Abstract
To fight resistances to radiotherapy, the understanding of escape mechanisms of tumor cells is crucial. The aim of this study was to identify phosphoproteins that are regulated upon irradiation. The comparative analysis of the phosphoproteome before and after irradiation brought nucleophosmin (NPM1) into focus as a versatile phosphoprotein that has already been associated with tumorigenesis. We could show that knockdown of NPM1 significantly reduces tumor cell survival after irradiation. NPM1 is dephosphorylated stepwise within 1 hour after irradiation at two of its major phosphorylation sites: threonine-199 and threonine-234/237. This dephosphorylation is not the result of a fast cell cycle arrest, and we found a heterogenous intracellular distribution of NPM1 between the nucleoli, the nucleoplasm, and the cytoplasm after irradiation. We hypothesize that the dephosphorylation of NPM1 at threonine-199 and threonine-234/237 is part of the immediate response to irradiation and of importance for tumor cell survival. These findings could make NPM1 an attractive pharmaceutical target to radiosensitize tumor cells and improve the outcome of radiotherapy by inhibiting the pathways that help tumor cells to escape cell death after gamma irradiation.
Introduction
Despite recent advancements in tumor therapy, the development of resistances and the recidivation of tumors remain a major challenge in cancer treatment. Tumor diseases represent the second most frequent cause of death in the Western world, and the predicted global burden is expected to surpass 20 million new cancer cases by 2025 compared with an estimated 14.1 million new cases in 2012 [1].
Radiotherapy is a very important part of the treatment regimen for cancer of different origins as it is noninvasive and not accompanied by an intense systemic toxicity such as chemotherapy [2]. Approximately 40% of all cancer patients who are cured received radiotherapy alone or in combination with other treatment options [3]. Unfortunately, the curative potential of radiotherapy is impeded by mechanisms of tumor radiation resistance that enable tumor cells to survive and repopulate. To reestablish radiosensitivity, different strategies can be pursued [4] which require an in-depth understanding of the radiation response of tumor cells to enable a targeted intervention.
The cell's fate after irradiation is determined by the DNA damage response which paves the way for either cell death or repair of the sustained damage. Posttranslational modifications — above all phosphorylation and dephosphorylation — play a crucial role in coordinating the DDR at different levels in the signal transduction cascade [5]. This confers special significance to the phosphoproteome in the light of the cellular response to irradiation.
Our proteome-wide analysis of the specific differences in protein phosphorylation before and after irradiation brought the multifunctional hub-protein nucleophosmin (NPM1 / B23 / NO38 / numatrin) into focus. NPM1 is a classical phosphoprotein that is regulated in manifold ways by phosphorylation and dephosphorylation. Around 10 of its phosphorylation sites have been characterized in greater detail [6], [7], [8], [9], [10], [11], about 20 phosphorylation sites have been found in high-throughput phosphoproteome studies [12], [13], and up to 40 sites have been predicted in silico [14]. NPM1 exerts a bunch of different functions. NPM1 is involved in ribosome biogenesis [15], maintenance of genomic stability [16] by securing correct centrosome duplication [17], DNA repair [18], chromatin remodeling as well as chaperoning of histones [19], [20], and regulation of cellular response to various stress stimuli [21] such as gamma irradiation [22], [23], UV irradiation [24], chemotherapeutics [25], heat shock [26], oxidative stress [27], and hypoxia [28].
Apart from posttranscriptional modifications and oligomerization [29], [30], [31], the subcellular localization of NPM1 plays a very important role for the regulation of the protein and the switch between its manifold functions and protein interaction partners. NPM1 mainly resides in the nucleoli, but upon cellular stress stimuli, it can be relocated into the nucleoplasm and the cytoplasm. The shuttling activity and the subcellular localization of NPM1 are essential for the protein to exert its correct cellular functions.
The involvement of NPM1 in the tumorigenesis of hematological malignancies is well demonstrated in several studies [32], [33], [34], and also the link between proliferation and malignant transformation in solid tumors has been drawn [35], [36], [37], [38]. In many solid tumors, the protein is overexpressed [39], [40], [41], [42], [43], [44], [45] even though also misregulation in the sense of reduced NPM1 levels has been reported [46], [47].
To gain deeper insights into the role of NPM1 in the response of tumor cells to irradiation, we evaluated the phosphorylation of NPM1 at four of its major phosphorylation sites that are serine-4, serine-125, threonine-199, and threonine-234/237. Additionally, we analyzed the subcellular localization of the protein and its influence on the survival of tumor cells after irradiation.
Materials and Methods
Tumor Cell Lines and Cell Culture
The non–small cell lung cancer (NSCLC) cell line A549 and the cervical cancer cell line HeLa were purchased from DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). The head and neck squamous cell carcinoma (HNSCC) cell line HNSCCUM-02T was previously established and characterized in our laboratory [48]. The identity of the cell lines was verified by STR analysis by the DSMZ. Cells were maintained in DMEM/Ham's F12 (Sigma-Aldrich, St. Louis, MO) supplemented with 5% FCS (fetal calf serum; Sigma-Aldrich, St. Louis, MO) and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin) at 37°C in 5% CO2.
Isoelectric Focusing (IEF), Two-Dimensional (2D) Gel Electrophoresis, and Mass Spectrometry (MS)
According to previous studies ([49] and therein), a single dose of 8 Gy is well suited to study the irradiation response of tumor cells. To study differences in the phosphoproteome after irradiation, the amount of phosphoproteins in cells 30 minutes after irradiation was compared to that in nonirradiated control cells. In brief, harvested cells were resuspended in lysis buffer (PhosphoProtein Purification Kit lysis buffer (Qiagen) with 0.25% CHAPS, phosphatase (PhosSTOP EASYpack Phosphatase inhibitor cocktail tablets, Roche, Grenzach-Wyhlen, Germany), and protease inhibitors (cOmplete Protease inhibitor cocktail tablets, Roche, Grenzach-Wyhlen, Germany) and sonicated. The phosphorylated proteins were isolated by IMAC-columns followed by ultracentrifugation with Roti-Spin MIDI (Carl Roth, Karlsruhe, Germany) and Vivaspin (Sartorius, Göttingen, Germany) centrifugal concentrators. Protein concentration was adjusted to 1.5 mg/ml, and 1/125 vol. 2.5 M DTT, 1/80 vol HED, and 1/200 vol IPG buffer, pH 4-7, were added for IEF. The procedures of IEF and MS have been described in detail before [50]. Shortly, for each sample, 360 μl lysate was applied to an 18-cm IPG-strip (pH 4-7, Immobiline Dry Strips, GE Healthcare Bio-Sciences, München, Germany). After focusing, strips were transferred to the top of a 14.5% SDS gel. Gels were run with 20 mA/gel for 45 minutes and then with 40 mA/gel for 3 hours. The gels were stained with SYPRO orange overnight. For scanning, we used a fluorescence scanner (Storm 840, Amersham Pharmacia, Freiburg, Germany) at a PMT voltage of 1000, resolution 100 μm. At least three biological replicates were performed in each case. Scanned gels were analyzed using the Delta 2D program (Decodon, Greifswald, Germany). Differentially expressed protein spots (normalized intensity values diverging by a factor of 1.5 or more) were analyzed by MS. Spots were trypsin-digested and spotted onto AnchorChip 600/384 T F targets (Bruker Daltonics, Bremen, Germany) using the dried droplet method. As energy-absorbing molecule, we used α-cyano-4-hydroxycinnamic acid (2 mg/ml in 50% ACN containing 0.2% TFA). Mass spectrometry analysis was performed using a MALDI-TOFTOF MS (Ultraflex II, Bruker Daltonics). The top 20 peaks were used for subsequent MS/MS analysis. Data processing of raw spectra and protein identification was performed using Bruker software (Flex analysis 2.4 and BioTools 3.1) and Mascot. As Mascot result parameters, we chose standard scoring and a significance threshold of P < .05 for protein / peptide identification.
RNA Interference and Colony-Forming Assay
To knock down NPM1, small interfering RNA (siRNA) transfection was performed according to the manufacturer's protocol using either NPM1 siRNA (Silencer Select siRNA, s9678, 5′->3′ sequence CUA UCU UUU CGG UUG UGA Att, Ambion Applied Biosystems, Darmstadt, Germany) or positive and negative controls (Silencer Select siRNA GAPDH and negative control #2). Knockdown of NPM1 was analyzed by Western blot (Figure 2B) at day 3, day 4, day 6, day 8, and day 10 after transfection. Thirty-six house (day 3) after siRNA transfection, when knockdown was fully established, tumor cells were detached, cell numbers were determined by Casy1, and cell suspensions were irradiated with 4 or 8 Gy, respectively, using a Cs137 source. Cell numbers were chosen according to pilot tests to reach between 10 and 600 colonies after 10 days, and the assay was performed as previously described [51]. Nonirradiated cultures were processed in parallel. Colony formation was assessed with the COLCOUNT system (Oxford Optronix Ltd., Abingdon, UK). Each experiment was performed in duplicates and repeated at least three times. The relative surviving fraction and the relative colony size were calculated.
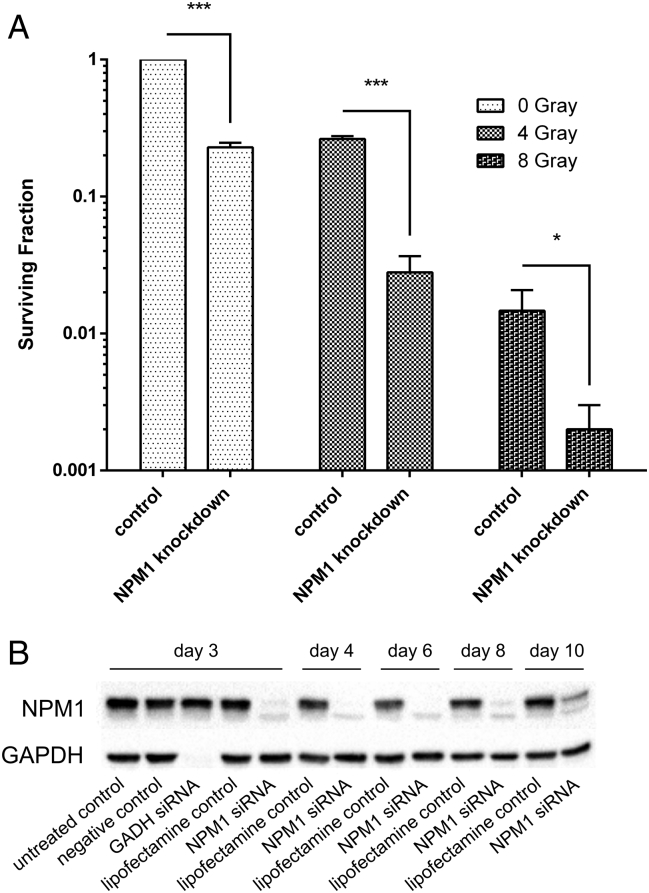
Figure 2.
Knockdown of NPM1 reduces tumor cell survival.
(A) The surviving fraction of A549 cells in a colony-forming assay was significantly reduced after knockdown of NPM1. Combined treatment with 4-Gy or 8-Gy gamma irradiation, respectively, with knockdown further reduced tumor cell survival significantly compared to cells which received irradiation alone. Shown are means ± SD, *P < .05, **P < .01, ***P < .001, unpaired t test, comparison between cells which received NPM1 knockdown and respective control groups as indicated, correction for multiple comparisons by Bonferroni, N = 3. (B) Knockdown of NPM1 was confirmed by Western blot analysis at day 3, 4, 6, 8, and 10 after transfection. At day 3, the knockdown was fully established, the cells were irradiated and seeded for the colony assay, and the knockdown lasted until day 10.
Irradiation and Fractionated Cell Lysis
For irradiation, tumor cell lines were isolated by tryptic digestion (Sigma-Aldrich, St. Louis, MO), and 1.8 × 104 cells/ml were seeded in culture dishes with a growth area of 25 cm2. After 24 hours, the medium was replaced by new medium, and the cells were cultured for additional 24 hours. Subsequently, cells were irradiated with 8 Gy using a Cs137 source and harvested directly 3, 10, 30, and 60 minutes after irradiation using a rapid fractionated lysis [52] with phosphatase (PhosSTOP EASYpack Phosphatase inhibitor cocktail tablets, Roche, Grenzach-Wyhlen, Germany) and protease inhibitors (cOmplete Protease inhibitor cocktail tablets, Roche, Grenzach-Wyhlen, Germany) included in the lysis buffer. To verify successful fractionation, control Western blots with antibodies against histone H3 (for the nuclear probes) and GAPDH (for the cytoplasmic probes) were performed.
Expression Analysis by SDS-PAGE and Western Blotting
Twenty micrograms of total protein was loaded onto 12% acrylamide gels and subjected to SDS-PAGE. Gels were transferred to Sequi-Blot PVDF Membrane (Bio-Rad, München, Germany) by semidry Western blotting procedure. We used the following antibodies: NPM antibody (Cell Signaling, Boston, MA), pNPM antibody phospho T199 (Cell Signaling, Boston, MA), pNPM antibody phospho T234/237 (Abcam, Cambridge, UK), pNPM antibody phospho S4 (Cell Signaling, Boston, MA), pNPM antibody S125 (Abcam, Cambridge, UK), anti-mouse IgG, HRP-linked Antibody (#7076 Cell Signaling), anti-rabbit IgG, HRP-linked Antibody (#7074 Cell Signaling) and GAPDH antibody (Abcam, Cambridge, UK) and histone H3 antibody (Cell Signaling, Boston, MA), as loading control and for verification of proper cell fractionation. Blots were developed by enhanced chemiluminescent substrate (Thermo Fisher Scientific, Rockford, IL) and documented using the ChemiDoc Imager (Bio-Rad, München, Germany) and evaluated with the Image Lab software (version 5.0 build 18, Bio-Rad). Band densities were normalized to those of the nonirradiated sample on each blot and expressed as percentage of that control. Each experiment was performed at least in triplicate.
Immunohistochemistry
For immunofluorescent staining, 2.5 × 104 cells were grown on cover slips in 6-well dishes. Twenty-four hours after seeding or transfection depending on the experiment, cells were washed 3 × 5 minutes with DPBS (Phosphate Buffered Saline; Sigma-Aldrich, St. Louis, MO) and fixated by incubation with 4% (w/v) paraformaldehyde in DPBS for 15 minutes, followed by another 3 × 5–minute washing step with DPBS. Cells were incubated with ice-cold methanol for 10 minutes, washed again for 3 × 5 minutes with DPBS, and blocked with DPBS with 5% BSA and 0.3% Triton-X100 for 1 hour to avoid unspecific binding. The following antibodies were used: Phospho-NPM (Thr199) Antibody (#3541 Cell Signaling), Mitotic Cells Antibody [8B3G] (ab8956 Abcam), Goat anti-Rabbit IgG (H + L) Highly CrossAdsorbed Secondary Antibody, Alexa Fluor 555, (A-21429 Invitrogen), and Goat anti-Mouse IgG (H + L) Highly CrossAdsorbed Secondary Antibody, Alexa Fluor 488, (A-11029 Invitrogen). The cover slips were transferred to microscopy slides, covered by VECTASHIELD Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA), and evaluated by wide field microscopy.
Cell Cycle Analysis
For cell cycle analysis, tumor cell lines were treated according to the corresponding protocol and isolated by tryptic digestion. Cells were washed with DPBS and finally suspended in 500 μl DPBS and 4.5 ml 70% ice-cold ethanol. After incubation for at least 2 hours at −20°C, cells were washed with washing buffer [0.2% (v/v) Triton X-100, 1% BSA in DPBS] and transferred to propidium iodide staining solution [0.1% (w/v) RNAse A, 5 μg/ml PI in DPBS]. The staining was evaluated with FACSCanto flow cytometer and analyzed with the ModFit LT software (Verity Software House, Topsham, ME).
Side-Directed Mutagenesis and Transfection
The side-directed mutagenesis was performed according to the Quik Change II L Sited-Directed Mutagenesis Kit (Agilent Technologies, Böblingen, Germany) using the pc3-NPM1-wt-GFP expression vector based on the pcDNA 3.1 plasmid (Invitrogen Life Technologies, Carlsbad, CA) which was a gift of Prof. Stauber (Molecular and Cellular Oncology, Department of Otorhinolaryngology, Head and Neck Surgery, University Medical Centre Mainz) and has previously been described [53]. Threonine or serine residues at the phosphorylation sites of NPM1 were replaced by alanine residues, which leave them phosphorylation insensitive. GFP-tagged NPM1 was transiently transfected into HeLa cells using the above described expression vector according to the jetPrime system (Polyplus, Illkirch, France).
Statistics
One-way ANOVA and unpaired t tests followed by Bonferroni correction for multiple comparisons and Welch correction for uneven variations, where applicable, were used to assess statistical significances. All calculations were performed using the software GraphPad Prism 6 for Windows, Version 6.01 (GraphPad Software, La Jolla, CA). Shown are mean values ± SD; P values < .05 are indicated as *P < .05, **P < .01, and ***P < .001.
Results
Regulation of NPM1 Postirradiation
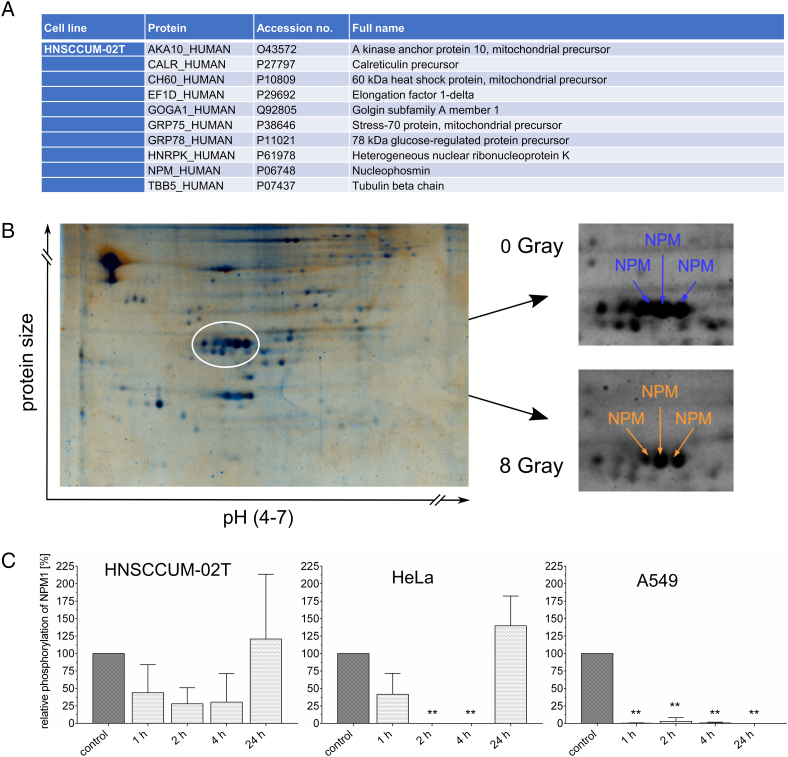
HNSCCUM-02T cells were treated with a single 8-Gy dosage of gamma irradiation, and changes of the phosphoproteome were studied 30 minutes after irradiation by means of 2D electrophoresis and MALDI-TOF-mass spectrometry. In total, we could identify 10 phosphoproteins of different protein families which were subjected to regulation upon irradiation (Figure 1A). Two of these proteins showed increased phosphorylation, and eight proteins were dephosphorylated following irradiation. Among them, the phosphoprotein nucleophosmin (NPM1) was very prominent (Figure 1B). In the 2D gel, several NPM1 containing spots could be identified. The NPM1 in the spots differed in the isoelectric point; thus, most probably, the spots correspond to differentially phosphorylated protein variants.
Figure 1.
Phosphoproteome analysis after gamma irradiation brings NPM1 into focus.
(A) Phosphoproteome analysis of HNSCCUM-02 T cells by 2D-electrophoresis and MALDI-TOF-mass spectrometry revealed 10 phosphoproteins regulated upon 8-Gy gamma irradiation within 30 minutes. NPM1 stood out as it was detected in several spots on the gel. (B) Two representative gels of lysates of HNSSCUM-02T cells. Blue represents the control gel, and orange represents the gel containing the cell lysates harvested after irradiation. (C) By 1D immunoblotting, we could show that NPM1 is dephosphorylated in HNSCCUM-02T, HeLa, and A549 cells within 1 to 4 hours after 8-Gy irradiation compared to untreated control cells (=100%). The dephosphorylation lasts for up to 24 hours depending on the cell line. Shown are means ± SD, *P < .05, **P < .01, ***P < .001, one-way ANOVA, comparison between untreated control cells and irradiated cells, correction for multiple comparisons by Bonferroni, N = 3. Representative Western blots are shown in Figure S3 in the supplementary data.
Dephosphorylation of NPM1 at Threonine-199 within 24 Hours after Irradiation
Western blot analysis showed NPM1 expression in all tumor cell lines under study. NPM1 expression was highest in HeLa cells, followed by HNSCCUM-02T cells and A549 cells (data not shown). NPM1 occurs in three different splicing forms: isoform 1 is considered the dominant form [54] which consists of 294 amino acids, and isoforms 2 and 3 are a little bit shorter with 265 and 259 amino acids [55]. In our Western blot experiments, we were able to detect one to three different bands in the gel, which were bound by the NPM1 antibody. In all three bands, NPM1 could be identified by MALDI analysis (data not shown). For our analysis, all three bands were evaluated together. To avoid falsification of the phosphorylation status, we worked strictly on ice with protease and phosphatase inhibitors included. Contrary to Koike et al. [23], Sekhar et al. [22], and Penthala et al. [56], we did not separate the fraction of the soluble proteins in the nucleoplasm from the insoluble proteins in the chromatin fraction according to Groisman et al. [57], but we obtained one nuclear fraction containing all proteins. Complete dissolution of all proteins was ensured by sonification and heating of the lysates prior to loading onto the gel.
Threonine-199 is one of the most important and well-characterized NPM1 phosphorylation sites. Therefore, we analyzed the phosphorylation at this site within 24 hours after irradiation with 8 Gy in A549, HeLa, and HNSCCUM-02T cells (Figure 1C). Representative blots are shown in Figure S3 in the supplementary data. A549 as well as HeLa cells bear wild-type p53, while HNSCCUM-02T cells bear a p53 mutation which results in unconditionally high p53 expression [58]. Hela cells represent a special case concerning their p53 level; they are affected by a mutation-independent reduction in p53 protein level due to the expression of the viral E6 protein [59]. Thus, the analysis covers different entities as well as differences in the p53 status. We found that NPM1 is dephosphorylated at threonine-199 shortly after irradiation in all three cell lines. In A549 cells, threonine-199 was completely dephosphorylated, and the dephosphorylation persisted for 24 hours. In HNSCCUM-02T cells, the dephosphorylation was less pronounced, and 24 hours after irradiation, the phosphorylation at threonine-199 returned to the basal level. In HeLa cells, the dephosphorylation was a little bit slower compared to A549 cells, reaching complete dephosphorylation after 2 hours, and phosphorylation was regained after 24 hours.
Depletion of NPM1 Diminishes Tumor Cell Survival after Irradiation
To tackle the question of the importance of NPM1 for the survival of tumor cells after irradiation, we performed a colony-forming assay with A549 cells as a representative cancer cell line, which showed the most pronounced NPM1 dephosphorylation after irradiation. This revealed that knockdown of NPM1 significantly reduced tumor cell survival when it was administered alone and in combination with 4-Gy or 8-Gy dosages of irradiation (Figure 2A). The surviving fraction of untreated tumor cells was set to 1.0, and the surviving fraction of tumor cells which were subjected to NPM1 knockdown and/or irradiation was related to that value. We could detect a significantly reduced tumor cell survival after NPM1 knockdown, and additional NPM1 knockdown further reduced tumor cell survival after irradiation with 4 or 8 Gy. Moreover, the colony size after depletion of NPM1 was reduced when NPM1 knockdown was administered alone (data not shown). NPM1 depletion after siRNA-mediated knockdown was confirmed by Western blot (Figure 2B).
Phosphorylation Pattern of NPM1 at Serine-4, Serine-125, Threonine-199, and Threonine-234/237 within 1 Hour after Irradiation
For the subsequent analysis of the phosphorylation status of NPM1 shortly after irradiation, we chose four of its most important phosphorylation sites: serine-4, serine-125, threonine-199, and threonine-234/237. A549 cells as well as HeLa cells, which both bear wild-type p53 but differ in their p53 expression level, were used. Cells were irradiated with 8 Gy, and cell lysates were harvested within 1 minute, 10 minutes, 30 minutes, and 1 hour after irradiation using a fast fractionated lysis protocol [52]. All cell lysates of each experiment (different compartments, different time points, and irradiated vs. control) were loaded on one gel to correct for differences between the single experiments, and the phosphorylation was expressed as relative phosphorylation in percent compared to the corresponding nonirradiated control in the corresponding compartment (Figure 3). Representative blots are shown in Figures S1 and S2 in the supplementary data.
Figure 3.
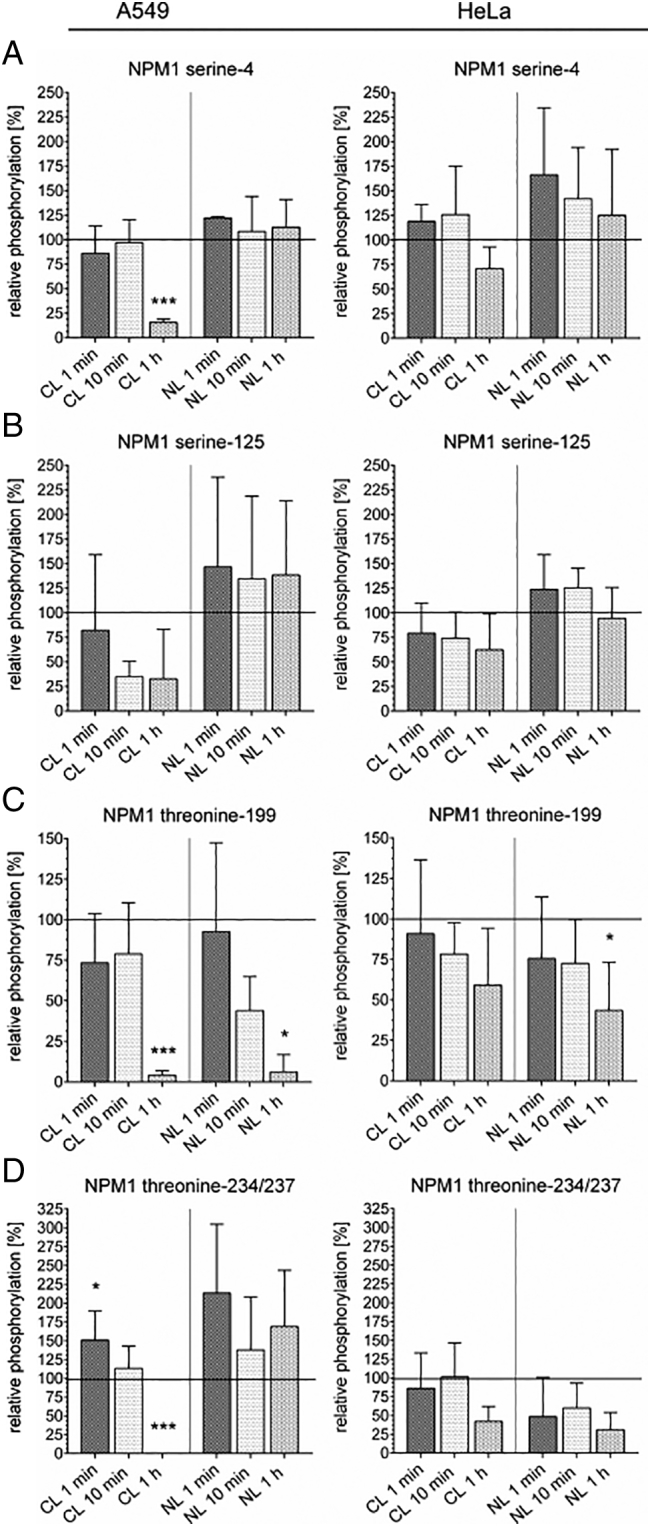
Phosphorylation pattern of NPM1 rapidly changes upon irradiation.
The phosphorylation of NPM1 after 8-Gy irradiation was tracked after 1 minute, 10 minutes, and 1 hour in the cytoplasm and in the nucleus of A549 and HeLa cells by Western blot analysis. NPM1 was significantly dephosphorylated at serine-4 in the cytoplasm and at threonine-199 in both compartments in A549 cells. Dephosphorylation at theonine-199 was also found in HeLa cells. At theorine-234/237, NPM1 was phosphorylated 1 minute after irradiation in the cytoplasm of A549 cells followed by a dephosphorylation after 1 hour. Shown are means ± SD, *P < .05, **P < .01, ***P < .001, one-way ANOVA, comparison between untreated control cells (=100%) and irradiated cells of the same compartment, correction for multiple comparisons by Bonferroni, N = 3. Representative Western blots are shown in the Figures S1 and S2 in the supplementary data.
At serine-4, we could show a significant dephosphorylation 1 hour after irradiation in the cytoplasm of A549 cells, while the phosphorylation in the nucleus at this site remained unchanged (Figure 3A). In HeLa cells, this dephosphorylation could not be confirmed due to major variability in the phosphorylation at this site between the biological replicates. At serine-125, there could be detected a slight dephosphorylation in the cytoplasm in A549 and in HeLa cells after irradiation, but the effect was not significant due to variations within the replicates (Figure 3B). Nevertheless, it was clearly visible that serine-125 just like serine-4 was dephosphorylated in the cytoplasm but not in the nucleus. At threonine-199, NPM1 was dephosphorylated in both cell lines in the nucleus as well as in the cytoplasm (Figure 3C). The dephosphorylation in A549 cells was more pronounced and faster than the one in HeLa cells, confirming the results in the long-duration measurement (Figure 1C). At threonine-234/237, there could be detected a very fast phosphorylation in the cytoplasm within 1 minute after irradiation in A549 cells followed by a dephosphorylation within 1 hour. The phosphorylation at threonine-234/237 in the nucleus was slightly elevated throughout the complete timespan compared to nonirradiated control cells. In HeLa cells, the analysis of phosphorylation was inconclusive: 1 hour after irradiation, there could be seen a slight dephosphorylation in the nucleus and the cytoplasm, but the effect did not reach significance (Figure 3D).
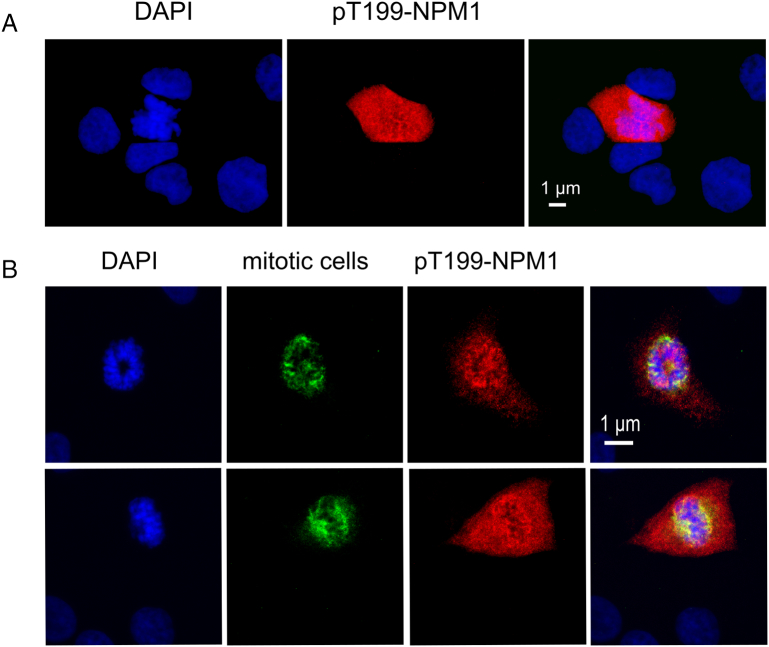
Mitotic Cells Exhibit High Level of NPM1 phosphorylated at Threonine-199
In the immunohistochemical staining of NPM1 phosphorylated at threonine-199 (pT199-NPM1), we observed some cells that exhibited a very strong staining for pT199-NPM1 in the whole cell (Figure 4A). We wondered what was special about these cells that they contained a huge amount of pT199-NPM1 while other cells did not. One first hint gave us the staining with DAPI of those cells: the DAPI staining was not as smooth as in other cells; the nucleus had no round shape but was rough and the staining uneven. This led us to a staining with an antibody against mitotic cells, and we could show that the cells with a strong staining for pT199-NPM1 were currently undergoing mitosis (Figure 4B).
Figure 4.
Phosphorylation of NPM1 at threonine-199 is high in mitotic cells.
Fluorescent staining for NPM1 phosphorylated at threonine-199 revealed that some A459 cells were especially high in phosphorylation (A). Staining with an antibody against mitotic cells revealed that those cells were currently undergoing mitosis (B).
Dephosphorylation of NPM1 at Threonine-199 is not Attributable to Cell Cycle Arrest
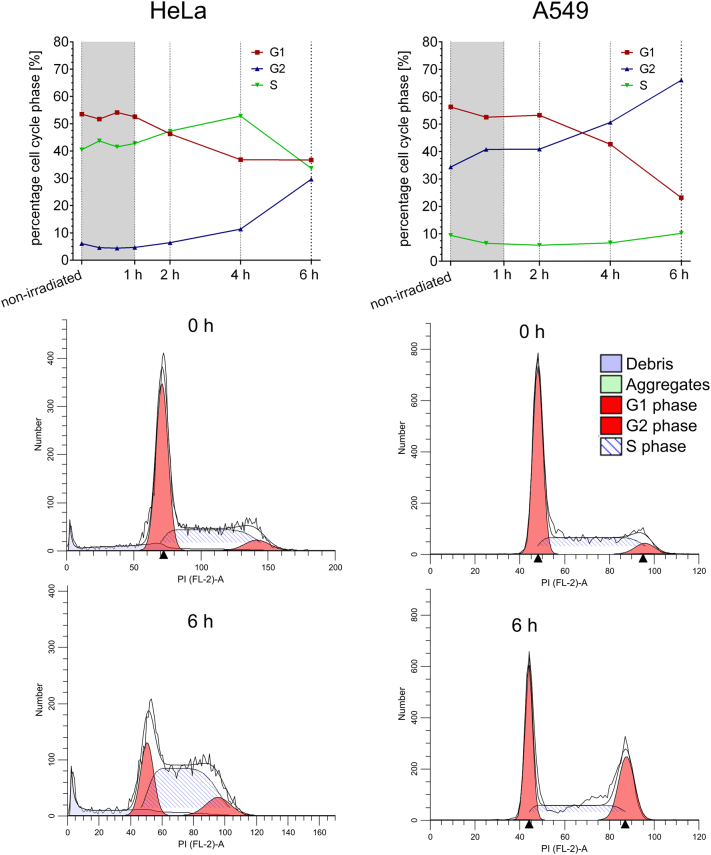
To evaluate a potential association between the dephosphorylation of NPM1 at threonine-199 and a reduction in mitotic cells after irradiation, we performed a cell cycle analysis (Figure 5). We could show that the cell cycle distribution within the first hour after irradiation remained constant in HeLa as well as in A549 cells. This means that within 1 hour after irradiation, the fractions of cells in G1, G2, or S phase did not change; thus, within this time scale, no cell cycle arrest is induced. Taking a closer look at the later time points, we could see that the percentage of cells in G1 phase was reduced and the percentage of cells in G2 phase rose within 6 hours after irradiation. This suggests a cell cycle arrest in G2 phase 6 hours postirradiation.
Figure 5.
Cell cycle arrest is induced in HeLa and A549 cells 6 hours after irradiation.
Within the first hour after irradiation with 8 Gy, the distribution between G1, S, and G2 phase of the cell cycle remained constant in A549 and HeLa cells. Six hours postirradiation, a cell cycle arrest in the G2 phase was initiated in both cell lines. The time course of cell cycle distribution is shown in the upper panel for HeLa cells on the left and for A549 cells on the right. Beneath there are shown exemplary cell cycle distributions after 0 hour and after 6 hours, which were depicted and evaluated by the ModFit LT software.
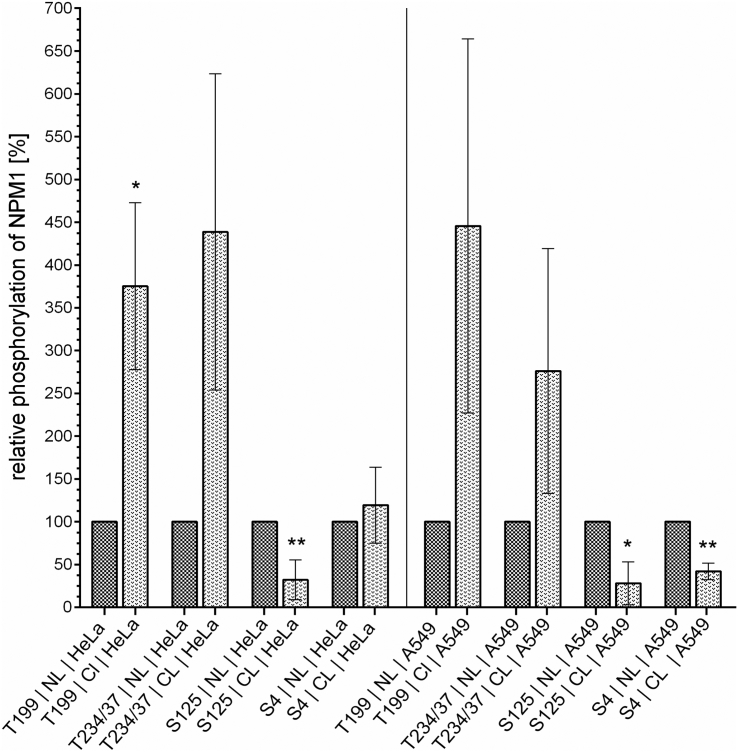
Phosphorylation of NPM1 Differs between Cellular Compartments
Several recent studies associated the phosphorylation status of NPM1 with its localization within the cell [9], [60] NPM1 is a predominantly nucleolar protein [15], but it is well established that the protein can undergo redistribution to the nucleoplasm and the cytoplasm upon different stimuli [24], [61], [62]. Thus, we analyzed the relative phosphorylation of NPM1 in the nucleus and in the cytoplasm at serine-4, serine-125, threonine-199, and threonine-234/237 in HeLa and in A549 cells (Figure 6). This revealed that NPM1 phosphorylation at threonine-199 and threonine-234/237 was more prominent in the cytoplasm than in the nucleus in both cell lines. By contrast, the phosphorylation at serine-125 was more pronounced in the nucleus. The analysis of the phosphorylation of serine-4 delivered inconclusive results: it was less prominent in the cytoplasm of A549 cells compared to the nucleus while being nearly equally phosphorylated in nucleus and cytoplasm in HeLa cells. Representative blots are shown in Figures S1 and S2 in the supplementary data (A549 and HeLa, 0 Gy = untreated cells).
Figure 6.
Phosphorylation of NPM1 differs between the cellular compartments.
The phosphorylation of NPM1 at threonine-199 and threonine- 234/237 was more pronounced in the cytoplasm than in the nucleus of HeLa cells and A549 cells. In contrast, phosphorylation of NPM1 at serine-125 could mainly be found in the nucleus. Phosphorylation of NPM1 at serine-4 was detected in both compartments, and it was dependent on the cell line in which part of the cell it was more prominent. Shown are means ± SD, *P < .05, **P < .01, ***P < .001, unpaired t test with Welch correction for uneven variations, comparison between expression in the nucleus (=100%) and expression in the cytoplasm, N = 3. Representative Western blots are shown in the Figures S1 and S2 in the supplementary data.
Heterogenous Localization of NPM1 after Irradiation
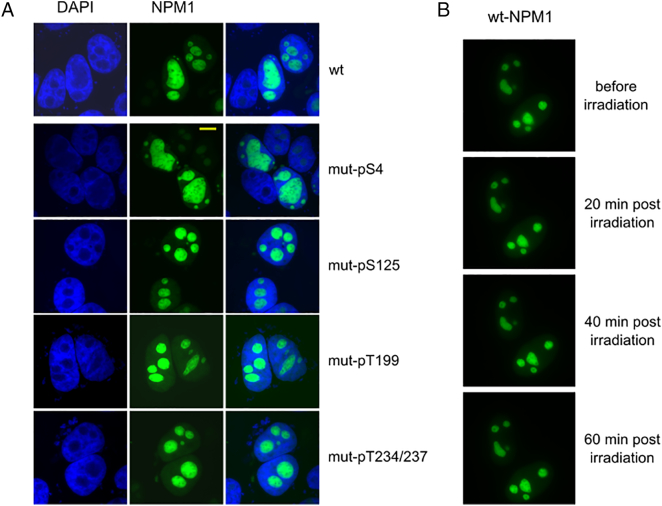
Next, we analyzed the dependence of the localization of NPM1 on its phosphorylation status. Thereto we transfected HeLa cells with a vector containing the NPM1 gene tagged with GFP. By side-directed mutagenesis, the phosphorylation sites serine-4, serine-125, threonine-199, or threonine-234/237, respectively, were replaced with alanine to make them phosphorylation insensitive. In the following, we analyzed whether the mutation of these phosphorylation sites had an influence on the distribution of the protein within the cell. This was not the case; irrespective of the mutation of the phosphorylation sites, the majority of the NPM1 protein still localized to the nucleoli (Figure 7A).
Figure 7.
Transfection with GFP-tagged NPM-1 to study subcellular distribution of the protein.
HeLa cells were transfected with a vector containing GFP-tagged NPM1 with mutations in different phosphorylation sites which rendered them phosphorylation insensitive. Still, the protein showed predominantly nucleolar localization (A). With the help of live cell imaging, it could be seen that GFP-tagged wt-NPM1 does not redistribute within the first hour after irradiation with 8 Gy (B).
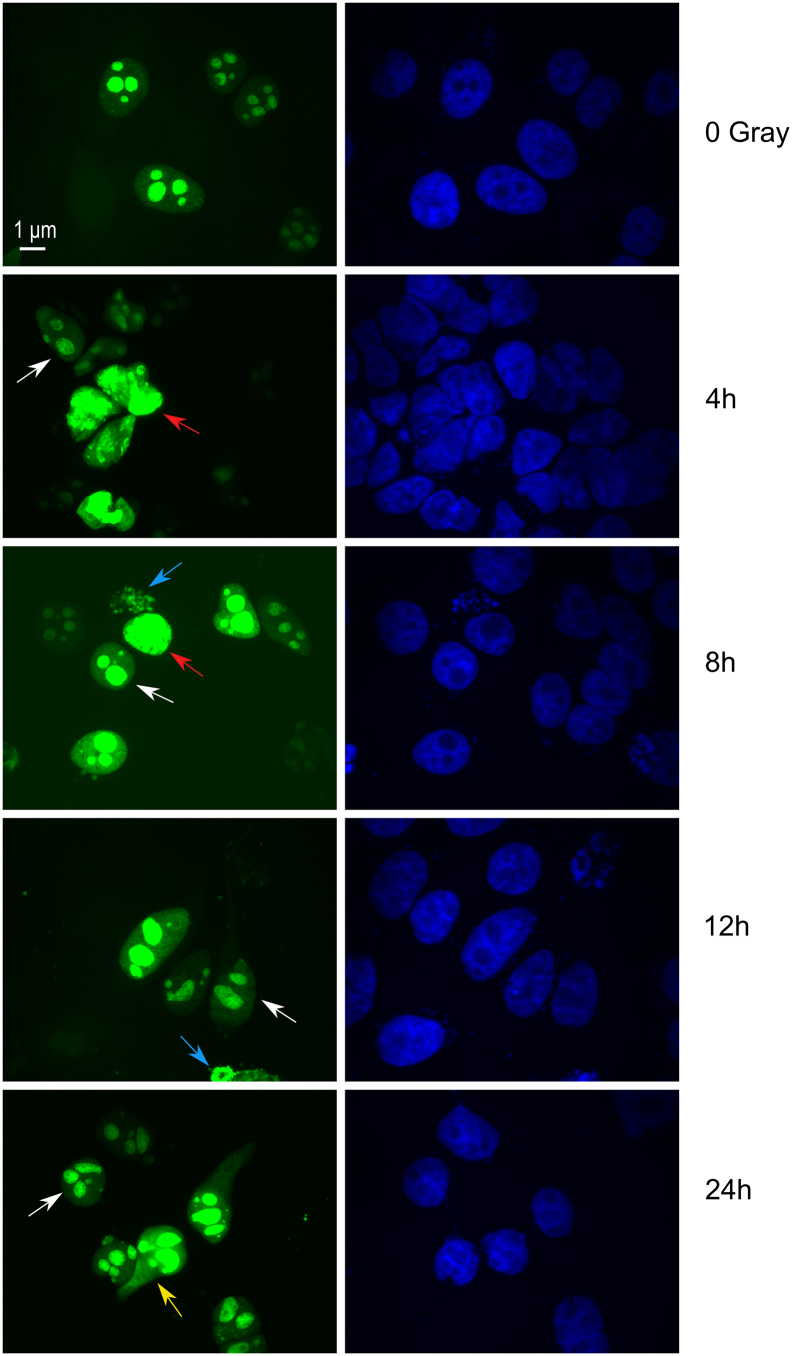
Moreover, we analyzed the distribution of NPM1 within 1 hour after irradiation with 8 Gy (Figure 7B). By live cell imaging, we could show that the cellular distribution of wild-type NPM1 did not change within 1 hour after irradiation. Next, we transfected cells with wild-type NPM1 and tracked its localization for 24 hours after irradiation (Figure 8). We observed a heterogenous reaction of the tumors cells to irradiation concerning the NPM1 distribution. Some cells maintained their predominant nucleolar localization of NPM1 (white arrows). In other cells, NPM1 lost its localization in the nucleolus and was distributed in the whole nucleus (red arrows). We also saw cells that were fragmented and most likely were undergoing cell death (blue arrows). In some cases, NPM1 could also be detected in the cytoplasm (yellow arrow).
Figure 8.
Localization of GFP-tagged NPM1 after irradiation was heterogenous following irradiation.
HeLa cells transfected with GFP-tagged NPM1 irradiated with 8 Gy showed a heterogenous distribution of the protein between the different cellular compartments. Many cells maintained their predominant nucleolar localization of NPM1 (white arrows). In some cells, NPM1 was redistributed in nucleoplasm (red arrows). A few cells were fragmented (blue arrows), and in some cases, NPM1 could be detected in the cytoplasm (yellow arrow). Shown are representative pictures of at least N = 3 independent experiments.
Discussion
We identified NPM1 as a very prominent protein in our analysis of the phosphoproteome after irradiation and could show that NPM1 is important for tumor cell survival after gamma irradiation. It came into focus because we could detect several different spots containing NPM1 which were lined up like pearls on a string. They differed in their isoelectric point which indicated that the NPM1 in the different spots differed in the phosphorylation pattern. NPM1 is a classical phosphoprotein which is regulated by phosphorylation and dephosphorylation to a great extent [9], [63], [64]. It is well established that NPM1 is involved in cellular proliferation and also in tumorigenesis [35], [65], and it is frequently overexpressed, mutated, or rearranged in tumor cells [33]. NPM1 has versatile cellular functions, and several of them are known to be relevant for malignant degeneration and progression, among them ribosomal biogenesis [15] and licensing of centrosomal duplication, thus assuring of genomic stability [17], DNA repair processes [18], [56], [66], [67], [68], and apoptosis [28], [69].
To check whether NPM1 is relevant for the survival of tumor cells after irradiation, we performed a colony-forming assay. We compared the ability of tumor cells to survive and form colonies after irradiation between cells expressing NPM1 and cells in which NPM1 was knocked down. After depletion of NPM1 by siRNA knockdown, we could see a significant decrease in survival rates compared to control cells. This was true for cells which received NPM1 knockdown alone as well as for those which additionally were irradiated with 4 or 8 Gy, respectively. Additionally, we could see a reduction of the colony size after knockdown of NPM1. This underlines that the protein is important for the survival and proliferation of tumor cells after irradiation. As NPM1 performs versatile functions within the cell, we had a closer look at its phosphorylation status after irradiation to understand its role in the response of tumor cells to irradiation.
Threonine-T199 is the best studied phosphorylation site of NPM1; thus, as a first step, we tracked the phosphorylation at this site within 24 hours after irradiation, and we found a dephosphorylation of the protein in all three cell lines under investigation. This dephosphorylation was initiated very fast, as it could be detected within 1 hour after irradiation, and depending on the cell line, it lasted for several hours. Therefore, we decided to have a closer look at the early phosphorylation or dephosphorylation events within the first hour after irradiation. Our detailed analysis of the phosphorylation and dephosphorylation events within 1 hour after irradiation revealed that NPM1 was dephosphorylated at threonine-199 and threonine-234/237 in A549 and in HeLa cells. The dephosphorylation was more pronounced in HeLa cells than in A549 cells, which may be attributable to the lower amount of total NPM1 in HeLa cells which enables the dephosphorylation to come into effect more rapidly which is also in line with the long-duration measurements. As NPM1 is known to directly and indirectly interact with the tumor suppressor p53 and its associated pathways [38], it is important to notice that A549 cells and HeLa cells both bear wild-type p53, but p53 expression in HeLa cells is diminished by the viral E6 protein to a nearly undetectable level [70]. The fact that dephosphorylation of NPM1 at threonine-199 and threonine-234/237 could be detected in both cell lines suggests that this cellular reaction is not dependent on p53 expression. Dephosphorylation of NPM1 at threonine-199 and threonine-234/237 was also reported following UV irradiation [18], where it was associated with DNA-damage repair pathways. Several studies were able to relate NPM1 and its phosphorylation at threonine-199 with DNA-damage repair pathways. For example, Koike et al. and Sekhar et al. [22], [23], [31], [56] postulated the recruitment of NPM1 to the sites of DNA damage and a critical role of the protein for their repair. Our study focused on phosphorylation status of NPM1 in the whole nucleus and in the cytoplasm and was able to underline the importance of the protein for the survival of tumor cells after irradiation. Altogether, there is growing evidence that NPM1 is highly relevant for DNA-damage repair pathways and as such is an attractive target for innovative antitumor agents.
Immunohistochemical staining of pT199-NPM1 revealed that phosphorylation at threonine-199 is particularly prominent in cells undergoing mitosis, which is in line with previous studies [9], [71]. This led us to the question of whether the observed dephosphorylation at threonine-199 is attributable to a cell cycle arrest which is accompanied by a reduction of the proportion of mitotic cells. Thus, we performed a cell cycle analysis, and we could show that there is no relevant change in the distribution of cells between G1, G2, and S phase within the first hour after irradiation. Thus, the dephosphorylation of threonine-199 could not be the consequence of a fast cell cycle arrest. We could see an arrest of the cell cycle in both cell lines, but it took longer to be initiated and was detectable after 6 hours postirradiation.
Several studies suggested that the phosphorylation status of NPM1 takes influence on the subcellular distribution of the protein [9], [72]. Therefore, we pursued the question whether the dephosphorylation of NPM1 after irradiation is accompanied by a relocation of the protein. First, we checked for each phosphorylation site under study in which compartment of the cell is was most prevalent. With the help of a fractionated cell lysis, we were able to show that threonine-199 and threonine-234/237 were mainly phosphorylated in the cytoplasm. In contrast, serine-125 was predominantly phosphorylated in the nucleus. The observation that pT199-NPM1 and pT234/237-NPM1 are mostly localized in the cytoplasm is congruent with studies showing that phosphorylation at these sides reduces the binding to nucleic acids and thus the affinity to the nucleus [9].
Next, we constructed a vector containing NPM1 tagged with GFP and expressed it in HeLa cells. Additionally, mutations in the NPM1 gene were introduced which made the protein phosphorylation-insensitive at serine-4, serine-125, theonine-199, or threonine-234/237, respectively. Contrary to our expectations, we could not see any relocation of NPM1 from the nucleoli into other parts of the cell when the individual phosphorylation sites were “turned off.” This supports the synergistic model of Negi et al. [9] which says that different phosphorylation sites take effect together and alternations of the subcellular distribution are only expected if several phosphorylation sites are mutated. Also, oligomerization of NPM1, which also influences distribution of the protein, is regulated by a stepwise process [63], [64], which supports that localization of NPM1 is influenced by several phosphorylation sites which act in cooperation and mutation of one of them is not sufficient to disrupt normal localization. We cannot completely rule out that the NPM1 tagged with GFP partially formed heterooligomers with the native NPM1, and thus, the localization of the mutated NPM1 was distorted. Nevertheless, transfection with the mutated NPM1 led to a clear increase in the protein level by a factor of 2.6 on average; thus, it is likely that some effect in the localization of the protein should have been detectable if there had been one.
Finally, we studied the distribution of NPM1 within the different cellular compartments after irradiation. Within the first hour after irradiation, we could not observe any dislocation of the protein by live cell imaging. Within the following hours, we saw different cellular reactions upon irradiation within the cell population. Some cells conserved the predominant nucleolar localization of NPM1; in others, the protein was redistributed in the whole nucleus, and in some cells, an increase in cytoplasmic NPM1 could be detected. This shows that the reaction upon irradiation concerning NPM1 is not uniform, and there could not be detected a clear redistribution of NPM1 to the nucleoplasm as it was suggested in case of the response to UV irradiation [21], [24], [73].
All in all, we could show that NPM1 is important for the survival of tumor cells following irradiation, as knockdown of the protein reduced the surviving fraction and the colony size of tumor cells in a colony-forming assay. Additionally, we could demonstrate that NPM1 is dephosphorylated at threonine-199 and threonine-234/237 within 1 hour after gamma irradiation, which is in line with similar findings made observing cellular reaction to UV irradiation [18]. This dephosphorylation could not be associated either with a fast cell cycle arrest or with a uniform redistribution of the protein within the cell. We found a heterogenous response of the tumor cells upon irradiation concerning the NPM1 distribution. This study suggests that NPM1 is regulated by a complex system of phosphorylation and dephosphorylation to switch between the versatile functions of the protein. We hypothesize that NPM1 and its phosphorylation status after irradiation are relevant for the immediate early response of tumor cells upon irradiation and their ability to repair sustained damage and survive the treatment. Thus, we think that further investigation of the cooperative activity of several phosphorylation sites of NPM1 would be rewarding to gain an in-depth understanding of their role in the early irradiation response of tumor cells in the light of irradiation resistances that frequently hamper the success of radiotherapy.
Funding
This work was funded by a grant provided by the Foundation Tumor Research Head and Neck, Wiesbaden, Germany.
Acknowledgments
Acknowledgements
We thank Katrin Gellner for her contribution to experiments within the scope of her doctoral thesis as well as Julia Heim and Cinja Kappel for their contribution as part of their master theses. Furthermore, we are greatly indebted to Simone Mendler and Karin Benz for their excellent technical assistance.
Conflicts of interest statement
None declared.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.10.015.
Appendix A. Supplementary data
Suppl. Fig. S1: Representative Western blots of the short-term measurements of the phosphorylation status of NPM1 in A549 cells after irradiation.
In Figure S1 are shown four representative Western blots that document the phosphorylation of NPM1 in A549 cells at threonine-199, threonine-234/237, serine-4, and serine-125 in nonirradiated control cells and 1 minute, 10 minutes, and 1 hour after irradiation with 8 Gy, as well as the total amount of NPM1 (last row). Each probe (treatment + time point) consists of one nuclear lysate (NL), one total lysate (TL), and one cytoplasmic lysate (CL), and the images are not cropped; all depicted probes were run on one gel.
Suppl. Fig. S2: Representative Western blots of the short-term measurements of the phosphorylation status of NPM1 in HeLa cells after irradiation.
In Figure S2 are shown four representative Western blots that document the phosphorylation of NPM1 in HeLa cells at threonine-199, threonine-234/237, serine-4, and serine-125 in nonirradiated control cells and 1 minute, 10 minutes, and 1 hour after irradiation with 8 Gy, as well as the total amount of NPM1 (last row). Each probe (treatment + time point) consists of one nuclear lysate (NL), one total lysate (TL), and one cytoplasmic lysate (CL), and the images are not cropped; all depicted probes were run on one gel.
Suppl. Fig. S3: Representative Western blots of the long-term measurements of the phosphorylation of NPM1 at threonine-199 in A549, HeLa, and HNSCCUM-02T cells after irradiation.
In Figure S3 are shown three representative Western blots that document the phosphorylation of NPM1 at position threonine-199 in HNSCCUM-02T, HeLa, and A549 cells. The phosphorylation of nonirradiated control cells was compared to the one in cells irradiated with 8 Gy at the indicated time points. In the first row, the phosphorylation of NPM1 at threonine-199 is shown, and in the second row, the total amount of NPM1 is shown. All images are not cropped, and all depicted probes were run on one gel.
References
- 1.Stewart BW, Wild CP. 2015. World Cancer Report 2014. [Google Scholar]
- 2.Deorukhkar A, Krishnan S. Targeting inflammatory pathways for tumor radiosensitization. Biochem Pharmacol. 2010;80(12):1904–1914. doi: 10.1016/j.bcp.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma RA, Plummer R, Stock JK, Greenhalgh TA, Ataman O, Kelly S, Clay R, Adams RA, Baird RD, Billingham L. Clinical development of new drug-radiotherapy combinations. Nat Rev Clin Oncol. 2016;13(10):627–642. doi: 10.1038/nrclinonc.2016.79. [DOI] [PubMed] [Google Scholar]
- 4.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11(4):239–253. doi: 10.1038/nrc3007. [DOI] [PubMed] [Google Scholar]
- 5.Huen MS, Chen J. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 2008;18(1):8–16. doi: 10.1038/cr.2007.109. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Shi X, Paddon H, Hampong M, Dai W, Pelech S. B23/nucleophosmin serine 4 phosphorylation mediates mitotic functions of polo-like kinase 1. J Biol Chem. 2004;279(34):35726–35734. doi: 10.1074/jbc.M403264200. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton G, Abraham AG, Morton J, Sampson O, Pefani DE, Khokorenkova S, Grawenda A, Papaspyropoulos A, Jamieson N, McKay C. AKT regulates NPM dependent ARF localization and p53mut stability in tumors. Oncotarget. 2014;5(15):6142–6167. doi: 10.18632/oncotarget.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M, Dereli-Oz A, Kocylowski M, Pateras IS, Evangelou K. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol. 2013;15(8):967–977. doi: 10.1038/ncb2795. [DOI] [PubMed] [Google Scholar]
- 9.Negi SS, Olson MOJ. Effects of interphase and mitotic phosphorylation on the mobility and location of nucleolar protein B23. J Cell Sci. 2006;119(17):3676–3685. doi: 10.1242/jcs.03090. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Budhu A, Forgues M, Wang XW. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat Cell Biol. 2005;7(8):823–830. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- 11.Okuwaki M, Tsujimoto M, Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol Biol Cell. 2002;13(6):2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. PNAS. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 14.Ramos-Echazábal G, Chinea G, García-Fernández R, Pons T. In silico studies of potential phosphoresidues in the human nucleophosmin/B23: its kinases and related biological processes. J Cell Biochem. 2012;113(7):2364–2374. doi: 10.1002/jcb.24108. [DOI] [PubMed] [Google Scholar]
- 15.Lindström MS. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int. 2011;2011(195209):1–16. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437(7055):147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 17.Okuda M. The role of nucleophosmin in centrosome duplication. Oncogene. 2002;21(40):6170–6174. doi: 10.1038/sj.onc.1205708. [DOI] [PubMed] [Google Scholar]
- 18.Lin CY, Tan BC-M, Liu H, Shih C-J, Chien K-Y, Lin C-L, Yung BY-M. Dephosphorylation of nucleophosmin by PP1beta facilitates pRB binding and consequent E2F1-dependent DNA repair. Mol Biol Cell. 2010;21(24):4409–4417. doi: 10.1091/mbc.E10-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2012;506(3):272–276. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- 20.Swaminathan V, Kishore AH, Febitha KK, Kundu TK. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol Cell Biol. 2005;25(17):7534–7545. doi: 10.1128/MCB.25.17.7534-7545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22(22):6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekhar KR, Reddy YT, Reddy PN, Crooks PA, Venkateswaran A, McDonald WH, Geng L, Sasi S, van der Waal RP, Roti JLR. The novel chemical entity YTR107 inhibits recruitment of nucleophosmin to sites of DNA damage, suppressing repair of DNA double-strand breaks and enhancing radiosensitization. Clin Cancer Res. 2011;17(20):6490–6499. doi: 10.1158/1078-0432.CCR-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koike A, Nishikawa H, Wu W, Okada Y, Venkitaraman AR, Ohta T. Recruitment of phosphorylated NPM1 to sites of DNA damage through RNF8-dependent ubiquitin conjugates. Cancer Res. 2010;70(17):6746–6756. doi: 10.1158/0008-5472.CAN-10-0382. [DOI] [PubMed] [Google Scholar]
- 24.Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala P, Meek D, Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5(5):465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 25.Poletto M, Lirussi L, Wilson DM, Tell G. Nucleophosmin modulates stability, activity and nucleolar accumulation of base excision repair proteins. Mol Biol Cell. 2014;25(10):1641–1652. doi: 10.1091/mbc.E13-12-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanderwaal RP, Maggi LB, Weber JD. Nucleophosmin redistribution following heat shock: a role in heat-induced radiosensitization in heat-induced radiosensitization. Cancer Res. 2009;69(16):6454–6462. doi: 10.1158/0008-5472.CAN-08-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu G-Y, Shi J-X, Shi S-L, Liu F, Rui G, Li X, Gao L-B, Deng X-L, Li Q-F. Nucleophosmin regulates intracellular oxidative stress homeostasis via antioxidant PRDX6. J Cell Biochem. 2017;118(12):4697–4707. doi: 10.1002/jcb.26135. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Zhang X, Sejas DP, Bagby GC, Pang Q. Hypoxia-induced nucleophosmin protects cell death through inhibition of p53. J Biol Chem. 2004;279(40):41275–41279. doi: 10.1074/jbc.C400297200. [DOI] [PubMed] [Google Scholar]
- 29.Qi W, Shakalya K, Stejskal A, Goldman A, Beeck S, Cooke L, Mahadevan D. NSC348884, a nucleophosmin inhibitor disrupts oligomer formation and induces apoptosis in human cancer cells. Oncogene. 2008;27(30):4210–4220. doi: 10.1038/onc.2008.54. [DOI] [PubMed] [Google Scholar]
- 30.Jian Y, Gao Z, Sun J, Shen Q, Feng F, Jing Y, Yang C. RNA aptamers interfering with nucleophosmin oligomerization induce apoptosis of cancer cells. Oncogene. 2009;28(47):4201–4211. doi: 10.1038/onc.2009.275. [DOI] [PubMed] [Google Scholar]
- 31.Sekhar KR, Benamar M, Venkateswaran A, Sasi S, Penthala NR, Crooks PA, Hann SR, Geng L, Balusu R, Abbas T. Targeting nucleophosmin 1 represents a rational strategy for radiation sensitization. Int J Radiat Oncol Biol Phys. 2014;89(5):1106–1114. doi: 10.1016/j.ijrobp.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naoe T, Suzuki T, Kiyoi H, Urano T. Nucleophosmin: a versatile molecule associated with hematological malignancies. Cancer Sci. 2006;97(10):963–969. doi: 10.1111/j.1349-7006.2006.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colombo E, Alcalay M, Pelicci PG. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene. 2011;30(23):2595–2609. doi: 10.1038/onc.2010.646. [DOI] [PubMed] [Google Scholar]
- 34.Sportoletti P, Grisendi S, Majid SM, Cheng K, Clohessy JG, Viale A, Teruya-Feldstein J, Pandolfi PP. Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood. 2008;111(7):3859–3862. doi: 10.1182/blood-2007-06-098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan WY, Liu QR, Borjigin J, Busch H, Rennert OM, Tease LA, Chan PK. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry. 1989;28(3):1033–1039. doi: 10.1021/bi00429a017. [DOI] [PubMed] [Google Scholar]
- 36.Feuerstein N, Chan PK, Mond JJ. Identification of numatrin, the nuclear matrix protein associated with induction of mitogenesis, as the nucleolar protein B23: implications for the role of the nucleus in early transduction of mitogenic signals. J Biol Chem. 1988;263(22):10608–10612. [PubMed] [Google Scholar]
- 37.Feuerstein N, Spiegel S, Mond JJ. The nuclear matrix protein, numatrin (B23) is associated with growth factor-induced mitogenesis in Swiss 3T3 fibroblasts and with T lymphocyte proliferation stimulated by lectins and anti-T cell antigen receptor antibody. J Cell Biol. 1988;107(5):1629–1642. doi: 10.1083/jcb.107.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6(7):493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Sun Z, Liu K, Qiu W, Yao R, Feng T, Xin C, Yue L. Prognostic significance of the co-expression of nucleophosmin and trefoil factor 3 in postoperative gastric cancer patients. Mol Clin Oncol. 2014;2(6):1055–1061. doi: 10.3892/mco.2014.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y-F, Zhang X-Y, Yang M, He Z-H, Peng N-F, Xie S-R, Xie Y-F. Prognostic role of nucleophosmin in colorectal carcinomas. Asian Pac J Cancer Res. 2014;15(5):2021–2026. doi: 10.7314/apjcp.2014.15.5.2021. [DOI] [PubMed] [Google Scholar]
- 41.Subong ENP, Shue MJ, Epstein JI, Briggman JV, Chan PK, Partin AW. Monoclonal antibody to prostate cancer nuclear matrix protein (PRO:4-216) recognizes nucleophosmin/B23. Prostate. 1999;39(4):298–304. doi: 10.1002/(sici)1097-0045(19990601)39:4<298::aid-pros11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Yuan G, Zhao B, Zhao Y, Qiu Y. High expression of B23 is associated with tumorigenesis and poor prognosis in bladder urothelial carcinoma. Mol Med Rep. 2017;15(2):743–749. doi: 10.3892/mmr.2016.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernard K, Litman E, Fitzpatrick JL, Shellman YG, Argast G, Polvinen K, Everett AD, Fukasawa K, Norris DA, Ahn NG. Functional proteomic analysis of melanoma progression. Cancer Res. 2003;63(20):6716–6725. [PubMed] [Google Scholar]
- 44.Gollapalli K, Ghantasala S, Atak A, Rapole S, Moiyadi A, Epari S, Srivastava S. Tissue proteome analysis of different grades of human gliomas provides major cues for glioma pathogenesis. OMICS. 2017;21(5):275–284. doi: 10.1089/omi.2017.0028. [DOI] [PubMed] [Google Scholar]
- 45.Yun J-P, Miao J, Chen GG, Tian Q-H, Zhang C-Q, Xiang J, Fu J, Lai PBS. Increased expression of nucleophosmin/B23 in hepatocellular carcinoma and correlation with clinicopathological parameters. Br J Cancer. 2007;96(3):477–484. doi: 10.1038/sj.bjc.6603574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leal MF, Mazzotti TKF, Calcagno DQ, Cirilo PDR, Martinez MC, Demachki S, Assumpcao PP, Chammas R, Burbano RR, Smith MC. Deregulated expression of Nucleophosmin 1 in gastric cancer and its clinicopathological implications. BMC Gastroenterol. 2014;14(9):1–10. doi: 10.1186/1471-230X-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karhemo P-RR, Rivinoja A, Lundin J, Hyvönen M, Chernenko A, Lammi J, Sihto H, Lundin M, Heikkilä P, Joensuu H. An extensive tumor array analysis supports tumor suppressive role for nucleophosmin in breast cancer. Am J Pathol. 2011;179(2):1004–1014. doi: 10.1016/j.ajpath.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welkoborsky H-J, Jacob R, Riazimand SH, Bernauer HS, Mann WJ. Molecular biologic characteristics of seven new cell lines of squamous cell carcinomas of the head and neck and comparison to fresh tumor tissue. Oncology. 2003;65(1):60–71. doi: 10.1159/000071206. [DOI] [PubMed] [Google Scholar]
- 49.Drigotas M, Affolter A, Mann WJ, Brieger J. Reactive oxygen species activation of MAPK pathway results in VEGF upregulation as an undesired irradiation response. J Oral Pathol Med. 2013;42(8):612–619. doi: 10.1111/jop.12056. [DOI] [PubMed] [Google Scholar]
- 50.Thiel UJE, Feltens R, Adryan B, Gieringer R, Brochhausen C, Schuon R, Fillies T, Grus F, Mann WJ, Brieger J. Analysis of differentially expressed proteins in oral squamous cell carcinoma by MALDI-TOF MS. J Oral Pathol Med. 2011;40(5):369–379. doi: 10.1111/j.1600-0714.2010.00982.x. [DOI] [PubMed] [Google Scholar]
- 51.Affolter A, Fruth K, Brochhausen C, Schmidtmann I, Mann WJ, Brieger J. Activation of mitogen-activated protein kinase extracellular signal-related kinase in head and neck squamous cell carcinomas after irradiation as part of a rescue mechanism. Head Neck. 2011;33(10):1448–1457. doi: 10.1002/hed.21623. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki K, Bose P, Leong-Quong RYY, Fujita DJ, Riabowol K. REAP: a two minute cell fractionation method. BMC Res Notes. 2010;3(294) doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bier C, Knauer SK, Docter D, Schneider G, Krämer OH, Stauber RH. The Importin-alpha/nucleophosmin switch controls Taspase1 protease function. Traffic. 2011;12(6):703–714. doi: 10.1111/j.1600-0854.2011.01191.x. [DOI] [PubMed] [Google Scholar]
- 54.Wang D, Umekawa H, Olson M. Expression and subcellular locations of two forms of nucleolar protein B23 in rat tissues and cells. Cell Mol Biol Res. 1993;39(1):33–42. [PubMed] [Google Scholar]
- 55.Frehlick LJ, Eirín-López JM, Ausió J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29(1):49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- 56.Penthala NR, Crooks PA, Freeman ML, Sekhar KR. Development and validation of a novel assay to identify radiosensitizers that target nucleophosmin 1. Bioorg Med Chem. 2015;23(13):3681–3686. doi: 10.1016/j.bmc.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groisman R, Polanowska J, Kuraoka I, Sawada J-i, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell Biochem Biophys. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 58.Wiesmann N, Strozynski J, Beck C, Zimmermann N, Mendler S, Gieringer R, Schmidtmann I, Brieger J. Knockdown of hnRNPK leads to increased DNA damage after irradiation and reduces survival of tumor cells. Carcinogenesis. 2017;38(3):321–328. doi: 10.1093/carcin/bgx006. [DOI] [PubMed] [Google Scholar]
- 59.Leroy B, Girard L, Hollestelle A, Minna JD, Gazdar AF, Soussi T. Analysis of TP53 mutation status in human cancer cell lines: A reassessment. Hum Mutat. 2014;35(6):756–765. doi: 10.1002/humu.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hisaoka M, Nagata K, Okuwaki M. Intrinsically disordered regions of nucleophosmin/B23 regulate its RNA binding activity through their inter- and intra-molecular association. Nucleic Acids Res. 2013;42(2):1180–1195. doi: 10.1093/nar/gkt897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yun J-P, Chew EC, Liew C-T, Chan JYH, Jin M-L, Ding M-X, Fai YH, Li HKR, Liang X-M, Wu Q-L. Nucleophosmin/B23 is a proliferate shuttle protein associated with nuclear matrix. J Cell Biochem. 2003;90(6):1140–1148. doi: 10.1002/jcb.10706. [DOI] [PubMed] [Google Scholar]
- 62.Zatsepina OV, Rousselet A, Chan PK, Olson MOJ, Jordan EG. The nucleolar phosphoprotein B23 redistributes in part to the spindle poles during mitosis. J Cell Sci. 1999;112(4):455–466. doi: 10.1242/jcs.112.4.455. [DOI] [PubMed] [Google Scholar]
- 63.Mitrea DM, Kriwacki R. Cryptic disorder: an order-disorder transformation regulates the function of nucleophosmin. Pac Symp Biocomput. 2012:152–163. [PubMed] [Google Scholar]
- 64.Mitrea DM, Grace CR, Buljan M, Yun M-K, Pytel NJ, Satumba J, Nourse A, Park C-G, Madan Babu M, White SW. Structural polymorphism in the N-terminal oligomerization domain of NPM1. PNAS. 2014;111(12):4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kozakai Y, Kamada R, Furuta J, Kiyota Y, Chuman Y, Sakaguchi K. PPM1D controls nucleolar formation by up-regulating phosphorylation of nucleophosmin. Sci Rep. 2016;6 doi: 10.1038/srep33272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu MH, Yung BYM. UV stimulation of nucleophosmin/B23 expression is an immediate-early gene response induced by damaged DNA. J Biol Chem. 2002;277(50):48234–48240. doi: 10.1074/jbc.M206550200. [DOI] [PubMed] [Google Scholar]
- 67.Wu MH, Chang JH, Chou CC, Yung Benjamin YM. Involvement of nucleophosmin/B23 in the response of HeLa cells to UV irradiation. Int J Cancer. 2002;97(3):297–305. doi: 10.1002/ijc.1606. [DOI] [PubMed] [Google Scholar]
- 68.Poletto M, Malfatti MC, Dorjsuren D, Scognamiglio PL, Marasco D, Vascotto C, Jadhav A, Maloney DJ, Wilson DM, Simeonov A. Inhibitors of the apurinic/apyrimidinic endonuclease 1 (APE1)/nucleophosmin (NPM1) interaction that display anti-tumor properties. Mol Carcinog. 2015;55(5):688–704. doi: 10.1002/mc.22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ando K, Parsons MJ, Shah RB, Charendoff CI, Paris SL, Liu PH, Fassio SR, Rohrman BA, Thompson R, Oberst A. NPM1 directs PIDDosome-dependent caspase-2 activation in the nucleolus. J Cell Biol. 2017;216(6):1795–1810. doi: 10.1083/jcb.201608095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wesierska-Gadek J, Schloffer D, Kotala V, Horky M. Escape of p53 protein from E6-mediated degradation in HeLa cells after cisplatin therapy. Int J Cancer. 2002;101(2):128–136. doi: 10.1002/ijc.10580. [DOI] [PubMed] [Google Scholar]
- 71.Peter M, Nakagawa J, Dorée M, Labbé JC, Nigg EA. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell. 1990;60(5):791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- 72.Hisaoka M, Ueshima S, Murano K, Nagata K, Okuwaki M. Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Mol Cell Biol. 2010;30(20):4952–4964. doi: 10.1128/MCB.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kurki S, Latonen L, Laiho M. Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization. J Cell Sci. 2003;116(Pt 19):3917–3925. doi: 10.1242/jcs.00714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. S1: Representative Western blots of the short-term measurements of the phosphorylation status of NPM1 in A549 cells after irradiation.
In Figure S1 are shown four representative Western blots that document the phosphorylation of NPM1 in A549 cells at threonine-199, threonine-234/237, serine-4, and serine-125 in nonirradiated control cells and 1 minute, 10 minutes, and 1 hour after irradiation with 8 Gy, as well as the total amount of NPM1 (last row). Each probe (treatment + time point) consists of one nuclear lysate (NL), one total lysate (TL), and one cytoplasmic lysate (CL), and the images are not cropped; all depicted probes were run on one gel.
Suppl. Fig. S2: Representative Western blots of the short-term measurements of the phosphorylation status of NPM1 in HeLa cells after irradiation.
In Figure S2 are shown four representative Western blots that document the phosphorylation of NPM1 in HeLa cells at threonine-199, threonine-234/237, serine-4, and serine-125 in nonirradiated control cells and 1 minute, 10 minutes, and 1 hour after irradiation with 8 Gy, as well as the total amount of NPM1 (last row). Each probe (treatment + time point) consists of one nuclear lysate (NL), one total lysate (TL), and one cytoplasmic lysate (CL), and the images are not cropped; all depicted probes were run on one gel.
Suppl. Fig. S3: Representative Western blots of the long-term measurements of the phosphorylation of NPM1 at threonine-199 in A549, HeLa, and HNSCCUM-02T cells after irradiation.
In Figure S3 are shown three representative Western blots that document the phosphorylation of NPM1 at position threonine-199 in HNSCCUM-02T, HeLa, and A549 cells. The phosphorylation of nonirradiated control cells was compared to the one in cells irradiated with 8 Gy at the indicated time points. In the first row, the phosphorylation of NPM1 at threonine-199 is shown, and in the second row, the total amount of NPM1 is shown. All images are not cropped, and all depicted probes were run on one gel.