Abstract
Various commercial assays have recently been developed for detecting glutamate dehydrogenase (GDH) and/or toxin A/B to diagnose Clostridioides difficile infection (CDI). We compared the performance of two assays for the simultaneous detection of C. difficile GDH and toxin A/B, using 150 stool samples: C. DIFF QUIK CHEK COMPLETE (QCC; TechLab, Blacksburg, VA, USA) and RIDASCREEN Clostridium difficile GDH (RC-GDH) and Toxin A/B (RC-Toxin A/B; R-Biopharm, Darmstadt, Germany). For GDH detection, QCC and RC-GDH showed satisfactory sensitivity (95.7% and 94.3%, respectively) and specificity (92.5% and 93.8%, respectively) compared with C. difficile culture. For toxin A/B detection, QCC showed higher sensitivity than RC-Toxin A/B (60.0% vs 33.3%, P<0.001) compared with toxigenic C. difficile culture. When the results of QCC or RC-GDH+RC-Toxin A/B were used as the first step of a two-step algorithm for diagnosing CDI, QCC permitted more accurate discrimination than RC of positive or negative results for CDI (77.3% and 65.3%, respectively). QCC is useful for the simultaneous detection of C. difficile GDH and toxin A/B as a part of the two-step algorithm for diagnosing CDI.
Keywords: Clostridioides difficile, Glutamate dehydrogenase, Toxin A/B, Performance, Algorithm
Clostridioides difficile is the major causative agent of healthcare-associated diarrhea [1,2,3]. Cell cytotoxicity assays and toxigenic C. difficile culture (TC) have been regarded as the gold standards for diagnosing C. difficile infection (CDI) [4,5]. However, a multiple-step algorithm for CDI diagnosis that includes a glutamate dehydrogenase (GDH) assay and toxin A/B assay as a screening test, along with a nucleic acid amplification test (NAAT), has recently been recommended [6,7].
Several commercial assays have recently been developed for detecting GDH and/or toxin A/B. C. DIFF QUIK CHEK COMPLETE (QCC; TechLab, Blacksburg, VA, USA), a lateral flow membrane immunoassay, tests for both GDH (QCC-GDH) and toxin A/B (QCC-Toxin A/B) simultaneously in one cartridge. RIDASCREEN (RC; R-Biopharm, Darmstadt, Germany) is a widely used enzyme immunoassay performed using a 96-well microwell plate per batch for RIDASCREEN Clostridium difficile GDH (RC-GDH) and RIDASCREEN Clostridium difficile Toxin A/B (RC-Toxin A/B) separately. We compared the performance of these two assays for the simultaneous detection of GDH and toxin A/B.
We used 150 stool samples submitted to the clinical microbiology laboratory at Samsung Medical Center, Seoul, Korea, from April 2017 to May 2017, including 101 consecutively collected and 49 preselected samples. This study was approved by the Institutional Review Board (IRB) of Samsung Medical Center (IRB No. SMC 2018-03-050). After routine testing, residual samples were stored at 2–8℃ for processing within 72 hours and frozen at −70℃ for further processing and evaluation.
TC was performed to confirm the presence of C. Difficile. All stool samples were inoculated on a chromogenic agar plate (CHROMID C. difficile agar; BioMérieux, Marcy l'Etoile, France) and incubated anaerobically at 35℃ for 24–48 hours. Gray to black colonies grown on the agar were investigated by Gram staining and tested for the production of proline-aminopeptidase using a PRO disk K1532B (Key Scientific Products, Inc., Stamford, TX, USA) [8]. Next, toxin gene PCR was performed with all C. difficile isolates. Briefly, DNA was extracted using more than five colonies by the MagNA Pure 96 kit (Roche Diagnostics, Mannheim, Germany), according to the manufacturer's protocol. Multiplex PCR was performed targeting the tcdA (toxin A), tcdB (toxin B), and triose phosphate isomerase (tpi; a C. difficile-specific housekeeping gene) using NK2/NK3 (tcdA), NK104/NK105 (tcdB), and tpi primers, respectively [9,10,11].
All stool samples were tested for the presence of GDH antigen and toxin A/B by QCC and RC according to the manufacturers' instructions. In brief, for QCC, a 500-µL mixture comprising a 25-µL stool sample with diluent and conjugate (TechLab) was transferred to the device sample well. After incubation for 15 minutes at 20–25℃, wash buffer was added, followed by addition of the substrate (TechLab) to the reaction window. Results were read after 10 minutes. The presence of GDH and/or toxins was indicated by the appearance of a color bar in the appropriate detection zone. The RC-GDH and RC-Toxin A/B tests were carried out sequentially using separate reagents. A total of 100 µL of stool sample with biotinylated anti-GDH and toxin A/B antibodies was transferred to each sample well and incubated for 60 minutes at 20–25℃. After washing with washing buffer five times, streptavidin poly-peroxidase conjugates were added and then incubated for 30 minutes. A fter washing, the substrates were added, followed by 15-minute incubation and the addition of a stop reagent. The concentrations of GDH and toxin A/B were measured at a dual wavelength of 450/630 nm using an automated immunoassay system, GEMINI (STRATEC Biomedical, Birkenfeld, Germany) [12].
The sensitivity and specificity of each enzyme immunoassay for GDH were calculated against the results of C. difficile culture, and those for toxin A/B were calculated against the results of TC as a reference method. McNemar's test was used to compare sensitivity and specificity between QCC and RC. Cohen's kappa was computed to evaluate the inter-assay agreement between QCC and RC (agreement:<0.4, poor; 0.4–0.75, fair to good; >0.75, excellent). Analyses were performed using SPSS version 24 (IBM Corp., Armonk, NY, USA). P<0.05 was considered statistically significant. We also investigated the performance of two-step algorithm for diagnosis of CDI by conducting simulations with the results of QCC and RC-GDH and Toxin A/B.
Among the 150 stool samples, 70 C. difficile isolates were obtained in culture, 60 of which (85.7%) were toxigenic C. difficile. For GDH, 73 and 71 samples were positive according to QCC-GDH and RC-GDH, respectively (Table 1). QCC-GDH and RC-GDH showed overall excellent agreement. The positive and negative percent agreement was 95.8% (95% confidence interval [CI], 87.3–98.9%) and 93.7% (95% CI, 85.2–97.6%), respectively. Discordant results between QCC-GDH and RC-GDH were observed for eight samples. A total of five results were QCC-GDH-positive and RC-GDH-negative, and two of them were confirmed to be positive on C. difficile culture. Of the three QCC-GDH-negative and RC-GDH-positive results, one was confirmed to be positive by C. difficile culture. In comparison with C. difficile culture, QCC-GDH and RC-GDH assays showed comparable diagnostic sensitivity and specificity (Table 1).
Table 1. Performance of C. DIFF QUIK CHEK COMPLETE– GDH and RIDASCREEN Clostridium difficile GDH compared with Clostridioides difficile culture.
| Test | Result | C. difficile culture | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Kappa (95% CI) | |
|---|---|---|---|---|---|---|
| Positive (N=70) | Negative (N=80) | |||||
| C. DIFF QUIK CHEK COMPLETE–GDH | Positive | 67 | 6 | 95.7 (87.2–98.9) | 92.5 (83.8–96.9) | 0.89 (0.82–0.97) |
| Negative | 3 | 74 | ||||
| RIDASCREEN Clostridium difficile GDH | Positive | 66 | 5 | 94.3 (85.3–98.2) | 93.8 (85.4–97.7) | |
| Negative | 4 | 75 | ||||
Abbreviations: CI, confidence interval; GDH, glutamate dehydrogenase.
For toxin A/B detection, 41 and 21 samples were positive according to QCC and RC-Toxin A/B, respectively (Table 2). QCC and RC-Toxin A/B showed overall fair to good agreement. The positive and negative percent agreement was 95.3% (95% CI, 74.1–99.8%) and 83.7% (95% CI, 76.0–89.4%), respectively. In comparison with TC, the QCC-Toxin A/B assay showed higher sensitivity than the RC-Toxin A/B assay (P<0.001); however, specificity did not significantly differ between the two assays (P=0.125; Table 2).
Table 2. Performance of C. DIFF QUIK CHEK COMPLETE–Toxin A/B and RIDASCREEN Clostridium difficile Toxin A/B compared with toxigenic Clostridioides difficile culture.
| Test | Result | Toxigenic culture | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Kappa (95% CI) | |
|---|---|---|---|---|---|---|
| Positive (N=60) | Negative (N=90) | |||||
| C. DIFF QUIK CHEK COMPLETE–Toxin A/B | Positive | 36 | 5 | 60.0 (46.5–72.2) | 94.4 (86.9–97.9) | 0.56 (0.41–0.71) |
| Negative | 24 | 85 | ||||
| RIDASCREEN Clostridium difficile Toxin A/B | Positive | 20 | 1 | 33.3 (22.0–46.8) | 98.9 (93.1–99.9) | |
| Negative | 40 | 89 | ||||
Abbreviation: CI, confidence interval.
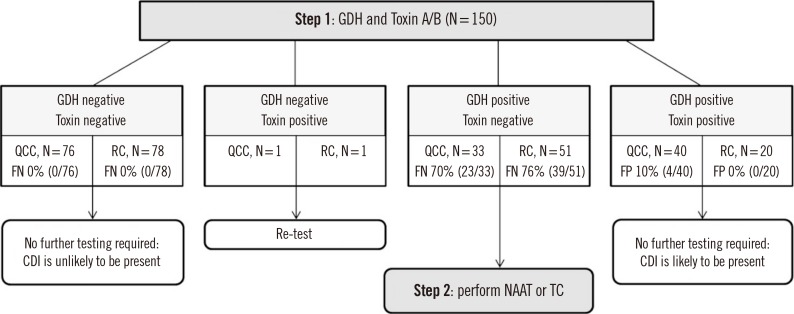
In the two-step algorithm for diagnosis (Fig. 1) [6,13], when QCC was used as an initial screening test, no further tests were needed for 116 of 150 (77.3%) samples. When RC-GDH and Toxin A/B were used as an initial screening test, no further tests were needed in 98 of 150 (65.3%) samples.
Fig. 1. Two-step algorithm for diagnosis of toxigenic Clostridioides difficile infection by applying QCC or RC-GDH and Toxin A/B.
Abbreviations: QCC, C. DIFF QUIK CHEK COMPLETE, QUIK-CHEK COMPLETE; RC, RIDASCREEN Clostridium difficile; FP, false positive; FN, false negative; CDI, Clostridioides difficile infection; GDH, glutamate dehydrogenase; NAAT, nucleic acid amplification test; TC, toxigenic Clostridioides difficile culture.
Overall, QCC-GDH and RC-GDH showed satisfactory performance in detecting GDH, with sensitivities and specificities in the range of previous reports (81.0–100% and 82.0–94.8%, respectively) [14,15]. QCC-Toxin A/B showed a sensitivity of 60%, which accords with previous reports of a wide range of sensitivity, from 29% to 79% [6,13]. However, the sensitivity of RC-Toxin A/B was relatively low at 33.3%, below that of previous reports showing a narrow range of 48–67%, despite the same test procedure [12,13,16,17]. One GDH-negative but toxin A/B-positive sample was identified by both QCC and RC. Since this sample was determined to be negative by TC, it was designated as a toxin A/B false-positive result. The most likely explanation for this discrepancy is cross-reactivity to toxins formed by other clostridial species, such as C. sordellii, which produce toxins that are antigenically similar to those of C. difficile [18].
Because no single test is suitable as a stand-alone test, the use of a multiple-step algorithm for CDI diagnosis is recommended in clinical laboratories [6,19]. Simultaneous detection of both GDH and toxin A/B is less time-consuming than conventional TC and has the advantage of being able to quickly and accurately discriminate negative cases as a screening test. The use of QCC and RC as the first step in a two-step algorithm eliminated the need for secondary testing in 77.3% and 65.3%, of the samples, respectively. In particular, QCC has the advantage that samples can be analyzed individually and do not need to be batched. However, RC is suitable for high-throughput batch testing in laboratories that require analyses of large numbers of samples.
In summary, as QCC-Toxin A/B is significantly more sensitive than RC-Toxin A/B for identifying toxin producers, QCC is useful for simultaneous detection of GDH and toxin A/B in the first step of the two-step algorithm for diagnosing CDI.
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: There are no potential conflicts of interest relevant to this article.
References
- 1.Makristathis A, Zeller I, Mitteregger D, Kundi M, Hirschl AM. Comprehensive evaluation of chemiluminescent immunoassays for the laboratory diagnosis of Clostridium difficile infection. Eur J Clin Microbiol Infect Dis. 2017;36:1253–1259. doi: 10.1007/s10096-017-2916-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin BM, Yoo SM, Shin WC. Evaluation of Xpert C. difficile, BD MAX Cdiff, IMDx C. difficile for Abbott m2000, and Illumigene C. difficile assays for direct detection of toxigenic Clostridium difficile in stool specimens. Ann Lab Med. 2016;36:131–137. doi: 10.3343/alm.2016.36.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mori N, Takahashi T. Characteristics and immunological roles of surface layer proteins in Clostridium difficile. Ann Lab Med. 2018;38:189–195. doi: 10.3343/alm.2018.38.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Planche T, Wilcox M. Reference assays for Clostridium difficile infection: one or two gold standards? J Clin Pathol. 2011;64:1–5. doi: 10.1136/jcp.2010.080135. [DOI] [PubMed] [Google Scholar]
- 5.Debast SB, Bauer MP, Kuijper EJ European Society of Clinical Microbiology and Infectious Diseases. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2014;20:1–26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 6.Crobach MJ, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, et al. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2016;22:S63–S81. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 7.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin Infect Dis. 2018;66:987–994. doi: 10.1093/cid/ciy149. [DOI] [PubMed] [Google Scholar]
- 8.Park KS, Ki CS, Lee NY. Isolation and identification of Clostridium difficile using ChromID C. difficile medium combined with Gram staining and PRO disc testing: a proposal for a simple culture process. Ann Lab Med. 2015;35:404–409. doi: 10.3343/alm.2015.35.4.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terhes G, Urbán E, Sóki J, Hamid KA, Nagy E. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J Clin Microbiol. 2004;42:4316–4318. doi: 10.1128/JCM.42.9.4316-4318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004;42:5710–5714. doi: 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, et al. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36:2178–2182. doi: 10.1128/jcm.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanpoucke H, De Baere T, Claeys G, Vaneechoutte M, Verschraegen G. Evaluation of six commercial assays for the rapid detection of Clostridium difficile toxin and/or antigen in stool specimens. Clin Microbiol Infect. 2001;7:55–64. doi: 10.1046/j.1469-0691.2001.00141.x. [DOI] [PubMed] [Google Scholar]
- 13.Chung HS, Lee M. Evaluation of the performance of C. DIFF QUIK CHEK COMPLETE and its usefulness in a hospital setting with a high prevalence of Clostridium difficile infection. J Investig Med. 2017;65:88–92. doi: 10.1136/jim-2016-000231. [DOI] [PubMed] [Google Scholar]
- 14.Swindells J, Brenwald N, Reading N, Oppenheim B. Evaluation of diagnostic tests for Clostridium difficile infection. J Clin Microbiol. 2010;48:606–608. doi: 10.1128/JCM.01579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ota KV, McGowan KL. Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C. difficile toxin enzyme immunoassays with C. difficile nucleic acid amplification testing increase diagnostic yield in a tertiary pediatric population. J Clin Microbiol. 2012;50:1185–1188. doi: 10.1128/JCM.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattner F, Winterfeld I, Mattner L. Diagnosing toxigenic Clostridium difficile: new confidence bounds show culturing increases sensitivity of the toxin A/B enzyme immunoassay and refute gold standards. Scand J Infect Dis. 2012;44:578–585. doi: 10.3109/00365548.2012.655772. [DOI] [PubMed] [Google Scholar]
- 17.Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211–3217. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toltzis P, Nerandzic MM, Saade E, O'Riordan MA, Smathers S, Zaoutis T, et al. High proportion of false-positive Clostridium difficile enzyme immunoassays for toxin A and B in pediatric patients. Infect Control Hosp Epidemiol. 2012;33:175–179. doi: 10.1086/663706. [DOI] [PubMed] [Google Scholar]
- 19.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]