Abstract
With the advent of next-generation sequencing (NGS), targeted sequencing is now contributing to decision making for which chemotherapeutics to administer to cancer patients, especially in refractory and metastatic cancer. Given that most patients with refractory cancer develop resistance to chemotherapy and have few treatment options, we performed NGS test to evaluate the efficacy and clinical feasibility of NGS-based targeted anticancer therapy. We used a gene panel for capturing target regions covering 83 cancer-related genes. A total of 25 refractory metastatic solid tumor patients were enrolled in this study. Among the 25 patients, 7 had FDA-approved drug-responsive or -resistant alterations. However, the effectiveness of targeted therapy was assessed by follow-up in three patients (12%). These included crizotinib for ALK-EML4 fusion in a malignancy of undefined origin patient and everolimus for AKT3 amplification in a uterine sarcoma patient. In addition, we identified a KRAS codon 146 mutation (A146V), which is associated with resistance to anti-EGFR, in a cetuximab-resistant colon cancer patient with wild-type KRAS exons 2 and 12, and EGFR amplification. He received bevacizumab therapy. All three patients showed partial response after targeted therapy. Furthermore, we characterized KRAS A146V biologically using colon cancer cells. In conclusion, this study suggests that targeted therapy based on NGS test may be a good choice for improving the care of patients with refractory solid tumors.
Introduction
Cancer is one of the leading causes of death worldwide. According to cancer statistics from the National Cancer Institute, in 2012, there were 14 million new cases and 8.2 million cancer-related deaths worldwide, and the number of cancer cases is projected to increase globally by 50% in 2030 [1]. Despite advances in chemotherapeutic treatments, most patients with refractory cancer develop resistance to these therapies and have few treatment options.
Next-generation sequencing (NGS) technologies have led to a comprehensive understanding of cancer genomes and are increasingly being used for both clinical and research applications. NGS testing of clinically relevant cancer-related genes has enabled cancer patients to access targeted therapies and predict response to treatments. Many research groups have established NGS testing for treatment decision making, and currently active clinical trials, such as umbrella and basket trials, have shown that the use of NGS testing results in better outcomes for patients than those in patients not using it [2], [3], [4].
Therefore, this study was undertaken to assess whether NGS testing is reliable and useful for decision making in the clinic for patients with refractory solid tumors in our institute. In this pilot study, 25 patients were enrolled, and their tissues were sequenced using a panel of 83 cancer-related genes.
Methods
Patients
Patients with refractory solid tumors were enrolled in this study. The study was approved by the institutional review board of Pusan National University Hospital. All study participants provided written informed consent before being enrolled in the study. The criteria for participation included the following: patients with pathologically confirmed cancer; patients with recurrent/metastatic solid tumors who did not respond to standard therapy; patients ≥18 years old; Eastern Cooperative Oncology Group performance status of 0, 1, or 2; presence of enough tumor tissue for targeted sequencing; measurable disease in accordance with the Response Evaluation Criteria version 1.1; life expectancy ≥3 months; adequate bone marrow function (neutrophils ≥1.5 × 109/l, platelets ≥100 × 109/l; Hb >10 g/dl); adequate liver function [aspartate aminotransferase/alanine aminotransferase ≤5× upper limit of normal (ULN), bilirubin ≤2× ULN, albumin >25 g/l]; and adequate renal function (creatinine ≤2× ULN). The exclusion criteria were alcohol or substance abuse; pregnant or breast-feeding women; patients with cardiac, renal, or hepatic dysfunction; patients with infectious, neurological, or psychiatric disorders that may affect the study's results; and patients with uncontrollable elevation of intracranial pressure.
Targeted Sequencing
A targeting panel was used to capture the target regions of 83 cancer-related genes for the detection of single nucleotide variants (SNVs), insertion/deletions (INDELs), and copy number variations (CNVs), including all coding exons of the following 72 genes: ABL1, AKT1, AKT2, AKT3, ALK, APC, ARID1A, ARID1B, ARID2, ATM, AURKA, AURKB, BCL2, BRAF, BRCA1, BRCA2, CDH1, CDK4, CDK6, CDKN2A, CSF1R, CTNNB1, DDR2, EGFR, EPHB4, ERBB2, ERBB3, ERBB4, EWSR1, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAS, GNAQ, HNF1A, HRAS, IDH1, IDH2, IGF1R, ITK, JAK1, JAK2, JAK3, KDR, KIT, KRAS, MDM2, MET, MLH1, MPL, MTOR, NF1, NOTCH1, NPM1, NRAS, NTRK1, PDGFRA, PDGFRB, PIK3CA, PIK3R1, PTCH1, PTCH2, PTEN, PTPN11, RB1, RET, ROS1, SMAD4, SMARCB1, SMO, SRC, STK11, SYK, TERT, TMPRSS2, TOP1, TP53, and VHL. Additionally, some introns of the following five genes were included for the detection of gene fusions: ALK, RET, ROS1, EWSR1, and TMPRSS2 (SureSelect, Agilent, Inc., USA). Two hundred to 500 ng of genomic DNA extracted from FFPE of cancer patients was prepared to construct libraries using the SureSelect targeting panel according to the manufacturer's protocol. Briefly, genomic DNA samples were randomly fragmented by Covaris (Covaris, Inc., USA), followed by adapter ligation, purification, hybridization, and PCR. Captured libraries were analyzed in the Agilent 2100 Bioanalyzer to estimate the quality of nucleotides and were loaded onto the Illumina HiSeq2500 instrument (TheragenEtex Bio Institute, Suwon, Korea) according to the manufacturer's recommendations. Raw image files were processed in the HCS1.4.8 software for base-calling using default parameters, and the sequences of each individual were generated as 101-bp paired-end reads.
Analytical Methods
For NGS data preprocessing, sequence reads were aligned to the human genome (hg19) using BWA-MEM [5]. To generate the analysis-ready Binary Alignment Map (BAM), the overall preprocessing steps, including removal of duplicates, local realignment, and recalibration, were performed using GATK Best Practice (Broad Institute) [6]. For variant discovery (SNVs and INDELs), we used three open-source callers (UnifiedGenotyper [7], LoFreq [8], and SNVer [9]) and Samsung SDS's in-house callers. CNVs and translocations were discovered using in-house callers developed by Samsung SDS. SNVs and INDELs were filtered using germ-line mutations and false-positive filters. SNVs with variant allele frequency ≥5% and INDELs ≥10% were selected for this study. CNVs were analyzed using the depth of coverage for each target region between tumor and preprocessed normal data. To calculate absolute copy number, tumor purity and ploidy were estimated using a statistical model consisting of log2 ratio values and SNV variant allele frequency values. As a cutoff value, copy number (CN) ≥7 and CN = 0 were used for amplification and homozygous deletion, respectively. For detection of translocations, a paired-end mapping analysis and a split-alignment analysis were performed. All discordant read-pairs with abnormal insert size or orientation were screened, and soft-clipping information of the split-reads was investigated as evidence of genomic rearrangements. The confidence cutoff value for translocations was a split-read support count ≥3.
Site-Directed Mutagenesis and Transfection
A KRAS A146 mutation was engineered into the pLenti-C-mGFP-P2A-puro vector using the QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, CA, USA) according to the manufacturer's instructions. The constructs were verified by Sanger sequencing. Lentivirus stocks were produced using the Virapower lentiviral packaging mix and the 293FT cell line according to the manufacturer's protocol (Invitrogen, CA, USA). HT29 cells were grown to 50% confluence and incubated for 24 hours in a 1:1 dilution of virus:media with 5 μg/ml Polybrene. After a 24-hour recovery period in complete media without virus, polyclonal stable cell lines were selected and maintained in media containing 5 μg/ml puromycin. Cells expressing green fluorescent protein were observed under the microscope, and mutations were verified by Sanger sequencing.
Measurement of Cell Viability
Cell viability was evaluated using an MTT assay. After washing the cells, culture medium containing 0.5 mg/ml MTT was added to each well. The cells were incubated for 2 hours at 37°C, the supernatant was removed, and the formazan crystals formed in viable cells were solubilized with 0.11 ml of dimethylsulfoxide. A 0.1-ml aliquot of each sample was then transferred to 96-well plates, and the absorbance of each well was measured at 570 nm using a spectrophotometer (Hewlett-Packard, Agilent Technologies, USA). Data are expressed as a percentage of control measured in the absence of paclitaxel.
Western Blotting
Cells were harvested at various time points after paclitaxel treatment and disrupted in lysis buffer (1% Triton X-100, 1 mM EGTA, 1 mM EDTA, 10 mM Tris–HCl, pH 7.4, and protease inhibitors). Cell debris was removed by centrifugation at 10,000×g for 10 minutes at 4°C. The resulting supernatant proteins were separated using SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with 5% nonfat dried milk at room temperature for 30 minutes and incubated with anti-MEK, anti-ERK, anti-phosphorylated MEK, anti-phosphorylated ERK (Cell Signaling Technology, MA, USA), and anti-GAPDH. The membranes were then washed and incubated with horseradish peroxidase–conjugated secondary antibody. Signals were visualized using enhanced chemiluminescence (Amersham; Buckinghamshire, UK).
Results
Patient Characteristics
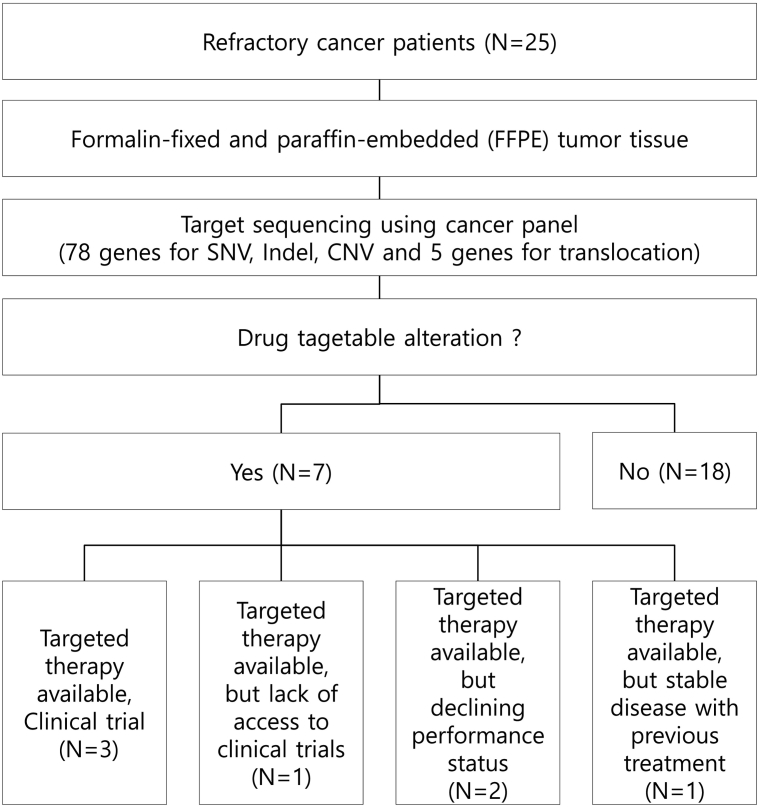
For this study, we enrolled 25 refractory metastatic solid tumor patients treated with conventional treatments (Figure 1). FFPE samples from those patients were used for targeted sequencing and analyzed. Their baseline characteristics are described in Table 1. The most common tumor types were uterine sarcoma (20%, n = 5) and breast carcinoma (16%, n = 4), followed by malignancy of undefined origin (MUO) (12%, n = 3), renal cell carcinoma (8%, n = 2), and neuroendocrine tumor (8%, n = 2). The median age at diagnosis was 51.9 years (range 22-72). Eighteen patients had metastatic disease at diagnosis.
Figure 1.
Schematic representation of the study design. The process started with targeted sequencing using FFPE samples from 25 refractory cancer patients. All patients underwent genomic sequencing. The number of patients who actually received matched therapy according to genomic alterations was three.
Table 1.
Characteristics of the 25 Study Patients
| Characteristics | Value |
|---|---|
| Age (year) | |
| Median | 51.9 |
| Range | 22–72 |
| Sex | |
| Male | 6 |
| Female | 19 |
| ECOG performance status score | |
| 0 | 13 |
| 1 | 8 |
| 2 | 3 |
| 3 | 1 |
| Metastatic disease at initial diagnosis | 18 |
| Received prior treatments (range, 1-6) | |
| 1 | 12 |
| 2 | 6 |
| 3 | 3 |
| 4 | 2 |
| 5 | 1 |
| 6 | 1 |
| Diagnosis | |
| Uterine sarcoma | 5 |
| Breast carcinoma | 4 |
| Malignancy or undefined origin | 3 |
| Renal cell carcinoma | 2 |
| Neuroendocrine tumor | 2 |
| Cholangiocarcinoma | 1 |
| Colon carcinoma | 1 |
| Tongue carcinoma | 1 |
| Leiomyosarcoma | 1 |
| Liposarcoma | 1 |
| Malignant peripheral nerve sheath tumor | 1 |
| Pleomorphic myogenic sarcoma | 1 |
| Cervical cancer | 1 |
| Uterus neuroendocrine carcinoma | 1 |
ECOG, Eastern Cooperation Oncology Group.
Sequencing Results
All patients underwent genomic sequencing owing to the high DNA concentrations and tumor cellularity of samples. The median time from FFPE to completion and final analysis of NGS patient data was approximately 3-4 weeks. Mean coverage was 1840.98×, with 98.23% over 100× (Supplemental Table 1). Six out of 25 cases had CNV in targetable genes. The most frequently detected amplifications were in PDGFRB (Table 2). A translocation was detected in only one case, which was the ALK-EML fusion gene.
Table 2.
List of Drug-Targetable Alterations in Seven Patients
| ID | Cancer Type | Gene | Mutation Type | Targeted Drug |
|---|---|---|---|---|
| Patient 1 | MUO | ALK-EML | Fusion | Crizotinib |
| Patient 4 | Uterine sarcoma | PDGFRB | CNV, amplification | Sorafenib |
| Patient 10 | Leiomyosarcoma | PDGFRB | CNV, amplification | Sorafenib |
| Patient 11 | Liposarcoma | AKT2 | CNV, amplification | Everolimus |
| Patient 14 | Uterine sarcoma | AKT3 | CNV, amplification | Everolimus |
| Patient 21 | Breast cancer | AKT1 | CNV, amplification | Everolimus |
| Patient 22 | Colon cancer |
EGFR KRAS |
CNV, amplification SNV |
Bevacizumab |
Molecularly Targeted Therapies
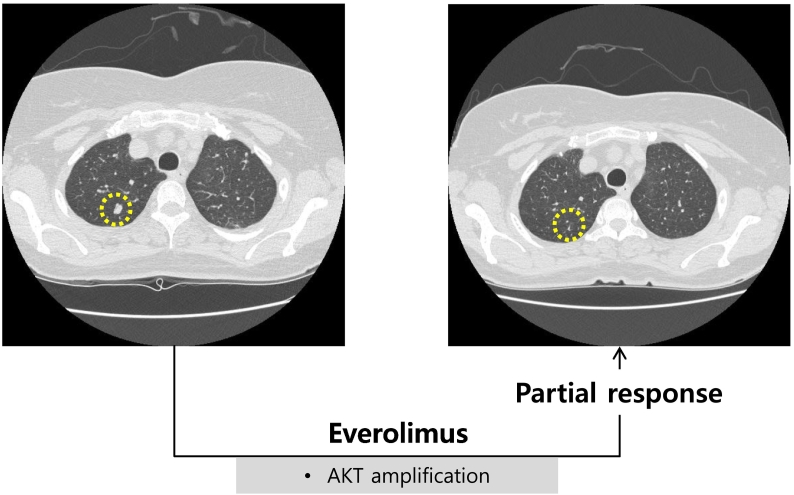
Among the 25 patients, seven patients had at least one molecular alteration matching one of the available targeted agents. However, targeted therapy was guided in only three patients (12%) on the basis of sequencing results because of lack of access to clinical trials (n = 1), declining clinical state and performance status (n = 2), and/or stable disease with previous treatment (n = 1) (Figure 1). Patient 1 with MUO had ALK-EML4 fusion and achieved partial response to crizotinib (Table 3). Before crizotinib therapy, the patient received several individual regimens of chemotherapy including 5-fluorouracil/cisplatin and paclitaxel/cisplatin but finally progressed to all regimens. Patient 14 with uterine sarcoma had amplifications of AKT3, BRAF, and EGFR (Table 3). He had multiple lung metastases. Adriamycin was initiated as a palliative chemotherapy. Subsequently, ifosfamide, gemcitabine, and docetaxel were administered as palliative therapy. Despite continual therapy, pulmonary metastasis progressed. On the basis of NGS test results, this patient was treated with everolimus for AKT3 amplification. As shown in Figure 2, patient 14 showed clinical improvement with partial responses to everolimus. CT scans of patient 14 before and after everolimus-based therapy showed a decreased tumor size. Treatment was discontinued at 5 months after disease progression was confirmed.
Table 3.
Outcomes of Targeted Therapy
| Patient ID | Tumor Type | Targeted Mutation | Other Mutation | Drug | Best Response |
|---|---|---|---|---|---|
| Patient 1 | MUO | ALK-EML4 fusion | Crizotinib | PR | |
| Patient 14 | Uterine sarcoma | AKT3 amplification | BRAF, EGFR amplification | Everolimus | PR |
PR, partial response.
Figure 2.
CT scans from patient 14, a 62-year-old woman with uterine sarcoma treated with everolimus. The pretreatment CT image (left panel) shows multiple lesions. The follow-up CT image (right panel) was obtained after 3 months of everolimus treatment and shows decreased lesion size in multiple lesions.
Mechanisms of Resistance to Targeted Therapies
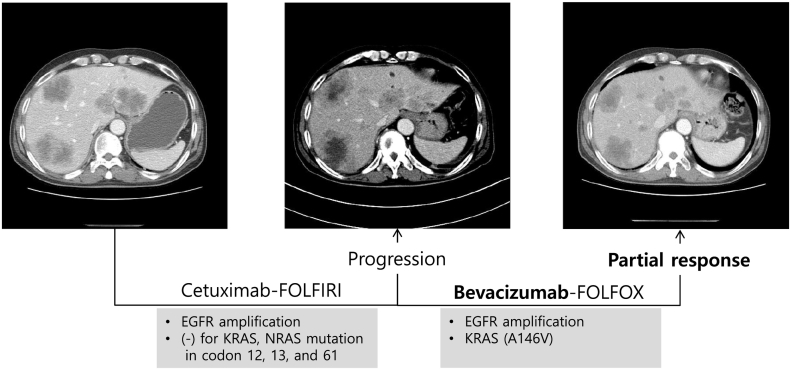
We also detected mutations associated with drug resistance. Patient 22 had wild-type KRAS codons 12 and 13 and EGFR amplification (Table 4). He had been treated with a combination of cetuximab and folinic acid, fluorouracil, and irinotecan (FOLFIRI), based on the results of previous molecular diagnostic tests, but showed resistance to EGFR-targeted therapy. Targeted sequencing of the tumor from this patient revealed a KRAS A146V mutation, which has been shown to confer resistance to anti-EGFR therapy. Based on this result, he was treated with combined bevacizumab and folinic acid, fluorouracil, and oxaliplatin (FOLFOX) and had a partial response. His CT scan images showed that the lesions of liver metastases that were increased after treatment with cetuximab and FOLFIRI significantly reduced in size after bevacizumab and FOLFOX therapy (Figure 3).
Table 4.
Mechanism of Resistance to Targeted Therapy
| Patient ID | Tumor Type | Test | Targeted Mutation | Other Mutation | Drug | Best Response |
|---|---|---|---|---|---|---|
| Patient 22 | Colon cancer | Molecular Diagnostic test | EGFR amplification | Cetuximab | PD | |
| Targeted Sequencing (NGS) |
EGFR amplification |
KRAS (A146V) |
Bevacizumab | PR |
PD, progressive disease; PR, partial response.
Figure 3.
CT scans from patient 22, a 55-year-old man with liver metastatic colon cancer. The CT image (middle panel) after therapy with cetuximab and FOLFIRI shows increased prevalence of liver metastases compared to that in the pretreatment CT image (left panel). The follow-up CT image (right panel) obtained after therapy with bevacizumab and FOLFOX shows attenuation of liver metastases.
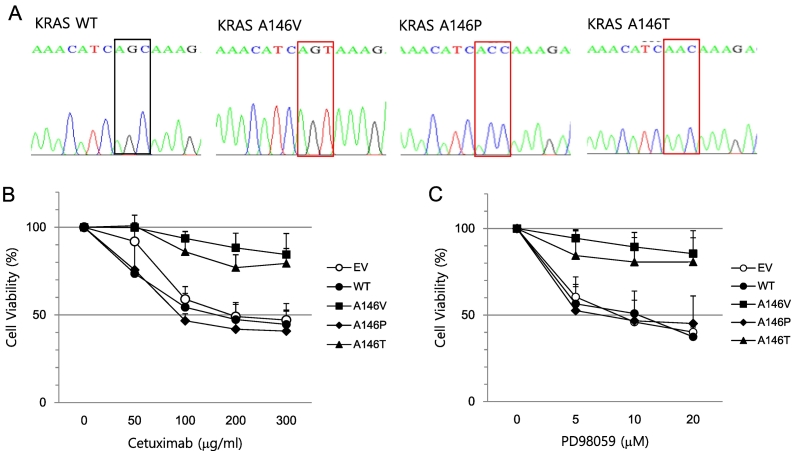
Next, to investigate the role of the KRAS A146V mutation in the resistance of colon cancer cells to anti-EGFR therapy, we used HT29 cells that did not harbor any activating KRAS mutations. HT29 cells were transfected with a lentiviral vector expressing the KRAS A146V mutation, as well as other well-known KRAS 146 mutations (A146T or A146P); the mutations were confirmed by Sanger sequencing (Figure 4A). Cell viability was higher in HT29 cells with KRAS A146V and A146T mutations after cetuximab treatment than in cells expressing wild-type KRAS and in control cells (Figure 4B). Moreover, to study the response of KRAS-mutant cells to MEK inhibition, we examined cell viability after treatment of KRAS-mutant cells with PD98059, an inhibitor of MEK1/2. Figure 4C shows that KRAS-mutant cells (A146V or T) were insensitive to PD98059. Together, these results suggest that colon cancer cells with the KRAS A146V mutation are resistant to cetuximab and MEK inhibition.
Figure 4.
Resistance to EGFR or MEK-targeted drugs in KRAS-mutant HT-29 cells. (A) Vectors expressing KRAS A146V, P, T, or wild type were introduced into HT-29 cells through lentiviral infection. Mutations were confirmed by Sanger sequencing. HT-29 cells expressing KRAS wild type, A146V, A146P, or A146T were treated with the indicated concentration of cetuximab (B) or PD98059 (C) for 24 hours. An MTT assay was performed to measure cell viability. Experiments were repeated three times, each with three replicates. Error bars indicate SEM.
Discussion
The use of NGS testing for clinical application has been spreading rapidly worldwide. The present pilot study was conducted to explore the feasibility of employing NGS-guided therapy in patients with refractory solid tumors. In this study, seven of 25 (28%) patients had molecular alterations matching one of the available targeted drugs. Of these, only three patients (12%) were enrolled in the genome-based clinical trial. In another pilot study in Korea, only a small number of patients (15%, 5/32) received NGS-based targeted therapy [10]. Although most patients were expected to have at least one drug-targetable mutation in their tumors, limiting factors included a lack of patient participation in clinical trials, poor performance status, and stable disease following previous treatment [10]. Access to trials and use of off-label drugs depend on the financial status of patients. To promote genome-based clinical trials, issues such as the lack of access to clinical-grade NGS testing and the limited number of targeted drugs must be resolved [10]. Most importantly, cancer patients need to be referred for possible clinical trial enrollment before their performance status deteriorates [11]. Thus, physicians must be encouraged to check patient availability for participation in trials. In addition, earlier NGS testing is needed so that more patients can receive targeted therapy.
It is reported that the main reason for failure of NGS testing is low cellularity and low DNA content in samples [12], [13]. We extracted DNA from a tumor region marked with a circle on the FFPE slide by the pathologist to enrich tumor cells. All patients enrolled in this study underwent successful NGS testing owing to sufficient amounts of DNA. Moreover, a high read depth is required for detection of variants of low frequency. In this study, the average coverage was 1840.98X, with 98.23% of the targeted base covered at over 100×. However, the turnaround time for testing took 3 to 4 weeks in this study. Faster turnaround times may help to better treat cancer patients.
Although only three patients received targeted therapy, the present study showed that NGS-based targeted therapy benefited the patients with refractory cancer. Partial response was achieved in all three patients receiving targeted therapy. The MUO patient with ALK-EML4 fusion received ALK inhibitor crizotinib treatment; however, this patient was not followed up because he transferred to another hospital. The uterine sarcoma patient with amplification of AKT3 was treated with everolimus, which targets mTOR downstream of PI3KCA and AKT. In addition, the colon cancer patient with wild-type KRAS codons 12 and 13 and EGFR amplification had been receiving a combination therapy of cetuximab and FOLFIRI prior to NGS testing. After the cancer relapsed, NGS testing was conducted using his tumor tissue, and the KRAS A146V mutation (which is known to cause resistance to cetuximab) was found. He then received a combination therapy of bevacizumab and FOLFOX, and showed partial response. These results are supported by clinical data suggesting that the use of bevacizumab (an anti-VEGF drug) after cetuximab-based treatment is effective [14]. Moreover, they are also supported by a previous preclinical study suggesting that anti-EGFR therapy–resistant colorectal cancer (CRC) cells upregulate VEGF levels and respond to antiangiogenic drugs [15].
KRAS mutations are found in about 30% to 50% of CRC patients, and the most frequent mutations are detected in approximately 40% of CRC patients in codons 12 and 13 [16], [17], [18]. Those are known as predictors of resistance to anti-EGFR therapy; therefore, KRAS mutation testing is necessary prior to administration of anti-EGFR therapy in CRC patients. Currently, KRAS codon 61 and 146 mutations have a frequency of 1%-4% and show resistance to anti-EGFR therapy in wild-type KRAS codon 12 and 13 patients [19]. In the case of patient 22, the KRAS codon 146 mutation, which was not included in prior KRAS mutation tests, was detected by NGS testing. The results highlighted the limitation of KRAS hotspot mutation testing and suggested that NGS testing is sufficient and necessary for detecting multiple mutations and for refractory cancer patients with low-frequency mutations.
KRAS 146 codon mutations, such as c.436G>A p.A146T, c.436G>C p.A146P, and c.437C>T p.A146V, were found in CRC [19]. In the present study, the A146V KRAS mutation was identified in the CRC patient with wild-type codons 12 and 13. Although clinical data have shown that those KRAS 146 mutations are associated with resistance to EGFR-targeted therapy (cetuximab) in CRC patients [20], [21], the molecular mechanism underlying the association of therapy resistance and those mutations is not well known. Janakiramam et al. reported that the KRAS A146T mutation increased RAS activity, and KRAS A146T-expressing xenografts were resistant to EGFR-targeted inhibition and sensitive to MEK inhibition [22]. However, the efficacy of MEK inhibitors in colon cancer patients with KRAS mutations has been modest [23], [24]. Consistent with this, MEK inhibition was ineffective in KRAS-mutant cells, even though our in vitro studies showing cell death inhibition in KRAS-mutant cells treated with cetuximab support that the KRAS A146V mutation is a predictor of resistance to EGFR-targeted inhibitors. These results demonstrate that MEK is not the key downstream pathway in those cells. In addition, our study showed that KRAS A146P mutant cells were sensitive to EGFR or MEK inhibitors, in contrast to cells with KRAS A146T/V. The characteristics of KRAS 146 mutations need to be elucidated further for the development of additional treatment options.
In the present study, we showed that NGS testing using a cancer gene panel allowed us to match patients to targeted therapies and provided shorter timelines for clinical trials than did those for testing single biomarkers. Although a very small number of patients were enrolled, our results demonstrate the feasibility and reliability of NGS testing for targeted therapies in refractory cancer patients, which may potentially help more patients achieve better outcomes. Recently, the clinical application of NGS has started under the medical insurance system in South Korea. This will increase NGS testing rates and also give physicians more options to better manage cancer patients with relevant mutations. Further functional studies on genetic variants will help elucidate their pathogenic role, which may have important clinical implications.
The following are the supplementary data related to this article.
Sequencing QC.
Footnotes
Funding:This study was supported by Busan Cancer Center Research Grant (2018), Pusan National University Hospital, Republic of Korea.
Declarations of Interest: None.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikchit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBCAN 2012. Int J Cancer. 2015;126:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Redig AJ, Jänne PA. Basket trials and the evolution of clinical trial design in an era of genomic medicine. J Clin Oncol. 2015;33:975–977. doi: 10.1200/JCO.2014.59.8433. [DOI] [PubMed] [Google Scholar]
- 3.Willyard C. 'Basket studies' will hold intricate data for cancer drug approvals. Nat Med. 2013;19:655. doi: 10.1038/nm0613-655. [DOI] [PubMed] [Google Scholar]
- 4.Kummar S, Williams PM, Lih CJ, Polley EC, Chen AP, Rubinstein LV, Zhao Y, Simon RM, Conley BA, Doroshow JH. Application of molecular profiling in clinical trials for advanced metastatic cancers. J Natl Cancer Inst. 2015;107:1–6. doi: 10.1093/jnci/djv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Drubin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philipparkis AA, Sivachenko Ay, Cibulskis K, Gabriel SB. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CCm Petric R, Hibberd ML, Nagarajan N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011;39:e132. doi: 10.1093/nar/gkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park HS, Lim AM, Kim A, Kim HR, Kwack KB, Lee MG, Kim JH, Moon YW. Pilot study of a next-generation sequencing-based targeted anticancer therapy in refractory solid tumors at a Korean Institution. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lara PN, Jr., Higdon R, Lim N, Kwan K, Tanaka M, Lau DH, Wun T, Welborn J, Meyers FJ, Christensen S. Prospective evaluation of cancer clinical trial accrual patterns: identifying potential barriers to enrollment. J Clin Oncol. 2001;19:1728–1733. doi: 10.1200/JCO.2001.19.6.1728. [DOI] [PubMed] [Google Scholar]
- 12.Tsimberidou AM, Wen S, Hong DS, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analyses. Clin Cancer Res. 2014;20:4827–4836. doi: 10.1158/1078-0432.CCR-14-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Tourneau C, Delord JP, Gonçalves A, Gavoille C, Dubot C, Isambert N, Campone M, Trédan O, Massiani MA, Mauborgne C. SHIVA investigators. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicenter, open-label, proof-of-concept, randomized, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 14.Heinemann V, Stintzing S. FOLFIRI with cetuximab or bevacizumab:FIRE-3-Authors' reply. Lancet Oncol. 2014;15:e583–e584. doi: 10.1016/S1470-2045(14)71036-8. [DOI] [PubMed] [Google Scholar]
- 15.Ciardiello F, Bianco R, Caputo R, Caputo R, Damiano V, Troiani T, Melisi D, De Cita F, De Plasido S, Bianco AR. Antitumor activity of ZD6474, a vascular endothelial growth factor receptor tyrosine kinase inhibitor, in human cancer cells with acquired resistance to antiepidermal growth factor receptor therapy. Clin Cancer Res. 2004;10:784–793. doi: 10.1158/1078-0432.ccr-1100-03. [DOI] [PubMed] [Google Scholar]
- 16.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858–862. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 18.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edkins S, O'Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 22.Jankiraman M, Vakinani E, Zeng Z, Pratilas C, Taylor B, Chitale D, Halilovic E, Wilson M, Huberman K, Filho J. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Migliardi G, Sassi F, Torti D, Galimi F, Zanella ER, Buscarino M, Ribero D, Muratore A, Massucco P, Pisacane A. Inhibition of MEK and PI3K/mTOR suppresses tumor growth but does not cause tumor regression in patient-derived xenografts of RAS-mutant colorectal carcinomas. Clin Cancer Res. 2012;18:2515–2525. doi: 10.1158/1078-0432.CCR-11-2683. [DOI] [PubMed] [Google Scholar]
- 24.Jänne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, Franke FA, Grinsted L, Zazulina V, Smith P. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled,phase 2 study. Lancet Oncol. 2013;14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing QC.