Abstract
Modifications to the Gram-positive bacterial cell wall play important roles in antibiotic resistance and pathogenesis, but the pathway for the d-alanylation of teichoic acids (DLT pathway), a ubiquitous modification, is poorly understood. The d-alanylation machinery includes two membrane proteins of unclear function, DltB and DltD, which are somehow involved in transfer of d-alanine from a carrier protein inside the cell to teichoic acids on the cell surface. Here, we probed the role of DltD in the human pathogen Staphylococcus aureus using both cell-based and biochemical assays. We first exploited a known synthetic lethal interaction to establish the essentiality of each gene in the DLT pathway for d-alanylation of lipoteichoic acid (LTA) and confirmed this by directly detecting radiolabeled d-Ala-LTA both in cells and in vesicles prepared from mutant strains of S. aureus. We developed a partial reconstitution of the pathway by using cell-derived vesicles containing DltB, but no other components of the d-alanylation pathway, and showed that d-alanylation of previously formed lipoteichoic acid in the DltB vesicles requires the presence of purified and reconstituted DltA, DltC, and DltD, but not of the LTA synthase LtaS. Finally, based on the activity of DltD mutants in cells and in our reconstituted system, we determined that Ser-70 and His-361 are essential for d-alanylation activity, and we propose that DltD uses a catalytic dyad to transfer d-alanine to LTA. In summary, we have developed a suite of assays for investigating the bacterial DLT pathway and uncovered a role for DltD in LTA d-alanylation.
Keywords: teichoic acid, acyltransferase, cell wall, Staphylococcus aureus (S. aureus), membrane protein, DLT pathway, MBOAT, pathway reconstitution, DltB
Introduction
Teichoic acids are found in the cell wall of almost all Gram-positive bacteria, and many pathogens from this group rely on these polymers to colonize and resist the defenses of their host (1). Two types of teichoic acids, lipoteichoic acid and wall teichoic acid, are made in these bacteria. In most organisms, the membrane-anchored lipoteichoic acid (LTA) 3 is synthesized outside the cell on a membrane anchor through a separate pathway from wall teichoic acid (WTA) (2). WTA is synthesized inside the cell on the undecaprenyl carrier lipid before it is flipped outside and transferred to peptidoglycan (3). Both of these polymers are modified with positively charged d-alanine residues outside the cell through a process that depends on the d-alanyl lipoteichoic acid (DLT) pathway (4). The d-alanine modification of lipoteichoic acid is ubiquitous among Gram-positive bacteria, which include the human pathogens Streptococcus pneumoniae (5), Enterococcus faecalis (6, 7), and Staphylococcus aureus (8–11). This modification modulates cell surface electrostatics as measured by cell-surface affinity for cytochrome c (12–16) and whole-cell electrophoretic mobility (17). Fluorescence polarization with S. aureus cells also showed an impact of d-alanylation on cell membrane fluidity (14). The change in charge of the cell wall from neutralization of polyanionic teichoic acids influences many cellular growth and homeostasis properties, including autolysis regulation (13, 18–23), growth at low pH (8, 10, 13, 22–25), metal ion binding (8, 26, 27), protein secretion (28), bacterial coaggregation (29), and biofilm formation (6, 23, 30). Furthermore, the d-alanine modification is important for pathogenesis, providing resistance to cationic antimicrobial peptides as well as antibacterial proteins induced during infection (12, 31, 32) (Table S1). S. aureus evasion from human neutrophils and colonization and virulence in mouse models were dependent on the d-alanine modification (30, 31). In an effort to stem the tide of antibiotic-resistant S. aureus infections, the DLT pathway has been suggested as a target for small molecule inhibitors, and some have already been identified (33, 34).
Lipoteichoic and wall teichoic acids become d-alanylated through a poorly understood process that depends on the dlt operon. LTA d-alanylation occurs first, and d-alanines are subsequently transferred to WTA (35, 36). The dlt operon encodes a set of proteins that activate d-alanine as a thioester in the cytoplasm before moving the d-alanine across the plasma membrane and onto teichoic acids (4, 36–39). Four genes constitute the core dlt operon, dltABCD, of which only two, dltA and dltC, have clearly defined functions (12, 37–39). DltA resembles adenylation domains found in nonribosomal peptide synthetases (40–43), and it has been shown that DltA transfers d-alanine in an ATP-dependent manner to DltC, a peptidyl carrier protein (38, 39, 44–47). Peptidyl carrier proteins are post-translationally modified with 4′-phosphopantetheine on a conserved serine residue (48), and the associated thiol functionality allows them to carry acyl groups via a labile thioester bond (49) and deliver their acyl groups to specific enzymes. DltC is proposed to present its d-alanyl thioester cargo to the putative acyltransferase DltB (see Fig. 1) (36, 38).
Figure 1.
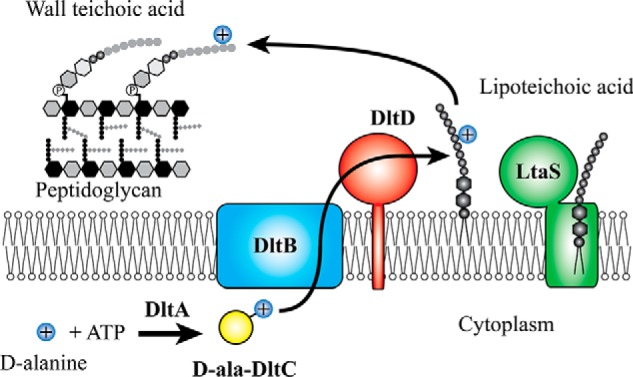
Model for teichoic acid d-alanylation by the DLT pathway. DltA-catalyzed DltC d-alanylation takes place in the cytoplasm at the expense of ATP. What happens to the thioesterified d-Ala-DltC is unclear, but the two predicted membrane-bound acyltransferases, DltB and DltD, are involved in the migration of the d-alanyl moiety across the membrane and onto lipoteichoic acid and wall teichoic acid. Lipoteichoic acid is a poly(glycerol phosphate) polymer that is biosynthesized on a diglucosyl diacylglycerol lipid anchor outside the cell by LtaS.
DltB is likely responsible for moving d-alanine across the plasma membrane (36, 39). DltB is a polytopic membrane protein containing multiple transmembrane helices. It belongs to a superfamily of membrane-bound O-acyltransferases (MBOATs) (50). MBOAT members have been shown to play roles in lipid (51) and sterol (52) biosynthesis and in acylation of peptides important for intracellular signaling, such as hedgehog, Wnt, and ghrelin (53). Ghrelin O-acyltransferase (GOAT), the best understood of the MBOATs, acts in the biosynthesis of ghrelin, a hormone that increases appetite (54). GOAT modifies proghrelin, a 28-amino-acid prohormone, on Ser-3 with an octanoyl group that is thought to come from octanoyl-CoA (55). In this reaction, the octanoyl-CoA, which is synthesized in the cytoplasm, is proposed to bind in the membrane-embedded active site of GOAT, and the peptide substrate, which is in the endoplasmic reticulum lumen, is proposed to enter the active site from the opposite side of the membrane (56). All known MBOATs are thought to use hydrophobic substrates as the acyl donor, the acyl acceptor, or both (50). Analogously, DltB may catalyze formation of a d-alanyl-lipid as an intermediate in the pathway (see Fig. 1). Indeed, DltB has been proposed to transfer d-alanine from the carrier protein to undecaprenyl phosphate, the carrier lipid for the biosynthesis of peptidoglycan and wall teichoic acid precursors (36, 39). Although this proposed intermediate has not yet been detected, an alternative intermediate, d-alanylated phosphatidylglycerol, has been identified in lipid extracts from Bacillus subtilis (57).
DltD is a single-pass membrane protein for which several functions have been proposed. These include hydrolyzing misacylated d-Ala-ACP (58), activating DltA catalysis (58), and transferring d-alanine from the undecaprenyl phosphate carrier lipid to teichoic acids outside the cell (38). Recent work showing that DltD is oriented outside the cell excludes a role for DltD in proofreading or activation of DltA (36), suggesting another function in the pathway.
In this report, we describe a new series of assays useful for addressing many questions that remain in the study of the DLT pathway. After establishing the essentiality of all four proteins in the pathway for LTA d-alanylation, we address the role of the last protein in the pathway, DltD. Based on active-site features required for DltD's function, we propose that it forms a covalent intermediate with d-Ala transferred from the acylated product of DltB, which may be a d-Ala-lipid or DltD itself; LTA then serves as the acyl acceptor for subsequent transfer of the d-Ala (Fig. 1).
Results
dltABCD constitutes the minimal dlt operon
We first used RT-PCR and bioinformatics to address which genes are part of the dlt operon in S. aureus and of those which are conserved in other Gram-positive bacteria. Upstream or downstream of the four recognized dlt genes are often found either a dltE or dltX gene, whose functions are unknown. In S. aureus, the DUF3687 family member, dltX, is positioned 16 bp upstream of dltA. dltX is itself preceded by a 50-bp predicted ORF with no obvious homologs, SAOUHSC_00867 (Fig. S1A). Using reverse transcription-PCR with primers spanning dltX-dltA and SAOUHSC_00867-dltX, we found that both of these ORFs are cotranscribed with the dltABCD operon (Fig. S1B). Although dltX was previously identified as being part of the operon and has been linked to DLT function in Bacillus thuringiensis (17, 27), these results identify an additional small ORF in the S. aureus dlt operon. It is unknown whether these ORFs encode peptides. We performed a genome-neighborhood network analysis (59) to determine the frequency with which any genes co-occur near dltABCD using dltD as the query (http://efi.igb.illinois.edu/efi-gnt/). 4 This analysis revealed that dltX is present in fewer than 60% of dltD-encoding organisms; dltE is more common but is not found universally (Fig. S1C). These small genes may encode regulatory factors, but the minimal conserved operon comprises only dltABCD. Following this analysis, we focused on the core components only.
dltABCD are each essential when wall teichoic acid biosynthesis is blocked
The DLT pathway becomes essential when WTA biosynthesis is prevented (60), which enabled us to use a synthetic lethal approach to evaluate the importance of each dlt gene for survival. Tunicamycin potently inhibits the first enzyme in the wall teichoic acid biosynthetic pathway, TarO, and can be used in agar plates to probe essentiality of genes when wall teichoic acids are depleted (Fig. 2A) (60–62). We previously reported that null mutants of dltA and dltD were nonviable in the absence of wall teichoic acids. Here, we tested all deletion mutants in DLT pathway genes on plates containing tunicamycin (Fig. 2B). The WT strain was unaffected by tunicamycin, whereas the dltA-, dltB-, dltC-, and dltD-null mutants all failed to grow on tunicamycin plates even though LTA production was robust (Fig. S2). Complementation with a plasmid expressing the dlt operon restored growth of each mutant on tunicamycin (Fig. 2B). Polar effects on transcription of dltC were excluded in control experiments (Fig. S3), and the requirement for functional DltB was independently confirmed using a small-molecule inhibitor (33). Thus, all four genes in the conserved dltABCD operon are necessary for survival in S. aureus when WTAs are not produced.
Figure 2.
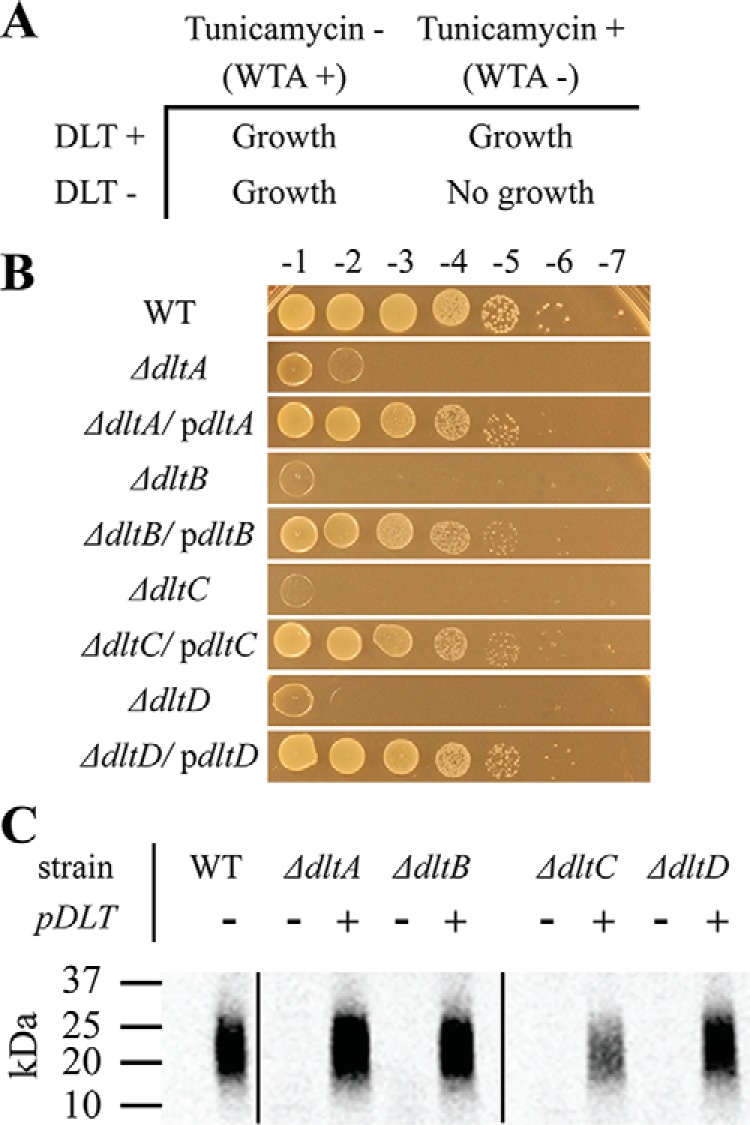
DltABCD are each necessary for LTA d-alanylation. The synthetic lethal interaction between the DLT pathway and WTA biosynthesis pathway allows for dlt selection. Our laboratory previously reported that the natural product, tunicamycin, is a potent inhibitor of TarO in WTA biosynthesis. A illustrates how either TarO inhibition or genetic DLT inactivation alone do not hinder growth, whereas tunicamycin treatment of a dlt-null mutant inhibits bacterial growth. WT S. aureus and null mutants of individual dlt genes were transformed with either plasmid-encoded dltABCD or empty vector. In B, growth on 0.4 μg/ml tunicamycin was used as a readout of DLT activity. The growth dependence of each null mutant on a plasmid-borne copy of the dlt operon demonstrates the essentiality of their respective dlt gene for teichoic acid d-alanylation. To directly detect changes in LTA d-alanylation, the same strains were fed d-[14C]alanine before SDS extraction, SDS-PAGE, and autoradiography as described under “In vivo LTA d-alanylation assay.” C shows the level of d-Ala-LTA produced in each mutant with or without complementation. d-Ala-LTA migrates as a smear between 20 and 25 kDa as compared with protein ladder.
dltABCD are necessary for d-[14C]alanine labeling of LTA in whole cells
The likely explanation for the tunicamycin sensitivity of dlt pathway mutants is that LTA d-alanylation is abolished. To confirm this, we extracted LTA from WT and mutant strains treated with d-[14C]alanine (33). We observed a complete lack of d-alanylated LTA in the DLT pathway mutants; complementation restored d-alanylated LTA (Fig. 2C). These studies establish that each gene in the dlt operon is required for LTA d-alanylation. A pathway reconstitution would thus require all four components.
Kinetics of DltA-catalyzed DltC d-alanylation
DltA orthologs from Lactococcus casei and B. subtilis were previously studied using a pyrophosphate exchange assay (38, 63). Here, we used a continuous, coupled-enzyme assay to monitor transfer of d-alanine to DltC by DltA. Loading of d-alanine onto DltC consumes ATP, which is regenerated at the expense of NADH, allowing for a convenient UV readout. We heterologously expressed and purified S. aureus DltA and DltC from Escherichia coli for in vitro assays. DltC expressed in E. coli was ∼50% post-translationally modified with 4′-phosphopantetheine based on conformationally sensitive native PAGE (Fig. S4). To fully form holoDltC, E. coli 4′-phosphopantetheinyltransferase, AcpS, was cloned, purified, and used to load phosphopantetheine onto apoDltC (Fig. S5; holoDltC is referred to below as DltC). ATP consumption depended on DltC, and no reaction was observed in the absence of DltC. Plots of the initial velocities against DltC concentration from triplicate assays were each fit to the Michaelis–Menten equation (all data plotted together in Fig. 3A), and kinetic constants were calculated (error reported as standard deviation): kcat of 2.45 ± 0.09 s−1, Km or 4 ± 1 μm, and kcat/Km of 6 ± 1 × 105 m−1 s−1. We confirmed that activity in the ATPase assay correlated with DltC d-alanylation by performing a time-course experiment with d-[14C]alanine at saturating DltC and analyzed it by SDS-PAGE (Fig. 3A, inset).
Figure 3.
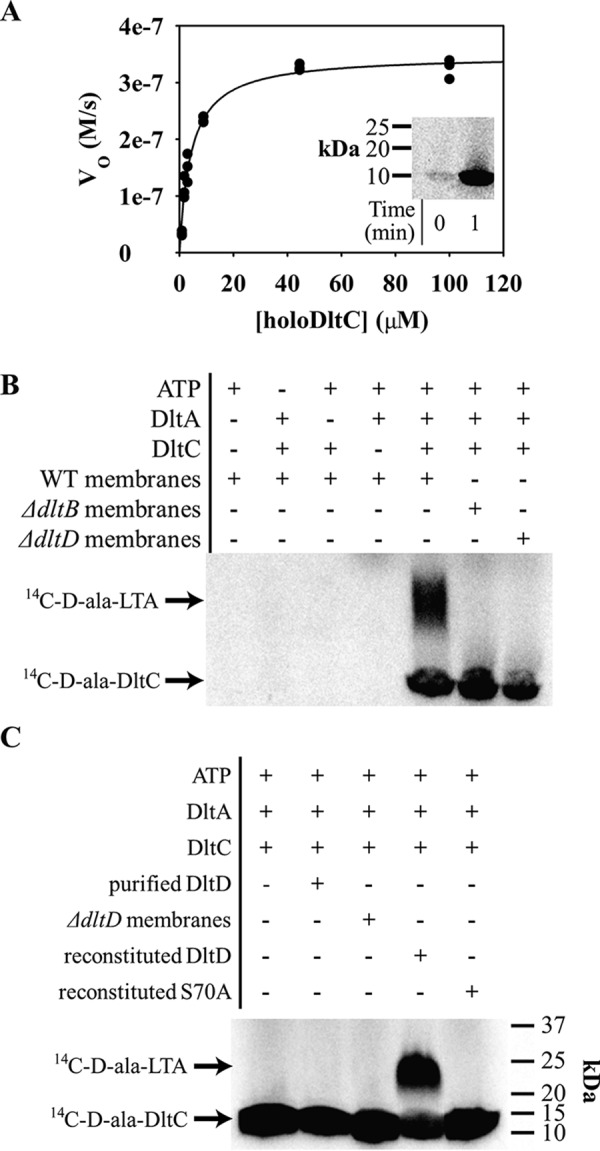
In vitro DLT activity assays show essentiality of each DLT protein for d-alanylation. A continuous ATP-hydrolysis assay for DltA-catalyzed DltC d-alanylation was developed using NADH turnover via a three-enzyme system for ATP regeneration (adenylate kinase, pyruvate kinase, and lactate dehydrogenase). A shows nonlinear curve fitting of V0 with varying concentrations of holoDltC plotted with the Michaelis–Menten equation. Three data sets are shown plotted together. DltA assay conditions were modified slightly for in vitro d-[14C]alanine incorporation into DltC (inset to A). Next, vesicles composed of fractionated membranes from S. aureus and dlt-null mutant cells were incubated with d-[14C]Ala-DltC. The left four lanes in B show that d-[14C]Ala-DltC was produced only when ATP, DltA, and DltC were all added, and no native DltC was contributed from the vesicle preparation. Using vesicles prepared with membranes from dltB- and dltD-null strains, the right three lanes of B show that d-[14C]Ala-LTA formation depends on both DltB and DltD. Lastly, the full-length DltD enzyme was purified and reconstituted into vesicles. The gel in C shows that d-[14C]Ala-LTA formation in vesicles from dltD-null membranes required DltD reconstitution and that an inactivated point mutant of DltD failed to support the same activity when reconstituted.
d-Alanine is transferred to LTA in vesicles containing DltB and DltD, but ongoing LTA synthesis is not required
Neuhaus and co-worker (38) previously demonstrated that radiolabeled d-alanine could be transferred from DltC to L. casei membranes, suggesting the formation of d-alanylated lipoteichoic acid. With d-[14C]Ala-DltC in hand, we set out to build on that work and develop a partial reconstitution of the S. aureus DLT pathway. Vesicles from WT, dltB-null, and dltD-null strains were prepared by homogenization of fractionated membranes, and the formation of d-alanylated LTA was analyzed under different reaction conditions. We found that the reaction depended on d-Ala-DltC formation and occurred only in vesicles prepared from WT membranes, whereas membranes derived from ΔdltB, ΔdltD, or ΔltaS (64) strains did not produce the same species (Fig. 3B and Fig. S6A). LTA d-alanylation was complemented in vesicles prepared from deletion mutants expressing a plasmid-borne dlt operon (Fig. S6A). These results demonstrate that d-[14C]Ala-DltC produced from recombinant protein can be transferred to LTA provided both DltB and DltD are present.
We next sought to test whether the LTA synthase, LtaS, was directly involved in LTA d-alanylation. Because LtaS uses phosphatidylglycerol to construct the LTA polymer, it is plausible that LTA could utilize a phosphatidylglycerol molecule that has been modified with d-alanine. To address this, the presence of full-length LtaS in DLT-active vesicles was tested by Western blotting. Gründling and co-workers (65) have shown that S. aureus LtaS is cleaved in vivo by the type I signal protease, SpsB, but an S218P substitution in LtaS prohibits this cleavage. Here, Western blot analysis of vesicles prepared from S. aureus expressing C-terminally myc-tagged LtaS from a plasmid failed to detect any protein. However, the same strain expressing a plasmid-encoded S218P mutant of C-terminally myc-tagged LtaS showed signal for the full-length myc-tagged LtaS protein in separate vesicle preparations (Fig. S7). SpsB cleavage of WT LtaS appeared to be complete during the vesicle preparation to the detection limits of Western blotting. Because the active site of LtaS requires the C-terminal soluble domain for activity, this result suggests that no active LtaS was present in the vesicles and that the d-Ala-LTA stems from the DLT enzymes in the assay.
A rough kinetic analysis of the partially reconstituted DLT pathway was performed to test whether the in vitro rate was consistent with the expected rate for the pathway in growing S. aureus. First, the dependence of product formation on the concentration of preformed d-[14C]Ala-DltC was determined. By fitting the d-[14C]Ala-LTA intensities from densitometry to a rectangular hyperbola, 32 μm d-[14C]Ala-DltC was calculated to yield half-maximal product formation in vitro (Fig. S8). This saturation behavior was not seen in previous work, but at ∼3 μm, the highest d-[14C]Ala-DltC concentration used would likely be in the linear range (38). Then, the rate of in vitro LTA d-alanylation was estimated from a time course at 67 μm d-[14C]Ala-DltC. By fitting intensities of d-[14C]Ala-LTA formation to a rectangular hyperbola and calculating the maximal product formation at infinite time, the rate of d-[14C]Ala-LTA formation showed a half-life of ∼50 min (Fig. S9). This rate is reasonably close to the 40-min doubling time of S. aureus at 30 °C. Whether 32 μm DltC is similar to the intracellular concentration of DltC is unknown. However, the similarity in the rate of the reconstituted pathway to the rate necessary for vegetative S. aureus growth suggested that the in vitro pathway was a reasonable model for the in vivo pathway.
Activity of purified, full-length DltD can be reconstituted in vesicles
We next tested whether we could reconstitute DltD's LTA d-alanylation activity by reincorporating heterologously expressed and purified DltD into vesicles of S. aureus membranes. DltD solubilized in 0.05% n-dodecyl β-d-maltoside (DDM) was mixed with membranes from the ΔdltD mutant. Lane 4 of Fig. 3C shows d-[14C]Ala-LTA produced from DltD-reconstituted vesicles treated with in vitro loaded d-[14C]Ala-DltC. For the DLT pathway model to be correct (Fig. 1), DltD and d-[14C]Ala-DltC would need to be on opposite sides of the vesicles to affect LTA d-alanylation. However, the inclusion of DDM (≥0.3 mm) from the DltD preparation is thought to allow the spontaneous incorporation of membrane proteins into both sides of vesicles (66). Here, the reconstituted DltD acted in the pathway after integration into the vesicle membranes because the DltD activity was retained in the vesicles after ultracentrifugation (Fig. S6B). LTA d-alanylation was produced from in vitro synthesized d-[14C]Ala-DltC because d-[14C]Ala-LTA formation depended on the addition of pure ATP, DltA, and DltC. Furthermore, substitution of WT DltD for an inactivated S70A mutant (see below) in the reconstitution assay failed to produce d-Ala-LTA (Fig. 3C, lane 5). Additionally, two separate tests of the essentiality of DltB for LTA d-alanylation in DltD-reconstituted membranes were performed. d-[14C]Ala-LTA was not produced when the DltB inhibitor amsacrine (33) was added to the DltD-reconstituted membranes. Also, no LTA d-alanylation was observed when DltD was reconstituted into vesicles prepared from membranes of the dltB-null mutant (Fig. S10). Thus, these data support the essentiality of both DltB and DltD for LTA d-alanylation in vitro.
DltD uses a catalytic dyad to catalyze d-alanyl O-acyl transfer
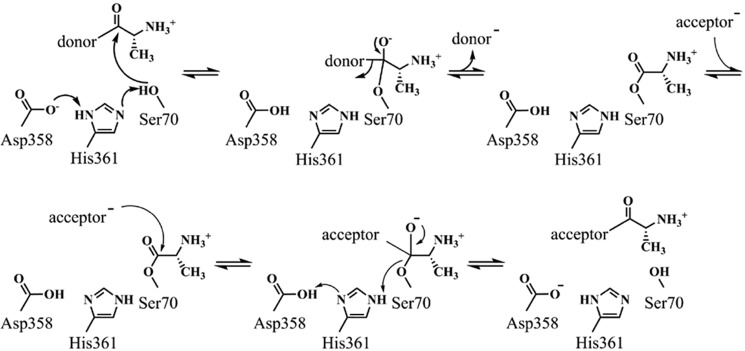
To further investigate the role of DltD in d-alanylation of LTA, we used a combination of bioinformatics and biochemistry to determine the importance of an apparent catalytic residue network in the DltD active site. DltD is a membrane protein with a single transmembrane helix predicted from residues 7 to 27 (Fig. S11). Little homology could be found from BLAST searches of DltD in public databases. However, comparison of the unpublished crystal structure of the DltD ortholog from S. pneumoniae (Protein Data Bank (PDB) code 3BMA) to all other structures in the Protein Data Bank using the DALI server (67) (http://ekhidna.biocenter.helsinki.fi/dali_server)4 reveals structural similarity to a wide range of flavodoxin-like (three-layered αβα sandwich-like) fold proteins, including many members of the SGNH-like family (IPR013830). Fig. 4 shows a homology model of S. aureus DltD calculated using Phyre 2 (68) (http://www.sbg.bio.ic.ac.uk/phyre2/html),4 which used PDB code 3BMA as a template; the two proteins share 20% sequence identity and 65% sequence similarity. Besides the SGNH hydrolases, DltD shows structural similarity to other SGNH-like proteins, including the known or predicted transferases AlgJ, AlgX, WssI, and PatB and the C-terminal domain of OatA (Fig. S12). In addition to sharing the αβα sandwich-like fold, these hydrolases and transferases each contain common active-site features, including a catalytic triad and an oxyanion hole. In S. aureus DltD, Ser-70, His-361, and Asp-358 comprise an apparent catalytic triad, and Gly-100 defines the oxyanion hole (Fig. 4B). The catalytic triad is a common motif in well-studied hydrolytic enzymes; the His and Asp residues work in tandem to activate a serine side chain for attack on a carbonyl (Scheme 1), and the oxyanion formed in the tetrahedral intermediate is stabilized by hydrogen bonds to the backbone amides of the serine nucleophile and a neighboring glycine. This half-reaction is shared with enzymes that use a catalytic triad to effect transfer reactions (69). For transferases, however, the next step involves attack of an organic acyl acceptor rather than water (Scheme 1).
Figure 4.
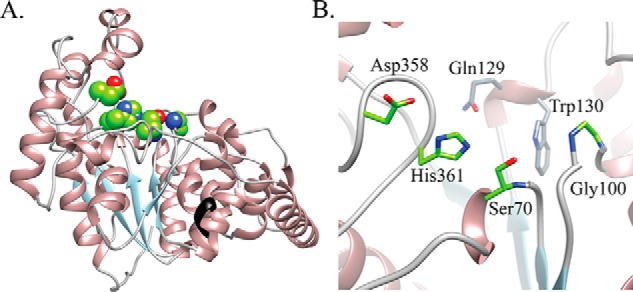
Homology model of S. aureus DltD demonstrates a Ser-His-Asp triad. The Phyre 2–calculated model of the S. aureus DltD is shown in A. An unpublished crystal structure of the ortholog from S. pneumoniae without the transmembrane helix was used as a template (PDB code 3BMA). The enzyme forms an αβα sandwich-like fold, and the black colored ribbon indicates where the N-terminal transmembrane helix would originate (before Glu-33). Structural similarity searches of the Protein Data Bank retrieved several families of SGNH hydrolase-like proteins (Fig. S11). The Ser-His-Asp triad (green sphere representation in A) in the DltD active site has been identified as a catalytic triad in SGNH hydrolases. In the active-site zoomed image in B, the amide backbones of Ser-70 and Gly-100 are well-positioned for oxyanion stabilization, and the conserved DltD residues Gln-129 and Trp-130 shown in gray appear to form the back of the pocket.
Scheme 1.
Proposed mechanism for acyl transfer via DltD's catalytic dyad.
Direct assays of DltD activity were hindered by a lack of knowledge of the d-alanyl acyl-donor substrate. We were unable to detect accumulation of radiolabeled lipids from methanol-chloroform extracts of d-[14C]alanine–fed S. aureus WT, ΔdltD, ΔltaS, or ΔdltDΔltaS cells (data not shown). In lieu of the native substrate, activity of DltD against the model hydrolase substrates p-nitrophenyl acetate, p-nitrophenyl butyrate, and p-nitrophenyl l-alanine was tested. However, none of these activated esters resulted in detectable DltD-dependent release of p-nitrophenol (data not shown). Therefore, LTA d-alanylation was used as a readout of DltD activity in the context of the full DLT pathway.
Our tunicamycin cell-based assay allowed testing of activity in site-directed mutants of the putative Dlt protein active sites (Fig. 5 and Fig. S13). Individual alanine-substitution mutants of the apparent catalytic triad residues showed that S70A and H361A DltD mutations exhibited the same inability to grow as observed for an empty vector in the dltD-null mutant when grown on tunicamycin (Fig. 5A). However, the D358A mutant showed an intermediate level of activity (Fig. 5A). Fig. 5B shows that protein expression levels of each of the mutants was consistent with WT. These findings are not unprecedented. An Asp-to-Ala mutant in E. coli thioesterase I (TAP), a homolog of DltD, retained 40% of WT activity, whereas the Ser and His mutants showed less than 1% (70). Furthermore, some SGNH hydrolase–like proteins are known to function with only a Ser-His dyad (71). Hence, although the active site of DltD contains an apparent catalytic triad, only the Ser-His dyad is essential for function.
Figure 5.
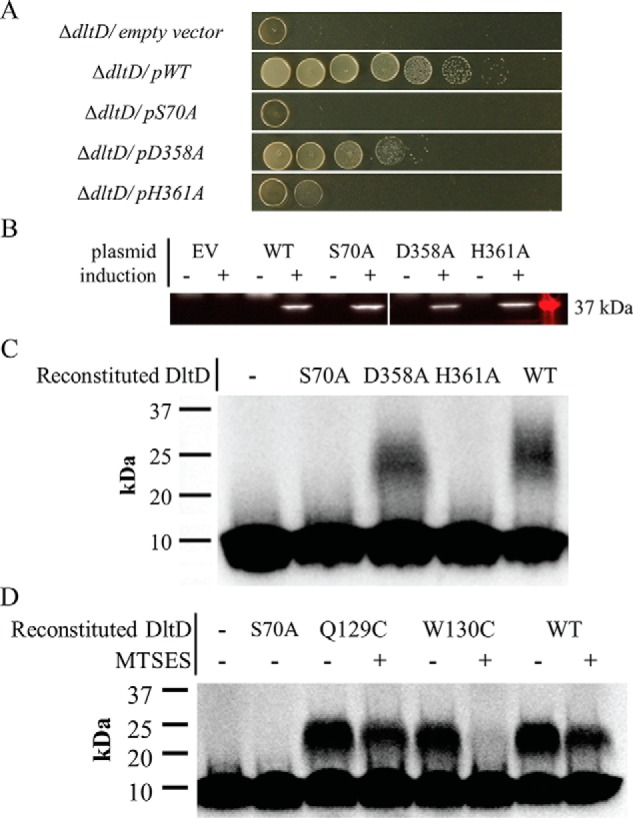
DltD-dependent LTA d-alanylation depends on an active-site pocket that includes Ser-70, Trp-130, His-361, and Asp-358. Site-directed alanine substitutions of DltD's apparent catalytic triad in an S. aureus expression vector were transformed into the S. aureus dltD-null strain. A shows a test of their activity via growth complementation in the presence of 0.4 μg/ml tunicamycin. Serial dilutions were spotted on agar plates from undiluted (left) to 10−7 diluted (right). Here, “−” indicates empty vector (EV), and WT, S70A, D358A, and H361A refer to the myc-dltD alleles expressed as part of plasmid-encoded dltABCD. Western blotting to the N-terminal myc tags fused to each DltD clone allowed confirmation of their expression (B). The tag showed no effect on DltD function (Fig. S15). Then, the mutant DltD enzymes were expressed in E. coli, purified, and reconstituted into vesicles made from S. aureus dltD-null membranes. C shows the result of in vitro LTA d-alanylation assay of these reconstituted DltD vesicles using d-[14C]Ala-DltC. Here, “−” refers to vesicles without DltD reincorporation. Lastly, in panel D, the Q129C and W130C were each tested for their ability to support LTA d-alanylation in vitro with or without the presence of thiol-reactive MTSES. Purified DltD WT and mutants were reconstituted into vesicles of membranes from the S. aureus dltD-null mutant. The reconstituted vesicles were pretreated with 5 mm MTSES where indicated with a “+.” The inactive S70A mutant-reconstituted vesicles as well as vesicles with no DltD added were run in the first two lanes for comparison.
The S70A mutant (see above) as well as D358A and H361A was purified for in vitro LTA d-alanylation assays in reconstituted vesicles. Fig. 5C shows results of assays with vesicles containing each of these mutant proteins (lanes 2–4) compared with WT DltD protein (lane 5) and blank vesicles (no DltD treatment; lane 1). S70A and H361A showed no activity in the form of d-[14C]Ala-LTA production, but again, D358A showed an intermediate level of activity.
Lastly, cysteine substitutions at dispensable residues in the DltD active site were constructed to test whether an active DltD mutant can be chemically inactivated. MTSES has been used as a way to quickly inactivate otherwise active cysteine-substituted mutant enzymes (72). Here, this chemical switch of activity was used to test whether mutations to the active-site pocket inactivate DltD through gross structural changes such as denaturation or via controlled changes to specific side chains. Many SGNH-like proteins contain an active-site asparagine juxtaposed across from the Ser-70 of the catalytic triad, i.e. the “N” of SGNH, but in DltD that asparagine is missing, and Gln-129 and Trp-130 lie roughly in its place (Fig. 4B). Gln-129 and Trp-130 were individually mutated to cysteine, and these mutants were found to have WT activity via tunicamycin growth complementation (Fig. S14) and the in vitro reconstitution assay (Fig. 5D, lanes 3 and 5). However, in vitro LTA d-alanylation assays in the presence of MTSES yielded a complete loss of activity in membrane vesicles with reconstituted W130C but not in WT or Q129C DltD-containing vesicles (Fig. 5D). Therefore, DltD function in the LTA d-alanylation can be chemically inhibited by targeting its active site.
Discussion
Although the dlt operon was implicated in d-alanylation of teichoic acids over 25 years ago, and the essentiality of several components of the dlt operon has previously been experimentally confirmed (12, 38, 39), the requirement of each Dlt protein for LTA d-alanylation has not been shown previously. This work is the first to demonstrate the essentiality of each of the four conserved proteins in the dlt operon for LTA d-alanylation using biochemistry.
The mechanism for teichoic acid d-alanylation by the DLT pathway is largely unknown. The role of DltA in the esterification of d-alanine onto DltC has been demonstrated with genetics or in vitro activity for homologs from many organisms, e.g. L. casei (37, 38, 44), B. subtilis (39, 63), Lactobacillus rhamnosus (45, 58), S. aureus (12), Streptococcus mutans (15, 24, 73), Streptococcus gordonii (29), S. pneumoniae (5), and Bacillus cereus (40). Although studies in many of these organisms clearly link DltC to the DLT pathway, the fate of d-Ala-DltC is not clear. Similarly, DltD in L. rhamnosus was shown to be essential for the DLT pathway using genetics (22, 58), but the hypothesis that DltD transfers d-alanine to LTA is untested. The in vitro activities reported for DltD from L. rhamnosus involving proofreading and activation of DltA (58) are likely artifacts given the more recent report that DltD faces outside the cell (36). Genetic work on dltB in S. mutans (24), dltD in Clostridium difficile (74), and both dltB and dltD in B. subtilis (39, 75) and S. aureus (12) link these genes to the DLT pathway. However, the possibility of polar effects in these mutant strains calls into question the causality of each gene's mutation to the phenotypes measured. For these reasons, the entire DLT pathway following formation of d-Ala-DltC by DltA remains unknown, and a biochemical approach is needed.
Biochemical experiments aimed at testing the roles of each of the DLT proteins are difficult for several reasons. First, the last two proteins in the pathway are embedded in the plasma membrane. Second, analyzing large d-alanylated polymers poses challenges absent for reactions that produce small, nonpolymeric products. Finally, d-Ala-intermediates may form in the pathway but have not been identified. To overcome these obstacles, we utilized the full DLT pathway using d-[14C]alanylation of LTA in vivo and in vitro to detect activity of individual components. This work focused on LTA rather than WTA d-alanylation because it is clear from previous studies that d-alanine is added to LTA before WTA (35, 36). Genetic and whole-cell labeling experiments with null mutants of each of the four individual dlt genes demonstrated that each was essential for LTA d-alanylation in vivo. A partial reconstitution in which DltA, DltC, and DltD are added to vesicles containing DltB and preformed LTA provides a more controlled analysis platform. By omitting components and using vesicles prepared from dltB- and ltaS-null strains, we confirmed that all four DLT components and LTA, but not LtaS itself, are required for LTA d-alanylation. The results here are the first to show that DltD acts directly in LTA d-alanylation.
We used our in vivo and in vitro assays to probe whether DltD is an enzyme as suggested by the presence of an apparent catalytic triad and homology to some characterized enzymes. We identified Ser-70 and His-361 as essential for d-alanylation activity and further showed that chemical modification of a proximal active-site residue, W130C, in an otherwise active protein abolishes d-alanylation. The essentiality of this apparent active site to the role of DltD in LTA d-alanylation, together with the requirement for an O-acyltransferase in the DLT pathway, suggests that DltD acts enzymatically to transfer d-alanine ester to LTA.
A well-known example of acyl transfer using a catalytic serine is malonyl-ACP transacetylase (FabD), an enzyme involved in fatty acid biosynthesis (76). FabD accepts malonate from a malonyl-CoA donor and transfers it to ACP. Malonate is loaded on an active-site serine, a transfer reaction facilitated by a histidine in the active site (77). This enzyme-bound acyl intermediate is then attacked by the sulfhydryl group of holo-ACP, regenerating FabD. We propose that the DltD-catalyzed reaction is analogous to this general mechanism in which a d-Ala-DltD acyl intermediate is formed followed by nucleophilic attack by the 2-hydroxyl group of a glycerol from LTA on this acyl-enzyme intermediate to regenerate the enzyme and produce d-Ala-LTA (Scheme 1).
The d-Ala-ester donor that presumably loads Ser-70 with d-alanine is still unknown (4). Currently, there are two hypotheses concerning the loading of d-Ala onto DltD. One hypothesis is that undecaprenyl phosphate is used as the carrier lipid to form a d-Ala-phosphoester (36, 38, 39); however, acyl transfer to form a d-Ala-phosphoester from a d-Ala-thioester is thermodynamically unfavorable. Alternatively, DltB could use one of the more abundant phospholipids in S. aureus membranes, such as phosphatidylglycerol (78), to form a d-Ala-ester–linked lipid intermediate that could then be used as a substrate by DltD. The plausibility of a d-Ala-lipid species in the DLT pathway is supported by analysis of DltB homologs from the MBOAT family, which directly acylate membrane components such as cholesterol (acyl-CoA:cholesterol acyltransferase) (79), triglycerides (diglyceride acyltransferase) (51), and phosphatidylinositol (MBOAT7) (80). Previously, Atila and Luo detected d-Ala-phosphatidylglycerol from B. subtilis, but they lacked a specific test for the species' dependence on the dlt operon (57). Attempts in this work to observe d-Ala-phosphatidylglycerol in S. aureus failed, but this negative result does not establish the absence of such a species in S. aureus.
A second hypothesis is that d-Ala is directly transferred from the phosphopantetheine arm of DltC onto DltD. This would require the active site of DltD, which contains the serine nucleophile, to access the active site of DltB where this thioester would be presented. The DltB active site is formed by a set of conserved residues on predicted transmembrane helices that are embedded in the membrane. DltB would facilitate transfer via conserved histidine residues, and DltD would then transfer d-Ala onto LTA. Our cell-based assay for tunicamycin growth complementation confirmed the necessity of the conserved MBOAT residues His-294 and His-341 to DltB function, implying that DltB follows a similar enzymatic mechanism as other MBOATs for acyl transfer (Fig. S13). Several MBOAT proteins require a DltD homolog for acyl transfer to various extracellular polymers, including PatAB (peptidoglycan), WssHI (cellulose), and AlgIJ (alginate) (69). The evolutionary link between DltB and DltD homologs for acylation of extracellular polymers is consistent with our proposal that DltD acts directly in LTA acylation but does not clarify whether DltD is acylated by a lipid intermediate or the DltC-bound thioester. Further work is required to resolve this issue, and the reconstitution described here may help answer this question.
Experimental procedures
Materials
Ampicillin, chloramphenicol, kanamycin, neomycin, erythromycin, tunicamycin, anhydrotetracycline, isopropyl thiogalactoside (IPTG), lysostaphin, d-[1-14C]alanine (American Radiolabeled Chemicals, 0341A), β-nicotinamide adenine dinucleotide (NADH; Sigma-Aldrich, 43420), phosphoenolpyruvate (Sigma-Aldrich, P0564), adenylate kinase (Sigma-Aldrich, M3003), pyruvate kinase (Sigma-Aldrich, P9136), and lactate dehydrogenase (Sigma-Aldrich, 59747) were obtained from the indicated sources. All reagents were purchased at the highest quality available.
S. aureus gene deletion
The ORFs for dltA and dltD were previously deleted from S. aureus strain Newman (60). The procedure for allelic replacement of dltD with a kanamycin resistance cassette was repeated for dltB and dltC (60, 81).
Cloning and site-directed mutagenesis
dltA, dltC, and dltD were each cloned from S. aureus RN4220 into an E. coli expression plasmid, pET15b. The primary structures of these gene products are identical across lab strains, e.g. RN4220 and HG003, and methicillin-resistant S. aureus strains common in the United States, e.g. MW2 and USA300. DltA and DltD were amplified using primers PMW56-PMW57 and PMW60-PMW61, respectively (see Table S1 for primer sequences). The PCR products were cloned into pET15b-NdeI/BamHI (NdeI- and BamHI-digested pET15b) via a TaKaRa Infusion kit (catalog number 638917). An N-terminally truncated form of DltD, DltDtrunc, lacking the first 27 residues of the protein that contain the transmembrane helix was also cloned into pET15b-NdeI/BamHI via the InFusion kit using primers PMW62-PMW61. DltC was cloned via T4 DNA ligation into pET15b-NheI/BamHI following amplification with primers PMW14-PMW8 and similar digestion. Lastly, the E. coli acpS gene for holo-[acyl-carrier-protein] synthase from E. coli BL21(DE3) was also cloned via the InFusion kit into pET15b-NdeI/BamHI using primers PMW63-PMW64.
For expression in S. aureus, the dltABCD operon was cloned using primers SM61-SM64 and SM148-SM149, respectively, into both pLOW-SalI/BamHI and pTP63-KpnI/BlpI via T4 DNA ligation. dltD was also cloned individually into pTP63-KpnI/BlpI with primers PMW181-SM149. Using the InFusion kit, ltaS was cloned into pTP63-KpnI/BlpI with either an N-terminal myc tag (primers PMW214, -215, and -216) or a C-terminal myc tag (primers PMW217, -218, and -219). The construction required three primers and two PCRs because of the length of the myc tag and ribosome-binding site being added.
Insertion of a myc tag at the N terminus of dltD in pTP63-DLT was achieved by overlap extension PCR (82). The dltABCD operon was amplified in two halves from pTP63-DLT using primers SM148-PMW143 and PMW142-SM149 in two separate reactions. These two PCR products, which anneal at the myc tag following the dltC gene and at the 5′-end of the dltD gene, were used in a megaprimer PCR with primers SM148-SM149 to produce the full DLT(myc-dltD). This final PCR product was cloned back into pTP63-KpnI/BlpI via T4 DNA ligase. Fusing dltD with a C-terminal myc tag in the pTP63-DLT plasmid only required amplification with SM148-PMW144 and religation with pTP63-KpnI/BlpI via T4 DNA ligase.
Site-directed mutagenesis was carried out in dltB, dltD, and ltaS via overlap extension PCR (82). In short, point mutations were created by amplifying individual genes (pET15b-dltD and pTP63-ltaS-myc) or the entire dltABCD operon (pTP63-DLT(DltBmyc) and pTP63-DLT(mycDltD)) in two halves in separate PCRs where the two PCR products anneal at the site to be changed in a subsequent megaprimer PCR reaction as described above. Primers for each mutation were the same regardless of the destined plasmid and can be found in Table S1. For heterologous expression and purification, the following DltD mutants were created in pET15b-dltD: S70A, E71A, E71N, E71Q, Q129A, Q129C, W130A, W130C, D358A, and H361A. For expression in S. aureus, the following mutations were made in pTP63-DLT(myc-DltD): S70A, S70V, S70C, E71A, E71N, E71Q, Q129A, Q129C, W130A, W130C, D358A, D358C, and H361A. Also, for expression in S. aureus, the following mutations were made in pTP63-DLT(DltB-myc): H294A, H294N, and H341A. Lastly, for expression in S. aureus, the S218P mutation was created in the plasmid pTP63-ltaS-myc.
Protein purification
The pET15b clones were transformed into E. coli BL21(DE3) for protein expression. For protein expression, each strain was grown in 4 liters of Luria-Bertani broth with 100 μg/ml ampicillin for 40 h at 20 °C with 0.5 mm IPTG added after 16 h for induction. For the purification of DltA, DltC, and AcpS, harvested cells were stored at −80 °C before thawing and resuspending on ice in binding buffer (0.5 m NaCl, 20 mm Tris, pH 7.9, and 5 mm imidazole) with 5 mg of DNase added and lysing on an Avestin EmulsiFlex-C5. Insoluble cell debris was cleared twice at 10,000 × g for 30 min, and the supernatant was loaded onto a 5-ml Ni-Sepharose gravity column pre-equilibrated with binding buffer at 4 °C. The loaded column was washed with 10 ml of binding buffer with 60 mm imidazole before a stepwise gradient of imidazole (60, 100, 150, 200, 250, 300, 500, and 1000 mm), each in 3 ml of binding buffer, was run on the column. The fractions containing the desired protein were identified by SDS-PAGE and pooled before loading onto a Superdex 200 column. Protein was eluted from the Superdex 200 column using an isocratic elution via FPLC of 150 mm NaCl and 20 mm Tris, pH 7.9. Each protein was then dialyzed into a unique storage buffer before concentrating on an Amicon centrifugal filter and flash freezing in liquid nitrogen. The storage buffer was 5 mm NaCl and 20 mm Tris, pH 7.8, for DltA, and AcpS received the same with the addition of 10% glycerol.
For DltC, the pooled fractions from Superdex 200 purification were further treated with both thrombin and AcpS in one pot for hexahistidine tag removal and phosphopantetheinylation. The reaction in 10 ml was composed of 675 μm DltC, 5 μm His6-AcpS, 1.8 mm CoA, 2 mm DTT, 10 mm MgCl2, 500 units of thrombin (GE Healthcare), 75 mm NaCl, and 25 mm Tris, pH 7.8. The reaction was run at room temperature overnight. After confirmation of holoDltC formation by native PAGE with and without DTT, the holoDltC was dialyzed into 20 mm Tris, pH 7.8; concentrated on a 3-kDa molecular mass-cutoff centrifugal filter; and purified on a Mono Q column. HoloDltC was eluted from the Mono Q column with a gradient of NaCl from 0 to 1 m in 20 mm Tris, pH 7.8. Purified holoDltC was then dialyzed against DltC storage buffer (1 mm DTT in 30 mm bis-Tris, pH 6.6), concentrated on 3-kDa molecular-mass cutoff centrifugal filter, and flash frozen in liquid nitrogen.
DltD is a single-pass membrane protein, so the protein purification was altered from above. After protein expression in BL21(DE3) and crude lysate preparation as described above, the supernatant was cleared of cell debris at 5,000 × g for 10 min twice before ultracentrifugation at 100,000 × g for 30 min. The final pellet was transferred to a Dounce homogenizer and homogenized with binding buffer containing 1% Triton X-100. The homogenate was then ultracentrifuged at 100,000 × g for an additional 30 min. The remaining steps of the purification were the same as for DltA and AcpS except that 0.05% DDM was included in all of the buffers.
Tunicamycin complementation assay
S. aureus strain Newman and dlt-null mutants derived from Newman were each transformed with pLOW-DLT or empty vector. These strains were each grown in tryptic soy broth (TSB) with appropriate antibiotic overnight before normalization by A600 and serial dilution. Each dilution was spot-plated onto TSB agar plates containing 5 μg/ml erythromycin and 1 mm IPTG with or without 0.4 μg/ml tunicamycin. The plates were incubated at 30 °C for 24 h before imaging. For complementation of the dltD-null mutant with site-directed mutants of DltD, the pTP63-DLT(myc-dltD) plasmid was used, and site-directed mutants were derived from that plasmid. Again, Newman WT and the Newman dltD-null mutant carrying either empty vector or pTP63-DLT(myc-dltD) with WT or mutant copies of dltD were grown overnight in TSB with 10 μg/ml chloramphenicol (Cm). These strains were each normalized by A600 and serially diluted into TSB. Each serial dilution was spot-plated onto each of four TSB-agar plates: (a) 10 μg/ml Cm, (b) 0.4 μm anhydrotetracycline + 10 μg/ml Cm, (c) 0.4 μg/ml tunicamycin + 10 μg/ml Cm, and (d) 0.4 μm anhydrotetracycline + 0.4 μg/ml tunicamycin + 10 μg/ml Cm. All of the plates were incubated at 30 °C for 24 h before imaging.
In vivo LTA d-alanylation assay
Feeding experiments and SDS-PAGE analysis of LTA derived from radiolabeled cells were carried out as previously described with a few exceptions (33). S. aureus strain Newman and dlt-null mutants derived from Newman were grown to mid-log phase (A600 ∼0.6) in TSB with appropriate antibiotics at 30 °C. The cells were normalized to 1 ml of culture at an A600 equal to 0.6, and the resulting cells pellets were resuspended in 100 μl of diluted TSB (4-fold) at pH 6 (via HCl) with 10 mm glucose and 200 μg/ml d-cycloserine. The cell suspensions were incubated at 30 °C for 30 min before addition of 1 μl of 1.43 nCi/μl d-[14C]alanine and incubation at 30 °C for an additional 30 min. The cells were then pelleted and stored at −20 °C after removal of the supernatant. The pellets were then resuspended in 30 μl of SDS loading dye, boiled for 5 min, and pelleted for 2 min at 20,000 × g, and 20 μl were separated by 4–20% Tris/glycine SDS-PAGE. The resulting gel was dried and imaged by autoradiography using a phosphor screen and Typhoon FLA9500 imager.
In vitro DltC d-alanylation assay
The DltA-catalyzed ATP-hydrolysis reaction was monitored continuously in vitro via NADH turnover at 340 nm (ϵ340,NADH 6220 m−1 cm−1) using a coupled-enzyme assay (83) and a Cary 300 UV/visible spectrophotometer. The components of the assay included:147 nm DltA with 5 mm d-alanine, 12 mm NADH, 30 mm phosphoenolpyruvate, 0.5 mm DTT, 10 mm MgCl2, 50 mm NaCl, and 30 mm bis-Tris, pH 6.6, with coupling enzymes (0.03 unit/μl adenylate kinase, 0.02 unit/μl pyruvate kinase, and 0.06 unit/μl lactate dehydrogenase). The concentration of holoDltC was varied from 1.5 to 100 μm. The assays were first set up without DltA or ATP, and upon addition of ATP, the consumption of contaminating ADP and AMP was observed to completion (<1 min) before initiating the reaction with DltA. The dependence of the reaction rate on DltA and not the coupling enzymes was verified by ensuring that the rate was linear with DltA concentration and that the rate was not affected by 2-fold changes in coupling enzyme concentration. Curve fitting of the initial velocities with varying holoDltC concentration to the Michaelis–Menten equation was performed using the program SigmaPlot. Error was determined by standard deviation from calculated rate constants from the three independent sets of assays. The protocol for radiolabeling DltC can be found under “In vitro LTA d-alanylation assay.”
In vitro LTA d-alanylation assay
To supply the membrane-bound components of the DLT pathway, vesicles were prepared by homogenization of S. aureus membranes. Membranes were routinely prepared by fractionating S. aureus lysate from 1 liter of culture grown to mid-log phase (A600 of about 0.6) in TSB medium with appropriate antibiotic. In short, cell pellets from these cultures were stored at −80 °C for at least a day before thawing on ice and resuspension in 20 ml of digestion buffer (0.5 m sucrose, 10 mm MgCl2, and 100 mm Tris, pH 7.0). Aliquots of 100 μl of 10 mg/ml lysostaphin and 100 μl of 10 mg/ml lysozyme each were added, and the peptidoglycan digestion reaction was incubated at 37 °C for 2 h before pelleting the sucrose-stabilized protoplasts at 5000 × g for 10 min. The pellets were resuspended in 20 ml of lysis buffer (10 mm MgCl2, 0.2 mg/ml DNase, and 100 mm Tris, pH 7.8). Lysis of the cells began with addition of the hypotonic medium, but the suspension was passed through an Avestin Emulsi-Flex C5 for 4 min to ensure thorough lysis. Cell debris was pelleted at 5000 × g before the supernatant was ultracentrifuged at 100,000 × g for 30 min. The pellet was resuspended in membranes buffer (50 mm NaCl, 20% glycerol, and 30 mm bis-Tris, pH 6.6) using a Dounce homogenizer, and the resulting vesicle suspension was flash frozen as ∼25-μl pellets in liquid nitrogen. The vesicles were stored at −80 °C. The concentration of the vesicles was approximated via a bicinchoninic acid (BCA) total protein assay (Thermo Fisher, 23225) using a BSA standard curve.
For in vitro LTA labeling assays, vesicles corresponding to 30 μg of total membrane protein were routinely used. The vesicles were thawed on ice and added to an altered DltC labeling reaction (200 μm ATP, 5.3 μm DltA, 10 μm holoDltC, 5 mm tris(2-carboxyethyl)phosphine, 91 μm (55 mCi/mmol) d-[14C]alanine, 10.2 mm MgCl2, 50 mm NaCl, and 30 mm bis-Tris, pH 6.6). The final volume for these reactions was 20 μl. This mixture was incubated at 30 °C for 30 min before addition of SDS loading dye and immediate 4–20% SDS-PAGE. The gel was dried under vacuum and exposed to a phosphor storage screen. The screen was imaged on the Typhoon imager after ∼65 h of exposure.
Kinetic experiments with varying holoDltC concentration and time were also conducted. For these reactions, the DltC d-alanylation reaction was carried out to completion by setting up the above reaction with a higher concentration of d-alanine (0.5 mm; 8.9 mCi/mmol [14C]alanine) in the absence of vesicles and incubating at 30 °C for 15 min. The holoDltC concentrations used were 0, 8.3, 17, 33, 67, and 130 μm holoDltC. Then, 5 mm MTSES was added, and the reaction was incubated at room temperature for 3 min to inactivate DltA. Finally, the vesicles were added, and the above procedure was followed for incubation, SDS-PAGE, and autoradiography. For the time-course experiment, the same procedure was followed as for varying holoDltC concentration except that the DltC labeling at 70 μm holoDltC and subsequent DltA inactivation reaction were prepared in a single, scaled-up reaction, which was then aliquoted for addition of membranes. Besides a zero time point, the replicate reactions were removed from 30 °C after 5-, 15-, 30-, 45-, and 60-min incubation, and the reaction was quenched by addition of room temperature SDS loading dye, mixing, and then incubation on ice until all reactions could be separated by SDS-PAGE for autoradiography.
DltD-reconstitution LTA d-alanylation
Purified, full-length WT and site-directed mutants of DltD as well as vesicles from membranes of the S. aureus dltD-null mutant were prepared as described above. The in vitro LTA d-alanylation reaction described in the previous section was altered only to include a premixing, “reconstitution reaction” for the vesicles in which DltD protein in DltD storage buffer was pipetted several times with the vesicles and allowed to incubate at 30 °C for 5 min before aliquoting into assays. Vesicles corresponding to 36 μg of total membrane protein before addition of DltD were used, and 8 μm DltD was added to these in the 6-μl-total volume reconstitution reaction. After incubation, the DltD-reconstituted vesicles were added to 14 μl of d-[14C]Ala-DltC charging reaction and treated as described above for in vitro LTA d-alanylation.
Author contributions
B. M. W., J. P. S. M., L. M. M., C. R. V., and S. W. conceptualization; B. M. W., J. P. S. M., and L. M. M. data curation; B. M. W., J. P. S. M., L. M. M., and C. R. V. formal analysis; B. M. W. investigation; B. M. W., J. P. S. M., L. M. M., and S. W. writing-original draft; B. M. W. project administration; B. M. W., C. R. V., and S. W. writing-review and editing; S. W. supervision; S. W. funding acquisition.
Supplementary Material
Acknowledgment
We thank T. Pang (Department of Microbiology and Immunobiology, Harvard Medical School) for providing plasmids.
This work was supported by National Institutes of Health Grants R01AI099144, R21AI119892, and P01AI083214 (to S. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S16 and Tables S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- LTA
- lipoteichoic acid
- WTA
- wall teichoic acid
- DLT
- d-alanyl lipoteichoic acid
- Cm
- chloramphenicol
- ACP
- acyl carrier protein
- MTSES
- (2-sulfonatoethyl)methanethiosulfonate
- MBOAT
- membrane-bound O-acyltransferase
- GOAT
- ghrelin O-acyltransferase
- DDM
- n-dodecyl β-d-maltoside
- IPTG
- isopropyl thiogalactoside
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TSB
- tryptic soy broth.
References
- 1. Weidenmaier C., and Peschel A. (2008) Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6, 276–287 10.1038/nrmicro1861 [DOI] [PubMed] [Google Scholar]
- 2. Percy M. G., and Gründling A. (2014) Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu. Rev. Microbiol. 68, 81–100 10.1146/annurev-micro-091213-112949 [DOI] [PubMed] [Google Scholar]
- 3. Brown S., Santa Maria J. P. Jr., and Walker S. (2013) Wall teichoic acids of Gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 10.1146/annurev-micro-092412-155620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neuhaus F. C., and Baddiley J. (2003) A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 686–723 10.1128/MMBR.67.4.686-723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kovács M., Halfmann A., Fedtke I., Heintz M., Peschel A., Vollmer W., Hakenbeck R., and Brückner R. (2006) A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in Gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188, 5797–5805 10.1128/JB.00336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fabretti F., Theilacker C., Baldassarri L., Kaczynski Z., Kropec A., Holst O., and Huebner J. (2006) Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 74, 4164–4171 10.1128/IAI.00111-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wobser D., Ali L., Grohmann E., Huebner J., and Sakinc T. (2014) A novel role for D-alanylation of lipoteichoic acid of Enterococcus faecalis in urinary tract infection. PLoS One 9, e107827 10.1371/journal.pone.0107827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Archibald A. R., Baddiley J., and Heptinstall S. (1973) The alanine ester content and magnesium binding capacity of walls of Staphylococcus aureus H grown at different pH values. Biochim. Biophys. Acta 291, 629–634 10.1016/0005-2736(73)90468-9 [DOI] [PubMed] [Google Scholar]
- 9. Fischer W., and Rösel P. (1980) The alanine ester substitution of lipoteichoic acid (LTA) in Staphylococcus aureus. FEBS Lett. 119, 224–226 10.1016/0014-5793(80)80257-2 [DOI] [PubMed] [Google Scholar]
- 10. MacArthur A. E., and Archibald A. R. (1984) Effect of culture pH on the D-alanine ester content of lipoteichoic acid in Staphylococcus aureus. J. Bacteriol. 160, 792–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruhland G. J., and Fiedler F. (1990) Occurrence and structure of lipoteichoic acids in the genus Staphylococcus. Arch. Microbiol. 154, 375–379 10.1007/BF00276534 [DOI] [PubMed] [Google Scholar]
- 12. Peschel A., Otto M., Jack R. W., Kalbacher H., Jung G., and Götz F. (1999) Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410 10.1074/jbc.274.13.8405 [DOI] [PubMed] [Google Scholar]
- 13. Kristian S. A., Datta V., Weidenmaier C., Kansal R., Fedtke I., Peschel A., Gallo R. L., and Nizet V. (2005) D-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187, 6719–6725 10.1128/JB.187.19.6719-6725.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mishra N. N., Bayer A. S., Weidenmaier C., Grau T., Wanner S., Stefani S., Cafiso V., Bertuccio T., Yeaman M. R., Nast C. C., and Yang S.-J. (2014) Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: relative roles of mprF and dlt operons. PLoS One 9, e107426 10.1371/journal.pone.0107426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nilsson M., Rybtke M., Givskov M., Høiby N., Twetman S., and Tolker-Nielsen T. (2016) The dlt genes play a role in antimicrobial tolerance of Streptococcus mutans biofilms. Int. J. Antimicrob. Agents 48, 298–304 10.1016/j.ijantimicag.2016.06.019 [DOI] [PubMed] [Google Scholar]
- 16. Kang K.-M., Mishra N. N., Park K. T., Lee G.-Y., Park Y. H., Bayer A. S., and Yang S.-J. (2017) Phenotypic and genotypic correlates of daptomycin-resistant methicillin-susceptible Staphylococcus aureus clinical isolates. J. Microbiol. 55, 153–159 10.1007/s12275-017-6509-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamar R., Réjasse A., Jéhanno I., Attieh Z., Courtin P., Chapot-Chartier M.-P., Nielsen-Leroux C., Lereclus D., El Chamy L., Kallassy M., and Sanchis-Borja V. (2017) DltX of Bacillus thuringiensis is essential for D-alanylation of teichoic acids and resistance to antimicrobial response in insects. Front. Microbiol. 8, 1437 10.3389/fmicb.2017.01437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wecke J., Perego M., and Fischer W. (1996) D-Alanine deprivation of Bacillus subtilis teichoic acids is without effect on cell growth and morphology but affects the autolytic activity. Microb. Drug Resist. 2, 123–129 10.1089/mdr.1996.2.123 [DOI] [PubMed] [Google Scholar]
- 19. Wecke J., Madela K., and Fischer W. (1997) The absence of D-alanine from lipoteichoic acid and wall teichoic acid alters surface charge, enhances autolysis and increases susceptibility to methicillin in Bacillus subtilis. Microbiology 143, 2953–2960 10.1099/00221287-143-9-2953 [DOI] [PubMed] [Google Scholar]
- 20. Nakao A., Imai S., and Takano T. (2000) Transposon-mediated insertional mutagenesis of the D-alanyl-lipoteichoic acid (dlt) operon raises methicillin resistance in Staphylococcus aureus. Res. Microbiol. 151, 823–829 10.1016/S0923-2508(00)01148-7 [DOI] [PubMed] [Google Scholar]
- 21. Steen A., Palumbo E., Deghorain M., Cocconcelli P. S., Delcour J., Kuipers O. P., Kok J., Buist G., and Hols P. (2005) Autolysis of Lactococcus lactis is increased upon D-alanine depletion of peptidoglycan and lipoteichoic acids. J. Bacteriol. 187, 114–124 10.1128/JB.187.1.114-124.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perea Vélez M., Verhoeven T. L., Draing C., Von Aulock S., Pfitzenmaier M., Geyer A., Lambrichts I., Grangette C., Pot B., Vanderleyden J., and De Keersmaecker S. C. (2007) Functional analysis of D-alanylation of lipoteichoic acid in the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73, 3595–3604 10.1128/AEM.02083-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walter J., Loach D. M., Alqumber M., Rockel C., Hermann C., Pfitzenmaier M., and Tannock G. W. (2007) D-Alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100–23 results in impaired colonization of the mouse gastrointestinal tract. Environ. Microbiol. 9, 1750–1760 10.1111/j.1462-2920.2007.01292.x [DOI] [PubMed] [Google Scholar]
- 24. Boyd D. A., Cvitkovitch D. G., Bleiweis A. S., Kiriukhin M. Y., Debabov D. V., Neuhaus F. C., and Hamilton I. R. (2000) Defects in D-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182, 6055–6065 10.1128/JB.182.21.6055-6065.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Revilla-Guarinos A., Alcántara C., Rozès N., Voigt B., and Zúñiga M. (2014) Characterization of the response to low pH of Lactobacillus casei ΔRR12, a mutant strain with low D-alanylation activity and sensitivity to low pH. J. Appl. Microbiol. 116, 1250–1261 10.1111/jam.12442 [DOI] [PubMed] [Google Scholar]
- 26. Lambert P. A., Hancock I. C., and Baddiley J. (1975) Influence of alanyl ester residues on the binding of magnesium ions to teichoic acids. Biochem. J. 151, 671–676 10.1042/bj1510671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koprivnjak T., Mlakar V., Swanson L., Fournier B., Peschel A., and Weiss J. P. (2006) Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 188, 3622–3630 10.1128/JB.188.10.3622-3630.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hyyryläinen H.-L., Vitikainen M., Thwaite J., Wu H., Sarvas M., Harwood C. R., Kontinen V. P., and Stephenson K. (2000) D-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J. Biol. Chem. 275, 26696–26703 10.1074/jbc.M003804200 [DOI] [PubMed] [Google Scholar]
- 29. Clemans D. L., Kolenbrander P. E., Debabov D. V., Zhang Q., Lunsford R. D., Sakone H., Whittaker C. J., Heaton M. P., and Neuhaus F. C. (1999) Insertional inactivation of genes responsible for the D-alanylation of lipoteichoic acid in Streptococcus gordonii DL1 (Challis) affects intrageneric coaggregations. Infect. Immun. 67, 2464–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gross M., Cramton S. E., Götz F., and Peschel A. (2001) Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69, 3423–3426 10.1128/IAI.69.5.3423-3426.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collins L. V., Kristian S. A., Weidenmaier C., Faigle M., Van Kessel K. P., Van Strijp J. A., Götz F., Neumeister B., and Peschel A. (2002) Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 186, 214–219 10.1086/341454 [DOI] [PubMed] [Google Scholar]
- 32. Hunt C. L., Nauseef W. M., and Weiss J. P. (2006) Effect of D-alanylation of (lipo)teichoic acids of Staphylococcus aureus on host secretory phospholipase A2 action before and after phagocytosis by human neutrophils. J. Immunol. 176, 4987–4994 10.4049/jimmunol.176.8.4987 [DOI] [PubMed] [Google Scholar]
- 33. Pasquina L., Santa Maria J. P. Jr., McKay Wood B., Moussa S. H., Matano L. M., Santiago M., Martin S. E., Lee W., Meredith T. C., and Walker S. (2016) A synthetic lethal approach for compound and target identification in Staphylococcus aureus. Nat. Chem. Biol. 12, 40–45 10.1038/nchembio.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matano L. M., Morris H. G., Wood B. M., Meredith T. C., and Walker S. (2016) Accelerating the discovery of antibacterial compounds using pathway-directed whole cell screening. Bioorg. Med. Chem. 24, 6307–6314 10.1016/j.bmc.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koch H. U., Döker R., and Fischer W. (1985) Maintenance of D-alanine ester substitution of lipoteichoic acid by reesterification in Staphylococcus aureus. J. Bacteriol. 164, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reichmann N. T., Cassona C. P., and Gründling A. (2013) Revised mechanism of D-alanine incorporation into cell wall polymers in Gram-positive bacteria. Microbiology 159, 1868–1877 10.1099/mic.0.069898-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heaton M. P., and Neuhaus F. C. (1992) Biosynthesis of D-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the D-alanine-activating enzyme. J. Bacteriol. 174, 4707–4717 10.1128/jb.174.14.4707-4717.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heaton M. P., and Neuhaus F. C. (1994) Role of the D-alanyl carrier protein in the biosynthesis of D-alanyl-lipoteichoic acid. J. Bacteriol. 176, 681–690 10.1128/jb.176.3.681-690.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perego M., Glaser P., Minutello A., Strauch M. A., Leopold K., and Fischer W. (1995) Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis: identification of genes and regulation. J. Biol. Chem. 270, 15598–15606 10.1074/jbc.270.26.15598 [DOI] [PubMed] [Google Scholar]
- 40. Du L., He Y., and Luo Y. (2008) Crystal structure and enantiomer selection by D-alanyl carrier protein ligase DltA from Bacillus cereus. Biochemistry 47, 11473–11480 10.1021/bi801363b [DOI] [PubMed] [Google Scholar]
- 41. Yonus H., Neumann P., Zimmermann S., May J. J., Marahiel M. A., and Stubbs M. T. (2008) Crystal structure of DltA: implication for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains. J. Biol. Chem. 283, 32484–32491 10.1074/jbc.M800557200 [DOI] [PubMed] [Google Scholar]
- 42. Osman K. T., Du L., He Y., and Luo Y. (2009) Crystal structure of Bacillus cereus D-alanyl carrier protein ligase (DltA) in complex with ATP. J. Mol. Biol. 388, 345–355 10.1016/j.jmb.2009.03.040 [DOI] [PubMed] [Google Scholar]
- 43. Du L., and Luo Y. (2014) Thiolation-enhanced substrate recognition by D-alanyl carrier protein ligase DltA from Bacillus cereus. F1000Res. 3, 106 10.12688/f1000research.4097.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Debabov D. V., Heaton M. P., Zhang Q., Stewart K. D., Lambalot R. H., and Neuhaus F. C. (1996) The D-alanyl carrier protein in Lactobacillus casei: cloning, sequencing, and expression of dltC. J. Bacteriol. 178, 3869–3876 10.1128/jb.178.13.3869-3876.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kiriukhin M. Y., and Neuhaus F. C. (2001) D-Alanylation of lipoteichoic acid: role of the D-alanyl carrier protein in acylation. J. Bacteriol. 183, 2051–2058 10.1128/JB.183.6.2051-2058.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Volkman B. F., Zhang Q., Debabov D. V., Rivera E., Kresheck G. C., and Neuhaus F. C. (2001) Biosynthesis of D-alanyl-lipoteichoic acid: the tertiary structure of apo-D-alanyl carrier protein. Biochemistry 40, 7964–7972 10.1021/bi010355a [DOI] [PubMed] [Google Scholar]
- 47. Zimmermann S., Pfennig S., Neumann P., Yonus H., Weininger U., Kovermann M., Balbach J., and Stubbs M. T. (2015) High-resolution structures of the D-alanyl carrier protein (Dcp) DltC from Bacillus subtilis reveal equivalent conformations of apo- and holo-forms. FEBS Lett. 589, 2283–2289 10.1016/j.febslet.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 48. Majerus P. W., Alberts A. W., and Vagelos P. R. (1965) Acyl carrier protein. IV. The identification of 4′-phosphopantetheine as the prosthetic group of the acyl carrier protein. Proc. Natl. Acad. Sci. U.S.A. 53, 410–417 10.1073/pnas.53.2.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nguyen C., Haushalter R. W., Lee D. J., Markwick P. R., Bruegger J., Caldara-Festin G., Finzel K., Jackson D. R., Ishikawa F., O'Dowd B., McCammon J. A., Opella S. J., Tsai S.-C., and Burkart M. D. (2014) Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature. 505, 427–431 10.1038/nature12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hofmann K. (2000) A superfamily of membrane-bound O-acyltransferases with implications for Wnt signaling. Trends Biochem. Sci. 25, 111–112 10.1016/S0968-0004(99)01539-X [DOI] [PubMed] [Google Scholar]
- 51. Cases S., Smith S. J., Zheng Y.-W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., Erickson S. K., and Farese R. V. Jr. (1998) Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. U.S.A. 95, 13018–13023 10.1073/pnas.95.22.13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zweytick D., Leitner E., Kohlwein S. D., Yu C., Rothblatt J., and Daum G. (2000) Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267, 1075–1082 10.1046/j.1432-1327.2000.01103.x [DOI] [PubMed] [Google Scholar]
- 53. Resh M. D. (2016) Fatty acylation of proteins: the long and the short of it. Prog. Lipid Res. 63, 120–131 10.1016/j.plipres.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Taylor M. S., Ruch T. R., Hsiao P.-Y., Hwang Y., Zhang P., Dai L., Huang C. R., Berndsen C. E., Kim M.-S., Pandey A., Wolberger C., Marmorstein R., Machamer C., Boeke J. D., and Cole P. A. (2013) Architectural organization of the metabolic regulatory enzyme ghrelin O-acyltransferase. J. Biol. Chem. 288, 32211–32228 10.1074/jbc.M113.510313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang J., Brown M. S., Liang G., Grishin N. V., and Goldstein J. L. (2008) Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 132, 387–396 10.1016/j.cell.2008.01.017 [DOI] [PubMed] [Google Scholar]
- 56. Barnett B. P., Hwang Y., Taylor M. S., Kirchner H., Pfluger P. T., Bernard V., Lin Y. Y., Bowers E. M., Mukherjee C., Song W.-J., Longo P. A., Leahy D. J., Hussain M. A., Tschöp M. H., Boeke J. D., et al. (2010) Glucose and weight control in mice with a designed ghrelin O-acyltransferase inhibitor. Science. 330, 1689–1692 10.1126/science.1196154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Atila M., and Luo Y. (2016) Profiling and tandem mass spectrometry analysis of aminoacylated phospholipids in Bacillus subtilis. F1000Res. 5, 121 10.12688/f1000research.7842.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Debabov D. V., Kiriukhin M. Y., and Neuhaus F. C. (2000) Biosynthesis of lipoteichoic acid in Lactobacillus rhamnosus: role of DltD in D-alanylation. J. Bacteriol. 182, 2855–2864 10.1128/JB.182.10.2855-2864.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gerlt J. A., Bouvier J. T., Davidson D. B., Imker H. J., Sadkhin B., Slater D. R., and Whalen K. L. (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037 10.1016/j.bbapap.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Santa Maria J. P. Jr., Sadaka A., Moussa S. H., Brown S., Zhang Y. J., Rubin E. J., Gilmore M. S., and Walker S. (2014) Compound-gene interaction mapping reveals distinct roles for Staphylococcus aureus teichoic acids. Proc. Natl. Acad. Sci. U.S.A. 111, 12510–12515 10.1073/pnas.1404099111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hancock I. C., Wiseman G., and Baddiley J. (1976) Biosynthesis of the unit that links teichoic acid to the bacterial wall: inhibition by tunicamycin. FEBS Lett. 69, 75–80 10.1016/0014-5793(76)80657-6 [DOI] [PubMed] [Google Scholar]
- 62. Campbell J., Singh A. K., Santa Maria J. P. Jr., Kim Y., Brown S., Swoboda J. G., Mylonakis E., Wilkinson B. J., and Walker S. (2011) Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6, 106–116 10.1021/cb100269f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. May J. J., Finking R., Wiegeshoff F., Weber T. T., Bandur N., Koert U., and Marahiel M. A. (2005) Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium's susceptibility to antibiotics that target the cell wall. FEBS J. 272, 2993–3003 10.1111/j.1742-4658.2005.04700.x [DOI] [PubMed] [Google Scholar]
- 64. Corrigan R. M., Abbott J. C., Burhenne H., Kaever V., and Gründling A. (2011) c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7, e1002217 10.1371/journal.ppat.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wörmann M. E., Reichmann N. T., Malone C. L., Horswill A. R., and Gründling A. (2011) Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J. Bacteriol. 193, 5279–5291 10.1128/JB.00369-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Knol J., Sjollema K., and Poolman B. (1998) Detergent-mediated reconstitution of membrane proteins. Biochemistry 37, 16410–16415 10.1021/bi981596u [DOI] [PubMed] [Google Scholar]
- 67. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Riley L. M., Weadge J. T., Baker P., Robinson H., Codée J. D., Tipton P. A., Ohman D. E., and Howell P. L. (2013) Structural and functional characterization of Pseudomonas aeruginosa AlgX: role of AlgX in alginate acetylation. J. Biol. Chem. 288, 22299–22314 10.1074/jbc.M113.484931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee L.-C., Lee Y.-L., Leu R.-J., and Shaw J.-F. (2006) Functional role of catalytic triad and oxyanion hole-forming residues on enzyme activity of Escherichia coli thioesterase I/protease I/phospholipase L1. Biochem. J. 397, 69–76 10.1042/BJ20051645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leščić Ašler I., Štefanić Z., Maršavelski A., Vianello R., and Kojić-Prodić B. (2017) Catalytic dyad in the SGNH hydrolase superfamily: in-depth insight into structural parameters tuning the catalytic process of extracellular lipase from Streptomyces rimosus. ACS Chem. Biol. 12, 1928–1936 10.1021/acschembio.6b01140 [DOI] [PubMed] [Google Scholar]
- 72. Sham L.-T., Butler E. K., Lebar M. D., Kahne D., Bernhardt T. G., and Ruiz N. (2014) MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222 10.1126/science.1254522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mazda Y., Kawada-Matsuo M., Kanbara K., Oogai Y., Shibata Y., Yamashita Y., Miyawaki S., and Komatsuzawa H. (2012) Association of CiaRH with resistance of Streptococcus mutans to antimicrobial peptides in biofilms. Mol. Oral Microbiol. 27, 124–135 10.1111/j.2041-1014.2012.00637.x [DOI] [PubMed] [Google Scholar]
- 74. McBride S. M., and Sonenshein A. L. (2011) The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 157, 1457–1465 10.1099/mic.0.045997-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bensaci M. F., and Takemoto J. Y. (2007) Syringopeptin SP25A-mediated killing of Gram-positive bacteria and the role of teichoic acid D-alanylation. FEMS Microbiol. Lett. 268, 106–111 10.1111/j.1574-6968.2006.00591.x [DOI] [PubMed] [Google Scholar]
- 76. Campbell J. W., and Cronan J. E. Jr. (2001) Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu. Rev. Microbiol. 55, 305–332 10.1146/annurev.micro.55.1.305 [DOI] [PubMed] [Google Scholar]
- 77. White S. W., Zheng J., Zhang Y.-M., and Rock C. O. (2005) The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831 10.1146/annurev.biochem.74.082803.133524 [DOI] [PubMed] [Google Scholar]
- 78. Beining P. R., Huff E., Prescott B., and Theodore T. S. (1975) Characterization of the lipids of mesosomal vesicles and plasma membranes from Staphylococcus aureus. J. Bacteriol. 121, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Spector A. A., Mathur S. N., and Kaduce T. L. (1979) Role of acylcoenzyme A: cholesterol O-acyltransferase in cholesterol metabolism. Prog. Lipid Res. 18, 31–53 10.1016/0163-7827(79)90003-1 [DOI] [PubMed] [Google Scholar]
- 80. Lee H.-C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., Gengyo-Ando K., Mitani S., and Arai H. (2008) Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell. 19, 1174–1184 10.1091/mbc.e07-09-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kato F., and Sugai M. (2011) A simple method of markerless gene deletion in Staphylococcus aureus. J. Microbiol. Methods 87, 76–81 10.1016/j.mimet.2011.07.010 [DOI] [PubMed] [Google Scholar]
- 82. Bryksin A. V., and Matsumura I. (2010) Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. BioTechniques 48, 463–465 10.2144/000113418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nørby J. G. (1988) Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 156, 116–119 10.1016/0076-6879(88)56014-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.