Abstract
The action mechanism of cyclopyrimorate, a novel herbicide for weed control in rice fields, was investigated. Cyclopyrimorate caused bleaching symptoms in Arabidopsis thaliana similar to those caused by existing carotenoid biosynthesis inhibitors, mesotrione and norflurazon. However, cyclopyrimorate treatment resulted in significant accumulation of homogentisate and a reduction in the level of plastoquinone. A metabolite of cyclopyrimorate, des-morpholinocarbonyl cyclopyrimorate (DMC), was detected in plants. These data suggested that cyclopyrimorate and/or DMC inhibit homogentisate solanesyltransferase (HST), a downstream enzyme of 4-hydroxyphenylpyruvate dioxygenase in the plastoquinone biosynthesis pathway. In vitro assays showed that A. thaliana HST was strongly inhibited by DMC and weakly by cyclopyrimorate, whereas other commercial bleaching herbicides did not inhibit HST. DMC derivatives showed a positive correlation between HST inhibition and in vivo bleaching activities. These results indicate that the target site of cyclopyrimorate and DMC is HST, a novel target site of commercial herbicides.
Keywords: carotenoid biosynthesis inhibitor, plastoquinone biosynthesis inhibitor, homogentisate solanesyltransferase, cyclopyrimorate, bleaching herbicide
Introduction
Herbicide-resistant weeds first became problematic in the USA and Europe in the 1970s and early 1980s.1) To date, weeds have evolved resistance to 23 of the 26 known herbicide sites of action and to 163 different herbicides.2) Weeds resistant to acetolactate synthase (ALS) inhibitor are the most problematic in wheat, corn, soybean, and rice production.1,2) Development of new herbicides with a novel mode of action, which have not been introduced in more than two decades, is needed to manage the evolution of weeds resistant to existing herbicides.3)
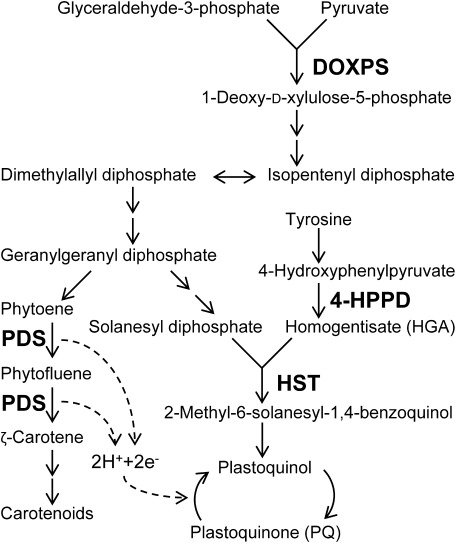
According to the Herbicide Resistance Action Committee classification, bleaching herbicides are categorized into four groups. Their target sites are related to carotenoid or plastoquinone (PQ) biosynthesis (Fig. 1). Most bleaching herbicides target phytoene desaturase (PDS) and 4-hydroxyphenylpyruvate dioxygenase (4-HPPD).4) PDS inhibitors block the desaturation of phytoene, which directly affects carotenoid biosynthesis.5) 4-HPPD inhibitors block the formation of homogentisate (HGA) in the biosynthesis pathway of PQ, a cofactor for PDS, and lead to indirect inhibition of its enzymatic activity.6,7) Because carotenoids protect plant cells from photooxidation, their loss leads to the destruction of chlorophyll8) and photobleaching symptoms.5) The bleaching herbicide clomazone inhibits the 1-deoxy-D-xylulose-5-phosphate synthase (DOXPS), which catalyzes the first committed step in the biosynthesis of isoprenoids, including carotenoids and PQ.9,10)
Fig. 1. Carotenoids and plastoquinone biosynthesis pathways. Target enzymes of inhibitors are indicated in bold. Two consecutive arrows indicate multi-step reactions. DOXPS, 1-deoxy-D-xylulose-5-phosphate synthase; PDS, phytoene desaturase; 4-HPPD, 4-hydroxyphenylpyruvate dioxygenase; HST, homogentisate solanesyltransferase.
It was reported in 2010 that haloxydine inhibits homogentisate solanesyltransferase (HST).11) HST catalyzes the prenylation and decarboxylation of HGA to form 2-methyl-6-solanesyl-1,4-benzoquinol in the PQ biosynthesis pathway. The in vitro assay systems were established with the HST gene of Chlamydomonas reinhardtii and Arabidopsis thaliana. Using the in vitro assay system of C. reinhardtii, haloxydine has been demonstrated to be a competitive inhibitor of homogentisate.11,12) Nevertheless, haloxydine has not yet been used commercially.
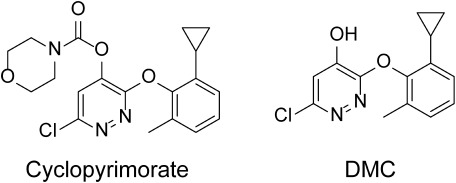
Cyclopyrimorate, 6-chloro-3-(2-cyclopropyl-6-methylphenoxy) pyridazin-4-yl morpholine-4-carboxylate, was invented by Mitsui Chemicals Agro Inc.(Fig. 2) and has proven to be highly effective against weeds in rice fields, including those resistant to ALS inhibitors. Cyclopyrimorate shows synergistic effects with 4-HPPD inhibitors, such as pyrazolynate.13–17) However, the target site of cyclopyrimorate is unknown. Identification of its target site will contribute to the establishment of novel strategies for the management of herbicide-resistant weeds. In this study, the levels of intermediates in PQ and carotenoid biosynthesis pathways were examined after cyclopyrimorate treatment. Escherichia coli cells expressing the A. thaliana HST gene were used to determine the HST inhibitory activities of various herbicides. Using des-morpholinocarbonyl cyclopyrimorate (DMC), a metabolite of cyclopyrimorate (Fig. 2), and its derivatives, we evaluated HST inhibitory activities and in vivo bleaching activities to verify their target site.
Fig. 2. Structures of cyclopyrimorate and its metabolite, des-morpholinocarbonyl cyclopyrimorate (DMC).
Materials and Methods
1. Chemicals and plant materials
Cyclopyrimorate, DMC and its derivatives, mesotrione (4-HPPD inhibitor),18) norflurazon (PDS inhibitor),19) and clomazone were synthesized at Agrochemicals Research Center, Mitsui Chemicals Agro, Inc. (Chiba, Japan). 2-Methyl-6-farnesyl-1,4-benzoquinol (MFBQH) was also synthesized there according to the procedures described previously.20,21) Haloxydine was purchased from Namiki Shoji Co., Ltd. (Tokyo, Japan). Hyponex was purchased from HYPONeX JAPAN Co., Ltd. (Osaka, Japan). Iron (III) chloride, L-ascorbic acid, and magnesium chloride hexahydrate were purchased from Wako Pure Chemical Industries Co., Ltd. (Osaka, Japan). L-Arabinose, sodium borohydride, and HGA were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Luria–Bertani broth (LB) was purchased from Becton, Dickinson and Company (NJ, USA). Farnesyl diphosphate and seeds of A. thaliana were purchased from Funakoshi Co., Ltd. (Tokyo, Japan). Seeds of Brassica juncea were purchased from Takii Seed Co., Ltd. (Kyoto, Japan). Seeds of Schoenoplectus juncoides were collected at the Agrochemicals Research Center, Mitsui Chemicals Agro, Inc. (Shiga, Japan).
2. Plant cultivation and herbicide treatment
Seeds of A. thaliana were sown on 10 mL of 1% agar media (w/v) containing 0.1% Hyponex (v/v) in a Petri dish (60 mm dia.×15 mm depth) and treated with 500 µL of herbicide solution containing 1% DMSO. Plates were incubated in a chamber with an 8 hr light (22°C)/16 hr dark (20°C) cycle for 4–5 days. Seeds of B. juncea were sown in a plastic pot (45 mm×55 mm) filled with an approximately 1 : 2 mixture of culture soil (NO3-N, PO4, K, Mg=550, 1500, 200, 100 mg/L, respectively; pH=6–7) and sieved and autoclaved soil collected from a field at the Agrochemicals Research Center, Mitsui Chemicals Agro, Inc. (Ibaraki, Japan). Immediately after sowing, 1 mL of herbicide solution containing 1% DMSO was added to the soil surface. Pots were incubated in a chamber with a 16 hr light (25°C)/8 hr dark (22°C) cycle for 4 days. Seeds of S. juncoides were sown on 10 mL of 0.5% agar media (w/v) containing 0.1% Hyponex (v/v) in a glass cylinder (35 mm×60 mm) and treated with 280 µL of herbicide solution containing 1% DMSO. Cylinders were incubated in a chamber with a 16 hr light (26°C)/8 hr dark (22°C) cycle for 4 days. For appropriate controls, 1% DMSO without the herbicides was applied to the seeds of each plant species.
3. HGA extraction and analysis
Whole plants of A. thaliana, leaves of B. juncea, and shoots of S. juncoides were homogenized in 500 µL of MeOH and 40 µL of 1 M L-ascorbic acid using a Biomasher II (Nippi Inc., Tokyo, Japan). After the addition of 500 µL of MeOH, the homogenate was centrifuged at 17,360×g for 5 min. Supernatants were dried in a centrifugal concentrator and dissolved in 400 µL of 50 mM KH2PO4–HCl buffer (pH 2.8). The solution was washed with 400 µL of n-hexane/ethyl acetate (6 : 4). After NaCl was added to saturation, the solution was extracted with 500 µL of ethyl acetate twice. Ethyl acetate solution was dried in a centrifugal concentrator, and the residue was resuspended in 100 µL of 50 mM KH2PO4–HCl buffer (pH 2.8). The solution was centrifuged at 17,360×g for 5 min, and the supernatant was subjected to HPLC analysis. The HPLC system consisted of a pump (LC-20AD), a UV detector (SPD-M20A), a fluorescence detector (RF-20Axs), and a column oven (CTO-20AC). Phase separation was performed on a reverse phase column (X-bridge, 4.6 mm ×15 mm, particle size 5 µm; Waters Co., MA, USA). The column oven temperature was maintained at 40°C. The mobile phase consisted of acetonitrile and 0.2% triethylamine-phosphoric acid buffer (pH 3.2) in a ratio of 0.5 : 99.5 (v/v). The flow rate was maintained at 1.4 mL/min, and the effluent was monitored using the fluorescence detector (Ex. 290 nm, Em. 330 nm). HGA was eluted with a retention time of 7.1 min. The concentration of HGA was determined using external standards and expressed on a fresh weight basis.
4. PQ extraction and analysis
Whole plants of A. thaliana, leaves of B. juncea, and shoots of S. juncoides were homogenized in 500 µL of ice-cold acetone and 5 µL of 0.5 M iron (III) chloride in EtOH using a Biomasher II. Samples were centrifuged at 17,360×g for 5 min, followed by the addition of 500 µL of water to the supernatant. The solution was extracted with 500 µL of n-hexane twice. The hexane solution was dried in a centrifugal concentrator, and the residue was resuspended in 100 µL of EtOH. The solution was centrifuged at 17,360×g for 5 min, and the supernatant was subjected to HPLC analysis. The column oven temperature was maintained at 40°C. MeOH was used as the mobile phase at a flow rate of 1.0 mL/min. The effluent was monitored using the UV detector at a wavelength of 254 nm. PQ was eluted with a retention time of 23.5 min. The concentration was expressed on a fresh weight basis.
5. Extraction and analysis of cyclopyrimorate and DMC from plants
Arabidopsis leaves were homogenized in 180 µL of 80% MeOH and 20 µL of 1 M L-ascorbic acid using a Biomasher II. Samples were centrifuged at 17,360×g for 5 min, followed by the addition of 50 µL of 50 mM KH2PO4–HCl buffer (pH 2.8) to 50 µL of the supernatant. Samples were centrifuged at 17,360×g for 5 min, and the supernatant was subjected to HPLC analysis. The column oven temperature was maintained at 40°C. The mobile phase consisted of acetonitrile and 0.2% triethylamine-phosphoric acid buffer (pH 3.2) in gradient mode as follows: 30%–100% acetonitrile in 20 min. The flow rate was maintained at 1.0 mL/min. The effluent was monitored using the UV detector. Cyclopyrimorate and DMC were eluted with a retention time of 13.8 min and 7.5 min at a wavelength of 272 nm and 277 nm, respectively. Concentrations of cyclopyrimorate and DMC were determined using external standards and expressed on a fresh weight basis.
6. Cloning and heterologous expression of A. thaliana HST
Cloning and heterologous expression of A. thaliana HST were performed as described.11,12,22) Briefly, peptide sequences of A. thaliana HST (accession number AT3G11945) lacking putative chloroplast targeting sequences were cloned into a pDEST15 vector (Invitrogen) according to the manufacturer’s instructions to generate a functional A. thaliana protein with an N-terminal glutathione S-transferase (GST) tag. The vectors were transformed into E. coli BL21-AI cells (Invitrogen) and incubated at 37°C in LB medium supplemented with ampicillin (100 µg/mL). Expression of A. thaliana HST was induced in exponentially growing cultures by adding L-arabinose to a final concentration of 0.2%. After 2 hr of induction, cells were harvested by centrifugation at 12,000×g at 4°C for 10 min and stored at −80°C. The frozen cells were resuspended in an extraction buffer (10 mM Tris–H2SO4, 1 mM phenylmethylsulfonylfluoride, pH 7.5) and homogenized with sonication on ice. The cells were centrifuged at 48,000×g at 4°C for 10 min. The supernatant was ultracentrifuged at 150,000×g at 4°C for 1 hr. The pellet was resuspended in a suspension buffer (5 mM Tris–H2SO4, 50% (v/v) glycerol, pH 7.5). Protein concentration was determined using the Bio-Rad assay (Bio-Rad Laboratories, Inc., CA, USA) according to the manufacturer's instructions. This protein suspension was adjusted to 15 mg/mL protein and used for in vitro HST assay.
7. In vitro HST assays
HST activity was measured using farnesyl diphosphate as the substrate as described previously.11,12,22) Eighty microliters of reaction mixture containing 50 mM tricine–NaOH buffer (pH 8.5), 25 µM homogentisate, 100 µM farnesyl diphosphate, 20 mM magnesium chloride, approximately 1.5 mg/mL protein suspension, and various concentrations of herbicides dissolved in 1% DMSO (or no-herbicide control) was incubated at 28°C for 30 min. In the case of cyclopyrimorate, the incubation time was shortened to 10 min to avoid the decomposition of cyclopyrimorate to DMC. After incubation, 80 µL of 0.05% sodium borohydride in EtOH and 32 µL of 0.1 M acetic acid were added to each sample. Subsequently, samples were centrifuged at 17,360×g for 5 min, and the supernatant was subjected to HPLC analysis. The column oven temperature was maintained at 40°C. The mobile phase consisted of acetonitrile and 0.1% acetic acid in a ratio of 65 : 35 (v/v). The flow rate was maintained at 1.0 mL/min, and the effluent was monitored using the fluorescence detector (Ex. 290 nm, Em. 330 nm). The product, MFBQH, was eluted with a retention time of 12.8 min and identified by LC-MS/MS. The inhibition rate was calculated as (1−T/C)*100, where C and T present the amount of MFBQH in control and herbicide treatment samples, respectively. The IC50 (half-inhibition concentration) values for HST were determined using a four-parameter logistic curve-fitting program (GraphPad Prism 6.00) with upper and lower constraints of 100% and 0% extension, respectively.
8. Determination of chlorophyll
Arabidopsis whole plants treated with various herbicide solutions were homogenized in dimethylformamide using a Biomasher II and stored at 4°C for 24 hr under dark conditions. Total chlorophyll content, including chlorophyll a and chlorophyll b, was determined using the following equation23):
where A646.8 and A663.8 represent the absorbance of extracts determined using a photometer at a wavelength of 646.8 and 663.8, respectively. The concentrations were expressed on a fresh weight basis. The EC50 (half-effective concentration) values for chlorophyll content reduction were determined using the four-parameter logistic curve-fitting program (GraphPad Prism 6.00) with upper and lower constraints of 100% and 0% extension, respectively. Bleaching activities were evaluated based on the EC50 values for chlorophyll.
Results
1. Bleaching symptoms of A. thaliana
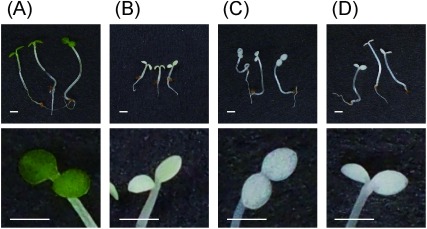
Five days after sowing, Arabidopsis seedlings grew normally with green cotyledons in the no-herbicide control (Fig. 3A), whereas leaves of seedlings treated with 500 ppm cyclopyrimorate solution were bleached (Fig. 3B). These bleaching symptoms were comparable to those observed with major carotenoid biosynthesis inhibitors, such as mesotrione and norflurazon (Fig. 3C and D). Furthermore, the lengths of shoots and roots treated by these herbicides were shorter than those of the control.
Fig. 3. Effect of herbicide treatment (500 ppm) on Arabidopsis seedlings. (A) Control, (B) cyclopyrimorate, (C) mesotrione, and (D) norflurazon. Scale bar=1 mm. The upper panel shows whole seedlings, whereas the lower panel shows the expanded leaves of each seedling. Images were taken 5 days after sowing.
2. Intermediates in the PQ biosynthesis pathway
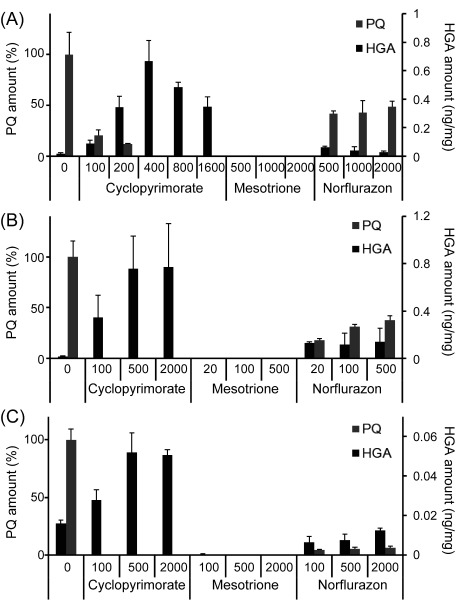
Five days after cyclopyrimorate treatment, the level of PQ in A. thaliana decreased in a concentration-dependent manner; no PQ was detected at cyclopyrimorate concentrations ≥400 ppm (Fig. 4A). Cyclopyrimorate treatment also resulted in the accumulation of HGA in A. thaliana. By contrast, no PQ or HGA was detected in plants treated with mesotrione. In the case of norflurazon, the changing pattern was somewhat similar to that of cyclopyrimorate, however PQ was still detected even at high concentrations, and the increase of HGA was moderate as compared with cyclopyrimorate. These changes in PQ and HGA were also observed in B. juncea and S. juncoides (Fig. 4B and C).
Fig. 4. The amount of plastoquinone (PQ) and homogentisate (HGA) in various plants 5 days after herbicide treatment. (A) A. thaliana seedlings, (B) B. juncea leaves, and (C) S. juncoides shoots. The concentration of each herbicide (ppm) is indicated along the X-axes. Data for PQ are normalized relative to the control, and data for HGA are expressed on a fresh weight basis. Values represent the mean±SD (n=3).
3. Metabolite of cyclopyrimorate
DMC was detected in Arabidopsis leaves 4 days after cyclopyrimorate treatment. The amount of DMC was nearly proportional to the treated concentration of cyclopyrimorate. The concentration of cyclopyrimorate was higher than that of DMC (Fig. 2 and Table 1).
Table 1. Concentrations of cyclopyrimorate and DMC in Arabidopsis leaves 4 days after cyclopyrimorate treatmenta)a).
| Treatment Cyclopyrimorate (ppm) | Concentration in leaves (pmol/mg fresh weight) | |
|---|---|---|
| DMC | Cyclopyrimorate | |
| 0 | 0 | 0 |
| 7.8 | 1.50±1.82 | 7.81±6.78 |
| 31.3 | 4.01±0.30 | 80.1±28.3 |
| 125 | 13.6±3.59 | 421±102 |
a) Values represent the mean±SD (n=3).
4. In vitro HST assays
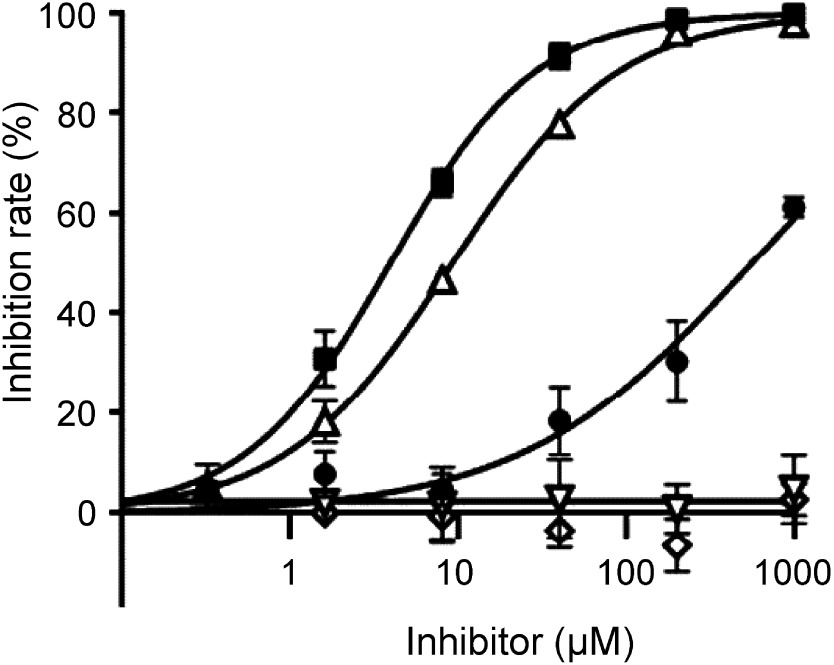
HST in crude E. coli extracts was weakly inhibited by cyclopyrimorate and strongly inhibited by DMC. DMC showed an IC50 of 3.93 µM and an inhibition rate of 99% at a concentration of 200 µM. The IC50 of haloxydine, a known HST inhibitor, was 9.19 µM, indicating that the inhibitory activity of DMC was higher than that of haloxydine. By contrast, mesotrione and norflurazon did not inhibit HST even at the concentration as high as 1 mM (Fig. 5).
Fig. 5. Effect of various herbicides on A. thaliana HST. ●, cyclopyrimorate; ■, DMC; △, haloxydine; ▽, mesotrione; ◇, norflurazon. Values represent the mean±SD (n=3).
5. Relationship between HST inhibitory activities and in vivo bleaching activities
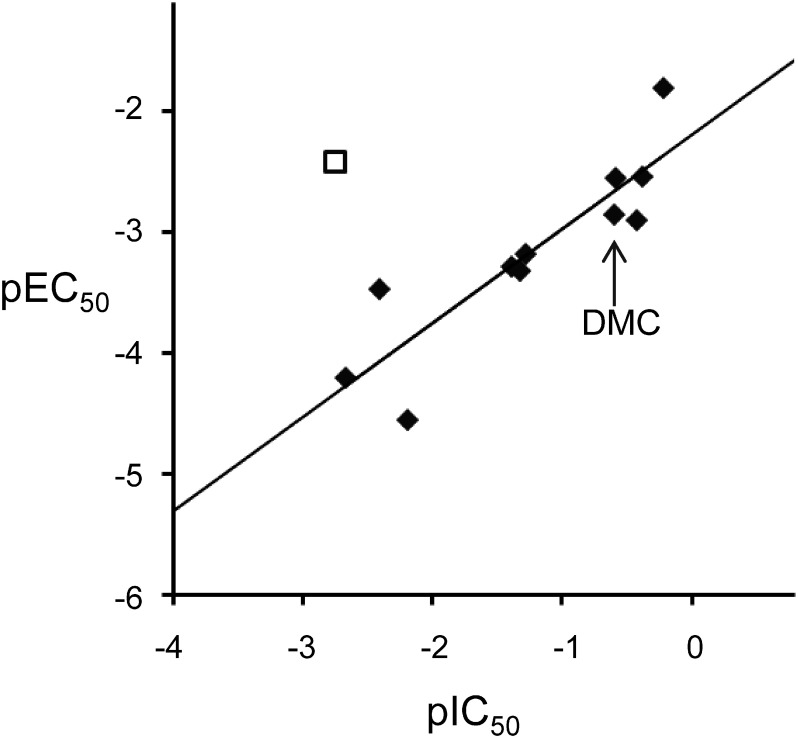
The inhibitory activities of cyclopyrimorate, DMC, and its derivatives (1–10) are listed in Table 2. DMC was one of the strongest HST inhibitors. Compounds possessing a methyl, an ethyl, or a bromo group at position X5 (DMC, 1, 2) showed high inhibitory activity, whereas those possessing a methoxy group or a hydrogen (3, 4) showed slightly lower inhibitory activity, suggesting that the presence of hydrophobic groups at position X5 may be important for HST inhibition. A similar tendency was also observed at positions X3 and X4; the introduction of a methyl group in X3 or X4 (5, 6) improved the inhibitory activity, whereas that of a hydroxyl group in X4 (7) decreased the inhibitory activity compared with that of DMC. By contrast, the introduction of a methyl group in X2 (8) slightly reduced the inhibitory activity. Additionally, positions A, X1, and R were more influential on the inhibitory activity of the compound. Changing a cyclopropyl substituent in X1 to a methyl group and a pyridazine ring to a pyridine ring (9) led to the lowest inhibitory activity. Compound 10 and cyclopyrimorate possessing a hydrogen and a morpholine-4-carbonyloxy group, respectively, at position R showed considerably low inhibitory activity. Compounds with low inhibitory activity exhibited weaker bleaching activity, except for cyclopyrimorate. In the case of DMC and its derivatives, a positive correlation (r2=0.78) was detected between the HST inhibitory activity (pIC50) and the in vivo bleaching activity (pEC50) (Fig. 6). However, cyclopyrimorate was an outlier in this correlation.
Table 2. Inhibitory activities of cyclopyrimorate, DMC, and its derivatives on HST and chlorophyll content.
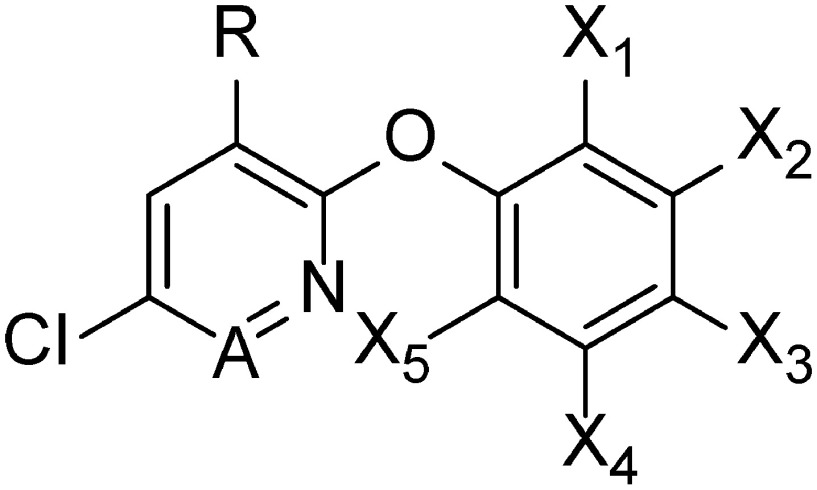 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | A | R | X1 | X2 | X3 | X4 | X5 | HST IC50 (μM) | Chlorophyll EC50 (μM) |
| Cyclopyrimorate | N | Morpholine-4-carbonyloxy | c-Pr | H | H | H | CH3 | 5.61×102 | 2.60×102 |
| DMC | N | OH | c-Pr | H | H | H | CH3 | 3.93 | 7.08×102 |
| 1 | N | OH | c-Pr | H | H | H | CH2CH3 | 2.39 | 3.52×102 |
| 2 | N | OH | c-Pr | H | H | H | Br | 3.85 | 3.56×102 |
| 3 | N | OH | c-Pr | H | H | H | OCH3 | 2.45×10 | 1.92×103 |
| 4 | N | OH | c-Pr | H | H | H | H | 2.13×10 | 2.10×103 |
| 5 | N | OH | c-Pr | H | CH3 | H | CH3 | 1.65 | 6.36×10 |
| 6 | N | OH | c-Pr | H | H | CH3 | H | 2.65 | 7.92×102 |
| 7 | N | OH | c-Pr | H | H | OH | CH3 | 1.57×102 | 3.60×104 |
| 8 | N | OH | c-Pr | CH3 | H | H | H | 1.93×10 | 1.52×103 |
| 9 | CH | OH | CH3 | H | H | H | CH3 | 4.63×102 | 1.62×104 |
| 10 | N | H | c-Pr | H | H | H | CH3 | 2.58×102 | 2.96×103 |
Fig. 6. Correlation between HST inhibitory activities and in vivo bleaching activities of herbicides. pIC50=log (1/IC50 (μM)) and pEC50=log (1/EC50 (μM)). ◆, DMC and its derivatives; □, cyclopyrimorate.
Discussion
Cyclopyrimorate exhibited bleaching symptoms similar to those caused by existing major carotenoid biosynthesis inhibitors, such as mesotrione or norflurazon (Fig. 3). Therefore, the target site of cyclopyrimorate was predicted to be an enzyme in the carotenoid or PQ biosynthesis pathway. Additionally, the shoots and roots of Arabidopsis seedlings treated with cyclopyrimorate were shorter than those of the control, which were similar to the dwarf phenotype of an A. thaliana mutant with the disrupted HST gene.24)
To narrow down the potential target sites of cyclopyrimorate, the levels of various intermediates in the PQ biosynthesis pathway were examined in Arabidopsis plants. The level of PQ decreased in a concentration-dependent manner of cyclopyrimorate, while HGA accumulated after cyclopyrimorate treatment (Fig. 4A). At or above 800 ppm cyclopyrimorate, the accumulated amount of HGA was lower than that at 400 ppm cyclopyrimorate, probably because the plants were damaged, and their HGA-synthesizing activity was weakened at higher cyclopyrimorate concentrations. Similar changes in the levels of intermediates were also observed in B. juncea and S. juncoides (Fig. 4B and C), which suggests that cyclopyrimorate inhibits HST, a downstream enzyme of 4-HPPD in the PQ biosynthesis pathway. Moreover, the pattern of change in HGA and PQ was similar between DMC and haloxydine, a known HST inhibitor (Supplemental Table S1).
The accumulation of HGA and the decrease of PQ in plants treated with norflurazon (Fig. 4) may be due to reduced expression of the geranylgeranyl diphosphate synthase (GGPS) gene. In an A. thaliana mutant with a disrupted PDS gene, the expression of GGPS was significantly lower than that in the wild-type.25) Geranylgeranyl diphosphate is a precursor of solanesyl diphosphate, which is a substrate of HST. It is, therefore, possible that lower GGPS activity indirectly caused by norflurazon results in a decrease in solanesyl diphosphate, which might further cause an indirect inhibition of HST and, subsequently, a little accumulation of HGA and decreased levels of PQ. Norflurazon completely bleached Arabidopsis leaves at a concentration of 500 ppm (Fig. 3D); however, PQ biosynthesis was not completely inhibited, indicating that the inhibition of PQ is not crucial for norflurazon to exhibit bleaching activity. By contrast, the inhibition of PQ biosynthesis by cyclopyrimorate treatment was closely associated with its bleaching activity (Fig. 4 and Supplemental Fig. S1).
Neither PQ nor HGA was detected in Arabidopsis plants after mesotrione treatment (Fig. 4). Similar trends have been observed before: the synthesis of HGA is inhibited by the 4-HPPD inhibitor, sulcotrione,26) and the level of PQ declines prior to that of β-carotene in Brassica kaber seedlings upon treatment with the 4-HPPD inhibitor, isoxaflutole.27) These effects of 4-HPPD inhibitors were different from those of cyclopyrimorate.
The action site of cyclopyrimorate could be distinguished from that of clomazone, a DOXPS inhibitor, by phytoene accumulation (Supplemental Fig. S2). It is known that phytoene accumulates in the presence of 4-HPPD and PDS inhibitors, whereas not in the presence of a DOXPS inhibitor.28,29) In this study, phytoene accumulation was observed with cyclopyrimorate, mesotrione, and norflurazon treatment, whereas not with clomazone treatment. These data suggest that the target site of cyclopyrimorate is not DOXPS. Based on changes in HGA, PQ, and phytoene levels, the action site of cyclopyrimorate was clearly distinguishable from those of the other bleaching herbicides except for haloxydine.
To confirm direct interaction between cyclopyrimorate and HST, we assayed HST activities in a cell-free system. This assay system, in which a radioactive substrate was used, has already been reported.11,12,22) Therefore, we developed alternative methods without the use of a radioactive isotope. Using these newly developed methods, the enzymatic reaction product, MFBQH, was quantified by HPLC with a fluorescence detection system, and the HST inhibitory activities of various herbicides were examined in a high-throughput manner. The data showed that cyclopyrimorate weakly inhibited HST (IC50=5.61×102 μM), whereas DMC strongly inhibited HST (IC50=3.93 µM), suggesting that DMC is the main active form of cyclopyrimorate. HST was not inhibited by the other bleaching herbicides except for haloxydine.
To verify the target site of cyclopyrimorate, we evaluated DMC and its derivatives for HST inhibitory activities and in vivo bleaching activities. The HST inhibition and bleaching activities showed a positive correlation r2=0.78, suggesting that the bleaching symptoms were attributable to the inhibition of HST. However, cyclopyrimorate was an outlier with the high bleaching activity and the low HST inhibitory activity, because cyclopyrimorate is metabolized to the highly active HST inhibitor, DMC.
Two intriguing questions remain: what is the mechanism of HST inhibition used by cyclopyrimorate, and why does cyclopyrimorate exhibit strong synergistic effects with 4-HPPD inhibitors? It is known that haloxydine is a competitive inhibitor of homogentisate and a non-competitive inhibitor of FPP for C. reinhardtii HST.11) Understanding the HST inhibitory mechanism of DMC will be valuable for the structural requirements to develop novel HST inhibitors. To address the synergistic effects of cyclopyrimorate and 4-HPPD inhibitors, it is possible that both HST and 4-HPPD function in the PQ biosynthesis pathway; therefore, two herbicides acting on sequential target sites may produce synergistic effects. However, detailed mechanisms need to be investigated in future studies.
In conclusion, it was indicated that the target site of cyclopyrimorate and DMC is HST. Cyclopyrimorate is potentially a highly effective herbicide for weed control in rice fields, especially those that are resistant to ALS inhibitors. Cyclopyrimorate will be on the market with a novel mode of action for the first time in over 20 years, thus offering a new strategy for the management of herbicide resistance.
Acknowledgments
We acknowledge S. Okaya for the organic synthesis of MFBQH and M. Kawashima for the LC-MS/MS analysis and J. Kadotani, H. Tamaru, K. Yoshino and T. Ando for helpful suggestions at Mitsui Chemicals Agro, Inc.
Supplementary Data
References
- 1).I. Heap: “Integrated Pest Management,” Springer, Netherlands, Chap. 12, pp. 281–301, 2014.
- 2).http://www.weedscience.org/ (Accessed 21 Nov., 2017)
- 3).S. O. Duke: Pest Manag. Sci. 68, 505–512 (2012). [DOI] [PubMed] [Google Scholar]
- 4).http://hracglobal.com/ (Accessed 21 Nov., 2017)
- 5).P. Boger and G. Sandmann: “Target Sites of Herbicide Action,” CRC Press, Boca Raton, FL, pp. 25–44, 1989.
- 6).M. P. Mayer, P. Beyer and H. Kleinig: Eur. J. Biochem. 191, 359–363 (1990). [DOI] [PubMed] [Google Scholar]
- 7).S. R. Norris, T. R. Barrette and D. Dellapenna: Plant Cell 7, 2139–2149 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).G. E. Bartley and P. A. Scolnik: Plant Cell 7, 1027–1038 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).C. Mueller, J. Schwender, J. Zeidler and H. K. Lichtenthaler: Biochem. Soc. Trans. 28, 792–793 (2000). [PubMed] [Google Scholar]
- 10).Y. Ferhatoglu and M. Barrett: Pestic. Biochem. Physiol. 85, 7–14 (2006). [Google Scholar]
- 11).R. Sadre, M. Frentzen, M. Saeed and T. Hawkes: J. Biol. Chem. 285, 18191–18198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).R. Sadre, J. Gruber and M. Frentzen: FEBS Lett. 580, 5357–5362 (2006). [DOI] [PubMed] [Google Scholar]
- 13).H. Tamaru, T. Sakamoto, K. Yoshino, N. Imamura, S. Saeki, T. Ando, S. Koda, Y. Tsukamoto, J. Kadotani, K. Ikemachi, K. Kitahara, C. Furuyama and R. Aoyama: Abstr. 26th Asian-Pacific Weed Science Society Conference, p. 190, 2017.
- 14).H. Matsumoto: ACS Symposium Series 892, 161–171 (2005). [Google Scholar]
- 15).H. Matsumoto, M. Mizutani, T. Yamaguchi and J. Kadotani: Weed Biol. Manage. 2, 39–45 (2002). [Google Scholar]
- 16).16) Y. Tsukamoto, H. Komai, J. Kadotani, K. Koi, S. Mio and H. Takeshiba (Sankyo Agro Inc.): Jpn. Kokai Tokkyo Koho JP 2004-262932 (2004) (in Japanese).
- 17).Y. Tsukamoto, H. Komai, J. Kadotani, K. Koi, S. Mio and H. Takeshiba (Sankyo Agro Inc.): Jpn. Kokai Tokkyo Koho JP 2004-002263 (2004) (in Japanese).
- 18).G. Mitchell, D. W. Bartlett, T. E. M. Fraser, T. R. Hawkes, D. C. Holt, J. K. Townson and R. A. Wichert: Pest Manag. Sci. 57, 120–128 (2001). [DOI] [PubMed] [Google Scholar]
- 19).G. Sandmann, I. E. Clarke, P. M. Bramley and P. Böger: J. Biosci. 39, 443–449 (1984). [Google Scholar]
- 20).K. Mori and T. Uno: Tetrahedron 45, 1945–1958 (1989). [Google Scholar]
- 21).K. Terashima, Y. Takaya and M. Niwa: Bioorg. Med. Chem. 10, 1619–1625 (2002). [DOI] [PubMed] [Google Scholar]
- 22).T. R. Hawkes, P. R. Drayton and R. Dale: (Syngenta Limited): Pat. WO 2010-029311 (2010).
- 23).R. J. Porra, W. A. Thompson and P. E. Kriedemann: Biochim. Biophys. Acta (BBA) Bioenerg. 975, 384–394 (1989). [Google Scholar]
- 24).Y. Chao, J. Kang, T. Zhang, Q. Yang, M. Y. Gruber and Y. Sun: PLoS ONE 9, 1–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).G. Qin, G. Qin, H. Gu, L. Ma, Y. Peng, X. W. Deng, Z. Chen and L. Qu: Cell Res. 17, 471–482 (2007). [DOI] [PubMed] [Google Scholar]
- 26).A. Schulz, O. Ort, P. Beyer and H. Kleinig: FEBS Lett. 318, 162–166 (1993). [DOI] [PubMed] [Google Scholar]
- 27).K. E. Pallett, J. P. Little, M. Sheekey and P. Veerasekaran: Pestic. Biochem. Physiol. 62, 113–124 (1998). [Google Scholar]
- 28).T. Soeda and T. Uchida: Pestic. Biochem. Physiol. 29, 35–42 (1987). [Google Scholar]
- 29).S. O. Duke, W. H. Kenyon and R. N. Paul: Weed Sci. 33, 786–794 (1985). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.