Key Points
Question
What molecular and clinical biomarkers can be used to better understand osimertinib mesylate resistance and develop treatment strategies?
Findings
In this cohort study of 143 patients who underwent tumor next-generation sequencing, loss of the EGFR T790M mutation was common on resistance to osimertinib and was associated with early treatment failure and development of a range of competing resistance mechanisms, some expected (MET amplification, small cell transformation) and some novel (acquired KRAS mutations, targetable gene fusions). Early changes in plasma EGFR mutation levels may indicate probable resistance patterns.
Meaning
Strategies to detect and target multiple coexistent resistance mechanisms will be needed to achieve more durable control of drug resistance in EGFR-mutant lung cancer.
Abstract
Importance
Osimertinib mesylate is used globally to treat EGFR-mutant non–small cell lung cancer (NSCLC) with tyrosine kinase inhibitor resistance mediated by the EGFR T790M mutation. Acquired resistance to osimertinib is a growing clinical challenge that is poorly understood.
Objective
To understand the molecular mechanisms of acquired resistance to osimertinib and their clinical behavior.
Design, Setting, and Participants
Patients with advanced NSCLC who received osimertinib for T790M-positive acquired resistance to prior EGFR tyrosine kinase inhibitor were identified from a multi-institutional cohort (n = 143) and a confirmatory trial cohort (NCT01802632) (n = 110). Next-generation sequencing of tumor biopsies after osimertinib resistance was performed. Genotyping of plasma cell-free DNA was studied as an orthogonal approach, including serial plasma samples when available. The study and analysis were finalized on November 9, 2017.
Main Outcomes and Measures
Mechanisms of resistance and their association with time to treatment discontinuation on osimertinib.
Results
Of the 143 patients evaluated, 41 (28 [68%] women) had tumor next-generation sequencing after acquired resistance to osimertinib. Among 13 patients (32%) with maintained T790M at the time of resistance, EGFR C797S was seen in 9 patients (22%). Among 28 individuals (68%) with loss of T790M, a range of competing resistance mechanisms was detected, including novel mechanisms such as acquired KRAS mutations and targetable gene fusions. Time to treatment discontinuation was shorter in patients with T790M loss (6.1 vs 15.2 months), suggesting emergence of pre-existing resistant clones; this finding was confirmed in a validation cohort of 110 patients with plasma cell-free DNA genotyping performed after osimertinib resistance. In studies of serial plasma levels of mutant EGFR, loss of T790M at resistance was associated with a smaller decrease in levels of the EGFR driver mutation after 1 to 3 weeks of therapy (100% vs 83% decrease; P = .01).
Conclusions and Relevance
Acquired resistance to osimertinib mediated by loss of the T790M mutation is associated with early resistance and a range of competing resistance mechanisms. These data provide clinical evidence of the heterogeneity of resistance in advanced NSCLC and a need for clinical trial strategies that can overcome multiple concomitant resistance mechanisms or strategies for preventing such resistance.
This cohort study examines mechanisms of acquired resistance to osimertinib in patients with non–small cell lung cancer and the associated clinical implications.
Introduction
Osimertinib mesylate is a third-generation tyrosine kinase inhibitor (TKI) targeting mutant epidermal growth factor receptor (EGFR) now available worldwide for the management of non–small cell lung cancer (NSCLC) carrying the EGFR (NCBI 1956) T790M resistance mutation after acquired resistance to prior EGFR TKI therapy. Patients treated in this setting experience a high response rate and median progression-free survival (PFS) of approximately 10 months.1,2 The clinical activity and favorable toxicity profile of osimertinib has led this drug to be studied broadly as a strategy for controlling and preventing drug resistance in EGFR-mutant NSCLC.3,4
With widespread clinical use of osimertinib, acquired resistance has become a clinical problem. A range of mechanisms of resistance has been described, including EGFR C797S mutations, MET amplification, and small cell transformation.5,6,7,8,9,10,11 The standard therapy after EGFR TKI options are exhausted is chemotherapy, such as the regimen of platinum/pemetrexed established in the IMPRESS trial, with a 34% response rate and 5.4-month median PFS.10 To improve on this outcome, a range of targeted treatment approaches is now in development to overcome resistance to osimertinib, an approach that requires an improved molecular and clinical understanding of what drives drug resistance. Genomic analysis of plasma cell-free DNA has been used to describe 2 new resistance mechanisms: acquired EGFR C797S mutations and loss of the T790M mutation.12 Herein, we report our study of resistance in a larger cohort to understand clinical and molecular indicators that might suggest a specific molecular mechanism of resistance and treatment approach.
Methods
Patients with advanced NSCLC treated with osimertinib were identified from institutional databases of 4 contributing cancer centers. Patients who received single-agent osimertinib for acquired resistance to prior EGFR TKI and were EGFR T790M-positive in either tumor or plasma were included. Patients from the first-in-human AURA trial13 were studied with the same inclusion criteria as a validation cohort; all patients signed informed consent forms for treatment in this protocol. Each site’s institutional review board gave approval with waiver of consent for the present analysis. The study and analysis were finalized on November 9, 2017.
Patients from the institutional cohort were eligible for resistance analysis based on the availability of tumor genotyping from a biopsy performed after development of osimertinib resistance. Patients who discontinued osimertinib due to toxic effects before progression were excluded. Genotyping was performed as part of clinical care or as a research test (with patient consent) using a Clinical Laboratory Improvement Amendments–approved next-generation sequencing (NGS) assay14; MET fluorescence in situ hybridization was also performed, when possible.
Plasma was collected in the AURA trial with patient consent. Patients from the AURA cohort were eligible for resistance analysis based on the availability of plasma genotyping results after development of osimertinib resistance. Plasma cell-free DNA was studied for EGFR driver (L858R and exon 19 del) and resistance mutations (T790M and C797S) using droplet digital polymerase chain reaction (PCR) or beads, emulsion, amplification, magnetics (BEAMing), as previously described.15,16 Patients with no detectable EGFR driver mutation in resistance plasma were excluded because there was no evidence of tumor DNA for analysis. In addition, some patients from the institutional cohort consented to serial plasma collection during treatment with osimertinib.
To study clinical outcomes of osimertinib, time to treatment discontinuation (TTD) was measured,17 defined as the time until the end of therapy for any reason. Patients were censored at the date that they were last known to have received therapy. Time to treatment discontinuation was studied rather than PFS because many patients received therapy as part of their standard clinical care, making it difficult to perform an objective analysis of radiographic progression. Furthermore, many patients can continue therapy beyond objective progression,18 such that TTD may more accurately capture the clinical benefit of therapy. A PFS analysis was included for the AURA cohort.
Statistical Analysis
The Kaplan-Meier method was used to estimate the TTD and PFS distributions. Fisher exact test was used to compare the distribution of categorical variables and Wilcoxon rank sum test was used to compare the distribution of continuous variables. Logistic regression modeling was used to estimate the association of time receiving therapy with the odds of T790M loss at the time of resistance. All P values are 2-sided and considered significant at the .05 level.
Results
We first studied a cohort of 143 patients from 4 institutions with EGFR-mutant NSCLC who were EGFR T790M-positive based on either a tumor or plasma genotyping assay before starting treatment with single-agent osimertinib. Forty-one patients (28 [68%] women) experienced progression and underwent a resistance biopsy adequate for genomic analysis (Figure 1). The median TTD for these patients was 8.0 months, which was lower than the expected PFS during osimertinib therapy.1 This TTD may represent an enrichment for patients with earlier acquired resistance—a common bias when studying patients developing therapy resistance.
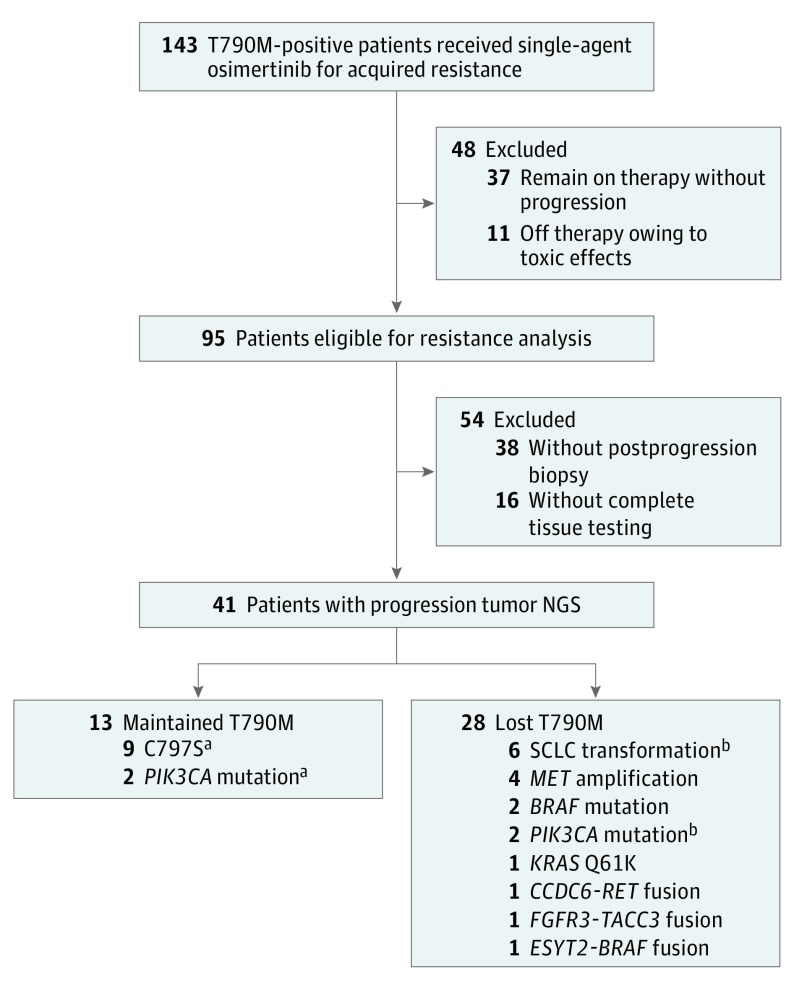
Figure 1. Patient Flowchart of Multi-institutional Cohort With Acquired Resistance to Osimertinib.
A total of 143 T790M-positive patients received single-agent osimertinib for EGFR-acquired resistance. Of these patients, 41 developed disease progression while receiving osimertinib and tumor genotyping during therapy, and tumor genotyping was completed on a resistance biopsy, revealing a range of competing resistance mechanisms in patients with T790M loss. NGS indicates next-generation sequencing.
aPatient with both EGFR C797S and PIK3CA.
bPatient with both PIK3CA and small-cell lung cancer transformation.
For all patients, tumor NGS was completed at resistance and an EGFR driver mutation was again detected (eTable 1 in the Supplement). Maintained EGFR T790M was detected in 13 patients (32%) and loss of T790M was seen in 28 patients (68%). Mutations of EGFR C797S were detected in 9 patients (22% overall; 69% of those with retained T790M), all in cis with a maintained T790M. Non-EGFR resistance mechanisms were identified in 19 patients (46%), with 17 (41%) occurring with T790M loss (Figure 1). Of the 28 patients with T790M loss, 13 (46%) developed resistance mechanisms that are well described after first-generation EGFR TKIs: 6 small-cell lung cancer transformation, 4 MET amplifications, 2 PIK3CA mutations (1 occurring with small cell transformation), and 2 BRAF mutations. Other patients with T790M loss developed unexpected resistance mechanisms, including RET, FGFR3, and BRAF fusions (1 each). Finally, 1 patient had a KRAS Q61K mutation detected on tumor NGS (allelic fraction [AF], 30%) in addition to the known EGFR 19 deletion mutation (AF, 31%) (Figure 2).
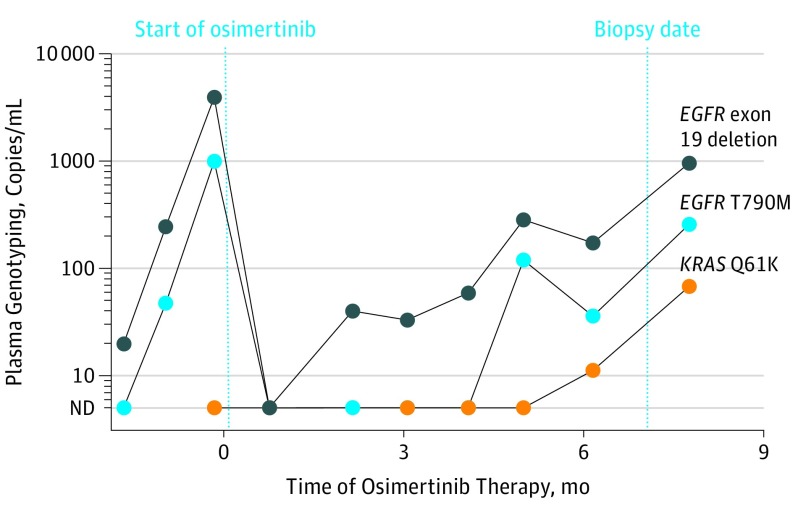
Figure 2. Acquired KRAS Mutation During Osimertinib Therapy.
In 1 patient with osimertinib resistance, next-generation sequencing of a tumor biopsy showed loss of T790M and an acquired KRAS Q61K mutation. Serial plasma genotyping confirmed the acquired KRAS mutation but also detected reemergence of the EGFR T790M mutation. ND indicates not detected.
In several cases, serial plasma genotyping performed retrospectively offered insight into the kinetics of osimertinib response and resistance heterogeneity (eFigure 1 in the Supplement). For example, in a patient who developed small cell transformation, plasma T790M levels dropped during osimertinib therapy while EGFR driver levels increased (eFigure 1A, top left in the Supplement), consistent with early progression after 6 weeks. In a patient with acquired EGFR C797S, an initial complete response in plasma was followed by emergence of the T790M plus 2 EGFR C797S variants (eFigure 1B, top left in the Supplement). Finally, in the patient with an acquired KRAS mutation, a new droplet digital PCR assay was developed to confirm the presence of the KRAS Q61K mutation, which was absent before therapy but gradually emerged with resistance (Figure 2); the T790M mutation reemerged in plasma but was not detected on tumor NGS. In several cases, plasma genotyping detected resistance mechanisms not detected on tumor NGS: 1 patient had a KRAS mutation (G12V) detected on plasma NGS at resistance that was not detected in the tumor, 1 patient with MET amplification detected in the tumor had T790M and an ALK rearrangement detected on plasma NGS, and a third patient with SCLC on biopsy had T790M and EGFR G724S detected in plasma (eTable 1 in the Supplement).
Patients developing loss of T790M were clinically similar to patients with maintained T790M aside from a predominance of women with T790M maintained (eTable 2A in the Supplement). We then explored the timing of osimertinib resistance, dividing patients based on maintained or lost EGFR T790M mutation in the resistance biopsy. Patients with T790M loss had a median TTD of 6.1 months, which was shorter than the median TTD of 15.2 months in patients with maintained T790M (log rank P = .01) (Figure 3). In 1 patient with a prolonged TTD of 23.4 months, NGS of a resistance biopsy showed an exon 19 deletion at a low AF (5%) and no detectable T790M; however, plasma droplet digital PCR detected the driver (AF, 47%) and both T790M (AF, 44%) and C797S (AF, 28%) mutations.
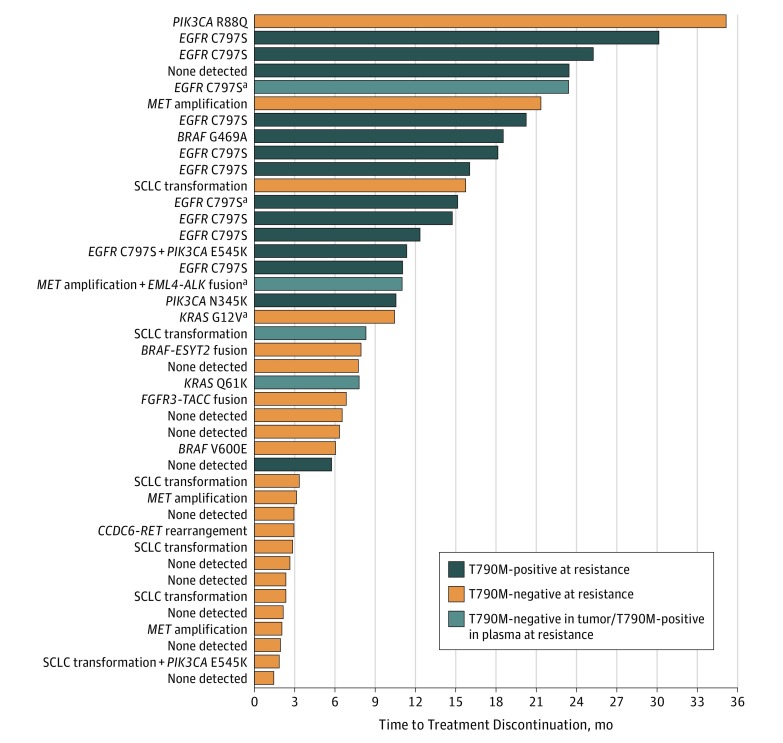
Figure 3. Early Time to Treatment Discontinuation (TTD) in 41 Patients With T790M Loss at Osimertinib Resistance.
Patients with loss of T790M in their resistance biopsy had a median TTD of 6.1 months. Patients with maintained T790M in their resistance biopsy had a median TTD of 15.2 months. Four patients had T790M detected in their plasma when tumor genotyping showed T790M loss. Details regarding resistance mechanisms detected are shown to the left of the bar graph.
aResistance mechanism detected in plasma but not tumor.
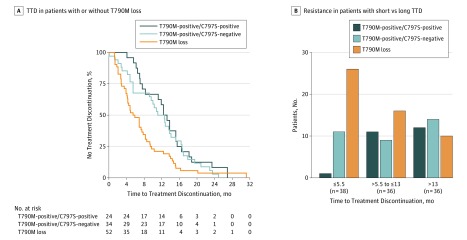
To validate our finding of early resistance in patients developing loss of T790M, we performed a retrospective analysis of patients in the AURA trial (eFigure 2 in the Supplement).1,19 Of 157 patients with plasma samples available after development of resistance, 110 had a driver EGFR mutation detected in plasma cell-free DNA and were deemed adequate for resistance analysis. Of these patients, 52 (47%) had loss of T790M and 58 (53%) maintained the T790M mutation (eTable 2B in the Supplement); 24 patients (22%) acquired EGFR C797S, all with maintained T790M. Patients with loss of T790M had a shorter median TTD (5.5 months) (Figure 4A) than patients with maintained T790M either without C797S (12.6 months) or with C797S (12.4 months) (log rank P = .006). Patients with loss of T790M similarly had a shorter PFS. With the cohort divided into tertiles, loss of T790M was seen in 26 of 38 patients (68%) with 5.5-months or less TTD; maintained T790M was seen in 26 of 36 patients (72%) with 13 months more TTD (Figure 4B).
Figure 4. Acquired Resistance to Osimertinib in a Validation Cohort (n = 110) From the AURA Trial.
A, This Kaplan-Meier curve shows that, in 110 patients, those with T790M loss had a median time to treatment discontinuation (TTD) of 5.5 months, which was lower than patients who were T790M-positive and C797S-negative (median, 12.6 months) or T790M-positive and C797S-positive (median, 12.4 months). B, A higher percentage of patients with short TTD (68%) had T790M loss; a higher percentage of patients with long TTD (72%) maintained T790M.
We studied strategies for anticipating loss of T790M using pretreatment features. Of the 19 patients with non-EGFR resistance mechanisms detected on biopsies after progression, 16 had pre-osimertinib resistance biopsies (eTable 1 in the Supplement). Among 6 individuals with small cell transformation, 1 had small cell differentiation detected on a biopsy that preceded osimertinib therapy by 8.5 months, although only adenocarcinoma was seen on a biopsy immediately preceding osimertinib therapy; 2 other patients showed RB1 loss on tumor NGS before osimertinib.20,21 Among 4 patients with MET amplification after osimertinib, 2 had pre-osimertinib MET fluorescence in situ hybridization, 1 of which showed focal MET amplification (6% of cells with 10 MET copies) in addition to the T790M mutation (AF, 7%); after treatment with osimertinib for 9 weeks, T790M was no longer detectable on rebiopsy, but MET amplification was seen in 94% of the cells (eFigure 3 in the Supplement). Pre-osimertinib NGS for the patient with the BRAF fusion (eFigure 4 in the Supplement) after osimertinib showed this fusion before therapy, coexistent with a T790M mutation (AF, 10%). Separately, 2 patients with early osimertinib failure and T790M loss in tumor had T790M detected in plasma but not in tumor before osimertinib treatment. We also studied the biologic outcome of genomic TP53 loss, which has been associated with earlier resistance on first-generation EGFR TKI.22,23 The TP53 mutational status was available for all 41 patients with resistance biopsies, and 26 individuals had a pathogenic TP53 mutation or 2-copy loss of TP53. Although these mutations were more common in patients with T790M loss (71% vs 46%), the difference was not statistically significant (P = .17).
Given the limited availability of matched pre- and post-osimertinib biopsies, we studied plasma genotyping to estimate the eventual pattern of resistance. Fifty patients from the AURA cohort had plasma genotyping performed before osimertinib administration and after development of resistance. The relative AF of T790M compared with the EGFR driver was calculated, as described previously,24 as a potential measure of T790M allelic prevalence. Pre- and post-osimertinib relative T790M AF were correlated (eFigure 5A in the Supplement) (Pearson correlation coefficient = 0.70; P < .001). However, when this association is studied in a dichotomous fashion, patients who develop T790M loss have only a slightly lower relative T790M AF before osimertinib (29% vs 38% median, P = .06) (Figure 5A). Generation of a receiver operating curve (eFigure 5B in the Supplement) revealed an area under the curve of 0.66, suggesting poor performance as a diagnostic test for estimating the probable loss of T790M.
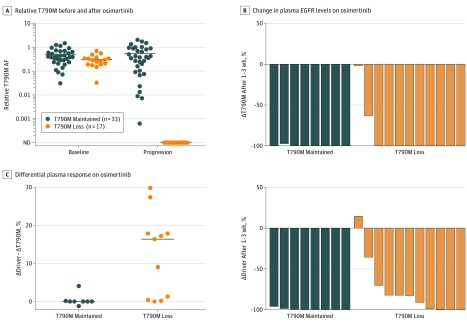
Figure 5. Estimating the Probability of T790M Loss From Baseline and Serial Plasma Samples.
A, Fifty patients from the AURA cohort had plasma genotyping performed before and after osimertinib treatment. Relative T790M allelic fraction (AF) was calculated as T790M AF/EGFR driver AF. Patients with T790M loss tended to have a lower relative T790M AF at baseline (29% vs 38% median, P = .06) than patients with T790M maintained, although the difference was modest. B, Measuring the relative change in plasma EGFR levels after 1 to 3 weeks of osimertinib therapy, both T790M loss and T790M maintained patients had decreases in T790M levels (ΔT790M). Patients with T790M maintained had a larger decrease in EGFR driver levels (ΔDriver, P = .01). C, In analysis of the differential plasma response between the EGFR driver and the T790M, patients had a greater T790M response than EGFR driver response compared with those who maintained T790M (median difference, 16% vs 0%; P = .003). ND indicates not detectable.
We then studied serial plasma genotyping to assess whether early response in plasma could indicate the eventual pattern of resistance. Nineteen patients from the institutional cohort had an early response plasma specimen (after 1-3 weeks) and detectable tumor DNA before treatment and at development of resistance.25 In the plasma samples drawn at resistance, 8 patients had maintained T790M and 11 had T790M loss. Study of the relative change in plasma EGFR mutation concentration after 1 to 3 weeks of osimertinib therapy showed a decrease in plasma T790M levels (ΔT790M) both in patients with maintained T790M and T790M loss (median 100% decrease for both) (Figure 5B). Evaluation of the relative change in EGFR driver levels (ΔDriver) demonstrated a larger decrease for patients with maintained T790M vs those who lost T790M (median, 100% vs 83% decrease, P = .01). We then analyzed the differential plasma response between the EGFR driver and T790M (ΔDriver – ΔT790M) and found that this difference was significant between patients who went on to have T790M loss or T790M maintained at resistance (median difference, 16% vs 0%; P = .003) (Figure 5C). Eight of 9 patients with a greater decrease in T790M than in driver EGFR mutation (ΔDriver – ΔT790M>1%) went on to develop loss of T790M at resistance.
Discussion
Osimertinib has become the first targeted therapy to receive regulatory approval for the treatment of solid tumors harboring a specific resistance mechanism. Although osimertinib is expected to move into the first-line setting for treatment of EGFR-mutant NSCLC,4 the challenge of detecting and targeting specific resistance mechanisms has relevance across a range of targetable genotypes where treatment of drug resistance is under active investigation.26,27
Our data provide clinical evidence of the influence of heterogeneity on treatment outcomes with osimertinib and suggest that there may be 2 types of T790M-positive resistance. In some patients, the T790M mutation represents the dominant resistance mechanism, and these patients will have a more durable response to treatment. In patients with heterogeneous mechanisms of resistance, T790M-mutant subclones coexist with subclones harboring distinct resistance mechanisms; targeting T790M alone is likely to result in transient benefit. What is currently unclear is how to determine whether there is a dominant or minor T790M-positive population. Some prior studies have found that quantification of the relative T790M AF is associated with the degree of response to third-generation EGFR TKIs, although this association has not been consistent across published reports.24,28,29 In our analysis, we studied whether such a calculation of relative T790M AF in plasma could indicate probable subsequent loss of T790M, and our data did not reveal a clear predictive ability. Instead, serial plasma monitoring may offer insight into subsequent resistance patterns.
The overgrowth of non-T790M resistance mechanisms highlights that coexistence of multiple resistance mechanisms is a reality with clinical implications—some resistance mechanisms are not acquired during osimertinib therapy but instead coexist with T790M. For example, in 1 case positive for T790M prior to osimertinib, MET was amplified in just 6% of cells; with osimertinib treatment, this subclone became the dominant resistance mechanism, detectable in 94% of cells (eFigure 3 in the Supplement). The finding of multiple resistance mechanisms has been described previously. Yu et al30 identified multiple resistance mutations in 4% of 155 patients with EGFR-mutant lung cancer and acquired resistance. Yet, the increased use of PCR assays for T790M detection means that coexistent resistance mechanisms may go undetected, although emerging data suggest that plasma NGS approaches can detect, in a subset of cases, resistance mechanisms that coexist with T790M.31,32 Further prospective study is needed to determine whether multigene analyses of tumor or plasma could detect competing resistance mechanisms reliably enough to allow early delivery of appropriate osimertinib-based combination therapies.
Loss of T790M is a potentially confusing clinical state that could be misinterpreted as resensitization to first-line EGFR TKIs. Our data do not support this hypothesis; most of the time, loss of T790M is associated with development of alternative competing resistance mechanisms (eFigure 6 in the Supplement). Many of these mechanisms, such as small cell transformation and MET amplification, have been reported in T790M-negative acquired resistance.33,34 However, we also have identified a novel finding of an acquired KRAS mutation, which was confirmed using tumor NGS and plasma droplet digital PCR. The complex variety of resistance mutations seen in patients with loss of T790M, including several acquired fusion genes, highlights the need for better strategies to prevent or delay the emergence of resistance. Because loss of T790M is associated with early resistance, this phenomenon could be more evident in usual clinical care than prospective cohorts would suggest; indeed, it was seen in 68% of patients in our post hoc institutional cohort, but 47% of patients in the prospective AURA cohort (P = .03).
Our study has potentially important implications in the care of patients who develop resistance to osimertinib. There is a wide range of osimertinib combination therapies now in clinical trials, but selecting the right approach will require insight into disease biology. Patients who develop early resistance to osimertinib likely have competing resistance mechanisms in other tumor subclones, and patients who develop late resistance are more likely to have maintained T790M and acquired C797S; different targeted therapies could be considered for these biologically varying populations. New clinical trials aiming to target patients with T790M-positive resistance and acquired resistance to osimertinib must consider which type of resistance is being targeted. Repeated testing for T790M would be an intuitive approach to stratify these 2 biologically distinct types of osimertinib resistance. Another strategy would be to improve the efficacy of osimertinib by developing combination approaches that prevent the development of resistance.35,36 However, the complex range of resistance mechanisms seen after osimertinib treatment make it difficult for any single combination approach to significantly delay osimertinib resistance overall.
Limitations
The limitations of our study must be acknowledged. Genomic analysis of resistance biopsies was performed using available clinical NGS assays rather than a single central assay, and pre-osimertinib NGS was available on only a subset of patients. To compensate for the limited numbers of patients receiving resistance biopsies, we performed plasma genotyping on a clinical trial cohort to validate our findings of different clinical behaviors in different types of osimertinib resistance. Loss of T790M was more common in our institutional cohort compared with the trial cohort—we suspect that this is because a retrospective institutional cohort will enrich data for patients presenting with early resistance, which is more often loss of T790M. Even the trial cohort may be falsely enriched for T790M loss because patients who are still receiving therapy without resistance may be enriched for C797S, which is seen more in late resistance. The development of therapies targeting the EGFR C797S resistance mutation will need to consider the possibility that C797S will be a late resistance mechanism on osimertinib, and extra time may be needed to identify these patients.37
Conclusions
Our data indicate that EGFR T790M is a key biomarker not only for indicating the probability of osimertinib sensitivity but also for understanding the biology of osimertinib resistance. Retesting for T790M after osimertinib failure is an important step for guiding patients to appropriate subsequent treatment strategies and relevant clinical trials. Our data indicate that drug resistance in some patients is genetically heterogeneous, impairing the effectiveness of targeted therapies. These findings must be considered as strategies are developed to treat resistance to newer first-line targeted therapies (eg, osimertinib, alectinib)4,38 with their unique resistance mechanisms. If we wish to achieve more durable outcomes, early interventions may be needed, such as combination targeted therapies, to prevent the development of drug resistance.
eTable 1. Pre- and Post-Osimertinib Tumor and Plasma Genotyping of 41 Institutional Cohort Patients
eFigure 1. Serial Plasma Genotyping of Patients Treated With Osimertinib
eTable 2. Clinical Characteristics of Osimertinib Resistance Cohorts
eFigure 2. Acquired Resistance to Osimertinib in a Validation Cohort From the AURA Trial
eFigure 3. Patient With MET Amplification Detected via FISH Pre- and Post-Osimertinib
eFigure 4. Patient With BRAF Fusion Detected on NGS Pre- and Post-Osimertinib
eFigure 5. 50 Patients From the AURA Cohort With Plasma Genotyping Before and After Osimertinib
eFigure 6. Molecular Subtypes of Acquired Resistance to Osimertinib
References
- 1.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor–resistant non–small-cell lung cancer. N Engl J Med. 2015;372(18):1527-1534. doi: 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 2.Mok TS, Wu YL, Ahn MJ, et al. ; AURA3 Investigators . Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376(7):629-640. doi: 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oxnard GR, Ramalingam SS, Ahn M-J, et al. Preliminary results of TATTON, a multi-arm phase Ib trial of AZD9291 combined with MEDI4736, AZD6094 or selumetinib in EGFR-mutant lung cancer. J Clin Oncol. 2015;33(15 suppl):2509.26150443 [Google Scholar]
- 4.Soria JC, Ohe Y, Vansteenkiste J, et al. ; FLAURA Investigators . Osimertinib in untreated EGFR-mutated advanced non–small-cell lung cancer. N Engl J Med. 2018;378(2):113-125. doi: 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 5.Ou SI, Agarwal N, Ali SM. High MET amplification level as a resistance mechanism to osimertinib (AZD9291) in a patient that symptomatically responded to crizotinib treatment post-osimertinib progression. Lung Cancer. 2016;98:59-61. doi: 10.1016/j.lungcan.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 6.Planchard D, Loriot Y, André F, et al. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann Oncol. 2015;26(10):2073-2078. doi: 10.1093/annonc/mdv319 [DOI] [PubMed] [Google Scholar]
- 7.Ahn S, Hwang SH, Han J, et al. Transformation to small cell lung cancer of pulmonary adenocarcinoma: clinicopathologic analysis of six cases. J Pathol Transl Med. 2016;50(4):258-263. doi: 10.4132/jptm.2016.04.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho CC, Liao WY, Lin CA, Shih JY, Yu CJ, Chih-Hsin Yang J. Acquired BRAF V600E mutation as resistant mechanism after treatment with osimertinib. J Thorac Oncol. 2017;12(3):567-572. doi: 10.1016/j.jtho.2016.11.2231 [DOI] [PubMed] [Google Scholar]
- 9.Yu HA, Tian SK, Drilon AE, et al. Acquired resistance of EGFR-mutant lung cancer to a T790M-specific EGFR inhibitor: emergence of a third mutation (C797S) in the EGFR tyrosine kinase domain. JAMA Oncol. 2015;1(7):982-984. doi: 10.1001/jamaoncol.2015.1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacher AG, Jänne PA, Oxnard GR. Management of acquired resistance to epidermal growth factor receptor kinase inhibitors in patients with advanced non–small cell lung cancer. Cancer. 2014;120(15):2289-2298. doi: 10.1002/cncr.28723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res. 2016;22(19):4837-4847. doi: 10.1158/1078-0432.CCR-15-1915 [DOI] [PubMed] [Google Scholar]
- 12.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21(6):560-562. doi: 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ClinicalTrials.gov AZD9291 First Time in Patients Ascending Dose Study. NCT01802632. https://clinicaltrials.gov/ct2/show/ NCT01802632. Accessed June 11, 2018.
- 14.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1(19):e87062. doi: 10.1172/jci.insight.87062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paweletz CP, Sacher AG, Raymond CK, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res. 2016;22(4):915-922. doi: 10.1158/1078-0432.CCR-15-1627-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. BEAMing up for detection and quantification of rare sequence variants. Nat Methods. 2006;3(2):95-97. doi: 10.1038/nmeth850 [DOI] [PubMed] [Google Scholar]
- 17.Kehl K, Gong YUT, Kalyani PM, Oxnard G, Blumenthal GM. Time to treatment discontinuation (TTD) as a pragmatic endpoint in metastatic non–small cell lung cancer (mNSCLC): a pooled analysis of 8 trials [abstract]. J Clin Oncol. 2018;36(suppl):9064. [Google Scholar]
- 18.Lo PC, Dahlberg SE, Nishino M, et al. Delay of treatment change after objective progression on first-line erlotinib in epidermal growth factor receptor-mutant lung cancer. Cancer. 2015;121(15):2570-2577. doi: 10.1002/cncr.29397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in pretreated T790M-positive advanced non-small-cell lung cancer: AURA study phase II extension component. J Clin Oncol. 2017;35(12):1288-1296. doi: 10.1200/JCO.2016.70.3223 [DOI] [PubMed] [Google Scholar]
- 20.Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JK, Lee J, Kim S, et al. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol. 2017;35(26):3065-3074. doi: 10.1200/JCO.2016.71.9096 [DOI] [PubMed] [Google Scholar]
- 22.VanderLaan PA, Rangachari D, Mockus SM, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: correlation with clinical outcomes. Lung Cancer. 2017;106:17-21. doi: 10.1016/j.lungcan.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aisner DL, Sholl LM, Berry L, et al. ; LCMC2 investigators. . The impact of smoking and TP53 mutations in lung adenocarcinoma patients with targetable mutations—the Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res. 2018;24(5):1038-1047. doi: 10.1158/1078-0432.CCR-17-2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non–small-cell lung cancer. J Clin Oncol. 2016;34(28):3375-3382. doi: 10.1200/JCO.2016.66.7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2(8):1014-1022. doi: 10.1001/jamaoncol.2016.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bahcall M, Sim T, Paweletz CP, et al. Acquired METD1228V mutation and resistance to MET inhibition in lung cancer. Cancer Discov. 2016;6(12):1334-1341. doi: 10.1158/2159-8290.CD-16-0686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: precision medicine takes on drug resistance. Cancer Discov. 2017;7(2):137-155. doi: 10.1158/2159-8290.CD-16-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 2015;5(7):713-722. doi: 10.1158/2159-8290.CD-15-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol. 2017;28(4):784-790. [DOI] [PubMed] [Google Scholar]
- 30.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240-2247. doi: 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. doi: 10.1038/ncomms11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol. 2018;29(4):1049-1055. doi: 10.1093/annonc/mdy005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011;17(5):1169-1180. doi: 10.1158/1078-0432.CCR-10-2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusan M, Li K, Li Y, et al. Suppression of adaptive responses to targeted cancer therapy by transcriptional repression. Cancer Discov. 2018;8(1):59-73. doi: 10.1158/2159-8290.CD-17-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tricker EM, Xu C, Uddin S, et al. Combined EGFR/MEK inhibition prevents the emergence of resistance in EGFR-mutant lung cancer. Cancer Discov. 2015;5(9):960-971. doi: 10.1158/2159-8290.CD-15-0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534(7605):129-132. doi: 10.1038/nature17960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters S, Camidge DR, Shaw AT, et al. ; ALEX Trial Investigators . Alectinib versus crizotinib in untreated ALK-positive non–small-cell lung cancer. N Engl J Med. 2017;377(9):829-838. doi: 10.1056/NEJMoa1704795 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Pre- and Post-Osimertinib Tumor and Plasma Genotyping of 41 Institutional Cohort Patients
eFigure 1. Serial Plasma Genotyping of Patients Treated With Osimertinib
eTable 2. Clinical Characteristics of Osimertinib Resistance Cohorts
eFigure 2. Acquired Resistance to Osimertinib in a Validation Cohort From the AURA Trial
eFigure 3. Patient With MET Amplification Detected via FISH Pre- and Post-Osimertinib
eFigure 4. Patient With BRAF Fusion Detected on NGS Pre- and Post-Osimertinib
eFigure 5. 50 Patients From the AURA Cohort With Plasma Genotyping Before and After Osimertinib
eFigure 6. Molecular Subtypes of Acquired Resistance to Osimertinib