Study.
(a) Conventional eQTL plot between TMM normalizedARSB expression and dominant coded genotype of the rs7732088 SNP selected as representative of 7 SNPs. (b) Conditional eQTL plot showing interaction effect between genotype and oxygen effect. The dotted-line and error bar represent mean±SEM. (c) Same data as in part (b), with oxygen treatment plotted on the x-axis. (d) Longitudinal gene expression pattern of the ARSB gene. There was no significant interaction (P= 0.20). (e) ARSB gene expression at screening and 12-month follow-up. There was no significant differential expression based on oxygen treatment at either time point.
Longitudinal transcriptome analysis in LOTT to detect genes affected by oxygen treatment
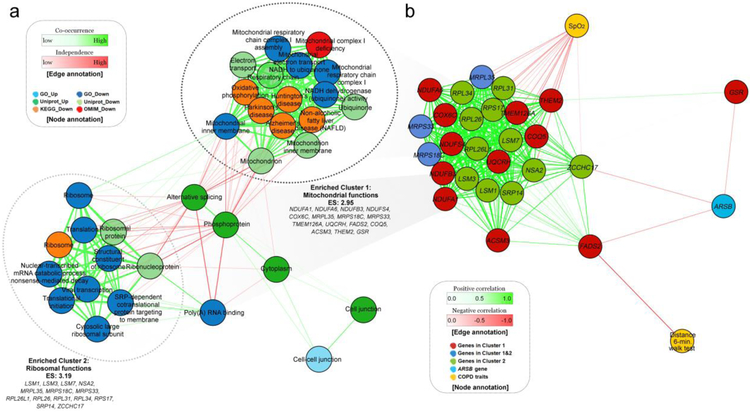
In a longitudinal analysis using a mixed effects model, 192 genes were differentially expressed between oxygen treatment groups at a nominal P<0.01; 80 and 112 genes were up- and down-regulated, respectively. Functional enrichment analysis was performed using DAVID [16]; these genes were enriched in 33 functional terms at false discovery rate (FDR) adjusted P < 0.1 (Supplementary Table S7). A network-based clustering analysis of the observed functional terms was performed, identifying two large clusters enriched in down-regulated genes (Figure 5). In Cluster 1 (Enrichment score: 2.95), functional terms were related to mitochondrial function, such as Mitochondrion inner membrane (Padj: 1.79e-03), Electron transport (1.98e-03), Respiratory chain (2.12e-03), Mitochondrion (3.63e-03), and 12 other terms. These terms of Cluster 1 were based on 15 down-regulated genes in the oxygen group (Figure 5 and Supplementary Figure S3). In Cluster 2 (Enrichment score: 3.19), 11 functional terms were related to ribosomes, such as Ribonucleoprotein (5.63e-09) and Ribosomal protein (3.12e-04). A total of 14 down-regulated genes belonged to this cluster (Figure 5). Three overlapping genes, MRPL35, MRPS18C, and MRPS33, were observed in both clusters, and these genes encoded mitochondrial ribosomal proteins. For this reason, we further investigated the co-expression pattern of these 26 genes in two clusters. We visualized a correlation-based network additionally including the ARSB gene and two COPD-related traits (resting oxygen saturation and 6-minute walk distance), and found that three genes were negatively correlated with ARSB (−0.344±0.005) (Figure 5). The rest of the 23 genes in Clusters 1 and 2 were positively correlated with each other (0.764±0.006) and negatively correlated with resting oxygen saturation (Figure 5, Supplementary Table S8). The FADS2 gene was negatively correlated with 6 minute walk distance (Padj= 0.026). ARSB expression was not significantly affected by oxygen therapy in the longitudinal analysis (P=0.20 for interaction between oxygen therapy and time and P= 0.061 at 12-month follow-up) (Figure 4de), and was not associated with resting oxygen saturation (P=0.95) or 6 minute walk distance (P=0.11).
Figure 5. Network plots of LOTT gene expression analysis.
(a) Network plot for DAVID functional enrichment analysis using up- and down-regulated genes in the oxygen group in longitudinal gene expression analysis. Two functional clusters were observed in down-regulated genes at FDR adjusted P < 0.1. (b) Correlation-based network of co-expressed genes and COPD related traits.
Validation in the COPDGene study
Using COPDGene whole blood RNA-sequencing data[14], we tested for differential expression of these 26 genes based on self-reported supplemental oxygen use adjusted for covariates including GOLD spirometry grade. After we combined the evidence from the results based on LOTT and those based on COPDGene, 25 of the 26 genes were significant at FDR adjusted P<0.05 (Table 3). The ARSB gene was not differentially expressed in LOTT, but was significant in the RNA-seq analysis of COPDGene (P=0.03). The combined evidence from LOTT and COPDGene showed that 4 of these genes were significantly associated with resting SpO2 (Supplementary Table S9). The association between FADS2 expression and 6-minute walk distance was not replicated in COPDGene data. SNPs near ARSB, tested using rs7732088 as a proxy, were not associated with resting SpO2, 6-minute walk distance, SGRQ score, FEV1, DLCO or chest CT scan measures of emphysema.
Table 3.
Meta-analysis of gene expression affected by oxygen therapy in LOTT and COPDGene
| Gene symbol |
P-value LOTT |
P-value COPDGene |
Combined P-value* |
Adj. Combined P-value*† |
|---|---|---|---|---|
| NDUFA1 | 1.37e-03 | 3.15e-01 | 3.89e-03 | 5.82e-03 |
| NDUFA6 | 4.42e-04 | 1.01e-02 | 6.92e-05 | 3.74e-04 |
| NDUFB3 | 5.87e-03 | 1.92e-02 | 9.41e-04 | 2.31e-03 |
| NDUFS4 | 1.47e-03 | 4.63e-03 | 9.58e-05 | 4.31e-04 |
| COX6C | 2.31e-03 | 2.54e-04 | 1.52e-05 | 2.78e-04 |
| MRPL35 | 4.49e-03 | 1.41e-01 | 4.45e-03 | 5.82e-03 |
| MRPS18C | 3.60e-03 | 1.25e-02 | 4.39e-04 | 1.32e-03 |
| MRPS33 | 9.27e-03 | 1.61e-02 | 1.19e-03 | 2.68e-03 |
| TMEM126A | 8.21e-03 | 4.66e-02 | 2.69e-03 | 4.84e-03 |
| UQCRH | 7.20e-03 | 4.34e-02 | 2.26e-03 | 4.69e-03 |
| FADS2 | 3.23e-03 | 1.07e-01 | 2.60e-03 | 4.84e-03 |
| COQ5 | 3.33e-03 | 7.25e-03 | 2.64e-04 | 8.90e-04 |
| ACSM3 | 4.97e-03 | 6.45e-01 | 3.38e-02 | 3.51e-02 |
| THEM2 | 8.29e-03 | 7.32e-02 | 4.04e-03 | 5.82e-03 |
| GSR | 1.23e-02 | 4.87e-02 | 3.94e-03 | 5.82e-03 |
| LSM1 | 5.84e-03 | 2.53e-04 | 2.99e-05 | 2.78e-04 |
| LSM3 | 5.09e-03 | 1.06e-02 | 5.09e-04 | 1.38e-03 |
| LSM7 | 7.24e-03 | 9.26e-02 | 4.47e-03 | 5.82e-03 |
| NSA2 | 9.86e-03 | 1.57e-01 | 9.53e-03 | 1.07e-02 |
| RPL26L1 | 7.23e-03 | 1.47e-01 | 6.92e-03 | 8.12e-03 |
| RPL26 | 3.84e-03 | 1.02e-03 | 6.23e-05 | 3.74e-04 |
| RPL31 | 8.91e-03 | 1.27e-03 | 1.43e-04 | 5.53e-04 |
| RPL34 | 2.70e-03 | 5.74e-04 | 3.09e-05 | 2.78e-04 |
| RPS17 | 7.25e-03 | 8.00e-01 | 7.95e-02 | 7.95e-02 |
| SRP14 | 9.88e-03 | 7.04e-02 | 4.53e-03 | 5.82e-03 |
| ZCCHC17 | 9.80e-03 | 8.47e-02 | 5.32e-03 | 6.53e-03 |
| ARSB | 2.04e-01 | 3.07e-02 | 3.10e-02 | 3.35e-02 |
Significant result at 5% significance level is shown in bold.
Combined P-values using weighted Z-method
Adjusted P-values using Benjamini and Hochberg’s method
DISCUSSION
In the LOTT pharmacogenomics study, we tested two hypotheses and two main results were observed which support the heterogeneity of the effects of oxygen among COPD patients with moderate hypoxemia at rest and/or with exercise. The first outcome was derived from the hypothesis that genetic variation and gene expression may influence the effect of long-term oxygen treatment. The second result was derived from hypothesis that oxygen-responsive genes may be differentially expressed in COPD patients treated with long-term oxygen.
We found an LD block including 7 SNPs on chromosome 5q14.1 was differentially associated with the time-to-event of death or hospitalization based on oxygen treatment and that the expression of the ARSB gene, located near these 7 SNPs, showed similar effects on the LOTT primary outcome. In blood samples from the larger COPDGene study, we confirmed these SNPs influence the levels of ARSB gene expression conditional on supplemental oxygen use. This result show these SNPs could conditionally cis-regulate the expression level of ARSB, thereby extending or shortening the time to death or hospitalization in COPD patients. Previous work supports ARSB as an oxygen-responsive gene. The expression levels of ARSB were found to be decreased in response to hypoxemia in human bronchial or colon epithelial cells and in mouse model experiments [19, 20]. ARSB deficiency led to increased cellular sulfated glycosaminoglycans (GAGs), extracellular acidification rate, and lactate production, and decreased mitochondrial membrane potential, oxygen consumption rate, mitochondrial complex I activity, and activity of NADH and NADPH [20] Mitochondria are highly sensitive to these changes including the loss of mitochondrial membrane potential, resulting in accumulation of reactive oxygen species (ROS), which cause lung inflammation [21, 22]. Additionally, recessive mutations in ARSB are the cause of Mucopolysaccharidosis type VI, which has been associated with pulmonary hypertension [23].
Our findings are consistent with these prior papers in cell and animal models [19, 20]. COPD patients with moderate hypoxemia and risk alleles not treated with oxygen showed downregulated ARSB, which may result in mitochondrial dysfunction, ROS accumulation and ultimately death or hospitalization events. On the other hand, if COPD patients received oxygen therapy, the gene-expression level of ARSB increases, improving survival and reducing hospitalization events. In COPD patients homozygous for major allele at these SNPs, oxygen treatment did not affect ARSB expression, and there is no difference in survival or hospitalization regardless of oxygen therapy.
The second outcome of this study focused on the transcriptome changes affected by oxygen therapy at one year. We found 26 downregulated genes which were enriched in two functional clusters; all of these genes were involved in mitochondrial function. In addition, these genes showed a very strong co-expression pattern and a negative association with resting oxygen saturation. The expression pattern of these mitochondrial-related genes was downregulated at one year in patients with COPD randomized to oxygen and upregulated in patients without oxygen. Six of the observed genes were involved in oxidative phosphorylation, including cytochrome c oxidase subunit 6C (COX6C), ubiquinol-cytochrome c reductase hinge protein (UQCRH) and four NADH dehydrogenase (ubiquinone) subunits of the mitochondrial respiratory chain complex (NDUFA1, NDUFA6, NDUFB3, and NDUFS4). Impaired oxidative phosphorylation is important in multiple aging-related diseases such as Parkinson's disease, Alzheimer's disease, and chronic inflammatory diseases like COPD [24-29]. Upregulation of oxidative phosphorylation can lead to increase in ROS. Recent studies have begun to address mitochondrial dysfunction as a mechanism in COPD pathogenesis [30-32]. In muscle biopsies from COPD patients, cytochrome oxidase activity and mitochondrial gene expression were inversely related to resting PaO2 [28]. In our results, COX6C and co-expressed oxidative phosphorylation genes were upregulated in control subjects, who would be assumed to have chronically lower PaO2 compared to oxygen treated subjects. These effects were confirmed in the COPDGene RNA-seq data analysis. Both hypoxemia and short term use of supplemental oxygen may increase oxidative stress [33]; however, long-term oxygen may actually reduce these effects.
Based on the observations from two different analyses, we speculate that the ARSB gene and 26 mitochondrial related genes may be connected with each other through a mechanism related to mitochondrial dysfunction. Low expression of ARSB and overexpression of oxidative phosphorylation genes in the absence of oxygen treatment may all suggest overproduction of ROS. The co-expression network demonstrated ARSB and three mitochondrial-function related genes zinc finger CCHC-type containing 17 (ZCCHC17), fatty acid desaturase 2 (FADS2), and glutathione-disulfide reductase (GSR) were negatively correlated, whereas ZCCHC17 was positively correlated with other mitochondrial genes.
This study has several limitations. Although this study was based in LOTT, the largest randomized trial of oxygen therapy to date, the sample size was limited for a genetics study. Therefore, we were not able to achieve the strict threshold of p<5e-8 which defines genomewide significance. Instead, we used a more liberal p<1e-5 in the time to event analysis, with validation of results using gene expression data in LOTT. In the absence of another randomized trial for replication, multi-omics data from the COPDGene study was used for validation and eQTL analysis. Since COPDGene is an observational study, there will be confounding between oxygen use and COPD severity [34], which was considered as a covariate in the analyses. Subjects were enrolled in LOTT based on moderate hypoxemia at rest and/or exercise, and were randomized to oxygen full time or with exercise and during sleep, respectively. As in the main LOTT trial [6], both groups were combined in our analysis. Because of the limited sample size, we were not able to stratify subjects to determine if the pharmacogenetic effects were different based on enrollment criteria and oxygen prescription. The genetic and gene expression associations, including possible eQTL effects of SNPs on ARSB expression should be confirmed experimentally in future studies. Despite these limitations, we have found consistent effects in the LOTT study and the independent sample from COPDGene. Furthermore, experimental confirmation of the relationship between ARSB and hypoxemia in cell and animal models suggests that our results are valid [19, 20].
Conclusions
We found that SNPs near the ARSB gene and reduced ARSB gene expression were associated with differential effects of long-term oxygen treatment in COPD patients with moderate hypoxemia. This suggests that ARSB genotypes or gene expression levels could potentially serve as a biomarker to personalize prescription of long-term oxygen in COPD patients with moderate hypoxemia at rest and/or with exercise, although more studies will be required to determine the clinical utility of this biomarker. In the absence of oxygen treatment, mitochondrial genes were upregulated, which supports the importance of mitochondrial dysfunction in the progression of COPD. The results of this study provide a step forward in understanding the molecular effects of supplemental oxygen in COPD and the heterogeneity of response to long-term oxygen treatment.
Supplementary Material
Key message.
SNPs and expression of ARSB are associated with response to long-term oxygen in COPD
The ARSB SNPs were expression quantitative trait loci depending on oxygen therapy
Genes differentially expressed by long-term oxygen were enriched in mitochondrial functions
This suggests a potential biomarker to personalize use of long-term oxygen in COPD
Acknowledgements
Members of the LOTT Research Group and the COPDGene Study investigators are listed in the online supplement.
Funding
Funded by NIH grants R01HL094635, R01HL125583, R01HL130512, U01HL089897, U01HL089856, R01HL124233.
LOTT was supported by NHLBI contracts HHSN268200736183C, HHSN268200736184C, HHSN268200736185C, HHSN268200736186C, HHSN268200736187C, HHSN268200736188C, HHSN268200736189C,
HHSN268200736190C, HHSN268200736191C, HHSN268200736192C,
HHSN268200736193C, HHSN268200736194C, HHSN268200736195C,
HHSN268200736196C, HHSN268200736197C, Y1-HR-7019-01, and Y1-HR-8076-01, and the Centers for Medicare and Medicaid Services.
The COPDGene® project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens and Sunovion.
List of abbreviations
- COPD
Chronic obstructive pulmonary disease
- LOTT
Long-term Oxygen Treatment Trial
- SNP
Single nucleotide polymorphism
- GOLD
Global Initiative for Obstructive Lung Disease
- BMI
Body mass index
- eQTL
Expression quantitative trait loci
- LD
Linkage disequilibrium
- FDR
False discovery rate
- GAGs
Glycosaminoglycans
- ROS
reactive oxygen species
Footnotes
Conflict of interest
Dr. Criner reports grants from Boehringer- Ingelheim, grants from Novartis, grants from Astra Zeneca, grants from Respironics, grants from MedImmune, grants from Actelion, grants from Forest, grants from Pearl, grants from Ikaria, grants from Aeris, grants from PneumRx, grants from Pulmonx, other from HGE Health Care Solutions, Inc, other from Amirall, other from Boehringer- Ingelheim, other from Holaira, outside the submitted work. Dr. Dransfield reports grants from NHLBI, during the conduct of the study; grants from Department of Defense, personal fees and other from Boehringer Ingeheim, personal fees and other from GlaxoSmithKline, other from Novartis, personal fees and other from AstraZeneca, other from Yungjin, other from PneumRx/BTG, other from Pulmonx, personal fees from Genentech, personal fees and other from Boston Scientific, outside the submitted work. Dr. Fuhlbrigge has received grant support from the NHBLI, AHRQ and PCORI related to asthma and COPD studies. She also has received personal fees from GSK, AstraZeneca and Icon Medical Imaging for consulting on investigations in asthma and COPD. Dr. Scholand reports other from Boehringer Ingelheim, other from Genentech, other from Fibrogen, other from Global Blood Therapeutics, outside the submitted work. In addition, Dr. Scholand has a patent Apparatus, Compositions and Methods for Assessment of Chronic Obstructive Pulmonary Disease Progression among Rapid and Slow Decline Conditions issued. Dr. Castaldi reports grants from GSK, personal fees from GSK, outside the submitted work. Dr. Cho reports grants from NHLBI, grants from GSK, during the conduct of the study; grants from Alpha-1 Foundation, outside the submitted work. Dr. Silverman reports grants from NIH, during the conduct of the study; personal fees from Novartis; and grant and travel support from GlaxoSmithKline, outside the submitted work. Dr. Hersh reports grants from National Institutes of Health, during the conduct of the study; personal fees from Mylan, personal fees from AstraZeneca, personal fees from Concert Pharmaceuticals, personal fees from 23andMe, grants from Novartis, grants from Boehringer-Ingelheim, outside the submitted work.
Drs. Seo, Qiu, Bailey, Reilly, Parker, Saferali, Yun, Crapo, Beaty and Mr. Chase report no competing interests.
REFERENCES
- 1.Nocturnal Oxygen Therapy Trial Group (1980) Continuous or nocturnal oxygen therapy in hypoxemic chronic obstructive lung disease: a clinical trial. Ann Intern Med 93: 391–398 [DOI] [PubMed] [Google Scholar]
- 2.Medical Research Council Working Party (1981) Long term domiciliary oxygen therapy in chronic hypoxic cor pulmonale complicating chronic bronchitis and emphysema. Lancet 1: 681–686 [PubMed] [Google Scholar]
- 3.Gorecka D, Gorzelak K, Sliwinski P, Tobiasz M, Zielinski J (1997) Effect of long-term oxygen therapy on survival in patients with chronic obstructive pulmonary disease with moderate hypoxaemia. Thorax 52: 674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaouat A, Weitzenblum E, Kessler R, Charpentier C, Enrhart M, Schott R, Levi-Valensi P, Zielinski J, Delaunois L, Cornudella R, et al. (1999) A randomized trial of nocturnal oxygen therapy in chronic obstructive pulmonary disease patients. Eur RespirJ 14: 1002–1008 [DOI] [PubMed] [Google Scholar]
- 5.Drummond MB, Blackford AL, Benditt JO, Make BJ, Sciurba FC, McCormack MC, Martinez FJ, Fessler HE, Fishman AP, Wise RA (2008) Continuous oxygen use in nonhypoxemic emphysema patients identifies a high-risk subset of patients: retrospective analysis of the National Emphysema Treatment Trial. Chest 134: 497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Long-Term Oxygen Treatment Trial Research G (2016) A randomized trial of long-term oxygen for COPD with moderate desaturation. N Engl J Med 2016: 1617–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald M-LN, Cho MH, Sørheim l-C, Lutz SM, Castaldi PJ, Lomas DA, Coxson HO, Edwards LD, MacNee W, Vestbo J, et al. (2014) Common genetic variants associated with resting oxygenation in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 51: 678–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhandari V, Bizzarro MJ, Shetty A, Zhong X, Page GP, Zhang H, Ment LR, Gruen JR (2006) Familial and genetic susceptibility to major neonatal morbidities in preterm twins. Pediatrics 117: 1901–1906 [DOI] [PubMed] [Google Scholar]
- 9.Cooke RW, Drury JA, Mountford R, Clark D (2004) Genetic polymorphisms and retinopathy of prematurity. Invest Ophthalmol Vis Sci 45: 1712–1715 [DOI] [PubMed] [Google Scholar]
- 10.Bizzarro MJ, Hussain N, Jonsson B, Feng R, Ment LR, Gruen JR, Zhang H, Bhandari V (2006) Genetic susceptibility to retinopathy of prematurity. Pediatrics 118: 1858–1863 [DOI] [PubMed] [Google Scholar]
- 11.Cho HY, Kleeberger SR (2007) Genetic mechanisms of susceptibility to oxidative lung injury in mice. Free Radic Biol Med 42: 433–445 [DOI] [PubMed] [Google Scholar]
- 12.Hopkins RO, Weaver LK, Valentine KJ, Mower C, Churchill S, Carlquist J (2007) Apolipoprotein E genotype and response of carbon monoxide poisoning to hyperbaric oxygen treatment. Am J Respir Crit Care Med 176: 1001–1006 [DOI] [PubMed] [Google Scholar]
- 13.Cho MH, McDonald M-LN, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, DeMeo DL, Sylvia JS, Ziniti J, Laird NM (2014) Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med 2: 214–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker MM, Chase RP, Lamb A, Reyes A, Saferali A, Yun JH, Himes BE, Silverman EK, Hersh CP, Castaldi PJ (2017) RNA sequencing identifies novel non-coding RNA and exon-specific effects associated with cigarette smoking. BMC Med Genomics 10: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo M, Qiu W, Bailey W, Criner G, Dransfield M, Fuhlbrigge A, Reilly J, Scholand M, Silverman E, Hersh C (2018) Genomics and Response to Long-Term Oxygen Treatment in COPD: The LOTT Pharmacogenomics Ancillary Study[abstract], Am J Respir Crit Care Med 2018;197:A743. [Google Scholar]
- 16.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo M, Kim K, Yoon J, Jeong JY, Lee H-J, Cho S, Kim H (2016) RNA-seq analysis for detecting quantitative trait-associated genes. Sci Rep 6: 24375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS (2001) Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med 163: 1256–1276 [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharyya S, Tobacman JK (2012) Hypoxia reduces arylsulfatase B activity and silencing arylsulfatase B replicates and mediates the effects of hypoxia. PloS one 7: e33250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharyya S, Feferman L, Tobacman JK (2016) Restriction of aerobic metabolism by acquired or innate arylsulfatase B deficiency: A new approach to the Warburg effect. Sci Rep 6: 32885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Wang X, Hu D (2017) Mitochondrial alterations during oxidative stress in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis 12: 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerner CA, Sundar IK, Rahman I (2016) Mitochondrial redox system, dynamics, and dysfunction in lung inflammaging and COPD. Int J Biochem Cell Biol 81: 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golda A, Jurecka A, Gajda K, Tylki-Szymahska A, Lalik A (2015) Human pulmonary artery endothelial cells in the model of mucopolysaccharidosis VI present a prohypertensive phenotype. Mol Genet Metab Rep 3: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramaniam SR, Chesselet M-F (2013) Mitochondrial dysfunction and oxidative stress in Parkinson's disease. Prog Neurobiol 106: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manczak M, Park BS, Jung Y, Reddy PH (2004) Differential expression of oxidative phosphorylation genes in patients with Alzheimer′s disease. Neuromolecular Med 5: 147–162 [DOI] [PubMed] [Google Scholar]
- 26.Cui H, Kong Y, Zhang H (2012) Oxidative stress, mitochondrial dysfunction, and aging. J Signal Transduct 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang M-J, Shadel GS (2016) A mitochondrial perspective of chronic obstructive pulmonary disease pathogenesis. Tuberc Respir Dis 79: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauleda J, Garcia-Palmer F, Wiesner RJ, Tarraga S, Harting I, Tomás P, Gomez C, Saus C, Palou A, Agusti AG (1998) Cytochrome oxidase activity and mitochondrial gene expression in skeletal muscle of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 157: 1413–1417 [DOI] [PubMed] [Google Scholar]
- 29.Granata S, Zaza G, Simone S, Villani G, Latorre D, Pontrelli P, Carella M, Schena FP, Grandaliano G, Pertosa G (2009) Mitochondrial dysregulation and oxidative stress in patients with chronic kidney disease. BMC Genomics 10: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cloonan SM, Glass K, Laucho-Contreras ME, Bhashyam AR, Cervo M, Pabón MA, Konrad C, Polverino F, Siempos II, Perez E (2016) Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat Med 22: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angara S, Bhandari V (2013) Targeting mitochondrial dysfunction in lung diseases: emphasis on mitophagy. Front Physiol 4: 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue L, Yao H (2016) Mitochondrial dysfunction in inflammatory responses and cellular senescence: pathogenesis and pharmacological targets for chronic lung diseases. Br J Pharmacol 173: 2305–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troosters T (2004) Oxygen: the good, the bad, and the necessary… Thorax 59: 1005–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim DK, Jacobson FL, Washko GR, Casaburi R, Make BJ, Crapo JD, Silverman EK, Hersh CP (2011) Clinical and radiographic correlates of hypoxemia and oxygen therapy in the COPDGene study. Respir Med 105: 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.