1. Introduction
One of the major neuropeptides of the hypothalamic-pituitary-adrenal (HPA) axis during the stress adaptation response is arginine vasopressin (AVP) / arginine vasotocin (AVT), the latter a non-mammalian homologue of AVP (Antoni, 1993; Lightman, 2008; Mason, Hassan, Chacko, & Thompson, 2002; Murat et al., 2012; Nagarajan, Tessaro, Kang, & Kuenzel, 2014; Gilchriest et al., 2000). The sustained adaptation of the HPA axis appears to occur with a reduction of corticotropin-releasing hormone (CRH) and a concomitant augmentation of AVP during chronic stress (Lightman, 2008; Goncharova, 2013). The effect(s) of AVP/AVT on adrenocorticotropic hormone (ACTH) secretion from anterior pituitary (AP) corticotropic cells (corticotropes) has been investigated and the V1b receptor (V1bR) is the most abundant and plays a key role during acute and chronic stress in mammals(Autelitano, Lundblad, Blum, & Robert, 1989; Hernando, Schoots, Lolait, & Burbach, 2001; Sugimoto et al., 1994; Koshimizu et al., 2012).
In birds, four subtype vasotocin receptors (VT1R-VT4R) have been identified for the ligand AVT (Cornett, Kang, & Kuenzel, 2013). VT2R, the avian homologue of the mammalian vasopressin 1b receptor, has been shown located on corticotropes in the AP (Jurkevich, Berghman, Cornett, & Kuenzel, 2005; Jurkevich, Berghman, Cornett, & Kuenzel, 2008; Selvam et al., 2013). The VT4R, avian homologue of the mammalian vasopressin 1a receptor, has likewise been shown to occur in corticotropes and both avian VTRs have shown significant changes in gene expression following a psychogenic stressor, immobilization (Selvam et al., 2013; Kang & Kuenzel, 2014; Kuenzel, Kang, & Jurkevich, 2013; Jayanthi et al., 2014). Due to similar function and significant sequence homology to the mammalian V1b and V1a receptors, the VT2R and VT4R were therefore renamed the avian V1bR and V1aR, respectively (Kuenzel et al., 2016). When chickens were subjected to immobilization stress or nutritional stress, a significant change in AP pro-opiomelanocortin (POMC) heteronuclear (hn) RNA occurred (Kang & Kuenzel, 2014; Jayanthi et al., 2014; Nagarajan, Kang, & Kuenzel, 2017). The subsequent secretion of ACTH activated adrenal production of the avian stress hormone corticosterone (CORT) (Selvam et al., 2013; Kuenzel, Kang, & Jurkevich, 2013). In addition, the avian V1aR has recently been shown to play a role(s) in the neuroendocrine regulation of food intake in the brain of chickens, Gallus gallus (Jayanthi et al., 2014; Kuenzel et al., 2016), suggesting that the avian V1aR in the brain not only functions in the neuroendocrine axis in regulating the stress response but may also function to suppress feeding by partially inhibiting the full orexigenic response initiated by neuropeptide Y (NPY).
The relationship between structure and functional activity of the AVP/AVT receptors was found in several studies using three-dimensional (3D) homology modelling techniques, chimeric receptor approaches and site-directed mutagenesis (Acharjee et al., 2004; Cho et al., 2007; Cotte et al., 2000; Czaplewski, Kazmierkiewicz, & Ciarkowski, 1998a; Czaplewski, Kazmierkiewicz, & Ciarkowski, 1998b; Hausmann et al., 1996; Mahlmann et al., 1994; Mouillac et al., 1995a). Our recent study, using in silico 3D modelling/docking analyses of the chicken V1aR (cV1aR) and an in vitro chicken AP cell culture study (Jayanthi et al., 2014), successfully identified a mammalian V1aR antagonist SR-49059 as a most effective antagonist of the avian V1aR compared to other candidate blockers including the Manning compound (Baran, Sklar, & Adkins-Regan, 2016; Goodson & Evans, 2004).
In this study, we used 3D homology model of the cV1aR and chicken V1bR (cV1bR), built using a common modelling template and performed docking analyses with a series of known effective mammalian antagonists. An in vitro primary AP cell culture was used to examine the effect(s) of antagonists on the AP stress response and the specificity of selected antagonists to attenuate expression of the cV1aR and cV1bR gene.
2. Material and Methods
2.1. Ethics Statement
The care and experimental use of animals were approved by the University of Arkansas Institutional Animal Care and Use Committee (Protocol number 16043). Animals were maintained according to a standard management program at the Poultry Farm, University of Arkansas. The procedures for animal management, and sampling followed the standard operation protocols of our laboratory.
2.2. Animals and Materials
One-day old male chicks (Cobb 500) were obtained from a commercial hatchery and raised in an environmentally controlled room and fed a standard starter diet ad libitum. Birds were maintained on continuous light (L) with no dark (D) periods (LD 24:0) for 3 days, and then moved to a long-day photoperiod of LD 16:8. The selected mammalian V1bR antagonists (SSR- 149415, Manning compound, L-368899) and V1aR antagonist (SR-49059) were purchased from Sigma-Aldrich (St. Louis, MO, USA), and AVT and CRH were purchased from Bachem Americas Inc. (Torrance, CA, USA).
2.3. Homology modelling and molecular docking of cV1aR and cV1bR with ligands
Amino acid sequences of the cV1aR (419 AAs) and cV1bR (425 AAs) were obtained from the Uni-Prot database (ID: A8CWP8, Q90YN1). Basic Local Alignment Search Tool for Protein (BlastP) and FUGUE algorithm search tools were employed to identify the suitable template candidate available in the Protein Data Bank (PDB) for building the 3D homology model (Altschul et al., 1997; Berman et al., 2000; Shi, Blundell, & Mizuguchi, 2001). Due to a high percentage of cV1bR sequence identity (~50%) to cV1aR, similar hits were considered as potential templates. Nociceptin/Orphanin FQ (N/OFQ) Opioid receptor (PDB ID: 4EA3) was selected as a template for model building. Similarities and differences between cV1aR and cV1bR sequences were evaluated by subjecting the sequences to EMBOSS-Needle pairwise alignment tool based on the Needleman-Wunsch algorithm available at EMBL-EBI portal (Needleman & Wunsch, 1970). 3D homology models of cV1aR and cV1bR were constructed by using the Composer tool available in the Bioploymer module of the SYBYL-X suite. The 3D homology model was subjected to multi-step energy minimization protocol for obtaining a stable conformation of the receptor molecule using Tripos force field steepest descent followed with Powell algorithm with a conjugate gradient. In the first step of the minimization process, hydrogen atoms were uniformly added to the entire receptor molecule. Positions of the hydrogen atom were refined keeping the receptor molecule in rigid conformation. Geometric optimization of the entire molecule was performed with total flexibility of the receptor molecule. Simulated annealing of the modeled receptor structure was carried with periodic boundary conditions for a total time of 1 nano-second with an increment of 20 pico-second interval. Stereo-chemical quality of the model was verified by submitting the homology model to protein structure validation software suite (PSVS) to visualize the backbone dihedral angles (phi and psi) of amino acids as well the distribution of the amino acid data points on a Ramachandran plot (Bhattacharya, Tejero, & Montelione, 2007). All ligand molecules (agonists and antagonists) were built using the sketch tool available on the SYBYL-X suite. Antagonists were screened using the Autodock Vina molecular docking program using the previously published protocols (Kuenzel et al., 2016; Trott & Olson, 2010). The best docking conformation, with the lowest docking score (ΔG binding), was selected for ranking. Protein-ligand conformations, including bond lengths and hydrogen bonds, were analyzed and presented using the PyMOL visualization software (http://pymol.sourceforge.net).
2.4. Primary AP cell culture
Primary AP cells from 5–6 week old male chickens were obtained using a modified trypsin/neuraminidase procedure, as described previously (Jayanthi et al., 2014; Fehrer, Silsby, Behnke & el Halawani, 1985; Kang et al., 2004). Briefly, birds were killed by cervical dislocation. The AP gland was quickly isolated from the head region and placed in a Krebs—Ringer bicarbonate (KRB; pH7.4) buffer solution supplemented with amino acids (Eagle’s minimum essential amino acids; Difco, Walkersville, MD), 0.3mg/ml L-glutamine (Sigma Chemical, St. Louis, MO), 2.5mg/ml α-D(+) glucose (grade III Sigma), 3mg/ml bovine serum albumin (BSA, fraction V; Sigma), and 0.1mg/ml gentamycin sulfate (Sigma). The pituitary fragments were enzymatically digested with 1mg/ml trypsin (bovine pancreas type III; Sigma) and 2 μg/ml deoxyribonuclease I (DNase I, type I; Sigma) for 15min at 37 °C in a shaking water bath. Dispersed AP cells were maintained at 39 °C in a humidified 5% CO2 / 95% air incubator for 4 days. Cell viability (85–94%) was determined by trypan blue dye exclusion and quantified using a hemocytometer. A half-million dispersed AP cells (0.5 × 105) were treated with AVT/CRH (1.0/0.1nM; Bachem Americas Inc. Torrance, CA USA) for 6 hours as previously described as an in vitro stressor (Kang & Kuenzel, 2014; Jayanthi et al., 2014), because the combination of AVT/CRH (1.0/0.1 nM) for 6 hours was found to exert maximum effect to stimulate POMC hnRNA expression as a stress marker gene. A critical reason that we used the AVT/CRH combination was to use the most biologically relevant in vitro model based on our previous results (Kang & Kuenzel, 2014; Jayanthi et al., 2014). Two selected antagonists, SSR-149415 and L-368899, for cV1bR were pre-treated 30 min before AVT/CRH treatment with different concentrations (1, 10, and 100 pM and 1 nM). The selected dose (10 pM) of cV1bR antagonists (SSR-149415 and L-368899) and cV1aR antagonist (SR-49059) and Manning compound (Table 1 in Jayanthi et al., 2014) was pretreated to verify the effect of co-treatment of combination of V1aR and V1bR antagonists in the AP stress response. At the end of the incubation period, AP cells were collected by centrifugation, washed with 2 ml of phosphate-buffered saline (PBS), and subsequently dissolved in 1 ml Trizol® reagent (Life Technologies, Palo Alto, CA, USA) and frozen at −80°C.
Table 1.
Comparison of the interacting residues in the cV1aR and cV1bR with ligands.
| Receptor | Ligand | Interacting Residues on Receptors |
|---|---|---|
| cVlaR | Vasotocin | Q115, L116, K135, Q138, Q192, W211, W226, F310, Q313, T332 |
| SR-49059 | L116, K135, Q138, Q192, W211 F310, Q313, S316, T332, A336 | |
| SSR-149415 | L116, V120, K135, Q138, F310, Q313, T332 | |
| Manning | L116, K135, Q138, F309, Q313, T332 | |
| cVlbR | Vasotocin | R109, K121, Q124, C196, W197, W212, F310, Q314, F333 |
| SR-49059 | K121, Q124, C196, W197, F310, Q314, S317, D330, M337 | |
| SSR-149415 | K121, Q124, C196, W197, F310, Q314, S317, D330, F333, M337 | |
| Manning | E47, R109, K121, Q124, Q178, D195, C196, W197, A198, W212, F310, Q314, S317, D322, F333, T334, M337 |
2.5. Real-time RT-PCR for cV1aR, cV1bR , cCRH-R1, and cCRH-R2 mRNA and POMC hnRNA
Total RNA was extracted from cultured AP cells using TRIzol® reagent (Invitrogen Life Technologies, Palo Alto, CA, USA) following the DNase I treatment (Invitrogen, Carlsbad, CA, USA, 1 U/μg RNA) and purification of total RNA by RNeasy mini kit (Qiagen, Valencia, CA, USA). The RNA quality and quantity were determined using agarose gel electrophoresis and NanoDrop 1000 (Thermo Scientific, Wilmington, DE, USA). Two micrograms of total RNA were converted into cDNA with oligo (dT)16 primer and SuperScript III reverse transcriptase (Invitrogen Grand Island, NY, USA), as previously described (Kang & Kuenzel, 2014; Kuenzel, Kang, & Jurkevich, 2013; Jayanthi et al., 2014). A portion (1 μl) of the cDNA was subjected to a quantitative real-time PCR (qRT-PCR) using an ABI prism 7500 system (Applied Biosystems LLC, Foster, CA, USA) with Power SYBR Green PCR Master Mix (Invitrogen Grand Island, NY, USA). Real-time qRT-PCR was performed using 1 cycle at 95 °C for 5 min, 40 cycles at 95 °C for 30 s, 56–60 °C for 1 min [annealing temperature: 56 °C for V1aR: 54 °C for V1bR: 59 °C for POMC hnRNA] and 1 cycle at 72 °C for 10 min. Chicken glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β-actin were used as internal controls (annealing temperature: GAPDH − 58 − 60 °C; β-actin − 54–56 °C). Dissociation curves were constructed at the end of amplification for validating the quality of the data. All qRT-PCR experiments were performed in triplicate and the values of average cycle threshold (Ct) were determined and Delta-Ct scores for gene transcripts in each sample were normalized using Delta-Ct scores for GAPDH / β-actin and expressed as the fold change in gene expression using the equation, 2-ΔΔCt. The NCBI accession numbers, PCR product size and primer sequences used in the present study were: cV1aR [NM001110438, 137 bp (V1aR-F: 5-GGT TGC AGT GTT TTC AGA GTC G-3; V1aR-R: 5-CAA GAT CCG CAC CGT CAA G-3)], cV1bR [AY008272, 129 bp (V1bR-F: 5-CTT CAG CAT GCA GAT GTG GT-3; V1bR-R: 5-AAC ATG TAG ATG CAG GGG TTG-3)], CRH-R1 [NM_204321, 141 bp (CRH-R1-F: 5-CCCTGCCCCGAGTATTTCTA-3; CRH-R1-R: 5- CTTGCTCCTCTTCTCCTCACTG-3)], CRH-R2 [NM_204454, 129 bp (CRH-R2-F: 5- GCAGTCTTTTCAGGGTTTCTTTG-3; CRH-R2-R: 5-CGGTGCCATCTTTTCCTGG-3)], hnRNA of POMC [NM_001031098, 141bp (hnPOMC-F: Intro 1 forward primer 5- ATT TTA CGC TTC CAT TTC GC-3; hnPOMC-R: Exon 2 reverse primer 5- AAT GGC TCA TCA CGT ACT TGC-3)], GAPDH [NM204305, 128 bp (GAPDH-F: 5-CTT TGG CAT TGT GGA GGG TC-3; GAPDH-R: 5-ACG CTG GGA TGA TGT TCT GG-3)], β-actin [L08165, 158 bp (Actin-F: 5- CAC AAT GTA CCC TGG CAT TG-3; Actin-R: 5-ACA TCT GCT GGA AGG TGG AC-3)].
2.6. Statistical Analyses
Statistical analyses were performed using JMP® 11.0 (SAS Institute Inc., NC). A normal distribution was first tested and subsequently differences among the groups were analyzed using one-way analysis of variance (ANOVA) followed by mean comparison using the Tukey’s HSD test at a significance level of p < 0.05. Multiple comparisons of group means by Tukey’s HSD test were used to evaluate relative changes of each VT receptor gene expression and POMC hnRNA. Data are presented as the mean ± SEM. A probability level of p < 0.05 was considered statistically significant.
3. Results
3.1. Homology modelling and validation of the 3D structural models of cV1aR and cV1bR
N/OFQ Opioid receptor was identified as the best hit to be used as a template for building the homology model for the cV1bR (Jayanthi et al., 2014). Template to target sequence identity was 26% with a query coverage of 72%. This template was considered to be satisfactory to generate a viable homology model. Overlay of the homology models of the template and the target yielded a root-mean-square deviation (RMSD) value of 0.72Å which is well within the agreement of ≤ 2Å difference. Similarly, due to the significant percentage of sequence identity between the cV1aR and cV1bR (48%), a final 3D homology model was built using the same template. The homology models of cV1aR and cV1bR were built using the template 4EA3, and showed extremely high structural similarities in the seven transmembrane segments between the two models (Fig 1A and 1B). Validation of the model by the Protein Structure Validation Suite program (http://psvs-1.5-dev.nesg.org) showed that the structure of the cV1bR built satisfied all the necessary criteria for performing the docking analysis.
Figure 1. Three-dimensional (3D) homology structures and docking of vasotocin onto the modelled structures of the cV1aR and cV1bR.
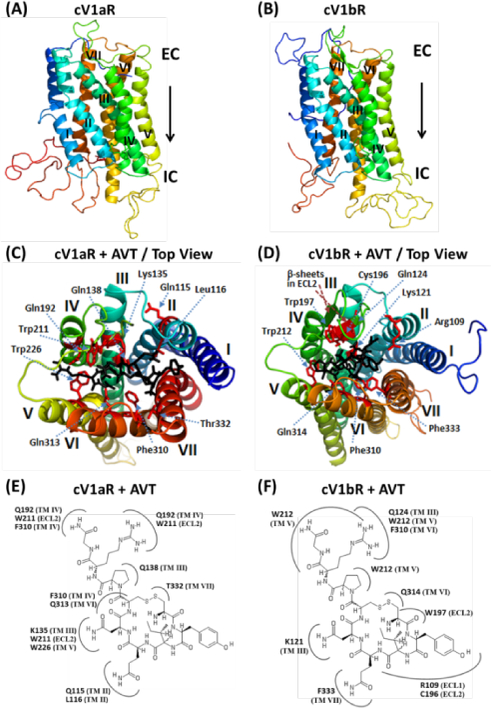
(A) and (B). Homology models of cV1aR and cV1bR built using the template opioid receptor (PDB ID: 4EA3). Seven transmembrane helices (TM-I-VII), each shown with a different spectral color are labelled with Roman numbers. EC – Extracellular side and IC – intracellular side of the receptor. (C) and (D). Top views of the binding amino acid residues (shown as sticks: magenta) of the modelled cV1aR and cV1bR structures with vasotocin (black). The residues are numbered according to their position in the primary sequence of the cV1aR and cV1bR (see Fig 3). The seven TM helices are labelled in different colors. Two-dimensional schematic views showing interactions of AVT and amino acid residues of the cV1aR and cV1bR structural models (E, F). All of the residues potentially interacting with the different parts of AVT are shown. Numbering of the residues and of the each TM is equivalent to that used in Fig 3. ECL- extracellular loop.
3.2. Differential ligand interacting residues on the VT receptors and binding affinities
Docking analyses of the agonist AVT with cV1aR and cV1bR models showed unique interacting 10 and 9 amino acid resides on cV1aR and cV1bR models, respectively (Table 1, Figs 1C,E and 1D,F). Based on the literature and also from our previous study (Jayanthi et al., 2014), a list of potential mammalian V1bR antagonists was screened against the cV1aR and cV1bR structures. Nine ligands with the highest binding affinity are shown in Table 2 and 3. From these ligands, two antagonists, SR-49059 and SSR-149415, were compared in the pose conformations and schematic views of docking models with cV1aR and cV1bR and summarized (Fig 2). As observed in a cV1aR structure, built using 4EA3 as a template, the binding pocket for agonists and antagonists was observed to be located closer to the extracellular side of the receptor (Fig 3). A closer look at the binding sites of the ligands with the cV1aR and cV1bR showed common and unique amino acid residues on the receptors that interact with the ligand (Table 1, Fig 3). Compound SR-49059 was found to have the highest binding affinity with cV1aR (−9.0 kcal/mol) (Table 2, Fig 4A). Molecular docking results were evaluated based on ligand-protein interactions including properties such as the steric, electrostatic and the intramolecular energy. Some of the screened ligands showed strong binding affinity to the cV1aR and cV1bR as evidenced by the high negative ΔGbinding values (Table 2 and 3). For cV1bR, the Manning compound showed the highest binding affinity with a binding energy of −9.2kcal/mol and ranked at the top among the list of candidate compounds screened (Table 3, Fig 4A). For cV1aR, the Manning compound showed the binding affinity with a binding energy of −7.4 kcal/mol (Table 2).
Table 2.
List of ligands and their in silico binding affinity to the cV1aR.
| SL. No. | Ligand | ΔGbinding (kcal/mol) |
|---|---|---|
| 1 | SR-49059((2S)-l-[(2R,3S)-5-chloro-3-(2-chlorophenyl)-l-3,4-dimethoxyphenyl)sulfonyl-3-hydroxy-2H-indole-2-carbonyl]pyrrolidine-2-carboxamide) | −9.0 |
| 2 | SSR-149415;((2S,4R)-l-[5-Chloro-l-[(2,4-dimethoxyphenyl)sulfonyl]-3-(2-methoxy-phenyl)-2- oxo-2,3-dihydro-lH-indol-3-yll-4-hydroxy-N,N-dimethyl-2-pyrrolidine carboxamide) | −8.8 |
| 3 | 3rd Compound of Table 10 in Fabio et al., 2010; Sanofi compound derivative by TaisltoiPharm. Co. | −8.8 |
| 4 | Compound 2h of Table 9 in Fabio et al., 2010; l-(oxo-4H-quinazolin-3-yl) acetamide derivative | −8.5 |
| 5 | Compound 12j in Napier et al., 2011; 2-(4-oxo-2-aryl-quinazolin-3(4H)-yl) acetamide derivative | −8.3 |
| 6 | 4th Compound of Table 10 in Fabio et al., 2010; Sanofi compound derivative by Taisho Pharm. Co. | −8.0 |
| 7 | Compound 30P of Table 3 in Scott et al., 2009; Tetrahydroquinoline sulfonamide derivative | −7.9 |
| 8 | L-368,899; l-((7,7-Dimethyl-2(S)-(2(S)-amino-4(methylsulfonyl)butyramido) bicyclo[2,2,l] heptan-l(S)-yl)methylsulfonyl)-4-(2-methylphenyl)piperazinehydrochloride | −7.6 |
| 9 | Manning compound; (d(CH2)51,Tyr(Me)2,Arg8)-Vasopressin | −7.4 |
| 10 | [Arg8]-Vasotocin | −7.2 |
The more negative score represent the higher binding affinity
Table 3.
List of ligands and their in silico binding affinity to the cV1bR.
| SL. No. | Ligand | ΔGbinding (kcal/mol) |
|---|---|---|
| 1 | Manning compound; (d(CH2)51(Me)2,Arg8)-Vasopressin | −9.2 |
| 2 | 3rd Compound of Table 10 in Fabio et al., 2010; Sanofi compound derivative by Taisho Pharm. Co. | −8.9 |
| 3 | Compound 2h of Table 9 in Fabio et al., 2010; l-(oxo-4H-quinazolin-3-yl) acetamide derivative | −8.8 |
| 4 | 4th Compound of Table 10 in Fabio et al., 2010; Sanofi compound derivative by Taisho Pharm. Co. | −8.3 |
| 5 | Compound 12j in Napier et al., 2011; 2-(4-oxo-2-aryl-quinazolin-3(4H)-yl) acetamide derivative | −8.3 |
| 6 | SR-49059((2S)-l-[(2R,3S)-5-chloro-3-(2-chlorophenyl)-l-3,4-dimethoxyphenyl)sulfonyl-3-hydroxy-2H-indole-2-carbonyl]pyrrolidine-2-carboxamide) | −7.9 |
| 7 | SSR-149415;((2S,4R)-l-[5-Chloro-l-[(2,4-dimethoxyphenyl)sulfonyl]-3-(2-methoxy-phenyl)-2- oxo-2,3-dihydro-lH-indol-3-yll-4-hydroxy-N,N-dimethyl-2-pyrrolidine carboxamide) | −7.9 |
| 8 | Compound 30P of Table 3 in Scott et al., 2009; Tetrahydroquinoline sulfonamide derivative | −7.6 |
| 9 | L-368,899; l-((7,7-Dimethyl-2(S)-(2(S)-amino-4(methylsulfonyl)butyramido) bicyclo[2,2,l] heptan-l(S)-yl)methylsulfonyl)-4-(2-methylphenyl)piperazine hydrochloride | −7.3 |
| 10 | [Arg8]-Vasotocin | −7.3 |
The more negative score represent the higher binding affinity
Figure 2. Close-up and two-dimensional (2D) schematic views of the interacting amino acid residues of cV1aR and cV1bR with SR-49059 and SSR-149415.
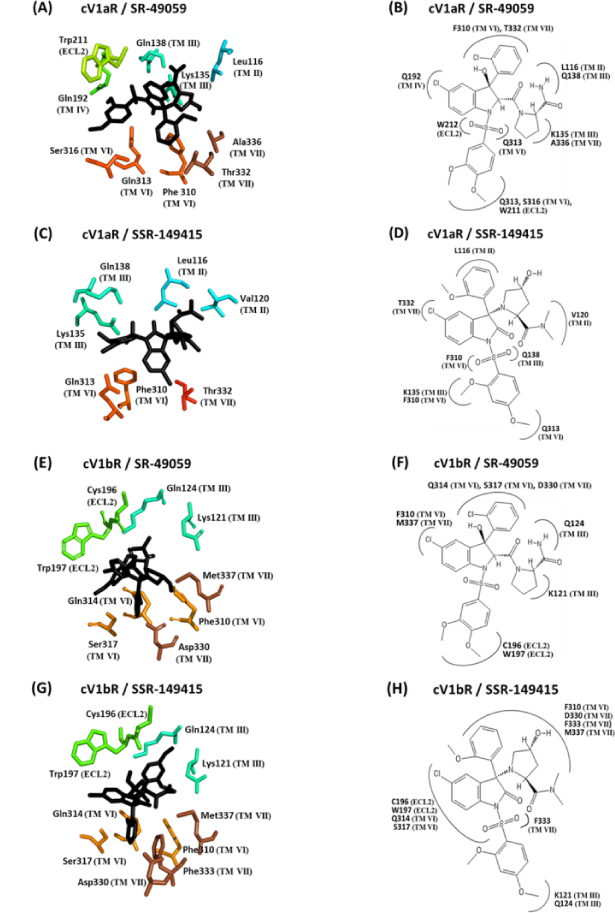
(A), (C), (E), and (G); Antagonists are shown as sticks in black color and the amino acid residues involved in the interaction are shown in different colors according to the transmembrane helices (TM) or extracellular loop (ECL). The residues are numbered according to their position in the primary sequence (see Fig 3). The seven TM helices are shown in different colors. (B), (D), (F), and (H); 2D schematic view of the interaction models with antagonists. All of the amino acid residues potentially interacting with the different parts of the antagonists are shown. Numbering of the residues and of the TM is equivalent to that used in Fig 3.
Figure 3. Two-dimensional schematic diagrams of cV1aR (A) and cV1bR (B) showing the amino acid residues involved in interactions with agonist and antagonists.
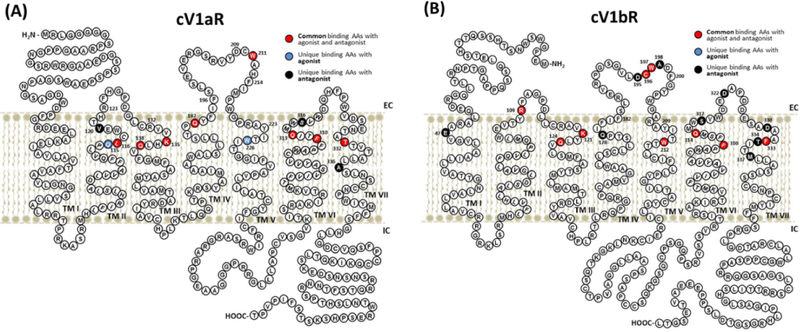
Common binding sites with agonist and antagonists in the modelled structures are shown in red and unique binding AAs with agonist and antagonists are shown in blue and dark purple coloration, respectively.
Figure 4. (A). Comparison of binding affinity of SR-49059, SSR-149415, and the Manning compound with cV1aR and cV1bR.
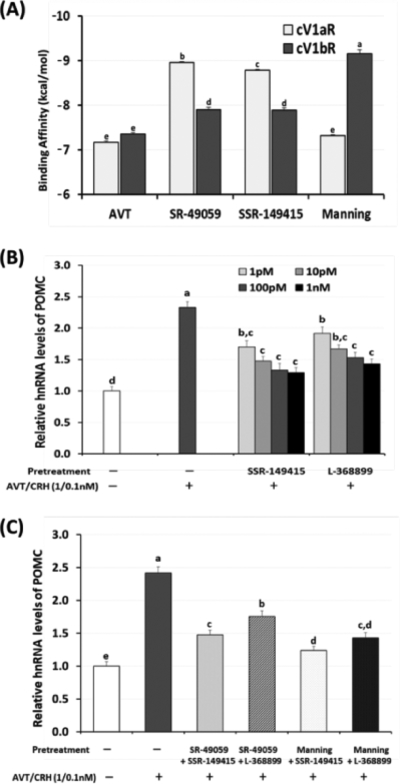
Docking analysis of ligands (agonist AVT and three VT antagonists) with the modeled cV1aR and cV1bR structures was performed to compare the binding affinity of each ligand to the receptors. Data (mean ± SEM) were obtained from twenty repetitions and are presented as the negative values of ΔGbinding (kcal/mol). (B). Dose-dependent inhibiting effect of two selected cV1bR antagonists, SSR-149415 and L-368899 on POMC hnRNA expression in AP cells following AVT/CRH treatments (1.0/0.1 nM, 6 hrs). The selected antagonists were applied 30 min before AVT/CRH treatments using 1, 10, 100, and 1000 pM concentrations to the AP cells. Total RNAs from AP cells were used for real-time RT-PCR of POMC hnRNA expression. (C). Combined inhibitory effects of selected antagonists on POMC hnRNA expression in AP cells. Selected antagonists for cV1aR and cV1bR (SR-49059/SSR-149415, SR-49059/L-368899, Manning/SSR-149415, and Manning/L-368899) were tested for 30 min followed by AVT/CRH (1.0 / 0.1 nM) administration. Six hours later, total RNAs were extracted and real time RT-PCR was performed for POMC hnRNA expression. Data were set as the fold changes of relative expression levels using the ΔΔCt method with GAPDH and β-actin as internal controls.
The binding affinities of SR-49059 and SSR-149415 with the cV1aR were higher than that observed with the cV1bR (p<0.05, Fig 3A). The binding affinity of the Manning compound with the cV1bR was higher than that observed with the cV1aR (p <0.05, Fig 4A). There was no significant difference between the binding affinities of the Manning compound and AVT with the cV1aR (p>0.05, Fig 4A).
3.3. Antagonistic effect of the selected cV1bR antagonists and the combined effects of cV1aR and cV1bR antagonists on the POMC hnRNA expression in AP cells
In primary AP cells, the dose-dependent antagonistic effects of two selected cV1bR antagonists, SSR-149415 and L-368899, were investigated (Fig 4B) as these two compounds were available commercially. Results showed that pre-treatment of AP cells with antagonists (1 pM) 30 min before AVT/CRH stimulation (1.0/0.1 nM), significantly reduced POMC hnRNA expression about 19 % (SSR-149415) and 10 % (L-368899) compared to AVT/CRH induced POMC hnRNA expression (Fig 4B). In higher dose (10, 100 pM and 1 nM) treatments, the two compounds inhibited POMC hnRNA expression resulting in a maximum decrease of 42 % (SSR- 149415, 1 nM) and 33 % (L-368899, 1 nM) compared to positive controls (AVT/CRH induced POMC hnRNA expression levels set at 100 %). The antagonistic effect of SSR-149415 pretreatment in different doses was not significantly different, but the L-368899 treatment showed significant differences at doses of 100 pM and greater.
Combined effects of cV1aR and cV1bR antagonists, therefore, were tested in 10 pM concentration of two compounds with four different combinations of V1aR and V1bR antagonists (SR-49059/SSR-149415, SR-49059/L-368899, Manning/SSR-149415, and Manning/L-368899). Co-treatment of SR-49059 with two selected V1bR antagonists, SSR-149415 or L-368899, decreased POMC hnRNA expression by 39 % or 27 %, respectively, compared to controls stimulated by AVT/CRH (positive controls set at 100 %). Co-treatment of the Manning compound with SSR-149415 or L-368899 decreased POMC hnRNA expression by 49 % or 41 %, respectively, compared to positive controls (Fig 4C).
3.4. Differential effect of selected antagonists on cV1aR, cV1bR, cCRH-R1, and cCRH-R2 gene expression in AP cells
Based upon their high binding affinities for the cV1aR and cV1bR (Table 2 and 3), the Manning compound was selected as a potential avian V1bR antagonist, and tested its effect to change gene expression of their respective target receptors with SR-49059, a selective cV1aR antagonist, (Jayanthi et al., 2014). Co-treatment of AVT / CRH (1.0/0.1 nM) was validated in our previous studies as an optimized in vitro stress in the primary AP cells (Jayanthi et al., 2014; Kang & Kuenzel, 2014). Stimulation of AP cells with AVT/CRH (10.0/0.1 nM, 6 hours) resulted in decreased expression levels of both the cV1aR and cV1bR mRNA (Fig 5A).
Figure 5. (A). Antagonistic effects of SR-49059 and Manning compound on cV1aR and cV1bR mRNA expression.
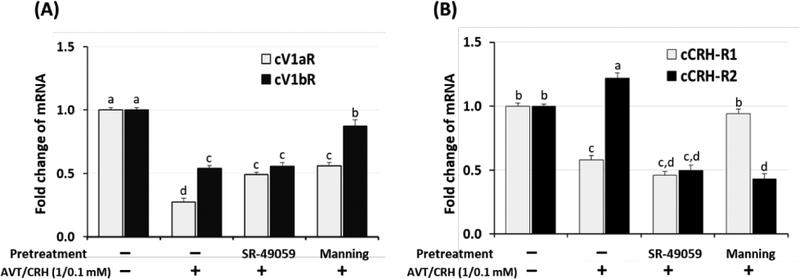
Dispersed chicken primary AP cells (0.5 × 106) were pretreated with SR-49059 (100 pM) or the Manning compound (100 pM) for 30 min followed by AVT/CRH (1.0 / 0.1 nM) treatments. Six hours later, total RNAs were extracted and real time RT-PCR was performed for cV1aR and cV1bR mRNA expression. Data were set as the fold changes of relative expression levels using the ΔΔCt method with GAPDH and β-actin as internal controls. Different lower-case letters above the bars denote significant differences (P<0.05) among groups. (B). Antagonistic effects of SR-49059 and Manning compound on cCRH-R1 and cCRH-R2 mRNA expression. Dispersed chicken primary AP cells (0.5 × 106) were pretreated with SR-49059 (100 pM) or Manning (100 pM) for 30 min followed by AVT/CRH (1.0 / 0.1 nM) treatments. Six hours later, total RNAs were extracted and real time RT-PCR was performed for cCRH-R1 and cCRH-R2 mRNA expression. Data were set as the fold changes of relative expression levels using the ΔΔCt method with GAPDH and β-actin as internal controls. Different lowercase letters above the bars denote significant differences (P<0.05) among groups.
The expression level of the cV1aR mRNA decreased to 27 % compared to controls (p < 0.05) by in vitro AP stress treatments (AVT/CRH). This result was significantly different from the decrease in cV1bR mRNA expression, shown to be 54 % compared to controls (p < 0.05). Pre-treatment of SR-49059 before AVT/CRH stimulation changed the down-regulated expression of cV1aR mRNA from 27 % to 49 % compared to controls (p < 0.05). However, pre-treatment of SR-49059 did not significantly change the down-regulated expression level of the cV1bR mRNA. Pre-treatment of the Manning compound significantly changed the down-regulation of both the cV1aR and cV1bR mRNA expression. Down-regulation of cV1aR and cV1bR mRNA expression levels changed from 27 % to 56 % and from 54 % to 87 %, respectively, compared to controls by pretreatment with the Manning compound.
The expression level of the cCRH-R1 mRNA decreased to 58 % and cCRH-R2 mRNA increased to 122 % compared to controls (p < 0.05) by in vitro AP stress treatments (AVT/CRH) (Fig. 5B). Pre-treatment of SR-49059 did not significantly change the down-regulated expression of the cCRH-R1, but significantly inhibited the cCRH-R2 expression from 122 % to 50 % compared to controls. Pre-treatment of the Manning compound significantly changed the cCRH-R1 expression from 58 % to 94 % and cCRH-R2 expression from 122 % to 43 % compared to controls.
4. Discussion
In the present study, we identified the most effective antagonists for the avian V1aR and V1bR using in silico molecular docking / binding studies. Using chicken AP cells, the in vitro effects of the in silico selected antagonists were tested and the effect of co-treatment with the previously found avian V1aR antagonist, SR-49059, was examined. Previous studies addressing the avian vasotocin receptors (VTRs) and their expression in AP cells revealed that two major VT receptors, the avian V1aR and V1bR, showed differential responses to an imposed in vivo acute and chronic psychological stress (Selvam et al., 2013; Kang & Kuenzel, 2014; Kuenzel, Kang, & Jurkevich, 2013). Unlike mammals, the presence of two VTR subtypes in corticotropes and their differential response in AP glands following restraint stress led us to initiate a study to identify receptor subtype-specific antagonists to the avian V1bR.
4.1. Homology modelling, validation of the 3D structural model, and ligand binding of the cV1aR and cV1bR
Our previous 3D modelling / docking studies of cV1aR indicated that the presence of the N-terminal, intracellular and extracellular loops and C-terminal amino acid sequences appeared to influence the binding interface of the peptide agonists and peptide / non-peptide antagonists (Jayanthi et al., 2014). However, the presence of loops did not appear to affect the relative binding affinity ranking of the peptide antagonists to the cV1aR. Nonetheless, the modelling/docking studies, coupled with in vitro AP cell culture studies enabled us to identify the most effective avian V1aR antagonist, SR-49059 (Jayanthi et al., 2014). In the current study, therefore, we followed similar protocols for the modelling / docking studies. A BLAST search using the cV1aR and cV1bR amino acid sequences, as queries against the Protein Data Bank (PDB), resulted in seven hits with a max score > 75 %. The N/OFQ opioid receptor (PDB: 4EA3) was selected as the template for homology modelling due to its optimum score. Interestingly, the secondary structural regions, consisting of the seven transmembrane helices, were highly conserved between the template and targets (cV1bR) and were well-aligned with a backbone RMSD of 0.72 A, satisfying the criteria for generating a viable homology model. In the case of membrane receptors, the binding pocket is invariably located inside the TM helices. Therefore, conservation in this region is an important criterion for generating an accurate homology model which in turn forms the reliable basis for performing the docking studies.
4.2. Extracellular binding pocket of cVTRs with ligands and differential arrangement of transmembrane domains
The current study showed extracellular surface ligand binding pockets on both the cV1aR and cV1bR using the template, N/OFQ opioid receptor. The opioid receptor was chosen due to its best overall coverage and sequence homology to the cV1aR and cV1bR in the BLAST search. The N/OFQ opioid receptor belongs to the superfamily of Rhodopsin-like receptors containing peptide-GPCRs that bind agonists of a wide range of sizes having binding cavities located near the receptor surface (Krumm & Grisshammer, 2015; Barwell et al., 2012; Conner et al., 2007). The binding sites on cV1aR and cV1bR for the agonist AVT are located close to or on the receptor surface including a few other amino acids located in the extracellular loops (Fig 3). Both the agonist and the peptide/non-peptide antagonists for cV1aR and cV1bR have common and unique binding sites, suggesting that the antagonists can be expected to show inhibition kinetics similar to those exhibited by competitive inhibitors (Table 1, Fig 3). The apparent difference was noted on the arrangements of transmembrane domains between cV1aR and cV1bR, showing the wider outer mouth of cV1aR than that of cV1bR when AVT interacted with receptors (Fig 1C and 1D). Agonist-induced conformational changes at the ligand-binding pocket of GPCRs are largely receptor specific due to the different chemical nature of the agonists, and were suggested to cause the differential re-arrangement of the intracellular face of the receptor required for G protein binding and activation (Deupi & Standfuss, 2011; White et al., 2012). Therefore, the different binding affinity between antagonists and receptors (Table 2 and 3, Fig. 4A) may cause unique conformational change of transmembrane domains, and induce differential arrangement of the intracellular face of the receptors which might be required for G protein binding and activation in the avian AP gland.
4.3. Role of extracellular residues of cVTRs for ligand binding, particularly charged amino acids
The molecular basis underlying ligand-receptor interactions has been extensively studied in the mammalian AVP receptor family (Mouillac et al., 1995a; Conner et al., 2007; Hawtin et al., 2006; Mouillac et al., 1995b). In avian species, Gln (Q) residues in TM II, III, IV, VI (Q115, Q138, Q192, Q313) and a Lys (K) in TM III (K135) of the cV1aR and Q residues in TM III, VI (Q124, Q314) and a K in TM III (K121) of the cV1bR interacted with the agonist AVT. Some of the Q residues (Q138 and Q313 of cV1aR and Q124 and Q314 of cV1bR) interact with antagonists. Q residues (Q115 of cV1aR and Q178 of cV1bR) are unique to agonist AVT binding (Q115 of cV1aR) or antagonist binding (Q178 of cV1bR with the Manning compound). Similarly, a mutagenesis study on the human V1aR (Mouillac et al., 1995b) showed that critical amino acids involved in agonist binding were Gln in TM II, IV (Q108, Q185), and a Lys that is located in the spatial vicinity of TM III (K128). Substitution of these critical amino acids with Ala, a neutral amino acid, (Q108A, Q185A, and K128A), significantly reduced binding affinity of agonists. Interestingly, these substitutions only caused minor differences in the binding affinities of an antagonist, indicating similarity of ligand-receptor binding interactions between the avian V1aR and human V1aR.
In general, the roles of extracellular residues of GPCRs have not been as well defined compared to residues in TMs (Hawtin et al., 2006; Wheatley et al., 2007; Wheatley et al., 2012). Recently, the role of charged residues in extracellular loops of GPCRs, which are conserved in a subfamily of peptide-GPCRs, was extensively examined using mutagenesis and by molecular modelling studies on the mammalian GPCRs, including the rat V1aR (Hawtin et al., 2006; Mouillac et al., 1995a; Tahtaoui et al., 2003; Unal, Jagannathan, Bhat, & Karnik, 2010; Kmiecik, Kamroz, & Kolinski, 2014; Nguyen et al., 2016; Serradeil-Le Gal et al., 2005). Arg116 (R116) in the first extracellular loop (ECL1), R125 at the top of TM III, and Asp204 (D204; in ECL2) of the rat V1aR were found to be important for agonist binding and/or receptor activation (Hawtin et al., 2006). In the cV1aR, we found similar residues including R123 in the ECL1, R132 near the extracellular region of TM III and as well as D209 in the ECL2. Interestingly, analysis of the receptor/agonist binding interface indicated that these residues of cV1aR did not interact with the ligands (Table 1, Fig 3A). In contrast, data for the cV1bR indicated that R109 in the ECL1 was interacting with the agonist AVT and peptide antagonist Manning compound. In addition, D195 in the ECL2 of the cV1bR was only interacting with the antagonist Manning compound that displayed the highest binding affinity with cV1bR in the current study (Table 3, Fig 3B).
4.4. Role of extracellular residues of cVTRs for ligand binding, particularly aromatic amino acids
ECL2 of GPCRs has attracted increased interest, because the x-ray structure of bovine rhodopsin revealed that ECL2 projects into the binding crevice within the TM bundle (Barwell et al., 2012; Hawtin et al., 2006). In mammals, analysis of residues in or close to ECL2 by mutagenesis revealed that four aromatic residues Phe189 (F189), Trp206 (W206), Phe209 (F209), and Tyr218 (Y218) appear to be important for agonist binding and receptor activation (Conner et al., 2007). These aromatic resides are highly conserved throughout the neurohypophysial hormone sub-family of peptide-GPCRs (Conner et al., 2007). Specifically, the highly conserved aromatic residues W206 and F209 were suggested to project into the binding crevice, and F189 and Y218, located at the extreme ends of ECL2 were suggested to have a critical role for determining the position of the ECL2 cap over the binding crevice in the human V1aR (Conner et al., 2007; Hawtin et al., 2006). In the cV1aR, W211 and F214 in its ECL2 are spaced 3AAs apart, similar to the spacing of W197 and F200 in the ECL2 of the cV1bR. Note that this spacing is identical with the human spacing in its V1aR and V1bR (Hawtin et al., 2006). Similarly the extreme ends of the human ECL2, F189 and Y218 are separated by 29 AAs (Hawtin et al., 2006). The identical spacing of the cV1aR (F194, Y223) and cV1bR (F180, Y209) at the extreme ends of the ECL2 for both avian receptors can be seen (Fig 3), suggesting an important role for the conserved aromatic residues in agonist binding in avian species as well.
4.5. Differential role of conserved Cys-Trp (CW) motif in ECL2 of cVTRs
In rhodopsin, class A GPCRs, ECL2 is usually the longest ECL and comprises two β-sheets which form a twisted β-hairpin that projects down into the TM bundle (Wheatley et al., 2012). In the ECL2 of cV1bR, there are two β -sheets and a highly conserved Cys -Trp (CW) motif that resides in the middle of the second β-sheet. The CW motif is a common binding site with agonist AVT and antagonists (SR-49059, SSR-149415, and the Manning compound), indicating the significance of the CW motif of the ECL2 in avian species. In mammals, including humans, the CW motif in the ECL2 of the V1aR and V1bR is conserved (Conner et al 2007; Hawtin et al., 2006). In this conserved CW motif, cV1aR showed the interaction of Trp211 (W211) with AVT and SR-49059. In a more extensive analysis the cV1bR showed the interaction of C196 and W197 with AVT and three tested antagonists (one peptide and two non-peptides), likewise indicating a critical role of the CW motif in ECL2 in avian species. In the interaction of cV1bR with the Manning compound, two adjacent conserved Asp195 (D195) and Ala198 (A198) were involved in the interaction (Table 1, Fig 3B), perhaps contributing to the highest binding affinity shown by the Manning compound with cV1bR obtained in the docking study. These results may indicate that the differences in the interaction(s) between the agonist/antagonists and the conserved CW motif in ECL2 of cV1aR and cV1bR probably accounts for the observed differences in specific ligand recognition. The conserved Cys residue in the middle of ECL2 (C210 in cV1aR and C196 in cV1bR) was suggested as the reference point for tertiary folding of ECL2 (Wheatley et al., 2007; Unal, Jagannathan, Bhat, & Karnik, 2010). Importantly, it has been shown that Cys (C205) in the mammalian ECL2 can form a disulfide bond with C124 near the extracellular region of TM III, and consequently affecting the ECL2 cap over the binding pocket crevice involving TM III, TM IV and other TMs (Hawtin et al., 2006). Similarly, both cV1aR and cV1bR have a Cys near their extracellular border of their TM III suggesting strongly that a disulfide bond can form with their Cys residue in the ECL2 forming an ECL2 cap over the binding pocket crevice involving TM III. These results provide the possible structural requirements for the cV1aR and cV1bR to attain high-affinity binding for both the vasotocin agonist and effective antagonists examined in this study.
4.6. Antagonistic effect of the selected avian V1bR antagonists and the combined effect of V1aR and V1bR antagonists on POMC hnRNA expression in chicken AP cells
In our previous study, several mammalian V1aR antagonists were shown capable of attenuating the response of the avian V1aR, however, one emerged as the most effective blocker, SR-49059 (Jayanthi et al., 2014). The lack of information on effective blockers of the avian V1bR led to the current study. From the list of candidate avian V1bR antagonists tested in the present in silico study, we found several compounds showing a higher binding affinity (a lower, more negative docking score) to the cV1bR compared to its natural agonist, AVT (Table 3, Fig 4A). A comparison of binding affinities of SR-49059, SSR-149415 and Manning compound to AVT using the modeled cV1aR and cV1bR confirmed that SR-49059 showed significantly greater binding affinity with the cV1aR than cV1bR, hence supporting its functional role as an effective cV1aR antagonist (Fig. 4A).
In contrast, the Manning compound showed the lowest binding affinity with cV1aR and the highest binding affinity with cV1bR (Table 2 and 3, Fig 4A). However, this compound was originally described as a potent mammalian V1aR-selective antagonist, but recently found to be not selective in humans (Manning et al., 2012) or in mice (Busnelli et al., 2013), suggesting not useful for a selective V1aR antagonist in mammals. Therefore, two commercially available V1bR antagonists, SSR-149415 and L-368899, from our list of ligands for cV1bR (Table 3) were selected for testing the in vitro antagonistic effects using the established primary AP cell stress response system (Kang & Kuenzel, 2014; Jayanthi et al., 2014). As it is difficult to measure the dynamic spatial and temporal patterns of ACTH release and synthesis from primary AP cells owing to the accumulation of ACTH in cell culture media, we developed the assay measuring hnRNA of the POMC gene as a stress-induced activating marker in primary chicken AP cells. The optimum time and concentration of in vitro stress inducers, AVT/CRH, were established (Jayanthi et al., 2014). In primary AP cells, pretreatments of the two antagonists before the imposed AVT/CRH in vitro stress, significantly reduced hnRNA of POMC compared to stressed controls, indicating their antagonistic effect. We found a dose of 10 pM for SSR-149059 and L-368899 pretreatment to be effective for blocking of hnRNA expression of POMC expression (Fig. 4B) and used the 10 pM dose for each antagonist in the next study (Fig 4C). Administration of either a single or the combination of two distinct receptor antagonists to AP cells followed by measuring subsequent POMC hnRNA expression showed that both SSR-149415 and L-368899 were effective in attenuating POMC hnRNA expression. However, when each candidate was used along with the avian V1aR blocker, SR-49059, results showed that SSR-149415 was significantly more effective than L-368899 as a specific avian V1bR antagonist (Fig 4C) similar to what was found in mammals (Serradeil-Le Gal et al., 2005). The combined antagonistic effects of the selected V1bR antagonists with SR-49059 or Manning compound appear to be additive in the primary AP cell culture study.
4.7. Effect of V1aR and V1bR antagonists on their receptor expression, a test of receptor specificity, as well as CRH-R1 and CRH-R2 in AP primary cells
The Manning compound showed the best binding affinity with the cV1bR, but showed no significant difference of binding affinity with the cV1aR compared to AVT. Therefore, the specificity of two antagonists, SR-49059 as a selective cV1aR antagonist and Manning compound as a selective cV1bR antagonist, were tested using the in vitro AP cell culture system. Both cV1aR and cV1bR mRNA expression were down-regulated by in vitro stress AVT/CRH (1.0/0.1 nM) application to AP cells as previously reported (Kang & Kuenzel, 2014). The decrease of cV1aR mRNA compared to controls, which was significantly different from the cV1bR mRNA, indicates a differential effect of the natural agonist (AVT) on each receptor and its sensitivity in primary AP cell cultures. Pre-treatment of SR-49059 augmented expression of the cV1aR mRNA from 27 % to 49 % compared to controls, but did not change expression of the cV1bR mRNA. This result indicates the specificity of SR-49059 for the cV1aR. In contrast, the Manning compound changed the cV1aR and cV1bR expression from 27 % and 54 % to 56 % and 87 %, respectively, compared to controls following administration of AVT/CRH (Fig 5A). Therefore, SR-49059 appears to be specific to the cV1aR, while the Manning compound appears to be promiscuous in blocking both the cV1aR and cV1bR.
Previous studies in avian and mammalian species indicate that V1bR and CRH-R1 mediate synergistic biological actions of vasopressin / vasotocin and CRH by heterodimerization and direct molecular interactions between V1bR and CRH-R1 (Young et al., 2007; Mikhailova et al., 2007; Murat et al., 2012; Cornett, Kang, & Kuenzel, 2013), indicating heterodimer formation between V1bR and CRH-R1 may be critical for activation of downstream signal transduction pathways and HPA axis adaptation. Young et al (2007) suggested that this heterodimerization between V1bR and CRH-R1 is not ligand dependent and does not influence the binding properties of these receptors. The results in the current study showed that both the expression of cCRH-R1 and cV1bR were not affected by SR-49059 pretreatment, while cV1aR was significantly up regulated. In contrast, cCRH-R2 expression was significantly down regulated and cCRH-R1 and cV1bR were up regulated by Manning compound pretreatment (Fig 5). The increased expression of cCRH-R1 and cV1bR provide supporting evidence for possible functional heterodimerization of V1bR and CRH-R1 in the AP gland of avian species (Mikhailova et al., 2007). Additionally, the antagonist SR-49059 appears to be specific for the cV1aR while the Manning compound lacks specificity.
5. Conclusion
Our results show that the specific molecular interaction of cV1aR and cV1bR with agonist and antagonists appears to be critical for their binding affinity and pharmacological properties. Data suggest that the antagonist, SR-49059 is specific for the avian V1aR while to date, SSR- 149415 is a more potent blocker of the avian V1bR, however it also shows significant binding affinity for the V1aR. As both compounds are non-peptides, their antagonistic effects will be more long lasting. In contrast the Manning compound is non-specific and appears to affect the avian V1aR, V1bR, CRH-R1 and CRH-R2. Being a peptide compound, its effect can be expected to be of a shorter duration. These findings are likely to provide useful leads for the continuing effort to develop pharmaceutical compounds capable of blocking or attenuating the AP stress response of non-mammalian species.
Acknowledgments
This research was supported by NSF Grant #0842937, Arkansas Biosciences Institute (ABI) Grant, and Arkansas Agricultural Experiment Station. We also want to thank the funding support from the Department of Energy (grant number DE-FG02–01ER15161), the National Institutes of Health/National Cancer Institute (NIH/NCI) (1 RO1 CA 172631) and the NIH through the COBRE program (P30 GM103450).
Footnotes
Author contributions
Conceived and designed the experiments: SK SJ TKSK WK. Performed the experiments: SK SJ GN. Analyzed the data: SK SJ. Contributed reagents/materials/analysis tools: SK TKSK WK. Wrote the paper: SK SJ TKSK WK.
Competing Interests: The authors declare that they have no competing interests.
References:
- Acharjee S, Do-Rego JL, Oh DY, Ahn RS, Choe H, Vaudry H, Kwon HB (2004). Identification of amino acid residues that direct differential ligand selectivity of mammalian and nonmammalian V1a type receptors for arginine vasopressin and vasotocin. Insights into molecular coevolution of V1a type receptors and their ligands. J Biol Chem, 279(52), 54445–54453 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, & Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res, 25(17), 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni FA (1993). Vasopressinergic control of pituitary adrenocorticotropin secretion comes of age. Front Neuroendocrinol, 14(2), 76–122 [DOI] [PubMed] [Google Scholar]
- Autelitano DJ, Lundblad JR, Blum M, & Roberts JL (1989). Hormonal regulation of POMC gene expression. Annu Rev Physiol, 51, 715–726. doi: 10.1146/annurev.ph.51.030189.003435 [DOI] [PubMed] [Google Scholar]
- Baran NM, Sklar NC, & Adkins-Regan E (2016). Developmental effects of vasotocin and nonapeptide receptors on early social attachment and affiliative behavior in the zebra finch. Horm Behav, 78, 20–31. doi: 10.1016/j.yhbeh.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwell J, Woolley MJ, Wheatley M, Conner AC, & Poyner DR (2012). The role of the extracellular loops of the CGRP receptor, a family B GPCR. Biochem Soc Trans, 40(2), 433–437. doi: 10.1042/BST20110726 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Bourne PE (2000). The Protein Data Bank. Nucleic Acids Res, 28(1), 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, & Montelione GT (2007). Evaluating protein structures determined by structural genomics consortia. Proteins 66(4), 778–795. doi: 10.1002/prot.21165 [DOI] [PubMed] [Google Scholar]
- Busnelli M, Bulgheroni E, Manning M, Kleinau G, & Chini B (2013). Selective and potent agonists and antagonists for investigating the role of mouse oxytocin receptors. J Pharmacol Exp Ther, 346(2), 318–327. doi: 10.1124/jpet.113.202994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Acharjee S, Moon MJ, Oh DY, Vaudry H, Kwon HB, & Seong JY (2007). Molecular evolution of neuropeptide receptors with regard to maintaining high affinity to their authentic ligands. Gen Comp Endocrinol, 153(1–3), 98–107 [DOI] [PubMed] [Google Scholar]
- Conner M, Hawtin SR, Simms J, Wootten D, Lawson Z, Conner AC, Wheatley M (2007). Systematic analysis of the entire second extracellular loop of the V(1a) vasopressin receptor: key residues, conserved throughout a G-protein-coupled receptor family, identified. J Biol Chem, 282(24), 17405–17412. doi: 10.1074/jbc.M702151200 [DOI] [PubMed] [Google Scholar]
- Cornett LE, Kang SW, & Kuenzel WJ (2013). A possible mechanism contributing to the synergistic action of vasotocin (VT) and corticotropin-releasing hormone (CRH) receptors on corticosterone release in birds. Gen Comp Endocrinol, 188, 46–53 [DOI] [PubMed] [Google Scholar]
- Cotte N, Balestre MN, Aumelas A, Mahe E, Phalipou S, Morin D, Mouillac B (2000). Conserved aromatic residues in the transmembrane region VI of the V1a vasopressin receptor differentiate agonist vs. antagonist ligand binding. Eur J Biochem, 267(13), 4253–4263 [DOI] [PubMed] [Google Scholar]
- Czaplewski C, Kazmierkiewicz R, & Ciarkowski J (1998a). Molecular modeling of the human vasopressin V2 receptor/agonist complex. J Comput Aided Mol Des, 12(3), 275–287 [DOI] [PubMed] [Google Scholar]
- Czaplewski C, Kazmierkiewicz R, & Ciarkowski J (1998b). Molecular modelling of the vasopressin V2 receptor/antagonist interactions. Acta Biochim Pol, 45(1), 19–26 [PubMed] [Google Scholar]
- DeLano WL The PyMOL Molecular Graphics System.DeLano Scientific, San Carlos, CA. http://pymol.sourceforge.net [Google Scholar]
- Deupi X, & Standfuss J (2011). Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr Opin Struct Biol, 21(4), 541–551. doi: 10.1016/j.sbi.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Fehrer SC, Silsby JL, Behnke EJ, & el Halawani ME (1985). Hypothalamic and serum factors influence on prolactin and luteinizing hormone release by the pituitary gland of the young turkey (Meleagris gallopavo). Gen Comp Endocrinol, 59(1), 73–81 [DOI] [PubMed] [Google Scholar]
- Gilchriest BJ, Tipping DR, Hake L, Levy A, & Baker BI (2000). The effects of acute and chronic stresses on vasotocin gene transcripts in the brain of the rainbow trout (Oncorhynchus mykiss). J Neuroendocrinol, 12(8), 795–801 [DOI] [PubMed] [Google Scholar]
- Goncharova ND (2013). Stress responsiveness of the hypothalamic-pituitary-adrenal axis: age-related features of the vasopressinergic regulation. Front Endocrinol (Lausanne), 4, 26. doi: 10.3389/fendo.2013.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson JL, & Evans AK (2004). Neural responses to territorial challenge and nonsocial stress in male song sparrows: segregation, integration, and modulation by a vasopressin V1 antagonist. Horm Behav, 46(4), 371–381. doi: 10.1016/j.yhbeh.2004.02.008 [DOI] [PubMed] [Google Scholar]
- Hausmann H, Richters A, Kreienkamp HJ, Meyerhof W, Mattes H, Lederis K, . . . Richter D (1996). Mutational analysis and molecular modeling of the nonapeptide hormone binding domains of the. Proc Natl Acad Sci U S A, 93(14), 6907–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawtin SR, Simms J, Conner M, Lawson Z, Parslow RA, Trim J, Wheatley M (2006). Charged extracellular residues, conserved throughout a G-protein-coupled receptor family, are required for ligand binding, receptor activation, and cell-surface expression. J Biol Chem, 281(50), 38478–38488. doi: 10.1074/jbc.M607639200 [DOI] [PubMed] [Google Scholar]
- Hernando F, Schoots O, Lolait SJ, & Burbach JP (2001). Immunohistochemical localization of the vasopressin V1b receptor in the rat brain and pituitary gland: anatomical support for its involvement in the central effects of vasopressin. Endocrinology, 142(4), 1659–1668 [DOI] [PubMed] [Google Scholar]
- Jayanthi S, Kang SW, Bingham D, Tessaro BA, Suresh Kumar TK, & Kuenzel WJ (2014). Identification of antagonists to the vasotocin receptor sub-type 4 (VT4R) involved in stress by molecular modelling and verification using anterior pituitary cells. J Biomol Struct Dyn, 32(4), 648660. doi: 10.1080/07391102.2013.787025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkevich A, Berghman LR, Cornett LE, & Kuenzel WJ (2005). Characterization and immunohistochemical visualization of the vasotocin VT2 receptor in the pituitary gland of the chicken, Gallus gallus. Gen Comp Endocrinol, 143(1), 82–91. doi: 10.1016/j.ygcen.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Jurkevich A, Berghman LR, Cornett LE, & Kuenzel WJ (2008). Immunohistochemical characterization of chicken pituitary cells containing the vasotocin VT2 receptor. Cell Tissue Res, 333(2), 253–262. doi: 10.1007/s00441-008-0636-2 [DOI] [PubMed] [Google Scholar]
- Kang SW, Gazzillo LC, You S, Wong EA, & El Halawani ME (2004). Turkey prolactin gene regulation by VIP through 35-bp cis-acting element in the proximal promoter. Gen Comp Endocrinol, 138(2), 157–165 [DOI] [PubMed] [Google Scholar]
- Kang SW, & Kuenzel WJ (2014). Regulation of gene expression of vasotocin and corticotropin-releasing hormone receptors in the avian anterior pituitary by corticosterone. Gen Comp Endocrinol, 204, 25–32. doi: 10.1016/j.ygcen.2014.04.018 [DOI] [PubMed] [Google Scholar]
- Kmiecik S, Jamroz M, & Kolinski M (2014). Structure prediction of the second extracellular loop in G-protein-coupled receptors. Biophys J, 106(11), 2408–2416. doi: 10.1016/j.bpj.2014.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshimizu TA, Nakamura K, Egashira N, Hiroyama M, Nonoguchi H, & Tanoue A (2012). Vasopressin V1a and V1b receptors: from molecules to physiological systems. Physiol Rev, 92(4), 1813–1864. doi: 10.1152/physrev.00035.2011 [DOI] [PubMed] [Google Scholar]
- Krumm BE, & Grisshammer R (2015). Peptide ligand recognition by G protein-coupled receptors. Front Pharmacol, 6, 48. doi: 10.3389/fphar.2015.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzel WJ, Hancock M, Nagarajan G, Aman NA, & Kang SW (2016). Central effect of vasotocin 4 receptor (VT4R/V1aR) antagonists on the stress response and food intake in chicks given neuropeptide Y (NPY). Neurosci Lett, 620, 57–61. doi: 10.1016/j.neulet.2016.03.036 [DOI] [PubMed] [Google Scholar]
- Kuenzel WJ, Kang SW, & Jurkevich A (2013). Neuroendocrine regulation of stress in birds with an emphasis on vasotocin receptors (VTRs). Gen Comp Endocrinol, 190, 18–23. doi: 10.1016/j.ygcen.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Lightman SL (2008). The neuroendocrinology of stress: a never ending story. J Neuroendocrinol, 20(6), 880–884 [DOI] [PubMed] [Google Scholar]
- Mahlmann S, Meyerhof W, Hausmann H, Heierhorst J, Schonrock C, Zwiers H, Richter D (1994). Structure, function, and phylogeny of [Arg8]vasotocin receptors from teleost fish and toad. Proc Natl Acad Sci U S A, 91(4), 1342–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning M, Misicka A, Olma A, Bankowski K, Stoev S, Chini B, Guillon G (2012). Oxytocin and vasopressin agonists and antagonists as research tools and potential therapeutics. J Neuroendocrinol, 24(4), 609–628. doi: 10.1111/j.1365-2826.2012.02303.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D, Hassan A, Chacko S, & Thompson P (2002). Acute and chronic regulation of pituitary receptors for vasopressin and corticotropin releasing hormone. Arch Physiol Biochem, 110(1–2), 74 89 [DOI] [PubMed] [Google Scholar]
- Mikhailova MV, Mayeux PR, Jurkevich A, Kuenzel WJ, Madison F, Periasamy A, Cornett LE(2007). Heterooligomerization between vasotocin and corticotropin-releasing hormone (CRH) receptors augments CRH-stimulated 3’, 5’ - cyclic adenosine monophosphate production. Mol Endorinol 21(9), 2178–2188. doi: 10.1210/me.2007-0160 [DOI] [PubMed] [Google Scholar]
- Mouillac B, Chini B, Balestre MN, Elands J, Trumpp-Kallmeyer S, Hoflack J, Barberis C (1995a). The binding site of neuropeptide vasopressin V1a receptor. Evidence for a major localization within transmembrane regions. J Biol Chem, 270(43), 25771–25777 [DOI] [PubMed] [Google Scholar]
- Mouillac B, Chini B, Balestre MN, Jard S, Barberis C, Manning M, et al. (1995b). Identification of agonist binding sites of vasopressin and oxytocin receptors. Adv Exp Med biol, 395, 301–310 [PubMed] [Google Scholar]
- Murat B, Devost D, Andres M, Mion J, Boulay V, Corbani M, Guillon G (2012). V1b and CRHR1 receptor heterodimerization mediates synergistic biological actions of vasopressin and CRH. Mol Endocrinol, 26(3), 502–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan G, Kang SW, & Kuenzel WJ (2017). Functional evidence that the nucleus of the hippocampal commissure shows an earlier activation from a stressor than the paraventricular nucleus: Implication of an additional structural component of the avian hypothalamo-pituitary-adrenal axis. Neurosci Lett, 642, 14–19. doi: 10.1016/j.neulet.2017.01.064 [DOI] [PubMed] [Google Scholar]
- Nagarajan G, Tessaro BA, Kang SW, & Kuenzel WJ (2014). Identification of arginine vasotocin (AVT) neurons activated by acute and chronic restraint stress in the avian septum and anterior diencephalon. Gen Comp Endocrinol, 202, 59–68. doi: 10.1016/j.ygcen.2014.04.012 [DOI] [PubMed] [Google Scholar]
- Needleman SB, & Wunsch CD (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol, 48(3), 443–453 [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Baltos JA, Thomas T, Nguyen TD, Munoz LL, Gregory KJ, May LT (2016). Extracellular Loop 2 of the Adenosine A1 Receptor Has a Key Role in Orthosteric Ligand Affinity and Agonist Efficacy. Mol Pharmacol, 90(6), 703–714. doi: 10.1124/mol.116.105007 [DOI] [PubMed] [Google Scholar]
- Selvam R, Jurkevich A, Kang SW, Mikhailova MV, Cornett LE, & Kuenzel WJ (2013). Distribution of the Vasotocin Subtype Four Receptor (VT4R) in the Anterior Pituitary Gland of the Chicken, Gallus gallus, and its Possible Role in the Avian Stress Response. J Neuroendocrinol, 25(1), 56–66 [DOI] [PubMed] [Google Scholar]
- Serradeil-Le Gal C, Wagnon J 3rd, Tonnerre B, Roux R, Garcia G, Griebel G, & Aulombard A (2005). An overview of SSR149415, a selective nonpeptide vasopressin V(1b) receptor antagonist for the treatment of stress-related disorders. CNS Drug Rev, 11(1), 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Blundell TL, & Mizuguchi K (2001). FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol, 310(1), 243–257. doi: 10.1006/jmbi.2001.4762 [DOI] [PubMed] [Google Scholar]
- Sugimoto T, Saito M, Mochizuki S, Watanabe Y, Hashimoto S, & Kawashima H (1994). Molecular cloning and functional expression of a cDNA encoding the human V1b vasopressin receptor. J Biol Chem, 269(43), 27088–27092 [PubMed] [Google Scholar]
- Tahtaoui C, Balestre MN, Klotz P, Rognan D, Barberis C, Mouillac B, & Hibert M (2003). Identification of the binding sites of the SR49059 nonpeptide antagonist into the V1a vasopressin receptor using sulfydryl-reactive ligands and cysteine mutants as chemical sensors. J Biol Chem, 278(41), 40010–40019. doi: 10.1074/jbc.M301128200 [DOI] [PubMed] [Google Scholar]
- Trott O, & Olson AJ (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem, 31(2), 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S, Griffante C, & Aguilera G (2007). Dimerization between vasopressin V1b and corticotropin releasing hormone type 1 receptors. Cell Mol Neurobiol 27(4), 439–461. doi: 10.1007/s10571-006-9135-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal H, Jagannathan R, Bhat MB, & Karnik SS (2010). Ligand-specific conformation of extracellular loop-2 in the angiotensin II type 1 receptor. J Biol Chem, 285(21), 16341–16350. doi: 10.1074/jbc.M109.094870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley M, Simms J, Hawtin SR, Wesley VJ, Wootten D, Conner M, Parslow RA (2007). Extracellular loops and ligand binding to a subfamily of Family A G-protein-coupled receptors. Biochem Soc Trans, 35(Pt 4), 717–720. doi: 10.1042/BST0350717 [DOI] [PubMed] [Google Scholar]
- Wheatley M, Wootten D, Conner MT, Simms J, Kendrick R, Logan RT, Barwell J (2012). Lifting the lid on GPCRs: the role of extracellular loops. Br J Pharmacol, 165(6), 1688–1703. doi: 10.1111/j.1476-5381.2011.01629.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Grisshammer R (2012). Structure of the agonist-bound neurotensin receptor. Nature, 490(7421), 508–513. doi: 10.1038/nature11 [DOI] [PMC free article] [PubMed] [Google Scholar]


