Abstract
Purpose
Advancing maternal and paternal age leads to a decrease in fertility, and hence, many infertile couples opt for assisted reproductive technologies [ART] to achieve biological parenthood. One of the key determinants of achieving a live outcome of ART, embryo quality, depends on both the quality of the oocyte and sperm that have created the embryo. Several studies have explored the effect of oocyte parameters on embryo quality, but the effects of sperm quality on the embryo have not been comprehensively evaluated.
Method
In this review, we assess the effect of various genetic factors of paternal origin on the quality and development of the embryo.
Results
The effects of sperm aneuploidy, sperm chromatin structure, deoxyribonucleic acid [DNA] fragmentation, role of protamines and histones, sperm epigenetic profile, and Y chromosome microdeletions were explored and found to negatively affect embryo quality.
Conclusion
We propose that careful assessment of spermatozoal parameters is essential to achieve embryo development and a healthy live birth. However, the heterogeneity in test results and the different approaches of assessing a single sperm parameter highlight the need for more research and the development of standardized protocols to assess the role of sperm factors affecting embryo quality.
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1304-4) contains supplementary material, which is available to authorized users.
Keywords: Embryo, Paternal factors, DNA fragmentation, Sperm chromatin, Y chromosome microdeletions, Sperm epigenetics, Histone, Protamines, Methylation
Background
The number of couples seeking to have children is steadily increasing [1]. This is leading to an increase in the number of couples seeking infertility treatment and in particular, patients being treated at an older age. Compounding this phenomenon is that fertility declines with advancing maternal [1] and paternal age [2]. Presently, infertility affects 10–15% of reproductive aged couples worldwide [3]. Studies have suggested a trend towards a reduced sperm quality in the last years where about 30% of infertile couples show a paternal origin of infertility [4].
Male factor infertility refers to the inability of the male to cause pregnancy in a clinically sound female [5]. Male fertility can be affected by several factors that may be congenital, endocrinal, immunological, oncological, infectious, or lifestyle-based in nature [6]. It can be classified as non-idiopathic [known causes of male infertility include cryptorchidism, varicocele, hormonal imbalances, and chemotherapy] or idiopathic, where the cause of male infertility remains unknown. It is in such cases that genetic lesions such as karyotypic abnormalities [7], single gene mutations, and polymorphisms [8] or Y chromosome microdeletions [9] have been shown to be responsible. In recent times, a gray area of sperm biology including that of sperm DNA damage [10] and epigenetic or methylation anomalies [11] are also believed to be potential candidates responsible for infertility. All these genetic defects may interfere with the development of the male reproductive system and urogenital tract, arrest germ cell production and maturation, and lead to the production of non-functional spermatozoa, manifested phenotypically as various forms of male factor infertility [12].
ART are a gamut of infertility treatments used to achieve a pregnancy bypassing the natural process of conception, with in vitro fertilization [IVF] and intracytoplasmic sperm injection [ICSI] being the two commonly used technologies [4]. Since the introduction of ICSI in 1992 [13], its high rate of success has opened avenues to treating previously untreatable cases of both female and male factor infertility. However, after almost a quarter century of experience in this field, the rate of achieving clinical pregnancy using ART still remains lower than expected [14]. Many couples can undergo repeated attempts at ART before finally achieving a live birth [15] while encountering significant physical, emotional, and financial discomposure [16]. To address these issues, it is essential to comprehensively understand the plethora of factors that hang like the sword of Damocles over the outcome of ART. In this review, we discuss one of the most crucial factors affecting ART outcome: embryo quality, with emphasis, in particular, on how the sperm may impact it.
From zygote to implanted embryo—journey to the endometrium
The preliminary step of ART involves the fertilization of retrieved oocytes by sperm: a process that involves the direct interaction of the sperm and oocyte, fusion of their cell membranes, and a painstakingly intricate manipulation of the maternal and paternal genomes to create a totipotent zygote. This is followed by the male and female genomes forming pronuclei, whose subsequent breakdown signals the start of a series of ‘reductive’ mitotic divisions to form successively smaller cells. At the four-eight cell stage, the zygotic genome activates and the dependence on maternal mRNAs and proteins is eliminated [17]. Compaction at the eight-cell stage brings the dividing cells into close physical contact and cavitation at the 16- to 32-cell stage leads to the formation of a fluid-filled cavity marking the transition to the blastocyst stage. At this point, a clear delineation of the first two cell fate lineages in the embryo, the trophectoderm and the inner cell mass, can be observed [18]. This composite and harmonized cascade of events occurs over a span of 5 to 6 days and results in the mystifying metamorphosis of the zygote to an embryo [19].
Embryo quality
The transformations occurring in the zygote following fertilization are meticulously monitored by all ART clinics to determine “embryo quality”. Embryo quality is assessed on the basis of several parameters: morphological, developmental, genetic, and metabolic. These include measurement of the cleavage speed, kinetics and synchrony of division, the number, symmetry and spatial arrangement of blastomeres, the degree of blastomere fragmentation, the presence of multinucleation, compaction of blastomeres and extent of cytoplasmic fragmentation, the presence of a clear granular homogeneous cytoplasm, absence of vacuoles, and the non-aggregation of organelles [20]. Measured together, these parameters are able to confer the embryo a quality score, the decisive factor in defining the number of embryos to be transferred or frozen. In ART, embryo transfer is usually performed on either day 3 or 5, the latter ensures greater synchronicity of endometrial and embryonic development. Developmentally, day 5 embryos or blastocysts are associated with a better chance of pregnancy because they are believed to have a higher potential for implantation compared with cleavage stage [days 2–3] embryos [21]. A “top quality embryo” has been defined as having “no multinucleated blastomeres, ≤ 20% anucleated fragments and four or five blastomeres on day two, or seven or more blastomeres on day 3” [22].
The quality of an embryo, undoubtedly, is dependent on the quality of the oocyte that has created it. While studies have described the relationship between oocyte and embryo quality [23, 24, 25], several of the “embryo quality” parameters have also been shown to be influenced by paternal factors [26]. Clinically, it has been observed that the transfer of euploid embryos results in an implantation rate of approximately 60% while almost 40% embryos remain unaccounted for. Although some losses can be accounted for by endometrial factors, the residual lost embryos could be partly accounted for by paternal factors. Identifying the paternal factors affecting embryo quality is particularly important because ICSI has allowed infertile men with severe male factor to successfully sire offspring thus increasing the risk of vertical transmission of paternal genetic defects.
Association of the paternal genome to embryonic development has been reported by several groups [26, 27, 28]; significantly lower cleavage rates and blastocyst formation rates have been noted when frozen or morphologically abnormal sperm were used to fertilize oocytes in IVF [29] and lower blastocyst formation rates have been noted after ICSI [30]. However, the wide-ranging impact of spermatozoal factors on early embryo development is only still being deciphered [31, 32]. Could these spermatozoal factors be the Trojan horse that majorly impact ART outcomes? If yes, then an enhanced understanding of these factors could help optimize the existing therapeutic schemes and further improve the success and safety of ARTs. This review thus addresses the impact of paternal factors of genetic origin, such as sperm structure abnormalities, sperm DNA fragmentation, epigenetic defects in sperm, sperm aneuploidies, and Y chromosome microdeletions that affect embryo quality in ART [Fig. 1] [9].
Fig. 1.
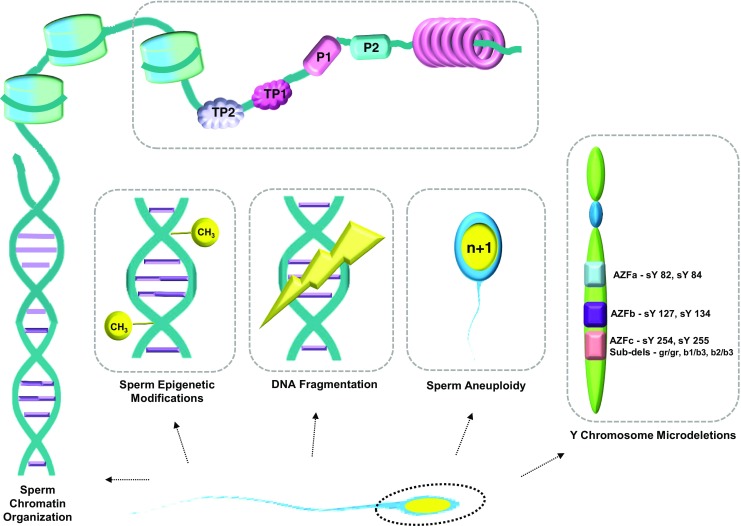
Genetic defects identified in spermatozoa
Paternal factors that contribute towards embryo development
Sperm chromatin structure
In contrast to the oocyte, the paternal contribution to the embryo is via a highly differentiated, transcriptionally inert cell with minimal cytoplasm (sperm). Human sperm are entrusted with the herculean task of ensuring that the paternal genome is delivered to the oocyte in a form that allows the developing embryo to access the genetic information for its development and also perpetuate the information to future generations [33]. A haploid sperm packages its DNA into a volume < 10% of a somatic cell nucleus. This astonishing level of genomic compaction is facilitated by the replacement of most of the sperm histones with smaller, highly basic, arginine, and cysteine-rich nucleoproteins called, protamines. Arginine permits for a higher degree of chromatin compaction by neutralizing the strong negative charges of the phosphate groups in the sperm DNA backbone while cysteine confers enhanced stability through intermolecular disulfide cross-links, thus forming the basic protamine packaging unit, called a toroid [34]. Protamination of sperm chromatin facilitates its efficient compaction into a small volume, bestows the spermatozoa an ergonomic nuclear shape to support its protracted transit in the male and female reproductive tracts, and protects the spermatozoa from physical or chemical genotoxic factors thus ensuring optimal genomic integrity.
So, by which mechanisms can sperm genomic integrity be breached? Abnormalities in sperm chromatin structure, for one, are known to alter chromatin configuration and cause DNA damage in the form of single-stranded or double-stranded DNA breaks. Sperm nucleoprotein defects such as altered histone to protamine conversion result in faulty DNA compaction. Sperm DNA damage can also be the consequence of defective spermatogenic machinery [35], the occurrence of apoptosis during spermatogenesis [36], or damage by exogenous sources such as reactive oxygen species [ROS] during transport in the male or female reproductive tract [37]. In cases where the oocyte is unable to repair the damage or in cases where the sperm chromatin perturbations are high, embryo development may also be impaired.
Currently, sperm chromatin integrity is assayed using several tests such as sperm chromatin structure assay [SCSA], sperm chromatin dispersion test [SCD], acridine orange test, aniline blue test [AB], and the toluidine blue test [TB]. Each of these tests relies on different principles and have different sensitivities and specificities [see Supplementary Table 1].
The quality of sperm chromatin is crucial, especially when one sperm is artificially selected to fertilize an oocyte as in the case of ART [38]. Evidence highlighting the role of chromatin organization in sperm came from observations that sperm which had not undergone the acrosome reaction, when used in ICSI, showed impaired decondensation of apical chromatin [39]. This delay in decondensation hindered the progression of the first mitotic division of the zygote, providing indirect evidence to explain the increase in sex chromosome aneuploidies observed in offspring after ICSI. Other reports have associated sperm with defective chromatin packing with significantly lower fertilization capacity following sub zonal insemination [40] or IVF [41].
An increased percentage of sperm with immature chromatin [as judged by its structure, condensation, and integrity] has been associated with lower fertilization rate and slower embryo development [42]. Sperm chromatin maturity has also been shown to play a pivotal role in the accomplishment of a pregnancy [43]; a chromatin maturity at the level of AB < 87% correlated significantly with the cleavage rate of the zygote [p = 0.022; r = 0.371] and chromatin integrity at the level of TB < 80% correlated with embryo formation [p = 0.048; r = 0,485] [44]. Another variation of sperm chromatin abnormality characterized by atypical packing of chromosome territories in sperm, aberrant positioning of chromosomes, or disturbed telomere-centromere interactions has also been reported in some infertile males and it is suggested that this abnormality may impact embryonic development [45].
However, several other studies have expressed uncertainty regarding the correlation between the sperm chromatin organization and embryo quality. A recent Polish study [46] failed to give credence to the association between sperm chromatin maturity and embryo development [measured as the number of blastomeres, symmetry, and cytoplasmic fragmentation]. Another study [47] reported that in IVF, the fertilization rate, percentage of cleaved zygotes, and embryos without fragmentation were similar in males with normal and abnormal sperm chromatin, except that sperm samples with both morphological defects and abnormal chromatin, displayed a reduced fertilization rate. Several other studies have also reported non-associative results [48, 49, 50]. Presently, the cause of such discrepant findings is hard to identify, but the lack of a single robust test that can serve as a gold standard for assessing chromatin integrity in the current clinical scenario may explain the variation in reported results. Each of the tests routinely employed to assess the integrity of sperm chromatin structure detect different aspects of chromatin abnormality.
Sperm protamine and histone levels
During the process of spermatogenesis, sperm chromatin undergoes a sperm-specific epigenetic mechanism that involves a major structural overhaul in DNA packaging brought about by the replacement of nucleohistones by protamines. The process involves breakdown and subsequent repair of double-stranded DNA to facilitate the replacement of hyperacetylated testis-specific histones in the round spermatids by transition nuclear proteins [TP1 and TP2] followed by ordered replacement of two types of phosphorylated protamines [P1 and P2] in the elongating spermatid stage [51] (Fig. 1). The new high-order chromatin packaging results in a global stop of transcription in the sperm.
In humans, both P1 and P2 are present in roughly equal quantities [52] but improper temporal regulation of these transcripts leads to altered expression of the mature proteins [53]. Infertile men can have altered P1/P2 ratios, increased histone to protamine ratios, or complete protamine deficiency [54, 55]. The incorrect distribution of histones and protamines can extend adverse effects on early embryo development [56].
A significant correlation has been shown to exist between sperm protamine messenger ribonucleic acid [mRNA] and protein levels and fertilization rate and embryo quality in couples undergoing IVF [57]. An altered P1/P2 ratio has been correlated with a negative impact on embryo quality and IVF outcome [58]. Most males with poor embryo quality [size of blastomeres, DNA fragmentation] show a P1/P2 ratio > 1 [59]. P2 deficiency has been shown to affect the development of embryos derived from P2-deficient spermatozoa [produced from knock-out protamine-deficient embryonic stem cells] with only 11% of P2-deficient embryos developing to the blastocyst stage while 86% remaining arrested in the 2–6 cell stages [60]. Low P1/P2 ratios have been reported to cause decreased fertilization [61] and implantation rates [62] in both IVF and ICSI [53].
It is thought that protamine deficiency affects sperm fertilizing capacity by bringing about premature chromosomal condensation [PCC]. During natural fertilization, sperm chromatin undergoes the histone to protamine replacement and suspends in the G1 cell cycle phase while the oocyte is suspended in metaphase II. PCC does not occur under these conditions since the meiosis promoting factors that induce PCC can act only on histones and not protamines. When the sperm penetrates the oocyte, sperm-associated oocyte activating factors are released from the acrosome resulting in the activation of meiosis promoting factors. During inactivation of meiosis promoting factors, protamines are replaced with histones and the oocyte terminates meiosis and enters G1 phase of the cell cycle becoming synchronized with the sperm. However, when a sperm carrying decreased levels of protamines enters an oocyte, the sperm undergoes PCC due to its high concentration of histones and this leads to failure of fertilization [57].
A correlation has also been found between sperm DNA fragmentation, altered P1/P2 ratios, and protamine concentration. Under normal conditions, approximately 10–15% of sperm chromatin remains linked to histones [63] but a decreased level or complete absence of protamines and greater persistence of histones during spermiogenesis can cause abnormal chromatin packing in the sperm head leading to DNA strand breaks in ejaculated sperm [64]. Protamine insufficiency-associated sperm DNA breakage as measured by the comet assay suggests that irreparable DNA damage is the chief cause of implantation failure in embryos derived from healthy eggs fertilized by protamine-compromised sperm [53]. However, another study suggests that increased sperm DNA fragmentation may be associated with both low and high P1/P2 ratios [65]. A meta-analysis of twelve studies found a significant association between protamine deficiency and DNA damage [n = 845; p < 0.001]. When the relationship between P1/P2 ratio and male fertility was analyzed using data from nine studies, a significantly higher protamine ratio was observed in sub-fertile men compared with healthy volunteers [n = 633 versus 453, respectively; P < 0.00001] [66].
A recent prospective cohort study in France [67] investigated the relationship between the proportion of sperm chromatin linked to remaining histone and ART outcome and found that the histone-to-protamine [HPR] ratio is prognostic of embryonic development up to the blastocyst stage. This group realized that when HPR values ranged between 6 and 26%, the probability of blastocyst formation was optimal and decreased when the HPR value was outside this range, regardless of whether IVF or ICSI was utilized. When sperm with altered HPR values is utilized in an ART procedure, it could result in desynchronized paternal genomic expression that alters embryo development until the blastocyst stage. Increased levels of histone in sperm chromatin have been shown to adversely affect the cleavage rate [p = 0.05] and embryo quality [p = 0.01] but not affect embryo production [68]. The histone content in sperm is related to its epigenetic modifications, and variations in the quantity of histones could disturb embryo development [67]. Even the replacement of histones by protamines has been shown to affect the epigenetic landscape in sperm. Aberrant protamine levels in oligozoospermic men have been correlated with an altered DNA methylation pattern at seven imprinted genes [KCNQ1OT1, MEST, SNRPN, PLAGL1, PEG3, H19, IGF2] [69].
Sperm epigenetic factors
Epigenetics refers to mechanisms that bring about gene regulation without altering the core DNA sequence. Epigenetic modifications permit the transformation of the same genome into several different transcriptomes in different cells of the organism. Sperm epigenetics is an emerging field of research in reproductive health and new studies have squashed the traditionally held belief that the sperm only delivers its genome to the oocyte at fertilization. In fact, an extensive genome-wide reprogramming of DNA methylomes, histone modifications, and chromatin accessibility have been reported to occur during early embryogenesis in mammals, facilitated by the distinct paternal-specific and maternal-specific epigenetic signatures carried by the newly developed embryo [70]. The sperm epigenome comprises its one-of-a-kind DNA methylation profile, presence of protamines and DNA-associated proteins, distinctive nucleosome distribution pattern, post-translational histone modifications and a milieu of stored RNAs, non-histone proteins and non-protamine proteins [71].
Genome-wide re-programming in the zygote occurs in two steps soon after fertilization. First, sperm protamines are exchanged for maternal nucleosomes [72] in a process called preimplantation reprogramming. This step involves extensive epigenetic modifications of the spermatozoal genome within the oocyte and subsequently bestows totipotency to the zygote [73]. The second step, called germline reprogramming, helps establish the sex-specific epigenetic signatures in the zygote [71].
The sperm is believed to be well equipped for supporting appropriate embryonic transcription, facilitated by the fraction of sperm genome that does not undergo the histone-protamine transition but retains a nucleosomal architecture. The histones in these nucleosomes carry epigenetic signatures that on fertilization are transferred to oocyte and consequently affect DNA access of transcription factors [74] and regulate gene expression [75] in the early embryo while also being transmitted to the offspring [76]. What is interesting to know is these retained nucleosomes are not randomly distributed in the paternal genome with studies showing enrichment of nucleosomes at loci of developmental importance and imprinted genes [55, 77, 78] and regions containing retrotransposable long and short interspersed nuclear elements [LINEs and SINEs, particularly LINE1] [79]. Further, a link between embryo DNA methylation dynamics and a post-fertilization function for sperm histones was reported in a study showing that DNA methylation-free regions in the early embryo correspond with nucleosome-rich regions in sperm chromatin [75].
Altered patterns of sperm DNA and histone methylation have been reported in infertile men [69]. Hypermethylation of the CREM promoter in spermatids resulting in an abnormal protamine 1: protamine 2 ratio has been associated with incomplete sperm chromatin compaction, reduced sperm motility, and male subfertility [58]. In a study of 63 men whose female partners were part of an IVF program, genome-wide hypermethylation of sperm DNA was associated with pregnancy failure [P < 0.05] [80]. Another study [81] where genome-wide sperm DNA methylation analysis was measured across > 485,000 sites in the genomes of IVF patients and normozoospermic fertile men indicated that sperm DNA methylation patterns could predict embryo quality during IVF. The researchers noted that although poor embryogenesis [developmental arrest or attrition at any point during embryonic development or poor implantation] could not be attributed to a few consistently altered cytosine-guanine dinucleotides, the genome-wide methylation profile of men in the poor-quality–embryo was inherently different.
Spermatozoa are also known to contain a plethora of mRNAs [82] and non-coding RNAs [ncRNAs], microRNAs [miRNAs] [83], small interfering RNAs [siRNAs] [26], and piwi-interacting RNAs [piRNAs] [84] that are transmitted to the oocyte [85]. Animal studies have shown that absence of sperm-delivered miRNAs in mice results in developmental delay in the zygote [86]. The prominent role of sperm-derived RNAs in the epigenetic regulation of gene expression in the early embryo and also adult life of offspring has been demonstrated in animal studies [87]. However, human studies in this direction are warranted.
DNA fragmentation
So far, we have looked at how the genomic architecture of the sperm can be alerted resulting in a poor organization of sperm chromatin. A disrupted state of sperm chromatin leaves the sperm vulnerable to DNA damage characterized by single- and double-stranded DNA breaks. DNA fragmentation in sperm can also occur by other mechanisms such as sperm chromatin remodeling during spermiogenesis, apoptosis during the process of spermatogenesis, ROS-induced post-testicular DNA fragmentation, movement of sperm through the seminiferous tubules and the epididymis, DNA fragmentation induced by endogenous caspases and endonucleases, radiotherapy or chemotherapy-induced DNA fragmentation, and DNA fragmentation caused by environmental toxicants [37]. Some commonly used techniques are the sperm chromatin structural assays [e.g., chromomycin A3, sperm chromatin structural assay—SCSA]; tests that directly assess DNA fragmentation [Terminal Deoxynucleotidyl Transferase dUTP Nick End Labelling] TUNEL, COMET assays]; and sperm nuclear matrix assays [e.g., sperm nuclear matrix stability assay, sperm chromatin dispersion test]. Each of these tests relies on different principles and has their own advantages and limitations [Supplementary Table 1].
It has long been known that samples with normal semen parameters express some level of DNA fragmentation [88] and not surprisingly, higher degrees sperm DNA fragmentation have been documented in infertile males [89]. Sperm DNA damage has been associated with high levels of ROS detected in the semen of approximately 25% of infertile men [90]. Another study that measured levels of the apoptotic marker protein, Fas, found that < 10% of apoptotic sperm exist in normozoospermic men whereas almost 60% of oligospermic men have > 10% of apoptotic sperm [91]. Studies have demonstrated that following chemotherapy, recovery of spermatogenesis and return to baseline aneuploidy levels can take a few months to several years after treatment has ceased [92]. Lifestyle factors such as obesity, smoking, and certain occupations have also been associated with increased levels of DNA damage [93]. However, sperm carrying fragmented DNA can successfully fertilize an oocyte [94] and achieve pregnancy with help of in vitro techniques [95]. It is thought that good quality metaphase II oocytes are able to overcome the negative effect of sperm DNA fragmentation by recruiting an army of DNA repair enzymes and anti-apoptotic proteins [96].
Several detailed meta-analyses have explored the clinical value of the sperm DNA fragmentation tests towards pregnancy outcomes [96, 97]. DNA fragmentation exceeding 30% with the SCSA test has been associated with a lower likelihood of fertilization through intrauterine inseminations [96] while one study suggests that a predictive threshold of 27% was required for a successful pregnancy via both IVF and/or ICSI when DNA fragmentation is assessed using SCSA [96].
A systematic review exploring the relationship between sperm DNA fragmentation [measured by TUNEL, SCSA, SCD, and COMET assays] and embryo quality [3] reported that 34% studies showed a significant association whereas 66% studies showed no significant relationship between these two parameters. The authors found a differential association between sperm DNA fragmentation and embryo quality when the studies were segregated into groups based on assay types [sperm DNA fragmentation detected by the alkaline comet assay had stronger association with poor embryo quality when compared to other assays]. This was attributed to the sensitivity of the comet assay, which measures both single- and double-stranded DNA breaks. Another systematic review [97] assessing the impact of sperm DNA fragmentation on embryo quality revealed a positive relation between DNA fragmentation and poor embryo quality in 12.5% in IVF studies and 42% in ICSI studies. The higher poor embryo quality rate in ICSI is thought to occur because this procedure permits fertilization with even highly DNA fragmented sperm while in IVF, the integrity of sperm DNA is closely monitored prior to the selection of sperm for fertilization thus reducing the probability of fertilization with a damaged sperm.
A study [98] evaluating the effects of DNA strand breaks in spermatozoa on embryo development in IVF reported of a negative correlation between blastocyst development and the percentage of TUNEL positivity in sperm. Reports also associate sperm DNA damage to lower cleavage rates [97] and arrest of embryonic development after the second or third cleavage [99, 98]. Faster dividing cleavage stage bovine embryos have been shown to be more likely to develop into blastocysts [100] and fast dividing embryos have been associated with higher pregnancy rates [101]. Thus, DNA fragmentation impacts both blastocyst formation rates and pregnancy rates.
A recent study [102] has identified statistically significant differences in three groups of DNA fragmented sperm [SCD ≤ 10%, SCD 11–20%, and SCD ≥ 21%]] with regard to day 3 good embryo formation, blastocyst formation, implantation rates, and clinical pregnancy rates in the males with SDF ≥ 21% as compared to the other two groups [P < 0.05], all undergoing IVF. This can be explained by the fact that embryonic genome activation occurs only after the second cell division, i.e., the 4–8 cell stage [17] and hence is bound to impact only the later stages of embryo development. However, Zheng et al. [103] studied DNA fragmentation effects in sperm from 215 men undergoing ART using an alkaline comet assay and reported a paternal effect at each stage of early embryonic development [peri-fertilization effect [fertilization rate], early paternal effect [embryonic days 1–2], late paternal effect [embryonic days 3–5], and implantation stage effect]. In fact, it has been reported that paternal DNA damage may present as chromosome aberrations soon after the first metaphase [104].
Tandara et al. 2014 have reported that a big halo formed in the halosperm test was the only independent predictor of optimal embryo quality [third day cumulative embryo score] while big halo and DNA fragmentation index [DFI] together were significant prognostic parameters for achieving pregnancy [105]. Osman et al. have used sensitivity analysis to demonstrate a statistically significant difference in live birth rates between low and high sperm DNA fragmentation when ICSI was used [RR 1.08, 95% CI 0.39 to 2.96; p = 0.88] [106].
However, few studies also report absence of association between sperm DNA fragmentation and fertilization, embryo development, cleavage, blastulation, implantation, and pregnancy rates [107, 108]. Although a negative correlation between the intensification of DNA fragmentation and the achievement of pregnancy in IVF has been reported [109] and ICSI [110], Sadeghi et al. [2011] did not find this sperm factor affecting the effectiveness of IVF [50]. Thus, it is challenging to summarize the effect of DNA fragmentation on embryo quality with several such conflicting results. When we collated data on the role of DFI on embryo quality in eight studies what we found noteworthy was that different studies used different methods to estimate DFI and each of these studies used different DFI cut-offs for their analysis suggesting a lack of consensus in the measurement of this parameter [Supplementary Table 2]. In fact, the different methods of sperm preparation before ICSI have also been shown to contribute towards the discrepancies reported by these researchers [111]. A recent review [112] even suggests that significant inter-observer and inter-laboratory variation during microscopy-based assays to detect sperm DNA fragmentation make it challenging to commercialize the assays.
Sperm aneuploidy
Abnormalities in sperm chromatin condensation have been reported to correlate to chromosomal aneuplodies in embryos [113, 114]. Chromosomal aneuploidy refers to an alteration in chromosomal number from the normal diploid chromosomal complement in somatic cells or haploid complement in gametes. Chromosomal aneuploidies are common in early human embryos [115] and strongly contribute to poor IVF outcomes [116, 117]. Although embryonic aneuploidies can arise from both the maternal or paternal genomes, in the case of aneuploid ICSI fetuses, some aneuploidies have been traced to arise from the infertile father’s sperm [118].
Indeed, all males produce some amount of aneuploid sperm but higher incidence of sperm aneuploidy and diploidy are observed in males with altered seminal parameters [119] or those with male factor infertility [7]. Males with oligoasthenoteratozoospermia [OAT] show an increased level of disomy, diploidy, and nullisomy as compared to their fertile counterparts while also giving rise to higher levels of numerically aneuploid embryos [120]. Males with abnormal karyotypes are predisposed to producing spermatozoa with an unbalanced chromosome complement; non-mosaic Klinefelter patients show approximately 6% sex chromosome aneuploidy [92]. Moreover, several other classes of males such as those harboring Y chromosome microdeletions [predisposed towards sex chromosome aneuploidies] [121], males with normal karyotypes, fertile males who have fathered offspring with paternally derived aneuploidy, males who have experienced recurrent pregnancy losses, and infertile oligospermic males are also at increased risk of producing chromosomally abnormal spermatozoa [122].
Genetic studies suggest that sperm chromosome abnormalities occur due to meiotic errors [123] caused by mutations in meiosis-specific genes involved in synaptic processes of DNA recombination and reparation [124] while other studies have identified mutations in genes that cause meiotic arrest [125]. Several other factors such as Y chromosome microdeletions, varicocele, chemotherapy, occupation- and lifestyle-based factors also negatively impact the efficiency of meiotic divisions during spermatogenesis. Another line of evidence suggests that spermatozoa containing large vacuoles exhibit greater chromosomal aneuploidies and diploidies [126]. All the above findings hold significance because oligozoospermic males would be the most common candidates for ARTs and the identification of sperm aneuploidy rates in these men could be considered as an appropriate supportive test before initiation of the reproductive strategems.
Sperm aneuploidy, earlier tested by karyotyping [hamster oocyte fertilization test], is currently screened by interphase FISH in human sperm nuclei or next-generation sequencing assays [127]. Several studies have analyzed the clinical repercussions of sperm chromosomal abnormalities on ARTs. Reports associate sperm aneuploidy and diploidy with repeated implantation failure and pregnancy loss after ICSI cycles [128], recurrent abortion [129], miscarriage [129], ICSI failure [128, 129], and higher aneuploidies rates in live births [130]. Nicopoullos et al. report significantly higher total aneuploidy levels [13, 18, 21 and sex chromosome; P = 0.01], aneuploidy rates in chromosome 18 [P = 0.01], the sex chromosomes [P = 0.05], and the composite total of chromosomes 18/X/Y [P = 0.005] in sperm ejaculates from men who failed to achieve a clinical pregnancy in ICSI [128]. Corroborating this finding, male partners of women suffering from recurrent pregnancy loss have been reported to show a greater percentage of sperm aneuploidy of the sex chromosomes and chromosomes 18 and 13/21 [1.04 vs. 0.38%; 0.18 vs. 0.03%; 0.26 vs. 0.08%] in case control studies [131, 132].
Aneuploidies in the developing embryo are believed to arise either due to an aneuploid sperm or oocyte or due to a mitotic gain, loss, or non-disjunction occurrence in the embryo leading to mosaicism [133]. A ‘mosaic’ embryo could also be the result of minor variations in spindle dynamics or due to anomalies in the embryonic centriole and centrosome, both of which are paternally inherited by the human embryo. Sperm centrosomal defects result in abnormal spindle formation and compromise early cell divisions in the human embryo leading to the early paternal effect. A genome-wide association study has identified single nucleotide polymorphisms in the sequence of PLK4, a key regulator of centriole duplication, to be associated with embryonic mitotic errors [134].
Several studies have also explored the effect of advanced paternal age on sperm aneuploidy rates. Two recent studies [135, 136] note that males above the age of 50 in IVF/ICSI cycles showed significantly more sperm with damaged DNA, low blastocyst development rate, higher global aneuploidy rates, and significantly greater number of embryos with trisomies [p ≤ 0.05]. Increasing paternal age leads to several alterations in the male endocrinal [decreased circulation of androgens, elevated levels of FSH] and reproductive phenotypes [alteration of testicular morphology and volume, changes in sperm production, and sperm characteristics and a significant increase in sperm DNA fragmentation] [99, 135]. This, coupled with the accumulation of DNA damage over years and the decreased capacity of germ cells to repair this damage, is what leads to a decline in sperm genome integrity leading to production of aneuploid sperm which translates to increased aneuploidy in embryos [137]. Similarly, in the case of chromosomal translocations, the high percentage of chromosomally unbalanced sperm is shown to translate to a high proportion of chromosomally unbalanced embryos [138].
Intriguingly, studies by Daughtry et al. report that diploid–aneuploid mosaic embryos show implantation rates comparable to euploid embryos and re-iterate that a blastocyst with up to 60% aneuploidy can result in a successful pregnancy outcome [139]. Another study [140] reports that if the number of aneuploid cells is < 40%, the embryo stands a 78% chance of developing to the blastocyst stage while a higher number of abnormal cells may be associated with a lower survival rate [33%] [141]. Thus, it appears that mosaic embryonic aneuploidy does not always terminate developmental potential. Bolton et al. suggest that only a small portion of euploid cells are necessary to sustain human foetal development [142]—a notion that is supported by the observation that frozen-thawed human embryos that have lost almost half of blastomeres during the cryopreservation procedure are still viable and result in live births [143].
Yq microdeletions
The Y chromosome is one of the smallest in the human genome, is specific to males, and represents around 2–3% of the haploid genome [144]. This chromosome is largely heterochromatic, gene-poor, and recombines with its homolog only at its pseudoautosomal regions [144]. The pseudoautosomal regions comprise only 5% of the Y chromosome, while the majority [63 Mb] of the Y chromosome is known as the male-specific Y or MSY region. Approximately two-thirds [41 Mb] of the MSY region is composed of three blocks of highly repetitive sequences called the azoospermia factor [AZF] regions which contain several genes that are critical for spermatogenesis and the development of male gonads [145].
The Y chromosome microdeletions are submicroscopic deletions that span several genes but are not large enough to be detected using conventional cytogenetic methods. These microdeletions occur precisely in Yq11, at the AZF region which is composed of three loci: AZFa, AZFb, and AZFc and are among the best known genetic causes of male infertility. Based on global data of > 30,000 Y chromosomes, the prevalence of the Y chromosome microdeletions in infertile men is estimated to be 7% [95% CL 6.74–6.79] [146]. These deletions are more common among men with non-obstructive azoospermia [NOA] [7–23%] compared with severe oligozoospermia [1–8%] males [9].
The Y chromosome microdeletions can be classical: where the complete AZFa, b, or c locus is deleted, respectively, partial: where only certain regions of the AZFa, b, or c loci are deleted or, combined: where two or more AZF loci are deleted simultaneously. Distinct histopathological phenotypes are correlated with the site of the microdeletion; the complete AZFa and AZFb deletions are associated with azoospermia [9] while the complete AZFc deletion is associated with hypospermatogenesis that leads to cryptozoospermia, oligozoospermia, or azoospermia [147]. The AZFc locus can also harbor partial deletions or sub-deletions called gr/gr, b1/b3, and b2/b3 which are associated to variable phenotypes, ranging from normo- to azoo-spermia [148].
The development of ARTs, together with testicular or epididymal sperm retrieval for azoospermic men, has allowed Y deleted males to father offspring using their own gametes. However, the effect of Y chromosome microdeletions and partial deletions of AZFc on embryo quality in ART remains controversial.
A study by Page et al. reported that spermatozoa retrieved from men harboring AZFc deletions were fertilization competent and when used for ICSI produced viable embryos and pregnancy at similar rates as spermatozoa of men without AZFc deletions [149]. One study [150] evaluated the ART outcomes of six males showing complete AZFc deletions and noted that a clinical pregnancy could be achieved in all cases. However, lower fertilization rates have been noted in azoospermic AZFc deleted males when compared to males with an intact Y chromosome [36 vs. 45%] when both groups of males underwent testicular sperm extraction [TESE] with subsequent ICSI cycles [151]. Another study [152] has also reported lower fertilization rates [55 vs. 71%, P < 0.001] with concomitant poorer embryo quality in AZFc deleted males with severe oligozoospermia who underwent ICSI cycles. A higher cleaved embryo rate [94.0 vs. 88.1%, P < 0.05] in AZFc deleted males versus controls [94.0 vs. 88.1%, P < 0.05] has been reported in one Chinese study [153]. Mateu et al. 2010 have reported a high percentage of abnormal embryos [aneuploidies or monosomy X] in males harboring complete AZFc microdeletions in conjunction with numeric chromosome abnormalities, as compared with oligozoospermic patients without Y chromosome microdeletions [154].
A report from our laboratory [155] has shown absence of difference in fertilization rate and embryo transfer rate but an increase in the numbers of grade III embryos [grade I embryos were defined as blastomeres of equal size without cytoplasmic fragments, grade II embryos were defined as blastomeres of equal size with minor cytoplasmic fragments or blebs, and grade III embryos were defined as blastomeres of unequal size with significant cytoplasmic fragmentation] in men with AZFc sub-deletions as compared to non-deleted controls. However, when a recent Turkish study [156] evaluated the effect of the partial AZFc sub-deletions on ICSI outcome by measuring embryo development and pregnancy rates, they failed to find any statistical correlations, suggesting that the AZFc sub-deletions may not play a significant role in ART outcomes. The heterogeneity in the genes deleted in the AZFc locus is thought to influence the fertility outcome in males harboring AZFc sub-deletions and could explain the incongruity in results.
Very few studies in the literature have elucidated the influence of Y chromosome microdeletions on embryo quality in ARTs. In Supplementary Table 3, data from 12 studies on 451 embryos suggested that these microdeletions predominantly lower fertilization rates and influence the quality of the embryo while not altering other embryo quality parameters. However, all sperm to be used in ARTs must be assessed for the presence of Y chromosome microdeletions to avoid the vertical transmission of these infertility causing Y chromosome defects and thus lead to perpetuation of infertility in future generations.
Summary and future directions
Our review highlights that several paternal factors are in play during development of the embryo and each of these factors contributes significantly towards embryo quality in ART [Fig. 2]. Effects may range from decreasing the fertilization rate, lowering the cleavage/blastocyst rate of the developing embryo, and even affecting implantation into the endometrium. Hence, careful analysis of spermatozoal factors in conjunction with assessment of the maternal contributory factors is essential to ensure good embryo quality and subsequently a healthy child.
Fig. 2.
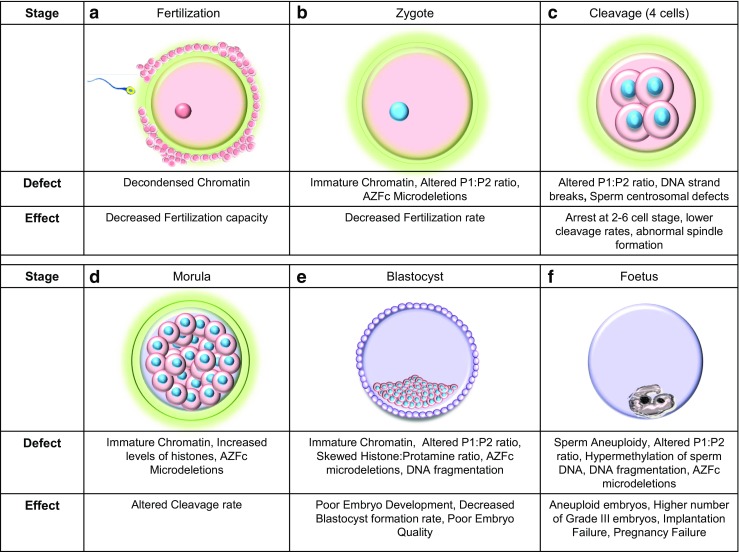
Genetic factors of paternal origin that impact embryo quality. Images a to e depict the stages in human embryonic development. a Structural defects of sperm chromatin can decrease the fertilization capacity of sperm. b The rate at which the sperm fertilizes the oocyte can be affected by sperm chromatin defects, alterations in the protamine ratios, and the [Azoospermia factor c] AZFc microdeletions in the Y chromosome. c At the 4 cell stage, the zygotic genome is activated and dependence on maternal genome is eliminated. Embryonic development may be arrested at this stage if the P1/P2 ratio is unbalanced. d The morula stage [16–32 cells] is characterized by a series of cleavage events that successively generate smaller cells. Structural chromatin defects and an increased level of histones can decelerate the cleavage rate with the presence of Y chromosome microdeletions can increase the cleavage rate. e The blastocyst stage that is characterized by the presence of the trophectoderm and the inner cell mass. A skewed histone to protamine ration [HPR] can decrease blastocyst formation rates, poor embryonic development is seen when the embryo is fertilized by sperm carrying chromatin defects and altered P1/P2 ratios. Poor quality embryos are noted when there is an altered protamine 1/protamine 2 ratio and the paternal Y chromosome harbors and AZFc microdeletions. In ART, embryo transfer is performed at this stage of embryonic development. f The growth of the fetus after implantation into the endometrium of the uterus. Presence of altered P1/P2 ratios can cause implantation failure while fertilization by aneuploid sperm, altered protamine ratios, and sperm epigenetic defects can lead to failure to establish pregnancy
Several studies have embarked on the odyssey of elucidating the role of sperm DNA damage in relation to embryo quality and on-going pregnancy. In a landmark study, Fernández-Gonzalez et al. performed ICSI in mouse using spermatozoa in which DNA damage had been induced by cryo-injury, to identify the long-term consequences of a damaged paternal genome on the health of offspring [157]. The authors reported several disturbing findings; the cryo-injury of sperm had led to gross DNA strand breaks and loss of telomeres, on fertilization by ICSI, these DNA-damaged sperm led to preimplantation embryo development and on birth, a reduced number of offspring per litter was noted. Methylation profiles of several epigenetically regulated genes were found to be altered and the ART offspring exhibited increased anxiety, lack of a habituation pattern, and a defective spatial memory [P < 0.05]. Moreover, 33% of females fathered with DNA-fragmented sperm presented some solid tumors in lungs and dermis and 20% of the mice died during the first 5 months of life, with 25% of the surviving animals showing premature aging symptoms. The authors suggested that although the oocytes had repaired parts of the fragmented DNA, the repair was incomplete, thus leading to long-term pathologies.
In terms of human studies, early publications have indicated that there were no increased health risks of children born via ART. However, all 25 studies in Hanson et al. [2005] [158] meta-analysis indicated an increased risk of birth defects. Studies from Europe have shown that ART children showed normal physical development up to 18 years with normal menarche and pubertal development. Studies from Australia show that the overall risk of any birth defect following ART was 8.3%, compared to the significantly lower, 5.8%, in spontaneously conceived children with the most common birth defects connected to ART being spina bifida, cerebral palsy, cleft palate and musculoskeletal, cardiovascular, and gastrointestinal conditions. Overall, the risk was found to be higher for ICSI compared to IVF and interestingly, use of frozen thawed embryos lessened the risk. However, it must be remembered here that these risks and anomalies cannot be pinned on defective sperm health alone.
The search for a marker of embryo quality which will permit the selection of the single best embryo for transfer continues to be a major challenge in ART with several contemporary and innovative approaches being ascertained with each passing day. While earlier research predominantly focused on assessing the impact of maternal factors affecting embryo quality, in recent times, the onus of contributing to better embryo quality is being equally shared by paternal factors as well.
At present, several sperm selection techniques such as density gradient centrifugation and swim-up are routinely used in most ART clinics with the aim of selecting the best sperm for fertilization. Unfortunately, not many of these techniques target the assessment of important sperm characteristics such as apoptosis, DNA integrity, membrane maturation, and ultrastructure, all of which contribute significantly towards improved embryo quality. Improving ART outcome by isolating mature, structurally intact and non-apoptotic spermatozoa with high DNA integrity remains an ongoing challenge with several methods based on surface charge [electrophoresis and zeta potential], apoptosis [magnetic cell sorting and glass wool], ultra-morphology [high magnification], or membrane maturity [hyaluronic acid binding] [159] being investigated.
Besides these methods, abnormalities in phospholipase c [PLC]-zeta, a sperm enzyme that mobilizes the Ca2+ oscillations to induce oocyte activation and embryo development, have also been associated with poor embryo quality [160, 161]. New data indicates that the replacement of defective centrosomes, which are responsible for specific forms of male infertility, with functional donor sperm centrosomes can restore normal functionality and thus ensure successful fertilization and embryo development [162]. The sperm cell has also been shown to contain various forms of RNA, e.g., mRNA, miRNA, siRNA [163] as well as more than 2000 proteins with unknown roles [164]. MicroRNA [miR]-34c is thought to initiate the first cleavage divisions in the mouse and represents a distinctive example of the impact of a single paternally derived miRNA on embryo development. Yuan et al. [165] produced zygotes from miRNA-depleted sperm [from Drosha conditional knockout mice] through ICSI and found that these zygotes resulted in embryos with reduced developmental potential, which could be recovered through the injection of small RNA from wild-type sperm. The potential predictive role of a novel sperm DNA-based marker of embryo quality, sperm telomere length [STL], is also being explored. Telomeres, the guanine-rich nucleoproteins found at the ends of chromosomes, shorten with each cell replication event [60–70 base pair per year] because of incomplete end replication but an enzyme called telomerase can synthesize de novo repeats to these chromosome ends. STL has been positively correlated with embryo morphology in IVF cycles and transplantable embryo rates [166].
With this myriad of novel parameters being identified what remains to be done is the development and validation of a robust and standardized panel of tests to assess the effects of the paternal genome on embryo quality.
Conclusion
The advent of ART and success of ICSI in particular has increased the chances of an abnormal spermatozoon being selected to fertilization and developing into an embryo. It was traditionally thought that the sperm was only a vessel to deliver the paternal genome to the oocyte, but recent advances have shown that the function of sperm extends immeasurably beyond this. Hence, it is of great essence to understand how abnormalities at different levels of paternal genomic organization may affect reproductive potential and ART outcome. Recent evidence suggests that spermatozoa containing chromatin structure anomalies, fragmented DNA, Y chromosome microdeletion, abnormal chromosome number, or an altered genetic imprint may be associated with impaired fertilization, embryogenesis, or embryonic development. Our review has discussed the possible mechanism and impact of these factors on fertilization and embryo quality comprehensively. However, the heterogeneity in test results and the different approaches of assessing a single sperm parameter highlight the urgent need to develop a standardized protocol to address the role of sperm factors affecting embryo quality and thus foster better reproductive healthcare.
The manuscript bears the NIRRH ID: REV/653/07–2018.
SC is a recipient of the ICMR postdoctoral grant.
Electronic supplementary material
(DOCX 35 kb)
Authors’ information
DS is Scientific Director at Boston IVF, Waltham MA 02451, USA.
SC is a post-doctoral fellow, Indian Council of Medical Research, National Institute for Research in Reproductive Health, Mumbai, India.
Authors’ contributions
DS contributed towards the design and structure of the manuscript. SC collected data and wrote the manuscript. Both authors approve the final manuscript.
Ethics approval and consent to participate
Not applicable as it is a review article.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Stacy Colaco, Email: stacy.colaco@gmail.com.
Denny Sakkas, Email: dsakkas@bostonivf.com.
References
- 1.CDC National Survey of Family Growth 2/6/15, CDC National Vital Statistics Reports 1/4/17.
- 2.De la Rochebrochard E, Thonneau P. Paternal age≥ 40 years: an important risk factor for infertility. Am J Obstet Gynecol. 2003;189:901–905. doi: 10.1067/s0002-9378(03)00753-1. [DOI] [PubMed] [Google Scholar]
- 3.Simon L, Emery BR, Carrell DT. Review: impact of sperm DNA damage in assisted reproduction. Best Pract Res Clin Obstet Gynaecol. 2017;44:38–56. doi: 10.1016/j.bpobgyn.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Chapuis A, Gala A, Ferrières-Hoa A, Mullet T, Bringer-Deutsch S, Vintejoux E, Torre A, Hamamah S. Sperm quality and paternal age: effect on blastocyst formation and pregnancy rates. Basic Clin Androl. 2017;27:2. doi: 10.1186/s12610-016-0045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpe RM. Sperm counts and fertility in men: a rocky road ahead: Science & Society Series on sex and science. EMBO Rep. 2012;13:398–403. doi: 10.1038/embor.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busetto GM, Agarwal A, Virmani A, Antonini G, Ragonesi G, Del Giudice F, Micic S, Gentile V, De Berardinis E. Effect of metabolic and antioxidant supplementation on sperm parameters in oligo-astheno-teratozoospermia, with and without varicocele: a double-blind placebo-controlled study. Andrologia. 2018;50:e12927. doi: 10.1111/and.12927. [DOI] [PubMed] [Google Scholar]
- 7.Templado C, Vidal F, Estop A. Aneuploidy in human spermatozoa. Cytogenet Genome Res. 2011;133:91–99. doi: 10.1159/000323795. [DOI] [PubMed] [Google Scholar]
- 8.Anifandis G, Markandona O, Dafopoulos K, Messini C, Tsezou A, Dimitraki M, Georgoulias P, Daponte A, Messinis I. Embryological results of couples undergoing ICSI-ET treatments with males carrying the single nucleotide polymorphism rs175080 of the MLH3 gene. Int J Mol Sci. 2017;18:314. doi: 10.3390/ijms18020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleiman SE, Yogev L, Lehavi O, Hauser R, Botchan A, Paz G, Yavetz H, Gamzu R. The likelihood of finding mature sperm cells in men with AZFb or AZFb-c deletions: six new cases and a review of the literature [1994–2010] Fertil Steril. 2011;95:2005–2012. doi: 10.1016/j.fertnstert.2011.01.162. [DOI] [PubMed] [Google Scholar]
- 10.Sakkas D, Ramalingam M, Garrido N, Barratt CL. Sperm selection in natural conception: what can we learn from mother nature to improve assisted reproduction outcomes? Hum Reprod Update. 2015;21:711–726. doi: 10.1093/humupd/dmv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurentino S, Borgmann J, Gromoll J. On the origin of sperm epigenetic heterogeneity. Reproduction. 2016;151:R71–R78. doi: 10.1530/REP-15-0436. [DOI] [PubMed] [Google Scholar]
- 12.WHO. Laboratory manual for the examination and processing of human semen . 5th ed. Geneva: World Health Organization; 2010. [Google Scholar]
- 13.Palermo G, Joris H, Devroey P, Van Steirteghem AC. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet. 1992;340:17–18. doi: 10.1016/0140-6736(92)92425-f. [DOI] [PubMed] [Google Scholar]
- 14.Sadeghi MR. Low success rate of ART, an illusion, a reality or simply a too high expectation? J Reprod Infertil. 2012;13:123. [PMC free article] [PubMed] [Google Scholar]
- 15.Malizia BA, Hacker MR, Penzias AS. Cumulative live-birth rates after in vitro fertilization. N Engl J Med. 2009;360:236–243. doi: 10.1056/NEJMoa0803072. [DOI] [PubMed] [Google Scholar]
- 16.Domar AD, Broome A, Zuttermeister PC, Seibel M, Friedman R. The prevalence and predictability of depression in infertile women. Fertil Steril. 1992;58:1158–1163. [PubMed] [Google Scholar]
- 17.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four-and eight-cell stages of preimplantation development. Nature. 1988;332:459. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 18.E. Charles. Embryogenesis. 2012. Human Embryogenesis. [Google Scholar]
- 19.Magli MC, Jones GM, Lundin K, Van den Abbeel E. Atlas of human embryology: from oocytes to preimplantation embryos. Hum Reprod 2012 ;27: i1. [DOI] [PubMed]
- 20.Cummins JM, Breen TM, Harrison KL, Shaw JM, Wilson LM, Hennessey JF. A formula for scoring human embryo growth rates in in vitro fertilization: its value in predicting pregnancy and in comparison with visual estimates of embryo quality. J In Vitro Fert Embryo Transf. 1986;3:284–295. doi: 10.1007/BF01133388. [DOI] [PubMed] [Google Scholar]
- 21.Wintner EM, Hershko-Klement A, Tzadikevitch K, Ghetler Y, Gonen O, Wintner O, Shulman A, Wiser A. Does the transfer of a poor quality embryo together with a good quality embryo affect the in vitro fertilization [IVF] outcome? J Ovarian Res. 2017;10:2. doi: 10.1186/s13048-016-0297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Royen E, Mangelschots K, De Neubourg D, Valkenburg M, Van de Meerssche M, Ryckaert G, Eestermans W, Gerris J. Characterization of a top quality embryo, a step towards single-embryo transfer. Hum Reprod. 1999;14:2345–2349. doi: 10.1093/humrep/14.9.2345. [DOI] [PubMed] [Google Scholar]
- 23.Lazzaroni-Tealdi E, Barad DH, Albertini DF, Yu Y, Kushnir VA, Russell H, Wu YG, Gleicher N. Oocyte scoring enhances embryo-scoring in predicting pregnancy chances with IVF where it counts most. PLoS One. 2015;10:e0143632. doi: 10.1371/journal.pone.0143632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cobo A, Coello A, Remohí J, Serrano J, de los Santos JM, Meseguer M. Effect of oocyte vitrification on embryo quality: time-lapse analysis and morphokinetic evaluation. Fertil Steril. 2017;108:491–497. doi: 10.1016/j.fertnstert.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 25.Demko ZP, Simon AL, McCoy RC, Petrov DA, Rabinowitz M. Effects of maternal age on euploidy rates in a large cohort of embryos analyzed with 24-chromosome single-nucleotide polymorphism-based preimplantation genetic screening. Fertil Steril. 2016;105:1307–1313. doi: 10.1016/j.fertnstert.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Parinaud J, Mieusset R, Vieitez G, Labal B, Richoilley G. Influence of sperm parameters on embryo quality. Fertil Steril. 1993;60:888–892. doi: 10.1016/s0015-0282(16)56292-x. [DOI] [PubMed] [Google Scholar]
- 27.Ménézo YJ, Sakkas D, Janny L. Co-culture of the early human embryo: factors affecting human blastocyst formation in vitro. Microsc Res Tech. 1995;32:50–56. doi: 10.1002/jemt.1070320105. [DOI] [PubMed] [Google Scholar]
- 28.Kumar M, Kumar K, Jain S, Hassan T, Dada R. Novel insights into the genetic and epigenetic paternal contribution to the human embryo. Clinics. 2013;68(S1):5–14. doi: 10.6061/clinics/2013(Sup01)02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janny L, Menezo YJ. Evidence for a strong paternal effect on human preimplantation embryo development and blastocyst formation. Mol Reprod Dev. 1994;38:36–42. doi: 10.1002/mrd.1080380107. [DOI] [PubMed] [Google Scholar]
- 30.Shoukir Y, Chardonnens D, Campana A, Sakkas D. Blastocyst development from supernumerary embryos after intracytoplasmic sperm injection: a paternal influence? Hum Reprod. 1998;13:1632–1637. doi: 10.1093/humrep/13.6.1632. [DOI] [PubMed] [Google Scholar]
- 31.Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet. 2005;6:633–642. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 32.Gòdia Marta, Swanson Grace, Krawetz Stephen A. A history of why fathers’ RNA matters†. Biology of Reproduction. 2018;99(1):147–159. doi: 10.1093/biolre/ioy007. [DOI] [PubMed] [Google Scholar]
- 33.Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod. 2010;16:30–36. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- 35.Sakkas D, Moffatt O, Manicardi GC, Mariethoz E, Tarozzi N, Bizzaro D. Nature of DNA damage in ejaculated human spermatozoa and the possible involvement of apoptosis. Biol Reprod. 2002;66:1061–1067. doi: 10.1095/biolreprod66.4.1061. [DOI] [PubMed] [Google Scholar]
- 36.Gorczyca W, Gong J, Darzynkiewicz Z. Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res. 1993;53:1945–1951. [PubMed] [Google Scholar]
- 37.Sakkas D, Alvarez JG. Sperm DNA fragmentation: mechanisms of origin, impact on reproductive outcome, and analysis. Fertil Steril. 2010;93:1027–1036. doi: 10.1016/j.fertnstert.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 38.Golan R, Shochat L, Weissenberg R, Soffer Y, Marcus Z, Oschry Y, Lewin LM. Evaluation of chromatin condensation in human spermatozoa: a flow cytometric assay using acridine orange staining. Mol Hum Reprod. 1997;3:47–54. doi: 10.1093/molehr/3.1.47. [DOI] [PubMed] [Google Scholar]
- 39.Sakkas D, Urner F, Bianchi PG, Bizzaro D, Wagner I, Jaquenoud N, Manicardi G, Campana A. Sperm chromatin anomalies can influence decondensation after intracytoplasmic sperm injection. Hum Reprod. 1996;11:837–843. doi: 10.1093/oxfordjournals.humrep.a019263. [DOI] [PubMed] [Google Scholar]
- 40.Bianchi Patrizia Grace, Manicardi Gian Carlo, Urner Françoise, Campana Aldo, Sakkas Denny. Chromatin packaging and morphology in ejaculated human spermatozoa: evidence of hidden anomalies in normal spermatozoa. Molecular Human Reproduction. 1996;2(3):139–144. doi: 10.1093/molehr/2.3.139. [DOI] [PubMed] [Google Scholar]
- 41.Auger J, Mesbah M, Huber C, Dadoune JP. Aniline blue staining as a marker of sperm chromatin defects associated with different semen characteristics discriminates between proven fertile and suspected infertile men. Int J Androl. 1990;13:452–462. doi: 10.1111/j.1365-2605.1990.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 42.Bounartzi T, Dafopoulos K, Anifandis G, Messini CI, Koutsonikou C, Kouris S, Satra M, Sotiriou S, Vamvakopoulos N, Messinis IE. Pregnancy prediction by free sperm DNA and sperm DNA fragmentation in semen specimens of IVF/ICSI-ET patients. Hum Fert. 2016;19:56–62. doi: 10.3109/14647273.2016.1157629. [DOI] [PubMed] [Google Scholar]
- 43.de Lamirande E, San Gabriel MC, Zini A. Human sperm chromatin undergoes physiological remodelling during in vitro capacitation and acrosome reaction. J Androl. 2012;33:1025–1035. doi: 10.2164/jandrol.111.015982. [DOI] [PubMed] [Google Scholar]
- 44.Asmarinah SA, Umar LA, Lestari SW, Mansyur E, Hestiantoro A, Paradowszka-Dogan A. Sperm chromatin maturity and integrity correlated to zygote development in ICSI program. Sys Biol Reprod Med. 2016;62:309–316. doi: 10.1080/19396368.2016.1210695. [DOI] [PubMed] [Google Scholar]
- 45.Zalensky A, Zalenskaya I. Organization of chromosomes in spermatozoa: an additional layer of epigenetic information? Biochem Soc Trans. 2007; 35:609–611. [DOI] [PubMed]
- 46.Gill K, Rosiak A, Gaczarzewicz D, Jakubik J, Kurzawa R, Kazienko A, Rymaszewska A, Laszczynska M, Grochans E, Piasecka M. The effect of human sperm chromatin maturity on ICSI outcomes. Hum Cell. 2018;29:1–2. doi: 10.1007/s13577-018-0203-4. [DOI] [PubMed] [Google Scholar]
- 47.Filatov M.V., Semenova E.V., Vorob'eva O.A., Leont'eva O.A., Drobchenko E.A. Relationship between abnormal sperm chromatin packing and IVF results. MHR: Basic science of reproductive medicine. 1999;5(9):825–830. doi: 10.1093/molehr/5.9.825. [DOI] [PubMed] [Google Scholar]
- 48.Bronet F, Martínez E, Gaytán M, Liñán A, Cernuda D, Ariza M, Nogales M, Pacheco A, San Celestino M, Garcia-Velasco JA. Sperm DNA fragmentation index does not correlate with the sperm or embryo aneuploidy rate in recurrent miscarriage or implantation failure patients. Hum Reprod. 2012;27:1922–1929. doi: 10.1093/humrep/des148. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z, Zhu L, Jiang H, Chen H, Chen Y, Dai Y. Sperm DNA fragmentation index and pregnancy outcome after IVF or ICSI: a meta-analysis. J Assist Reprod Genet. 2015;32:17–26. doi: 10.1007/s10815-014-0374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadeghi MR, Lakpour N, Heidari-Vala H, Hodjat M, Amirjannati N, Hossaini Jadda H, Binaafar S, Akhondi MM. Relationship between sperm chromatin status and ICSI outcome in men with obstructive azoospermia and unexplained infertile normozoospermia. Romanian J Morphol Embryol. 2011;52:645–651. [PubMed] [Google Scholar]
- 51.Balhorn Rod. A Clinician's Guide to Sperm DNA and Chromatin Damage. Cham: Springer International Publishing; 2018. Sperm Chromatin: An Overview; pp. 3–30. [Google Scholar]
- 52.Ward WS. Organization of sperm DNA by the nuclear matrix. Am J Clin Exp Urol. 2018;6:87. [PMC free article] [PubMed] [Google Scholar]
- 53.Aoki VW, Liu LH, Carrell DT. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod. 2005;20:1298–1306. doi: 10.1093/humrep/deh798. [DOI] [PubMed] [Google Scholar]
- 54.Carrell DT, Liu L. Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities of spermiogenesis. J Androl. 2001;22:604–610. [PubMed] [Google Scholar]
- 55.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carrell D. T., Hammoud S. S. The human sperm epigenome and its potential role in embryonic development. Molecular Human Reproduction. 2009;16(1):37–47. doi: 10.1093/molehr/gap090. [DOI] [PubMed] [Google Scholar]
- 57.Depa-Martynow M, Kempisty B, Jagodziński PP, Pawelczyk L, Jedrzejczak P. Impact of protamine transcripts and their proteins on the quality and fertilization ability of sperm and the development of preimplantation embryos. Reprod Biol. 2012;12:57–72. doi: 10.1016/s1642-431x(12)60077-1. [DOI] [PubMed] [Google Scholar]
- 58.Nanassy L, Liu L, Griffin J, T Carrell D. The clinical utility of the protamine 1/protamine 2 ratio in sperm. Protein Pept Lett. 2011;18:772–777. doi: 10.2174/092986611795713934. [DOI] [PubMed] [Google Scholar]
- 59.Nasr-Esfahani MH, Salehi MO, Razavi S, Mardani M, Bahramian H, Steger K, Oreizi F. Effect of protamine-2 deficiency on ICSI outcome. Reprod BioMed Online. 2004;9:652–658. doi: 10.1016/s1472-6483(10)61776-2. [DOI] [PubMed] [Google Scholar]
- 60.Cho C, Jung-Ha H, Willis WD, Goulding EH, Stein P, Xu Z, Schultz RM, Hecht NB, Eddy EM. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol Reprod. 2003;69:211–217. doi: 10.1095/biolreprod.102.015115. [DOI] [PubMed] [Google Scholar]
- 61.Mateo S, Gázquez C, Guimerà M, Balasch J, Meistrich ML, Ballescà JL, Oliva R. Protamine 2 precursors [pre-P2], protamine 1 to protamine 2 ratio [P1/P2], and assisted reproduction outcome. Fertil Steril. 2009;91:715–722. doi: 10.1016/j.fertnstert.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 62.Sakkas D, Shoukir Y, Chardonnens D, Bianchi PG, Campana A. Early cleavage of human embryos to the two-cell stage after intracytoplasmic sperm injection as an indicator of embryo viability. Hum Reprod. 1998;13:182–187. doi: 10.1093/humrep/13.1.182. [DOI] [PubMed] [Google Scholar]
- 63.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 64.D’Occhio MJ, Hengstberger KJ, Johnston SD. Biology of sperm chromatin structure and relationship to male fertility and embryonic survival. Anim Reprod Sci. 2007;101:1–7. doi: 10.1016/j.anireprosci.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 65.Simon L, Castillo J, Oliva R, Lewis SE. Relationships between human sperm protamines, DNA damage and assisted reproduction outcomes. Reprod BioMed Online. 2011;23:724–734. doi: 10.1016/j.rbmo.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 66.Ni K, Spiess AN, Schuppe HC, Steger K. The impact of sperm protamine deficiency and sperm DNA damage on human male fertility: a systematic review and meta-analysis. Andrology. 2016;4:789–799. doi: 10.1111/andr.12216. [DOI] [PubMed] [Google Scholar]
- 67.Fournier C, Labrune E, Lornage J, Soignon G, Giscard d'Estaing S, Guérin JF, Benchaib M. The impact of histones linked to sperm chromatin on embryo development and ART outcome. Andrology. 2018;6:436–445. doi: 10.1111/andr.12478. [DOI] [PubMed] [Google Scholar]
- 68.Tsuribe PM, Lima JN, Golim AM, Dell'Aqua PC, Issa JP, Gobbo CA. Assessment of sperm DNA in patients submitted the assisted reproduction technology procedures. JBRA Assist Reprod. 2016;20:17–22. doi: 10.5935/1518-0557.20160005. [DOI] [PubMed] [Google Scholar]
- 69.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–1733. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 70.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schagdarsurengin U, Steger K. Epigenetics in male reproduction: effect of paternal diet on sperm quality and offspring health. Nat Rev Urol. 2016;13:584. doi: 10.1038/nrurol.2016.157. [DOI] [PubMed] [Google Scholar]
- 72.Rodman TC, Pruslin FH, Hoffmann HP, Allfrey VG. Turnover of basic chromosomal proteins in fertilized eggs: a cytoimmunochemical study of events in vivo. J Cell Biol. 1981;90:351–361. doi: 10.1083/jcb.90.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okada Y, Yamaguchi K. Epigenetic modifications and reprogramming in paternal pronucleus: sperm, preimplantation embryo, and beyond. Cell Mol Life Sci. 2017;74:1957–1967. doi: 10.1007/s00018-016-2447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vavouri T, Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet. 2011;7:e1002036. doi: 10.1371/journal.pgen.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gannon JR, Emery BR, Jenkins TG, Carrell DT. The sperm epigenome: implications for the embryo. Adv Exp Med Biol. 2014;791:53–66. doi: 10.1007/978-1-4614-7783-9_4. [DOI] [PubMed] [Google Scholar]
- 76.Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, Trasler J. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350:aab2006. doi: 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]
- 77.Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod. 2011;26:2558–2569. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Samans B, Yang Y, Krebs S, Sarode GV, Blum H, Reichenbach M, Wolf E, Steger K, Dansranjavin T, Schagdarsurengin U. Uniformity of nucleosome preservation pattern in mammalian sperm and its connection to repetitive DNA elements. Dev Cell. 2014;30:23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 80.Benchaib M, Braun V, Ressnikof D, Lornage J, Durand P, Niveleau A, Guerin JF. Influence of global sperm DNA methylation on IVF results. Hum Reprod. 2005;20:768–773. doi: 10.1093/humrep/deh684. [DOI] [PubMed] [Google Scholar]
- 81.Aston KI, Uren PJ, Jenkins TG, Horsager A, Cairns BR, Smith AD, Carrell DT. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril. 2015;104:1388–1397. doi: 10.1016/j.fertnstert.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 82.Lelancette C, Miller D, Li Y, Krawetz SA. Paternal contributions: new functional insights for spermatozoal RNA. J Cell Biochem. 2008;104:1570–1579. doi: 10.1002/jcb.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan W, Morozumi K, Zhang J, Ro S, Park C, Yanagimachi R. Birth of mice after intracytoplasmic injection of single purified sperm nuclei and detection of messenger RNAs and microRNAs in the sperm nuclei. Biol Reprod. 2008;78:896–902. doi: 10.1095/biolreprod.107.067033. [DOI] [PubMed] [Google Scholar]
- 84.Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci U S A. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller D. Ensuring continuity of the paternal genome: potential roles for spermatozoal RNA in mammalian embryogenesis. Soc Reprod Fertil. 2007;65:373–389. [PubMed] [Google Scholar]
- 86.Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, Yeung WS. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A. 2012;109:490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 88.Evenson DP, Darzynkiewicz Z, Melamed MR. Relation of mammalian sperm chromatin heterogeneity to fertility. Science. 1980;210:1131–1133. doi: 10.1126/science.7444440. [DOI] [PubMed] [Google Scholar]
- 89.Oleszczuk K, Augustinsson L, Bayat N, Giwercman A, Bungum M. Prevalence of high DNA fragmentation index in male partners of unexplained infertile couples. Andrology. 2013;1:357–360. doi: 10.1111/j.2047-2927.2012.00041.x. [DOI] [PubMed] [Google Scholar]
- 90.Zini A, Libman J. Sperm DNA damage: importance in the era of assisted reproduction. Curr Opin Urol. 2006;16:428–434. doi: 10.1097/01.mou.0000250283.75484.dd. [DOI] [PubMed] [Google Scholar]
- 91.Varghese AC, du Plessis SS, Agarwal A. Male gamete survival at stake: causes and solutions. Reprod BioMed Online. 2008;17:866–880. doi: 10.1016/s1472-6483(10)60416-6. [DOI] [PubMed] [Google Scholar]
- 92.Tempest HG. Meiotic recombination errors, the origin of sperm aneuploidy and clinical recommendations. Syst Biol Reprod Med. 2011;57:93–101. doi: 10.3109/19396368.2010.504879. [DOI] [PubMed] [Google Scholar]
- 93.Zini A, Sigman M. Are tests of sperm DNA damage clinically useful? Pros and cons. J Androl. 2009;30:219–229. doi: 10.2164/jandrol.108.006908. [DOI] [PubMed] [Google Scholar]
- 94.Aitken J, Buckingham D, Krausz C. Relationships between biochemical markers for residual sperm cytoplasm, reactive oxygen species generation, and the presence of leukocytes and precursor germ cells in human sperm suspensions. Mol Reprod Dev. 1994;39:268–279. doi: 10.1002/mrd.1080390304. [DOI] [PubMed] [Google Scholar]
- 95.Erenpreiss J, Spano M, Erenpreisa J, Bungum M, Giwercman A. Sperm chromatin structure and male fertility:biological and clinical aspects. Asian J Androl. 2006;8:11–29. doi: 10.1111/j.1745-7262.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- 96.Meseguer M, Herrero J, Tejera A, Hilligsøe KM, Ramsing NB, Remohí J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 97.Zini A. Are sperm chromatin and DNA defects relevant in the clinic? Syst Biol Reprod Med. 2011;57:78–85. doi: 10.3109/19396368.2010.515704. [DOI] [PubMed] [Google Scholar]
- 98.Ruvolo G, Fattouh RR, Bosco L, Brucculeri AM, Cittadini E. New molecular markers for the evaluation of gamete quality. J Assist Reprod Genet. 2013;30:207–212. doi: 10.1007/s10815-013-9943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seli E, Gardner DK, Schoolcraft WB, Moffatt O, Sakkas D. Extent of nuclear DNA damage in ejaculated spermatozoa impacts on blastocyst development after in vitro fertilization. Fertil Steril. 2004;82:378–383. doi: 10.1016/j.fertnstert.2003.12.039. [DOI] [PubMed] [Google Scholar]
- 100.Fatehi AN, Bevers MM, Schoevers E, Roelen BA, Colenbrander B. Gadella BM. DNA damage in bovine sperm does not block fertilization and early embryonic development but induces apoptosis after the first cleavages. J Androl. 2006;27:176–188. doi: 10.2164/jandrol.04152. [DOI] [PubMed] [Google Scholar]
- 101.Knez J, Kovačič B, Vlaisavljević V. Comparison of embryo transfer strategies and assisted reproduction outcome in Slovenian and cross-border patients. Reprod BioMed Online. 2013;27:310–315. doi: 10.1016/j.rbmo.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 102.Van Montfoort AP, Dumoulin JC, Kester AD, Evers JL. Early cleavage is a valuable addition to existing embryo selection parameters: a study using single embryo transfers. Hum Reprod. 2004;19:2103–2108. doi: 10.1093/humrep/deh385. [DOI] [PubMed] [Google Scholar]
- 103.Zheng WW, Song G, Wang QL, Liu SW, Zhu XL, Deng SM, Zhong A, Tan YM, Tan Y. Sperm DNA damage has a negative effect on early embryonic development following in vitro fertilization. Asian J Androl. 2018;20:75. doi: 10.4103/aja.aja_19_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Simon L, Murphy K, Shamsi MB, Liu L, Emery B, Aston KI, Hotaling J, Carrell DT. Paternal influence of sperm DNA integrity on early embryonic development. Hum Reprod. 2014;29:2402–2412. doi: 10.1093/humrep/deu228. [DOI] [PubMed] [Google Scholar]
- 105.Tandara M, Bajić A, Tandara L, Bilić-Zulle L, Šunj M, Kozina V, Goluža T, Jukić M. Sperm DNA integrity testing: big halo is a good predictor of embryo quality and pregnancy after conventional IVF. Andrology. 2014;2:678–686. doi: 10.1111/j.2047-2927.2014.00234.x. [DOI] [PubMed] [Google Scholar]
- 106.Osman A, Alsomait H, Seshadri S, El-Toukhy T, Khalaf Y. The effect of sperm DNA fragmentation on live birth rate after IVF or ICSI: a systematic review and meta-analysis. Reprod BioMed Online. 2015;30:120–127. doi: 10.1016/j.rbmo.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 107.Larson-Cook KL, Brannian JD, Hansen KA, Kasperson KM, Aamold ET, Evenson DP. Relationship between the outcomes of assisted reproductive techniques and sperm DNA fragmentation as measured by the sperm chromatin structure assay. Fertil Steril. 2003;80:895–902. doi: 10.1016/s0015-0282(03)01116-6. [DOI] [PubMed] [Google Scholar]
- 108.Lin MH, Lee RK, Li SH, Lu CH, Sun FJ, Hwu YM. Sperm chromatin structure assay parameters are not related to fertilization rates, embryo quality, and pregnancy rates in in vitro fertilization and intracytoplasmic sperm injection, but might be related to spontaneous abortion rates. Fertil Steril. 2008;90:352–359. doi: 10.1016/j.fertnstert.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 109.Breznik BP, Kovačič B, Vlaisavljević V. Are sperm DNA fragmentation, hyperactivation, and hyaluronan-binding ability predictive for fertilization and embryo development in in vitro fertilization and intracytoplasmic sperm injection? Fertil Steril. 2013;99:1233–1241. doi: 10.1016/j.fertnstert.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 110.Jiang H, He RB, Wang CL, Zhu J. The relationship of sperm DNA fragmentation index with the outcomes of in-vitro fertilisation-embryo transfer and intracytoplasmic sperm injection. J Obstet Gynaecol. 2011;31:636–639. doi: 10.3109/01443615.2011.590910. [DOI] [PubMed] [Google Scholar]
- 111.Rougier N, Uriondo H, Papier S, Checa MA, Sueldo C, Sedó CA. Changes in DNA fragmentation during sperm preparation for intracytoplasmic sperm injection over time. Fertil Steril. 2013;100:69–74. doi: 10.1016/j.fertnstert.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 112.Selvam MK, Agarwal A. A systemic review on sperm DNA fragmentation in male factor infertility: laboratory assessment. Arab J Urol. 2018 Jan;17 [DOI] [PMC free article] [PubMed]
- 113.Morel F, Roux C, Bresson JL. Disomy frequency estimated by multicolour fluorescence in situ hybridization, degree of nuclear maturity and teratozoospermia in human spermatozoa. Reproduction. 2001;121:783–789. doi: 10.1530/rep.0.1210783. [DOI] [PubMed] [Google Scholar]
- 114.Ova’ri L, Sati L, Stronk J, Borsos A, Ward D, Cand Huszar G Double probing individual human spermatozoa: aniline blue staining for persistent histones and fluorescence in situ hybridization for aneuploidies. Fertil Steril. 2010;93:2255–2261. doi: 10.1016/j.fertnstert.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 115.Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, Scott RT. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril. 2014;101:656–663. doi: 10.1016/j.fertnstert.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 116.Fragouli E, Alfarawati S, Spath K, Wells D. Morphological and cytogenetic assessment of cleavage and blastocyst stage embryos. Mol Hum Reprod. 2013;20:117–126. doi: 10.1093/molehr/gat073. [DOI] [PubMed] [Google Scholar]
- 117.Kirkpatrick G, Ferguson KA, Gao H, Tang S, Chow V, Yuen BH, Ma S. A comparison of sperm aneuploidy rates between infertile men with normal and abnormal karyotypes. Hum Reprod. 2008;23:1679–1683. doi: 10.1093/humrep/den126. [DOI] [PubMed] [Google Scholar]
- 118.Mehdi M, Smatti B, Saad A, Guerin JF, Benchaib M. Analysis by fluorescence in situ hybridization [FISH] of the relationship between gonosomic aneuploidy and the results of assisted reproduction in men with severe oligozoospermia. Andrologia. 2006;38:137–141. doi: 10.1111/j.1439-0272.2006.00727.x. [DOI] [PubMed] [Google Scholar]
- 119.Calogero AE, De Palma A, Grazioso C, Barone N, Romeo R, Rappazzo G, D’Agata R. Aneuploidy rate in spermatozoa of selected men with abnormal semen parameters. Hum Reprod. 2001;16:1172–1179. doi: 10.1093/humrep/16.6.1172. [DOI] [PubMed] [Google Scholar]
- 120.Vozdova M, Heracek J, Sobotka V, Rubes J. Testicular sperm aneuploidy in non-obstructive azoospermic patients. Hum Reprod. 2012;27:2233–2239. doi: 10.1093/humrep/des115. [DOI] [PubMed] [Google Scholar]
- 121.O'Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertil Steril. 2010;93:1–2. doi: 10.1016/j.fertnstert.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 122.Chatziparasidou A, Christoforidis N, Samolada G, Nijs M. Sperm aneuploidy in infertile male patients: a systematic review of the literature. Andrologia. 2015;47(1):847–860. doi: 10.1111/and.12362. [DOI] [PubMed] [Google Scholar]
- 123.Sarrate Z, Blanco J, Anton E, Egozcue S, Egozcue J, Vidal F. FISH studies of chromosome abnormalities in germ cells and its relevance in reproductive counseling. Asian J Androl. 2005;7:227–236. doi: 10.1111/j.1745-7262.2005.00061.x. [DOI] [PubMed] [Google Scholar]
- 124.Baarends WM, van der Laan R, Grootegoed JA. DNA repair mechanisms and gametogenesis. Reproduction. 2001;121:31–39. doi: 10.1530/rep.0.1210031. [DOI] [PubMed] [Google Scholar]
- 125.Miyamoto T, Hasuike S, Yogev L, Maduro M, Ishikawa M, Westphal H, Lamb D. Azoospermia in patients heterozygous for a mutation in SYCP3. Lancet. 2003;362:1714–1719. doi: 10.1016/S0140-6736(03)14845-3. [DOI] [PubMed] [Google Scholar]
- 126.Perdrix A, Saidi R, Menard JF, Gruel E, Milazzo JP, Mace B, Rives N. Relationship between conventional sperm parameters and motile sperm organelle morphology examination [MSOME]. Int J Androl. Int J Androl. 2012;35:491–498. doi: 10.1111/j.1365-2605.2012.01249.x. [DOI] [PubMed] [Google Scholar]
- 127.Fragouli E. Next generation sequencing for preimplantation genetic testing for aneuploidy: friend or foe? Fertil Steril. 2018;109:606–607. doi: 10.1016/j.fertnstert.2018.01.028. [DOI] [PubMed] [Google Scholar]
- 128.Nicopoullos JD, Gilling-Smith C, Almeida PA, Homa S, Nice L, Tempest H, Ramsay JW. The role of sperm aneuploidy as a predictor of the success of intracytoplasmic sperm injection? Hum Reprod. 2007;23:240–250. doi: 10.1093/humrep/dem395. [DOI] [PubMed] [Google Scholar]
- 129.Rubio C, Gil-Salom M, Simon C, Vidal F, Rodrigo L, Minguez Y, Remohi J, Pellicer A. Incidence of sperm chromosomal abnormalities in a risk population: relationship with sperm quality and ICSI outcome. Hum Reprod. 2001;16:2084–2092. doi: 10.1093/humrep/16.10.2084. [DOI] [PubMed] [Google Scholar]
- 130.Devroey P. Van Steirteghem a. A review of ten years’ experience of ICSI. Hum Reprod Update. 2004;10:19–28. doi: 10.1093/humupd/dmh004. [DOI] [PubMed] [Google Scholar]
- 131.Ramasamy R, Scovell JM, Kovac JR, Cook PJ, Lamb DJ, Lipshultz LI. Fluorescence in situ hybridization detects increased sperm aneuploidy in men with recurrent pregnancy loss. Fertil Steril. 2015;103:906–909. doi: 10.1016/j.fertnstert.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Esquerré-Lamare C, Walschaerts M, Debordeaux LC, Moreau J, Bretelle F, Isus F, Karsenty G, Monteil L, Perrin J, Papaxanthos-Roche A, Bujan L. Sperm aneuploidy and DNA fragmentation in unexplained recurrent pregnancy loss: a multicenter case-control study. Basic Clin Androl. 2018;28:4. doi: 10.1186/s12610-018-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hassold T, Hunt P. To err [meiotically] is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 134.McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Fraser HB, Petrov DA. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science. 2015;348:235–238. doi: 10.1126/science.aaa3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.García-Ferreyra J, Luna D, Villegas L, Romero R, Zavala P, Hilario R, Dueñas-Chacón J. High aneuploidy rates observed in embryos derived from donated oocytes are related to male aging and high percentages of sperm DNA fragmentation. Clin Med Insights Reprod Health 2015;9: CMRH-S32769. [DOI] [PMC free article] [PubMed]
- 136.García-Ferreyra J, Hilario R, Dueñas J. High percentages of embryos with 21, 18 or 13 trisomy are related to advanced paternal age in donor egg cycles. JBRA Assist Reprod. 2018;22:26. doi: 10.5935/1518-0557.20180004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.El-Domyati MM, Al-Din AB, Barakat MT, El-Fakahany HM, Xu J, Sakkas D. Deoxyribonucleic acid repair and apoptosis in testicular germ cells of aging fertile men: the role of the poly [adenosine diphosphate-ribosyl] ation pathway. Fertil Steril. 2009;1(91):2221–2229. doi: 10.1016/j.fertnstert.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 138.Escudero T, Abdelhadi I, Sandalinas M, Munne S. Predictive value of sperm fluorescence in situ hybridization analysis on the outcome of preimplantation genetic diagnosis for translocations. Fertil Steril. 2003;79:1528–1534. doi: 10.1016/s0015-0282(03)00252-8. [DOI] [PubMed] [Google Scholar]
- 139.Daughtry BL, Rosenkrantz JL, Lazar NH, Fei SS, Redmayne N, Torkenczy KA, Adey A, Gao L, Park B, Nevonen KA, Carbone L. Chromosome Removal Via Cellular Fragmentation and Aneuploid Blastomere Exclusion in Primate Embryos. bioRxiv. 2018 :241851.
- 140.Manuelidis L. A view of interphase chromosomes. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- 141.Sandalinas M, Sadowy S, Alikani M, Calderon G, Cohen J, Munné S. Developmental ability of chromosomally abnormal human embryos to develop to the blastocyst stage. Hum Reprod. 2001;16:1954–1958. doi: 10.1093/humrep/16.9.1954. [DOI] [PubMed] [Google Scholar]
- 142.Bolton H, Graham SJ, Van der Aa N, Kumar P, Theunis K, Gallardo EF, Voet T, Zernicka-Goetz M. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun. 2016;7:11165. doi: 10.1038/ncomms11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zheng X, Liu P, Chen G, Qiao J, Wu Y, Fan M. Viability of frozen-thawed human embryos with one–two blastomeres lysis. J Assist Reprod Genet. 2008;25:281–285. doi: 10.1007/s10815-008-9224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Quintana-Murci L, Krausz C, McElreavey K. The human Y chromosome: function, evolution and disease. Forensic Sci Int. 2001;118:169–181. doi: 10.1016/s0379-0738(01)00387-5. [DOI] [PubMed] [Google Scholar]
- 145.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 146.Colaco S, Modi D. Genetics of the human Y chromosome and its association with male infertility. Reprod Biol Endo. 2018;16:14. doi: 10.1186/s12958-018-0330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Longepied G, Saut N, Aknin-Seifer I, Levy R, Frances AM, Metzler-Guillemain C, Guichaoua MR, Mitchell MJ. Complete deletion of the AZFb interval from the Y chromosome in an oligozoospermic man. Hum Reprod. 2010;25:2655–2663. doi: 10.1093/humrep/deq209. [DOI] [PubMed] [Google Scholar]
- 148.Navarro-Costa P, Pereira L, Alves C, Gusmão L, Proença C, Marques-Vidal P, Rocha T, Correia SC, Jorge S, Neves A, Soares AP. Characterizing partial AZFc deletions of the Y chromosome with amplicon-specific sequence markers. BMC Genomics. 2007;8:342. doi: 10.1186/1471-2164-8-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Page DC, Silber S, Brown LG. Men with infertility caused by AZFc deletion can produce sons by intracytoplasmic sperm injection, but are likely to transmit the deletion and infertility. Hum Reprod. 1999;14:1722–1726. doi: 10.1093/humrep/14.7.1722. [DOI] [PubMed] [Google Scholar]
- 150.Choi YM, Yoon JS, Hwang KR, Kim SH, Lee WD, Moon SY. Follicle-stimulating hormone receptor polymorphism and ovarian responses to controlled ovarian hyperstimulation for IVF-ET. Fertil Steril. 2004;82:S206–S207. doi: 10.1007/s10038-006-0005-5. [DOI] [PubMed] [Google Scholar]
- 151.Mulhall JP, Reijo R, Alagappan R, Brown L, Page D, Carson R, Oates RD. Azoospermic men with deletion of the DAZ gene cluster are capable of completing spermatogenesis: fertilization, normal embryonic development and pregnancy occur when retrieved testicular spermatozoa are used for intracytoplasmic sperm injection. Hum Reprod. 1997;12:503–508. doi: 10.1093/humrep/12.3.503. [DOI] [PubMed] [Google Scholar]
- 152.van Golde RJ, Wetzels AM, de Graaf R, Tuerlings JH, Braat DD, Kremer JA. Decreased fertilization rate and embryo quality after ICSI in oligozoospermic men with microdeletions in the azoospermia factor c region of the Y chromosome. Hum Reprod. 2001;16:289–292. doi: 10.1093/humrep/16.2.289. [DOI] [PubMed] [Google Scholar]
- 153.Liu XH, Qiao J, Li R, Yan LY, Chen LX. Y chromosome AZFc microdeletion may not affect the outcomes of ICSI for infertile males with fresh ejaculated sperm. J Assist Reprod Genet. 2013;30:813–819. doi: 10.1007/s10815-013-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mateu E, Rodrigo L, Martínez MC, Peinado V, Milán M, Gil-Salom M, Martínez-Jabaloyas JM, Remohí J, Pellicer A, Rubio C. Aneuploidies in embryos and spermatozoa from patients with Y chromosome microdeletions. Fertil Steril. 2010;94:2874–2877. doi: 10.1016/j.fertnstert.2010.06.046. [DOI] [PubMed] [Google Scholar]
- 155.Sen S, Ambulkar P, Hinduja I, Zaveri K, Gokral J, Pal A, Modi D. Susceptibility of gr/gr rearrangements to azoospermia or oligozoospermia is dependent on DAZ and CDY1 gene copy deletions. J Assist Reprod Genet. 2015;32:1333–1341. doi: 10.1007/s10815-015-0520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Beyaz CC, Gunes S, Onem K, Kulac T, Asci R. Partial deletions of Y-chromosome in infertile men with non-obstructive azoospermia and Oligoasthenoteratozoospermia in a Turkish population. in vivo. 2017;31:365–371. doi: 10.21873/invivo.11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fernández-Gonzalez R, Moreira P, Bilbao A, Jiménez A, Pérez-Crespo M, Ramírez MA, De Fonseca FR, Pintado B, Gutiérrez-Adán A. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc Natl Acad Sci U S A. 2004;101:5880–5885. doi: 10.1073/pnas.0308560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hansen M, Bower C, Milne E, de Klerk N, Kurinczuk J. J. Assisted reproductive technologies and the risk of birth defects—a systematic review. Hum Reprod. 2005;20:328–338. doi: 10.1093/humrep/deh593. [DOI] [PubMed] [Google Scholar]
- 159.Beck-Fruchter R, Shalev E, Weiss A. Clinical benefit using sperm hyaluronic acid binding technique in ICSI cycles: a systematic review and meta-analysis. Reprod BioMed Online. 2016;32:286–298. doi: 10.1016/j.rbmo.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 160.Heytens E, Parrington J, Coward K, Young C, Lambrecht ST, Yoon SY, Fissore RA, Hamer R, Deane CM, Ruas M, Grasa P. Reduced amounts and abnormal forms of phospholipase C zeta [PLCζ] in spermatozoa from infertile men. Hum Reprod. 2009;24:2417–2428. doi: 10.1093/humrep/dep207. [DOI] [PubMed] [Google Scholar]
- 161.Kashir J, Konstantinidis M, Jones C, Lemmon B, Chang Lee H, Hamer R, Heindryckx B, Deane CM, De Sutter P, Fissore RA, Parrington J. A maternally inherited autosomal point mutation in human phospholipase C zeta [PLCζ] leads to male infertility. Hum Reprod. 2011;27:222–231. doi: 10.1093/humrep/der384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schatten H, Sun QY. The functional significance of centrosomes in mammalian meiosis, fertilization, development, nuclear transfer, and stem cell differentiation. Environ Mol Mutagen. 2009;50:620–636. doi: 10.1002/em.20493. [DOI] [PubMed] [Google Scholar]
- 163.Cappallo-Obermann H, Schulze W, Jastrow H, Baukloh V, Spiess AN. Highly purified spermatozoal RNA obtained by a novel method indicates an unusual 28S/18S rRNA ratio and suggests impaired ribosome assembly. Mol Hum Reprod. 2011;17:669–678. doi: 10.1093/molehr/gar037. [DOI] [PubMed] [Google Scholar]
- 164.Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA. The presence, role and clinical use of spermatozoal RNAs. Reproductive Medicine Network Hum Reprod Update. 2013;19:604–624. doi: 10.1093/humupd/dmt031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yuan S, Schuster A, Tang C, Yu T, Ortogero N, Bao J, Zheng H, Yan W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development. 2016;143:635–647. doi: 10.1242/dev.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Balmori C, Varela E. Should we consider telomere length and telomerase activity in male factor infertility? Curr Opin Obstet Gynecol. 2018;30:197–202. doi: 10.1097/GCO.0000000000000451. [DOI] [PubMed] [Google Scholar]
- 167.Sedó CA, Bilinski M, Lorenzi D, Uriondo H, Noblia F, Longobucco V, Lagar VE, Nodar F. Effect of sperm DNA fragmentation on embryo development: clinical and biological aspects. JBRA Assisted Reproduction. 2017;21(4):343–350. doi: 10.5935/1518-0557.20170061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Haghpanah T, Salehi M, Ghaffari Novin M, Masteri Farahani R, Fadaei-Fathabadi F, Dehghani-Mohammadabadi M, Azimi H. Does sperm DNA fragmentation affect the developmental potential and the incidence of apoptosis following blastomere biopsy? Syst Biol Reprod Med. 2016;62:1–10. doi: 10.3109/19396368.2015.1103324. [DOI] [PubMed] [Google Scholar]
- 169.Duarte C, Núñez V, Wong Y, Vivar C, Benites E, Rodriguez U, Vergara C, Ponce J. Impact of the Z potential technique on reducing the sperm DNA fragmentation index, fertilization rate and embryo development. JBRA assisted reproduction. 2017;21:351. doi: 10.5935/1518-0557.20170055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xue LT, Wang RX, He B, Mo WY, Huang L, Wang SK, Mao XB, Cheng JP, Huang YY, Liu R. Effect of sperm DNA fragmentation on clinical outcomes for Chinese couples undergoing in vitro fertilization or intracytoplasmic sperm injection. J Int Med Res. 2016;44:1283–1291. doi: 10.1177/0300060516664240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gat I, Tang K, Quach K, Kuznyetsov V, Antes R, Filice M, Zohni K, Librach C. Sperm DNA fragmentation index does not correlate with blastocyst aneuploidy or morphological grading. PLoS One. 2017;12:e0179002. doi: 10.1371/journal.pone.0179002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Niu ZH, Shi HJ, Zhang HQ, Zhang AJ, Sun YJ, Feng Y. Sperm chromatin structure assay results after swim-up are related only to embryo quality but not to fertilization and pregnancy rates following IVF. Asian J Androl. 2011;13:862. doi: 10.1038/aja.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kihaile PE, Yasui A, Shuto Y. Prospective assessment of Y-chromosome microdeletions and reproductive outcomes among infertile couples of Japanese and African origin. J Exp Clin Assist Reprod. 2005;2:9. doi: 10.1186/1743-1050-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Peterlin B, Kunej T, Sinkovec J, Gligorievska N, Zorn B. Screening for Y chromosome microdeletions in 226 Slovenian subfertile men. Hum Reprod. 2002;17:17–24. doi: 10.1093/humrep/17.1.17. [DOI] [PubMed] [Google Scholar]
- 175.Tanaka A, Nagayoshi M, Tanaka I, Ikuma S, Miki T, Yamaguchi T, Kusunoki H, Watanabe S. Clinical outcome of treatments for azoospermia. Fertil Steril. 2015;104:e237–e238. [Google Scholar]
- 176.Sasamine K, Mizuta S, Nishiyama R, Yamaguchi K, Kitaya K, Matsubayashi H, Ishikawa T. Fertilization and embryonic development of azoospermia with AZFc microdeletion. Fertil Steril. 2015;104:e238. [Google Scholar]
- 177.Choi DK, Gong IH, Hwang JH, Oh JJ, Hong JY. Detection of Y chromosome microdeletion is valuable in the treatment of patients with nonobstructive azoospermia and oligoasthenoteratozoospermia: sperm retrieval rate and birth rate. Korean J Urol. 2013;54:111–116. doi: 10.4111/kju.2013.54.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhu Y, Wu T, Li G, Yin B, Liu H, Wan C, Zhang H, Zeng Y. The sperm quality and clinical outcomes were not affected by sY152 deletion in Y chromosome for oligozoospermia or azoospermia men after ICSI treatment. Gene. 2015;573:233–238. doi: 10.1016/j.gene.2015.07.051. [DOI] [PubMed] [Google Scholar]
- 179.Liu XY, Wang RX, Fu Y, Luo LL, Guo W, Liu RZ. Outcomes of intracytoplasmic sperm injection in oligozoospermic men with Y chromosome AZF b or AZF c microdeletions. Andrologia. 2017;49:e12602. doi: 10.1111/and.12602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 35 kb)


