Abstract
Premise of the Study
The production of banana (Musa spp.; Musaceae) plants is affected by various types of somaclonal variations (SV), including dwarfism. However, methods for specific detection of SV are still scarce. To overcome this, a metabolite‐based method for detection of dwarf variants was evaluated.
Methods
The gas chromatography–mass spectrometry (GC‐MS) metabolite profile of dwarf banana variants was investigated and compared to that of normal‐healthy (N) and cucumber mosaic virus (CMV)–infected plants using principal components analysis and partial least squares discriminant analysis (PLS‐DA).
Results
Significant differences among the sample groups were observed in 82 metabolites. Rhamnose was exclusively present in dwarf plants but allothreonine and trehalose were present in all but SV samples. Cellobiose was only detected in N plants, while 45 other metabolites, including methyl‐glucopyranoside, allopyranose, lactose, phenylalanine, and l‐lysine were detected in all but CMV‐infected samples. PLS‐DA models were able to detect SV, CMV, and N plants with 100% accuracy and specificity.
Discussion
The GC‐MS metabolite profile can be used for the rapid, specific detection of SV at early plant production stages. This is the first metabolite‐based characterization and detection of somaclonal variation in plants.
Keywords: cucumber mosaic virus, metabolomics, Musa, Musaceae, partial least squares regression, somaclonal variation
Metabolomics—the study of small metabolites in a system—has been successfully applied in the plant sciences to investigate plant development (Watanabe et al., 2013; Hu et al., 2016), characterize plant response to abiotic stresses (Caldana et al., 2011; Maruyama et al., 2014), and diagnosis plant diseases (Cevallos‐Cevallos et al., 2009, 2011). Among various metabolomics tools, the use of gas chromatography–mass spectrometry (GC‐MS) has been recommended for the assessment of plant varieties with different morphologies (Cevallos‐Cevallos et al., 2012), because metabolites have shown a more relevant relationship to plant phenotypes than genes (Fiehn, 2001; Yogendra et al., 2014). Therefore, metabolomics tools have the potential for characterizing morphological alterations in plants and detecting somaclonal variants.
Banana (Musa L. spp.; Musaceae) is one of the most cultivated crops worldwide and a major source of income for millions of people in many countries (Yeturu et al., 2016). The world's total production of banana was estimated at 113 million tons in 2016, with Ecuador standing out as the top banana‐exporting country (Food and Agriculture Organization of the United Nations, 2017). Banana plants are commonly produced by in vitro micropropagation techniques focusing on the generation of phenotypically uniform plantlets from a parent plant. However, phenotypic variations—known as somaclonal variations (SV)—are commonly observed in about 6% to 69% of the Musa spp. plants regenerated by tissue culture (Sahijram et al., 2003). Various types of atypical plant morphologies have been associated with SV, with dwarfism being the most common SV observed (Reuveni and Israeli, 1990).
Plants showing SV are commonly considered to be of inferior quality and reduced commercial value when compared to the parental clones (Sahijram et al., 2003; Oh et al., 2007), but improved characters of somaclonal variants have also been reported (Bairu et al., 2010). Dwarf variants of the ‘Williams’ banana cultivar can bear shorter fruits of atypical shapes (Israeli et al., 1991) but can show faster growth and flowering rates (Israeli et al., 1991) than normal plants. Additionally, the stocky build of dwarf bananas suggests a higher resistance to physical damages and advantages of cultivation convenience, field management, and labor savings when compared to normal clones (Ferrero‐Serrano and Assmann, 2016).
Detection of SV in Musa spp. has traditionally been carried out through visual examination of the plants. Dwarf off‐types have been detected by observing the plant height, leaf index (leaf length/width; Rodrigues et al., 1998), and pseudostem thickness (Oh et al., 2007) at the greenhouse production stage. However, banana morphological traits can also be affected by environmental factors (Bairu et al., 2010) and biotic stresses such as cucumber mosaic virus (CMV) infections (Yeturu et al., 2016), rendering visual diagnosis of SV unreliable. CMV can be transmitted by more than 80 aphid species and infect almost 1200 plant species including bananas (Zitter and Murphy, 2009), making CMV one of the most widespread banana viruses worldwide (Basavaraj et al., 2017). CMV can be detected wherever bananas are grown (Dheepa and Paranjothi, 2010), and Musa spp. plants infected by CMV can show symptoms resembling those of SV, including growth deficiency and leaf distortion (Yeturu et al., 2016), leading to problems of misdiagnosis between CMV and SV. Therefore, non‐visual methodologies need to be applied for the detection of SV in Musa spp. at early plant production stages, and the specificity of SV diagnosis methods must be validated against CMV‐infected plants.
Various DNA‐based methodologies have been proposed to assess genetic stability as a means to infer SV in micropropagated banana plants, including random‐amplified polymorphic DNA (RAPD) (Sheidai et al., 2008), inter‐simple sequence repeat (ISSR) (Ray et al., 2006), amplified fragment length polymorphism (AFLP) (Sahijram et al., 2003), and representational difference analysis (RDA) (Oh et al., 2007), among others. However, SV in clones showing polymorphic DNA was not phenotypically confirmed, and no association between DNA markers and alterations of visual traits was made.
Biochemical methods have also been suggested to detect dwarf banana variants, including the quantification of gibberellic acids (GAs) (Chen et al., 2016). However, GA is not the only endogenous signal that induces plant growth, and GA‐unresponsive dwarf variants have been reported (Sun, 2000). Moreover, the specificity of biochemical tests against CMV‐infected plants has not been evaluated. Specificity of biochemical methods for detection of various plant conditions can be significantly improved using a group of metabolites instead of a single marker (Cevallos‐Cevallos et al., 2009), but no metabolomics‐based characterization of banana dwarf variants can be found in the literature. Despite the importance of SV to the banana industry, reliable methods for the early and specific detection of SV in Musa spp. are still unavailable.
This research aimed to describe the GC‐MS metabolite profile of normal, dwarf, and CMV‐affected banana plants at the greenhouse establishment stage and propose a metabolomics‐based method for detection of dwarf variants.
METHODS
Plant material
Plants of the ‘Williams’ cultivar were obtained from a commercial banana propagation facility in Guayaquil, Ecuador, during 2015 and 2016. Plants in the greenhouse establishment phase (3–4 months) propagated from the same parental clone were selected for this study as suggested in previous reports (Rodrigues et al., 1998; Bairu et al., 2010). A total of 20 SV (dwarf variants), 16 normal‐healthy (N), and 15 CMV‐infected plants were collected. Only clones showing significantly shorter heights and smaller leaf indexes than standard normal plants were considered as dwarf variants (Israeli et al., 1991), whereas plants showing CMV symptoms, including yellow stripes on leaves, leaf distortion, and stunting of growth were preliminarily selected as putative CMV‐infected plants. The presence of CMV was then confirmed using a commercial triple antibody sandwich ELISA kit (Agdia, Elkhart, Indiana, USA) and CMV‐positive samples were selected for the study. The presence of CMV was also assessed in the SV and N samples yielding negative ELISA results. Plants were transported to our research facilities and kept at 28°C, 70% relative humidity with natural light (12 h) in a greenhouse, and watered every 48 h until analyzed (2 weeks).
Metabolite analysis
For metabolite characterization, small (5 × 5 cm) pieces were simultaneously extracted from the center of three different leaves from each plant using a scalpel while avoiding the midrib. Metabolite extraction, separation, and detection were executed on each leaf piece as described elsewhere (Cevallos‐Cevallos et al., 2011, 2012). Briefly, each leaf piece was ground under liquid nitrogen and 800 mg were mixed with 2 mL of an 8 : 1 : 1 methanol : water : chloroform solution followed by incubation at 7°C for 48 h. Extracts were then centrifuged at 21130 × g for 2 min using an Eppendorf 5424 microcentrifuge (Hamburg, Germany) and the pellet was discarded. Aliquots of 650 μL of each extract were transferred into 2‐mL vials and incubated with the cap open in a water bath at 50°C until dry. A total of 150 μL of N‐methyl‐N‐trimethylsilyltrifluoroacetamide (MSTFA) was added to the dried samples and incubated at 85°C for 90 min. Different amounts of MSTFA, different incubation temperatures, and different reaction times were tested but yielded lower numbers of detected peaks and poorer reproducibility, as reported in previous studies (Cevallos‐Cevallos et al., 2011) and validated in this research. The solution (1 μL) was then splitlessly injected into a GC‐MS. The injector was at 250°C, the initial oven temperature was 80°C held for 1 min, the temperature rate was 7°C/min, and the final temperature of 300°C was held for 5 min. Ultrapure helium was used as the carrier gas at 1 mL/min. The GC‐MS interface was set to 280°C, and after 8 min of solvent delay the scan was recorded with a frequency of 4 Hz. Data were acquired using ChemStation E.02.02 software (Agilent Technologies, Santa Clara, California, USA), and differentially expressed metabolites were putatively identified by MS spectra matching using two databases: the National Institute of Standards and Technology (NIST) Mass Spectral Library (NIST 11) and the Wiley Registry of Mass Spectral Data, 9th ed. (Wiley 9) (McLafferty, 2009). Metabolite identity was then confirmed by comparing the linear retention index of each compound with that of the pure standard using our internal database. All metabolites were quantified by estimating the peak area using the ChemStation software as suggested for untargeted analysis (Maridueña‐Zavala et al., 2017; Cevallos‐Cevallos et al., 2018). Preliminary GC‐MS runs showed that peaks were too close to each other throughout the chromatogram and the use of internal standards would have interfered with the chromatographic peaks. For this reason, normalization and quality control (QC) techniques not relying on internal standards were applied, including normalization to the total area (Cevallos‐Cevallos et al., 2012; Wu and Li, 2016) and QC by running the same sample after specific intervals (Warth et al., 2015; Maridueña‐Zavala et al., 2017). The quality of the GC‐MS runs was assessed by running one selected extract every five runs and estimating the variations in retention time and peak areas. Maximum acceptable coefficient of variation was 30% for a given metabolite in QC runs (Warth et al., 2015; Maridueña‐Zavala et al., 2017). Three replicates were run for each analysis.
Data analysis
Peak areas of each metabolite were used for data analysis. Peaks detected in less than one of six measurements were marked as potential metabolite carry‐over and were excluded from the analysis (Warth et al., 2015). Data were normalized to the total area and aligned using in‐house protocols based on metabolite comparisons of the MS spectra and retention times (Cevallos‐Cevallos et al., 2011). Differentially expressed metabolites were selected based on Student's t‐test comparisons of SV or CMV against N plants at the 0.05 significance level and reported as the logarithm base 2 of the fold change of each compound. Affected metabolic pathways were estimated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway database (Kanehisa and Goto, 2000; Maridueña‐Zavala et al., 2017) with Musa acuminata Colla as the model organism. Multivariate data analysis was carried out using XLSTAT 2018 (Addinsoft, Paris, France) and included principal component analysis (PCA) as well as partial least squares discriminant analysis (PLS‐DA) with jacknife cross‐validation and estimation of variable importance in the projection (VIP). Metabolites with VIP values above 2 (Steinfath et al., 2010) and the differentially expressed metabolites selected by Student's t‐test (Cevallos‐Cevallos et al., 2009) were considered to be potential biomarkers for SV.
For SV detection, three PLS‐DA models were created using the metabolite profile of 36 samples, and each model targeted the detection of the SV, CMV, or N sample class, respectively. Each model was built by assigning scores of 1 for samples belonging to the target class and zero for the other samples, and plants with model prediction values above 0.5 were assigned to the target class. The models were then validated using the remaining 15 plants.
RESULTS
A total of 466 metabolite peaks were detected in the GC‐MS runs, but Student's t‐test comparisons with N plants revealed that only 82 metabolites were differentially expressed (P < 0.05) in SV or CMV‐infected samples. Table 1 shows all the differentially expressed metabolites and the pathways potentially affected in SV and CMV‐infected plants.
Table 1.
Metabolites that were differentially expressed in dwarf (SV) and cucumber mosaic virus (CMV)–infected plants compared to normal‐healthy (N) samples.a
| Metabolite | Relative intensityb | Log2 FCc | Potential pathways | ||||
|---|---|---|---|---|---|---|---|
| N | SV | CMV | SV‐N | SV‐CMV | CMV‐N | ||
| Rhamnose | 0.00 ± 0.00a | 0.09 ± 0.01b | 0.00 ± 0.00a | >10 | >10 | ND | Fructose and mannose metabolism |
| d‐Mannitol | 0.27 ± 0.05a | 0.36 ± 0.07a | 0.15 ± 0.01b | 0.41 | 1.26 | −0.85 | Fructose and mannose metabolism |
| Fructose | 0.13 ± 0.06a | 0.79 ± 0.22b | 0.00 ± 0.00a | 2.57 | >10 | <−10 | Fructose and mannose metabolism, galactose metabolism |
| Fructopyranose | 1.44 ± 0.39a | 2.30 ± 0.36a | 0.00 ± 0.00b | 0.68 | >10 | <−10 | Fructose and mannose metabolism, galactose metabolism |
| Fructofuranose | 2.38 ± 0.36a | 2.96 ± 0.76a | 0.00 ± 0b | 0.32 | >10 | <−10 | Fructose and mannose metabolism, galactose metabolism |
| Glucose | 0.78 ± 0.13a | 0.93 ± 0.12a | 0.04 ± 0.01b | 0.27 | 4.50 | −4.24 | Starch and sucrose metabolism, galactose metabolism, fructose and mannose metabolism |
| Mannopyranose | 3.32 ± 0.81a | 4.01 ± 1.18a | 0.00 ± 0.00b | 0.27 | >10 | <−10 | Fructose and mannose metabolism, galactose metabolism |
| Sucrose | 0.63 ± 0.09a | 1.54 ± 0.45a | 5.16 ± 0.99b | 1.28 | −1.75 | 3.03 | Galactose metabolism (produced), starch and sucrose metabolism (consumed) |
| d‐Lactose | 0.50 ± 0.06a | 0.66 ± 0.09a | 0.00 ± 0.00b | 0.40 | >10 | <−10 | Galactose metabolism |
| Maltose | 0.17 ± 0.05a | 0.01 ± 0.00b | 0.00 ± 0.00b | −4.88 | >10 | <−10 | Starch and sucrose metabolism |
| Trehalose | 0.01 ± 0.01a | 0.00 ± 0.00a | 1.01 ± 0.15b | <−10 | <−10 | 6.63 | Starch and sucrose metabolism (consumed) |
| Cellobiose | 0.03 ± 0.01a | 0.00 ± 0.00b | 0.00 ± 0.00b | <−10 | ND | <−10 | Starch and sucrose metabolism |
| Linolenic acid | 1.27 ± 0.45a | 0.36 ± 0.08a | 0.00 ± 0.00b | −1.81 | >10 | <−10 | Biosinthesis of unsaturated fatty acids |
| Hexadecanoic acid | 0.14 ± 0.03a | 0.37 ± 0.05b | 1.27 ± 0.20c | 1.42 | −1.78 | 3.20 | Consumed by fatty acid degradation, produced by fatty acid enlongation |
| 9,12‐Octadecadienoic acid | 0.12 ± 0.03a | 0.09 ± 0.02a | 0.00 ± 0.00b | −0.36 | >10 | <−10 | Biosynthesis of unsaturated fatty acids |
| Octadecanoic acid | 0.98 ± 0.14a | 0.58 ± 0.12b | 0.02 ± 0.01c | −0.75 | 4.82 | −5.57 | Biosynthesis of unsaturated fatty acids |
| Phenylalanine | 0.20 ± 0.05a | 0.32 ± 0.06a | 0.00 ± 0.00b | 0.69 | >10 | <−10 | Biosynthesis of amino acids |
| Cystathionine | 0.02 ± 0.01a | 0.04 ± 0.01a,b | 0.00 ± 0.00b | 0.61 | >10 | <−10 | Biosynthesis of amino acids |
| l‐Isoleucine | 0.11 ± 0.02a | 0.09 ± 0.02a | 0.54 ± 0.04b | −0.26 | −2.56 | 2.30 | Valine, leucine, and isoleucine biosynthesis, biosynthesis of amino acids |
| l‐Leucine | 0.14 ± 0.03a | 0.11 ± 0.02a | 2.29 ± 0.49b | −0.37 | −4.43 | 4.06 | Valine, leucine, and isoleucine biosynthesis, biosynthesis of amino acids |
| l‐Lysine | 0.10 ± 0.02a | 0.08 ± 0.03a | 0.00 ± 0.00b | −0.41 | >10 | <−10 | Biosynthesis of amino acids, lysine biosynthesis |
| l‐Asparagine | 0.27 ± 0.10a | 0.08 ± 0.02a | 0.00 ± 0.00b | −1.70 | >10 | <−10 | Biosynthesis of amino acids, alanine, aspartate, and glutamate metabolism |
| Allothreonine | 0.00 ± 0.00a | 0.00 ± 0.00a | 0.18 ± 0.03b | <−10 | <−10 | 6.09 | Glycine, serine, and threonine metabolism |
| Butanoic acid | 0.14 ± 0.06a | 0.11 ± 0.03a | 0.00 ± 0.00b | −0.32 | >10 | <−10 | Butanoate metabolism |
| 2‐Butenedioic acid | 0.04 ± 0.01a | 0.03 ± 0.01a | 0.00 ± 0.00b | −0.34 | >10 | <−10 | Butanoate metabolism |
| Tyramine | 0.01 ± 0.00a | 0.02 ± 0.00a | 0.07 ± 0.01b | 1.38 | −2.05 | 3.43 | Tyrosine metabolism |
| Stigmasterol | 0.53 ± 0.13a | 0.19 ± 0.06b | 0.00 ± 0.00c | −1.44 | >10 | <−10 | Steroid biosynthesis |
| Ribitol | 0.02 ± 0.01a | 0.08 ± 0.04a | 0.38 ± 0.04b | 2.25 | −2.18 | 4.43 | Riboflavin metabolism, pentose glucoronate interconversions |
| Adenosine | 0.13 ± 0.03a | 0.08 ± 0.02a | 0.00 ± 0.00b | −0.75 | >10 | <−10 | Purine metabolism |
| Ribofuranose | 0.16 ± 0.04a | 0.18 ± 0.06a | 0.74 ± 0.09b | 0.20 | −2.01 | 2.21 | Pentose phosphate pathway |
| Methyl galactoside | 1.38 ± 0.62a | 2.53 ± 0.72a | 0.00 ± 0.00b | 0.88 | >10 | <−10 | NF |
| Methyl galactopyranoside | 0.02 ± 0.01a | 0.01 ± 0.00a | 0.19 ± 0.02b | −1.14 | −4.63 | 3.49 | NF |
| Allopyranose | 1.92 ± 0.78a | 5.36 ± 1.55a | 0.00 ± 0.00b | 1.48 | >10 | <−10 | NF |
| Allofuranose | 0.80 ± 0.16a | 1.39 ± 0.20b | 3.11 ± 0.46c | 0.81 | −1.16 | 1.97 | NF |
| l‐Threitol | 0.08 ± 0.01a | 0.12 ± 0.01a | 1.58 ± 0.19b | 0.64 | −3.76 | 4.40 | NF |
| Methyl glucofuranoside | 0.05 ± 0.02a | 0.40 ± 0.08b | 0.00 ± 0.00c | 3.04 | >10 | <−10 | NF |
| NI1 | 0.03 ± 0.03a | 0.24 ± 0.08b | 0.00 ± 0.00a | 2.97 | >10 | <−10 | NF |
| Ribonic acid | 0.04 ± 0.02a | 0.24 ± 0.06b | 0.49 ± 0.03c | 2.68 | −1.03 | 3.71 | NF |
| NI2 | 0.03 ± 0.01a | 0.11 ± 0.02b | 0.06 ± 0.02a,b | 1.90 | 0.89 | 1.01 | NF |
| Galactopyranosyl bromide, tetraacetate | 0.22 ± 0.07a | 0.81 ± 0.20b | 0.00 ± 0.00c | 1.88 | >10 | <−10 | NF |
| NI3 | 0.04 ± 0.01a | 0.07 ± 0.02a | 0.00 ± 0.00b | 0.85 | >10 | <−10 | NF |
| NI4 | 0.05 ± 0.02a | 0.09 ± 0.03a,b | 0.11 ± 0.01b | 0.84 | −0.34 | 1.18 | NF |
| NI5 | 2.09 ± 0.56a | 3.55 ± 0.85a,b | 4.06 ± 0.49b | 0.77 | −0.19 | 0.96 | NF |
| l‐Altrose | 2.03 ± 0.36a | 3.00 ± 0.45a | 0.07 ± 0.01b | 0.56 | 5.41 | −4.84 | NF |
| NI6 | 0.04 ± 0.01a | 0.07 ± 0.03a,b | 0.00 ± 0.00b | 0.56 | >10 | <−10 | NF |
| Glucopyranosiduronic acid | 0.33 ± 0.04a | 0.48 ± 0.04a | 0.25 ± 0.03b | 0.55 | 0.97 | −0.42 | NF |
| Propanoic acid | 0.34 ± 0.04a | 0.45 ± 0.08a,b | 0.55 ± 0.06b | 0.43 | −0.28 | 0.71 | NF |
| d‐glucosone | 0.17 ± 0.06a | 0.22 ± 0.06a | 2.04 ± 0.13b | 0.38 | −3.23 | 3.60 | NF |
| Hexopyranose | 3.52 ± 1.24a | 4.57 ± 0.95a | 0.00 ± 0.00b | 0.38 | >10 | <−10 | NF |
| Docosane | 0.05 ± 0.01a | 0.06 ± 0.02a | 0.29 ± 0.04b | 0.38 | −2.28 | 2.66 | NF |
| Butanedioic acid | 0.45 ± 0.07a | 0.57 ± 0.09a | 1.20 ± 0.07b | 0.34 | −1.08 | 1.42 | NF |
| NI7 | 2.38 ±0.36a | 2.96 ± 0.76a | 0.00 ± 0.00b | 0.32 | >10 | <−10 | NF |
| Galacturonic acid | 0.15 ± 0.03a | 0.18 ± 0.02a | 0.00 ± 0.00b | 0.27 | >10 | <−10 | NF |
| Phosphate | 0.49 ± 0.08a | 0.59 ± 0.07a | 0.00 ± 0.00b | 0.26 | >10 | <−10 | NF |
| Methyl glucopyranoside | 1.41 ± 0.31a | 1.69 ± 0.47a | 0.55 ± 0.15b | 0.26 | 1.63 | −1.37 | NF |
| Niacin | 0.06 ± 0.02a | 0.06 ± 0.03a,b | 0.00 ± 0.00b | 0.15 | >10 | <−10 | NF |
| Phosphoric acid propyl ester | 0.48 ± 0.15a | 0.49 ± 0.22a,b | 0.11 ± 0.07b | 0.05 | 2.21 | −2.17 | NF |
| Phenylethanolamine | 0.07 ± 0.02a | 0.07 ± 0.03a | 0.25 ± 0.02b | −0.06 | −1.81 | 1.75 | NF |
| NI10 | 11.59 ± 1.58a | 10.37 ± 1.33a | 0.00 ± 0.00b | −0.16 | >10 | <−10 | NF |
| 7,7′,8,8′,11,11′,12,12′,15,15′‐decahydro‐carotene | 0.33 ± 0.15a | 0.24 ± 0.11a | 0.00 ± 0.00b | −0.48 | >10 | <−10 | NF |
| 1‐benzopyran‐4‐one | 0.31 ± 0.09a | 0.22 ± 0.06a | 0.00 ± 0.00b | −0.51 | >10 | <−10 | NF |
| Quinic acid | 0.59 ± 0.11a | 0.42 ± 0.12a | 0.00 ± 0.00b | −0.51 | >10 | <−10 | NF |
| l‐Threonic acid | 3.77 ± 0.79a | 2.45 ± 0.45a,b | 1.60 ± 0.39b | −0.63 | 0.61 | −1.24 | NF |
| NI11 | 1.35 ± 0.33a | 0.85 ± 0.21a | 0.01 ± 0.00b | −0.67 | 6.62 | −7.30 | NF |
| 4‐O‐β‐Galactopyranosyl‐d‐mannopyranose | 0.06 ± 0.02a | 0.04 ± 0.00a | 0.00 ± 0.00b | −0.74 | >10 | <−10 | NF |
| Glyceryl‐glycoside | 2.42 ± 0.39a | 1.31 ± 0.19b | 0.00 ± 0.00c | −0.89 | >10 | <−10 | NF |
| NI12 | 0.12 ± 0.04a | 0.06 ± 0.02a | 0.00 ± 0.00b | −1.02 | >10 | <−10 | NF |
| NI13 | 0.15 ± 0.05a | 0.07 ± 0.04a,b | 0.00 ± 0.00b | −1.12 | >10 | <−10 | NF |
| Dihydro‐3,4‐dimethyl‐2(3H)‐furanone | 0.05 ± 0.01a | 0.02 ± 0.00b | 0.00 ± 0.00c | −1.17 | >10 | <−10 | NF |
| NI14 | 0.12 ± 0.05a | 0.05 ± 0.02a | 0.00 ± 0.00b | −1.22 | >10 | <−10 | NF |
| NI15 | 0.34 ± 0.08a | 0.14 ± 0.03b | 0.00 ± 0.00c | −1.29 | >10 | <−10 | NF |
| NI16 | 0.16 ± 0.04a | 0.06 ± 0.01b | 0.00 ± 0.00c | −1.39 | >10 | <−10 | NF |
| Ribono‐1,4‐lactone | 0.12 ± 0.02a | 0.04 ± 0.01b | 0.10 ± 0.03a,b | −1.45 | −1.19 | −0.26 | NF |
| NI17 | 0.23 ± 0.05a | 0.08 ± 0.02b | 2.42 ± 0.32c | −1.48 | −4.87 | 3.39 | NF |
| 3‐Penten‐2‐one | 0.21 ± 0.06a | 0.07 ± 0.02b | 0.00 ± 0.00c | −1.65 | >10 | <−10 | NF |
| NI18 | 0.05 ± 0.02a | 0.02 ± 0.01a,b | 0.00 ± 0.00b | −1.68 | >10 | <−10 | NF |
| Mannobiose | 4.03 ± 1.09a | 1.13 ± 0.55b | 15.40 ± 1.52c | −1.84 | −3.77 | 1.93 | NF |
| NI19 | 0.37 ± 0.11a | 0.09 ± 0.03b | 0.00 ± 0.00c | −2.03 | >10 | <−10 | NF |
| NI20 | 0.58 ± 0.17a | 0.13 ± 0.03b | 0.00 ± 0.00c | −2.20 | >10 | <−10 | NF |
| NI21 | 0.34 ± 0.15a | 0.05 ± 0.01a | 0.00 ± 0.00b | −2.88 | >10 | <−10 | NF |
| Tricosane | 0.00 ± 0.00a | 0.00 ± 0.00a | 0.69 ± 0.09b | ND | <−10 | >10 | NF |
| Ethyl‐d‐glucopyranoside | 0.00 ± 0.00a | 0.00 ± 0.00a | 0.02 ± 0.00b | ND | <−10 | >10 | NF |
NF = not found; NI = not identified.
Metabolites with zero area values were below the detection limit of the GC‐MS instrument.
Different letters represent significant differences in the relative intensities of each metabolite when comparing N, SV, and CMV samples.
Values represent the logarithm base 2 of the fold change (log2 FC) in metabolite intensity between two sample classes.
All SV samples were characterized by the presence of rhamnose as this metabolite was not detected in N or CMV‐infected plants. Similarly, only SV samples showed undetectable levels of allothreonine and trehalose. Significantly higher levels of nine other metabolites, including methyl glucofuranoside, ribonic acid, fructose, hexadecenoic acid, allofuranose, butanedioic acid, and glucopyranosiduronic acid, were also characteristic of SV when compared to N samples. Conversely, the levels of 13 metabolites were significantly lower in SV than N plants, including octadecanoic acid, glyceryl‐glycoside, dihydro‐3,4‐dimethyl‐2(3H)‐furanone, stigmasterol, ribono‐1,4‐lactone, 3‐penten‐2‐one, mannobiose, maltose, and cellobiose. Significant differences (P < 0.05) between SV and CMV‐infected samples were observed in 68 metabolites, including 47 upregulated and 21 downregulated in SV when compared to the CMV‐infected plants using Student's t‐tests. Among them, sugars and sugar derivatives such as rhamnose, fructose, methyl glucofuranoside, allopyranose, lactose, and galacturonic acid; amino acids including phenylalanine and l‐lysine; and organic acids like butanoic acid and 2‐butenedioic acid were among the significantly upregulated metabolites. Likewise, some sugars and sugar derivatives like ribonic acid, allofuranose, sucrose, mannobiose, and trehalose; organic acids like butanedioic and hexadecenoic acids; and amino acids such as allo‐threonine, leucine, and isoleucine were among the significantly downregulated metabolites.
Results of the Student's t‐tests showed significant differences (P < 0.05) in the levels of 76 metabolites, including 24 metabolites upregulated and 52 downregulated in CMV‐infected plants when compared to N plants. Among the upregulated metabolites, two metabolites (tricosane and ethyl‐d‐glucopyranoside) were exclusively detected in CMV samples. Other upregulated metabolites included sugars and sugar derivatives such as trehalose, ribitol, methyl galactopyranoside, sucrose, ribofuranose, allofuranose, and mannobiose; amino acids like allothreonine, leucine, and isoleucine; and organic acids such as hexadecenoic and butanedioic acids. Among the downregulated metabolites, 45 metabolites including sugars and sugar derivatives such as methyl glucofuranoside, cellobiose, methyl galactoside, fructopyranose, lactose, and galacturonic acid; amino acids like phenylalanine, l‐lysine, and asparagine; and organic acids like butanoic, 2‐butenedioic, 9,12‐octadecadienoic, and linolenic acids were only absent in plants infected with CMV. The disaccharide cellobiose was detected in N plants only.
Pathway mapping suggests a significant upregulation of the fructose and mannose metabolism in SV plants, but other metabolic pathways were significantly downregulated in the dwarf variants, including fatty acid degradation, starch and sucrose metabolism, biosynthesis of unsaturated fatty acids, and steroid biosynthesis pathways. Similarly, various pathways were significantly downregulated in CMV samples, including fructose and mannose metabolism, galactose metabolism, starch and sucrose metabolism, biosynthesis of unsaturated fatty acids, fatty acid degradation, biosynthesis of amino acids, butanoate metabolism, steroid biosynthesis, and purine metabolism, whereas other pathways were significantly upregulated, including valine, leucine, and isoleucine biosynthesis; glycine, serine, and threonine metabolism; tyrosine metabolism; and riboflavin metabolism, as well as the pentose phosphate pathway.
Multivariate PCA showed a clear grouping of the CMV‐infected plants and a partial grouping of the SV and N samples (Fig. 1A). Principal components (PC) 1 and 2 accounted for 30.74% of the total variation, and sample grouping occurred mostly on PC1. All CMV‐infected plants were characterized with high positive scores on PC1, whereas SV samples showed low negative score values. All N samples yielded highly negative score values in PC1. The metabolites with the highest absolute loading values in PC1 were sugars and sugar derivatives such as mannobiose, l‐altrose, galacturonic acid, hexopyranose, glucopyranosiduronic acid, d‐glucosone, and methyl galactopyranoside; organic acids including propanoic, butanedioic, octadecanoic, hexadecanoic, linolenic, and 2‐butenedioic acids; and other metabolites such as glyceryl‐glycoside and butanal (Fig. 1B).
Figure 1.
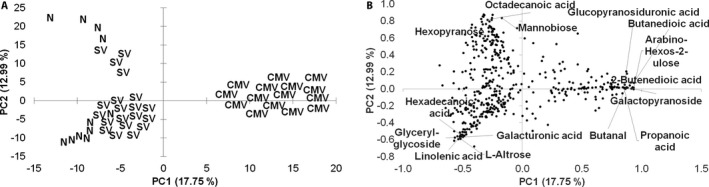
Score (A) and loading plot (B) of the principal components analysis (PCA) run on dwarf (SV), normal‐healthy (N), and cucumber mosaic virus (CMV)–infected plants. Values in parentheses represent the percentage of the variance explained by each component.
Prediction models for N, SV, and CMV sample classes were built using PLS‐DA. The models consisted of two PLS components and yielded total R 2 and Q 2 values of 0.87 and 0.71, respectively (Table 2). A 100% classification accuracy was observed in all samples used for calibration of each model, and the PLS scores plot showed a more marked grouping of SV, N, and CMV‐infected samples than PCA (Fig. 2A). The metabolites with the highest VIP scores (from 1.5 to 2.0) for the three models were sugars and sugar derivatives such as methyl glucopyranoside, glucopiranosiduronic acid, sucrose, gluconic acid, ribofuranose, mannitol, lactose, and galactopyranose; organic acids including butanedioic, 2‐butenedioic, and propanoic acids; amino acids like leucine, allo‐threonine, and serine; and the aldehyde butanal (Fig. 2B). However, none of the metabolites showed VIP values above 2 and were not considered among the potential biomarkers for SV. The potential of the three models combined for SV diagnostic was tested with 15 plants not previously used for model calibration, including five N, six SV, and four CMV‐infected plants. The models were 100% accurate in all validation tests (Fig. 3).
Table 2.
Quality values for the PLS‐DA prediction models
| Model | R 2 | Q 2 |
|---|---|---|
| CMV | 0.972 | 0.957 |
| N | 0.799 | 0.578 |
| SV | 0.838 | 0.599 |
| Total | 0.870 | 0.711 |
CMV = cucumber mosaic virus–infected plants; N = normal‐healthy plants; Q 2 = goodness of prediction obtained from the jacknife cross‐validation data; R 2 = coefficient of determination for multivariate analysis; SV = dwarf plants.
Figure 2.
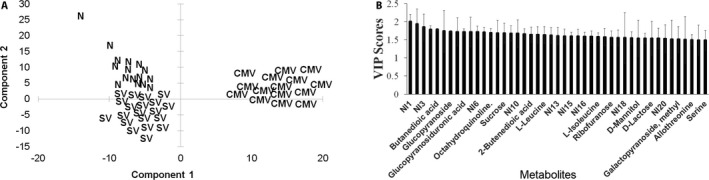
Scores (A) and variable importance in the projection (B) plots of the partial least squares discriminant analysis (PLS‐DA) run on dwarf (SV), normal‐healthy (N), and cucumber mosaic virus (CMV)–infected samples. NI = unidentified metabolites; VIP = variable importance in the projection. VIP bars are presented as mean and standard deviation of the metabolites with VIP scores of 1.5 or greater.
Figure 3.
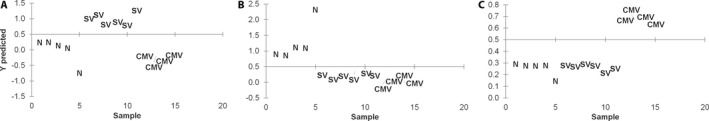
Validation of the SV (A), N (B), and CMV (C) models. SV = dwarf; N = normal‐healthy; CMV = cucumber mosaic virus–infected.
DISCUSSION
A novel metabolite‐based approach using PLS‐DA to detect SV in banana plants is presented, and a number of metabolite biomarkers of banana SV were discovered and validated. Metabolite biomarkers can be qualitative (presence/absence) or quantitative (abundance) (Cevallos‐Cevallos et al., 2009). Quantitative biomarkers can be selected based on VIP values (Steinfath et al., 2010) or differential expression assessment by Student's t‐test (Cevallos‐Cevallos et al., 2009), among other methods. Because VIP values were below 2, we used the P values obtained by Student's t‐tests to select potential quantitative biomarkers.
Among the qualitative biomarkers of SV, rhamnose can be produced in banana plants through the fructose and mannose pathway; derivatives of this metabolite are required for the synthesis of important cell wall components in plants (Oka et al., 2007). Therefore, the presence of this monosaccharide in SV samples only might have contributed to the thicker pseudostem observed in the dwarf variants when compared to the N or CMV‐infected plants (Oh et al., 2007; Ferrero‐Serrano and Assmann, 2016). Trehalose—another qualitative biomarker of SV plants—is produced in plants through the dephosphorylation of trehalose‐6‐phosphate (T6P) catalyzed by T6P phosphatase (TPP) (Grennan, 2007). T6P is considered an important signaling metabolite that is involved in the regulation of plant growth (O'Hara et al., 2013). Molecules of T6P are produced by T6P synthase (TPS), and alterations in this enzyme have been shown to inhibit plant growth and trehalose production (O'Hara et al., 2013). The undetectable levels of trehalose in SV plants suggest aberrations in TPP or TPS activity resulting in the accumulation or absence of T6P, respectively. Both the absence and excess of T6P have caused stunted growth in plants (O'Hara et al., 2013) and may have contributed to the dwarfism observed in the SV samples. Further research is needed to elucidate the T6P metabolism in SV plants. Similarly, allothreonine—undetected in SV plants only—is produced by the glycine, serine, and threonine metabolism pathway, but the role of this amino acid in plant development is still unclear.
Cellobiose was the only qualitative (presence) biomarker of N plants only. This disaccharide is an intermediary metabolite in the synthesis of cellulose, a main constituent of plant cell walls (Maleki et al., 2016). The deposition of cellulose in the cell wall is essential to plant growth, and deficient cellulose production has yielded dwarfed plant mutants (Taylor, 2008). The absence of cellobiose in SV and CMV‐infected plants suggests reduced cellulose synthesis that may have contributed to the stunted growth observed in the samples.
The CMV‐infected plants showed 47 qualitative biomarkers out of which 45 were absence biomarkers (Table 1). The only CMV presence biomarkers were tricosane and ethyl‐d‐glucopyranoside, and further research is needed to establish the role of both metabolites in CMV plant infection. Reduction in levels of various metabolites has previously been reported in plant infection (Cevallos‐Cevallos et al., 2012), and an overall reduced metabolism may be occurring in CMV‐infected plants. Particularly, the absence of l‐lysine might have contributed to the stunted growth of plants infected with the virus as this metabolite has been reported to participate in reactions associated with plant growth and development (Tomar et al., 2013).
The downregulated pathways observed in SV and CMV plants, including fatty acid degradation, starch and sucrose metabolism, biosynthesis of unsaturated fatty acids, and steroid biosynthesis pathways, suggest overall inhibited assimilatory processes in the plant samples (Rojas et al., 2014). Upregulated pathways specific to CMV‐infected plants included the metabolism of various amino acids (e.g., tyrosine, leucine, isoleucine, glycine, serine, and threonine) previously reported to accumulate in plants infected with pathogens. Furthermore, the upregulation of the pentose phosphate pathway—observed in the CMV plants—has been suggested to promote the generation of reactive oxygen species and pathogenesis‐related proteins in infected plants (Rojas et al., 2014).
A clear PCA and PLS‐DA class grouping was observed CMV‐infected samples, whereas separation between SV and N samples was less obvious, suggesting similarities in the GC‐MS metabolite profile of both groups. SV and N plants were not affected by biotic stresses as the CMV‐infected plants were, and biotic stress has been shown to yield a higher number of metabolic differences than those caused by abiotic conditions in plants (Cevallos‐Cevallos et al., 2011).
PLS‐DA with validation using additional data sets was selected for the development of prediction models because this method is not sensitive to multicollinearity (Palermo et al., 2009; Worley and Powers, 2012) and has been reported to yield better class separation than other techniques such as random forest (Gromski et al., 2015). Additionally, the three classes analyzed in this experiment prevent the use of procedures designed to solve a two‐class problem, including the support vector machines algorithm. PLS‐DA has yielded similar results than the commonly used principal component–discriminant function analysis (PC‐DFA), but the ability to rank the variables responsible for class separation is greater in PLS‐DA when compared to PC‐DFA (Gromski et al., 2015). The VIP scores were used for variable selection as this technique has outperformed other methods including PLS, PCR, and Lasso regression coefficients (Palermo et al., 2009).
The PLS‐DA prediction models were able to accurately classify the samples showing dwarf SV. Metabolite‐based PLS models have been successfully used to predict phenotypic traits in plants, including the susceptibility to black spot bruising and chip quality in potato tubers (Steinfath et al., 2010). To the best of our knowledge, this is the first report of a metabolite‐based PLS‐DA model for diagnosis of dwarf bananas. The proposed methodology could be directly applied in banana plant production facilities to detect dwarf variants and discriminate SV from CMV‐infected bananas before releasing the plants to the field as SV diagnosis using only visual symptoms can be highly unreliable. A symptoms‐based preselection of plants is required before this methodology is used. Further research is needed to assess the suitability of metabolomics‐based prediction models to detect SV in banana plants at the early production stages and before the first symptoms appear.
DNA markers have also been proposed to detect SV in banana plants, but these markers have not been shown to have any association with plant phenotypic variations. For instance, the use of RAPDs was able to detect 51.4% genetic variation in banana plants produced by tissue culture (Sheidai et al., 2008), but SV was not phenotypically confirmed and no association between DNA markers and alterations of visual traits was made. Similarly, an ISSR‐based study reported about 5.0% genetic variation in micropropagated Musa spp., but morphological variations were not observed in the generated clones (Ray et al., 2006). The PLS‐DA model proposed in this study represents the first SV diagnosis method at the phenotypic level.
The proposed models were also able to diagnose CMV infection in banana plants at the greenhouse establishment phase. Metabolite‐based PLS models have been used for prediction of plant pathogen infections, including the presence of Botrytis cinerea in commercial berry groves (Hong et al., 2012). Results show the potential of metabolite‐based PLS models to detect CMV in young banana plants, but further model development is needed for CMV detection in adult plants from commercial groves, as the disease mostly occurs in banana fields where various subgroups of the pathogen can be present (Yeturu et al., 2016).
In conclusion, the metabolite profile of plants with dwarf SV offered additional insights into banana dwarfism and provided a novel alternative for the specific detection of dwarf banana variants. Because of the observed metabolome–phenotype relationships, metabolite‐based detection of dwarf SV has the potential to become a superior SV diagnosis tool when compared to molecular‐based methods. This is the first metabolite‐based characterization and detection of somaclonal variation, showing the potential of metabolomics tools to understand and selectively detect phenotypic variations in plants.
AUTHOR CONTRIBUTIONS
J.M.C.‐C. conceived, designed, wrote, and approved the article. C.J. performed sample processing, data aquisition, and data pre‐treatment. M.G.M.‐Z. performed data validation and statistical analysis. M.J.M.‐M. performed data analysis and interpretation. D.E.O. reviewed the manuscript and obtained funding. J.A.F.‐C. designed the experiment, performed sampling, and interpreted the data.
ACKNOWLEDGMENTS
This project was supported by Vlaamse Interuniversitaire Raad–Universitaire Ontwikkelingssamenwerking (VLIR UOS; grant 2014RIP24).
Cevallos‐Cevallos, J. M. , Jines C., Maridueña‐Zavala M. G., Molina‐Miranda M. J., Ochoa D. E., and Flores‐Cedeno J. A.. 2018. GC‐MS metabolite profiling for specific detection of dwarf somaclonal variation in banana plants. Applications in Plant Sciences 6(11): e1194.
LITERATURE CITED
- Bairu, M. W. , Aremu A. O., and Staden J.. 2010. Somaclonal variation in plants: Causes and detection methods. Plant Growth Regulation 63(2): 147–173. [Google Scholar]
- Basavaraj, S. , Rangaswamy K. T., and Bhagyashree M.. 2017. Molecular characterization of CMV infecting banana from Karnataka based on complete coat protein gene sequence. International Journal of Current Microbiology and Applied Sciences 6(11): 3758–3763. [Google Scholar]
- Caldana, C. , Degenkolbe T., Cuadros‐Inostroza A., Klie S., Sulpice R., Leisse A., Steinhauser D., et al. 2011. High‐density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant Journal 67(5): 869–884. [DOI] [PubMed] [Google Scholar]
- Cevallos‐Cevallos, J. , Rouseff R., and Reyes‐De‐Corcuera J. I.. 2009. Untargeted metabolite analysis of healthy and Huanglongbing‐infected orange leaves by CE‐DAD. Electrophoresis 30(7): 1240–1247. [DOI] [PubMed] [Google Scholar]
- Cevallos‐Cevallos, J. M. , García‐Torres R., Etxeberria E., and Reyes‐De‐Corcuera J. I.. 2011. GC‐MS analysis of headspace and liquid extracts for metabolomic differentiation of citrus Huanglongbing and zinc deficiency in leaves of ‘Valencia’ sweet orange from commercial groves. Phytochemical Analysis 22(3): 236–46. [DOI] [PubMed] [Google Scholar]
- Cevallos‐Cevallos, J. M. , Futch D. B., Shilts T., Folimonova S. Y., and Reyes‐De‐Corcuera J. I.. 2012. GC–MS metabolomic differentiation of selected citrus varieties with different sensitivity to citrus huanglongbing. Plant Physiology and Biochemistry 53: 69–76. [DOI] [PubMed] [Google Scholar]
- Cevallos‐Cevallos, J. M. , Gysel L., Maridueña‐Zavala M. G., and Molina‐Miranda M. J.. 2018. Time‐related changes in volatile compounds during fermentation of bulk and fine‐flavor cocoa (Theobroma cacao) beans. Journal of Food Quality 2018: 1–14. [Google Scholar]
- Chen, J. , Xie J., Duan Y., Hu H., Hu Y., and Li W.. 2016. Genome‐wide identification and expression profiling reveal tissue‐specific expression and differentially‐regulated genes involved in gibberellin metabolism between Williams banana and its dwarf mutant. BMC Plant Biology 16(1): 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheepa, R. , and Paranjothi S.. 2010. Transmission of Cucumber Mosaic Virus (CMV) infecting banana by aphid and mechanical methods. Emirates Journal of Food and Agriculture 22(2): 117–129. [Google Scholar]
- Ferrero‐Serrano, Á. , and Assmann S. M.. 2016. The α‐subunit of the rice heterotrimeric G protein, RGA1, regulates drought tolerance during the vegetative phase in the dwarf rice mutant d1. Journal of Experimental Botany 67(11): 3433–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn, O. 2001. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comparative and Functional Genomics 2(3): 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations . 2017. FAOSTAT. Website http://www.fao.org/faostat/en/#data/QC [accessed 1 May 2018].
- Grennan, A. K. 2007. The role of trehalose biosynthesis in plants. Plant Physiology 144(1): 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromski, P. S. , Muhamadali H., Ellis D. I., Xu Y., Correa E., Turner M. L., and Goodacre R.. 2015. A tutorial review: Metabolomics and partial least squares‐discriminant analysis – a marriage of convenience or a shotgun wedding. Analytica Chimica Acta 879: 10–23. [DOI] [PubMed] [Google Scholar]
- Hong, Y. S. , Martinez A., Liger‐Belair G., Jeandet P., Nuzillard J.‐M., and Cilindre C.. 2012. Metabolomics reveals simultaneous influences of plant defence system and fungal growth in Botrytis cinerea–infected Vitis vinifera cv. Chardonnay berries. Journal of Experimental Botany 63(16): 5773–5785. [DOI] [PubMed] [Google Scholar]
- Hu, C. , Tohge T., Chan S.‐A., Song Y., Rao J., Cui B., Lin H., et al. 2016. Identification of conserved and diverse metabolic shifts during rice grain development. Scientific Reports 6(1): 20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli, Y. , Reuveni O., and Lahav E.. 1991. Qualitative aspects of somaclonal variations in banana propagated by in vitro techniques. Scientia Horticulturae 48(1–2): 71–88. [Google Scholar]
- Kanehisa, M. , and Goto S.. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research 28(1): 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki, S. S. , Mohammadi K., and Ji K.. 2016. Characterization of cellulose synthesis in plant cells. Scientific World Journal 2016: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maridueña‐Zavala, M. G. , Freire‐Peñaherrera A., Cevallos‐Cevallos J. M., and Peralta E. L.. 2017. GC‐MS metabolite profiling of Phytophthora infestans resistant to metalaxyl. European Journal of Plant Pathology 149(3): 563–574. [Google Scholar]
- Maruyama, K. , Urano K., Yoshiwara K., Morishita Y., Sakurai N., Suzuki H., Kojima M., et al. 2014. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiology 164(4): 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLafferty, F. W. 2009. Wiley Registry of Mass Spectral Data, with NIST 2008, p. 100 Wiley, New York, New York, USA. [Google Scholar]
- Oh, T. J. , Cullis M. A., Kunert K., Engelborghs I., Swennen R., and Cullis C. A.. 2007. Genomic changes associated with somaclonal variation in banana (Musa spp.). Physiologia Plantarum 129(4): 766–774. [Google Scholar]
- O'Hara, L. E. , Paul M. J., and Wingler A.. 2013. How do sugars regulate plant growth and development? New insight into the role of trehalose‐6‐phosphate. Molecular Plant 6(2): 261–274. [DOI] [PubMed] [Google Scholar]
- Oka, T. , Nemoto T., and Jigami Y.. 2007. Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP‐d‐glucose to UDP‐l‐rhamnose conversion. Journal of Biological Chemistry 282(8): 5389–5403. [DOI] [PubMed] [Google Scholar]
- Palermo, G. , Piraino P., and Zucht H. D.. 2009. Performance of PLS regression coefficients in selecting variables for each response of a multivariate PLS for omics‐type data. Advances and Applications in Bioinformatics and Chemistry 2(1): 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, T. , Dutta I., Saha P., Das S., and Roy S. C.. 2006. Genetic stability of three economically important micropropagated banana (Musa spp.) cultivars of lower Indo‐Gangetic plains, as assessed by RAPD and ISSR markers. Plant Cell, Tissue and Organ Culture 85(1): 11–21. [Google Scholar]
- Reuveni, O. , and Israeli Y.. 1990. Measures to reduce somaclonal variation in in vitro propagated bananas. Acta Horticulturae 275: 307–314. [Google Scholar]
- Rodrigues, P. H. V. , Tulmann Neto A., Cassieri Neto P., and Mendes B. M. J.. 1998. Influence of the number of subcultures on somoclonal variation in micropropagated Nanicão (Musa spp., AAA group). Acta Horticulturae 490: 469–473. [Google Scholar]
- Rojas, C. M. , Senthil‐Kumar M., Tzin V., and Mysore K. S.. 2014. Regulation of primary plant metabolism during plant‐pathogen interactions and its contribution to plant defense. Frontiers in Plant Science 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahijram, L. , Soneji J. R., and Bollamma K. T.. 2003. Analyzing somaclonal variation in micropropagated bananas (Musa spp.). In Vitro Cellular and Developmental Biology ‐ Plant 39(6): 551–556. [Google Scholar]
- Sheidai, M. , Aminpoor H., Noormohammadi Z., and Farahani F.. 2008. RAPD analysis of somaclonal variation in banana (Musa acuminate [sic] L.) cultivar Valery. Acta Biologica Szegediensis 52: 307–311. [Google Scholar]
- Steinfath, M. , Strehmel N., Peters R., Schauer N., Groth D., Hummel J., Steup M., et al. 2010. Discovering plant metabolic biomarkers for phenotype prediction using an untargeted approach. Plant Biotechnology Journal 8(8): 900–911. [DOI] [PubMed] [Google Scholar]
- Sun, T.‐P. 2000. Gibberellin signal transduction. Current Opinion in Plant Biology 3(5): 374–380. [DOI] [PubMed] [Google Scholar]
- Taylor, N. G. 2008. Cellulose biosynthesis and deposition in higher plants. New Phytologist 178(2): 239–252. [DOI] [PubMed] [Google Scholar]
- Tomar, P. C. , Lakra N., and Mishra S. N.. 2013. Cadaverine: A lysine catabolite involved in plant growth and development. Plant Signaling and Behavior 8(10): e25850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth, B. , Parich A., Bueschl C., Schoefback D., Neumann N. K. N., Kluger B., Schuster K., et al. 2015. GC–MS based targeted metabolic profiling identifies changes in the wheat metabolome following deoxynivalenol treatment. Metabolomics 11(3): 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M. , Balazadeh S., Tohge T., Erban A., Giavalisco P., Kopka J., Mueller‐Roeber B., et al. 2013. Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis . Plant Physiology 162(3): 1290–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley, B. , and Powers R.. 2012. Multivariate analysis in metabolomics. Current Metabolomics 1(1): 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , and Li L.. 2016. Sample normalization methods in quantitative metabolomics. Journal of Chromatography A 1430: 80–95. [DOI] [PubMed] [Google Scholar]
- Yeturu, S. , Méndez K., Garrido P., Serrano S., and Garrido A.. 2016. Serological and molecular identification of cucumber mosaic virus (CMV) infecting banana crops in Ecuador. Ecuador Es Calidad 3: 17–22. [Google Scholar]
- Yogendra, K. N. , Kushalappa A. C., Sarmiento F., Rodriguez E., and Mosquera T.. 2014. Metabolomics deciphers quantitative resistance mechanisms in diploid potato clones against late blight. Functional Plant Biology 42(3): 284–298. [DOI] [PubMed] [Google Scholar]
- Zitter, T. A. , and Murphy J. F.. 2009. Cucumber mosaic. Plant Health Instructor 10.1094/PHI-I-2009-0518-01. [DOI] [Google Scholar]


