Abstract
The transcription factors C/EBPα and PU.1 are upregulated by RANKL through activation of its receptor RANK during osteoclastogenesis and are critical for osteoclast differentiation. Herein we investigated the mechanisms underlying how C/EBPα and PU.1 regulate osteoclast differentiation in response to RANK signaling. We showed that C/EBPα or PU.1 overexpression could initiate osteoclastogenesis and upregulate the expressions of the osteoclast genes encoding the nuclear factor of activated T-cells, C1, cathepsin K, and tartrate-resistant acid phosphatase independently of RANKL. However, while PU.1 upregulated C/EBPα, C/EBPα could not upregulate PU.1. RANK has a unique cytoplasmic domain, 535IVVY538 motif, which is crucial for osteoclast differentiation. We demonstrated that mutational inactivation of RANK IVVY motif blocked osteoclast differentiation and significantly attenuated C/EBPα, but not PU.1, expression, indicating that RANK-IVVY-induced signaling is dispensable to PU.1 upregulation during osteoclastogenesis. However, C/EBPα or PU.1 overexpression failed to promote osteoclastogenesis in cells expressing mutated RANK IVVY motif. We noted that RANK-IVVY-motif inactivation significantly repressed osteoclast genes as compared with a vector control, suggesting that IVVY motif might also negatively regulate osteoclast inhibitors during osteoclastogenesis. Consistently, IVVY-motif inactivation triggered upregulation of RBP-J, a potent osteoclast inhibitor, during osteoclastogenesis. Notably, C/EBPα or PU.1 overexpression in cells expressing mutated RANK IVVY motif failed to control the deregulated RBP-J expression, resulting in repression of osteoclast genes. Accordingly, RBP-J silencing in the mutant cells rescued osteoclastogenesis with C/EBPα or PU.1 overexpression. In conclusion, we revealed that while PU.1 and C/EBPα are critical for osteoclastogenesis, they respond differently to RANKL-induced activation of RANK IVVY motif.
Keywords: C/EBPα, differentiation, IVVY motif, osteoclast, PU.1, RANK, RBP-J
1. Introduction
Bone is continuously remodeled through the balance activities of the osteoblasts, the bone-forming cells, and the osteoclasts, the bone resorbing cells (1,2). During skeletal remodeling and healthy states, bone resorption is synchronized by bone formation. However, in many bone diseases, the rate of bone resorption exceeds that of bone formation (3,4). As such, the osteoclast has been regarded as a key player in the bone loss stemming from various bone diseases (5). Osteoclasts are polykaryons that are originated from the macrophages upon stimulation by the macrophage colony-stimulating factor (M-CSF) and the receptor activator of NF-қB (RANK) ligand (RANKL) (6). M-CSF promotes the proliferation and survival of the bone marrow macrophages (BMMs), and RANKL, through its receptor RANK, mediates the differentiation of BMMs into osteoclasts. Specifically, activation of RANK by RANKL strongly upregulates the expressions of many crucial transcription factors, such as CCAAT/enhancer binding protein α (C/EBPα) (7), a member of the C/EBP family of transcription factors, and the spleen focus-forming virus proviral integration 1 (PU.1 also called Spi-1) (8), a member of the ETS family transcription factor. C/EBPα and PU.1 are both critical for osteoclastogenesis through induction or upregulation of osteoclast genes (6–8). Importantly, RANK has a specific motif within its cytoplasmic domain, 535IVVY538, which is essential for osteoclast formation by regulating gene expression (9–12).
C/EBPα is critical for hematopoiesis through its ability to induce the expressions of genes responsible for myeloid cell differentiation, including macrophages (13,14). Mice deficient in the C/EBPα gene die shortly after birth and exhibit defective granulocyte development as well as impaired homeostasis (15,16). We have recently demonstrated that newborn C/EBPα-deficient mice also display osteopetrosis due to impaired osteoclast development (7). Consistently, C/EBPα can induce the expressions of the osteoclast genes encoding nuclear factor of activated T-cells, C1 (NFATc1), cathepsin K (Ctsk), and tartrate-resistant acid phosphatase (TRAP) during osteoclast differentiation (17). Similarly, PU.1 is also important for the development of cells of the hematopoietic lineage, including macrophages (18–21). Mice deficient in the PU.1 gene die during embryonic development or shortly after birth (8). The PU.1-deficient mice also develop osteopetrosis from impaired osteoclast development (8). Furthermore, PU.1 is critical for the induction of NFATc1, Ctsk, and TRAP during osteoclast differentiation (22–24).
Although C/EBPα and PU.1 are both upregulated by RANKL and are also crucial for osteoclast differentiation by inducing gene expression (6–8,17), the mechanisms underlying how C/EBPα and PU.1 regulate osteoclast differentiation in response to RANK activation are unknown. Moreover, while the RANK IVVY motif is essential for induction of osteoclast genes including Ctsk and TRAP which are known to be regulated by C/EBPα and PU.1 (7,8,17), the roles of RANK IVVY motif in regulating the expressions of C/EBPα and PU.1 have not been investigated. The current study was aimed at investigating the roles of C/EBPα and PU.1 in mediating osteoclast differentiation in response to RANKL/RANK signaling by using a gain-of-function strategy in a RANK-IVVY motif dependent manner. The results provide an important insight into the roles of C/EBPα, PU.1, and RANK signaling in osteoclast differentiation.
2. Materials and Methods
2.1. Reagents
The chemicals were purchased from Sigma. Recombinant mouse RANKL (catalog no. 462-TEC) and M-CSF (catalog no. 416-ML) were obtained from R&D Systems. Anti-Human FAS activating antibody (α-FAS, catalog no. 05–201) was obtained from Millipore. Anti-FLAG antibody (catalog no. F1804) was from Sigma. Anti-β actin (catalog no. SC-81178) and anti-RBP-J (catalog no. SC-271128) antibodies were from Santa Cruz Biotechnology. Recombinant recognition sequence binding protein at the Jκ site (RBP-J) and Scramble shRNA lentiviral constructs were purchased from Sigma.
2.2. Plasmid Generation and Viral Transduction
The pMX-puro-3xFLAG-C/EBPα (FLAG-C/EBPα) and pMX-puro-3xFLAG constructs were generated in a previous study (17). The pMX-puro-3xFLAG-PU.1 (FLAG-PU.1) construct was generated by first amplifying the mouse PU.1 cDNA from the pSport6-PU.1 vector (Addgene). We then subcloned the amplified PU.1 cDNA in-frame with the 3xFLAG sequence into the pMx-puro-3xFLAG vector. The resulted construct was confirmed by sequencing. The pMX-puro-GFP (GFP), pMX-puro-FAS-RANK (FAS-RANK), and pMX-puro-FAS-mIVVY (FAS-mIVVY) vectors were generated and kindly provided by Dr. Xu Feng (University of Alabama at Birmingham) (9,25). The 293GPG retroviral packaging cell line was used for retrovirus generation as described previously (26). In brief, 293GPG cells were cultured in Dulbecco’s Modified Eagle Medium with 10% heat-inactivated fetal bovine serum, G418, tetracycline, penicillin/streptomycin, and puromycin before being transfected with pMX retroviral constructs using the calcium phosphate precipitation method. Retroviral supernatant was harvested at 2, 3, and 4 days post transfection. For the lentivirus generation, the RBP-J lentiviral vector or a Scramble shRNA lentiviral construct along with packaging vectors were co-transfected into HEK-293 cells using the calcium phosphate precipitation method. The lentiviral supernatant was collected at 60 hours post transfection. The viral supernatant was used to infect BMMs for osteoclastogenesis assays.
2.3. In Vitro Osteoclastogenesis Assays
BMMs were isolated from long bones of 4-to 6-week old C57BL/6 mice, and 5 × 104 cells/well in 24-well culture dishes were cultured in α-Minimal Essential Medium with 10% heat-inactivated fetal bovine serum and M-CSF (20 ng/ml) for 24 hours. Some cells were then directly differentiated into osteoclasts as indicated in individual experiments, and other cells were infected with a virus before being submitted to osteoclastogenesis assays as indicated in the related experiments (27,28). At the end of the assays, the cultures were stained for TRAP activity using a leukocyte acid phosphatase kit (catalog no. 387-A, Sigma) according to the instruction of the manufacturer to examine osteoclast formation. The assays were quantified by counting and/or accessing the size of the multinucleated TRAP-positive cells (more than three nuclei) in representative areas. The experiments involving mice were approved by The University of Alabama at Birmingham Institutional Animal Care and Use Committee. The osteoclastogenesis assays were carried in duplicate and repeated independently at least three times.
2.4. Western Blotting Analysis
Western blotting was carried out as described in a previous study (29). In brief, cells were cultured as indicated in the individual experiments before protein collection for gel electrophoresis. Membranes were washed, and enhanced chemiluminescence detection was carried using Luminata Forte HRP Substrate from Millipore. Membranes were visualized using a C-DiGit® Blot Scanner and Image Studio Software from Li-Cor. The Western blotting analysis was repeated independently at least three times using β actin as a loading control.
2.5. Quantitative Real-time PCR (qPCR) Analysis
qPCR analysis was performed as described in a previous study (30). In brief, cells were cultured as indicated in the individual experiments, and total RNA was collected using TRIzol reagent (Life Technologies). 1 μg of total RNA was transcribed into cDNA using the ProtoScript® First Strand cDNA Synthesis Kit (New England BioLabs) according to the instruction of the manufacturer. qPCR reactions were carried by utilizing the Fast SYBR® Green Master Mix reagent (Life Technologies) using hypoxanthine-guanine phosphoribosyl-transferase as an endogenous control for normalization. The qPCR analysis was repeated independently three times.
2.6. Reverse Transcription PCR (RT-PCR) Analysis
BMMs were cultured as indicated in the individual experiments, and total RNA was collected for cDNA synthesis as indicated above in 2.5. Gene amplification was carried using Taq DNA polymerase (catalog no E001, Novo Protein) (25). RT-PCR primers to detect the chimeric receptors (FAS-RANK and FAS-mIVVY) are 5′-ATGCTGGGCATCTGGACCCTCCTA-3′ for the Human FAS extracellular domain (Forward) and 5′-GAAGTCACAGCCCTCAGAATC-3′ for the mouse RANK intracellular domain (reverse). Primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), used as a loading control, are 5’-TCATTGAGAGCAATGCCAGC-3’ (Forward) and 5′-ACATCATCCCTGCATCCACTG-3’ (Reverse). The RT-PCR reaction was loaded on 2% agarose gel for electrophoretic analysis. The RT-PCR analysis was repeated independently three times.
2.7. Statistical Analysis
Data are reported as averages ±SD. Statistical significance was assessed using the Student’s t test. p values less than 0.05 were considered significant.
3. Results
3.1. C/EBPα or PU.1 can initiate osteoclastogenesis independently of RANKL.
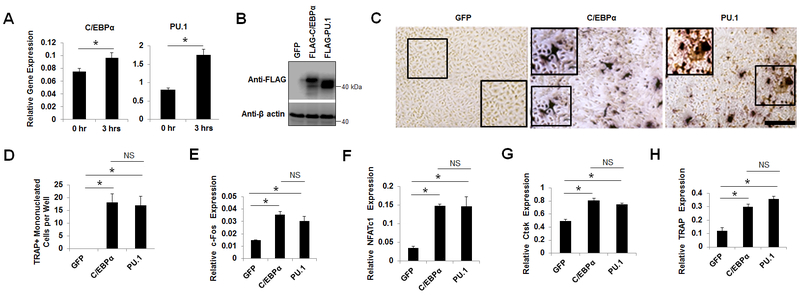
C/EBPα and PU.1 are critical for osteoclast formation both in vitro and in vivo (7,8,17,31). In order to examine the influence of C/EBPα and PU.1 on osteoclastogenesis, we first examined their roles in osteoclast lineage commitment. We stimulated BMMs, widely used as primary osteoclast precursors, with M-CSF plus RANKL for 0 or 3 hours and then accessed the expressions of C/EBPα and PU.1 by qPCR. The data showed that the combined stimulation of BMMs with M-CSF/RANKL could significantly upregulate C/EBPα and PU.1 (Fig. 1A), confirming previous reports that RANKL can upregulate C/EBPα and PU.1 very early during osteoclastogenesis (7,8). We then investigated the effects of C/EBPα or PU.1 overexpression in mediating osteoclast lineage commitment without RANKL stimulation (Fig. 1B-D). We have recently reported that C/EBPα overexpression could initiate osteoclastogenesis independently of RANKL (7,17). We confirmed this finding and showed that C/EBPα overexpression in BMMs, as confirmed by Western blotting (Fig. 1B), could generate TRAP-positive mononucleated cells independently of RANKL (Fig. 1C,D). Notably, we confirmed that, similarly to C/EBPα, PU.1 overexpression could also generate TRAP-positive mononucleated cells independently of RANKL (Fig. 1 B-D) (8,32). In confirming the abilities of C/EBPα and PU.1 to induce lineage commitment, we showed that C/EBPα or PU.1 overexpression could significantly induce the expressions of c-Fos (Fig. 1E), an early osteoclast transcription factor (33), and NFATc1 (Fig. 1F), a master transcriptional regulator of osteoclast differentiation, as compared to a GFP control (34). Importantly, we demonstrated that c-Fos or NFATc1 overexpression could not upregulate C/EBPα or PU.1 (Suppl. Fig. 1), confirming the previous studies that c-Fos and NFATc1 are target genes of C/EBPα and PU.1 during osteoclastogenesis (6–8). Moreover, C/EBPα or PU.1 overexpression could significantly induce the expressions of the osteoclast genes encoding Ctsk (Fig. 1G) and TRAP (Fig. 1H) as compared to a GFP control under the stimulation by M-CSF alone.
Fig. 1.
C/EBPα or PU.1 overexpression can initiate osteoclastogenesis independently of RANKL. A, analysis of C/EBPα and PU.1 expression in BMMs stimulated by M-CSF and RANKL for 0 hour or 3 hours by qPCR. B, analysis of gene overexpression in BMMs expressing a GFP control, FLAG-C/EBPα, or FLAG-PU.1 cultured with M-CSF for 4 days by Western blotting. C-H, BMMs expressing GFP, FLAG-C/EBPα (C/EBPα), or FLAG-PU.1 (PU.1) were cultured with M-CSF for 4 days. Some cells were submitted to TRAP staining to examine osteoclastogenesis (C) and then quantified for the number of TRAP-positive monucleated cells (D). Scale bar = 250 μm. The remaining cells were subjected to qPCR analysis for c-Fos (E), NFATc1 (F), Ctsk (G), or TRAP expression (H). Error bars show averages ± S.D. *, p < 0.05; NS, not significant..
Given that RANKL could transiently upregulate C/EBPα and PU.1 both of which could induce osteoclast lineage priming (Fig. 1), we examined the abilities of C/EBPα and PU.1 to upregulate each other independently of RANKL (Fig. 2). PU.1 overexpression significantly upregulated C/EBPα under stimulation by M-CSF alone as compared to a GFP control (Fig. 2A). However, C/EBPα overexpression failed to upregulate PU.1 in the absence of RANKL as compared to the GFP control (Fig. 2B). These results indicated that while overexpression of C/EBPα or PU.1 could similarly induce the lineage commitment, C/EBPα was a target gene of PU.1 during osteoclastogenesis.
Fig. 2.
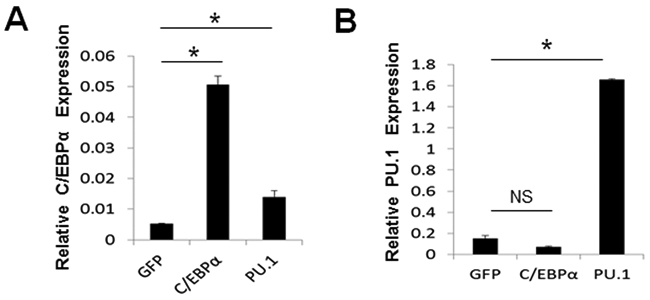
Analysis of the effects of C/EBPα or PU.1 in inducing the expressions of each other. A, analysis of C/EBPα expression in BMMs expressing a GFP control (GFP), FLAG-C/EBPα (C/EBPα), or FLAG-PU.1 (PU.1) cultured with M-CSF for 4 days by qPCR. B, analysis of PU.1 expression in BMMs expressing GFP, C/EBPα, or PU.1 cultured with M-CSF for 4 days. Error bars show averages ± S.D. *, p < 0.05. NS, not significant.
3.2. PU.1 generates more osteoclasts than C/EBPα from pre-committed BMMs.
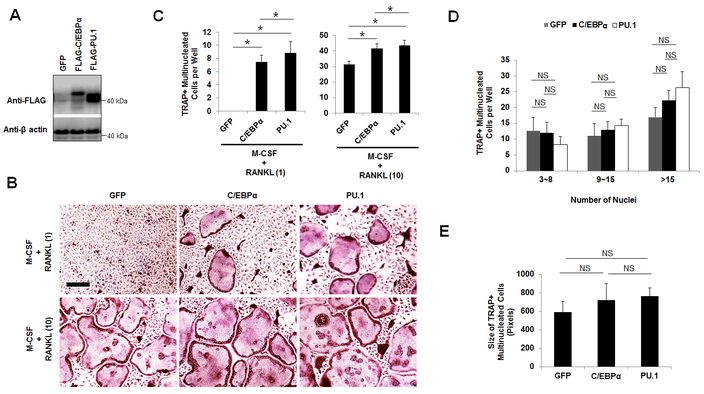
Next, we compared the roles of C/EBPα and PU.1 in mediating osteoclast differentiation which follows the lineage commitment by using the gain-of-function strategy (Fig. 3). Treatment of BMMs with low amount of RANKL was shown to be sufficient to promote lineage commitment but was unable to sustain osteoclast differentiation (35–37). Using this strategy, we overexpressed C/EBPα or PU.1 using a retrovirus and then treated the infected cells with M-CSF plus low amount (1 ng/ml) or optimum amount (10 ng/ml) of RANKL as determined in our previous assay to stimulate osteoclast differentiation with RANKL-evoked lineage priming (Fig. 3A,B) (17). Whereas C/EBPα or PU.1 overexpression could promote osteoclast differentiation with low RANKL doses, the PU.1 overexpressers generated significantly more osteoclasts than the C/EBPα overexpressers (Fig. 3B,C). Consistently, PU.1 could also generate more osteoclasts than C/EBPα in BMMs treated with optimum RANKL doses (Fig. 3B,C). However, we found that PU.1 overexpression did not influence the osteoclast size as compared with BMMs overexpressing C/EBPα or expressing the GFP control (Fig. 3D,E). The results indicated that PU.1 exhibited a stronger influence on osteoclast differentiation than C/EBPα, and showed PU.1 and C/EBPα displayed similar effects on OC size.
Fig. 3.
PU.1 overexpression generates more osteoclasts than C/EBPα overexpression from pre-committed BMMs. A, gene expression analysis from BMMs expressing a GFP control (GFP), FLAG-C/EBPα, or FLAG-PU stimulated by M-CSF for 4 days by Western blotting. B, TRAP staining for osteoclast differentiation from BMMs expressing GFP, FLAG-C/EBPα (C/EBPα), or FLAG-PU.1(PU.1) stimulated by M-CSF plus RANKL for 4 days. Scale bars = 200 μm. C, quantifications for B are shown. D and E, quantifications of the osteoclast size for B via the number of nuclei (D) and area (E) of TRAP-positive multinucleated cells. The numbers in parentheses show concentrations in nanograms per milliliter. Error bars show averages ± S.D. *, p < 0.05; NS, not significant.
3.3. C/EBPα and PU.1 respond differently to RANKL-induced activation of the RANK IVVY motif.
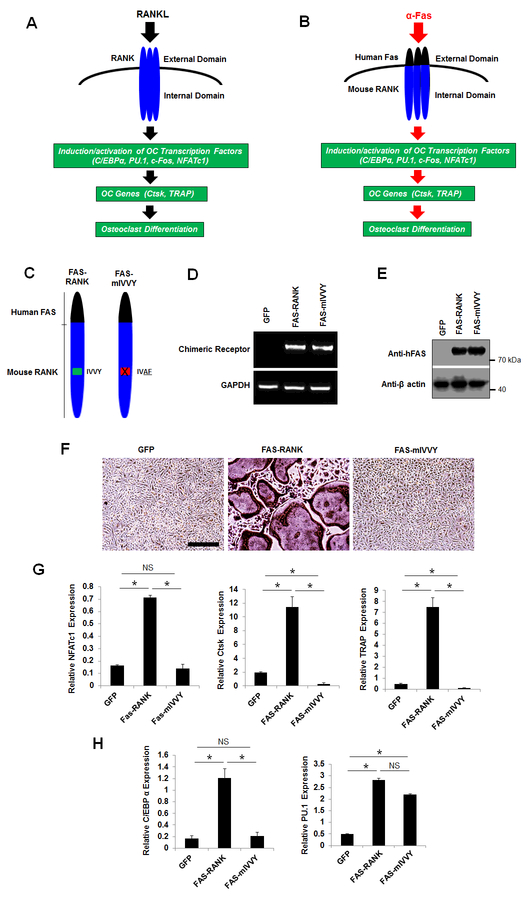
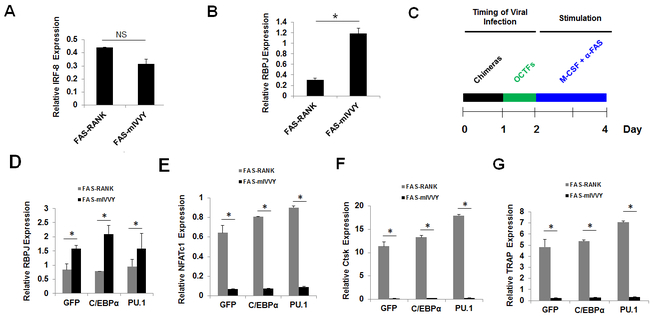
RANK contains an unique cytoplasmic domain, the IVVY motif, at the amino acids 535–538 that is essential for osteoclastogenesis by inducing gene expression (9–11). To gain more insight into the role of the RANK IVVY motif in osteoclastogenesis, we compared the influence of C/EBPα and PU.1 in osteoclast differentiation through RANKL-induced activation of the RANK IVVY motif (Fig. 4). RANK and FAS are both members of the tumor necrosis factor receptor superfamily, which are activated by ligand-induced receptor trimerization (38). Upon binding to RANK, RANKL triggers RANK trimerization and subsequently transduces intracellular signaling to drive gene expression and ultimately osteoclastogenesis (Fig. 4A). In order to delineate specific motifs within the RANK cytoplasmic domain that mediate osteoclastogenesis, Xu and colleagues developed a chimeric receptor system which consists of the Human FAS external domain linked to the transmembrane and cytoplasmic domains of mouse RANK (39). This chimeric receptor system can be specifically activated by a Human FAS activating antibody (α-FAS) which can only activate the Human FAS, but not the mouse FAS, external domain (9). The authors showed that treatment of BMMs expressing the chimeric receptor system with M-CSF and α-FAS could induce gene expression and promote osteoclastogenesis in a similar fashion as RANKL (Fig. 4B) (9,28,40). In investigating whether the RANK IVVY motif could regulate the expressions of C/EBPα and PU.1 during osteoclastogenesis, we used two chimeras (FAS-RANK and FAS-mIVVY) that were previously developed and validated by Xu and colleagues (9). FAS-RANK contains the Human FAS external domain linked to the transmembrane and cytoplasmic domains of normal mouse RANK, and FAS-mIVVY has the Human FAS external domain linked to the transmembrane and cytoplasmic domains of mouse RANK bearing an inactivating mutation in the IVVY motif (Fig. 4C). BMMs were infected with a retrovirus encoding FAS-RANK, FAS-mIVVY, or a GFP control before being submitted to gene expression analysis (Fig. 4D,E) and osteoclastogenesis assays (Fig. 4F). To confirm the expressions of FAS-RANK and FAS-mIVVY in the infected cells, we utilized both a RT-PCR strategy by designing a forward primer against the Human FAS extracellular domain and a reverse primer against the mouse RANK cytoplasmic domain (Fig. 4D) and Western blotting using an anti-Human FAS antibody (Fig. 4E). We confirmed that FAS-RANK and FAS-mIVVY were highly expressed in the BMMs expressing the chimeras but not in cells expressing the GFP control. Whereas treatment of the FAS-RANK expressers with M-CSF and α-FAS generated numerous osteoclasts, the FAS-mIVVY expressers failed to form osteoclast (Fig. 4F). Accordingly, the FAS-RANK, but not the FAS-mIVVY, expressers, could induce the expressions of NFATc1, Ctsk, and TRAP during osteoclast differentiation (Fig. 4G). These results confirmed the previous reports that the RANK IVVY motif is critical for osteoclastogenesis through induction of osteoclast genes (9,27). However, we found that the mutational inactivation of the RANK IVVY motif significantly repressed C/EBPα expression but exerted no significant effect on PU.1 expression (Fig. 4H). These results indicated that C/EBPα and PU.1 responded differently to RANK-IVVY signaling during osteoclastogenesis upon RANK activation.
Fig. 4.
Mutational inactivation of the RANK cytoplasmic 535IVVY538 motif blocks osteoclast differentiation and attenuates C/EBPα, but not PU.1, expression. A, a schematic of RANK activation by RANKL to induce osteoclast differentiation. B, a schematic of the chimeric receptor system which can be activated by α-FAS to promote osteoclast differentiation. C, schematics of FAS-RANK and FAS-mIVVY. D and E, analysis of the expressions of the chimeras in BMMs expressing a GFP control (GFP), FAS-RANK, or FAS-mIVVY cultured with M-CSF for 4 days by RT-PCR (D) or Western blotting (E). F, TRAP staining for osteoclast differentiation from BMMs expressing GFP, FAS-RANK, or FAS-mIVVY treated with M-CSF plus α-FAS (100 ng/ml) for 4 days. Scale bar = 250 μm. G and H, analysis of osteoclast genes from BMMs expressing GFP, FAS-RANK or FAS-mIVVY treated with M-CSF plus α-FAS (100 ng/ml) for 2 days by qPCR. Error bars show averages ± S.D. *, p < 0.05; NS, not significant.
3.4. C/EBPα or PU.1 fails to mediate osteoclast differentiation with inactivation of the RANK IVVY motif.
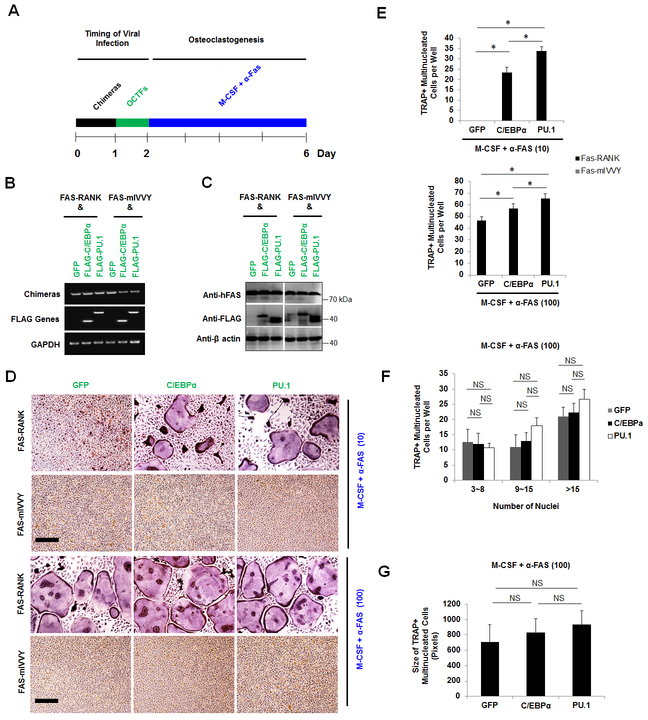
Next, we investigated whether PU.1 overexpression could rescue osteoclastogenesis in cells expressing the mutated RANK IVVY motif. Toward this end, BMMs were first infected with a retrovirus encoding FAS-RANK or FAS-mIVVY before being infected with another retrovirus encoding a GFP control, C/EBPα, or PU.1 (Fig. 5A). BMMs doubly expressing FAS-RANK or FAS-mIVVY plus the GFP control, C/EBPα, or PU.1, as confirmed by RT-PCR (Fig. 5B) and Western blotting (Fig. 5C), were stimulated with M-CSF plus α-FAS to promote osteoclast differentiation (Fig. 5D). These osteoclastogenic assays utilized 10 ng/ml and 100 ng/ml of α-FAS which were validated in previous studies to promote osteoclast differentiation with permissive and optimum activation of the chimeric receptor system, respectively (9,25,27,28,40). As expected, treatment of BMMs doubly expressing FAS-RANK plus the GFP control, C/EBPα, or PU.1 with 10 ng/ml or 100 ng/ml of α-FAS in the presence of M-CSF generated numerous osteoclasts (Fig. 5D). Consistently, we noted that the cells doubly expressing the FAS-RANK and PU.1 generated significantly more osteoclasts than the FAS-RANK and C/EBPα double expressers under both the permissive and optimum α-FAS stimulation (Fig. 5E). However, treatment of BMMs doubly expressing FAS-mIVVY plus the GFP control, C/EBPα, or PU.1 with the permissive and optimum doses of α-FAS in the presence of M-CSF formed no osteoclast (Fig. 5D,E), indicating that C/EBPα or PU.1 overexpression could not rescue osteoclastogenesis from the mutational inactivation of RANK IVVY motif. Moreover, similarly to the RANKL-induced osteoclast differentiation (Fig. 3), PU.1 overexpression did not influence the osteoclast size as compared with C/EBPα or the GFP control under stimulation by optimum α-FAS levels (Fig. 5F,G), further confirming that PU.1 and C/EBPα might not regulate osteoclast size. Collectively, the results indicated that mutational inactivation of the RANK IVVY motif blocked osteoclast differentiation, and overexpression of C/EBPα or PU.1 could not rescue osteoclast differentiation from the mutational inactivation of the RANK IVVY motif.
Fig. 5.
C/EBPα or PU.1 overexpression fails to mediate osteoclast differentiation with the mutational inactivation of the RANK cytoplasmic 535IVVY538 motif. A, a schematic of the experimental strategy. B and C, gene expression analysis from BMMs doubly expressing FAS-RANK or FAS-mIVVY plus a GFP control (GFP), FLAG-C/EBPα (C/EBPα), or FLAG-PU.1 (PU.1) cultured with M-CSF for 4 days by RT-PCR (B) and Western blotting (C). D, TRAP staining for osteoclast differentiation from BMMs doubly expressing FAS-RANK or FAS-mIVVY plus GFP, C/EBPα, or PU.1 treated with M-CSF and α-FAS for 4 days. Scale bar = 200 μm. E , quantifications for D are shown. F and G, quantifications of the osteoclast size for D via the number of nuclei (F) and area (G) of TRAP-positive multinucleated osteoclasts from FAS-RANK expressers treated with M-CSF and α-FAS (100 ng/ml). The numbers in parentheses show concentrations in nanograms per milliliter. Error bars show averages ± S.D. *, p < 0.05; NS, not significant.
3.5. Inactivation of the RANK cytoplasmic IVVY motif triggers RBP-J upregulation but exerts no significant effect on the expression of interferon regulatory factor 8 (IRF-8).
While C/EBPα or PU.1 overexpression could initiate osteoclastogenesis independently of RANKL (Fig. 1) and promote osteoclast differentiation from pre-committed BMMs (Fig. 3), C/EBPα or PU.1 overexpression failed to mediate osteoclast differentiation with inactivation of the RANK IVVY motif (Fig. 5). In our attempt to understand this discrepancy, we found that IVVY-motif inactivation led to significantly lower expressions of Ctsk and TRAP as compared with a GFP control (Fig. 4G). This observation suggested that the RANK IVVY motif, besides positively regulating the expressions of osteoclast genes, might also negatively regulate the expressions of osteoclast inhibitors during osteoclastogenesis. This is consistent with the notion that deregulation of potent negative regulators of osteoclastogenesis from the inactivation of the IVVY motif might negatively affect the ability of C/EBPα or PU.1 to promote osteoclast differentiation in cells expressing FAS-mIVVY.
Among the factors that can potently inhibit osteoclastogenesis, IRF-8 and RBP-J have been the most studied (41–47). Hence, we examined the role of RANK IVVY motif in regulating the expressions of IRF-8 and RBP-J during osteoclast differentiation. Mutational inactivation of RANK IVVY motif showed no significant effect on IRF-8 expression (Fig. 6A), but led to a significant increase in RBP-J expression (Fig. 6B), indicating that this RANK motif could negatively regulate RBP-J expression during osteoclastogenesis. It was recently shown that RBP-J inhibits osteoclastogenesis by suppressing the immunoreceptor tyrosine-based activation motif (ITAM)–associated receptor costimulatory signaling, which is critical for induction of osteoclast genes during osteoclast differentiation (48,49). We showed that overexpression of C/EBPα or PU.1 was unable to control the deregulated RBP-J expression from the inactivation of RANK IVVY motif as compared with normal RANK (Fig. 6C,D). To exclude the contribution of RANK signaling in regulating RBP-J expression upon the activation of the chimeric receptor system, we demonstrated that C/EBPα or PU.1 overexpression in BMMs not expressing the chimeric receptors exhibited no over effect on RBP-J expression (Suppl. Fig. 2). In addressing the molecular basis of the failure of C/EBPα or PU.1 overexpression to mediate osteoclast differentiation in the mutant cells, we revealed that C/EBPα or PU.1 overexpression in the FAS-mIVVY expressers significantly repressed NFATc1 (Fig. 6C,E), Ctsk (Fig. 6C,F), and TRAP (Fig. 6C,G) during osteoclast differentiation. Our data mimicked the reported role of RBP-J in repressing osteoclast genes during osteoclastogenesis (41). The results indicated that the inability of C/EBPα or PU.1 overexpression to rescue osteoclast differentiation in the context of RANK-IVVY-motif inactivation was due in part to the deregulated RBP-J expression, which negatively affected osteoclast gene expression.
Fig. 6.
Mutational Inactivation of the RANK cytoplasmic 535IVVY538 motif triggers RBP-J upregulation but represses osteoclast genes. A and B, analysis of the expressions of IRF-8 (A) and RBP-J (B) from BMMs expressing FAS-RANK or FAS-mIVVY stimulated by M-CSF plus α-FAS (100 ng/ml) for 2 days by qPCR. C, a schematic of the experimental strategy for D-G. D-G, BMMs doubly expressing FAS-RANK or FAS-mIVVY plus a GFP control (GFP), FLAG-C/EBPα (C/EBPα), or FLAG-PU.1 (PU.1) were cultured with M-CSF and α-FAS (100 ng/ml) for analyses of the expressions of the osteoclast inhibitor RBP-J (D) and the osteoclast genes NFATc1 (E), Ctsk (F) and TRAP (G) by qPCR. Error bars show averages ± S.D. *, p < 0.05; NS, not significant.
3.6. RBP-J silencing rescues osteoclastogenesis in cells expressing the mutated RANK IVVY motif.
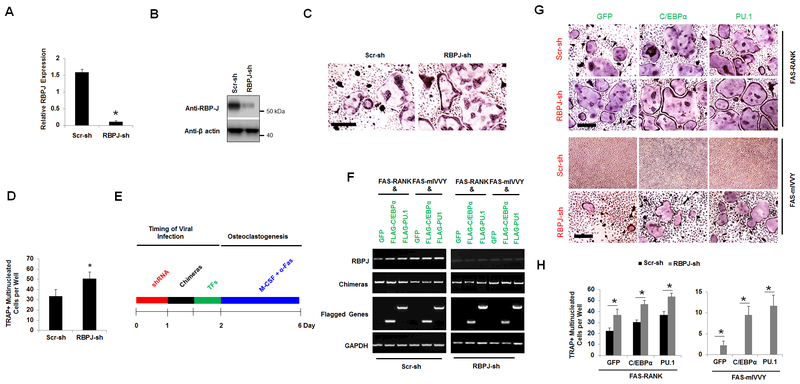
Finally, we investigated whether RBP-J silencing in BMMs expressing the mutated RANK IVVY motif could mediate osteoclastogenesis with C/EBPα or PU.1 overexpression (Fig. 7). We were able to knockdown the RBP-J gene efficiently in BMMs using a shRNA construct that was purchased from Sigma as accessed by qPCR (Fig. 7A) and Western blotting (Fig. 7B). RBP-J silencing significantly enhanced RANKL-induced osteoclastogenesis (Fig. 7C,D), confirming the established role of RBP-J as a strong inhibitor of osteoclast differentiation (50) (41). Next, in investigating whether RBP-J silencing could rescue osteoclastogenesis in cells doubly expressing the mutated RANK IVVY motif plus C/EBPα or PU.1, BMMs were first infected with a lentivirus encoding the RBP-J shRNA construct or a Scramble control to silence the RBP-J gene before being infected with a mixture of retrovirus encoding FAS-RANK or FAS-mIVVY plus FLAG-C/EBPα, FLAG-PU.1, or a GFP control (Fig. 7E,F). The data showed that RBP-J silencing in BMMs doubly expressing FAS-RANK plus C/EBPα, PU.1, or the GFP control significantly enhanced osteoclast differentiation as compared with cells triply expressing the Scramble control along with FAS-RANK plus FLAG-C/EBPα, FLAG-PU.1, or the GFP control (Fig. 7G,H). Notably, RBP-J silencing in the cells doubly expressing the mutated RANK IVVY motif plus the GFP control generated a few small TRAP-positive multinucleated cells as compared to the Scramble control cells doubly expressing the mutated RANK IVVY motif plus the GFP control (Fig. 7G,H). Notably, forced expression of C/EBPα or PU.1 in the cells doubly expressing the RBP-J shRNA construct and the mutated RANK IVVY motif generated more osteoclasts than cells triply expressing the RBP-J shRNA construct, the Fas-mIVVY, and the GFP control, but formed less osteoclasts than the cells triply expressing the RBP-J shRNA construct, normal RANK, and the transcription factors (Fig. 7G,H). Consistently, we noted that RBPJ-depleted BMMs doubly expressing Fas-RANK or Fas-mIVVY plus PU.1 generated more osteoclasts than the RBPJ-depleted BMMs doubly Fas-RANK or Fas-mIVVY plus C/EBPα or the GFP control. (Fig. 7H). In examining the effects of RBP-J silencing on the basal levels of C/EBPα and PU.1, we found that RBP-J silencing showed no significant effect on C/EBPα or PU.1 expression (Suppl. Fig. 3). Collectively, the results indicated that RBP-J silencing alone was sufficient to initiate osteoclastogenesis in the cells expressing the mutated RANK IVVY motif, but C/EBPα or PU.1 overexpression further enhanced osteoclast differentiation.
Fig. 7.
RBP-J silencing rescues osteoclastogenesis from inactivation of the RANK cytoplasmic 535IVVY538 motif. A and B, analysis of RBP-J knockdown in BMMs expressing a Scramble shRNA control (Scr-sh) or RBP-J shRNA (RBPJ-sh) construct cultured with M-CSF for 4 days by qPCR (A) and Western blotting (B). C, TRAP staining for osteoclast differentiation from BMMs expressing Scr-sh or RBPJ-sh treated with M-CSF and RANKL (10 ng/ml) for 4 days. Scale bar = 250 μm. D, quantification for C is shown. E, a schematic for the experimental strategy for F-H. F, analysis of gene expression from BMMs triply expressing the shRNA constructs (RBPJ-shRNA or Scr-sh), the chimeric receptor system (FAS-RANK or FAS-mIVVY), and the GFP control (GFP), FLAG-C/EBPα (C/EBPα), or FLAG-PU.1 (PU.1) treated with M-CSF for 4 days by RT-PCR. G, TRAP staining for osteoclast differentiation from BMMs triply expressers as in F treated with M-CSF and α-FAS (100 ng/ml) for 4 days. Scale bar = 200 μm. H, quantifications for G are shown. Error bars show averages ± S.D. *, p < 0.05.
4. Discussion
RANK signaling triggers upregulation of the transcription factors PU.1, C/EBPα, c-Fos, and NFATc1, which are essential for osteoclast formation (6). Whereas PU.1 is long known to be essential for osteoclastogenesis (8) (22), the role of C/EBPα in osteoclastogenesis has only recently been documented (7,17). Importantly, PU.1 or C/EBPα can upregulate c-Fos and NFATc1 during osteoclastogenesis, establishing C/EBPα and PU.1 as the earliest known osteoclast transcription factors (51). However, the mechanisms through which C/EBPα and PU.1 regulate osteoclast differentiation in response to RANK activation remain unknown. The current study sought to address this issue in vitro.
We confirmed that PU.1 and C/EBPα can mediate osteoclast lineage priming by inducing gene expression in a RANKL-independent manner (7). Notably, PU.1 can upregulate C/EBPα, but C/EBPα is unable to upregulate PU.1, establishing C/EBPα as a target gene of PU.1 during osteoclastogenesis. Our finding agrees with another study identifying C/EBPα as a target gene of PU.1 in granulocyte (52), but disagrees with other reports that C/EBPα can upregulate PU.1 during early myeloid cell fate decision (53,54). Nonetheless, we found that while C/EBPα and PU.1 could similarly initiate osteoclastogenesis independently of RANKL, PU.1 exhibited a stronger ability in promoting osteoclast differentiation than C/EBPα in pre-committed BMMs. Given that PU.1 and C/EBPα show similar abilities in inducing osteoclast genes, the stronger ability of PU.1 in mediating osteoclast differentiation than C/EBPα may not stem from its ability to upregulate C/EBPα. We believe that this may instead result from the ability of PU.1 or C/EBPα to induce different sets of genes, besides a common set of genes, during osteoclastogenesis. Our notion is underscored by a recent study demonstrating that C/EBPα and PU.1 exhibit distinct responses in the human acute leukemia HL-60 and NB4 cell lines (55).
In comparing the roles of C/EBPα and PU.1 in osteoclast differentiation, we examined their response to RANK signaling from activation of the IVVY motif which is essential for osteoclastogenesis (9–11) (56). We showed that RANK IVVY motif upregulates C/EBPα but is dispensable to PU.1 induction, indicating that C/EBPα and PU.1 respond differently to RANK-IVVY-induced signaling during osteoclastogenesis. The fact that C/EBPα is repressed with the inactivation of RANK IVVY motif despite normal PU.1 expression indicated that PU.1 may function with other unknown factors that are regulated by the RANK IVVY motif to upregulate C/EBPα during osteoclast differentiation. Nonetheless, we speculate that a different region within the RANK cytoplasmic domain is responsible for PU.1 upregulation during osteoclastogenesis. Studies have shown that RANK transduces two types of signaling pathways emanating from the IVVY motif, a TNF receptor associated factor (TRAF) independent site, and its TRAF-binding sites (9,27,57). RANK has three functional TRAF-binding motifs (369PFQEP373, 559PVQEET564, and 604PVQEQG609) that are as essential as the IVVY motif for osteoclastogenesis (57). Importantly, it was reported that the TRAF-binding sites and IVVY motif of RANK do not function independently but cooperate in mediating osteoclastogenesis (27). We believe that the TRAF-binding sites of RANK are likely to regulate PU.1 during osteoclastogenesis. Notably, the fact that RANK IVVY-motif inactivation fails to mediate osteoclastogenesis despite normal PU.1 expression indicates that the convergence of the IVVY and TRAF signaling pathways to induce a common set of genes (e.g Ctsk and TRAP) as well as the unique C/EBPα and PU.1 target genes is essential for osteoclast differentiation.
In further characterizing the influence of RANK-IVVY signaling on osteoclast differentiation, we hypothesized that C/EBPα, unlike PU.1, overexpression might rescue osteoclastogenesis from the IVVY-motif inactivation. We reasoned that C/EBPα overexpression in the context of RANK IVVY-motif inactivation should rescue the C/EBPα target genes in the presence of the PU.1 target genes to promote osteoclastogenesis. Consistently, PU.1 overexpression in cells expressing the mutated IVVY motif formed no osteoclast, indicating that repression of the C/EBPα target genes from IVVY-motif inactivation which caused C/EBPα downregulation impeded osteoclastogenesis despite PU.1 overexpression. However, C/EBPα overexpression in the mutant cells also formed no osteoclast despite normal PU.1 expression. In elucidating this discrepancy, we showed that PU.1 or C/EBPα overexpression failed to induce osteoclast genes in the context of RANK-IVVY-motif inactivation. We noted that RANK IVVY motif inactivation caused significantly lower Ctsk and TRAP expressions as compared with a vector control, suggesting that the RANK IVVY motif might also promote osteoclast differentiation by down-regulating osteoclast inhibitors. This assumption goes with the idea that RANK IVVY motif inactivation can also trigger upregulation of potent osteoclast inhibitors which affects the ability of C/EBPα or PU.1 to promote osteoclastogenesis.
We later revealed that RANK-IVVY-motif inactivation triggered RBP-J upregulation, a potent osteoclast inhibitor. Mechanistically, we demonstrated that C/EBPα or PU.1 overexpression failed to control the deregulated RBP-J expression from the IVVY-motif inactivation, leading to repression of osteoclast genes and inhibition of osteoclast differentiation. Osteoclastogenesis requires two essential cross-talk signaling induced by RANK activation and the ITAM-associated receptors (DAP12 and FcRγ) (48,49,58). Our finding is consistent with a recent study indicating that RBP-J suppresses the ITAM-mediated costimulatory signaling and limits the cross-talk between the ITAM and RANK signaling during osteoclastogenesis. Notably, RBP-J silencing partially rescues osteoclastogenesis from the inactivation of the IVVY motif. Whereas the IVVY motif can upregulate C/EBPα, but not PU.1, and downregulates RBP-J, C/EBPα or PU.1 overexpression only partially rescues osteoclast differentiation in RBP-J depleted cells expressing the mutated IVVY motif. This finding indicates that our understanding of the mechanism by which the IVVY motif mediates osteoclastogenesis remains incomplete (9–11).
On the basis of our findings, we propose a working model to, in conjunction with other studies (8,9,27,33), summarize the roles of transcription factors and RANK signaling in osteoclast differentiation (Fig. 8). The model expands on the findings of a recent study that has demonstrated that the IVVY motif and TRAF-binding sites of RANK are equally important for osteoclastogenesis (27). Hence, RANKL-induced activation of the TRAF-independent IVVY signaling pathway upregulates C/EBPα, which subsequently activates transcription factors (e.g. c-Fos and NFATc1) for induction of osteoclast genes (e. g. Ctsk and TRAP) and osteoclast differentiation (33,34). Unlike C/EBPα, PU.1 is likely to be regulated by the RANK TRAF-binding sites during osteoclastogenesis. PU.1 can then upregulate C/EBPα, c-Fos, and NFATc1 expressions to induce osteoclast genes and thereby promote osteoclastogenesis. However, we believe that C/EBPα is mainly regulated by the RANK IVVY motif during osteoclast differentiation. In the light of various studies reporting that transcription factors can function in complex with other factors to regulate gene expression for osteoclastogenesis (22,23,59), we anticipate that PU.1 and C/EBPα may induce a common set of osteoclast genes, including Ctsk, TRAP, and NFATc1. Moreover, C/EBPα and PU.1 may also induce a different set of genes that are also critical for osteoclastogenesis. Finally, the IVVY motif can negatively regulate the osteoclast inhibitor RBP-J in mediating osteoclast differentiation.
Fig. 8.
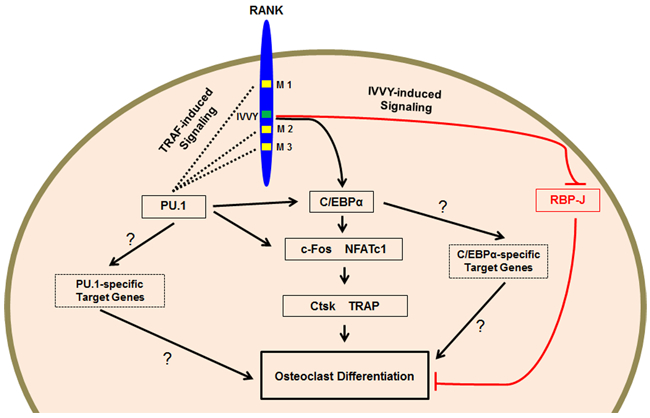
Proposed working model. RANK emits two types of signaling pathways from the IVVY motif and TRAF-binding sites that are equally important for osteoclastogenesis through activation of transcription factors for induction of osteoclast genes. RANK has three functional sites, 369PFQEP373 (M1), 559PVQEET564 (M2), and 604PVQEQG609 (M3), that can recruit TRAFs to transduce osteoclastogenic signaling. Moreover, RANK has a TRAF-independent 535IVVY538 motif (IVVY) that can also transduce signaling for osteoclastogenesis. Furthermore, osteoclastogenesis is likely to require two different but unique sets of genes that are specifically regulated by C/EBPα and PU.1, respectively. The question marks indicate unknown mechanisms.
In conclusion, whereas PU.1 can upregulate C/EBPα during the lineage commitment and exhibits a stronger osteoclastogenic potential than C/EBPα, the ability of PU.1 to upregulate C/EBPα may not be the primary factor responsible for its stronger ability in mediating osteoclast differentiation than C/EBPα. C/EBPα and PU.1 display different responses to RANKL/RANK signaling through activation of the IVVY motif. The IVVY motif mediates osteoclastogenesis by positively regulating osteoclast activators and negatively regulating osteoclast inhibitors. Our study provides an important insight into the mechanism underlying the responses of transcription factors to RANK signaling during osteoclatogenesis.
Supplementary Material
Acknowledgments
The authors thank Dr. Xu Feng for providing the chimeric receptor constructs and Dr. Jue Wang for her extensive reading and discussion of the manuscript. This study was supported by NIH grants R01-AR-044741 (YPL) and R01DE023813 (YPL).
List of abbreviations
- BMMs
bone marrow macrophages
- C/EBPα
CCAAT/enhancer binding protein-alpha
- Ctsk
cathepsin K
- FAS-RANK
a chimeric receptor with the Human FAS external domain linked to the normal mouse RANK transmembrane and cytoplasmic domains
- FAS-mIVVY
a chimeric receptor with the Human FAS external domain linked to the mouse RANK transmembrane and cytoplasmic domains bearing an inactivating mutation in the IVVY motif
- ITAM
immunoreceptor tyrosine-based activation motif
- IRF-8
interferon regulatory factor 8
- M-CSF
macrophage colony-stimulating factor
- NFATc1
nuclear factor of activated T-cells, C1
- RANK
receptor activator of NF-κB
- RANKL
receptor activator of NF-κB ligand
- RBP-J
recombinant recognition sequence binding protein at the Jκ site
- TRAP
tartrate-resistant acid phosphatase 5
- TRAF
TNF receptor associated factor
Footnotes
Conflict of interest: The authors have no conflicts of interest.
References
- 1.Ikeda K, and Takeshita S (2016) The role of osteoclast differentiation and function in skeletal homeostasis. J Biochem 159, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle WJ, Simonet WS, and Lacey DL (2003) Osteoclast differentiation and activation. Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 3.Feng X, and McDonald JM (2011) Disorders of bone remodeling. Annu Rev Pathol 6, 121–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soltanoff CS, Yang S, Chen W, and Li YP (2009) Signaling networks that control the lineage commitment and differentiation of bone cells. Crit Rev Eukaryot Gene Expr 19, 1–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers TJ (2000) Regulation of the differentiation and function of osteoclasts. J Pathol 192, 4–13 [DOI] [PubMed] [Google Scholar]
- 6.Huang W, Yang S, Shao J, and Li YP (2007) Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front Biosci 12, 3068–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Zhu G, Hao L, Wu M, Ci H, and Li YP (2013) C/EBPalpha regulates osteoclast lineage commitment. Proc Natl Acad Sci U S A 110, 7294–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tondravi MM, McKercher SR, Anderson K, Erdmann JM, Quiroz M, Maki R, and Teitelbaum SL (1997) Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature 386, 81–84 [DOI] [PubMed] [Google Scholar]
- 9.Xu D, Wang S, Liu W, Liu J, and Feng X (2006) A novel receptor activator of NF-kappaB (RANK) cytoplasmic motif plays an essential role in osteoclastogenesis by committing macrophages to the osteoclast lineage. J Biol Chem 281, 4678–4690 [DOI] [PubMed] [Google Scholar]
- 10.Taguchi Y, Gohda J, Koga T, Takayanagi H, and Inoue J (2009) A unique domain in RANK is required for Gab2 and PLCgamma2 binding to establish osteoclastogenic signals. Genes Cells 14, 1331–1345 [DOI] [PubMed] [Google Scholar]
- 11.Kim H, Choi HK, Shin JH, Kim KH, Huh JY, Lee SA, Ko CY, Kim HS, Shin HI, Lee HJ, Jeong D, Kim N, Choi Y, and Lee SY (2009) Selective inhibition of RANK blocks osteoclast maturation and function and prevents bone loss in mice. J Clin Invest 119, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jules J, Ashley JW, and Feng X (2010) Selective targeting of RANK signaling pathways as new therapeutic strategies for osteoporosis. Expert Opin Ther Targets 14, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nerlov C (2007) The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol 17, 318–324 [DOI] [PubMed] [Google Scholar]
- 14.Ye M, Zhang H, Amabile G, Yang H, Staber PB, Zhang P, Levantini E, Alberich-Jorda M, Zhang J, Kawasaki A, and Tenen DG (2013) C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol 15, 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, and Tenen DG (1997) Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A 94, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, and Darlington GJ (1995) Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269, 1108–1112 [DOI] [PubMed] [Google Scholar]
- 17.Jules J, Chen W, Feng X, and Li YP (2016) CCAAT/Enhancer-binding Protein alpha (C/EBPalpha) Is Important for Osteoclast Differentiation and Activity. J Biol Chem 291, 16390–16403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng X, Teitelbaum SL, Quiroz ME, Cheng SL, Lai CF, Avioli LV, and Ross FP (2000) Sp1/Sp3 and PU.1 differentially regulate beta(5) integrin gene expression in macrophages and osteoblasts. J Biol Chem 275, 8331–8340 [DOI] [PubMed] [Google Scholar]
- 19.Sheng Y, Ju W, Huang Y, Li J, Ozer H, Qiao X, and Qian Z (2016) Activation of wnt/beta-catenin signaling blocks monocyte-macrophage differentiation through antagonizing PU.1-targeted gene transcription. Leukemia [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Oevelen C, Collombet S, Vicent G, Hoogenkamp M, Lepoivre C, Badeaux A, Bussmann L, Sardina JL, Thieffry D, Beato M, Shi Y, Bonifer C, and Graf T (2015) C/EBPalpha Activates Pre-existing and De Novo Macrophage Enhancers during Induced Pre-B Cell Transdifferentiation and Myelopoiesis. Stem Cell Reports 5, 232–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoppe PS, Schwarzfischer M, Loeffler D, Kokkaliaris KD, Hilsenbeck O, Moritz N, Endele M, Filipczyk A, Gambardella A, Ahmed N, Etzrodt M, Coutu DL, Rieger MA, Marr C, Strasser MK, Schauberger B, Burtscher I, Ermakova O, Burger A, Lickert H, Nerlov C, Theis FJ, and Schroeder T (2016) Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 535, 299–302 [DOI] [PubMed] [Google Scholar]
- 22.Sharma SM, Bronisz A, Hu R, Patel K, Mansky KC, Sif S, and Ostrowski MC (2007) MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. J Biol Chem 282, 15921–15929 [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto M, Kogawa M, Wada S, Takayanagi H, Tsujimoto M, Katayama S, Hisatake K, and Nogi Y (2004) Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J Biol Chem 279, 45969–45979 [DOI] [PubMed] [Google Scholar]
- 24.Luchin A, Suchting S, Merson T, Rosol TJ, Hume DA, Cassady AI, and Ostrowski MC (2001) Genetic and physical interactions between Microphthalmia transcription factor and PU.1 are necessary for osteoclast gene expression and differentiation. J Biol Chem 276, 36703–36710 [DOI] [PubMed] [Google Scholar]
- 25.Jules J, Zhang P, Ashley JW, Wei S, Shi Z, Liu J, Michalek SM, and Feng X (2012) Molecular basis of requirement of receptor activator of nuclear factor kappaB signaling for interleukin 1-mediated osteoclastogenesis. J Biol Chem 287, 15728–15738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ory DS, Neugeboren BA, and Mulligan RC (1996) A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci U S A 93, 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jules J, Wang S, Shi Z, Liu J, Wei S, and Feng X (2015) The IVVY Motif and Tumor Necrosis Factor Receptor-associated Factor (TRAF) Sites in the Cytoplasmic Domain of the Receptor Activator of Nuclear Factor kappaB (RANK) Cooperate to Induce Osteoclastogenesis. J Biol Chem 290, 23738–23750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jules J, Shi Z, Liu J, Xu D, Wang S, and Feng X (2010) Receptor activator of NF-{kappa}B (RANK) cytoplasmic IVVY535–538 motif plays an essential role in tumor necrosis factor-{alpha} (TNF)-mediated osteoclastogenesis. J Biol Chem 285, 37427–37435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos MA, Faryabi RB, Ergen AV, Day AM, Malhowski A, Canela A, Onozawa M, Lee JE, Callen E, Gutierrez-Martinez P, Chen HT, Wong N, Finkel N, Deshpande A, Sharrow S, Rossi DJ, Ito K, Ge K, Aplan PD, Armstrong SA, and Nussenzweig A (2014) DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature 514, 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian F, Wu M, Deng L, Zhu G, Ma J, Gao B, Wang L, Li YP, and Chen W (2014) Core binding factor beta (Cbfbeta) controls the balance of chondrocyte proliferation and differentiation by upregulating Indian hedgehog (Ihh) expression and inhibiting parathyroid hormone-related protein receptor (PPR) expression in postnatal cartilage and bone formation. J Bone Miner Res 29, 1564–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaidi M, Blair HC, Moonga BS, Abe E, and Huang CL (2003) Osteoclastogenesis, bone resorption, and osteoclast-based therapeutics. J Bone Miner Res 18, 599–609 [DOI] [PubMed] [Google Scholar]
- 32.Tsuneto M, Tominaga A, Yamazaki H, Yoshino M, Orkin SH, and Hayashi S (2005) Enforced expression of PU.1 rescues osteoclastogenesis from embryonic stem cells lacking Tal-1. Stem Cells 23, 134–143 [DOI] [PubMed] [Google Scholar]
- 33.Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, and Wagner EF (1994) c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science 266, 443–448 [DOI] [PubMed] [Google Scholar]
- 34.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, and Taniguchi T (2002) Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 35.Jules J, and Feng X (2014) In vitro investigation of the roles of the proinflammatory cytokines tumor necrosis factor-alpha and interleukin-1 in murine osteoclastogenesis. Methods Mol Biol 1155, 109–123 [DOI] [PubMed] [Google Scholar]
- 36.Wei S, Kitaura H, Zhou P, Ross FP, and Teitelbaum SL (2005) IL-1 mediates TNF-induced osteoclastogenesis. J Clin Invest 115, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam J, Takeshita S, Barker JE, Kanagawa O, Ross FP, and Teitelbaum SL (2000) TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J Clin Invest 106, 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horowitz MC, Xi Y, Wilson K, and Kacena MA (2001) Control of osteoclastogenesis and bone resorption by members of the TNF family of receptors and ligands. Cytokine Growth Factor Rev 12, 9–18 [DOI] [PubMed] [Google Scholar]
- 39.Xu D, Shi Z, McDonald J, Pan G, Cao X, Yu X, and Feng X (2004) Development of a chimaeric receptor approach to study signalling by tumour necrosis factor receptor family members. Biochem J 383, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashley JW, McCoy EM, Clements DA, Shi Z, Chen T, and Feng X (2011) Development of cell-based high-throughput assays for the identification of inhibitors of receptor activator of nuclear factor-kappa B signaling. Assay Drug Dev Technol 9, 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Miller CH, Giannopoulou E, Hu X, Ivashkiv LB, and Zhao B (2014) RBP-J imposes a requirement for ITAM-mediated costimulation of osteoclastogenesis. J Clin Invest 124, 5057–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swarnkar G, Karuppaiah K, Mbalaviele G, Chen TH, and Abu-Amer Y (2015) Osteopetrosis in TAK1-deficient mice owing to defective NF-kappaB and NOTCH signaling. Proc Natl Acad Sci U S A 112, 154–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao B, Grimes SN, Li S, Hu X, and Ivashkiv LB (2012) TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med 209, 319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma J, Liu YL, Hu YY, Wei YN, Zhao XC, Dong GY, Qin HY, Ding Y, and Han H (2013) Disruption of the transcription factor RBP-J results in osteopenia attributable to attenuated osteoclast differentiation. Mol Biol Rep 40, 2097–2105 [DOI] [PubMed] [Google Scholar]
- 45.Wagner H (2010) Bone diseases: Interferon regulatory factor-8 suppresses osteoclastogenesis. Nat Rev Rheumatol 6, 73–74 [DOI] [PubMed] [Google Scholar]
- 46.Zhao B, Takami M, Yamada A, Wang X, Koga T, Hu X, Tamura T, Ozato K, Choi Y, Ivashkiv LB, Takayanagi H, and Kamijo R (2009) Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med 15, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swarnkar G, Shim K, Nasir AM, Seehra K, Chen HP, Mbalaviele G, and Abu-Amer Y (2016) Myeloid Deletion of Nemo Causes Osteopetrosis in Mice Owing to Upregulation of Transcriptional Repressors. Sci Rep 6, 29896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mocsai A, Humphrey MB, Van Ziffle JA, Hu Y, Burghardt A, Spusta SC, Majumdar S, Lanier LL, Lowell CA, and Nakamura MC (2004) The immunomodulatory adapter proteins DAP12 and Fc receptor gamma-chain (FcRgamma) regulate development of functional osteoclasts through the Syk tyrosine kinase. Proc Natl Acad Sci U S A 101, 6158–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koga T, Inui M, Inoue K, Kim S, Suematsu A, Kobayashi E, Iwata T, Ohnishi H, Matozaki T, Kodama T, Taniguchi T, Takayanagi H, and Takai T (2004) Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature 428, 758–763 [DOI] [PubMed] [Google Scholar]
- 50.Zhao B, and Ivashkiv LB (2011) Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res Ther 13, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asagiri M, and Takayanagi H (2007) The molecular understanding of osteoclast differentiation. Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 52.Burda P, Curik N, Kokavec J, Basova P, Mikulenkova D, Skoultchi AI, Zavadil J, and Stopka T (2009) PU.1 activation relieves GATA-1-mediated repression of Cebpa and Cbfb during leukemia differentiation. Mol Cancer Res 7, 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang D, D’Costa J, Civin CI, and Friedman AD (2006) C/EBPalpha directs monocytic commitment of primary myeloid progenitors. Blood 108, 1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeamans C, Wang D, Paz-Priel I, Torbett BE, Tenen DG, and Friedman AD (2007) C/EBPalpha binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood 110, 3136–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savickiene J, Treigyte G, Vistartaite G, Tunaitis V, Magnusson KE, and Navakauskiene R (2011) C/EBPalpha and PU.1 are involved in distinct differentiation responses of acute promyelocytic leukemia HL-60 and NB4 cells via chromatin remodeling. Differentiation 81, 57–67 [DOI] [PubMed] [Google Scholar]
- 56.Guerrini MM, Sobacchi C, Cassani B, Abinun M, Kilic SS, Pangrazio A, Moratto D, Mazzolari E, Clayton-Smith J, Orchard P, Coxon FP, Helfrich MH, Crockett JC, Mellis D, Vellodi A, Tezcan I, Notarangelo LD, Rogers MJ, Vezzoni P, Villa A, and Frattini A (2008) Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations. Am J Hum Genet 83, 64–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W, Xu D, Yang H, Xu H, Shi Z, Cao X, Takeshita S, Liu J, Teale M, and Feng X (2004) Functional identification of three receptor activator of NF-kappa B cytoplasmic motifs mediating osteoclast differentiation and function. J Biol Chem 279, 54759–54769 [DOI] [PubMed] [Google Scholar]
- 58.Humphrey MB, and Nakamura MC (2016) A Comprehensive Review of Immunoreceptor Regulation of Osteoclasts. Clin Rev Allergy Immunol 51, 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crotti TN, Sharma SM, Fleming JD, Flannery MR, Ostrowski MC, Goldring SR, and McHugh KP (2008) PU.1 and NFATc1 mediate osteoclastic induction of the mouse beta3 integrin promoter. J Cell Physiol 215, 636–644 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.