Abstract
Introduction
Currently, there are no tools that can accurately predict which patients with mild cognitive impairment (MCI) will progress to Alzheimer's disease (AD). Texture analysis uses image processing and statistical methods to identify patterns in voxel intensities that cannot be appreciated by visual inspection. Our main objective was to determine whether MRI texture could be used to predict conversion of MCI to AD.
Methods
A method of 3-dimensional, whole-brain texture analysis was used to compute texture features from T1-weighted MR images. To assess predictive value, texture changes were compared between MCI converters and nonconverters over a 3-year observation period. A predictive model using texture and clinical factors was used to predict conversion of patients with MCI to AD. This model was then tested on ten randomly selected test groups from the data set.
Results
Texture features were found to be significantly different between normal controls (n = 225), patients with MCI (n = 382), and patients with AD (n = 183). A subset of the patients with MCI were used to compare between MCI converters (n = 98) and nonconverters (n = 106). A composite model including texture features, APOE-ε4 genotype, Mini-Mental Status Examination score, sex, and hippocampal occupancy resulted in an area under curve of 0.905. Application of the composite model to ten randomly selected test groups (nonconverters = 26, converters = 24) predicted MCI conversion with a mean accuracy of 76.2%.
Discussion
Early texture changes are detected in patients with MCI who eventually progress to AD dementia. Therefore, whole-brain 3D texture analysis has the potential to predict progression of patients with MCI to AD.
Keywords: Alzheimer's disease, Mild cognitive impairment, Texture, MRI, ADNI
1. Introduction
Alzheimer's disease (AD) is a neurodegenerative disorder that is characterized by progressive cognitive and functional deficits. It is associated with the accumulation of amyloid and tau proteins in the brain and is the most common cause of dementia, accounting for nearly 70% of dementia cases [1]. According to the Alzheimer's Association, the estimated financial burden of AD on the Canadian and United States economy is 10.4 and 236 billion dollars per annum, respectively [2]. In addition to the financial burden, caregivers of patients with AD also have one of the highest prevalence of emotional stress and depression [3]. This unequivocally highlights the need for the development of therapy to target this disease.
While research into AD, vis-à-vis development of animal models to decipher the pathophysiology of this disease and to test novel therapeutics, has yielded a plethora of new data, this has not readily translated to successful therapeutics for this disease. One reason for this includes the inconsistency in diagnosing individuals early on in their disease course when interventions may have a greater chance of being effective before the condition reaches a state of irreversible degeneration. Establishing the initial diagnosis of AD in a patient at an earlier stage (“prodromal AD”) may be facilitated by recognition of the syndrome of mild cognitive impairment (MCI) [4], [5]. Present studies show that up to 20% of individuals with MCI will progress to dementia per year [4], [6], [7]. Hence, the establishment of biomarkers to identify individuals with MCI who will convert to dementia is a critical step in the development of novel therapeutics.
One particular area that has received extensive investigation is neuroimaging. For instance, MRI measurements of volume reduction in multiple brain regions and in cortical thickness have been predictive of those who will convert from MCI to AD [8], [9], [10], [11], [12]. More specifically, the hippocampus has been extensively studied and hippocampal volume and shape have predictive value in conversion [12], [13], [14]. Similar predictions for conversion have also been seen in other imaging modalities including diffusion tensor imaging [15], magnetic resonance spectroscopy [16], fluorodeoxyglucose positron emission tomography [12], [17], amyloid imaging [18], [19], and positron emission tomography acetylcholinesterase activity [20].
However, despite these various studies, the low accuracy with which clinicians have been able to predict MCI to AD conversion has limited its use in daily clinical practice. This is due in part to the lack of access to some of these technologies at smaller health centers (i.e., radioactive tracers). Another hurdle is that some of these diagnostic tools cannot be used on a mass scale to benefit a large population because of the various barriers associated with their integration into large-scale analysis (i.e., cost, time required for analysis, lack of expertise for manual analysis etc.).
In this study, we used an automated whole-brain texture analysis [21], [22]. We used this technology to compare MRI texture in normal controls (NC), patients with MCI, and patients with AD. We further tested whether MRI texture could be used to predict MCI conversion to AD. We predict that MRI texture will be different in NC, patients with MCI, and patients with AD. We further predict that these MRI texture changes can be used to classify MCI converters (MCI-C) from nonconverters (MCI-NC).
2. Materials and methods
2.1. Alzheimer's Disease Neuroimaging Initiative
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by principal investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD.
2.2. Subjects
A total of 790 ADNI-1 participants were included in this study including 225 NC, 382 patients with MCI, and 183 patients with AD. NC were identified as having Mini-Mental Status Examination (MMSE) scores of 24–30, clinical dementia rating scores of zero, and no diagnosis of depression, MCI, or dementia. Participants with MCI were defined as having MMSE scores of 24–30, a memory complaint verified by a partner, objective memory loss measured by Wechsler Memory Scale Logical Memory II, and a clinical dementia rating of 0.5. Participants with AD were defined as having an MMSE score of 20–26, clinical dementia rating of 0.5 to 1.0, and meeting NINCDS/ADRDA criteria for probable AD. These criteria have previously been verified for reliability and validity [23].
2.3. MRI data
MRI data consisted of the baseline 3D volumetric T1-weighted scans acquired at 1.5T from ADNI-1. MRI data acquisition techniques were standardized across different sites according to ADNI protocol (http://adni.loni.usc.edu/methods/documents/mri-protocols/). Clinical and MRI data were downloaded in July 2016.
2.4. Hippocampal volume
Hippocampal and inferior lateral ventricle volumes were extracted from T1 MRI scans using an automated method with FreeSurfer 6.0 and was aided with cBRAIN high-performance computing cluster to enable distributed computing [24], [25]. This method has been previously described and compared with other automated methods of segmentation [26]. FreeSurfer 6.0 image analysis suite (http://surfer.nmr.mgh.harvard.edu/) follows a standardized pipeline for volumetric analysis. Details of this have been previously discussed in detail [25]. In brief, this multistep pipeline includes motion correction, automated Talairach transformation, first normalization of voxel intensities, removal of the skull, linear volumetric registration, intensity normalization, nonlinear volumetric registration, volumetric labeling, second normalization of voxel intensities, and white matter segmentation. Hippocampal volumes were normalized based on the calculation of hippocampal occupancy (HOC = hippocampal volume divided by hippocampal volume plus volume of inferior lateral ventricle). This has previously been shown to perform better on both discrimination and predictive accuracy as opposed to raw hippocampal volume [27].
2.5. Three-dimensional texture analysis
For a detailed discussion of the methods for 3-dimensional voxel-based texture analysis (VGLCM-TOP-3D) of MR images, see Maani et al. [21]. In brief, this novel method of 3D voxel-based texture analysis calculates a texture feature at each voxel by averaging the texture feature calculated in three orthogonal planes at the voxel. A 3D texture map is generated from each 3D T1 data set for each feature. Eight texture features that had previously been shown to distinguish between nondementia and dementia patients were utilized in this study. These features include autocorrelation (autoc), correlation matlab (corrm), dissimilarity (dissi), energy (energ), homogeneity matlab (homom), sum of squares: variance (sosvh), sum average (savgh), and sum entropy (senth). Analysis was aided with the use of this toolbox on WestGrid Cluster distributed computing (https://www.westgrid.ca/) and Compute Canada (https://www.computecanada.ca/) clusters.
2.6. Statistical analysis
Texture maps of subjects' baseline scans were compared between groups (NC, MCI, AD) in SPM8 with an F-test. Within the MCI group, texture maps of subjects who eventually transitioned to AD (“converters”, MCI-C) were compared with those who did not (“nonconverters”, MCI-NC). Group comparisons incorporated a familywise error correction for multiple comparisons at P < .05 with reporting of clusters greater than 30 significant voxels. If the results did not survive correction, then results are reported at an uncorrected P < .001 with a minimum cluster size of 20. Texture values were extracted from statistically significant regions with MarsBaR region of interest toolbox for SPM version 0.44 [28].
To determine the performance of the various texture and clinical variables and their ability to distinguish NC from patients with AD, as well as classify conversion status, we used receiver operating characteristic curves. The resulting area under curve (AUC) was used to determine the capability for diagnosis and prognostication. The significance of an AUC was determined using DeLong test.
The MCI group was randomly divided into both a training set (∼75% of the participants) and a trial set (∼25%). This was iterated 10 times to provide 10 unique training and test groups. The training set was used to generate a binary logistic regression model, using MRI texture, HOC, and clinical data as variables. The generated logistic regression formula was then applied to the trial set and these values were then compared with the Youden index determined from the training set. This was used to determine predicted conversion, which was then compared with actual conversion status to determine its accuracy.
One-way analysis of variance, independent sample t-test, as well as chi-squared test were used to determine statistical differences between various clinical variables between baseline (NC, MCI, AD) and converter (MCI-NC, MCI-C) groups. Statistical significance was defined as P < .05.
All statistics were done with IBM SPSS Statistics Version 20 (IBM Corp. Released 2011. IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp.) or MedCalc Version 14 (MedCalc Software bvba, Ostend, Belgium; https://www.medcalc.org; 2016).
3. Results
Baseline demographics including age, sex, MMSE scores, education level, and APOE-ε4 genotype, and calculated HOC are shown in Table 1. All baseline variables were significantly different between the three groups based on one-way analysis of variance or chi-square analysis with the exception of age.
Table 1.
Baseline demographics
| Variables at baseline | Diagnosis group |
P value | ||
|---|---|---|---|---|
| Normal control (n = 225) | MCI (n = 382) | AD (n = 183) | ||
| Age (mean ± SD) | 75.8 ± 5.0 | 74.6 ± 7.4 | 75.4 ± 7.6 | .107 |
| Sex (% male; M/F) | 51.6 (116/109) | 64.1 (245/137) | 53.0 (97/86) | <.01 |
| MMSE score | 29.1 ± 1.0 | 27.0 ± 1.8 | 23.3 ± 2.2 | <.001 |
| Education (years) | 16.1 ± 2.8 | 15.7 ± 3.0 | 14.6 ± 3.2 | <.001 |
| Hippocampal occupancy | 0.824 ± 0.084 | 0.751 ± 0.121 | 0.666 ± 0.136 | <.001 |
| % with 1|2 APOE-ε4 alleles | 24.0|2.2 | 42.7|12.0 | 47.0|18.6 | <.001 |
Abbreviations: MCI, mild cognitive impairment; AD, Alzheimer's disease; SD, standard deviation; MMSE, Mini-Mental Status Examination; APOE, apolipoprotein E.
3.1. 3D texture analysis of T1 MRI scans reveals distinct texture differences between NC, MCI, and AD
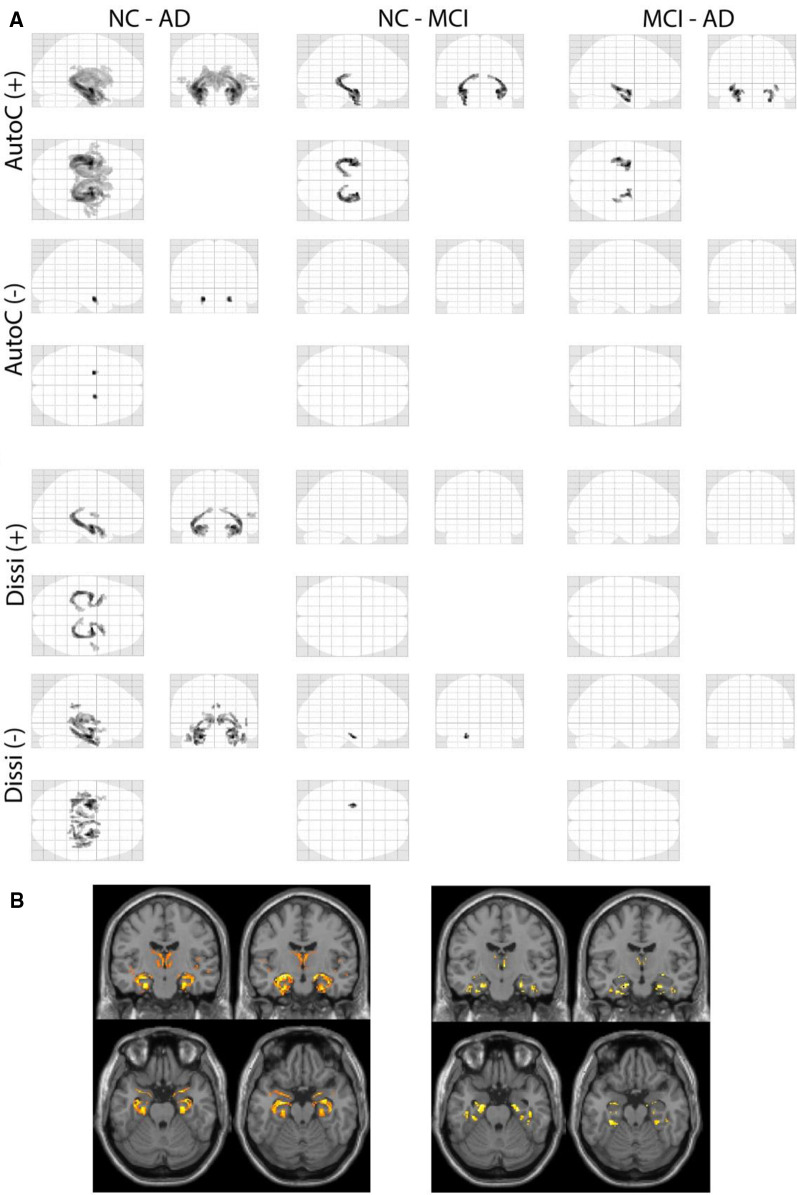
Baseline MRI T1 images were processed and 3D whole-brain texture maps for eight different features were calculated for each scan using VGLCM-TOP-3D. Statistical maps revealed differences in texture value between all groups for all eight textures (Fig. 1). Texture changes for different features have areas of overlap, but also unique areas. In particular, these areas include the bilateral hippocampus, medial temporal lobe, amygdala, and inferior parietal lobe.
Fig. 1.
Statistical maps of significant regions of texture differences between the subgroups. Two representative textures are shown (A: autocorrelation, Autoc; dissimilarity; Dissi). Areas of (+) higher texture value and (−) lower texture value when comparing the first to the second group are shown. Familywise error <0.05 and cluster size >30. Results are superimposed on MRI T1 templates for autoc (B, left) and dissi (B, right). Abbreviations: NC, normal controls; MCI, mild cognitive impairment; AD, Alzheimer's disease.
3.2. Texture changes between NC and patients with AD
To determine the classification of NC from patients with AD using texture alone within our sample group, we extracted the texture value for the significant regions identified in each of the eight texture features (Table 2). All eight texture features revealed regions with higher and lower texture values when comparing these two groups except for energy and homogeneity. We calculated a binary logistic regression model for each individual texture variable and using receiver operating characteristic curves, we determined their AUC (Table 2). The AUC values ranged from 0.722 to 0.866. When all texture features and HOC were combined into a single classification model using binary logistic regression, the AUC was 0.930 (sensitivity 83.1%, specificity of 92.0%). This model was statistically significant compared with using HOC score alone (AUC 0.843, sensitivity 77.6%, specificity 79.6%). The AUC of texture (0.928) was also statistically significant compared with HOC.
Table 2.
Texture and hippocampal volume in distinguishing normal controls (NC) from patients with Alzheimer's disease (AD)
| Variable | Texture value |
ROC analysis |
95% CI | Sens (%) | Spec (%) | P value | |
|---|---|---|---|---|---|---|---|
| NC (n = 225) | AD (n = 183) | AUC | |||||
| Autoc (+) | 42.6 ± 4.5 | 35.1 ± 6.2 | 0.837 | 0.797–0.871 | 75.4 | 78.7 | <.0001 |
| Autoc (−) | 42.0 ± 3.1 | 45.8 ± 3.3 | 0.809 | 0.767–0.846 | 66.1 | 85.3 | <.0001 |
| Corrm (+) | 69.7 ± 16.4 | 45.1 ± 19.6 | 0.828 | 0.788–0.864 | 74.9 | 76.9 | <.0001 |
| Corrm (−) | 77.7 ± 12.0 | 88.8 ± 14.3 | 0.722 | 0.676–0.765 | 74.3 | 61.3 | <.0001 |
| Dissi (+) | 32.6 ± 9.1 | 18.7 ± 10.8 | 0.828 | 0.788–0.864 | 77.1 | 76.9 | <.0001 |
| Dissi (−) | 27.6 ± 4.0 | 33.6 ± 4.9 | 0.825 | 0.784–0.869 | 77.6 | 71.6 | <.0001 |
| Energ (+) | 45.6 ± 8.3 | 32.1 ± 9.0 | 0.866 | 0.830–0.898 | 81.4 | 77.8 | <.0001 |
| Homom (+) | 124.2 ± 12.0 | 99.5 ± 19.0 | 0.866 | 0.829–0.897 | 76.0 | 84.9 | <.0001 |
| Savgh (+) | 133.1 ± 13.1 | 108.9 ± 19.4 | 0.850 | 0.812–0.883 | 78.1 | 79.1 | <.0001 |
| Savgh (−) | 146.2 ± 5.7 | 153.2 ± 5.8 | 0.815 | 0.774–0.852 | 66.1 | 86.7 | <.0001 |
| Senth (+) | 121.1 ± 26.2 | 76.6 ± 34.5 | 0.840 | 0.800–0.874 | 74.3 | 80.9 | <.0001 |
| Senth (−) | 120.0 ± 12.8 | 138.0 ± 13.8 | 0.825 | 0.785–0.861 | 77.6 | 75.1 | <.0001 |
| Sosvh (+) | 83.8 ± 9.9 | 67.6 ± 13.4 | 0.836 | 0.796–0.870 | 76.0 | 78.2 | <.0001 |
| Sosvh (−) | 84.3 ± 6.3 | 92.5 ± 6.8 | 0.818 | 0.777–0.854 | 63.4 | 90.2 | <.0001 |
| Texture (all features) | 0.928 | 0.898–0.951 | 88.0 | 84.9 | <.0001 | ||
| HOC | 0.843 | 0.804–0.877 | 77.6 | 79.6 | <.0001 | ||
| Texture (all features) + HOC | 0.930 | 0.901–0.953 | 83.1 | 92.0 | <.0001 | ||
Abbreviations: ROC, receiver operating characteristic; AUC, area under curve, CI, confidence interval; Sens, sensitivity; Spec, specificity; HOC, hippocampal occupancy.
3.3. Texture differences in participants with MCI predicted conversion to AD within 36 months
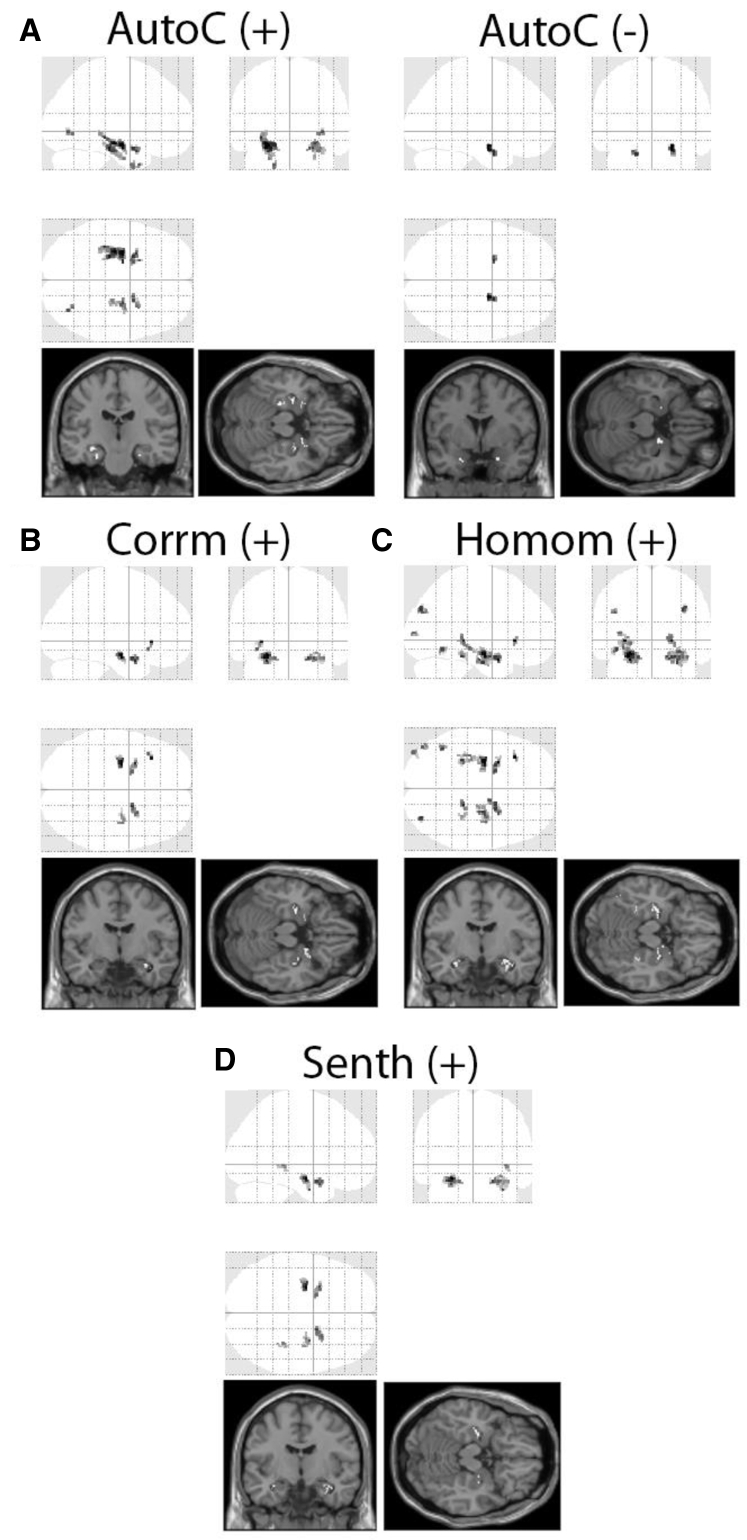
To address the main objective of our study, participants who were diagnosed as MCI at the baseline were further divided into two categories: those who remained as MCI at 36 months (n = 106; MCI-NC, nonconverters) versus those who converted to AD within 36 months (n = 98; MCI-C, converters) (Supplementary Table 1). Only patients with MCI who had follow-up for the full 36 months or converted within the 36 months were included in this analysis (n = 204). Baseline T1 MRI scans of these patients with MCI were selected from the data set and 3D texture maps were generated (Supplementary Fig. 1). A voxelwise comparison was conducted between MCI-NC and MCI-C, revealing significant texture changes in the mesial temporal region with some involvement of the parietal lobes (Fig. 2). In particular, we found that MCI-NC had higher texture values for the features autocorrelation, correlation, homogeneity, and sum of entropy. A region of lower texture value for texture feature autocorrelation was identified in the right mesial temporal region in MCI-C compared with MCI-NC. Next, a binary logistic regression model was used to test the ability of each texture feature to predict MCI to AD conversion (Table 3). Their individual AUCs ranged from 0.709 to 0.794, which were higher than HOC alone (AUC 0.655). The predictive ability was improved with a model combining all five texture features (AUC 0.825).
Fig. 2.
Statistical maps showing significant areas of texture change at the baseline between MCI nonconverters (MCI-NC) and converters (MCI-C) in the training group. Autocorrelation (Autoc) had areas of both higher (A, left, +) and lower (A, right, −) texture values in MCI-NC when compared with MCI-C, whereas correlation (B, Corrm), homogeneity (C, Homom), and sum of entropy (D, Senth) only revealed higher texture value for MCI-NC. Voxel clusters >20 at P < .001, uncorrected were designated as significant.
Table 3.
Texture, hippocampal occupancy, clinical variables, and the resulting area under curve in distinguishing mild cognitive impairment nonconverters (MCI-NC) from MCI converters (MCI-C)
| Variable | Texture value |
ROC analysis |
95% CI | Sens (%) | Spec (%) | P value | |
|---|---|---|---|---|---|---|---|
| Non-converters | Converters | AUC | |||||
| Autoc (+) | 41.8 ± 5.2 | 36.3 ± 6.0 | 0.753 | 0.688–0.810 | 73.5 | 69.8 | <.0001 |
| Autoc (−) | 42.1 ± 3.5 | 44.8 ± 3.1 | 0.709 | 0.641–0.770 | 82.7 | 49.1 | <.0001 |
| Corrm | 53.4 ± 18.2 | 34.9 ± 17.4 | 0.774 | 0.710–0.829 | 83.7 | 62.3 | <.0001 |
| Homom | 108.8 ± 16.6 | 88.0 ± 18.5 | 0.794 | 0.732–0.848 | 64.3 | 80.2 | <.0001 |
| Senth | 94.5 ± 37.8 | 57.6 ± 35.2 | 0.762 | 0.697–0.819 | 81.6 | 63.2 | <.0001 |
| HOC | 0.655 | 0.585–0.720 | 64.3 | 64.2 | <.0001 | ||
| Texture | 0.825 | 0.766–0.875 | 71.4 | 79.3 | <.0001 | ||
| Texture + HOC | 0.881 | 0.828–0.922 | 87.8 | 75.5 | <.0001 | ||
| Texture + HOC + APOE-ε4 | 0.895 | 0.845–0.933 | 90.8 | 73.6 | <.0001 | ||
| Texture + HOC + APOE-ε4 + MMSE + Sex | 0.905 | 0.856–0.941 | 86.7 | 83.0 | >.0001 | ||
Abbreviations: ROC, receiver operating characteristic; AUC, area under curve, CI, confidence interval; Sens, sensitivity; Spec, specificity; MMSE, Mini-Mental Status Examination; HOC, hippocampal occupancy.
To further characterize the utility of MRI texture in conversion discrimination, we next asked the question of whether the inclusion of additional clinical variables to texture could improve prognostic ability. Our binary logistic regression model determined that among the clinical variables, only APOE-ε4 provided a statistical significance of P < .05. In this composite model including texture, HOC, and APOE-ε4, an AUC of 0.895 was achieved with a sensitivity of 90.8% and a specificity of 73.6%. A final model which also included MMSE score and sex had a further increase in AUC to 0.905 and a sensitivity and specificity of 86.7% and 83.0%, respectively (Table 3). Pairwise comparison of receiver operating characteristic curves using DeLong test showed statistical significance with P < .05 for all comparisons with the exception of (Texture + HOC) compared with (Texture + HOC + APOE-ε4) and (Texture + HOC + APOE-ε4) compared with (Texture + HOC + APOE-ε4 + MMSE + Sex).
Next, the MCI group was further divided into a training set (∼75%) and a trial set (∼25%). We randomly generated a total of 10 of these sets, with each training set having a total of n = 80 MCI-NC and n = 74 MCI-C, whereas the trial set had a total of n = 26 MCI-NC and n = 24 MCI-C. Independent sample t-test and chi-square analysis showed no statistical difference between the baseline demographics in the training and trial sets in each iteration. Next, ten binary logistic regression models were determined, corresponding to one for each training set, achieving AUC ranging from 0.896 to 0.930 (Table 4).
Table 4.
Area under curve in ten separate trials of randomly splitting training (75%) and trail data (25%) and the resulting accuracy of prediction in the trial set
| Trial | ROC analysis |
95% CI | Sens (%) | Spec (%) | P value | Accuracy (%) in trial set |
|---|---|---|---|---|---|---|
| AUC | ||||||
| 1 | 0.925 | 0.871–0.961 | 86.5 | 87.5 | <.0001 | 68 |
| 2 | 0.896 | 0.837–0.940 | 78.4 | 90.0 | <.0001 | 84 |
| 3 | 0.899 | 0.840–0.942 | 85.1 | 85.0 | <.0001 | 82 |
| 4 | 0.921 | 0.867–0.958 | 94.6 | 78.8 | <.0001 | 78 |
| 5 | 0.910 | 0.854–0.950 | 89.2 | 85.0 | <.0001 | 76 |
| 6 | 0.906 | 0.849–0.947 | 89.2 | 81.3 | <.0001 | 84 |
| 7 | 0.917 | 0.861–0.955 | 86.5 | 87.5 | <.0001 | 70 |
| 8 | 0.930 | 0.878–0.965 | 89.2 | 87.5 | <.0001 | 66 |
| 9 | 0.904 | 0.846–0.946 | 89.2 | 80.0 | <.0001 | 82 |
| 10 | 0.906 | 0.849–0.947 | 85.1 | 85.0 | <.0001 | 72 |
Abbreviations: ROC, receiver operating characteristic; AUC, area under curve, CI, confidence interval; Sens, sensitivity; Spec, specificity.
To test our discriminant model, we then applied these binary logistic regression models to their corresponding trial sets to determine their accuracy for distinguishing between MCI-NC and MCI-C. The composite model including texture, HOC, APOE-ε4 genotype, MMSE score, and sex had a mean accuracy of 76.2 ± 6.8% in the 10 trials (Table 4).
4. Discussion
The main objective of this study was to characterize cerebral degeneration in vivo using MRI-based texture analysis and to determine whether MRI texture could be used to predict conversion of MCI to AD. We used a whole-brain texture analysis (VGLCM-TOP-3D) to compute eight different texture features from T1-weighted MR images. As we hypothesized, MRI texture was different between NC and patients with AD and could further be used to predict MCI-C from nonconverters. Specifically, MRI texture changes were seen in all eight texture features between NC and patients with AD and in a combined model had an AUC of 0.928. This was superior to HOC, which had an AUC of 0.843. Furthermore, we determined areas of significant texture change between MCI-NC and MCI-C, which alone accounted for an AUC of 0.825, which was higher than HOC at 0.655. The model that yielded the greatest AUC (0.905) incorporated texture, HOC, APOE-ε4, MMSE, and sex. We further tested this model on 10 separate data splits, each with 75% training and 25% trial and found accuracy in predicting MCI-C and MCI-NC in the trial group of 76.2%.
In this study, we included hippocampal volume (expressed as the HOC score), a previously well-defined biomarker of disease to compare with MRI texture in distinguishing the various AD states [29]. This score has previously been shown to be superior to raw hippocampal volume alone in distinguishing NC and patients with AD [27]. Previous studies have demonstrated that hippocampal volume can be used to classify NC from patients with AD, reaching AUC levels of 0.75 to 0.887 using whole hippocampal volume and 0.81 to 0.895 when using subsegments of the hippocampus [30], [31]. This is comparable with our own hippocampal volume segmentations, which achieved an AUC score of 0.843. When we combined texture and HOC, this achieved the greatest AUC of 0.930.
The use of hippocampal volume to distinguish MCI-NC from MCI-C has been demonstrated in multiple previous studies. In particular, a meta-analysis of these various studies concluded that atrophy of mesial temporal lobe structures, specifically the hippocampus and parahippocampal gyrus in the left hemisphere, had the highest predictive value for conversion [32]. Texture changes in these regions were also identified with our texture analysis, in addition to also identifying changes in the right mesial temporal lobe. Khan et al. [31] were able to use hippocampal volume to accurately identify 76.7% of all MCI-C and 50.1% of all MCI-NC. When they used subfield analysis, selecting for the subiculum, the accuracy of identifying all MCI-C increased to 81.1%, with a marginal decrease to 49.0% in MCI-NC. Furthermore, using an automated temporal lobe atrophy assessment, Chincarini et al. [14] were able to achieve an AUC of 0.81 when classifying MCI-NC from MCI-C. We show in our study that using HOC alone achieved an AUC of 0.655. The lower AUC and accuracy may be related to the heterogeneity of our MCI group, which contained participants with MCI-C showing conversion at any time point at 12, 24, and 36 months. This is in contrast to the data set used in the study by Khan et al. and Chincarini et al., which included MCI-C up to 12 months and 24 months, respectively. Despite this heterogeneity, MRI texture proved superior to hippocampal volume in both its ability to distinguish NC from patients with AD (AUC 0.928) and MCI-NC from MCI-C (AUC 0.825).
Our method of texture analysis is not confined to the use of a priori determined regions of interest, thus allowing for the discovery of novel regions of change. For instance, previous VBM studies showed that amnestic patients with MCI who converted to AD had significant clusters of gray matter volumetric reduction in the left hippocampus and parahippocampal gyrus. In our present study, we determined hippocampal and parahippocampal texture changes were also present in the right hemisphere. This suggests the possibility that texture change may precede the development of atrophy. However, a specific study designed to longitudinally compare volume changes with texture changes will need to be untaken to further verify this hypothesis. This is in keeping with previous studies on the ADNI data set that demonstrated the utility of hippocampal texture alone to act as a predictor of MCI to AD conversion [33]. Sorensen et al. showed that texture had the ability to predict conversion to dementia independent of volume, with an AUC of 0.740 to 0.742 in those who converted within 12 and 24 months, respectively. Previous studies from Chincarini et al. using texture features from defined volumes of interest with random forest classifiers have also shown that texture can be used to distinguish NC, MCI, and AD groups and MCI-C from MCI-NC. They were able to achieve an AUC of 0.74 with a classification index for the latter [34]. However, given that these previous study also used a region of interest analysis, they were constrained by the a priori selection bias. Our present study was able to determine more specific regions of texture change. In addition, our work builds on original investigations completed by Maani et al., which showed mesial temporal lobe changes in a smaller population of 30 NC and 30 patients with AD from the OASIS (http://www.oasis-brains.org/) database where similar regions of change were identified. However, our present study used a larger population and included patients with MCI in the analysis.
A previous study using a single hippocampal texture with predefined hippocampus region of interest along with hippocampal volume, shape, and cortical thickness resulted in an accuracy prediction on the Australian Imaging, Biomarker & Lifestyle Flagship Study of Aging of 63% after being trained on the ADNI data set [35]. Our present study utilizes random splitting of our data set into both a training and trial group to generate models and test on a trial group not involved in the creation of the model. While these results are not as robust as those seen in the study by Sorensen et al. where the model was tested on an independent test set, we note that our model has average accuracy of 76.2% when tested over 10 times. Future studies would involve testing this model on an independent data set.
There are some limitations to our present study in terms of their applicability to the general population. First, the ADNI cohort cannot be generalized to the normal population given the patient recruitment was targeted toward clinical trials in patients with AD. Second, the baseline demographics of these sample patients do not fit with the actual demographics of the broader population. We do also note that although we verified our model using multiple iterations of splitting our data, it was not verified on an independent population. Regardless, this study shows that (1) there are texture differences between NC, patients with MCI, and patients with AD and (2) MRI texture changes may be a viable biomarker in predicting which patients with MCI may convert to AD.
While previous methods of texture analysis and toolboxes have been limited to analyzing region of interest and therefore require a priori analysis, the utilization of VGLCM-TOP-3D in our study allowed for a hypothesis-free analysis [36], [37]. This is the first study to our knowledge that has used a 3D whole-brain analysis to identify novel texture changes in distinguishing NC from patients with AD and MCI-NC from MCI-C. In conclusion, the resulting ability to select patients at highest risk for conversion to AD from MCI will result in a more homogenous population of research participants, enabling more clear therapeutic studies as well as targeting those patients with the greatest benefit.
Research in context.
-
1.
Systematic review: In the review of literature, the authors found no articles using whole-brain MRI texture as a means to predict mild cognitive impairment (MCI) conversion to Alzheimer's disease (AD).
-
2.
Interpretation: MRI texture is different between normal control and patient with AD. Differences in MRI texture were also found between MCI converters and nonconverters. Using a model that incorporates MRI texture and clinical data allowed for an increased accuracy of predicting MCI to AD conversion.
-
3.
Future directions: Future work should be geared toward correlating MRI texture with tissue pathology. Furthermore, the incorporation of additional clinical markers with MRI texture has the potential for the development of a more accurate predictive model for MCI to AD conversion.
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
The authors have declared that no conflict of interest exists.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2018.09.002.
Supplementary data
References
- 1.Apostolova L.G. Alzheimer Disease. Continuum (Minneap Minn) 2016;22:419–434. doi: 10.1212/CON.0000000000000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers L.W., Bancej C., McDowell I. The Alzheimer Society Canada in Collaboration with Public Health Agency of Canada; Toronto, ON, Canada: 2016. Prevalence and Monetary Costs of Dementia in Canada. [Google Scholar]
- 3.Alzheimer's Association . 2016. 2016 Alzheimer's Disease Facts and Figures. Available at: https://www.alz.org/media/HomeOffice/Facts%20and%20Figures/facts-and-figures.pdf. [DOI] [PubMed] [Google Scholar]
- 4.Petersen R.C., Smith G.E., Waring S.C., Ivnik R.J., Tangalos E.G., Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 5.Petersen R.C. Mild Cognitive Impairment. Continuum (Minneap Minn) 2016;22:404–418. doi: 10.1212/CON.0000000000000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganguli M., Dodge H.H., Shen C., DeKosky S.T. Mild cognitive impairment, amnestic type: an epidemiologic study. Neurology. 2004;63:115–121. doi: 10.1212/01.wnl.0000132523.27540.81. [DOI] [PubMed] [Google Scholar]
- 7.Langa K.M., Levine D.A. The diagnosis and management of mild cognitive impairment: a clinical review. JAMA. 2014;312:2551–2561. doi: 10.1001/jama.2014.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.deToledo-Morrell L., Stoub T.R., Bulgakova M., Wilson R.S., Bennett D.A., Leurgans S. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging. 2004;25:1197–1203. doi: 10.1016/j.neurobiolaging.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Devanand D.P., Liu X., Tabert M.H., Pradhaban G., Cuasay K., Bell K. Combining early markers strongly predicts conversion from mild cognitive impairment to Alzheimer's disease. Biol Psychiatry. 2008;64:871–879. doi: 10.1016/j.biopsych.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack C.R., Jr., Petersen R.C., Xu Y.C., O'Brien P.C., Smith G.E., Ivnik R.J. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Risacher S.L., Saykin A.J., West J.D., Shen L., Firpi H.A., McDonald B.C. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosconi L., Perani D., Sorbi S., Herholz K., Nacmias B., Holthoff V. MCI conversion to dementia and the APOE genotype: a prediction study with FDG-PET. Neurology. 2004;63:2332–2340. doi: 10.1212/01.wnl.0000147469.18313.3b. [DOI] [PubMed] [Google Scholar]
- 13.Achterberg H.C., van der Lijn F., den Heijer T., Vernooij M.W., Ikram M.A., Niessen W.J. Hippocampal shape is predictive for the development of dementia in a normal, elderly population. Hum Brain Mapp. 2014;35:2359–2371. doi: 10.1002/hbm.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chincarini A., Bosco P., Gemme G., Esposito M., Rei L., Squarcia S. Automatic temporal lobe atrophy assessment in prodromal AD: Data from the DESCRIPA study. Alzheimers Dement. 2014;10:456–467. doi: 10.1016/j.jalz.2013.05.1774. [DOI] [PubMed] [Google Scholar]
- 15.Kantarci K., Petersen R.C., Boeve B.F., Knopman D.S., Weigand S.D., O'Brien P.C. DWI predicts future progression to Alzheimer disease in amnestic mild cognitive impairment. Neurology. 2005;64:902–904. doi: 10.1212/01.WNL.0000153076.46126.E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metastasio A., Rinaldi P., Tarducci R., Mariani E., Feliziani F.T., Cherubini A. Conversion of MCI to dementia: Role of proton magnetic resonance spectroscopy. Neurobiol Aging. 2006;27:926–932. doi: 10.1016/j.neurobiolaging.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Herholz K., Nordberg A., Salmon E., Perani D., Kessler J., Mielke R. Impairment of neocortical metabolism predicts progression in Alzheimer's disease. Dement Geriatr Cogn Disord. 1999;10:494–504. doi: 10.1159/000017196. [DOI] [PubMed] [Google Scholar]
- 18.Forsberg A., Engler H., Almkvist O., Blomquist G., Hagman G., Wall A. PET imaging of amyloid deposition in patients with mild cognitive impairment. Neurobiol Aging. 2008;29:1456–1465. doi: 10.1016/j.neurobiolaging.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 19.Ben Bouallegue F., Mariano-Goulart D., Payoux P., Alzheimer's Disease Neuroimaging Initiative (ADNI) Comparison of CSF markers and semi-quantitative amyloid PET in Alzheimer's disease diagnosis and in cognitive impairment prognosis using the ADNI-2 database. Alzheimers Res Ther. 2017;9:32. doi: 10.1186/s13195-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herholz K., Weisenbach S., Kalbe E., Diederich N.J., Heiss W.D. Cerebral acetylcholine esterase activity in mild cognitive impairment. Neuroreport. 2005;16:1431–1434. doi: 10.1097/01.wnr.0000177007.21732.7b. [DOI] [PubMed] [Google Scholar]
- 21.Maani R., Yang Y.H., Kalra S. Voxel-based texture analysis of the brain. PLoS One. 2015;10:e0117759. doi: 10.1371/journal.pone.0117759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maani R., Yang Y.H., Emery D., Kalra S. Cerebral degeneration in amyotrophic lateral sclerosis revealed by 3-dimensional texture analysis. Front Neurosci. 2016;10:120. doi: 10.3389/fnins.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blacker D., Albert M.S., Bassett S.S., Go R.C., Harrell L.E., Folstein M.F. Reliability and validity of NINCDS-ADRDA criteria for Alzheimer's disease. The National Institute of Mental Health Genetics Initiative. Arch Neurol. 1994;51:1198–1204. doi: 10.1001/archneur.1994.00540240042014. [DOI] [PubMed] [Google Scholar]
- 24.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 26.Zandifar A., Fonov V., Coupe P., Pruessner J., Collins D.L., Alzheimer's Disease Neuroimaging Initiative (ADNI) A comparison of accurate automatic hippocampal segmentation methods. NeuroImage. 2017;155:383–393. doi: 10.1016/j.neuroimage.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 27.Heister D., Brewer J.B., Magda S., Blennow K., McEvoy L.K., Alzheimer's Disease Neuroimaging Initiative (ADNI) Predicting MCI outcome with clinically available MRI and CSF biomarkers. Neurology. 2011;77:1619–1628. doi: 10.1212/WNL.0b013e3182343314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brett M, Anton J, Valabregue R, Poline J. Region of interest analysis using an SPM toolbox. 8th International Conference on Functional Mapping of the Human Brain. Sendai, Japan. 2002.
- 29.Jack C.R., Jr., Barkhof F., Bernstein M.A., Cantillon M., Cole P.E., Decarli C. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer's disease. Alzheimers Dement. 2011;7:474–485.e4. doi: 10.1016/j.jalz.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller S.G., Schuff N., Yaffe K., Madison C., Miller B., Weiner M.W. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer's disease. Hum Brain Mapp. 2010;31:1339–1347. doi: 10.1002/hbm.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan W., Westman E., Jones N., Wahlund L.O., Mecocci P., Vellas B. Automated hippocampal subfield measures as predictors of conversion from mild cognitive impairment to Alzheimer's disease in two independent cohorts. Brain Topogr. 2015;28:746–759. doi: 10.1007/s10548-014-0415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira L.K., Diniz B.S., Forlenza O.V., Busatto G.F., Zanetti M.V. Neurostructural predictors of Alzheimer's disease: a meta-analysis of VBM studies. Neurobiol Aging. 2011;32:1733–1741. doi: 10.1016/j.neurobiolaging.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Sorensen L., Igel C., Liv Hansen N., Osler M., Lauritzen M., Rostrup E. Early detection of Alzheimer's disease using MRI hippocampal texture. Hum Brain Mapp. 2016;37:1148–1161. doi: 10.1002/hbm.23091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chincarini A., Bosco P., Calvini P., Gemme G., Esposito M., Olivieri C. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer's disease. NeuroImage. 2011;58:469–480. doi: 10.1016/j.neuroimage.2011.05.083. [DOI] [PubMed] [Google Scholar]
- 35.Sorensen L., Igel C., Pai A., Balas I., Anker C., Lillholm M. Differential diagnosis of mild cognitive impairment and Alzheimer's disease using structural MRI cortical thickness, hippocampal shape, hippocampal texture, and volumetry. Neuroimage Clin. 2017;13:470–482. doi: 10.1016/j.nicl.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freeborough P.A., Fox N.C. MR image texture analysis applied to the diagnosis and tracking of Alzheimer's disease. IEEE Trans Med Imaging. 1998;17:475–479. doi: 10.1109/42.712137. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Yu C., Jiang G., Liu W., Tong L. 3D texture analysis on MRI images of Alzheimer's disease. Brain Imaging Behav. 2012;6:61–69. doi: 10.1007/s11682-011-9142-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.