Abstract
The histone transmethylase complex comprising WD repeat domain 77 (WDR77) and protein arginine methyltransferase 5 (PRMT5) catalyzes dimethylation of H4R3 (H4R3me2) and drives cancer cell proliferation and migration, but its regulation is not fully understood. Here, we report that sirtuin 7 (SIRT7) directly deacetylates WDR77 and that this deacetylation interferes with the WDR77–PRMT5 interaction and suppresses proliferation of human colon cancer HCT116 cells. Using co-expression in HEK293T cells and co-immunoprecipitation assays, we observed that SIRT7 deacetylates WDR77 at Lys-3 and Lys-243, which reduced of WDR77's interaction with PRMT5. More importantly, this reduction suppressed the transmethylase activity of the WDR77/PRMT5 complex, resulting in a reduction of the H4R3me2 modification. Rescue of the WDR77-KO HCT116 cells with a WDR77–2KR (K3R and K243R) variant yielded cell migration and proliferation rates that were significantly lower than those of WDR77-KO HCT116 cells rescued with WT WDR77. In summary, SIRT7 is a major deacetylase for WDR77, and SIRT7-mediated deacetylation of WDR77 at Lys-3 and Lys-243 weakens the WDR77–PRMT5 interaction and activity and thereby suppresses growth of cancer cells.
Keywords: histone methylation, cell proliferation, post-translational modification (PTM), colon cancer, epigenetics, H4R3me2, protein acetylation, SIRT7, transmethylase, WDR77
Introduction
Sirtuins are a family of NAD+-dependent deacetylases, also identified as class III histone deacetylases (1). Mammalian sirtuins comprise seven members (SIRT1–SIRT7) that participate in many cellular functions encompassing cellular proliferation, metabolism, genomic stability, and stress responses (2). Sirtuins have also been associated with several human diseases and disorders, including cancers, aging, and aging-related disorders, such as hearing-loss and cardiac dysfunction, and metabolic diseases, such as diabetes and fatty liver disease (3). Interestingly, Sirtuins appear to play a dual role in cancer. In addition to protecting organisms from tumors by increasing genomic stability and limiting cellular replicative lifespan, Sirtuins can also induce tumorigenesis by promoting cell survival under stress and enhancing uncontrolled cell division (4).
Of the seven Sirtuin members, SIRT7 has remained the least understood. Only a handful of deacetylation targets have been reported, including p53, NRF1, GABP-β1, H3K18ac, PAF53, U3–55K, and Smad4 (5–11). SIRT7 may have many more potential substrates according to a study using stable isotope labeling by amino acids in cell culture (SILAC)-coupled quantitative MS (12). SIRT7 plays critical roles in rDNA transcription, protein synthesis, chromatin remodeling, cellular survival, and lipid metabolism (13, 14). It is also reported to be highly associated with tumor progression. Evidence also indicates that SIRT7 may act as an oncogene, as its expression is elevated in several human cancers (15, 16). It also selectively deacetylates H3K18Ac at the promoters of a network of genes with links to tumor suppression (6). Interestingly, SIRT7 is reported to be significantly down-regulated in breast cancer lung metastases in humans and mice and predicts metastasis-free survival (11). Much more remains to be explored regarding the role of SIRT7 in tumor progression.
WDR77 is a WD-repeat protein that can interact with protein arginine methyltransferase 5 (PRMT5) 2 and form a higher-order tetrameric complex while acting independently as an androgen receptor cofactor (p44) that is overexpressed in prostate cancer cells (17). The WDR77/PRMT5 complex catalyzes the methylation of arginines on histones and other proteins. H4R3me2 is an important target of WDR77/PRMT5, but its function and regulation have yet to be fully elucidated. Elevated PRMT5 and WDR77 expression is observed in many cancers and is often correlated with enhanced tumor growth and poor disease prognosis (18–20).
In this study, we demonstrate that SIRT7 can directly deacetylate WDR77 and influence the interaction with PRMT5. More importantly, decreased WDR77–PRMT5 interactions leads to the down-regulation of H4R3me2 levels and can thus restrain cancer cell proliferation.
Results
SIRT7 interacts with WDR77
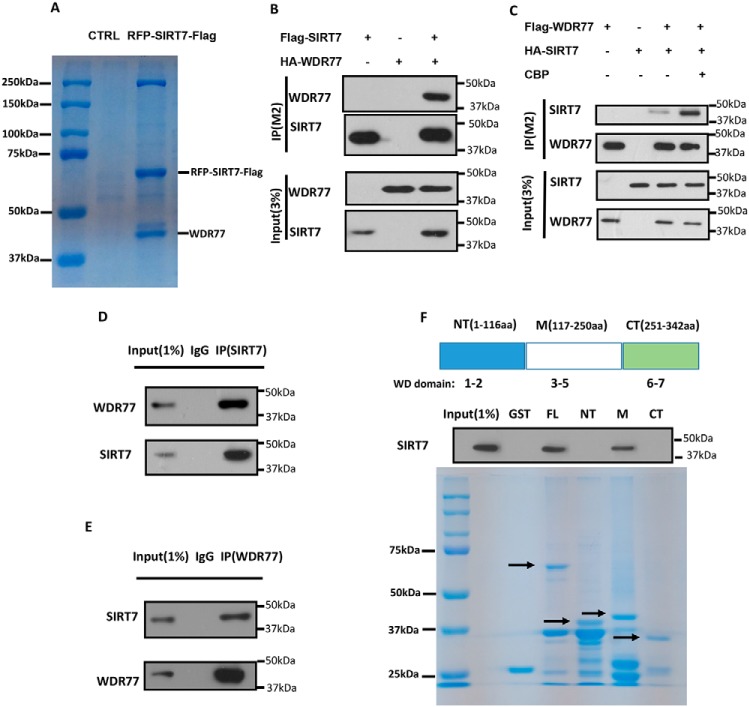
Relatively few SIRT7 targets have been reported in literature. We therefore searched for more potential targets of SIRT7 through purification and MS and found that WDR77 may be a new target of SIRT7 (Fig. 1A). To verify the interaction, we overexpressed tagged SIRT7 and WDR77 in HEK293T cells and performed co-immunoprecipitation assays to confirm the interaction between these two proteins. SIRT7 can co-immunoprecipitate WDR77 (Fig. 1B), and vice versa (Fig. 1C). Interestingly, the interaction of these two proteins was enhanced by the co-expression of histone acetyltransferase CBP (Fig. 1C). The SIRT7–WDR77 interaction was further confirmed by the co-immunoprecipitation of endogenous SIRT7 and WDR77 (Fig. 1, D and E). We also confirmed the SIRT7 and WDR77 interaction by GST pulldown assay. We first generated GST fusion proteins for the assay. WDR77 was divided into 3 segments according to its domains: N terminus (NT, 1–116 aa), central domain (M, 117–250 aa), and C terminus (CT, 251–342 aa). SIRT7 can clearly be pulled down by GST-WDR77 full-length and central domains, but not the N-terminal and C-terminal domains (Fig. 1F), indicating that SIRT7 interacts with WDR77 at its central domain. Taken together, these results demonstrated that SIRT7 interacts with WDR77 both in vivo and in vitro.
Figure 1.
WDR77 interacts with SIRT7 in vivo and in vitro. A, immunopurification and MS analysis of SIRT7-associated proteins. Co-precipitated proteins were analyzed by 10% SDS-PAGE and Coomassie Blue staining. The protein bands were cut and analyzed by MS. B and C, SIRT7 interacts with WDR77 in vivo, and CBP can enhance the interaction. Western blotting of whole cell extracts and immunoprecipitates with the anti-FLAG M2 beads (IP/M2) from cells were transfected with or without tagged SIRT7-tagged WDR77/CBP as indicated and detected with anti-HA and anti-FLAG antibodies. D and E, endogenous SIRT7 interacts with WDR77 in vivo. Whole cell lysates were immunoprecipitated with control IgG, anti-SIRT7, or anti-WDR77 antibody, and the precipitated proteins were detected with anti-WDR77 or anti-SIRT7 antibody, respectively. F, SIRT7 interacts with WDR77 in vitro. GST fusion proteins were generated for full-length WDR77 (FL), N-terminal (NT), central (M), and C-terminal (CT) WDR77, and GST pulldown assays were carried out as described under “Experimental procedures.”
WDR77 is acetylated at lysines 3 and 243
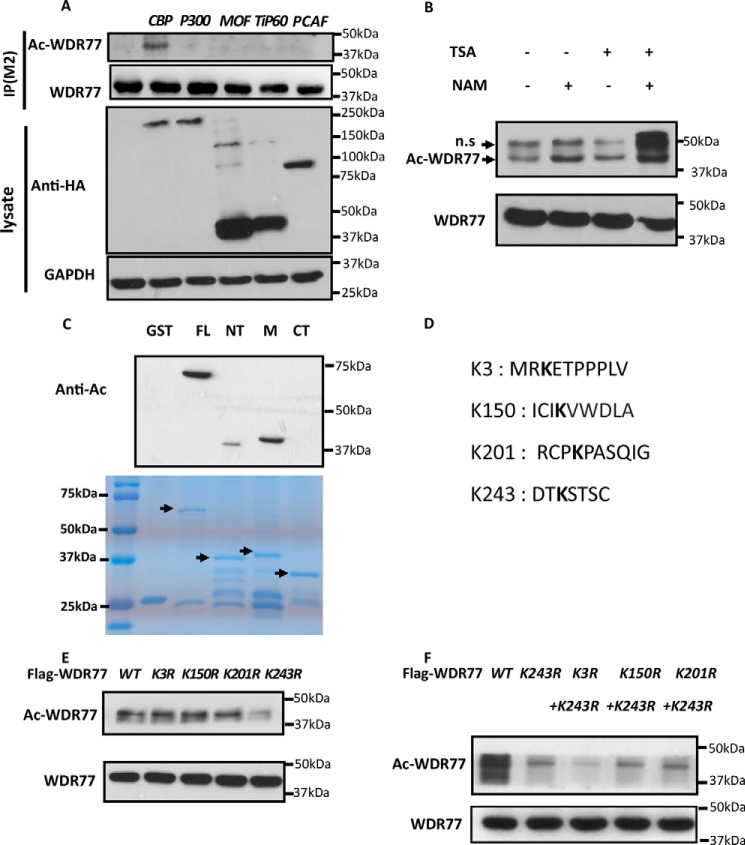
The interaction between WDR77 and SIRT7 prompted us to consider whether WDR77 is a novel target of SIRT7, especially considering that the interaction is enhanced in the presence of CBP (Fig. 1C). Therefore, we first examined whether WDR77 was acetylated. We co-transfected FLAG-WDR77 with different acetyltransferases in HEK293T cells and incubated the cell lysates with M2 beads to immunoprecipitate WDR77. WDR77 acetylation was detected only when co-transfected with CBP (Fig. 2A), consistent with the results of the interaction studies (Fig. 1C). WDR77 acetylation was more significant in cells treated with nicotinamide than in cells treated with trichostatin A (TSA) (Fig. 2B), suggesting that Sirtuins could be the major deacetylase for WDR77. Using the GST fusion proteins described in Fig. 1F, we performed the in vitro acetylation assay. The results indicated that WDR77 was primarily acetylated at the central region, although weak acetylation was also detected in the N-terminal region (Fig. 2C).
Figure 2.
WDR77 can be acetylated by CBP at lysine 3 and lysine 243. A, HEK293T cells were co-transfected with plasmids containing FLAG-WDR77 and different HA-tagged acetyltransferases, CBP, p300, MOF, Tip60, or P300/CBP-associated factor (PCAF). Whole cell lysates were immunoprecipitated with M2 beads and analyzed by Western blotting with anti-acetylated lysine, anti-FLAG, anti-HA, and anti-GAPDH antibodies. B, HEK293T cells were transfected with FLAG-WDR77 for 24 h and incubated with or without 1 μm TSA and/or 5 mm nicotinamide (NAM), as indicated, for an additional 6 h. An in vivo acetylation assay and Western blot analysis were then performed. C, four types of GST-WDR77 fusion proteins were used for in vitro acetylation assays. Top panel, Western blotting data. Bottom panel, gel code blue staining. D, K represents potential acetylation sites in WDR77 analyzed by MS. E and F, HEK293T cells were transfected with WT or the indicated Lys to Arg mutant FLAG-tagged WDR77 constructs for 24 h and incubated with 1 μm TSA and 5 mm nicotinamide for an additional 6 h. The levels of acetylation and total WDR77 protein were detected after anti-FLAG immunoprecipitation.
To identify the major acetylation sites of WDR77, we purified the acetylated WDR77 from HEK293T cells co-transfected with WDR77 and CBP and performed MS assay. Lysine residues 3, 150, 201, and 243 were detected in the peptides with acetylated K (Fig. 2D), consistent with the in vitro acetylation assay (Fig. 2C). By mutating these 4 lysines to arginine (K3R, K150R, K201R, K243R), we were able to observe significantly reduced acetylation levels for the K243R mutant (Fig. 2E). Because the N-terminal of WDR77 also showed an acetylation signal in the in vitro acetylation assay (Fig. 2C), we generated a construct containing K3R and K243R (2KR), which showed even lower acetylation levels than those of K243R (Fig. 2F). Taken together, these results indicated that WDR77 is acetylated by CBP both in cells and in vitro and that both lysine 3 and lysine 243 are the major acetylation sites of WDR77.
WDR77 is deacetylated by SIRT7
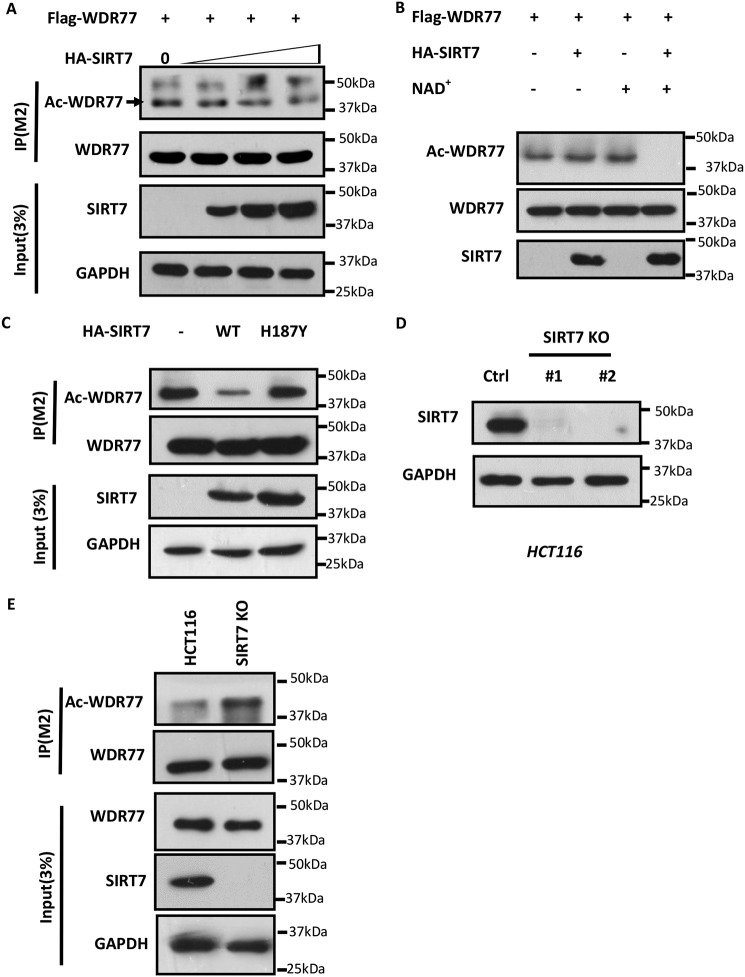
We then explored the possibility that SIRT7 deacetylates WDR77. FLAG-WDR77 and different HA-SIRT7 plasmid amounts were co-transfected into HEK293T cells. Western blotting showed that WDR77 acetylation levels decreased with increasing amounts of SIRT7 transfection (Fig. 3A). WDR77 deacetylation by SIRT7 was further confirmed by an in vitro deacetylation assay. We purified and incubated acetylated WDR77 under different conditions. The results revealed that WDR77 was deacetylated only in the presence of both SIRT7 and NAD+, as SIRT7 is a NAD+-dependent deacetylase (Fig. 3B). To further investigate whether WDR77 deacetylation depends on SIRT7 catalytic activity, we co-transfected WDR77 with SIRT7-WT or SIRT7-H187Y, which lacks catalytic activity, and analyzed WDR77 acetylation levels by Western blotting. Although SIRT7-WT was able to deacetylate WDR77, SIRT7-H187Y could not deacetylate WDR77 (Fig. 3C). To provide further insight into the role of SIRT7, we generated SIRT7 knockout cells using CRISPR-Cas9 technology (Fig. 3D) and overexpressed FLAG-WDR77 with CBP in both HCT116 WT and HCT116 SIRT7-KO cells. Western blotting revealed that WDR77 acetylation levels were significantly increased in SIRT7-KO cells compared with SIRT7-WT cells (Fig. 3E). Thus, WDR77 is a novel target of SIRT7, which can deacetylate WDR77 both in cells and in vitro, dependent on SIRT7 catalytic activity. Conversely, depletion of SIRT7 in cells enhanced WDR77 acetylation.
Figure 3.
Sirt7 deacetylates WDR77 in vivo and in vitro. A, HEK293T cells were transfected with FLAG-WDR77 alone or with increasing amounts of HA-SIRT7 plasmid, followed by deacetylation assays. B, in vitro deacetylation assay for WDR77. FLAG-WDR77 and HA-SIRT7 were purified from HEK293T cells, followed by in vitro deacetylation assays, in the presence of NAD or not. C, HEK293T cells were transfected with FLAG-WDR77 and empty vector or with HA-SIRT7 (WT) or HA-SIRT7-H187Y (H187Y), followed by deacetylation assays. D, HCT116-SIRT7-KO cells generated by CRISPR-CAS9 were analyzed by Western blotting for SIRT7 expression. E, HCT116-WT cells or HCT116-SIRT7-KO cells were transfected with FLAG-WDR77, followed by an acetylation assay.
Deacetylation of WDR77 influences the interaction of WDR77 and PRMT5
As an important component of the WDR77/PRMT5 complex, WDR77 mediates interactions with binding partners and substrates through its interaction with PRMT5 to form an atypical heterooctameric complex (21). Moreover, WDR77 is reported to interact with PRMT5 through both its N-terminal (Trp-44) and middle region (Phe-289) (22), which spans our identified acetylation sites (Lys-3 and Lys-243). Thus, we investigated whether WDR77 deacetylation affects the interaction with PRMT5. We overexpressed HA-PRMT5 with FLAG-WDR77-WT or FLAG-WDR77–2KR in HEK293T cells and co-immunoprecipitated FLAG-WDR77-WT and FLAG-WDR77–2KR using M2 beads. Western blotting revealed that PRMT5 was pulled down more weakly by WDR77–2KR than by WDR77-WT (Fig. 4A). Endogenous PRMT5 also interacted less strongly with FLAG-WDR77 when we overexpressed HA-tagged SIRT7 in HEK293T cells (Fig. 4B), consistent with the results of the WDR77 deacetylation by SIRT7. Conversely, endogenous WDR77 interacted more tightly with endogenous PRMT5 when SIRT7 was knocked out (Fig. 4C). Thus, the interaction of WDR77 with PRMT5 was affected by WDR77 deacetylation, which is regulated by SIRT7.
Figure 4.
WDR77 deacetylation decreases the WDR77/PRMT5 interaction. A, immunoprecipitation (IP) analysis of the interaction between PRMT5 and WT or mutant WDR77. HEK293T cells were transfected with HA-PRMT5 either alone (lane 2) or with FLAG-WDR77-WT (lane 3) or FLAG-WDR77–2KR (K3R and K243R) (lane 4), or FLAG-WDR77-WT alone (lane 1). Whole cell extracts and immunoprecipitates with anti-FLAG M2 beads (IP/M2) were analyzed by Western blotting. B, immunoprecipitation analysis of the interaction between endogenous PRMT5 and FLAG-WDR77 with or without HA-SIRT7. C, whole cell lysates from HCT116-WT or HCT116-SIRT7-KO cells were immunoprecipitated with control IgG or anti-WDR77 antibody, and the precipitated proteins were detected using anti-WDR77 and anti-PRMT5 antibodies, respectively.
WDR77 deacetylation influences cancer cell proliferation by altering WDR77/PRMT5 complex activity
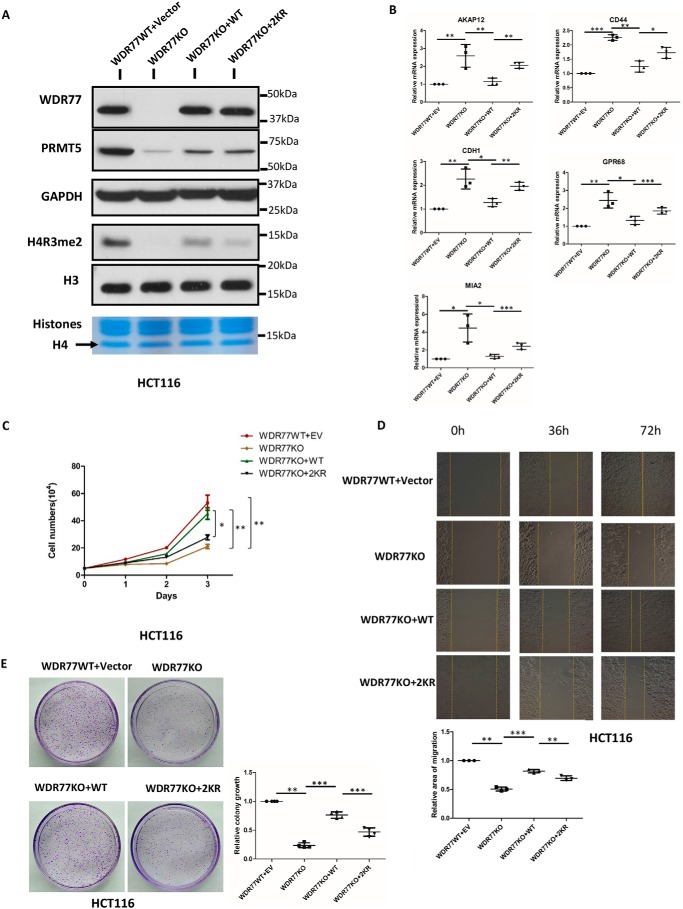
The effect of SIRT7 on the WDR77–PRMT5 interaction prompted us to further explore the enzymatic activity of this complex. We first generated WDR77-knockout HCT116 cells by CRISPR-Cas9. PRMT5 and H4R3me2 were both down-regulated (Fig. 5A). When stably re-expressing WDR77-WT and WDR77–2KR in WDR77-KO cells, we observed a significant difference in the enzymatic activity of the WDR77/PRMT5 complex by measuring the levels of H4R3me2, a known target of this complex. Although the expression levels of WDR77-WT and WDR77–2KR were almost the same, WDR77–2KR/PRMT5 showed much less activity than WDR77-WT/PRMT5 (Fig. 5A).
Figure 5.
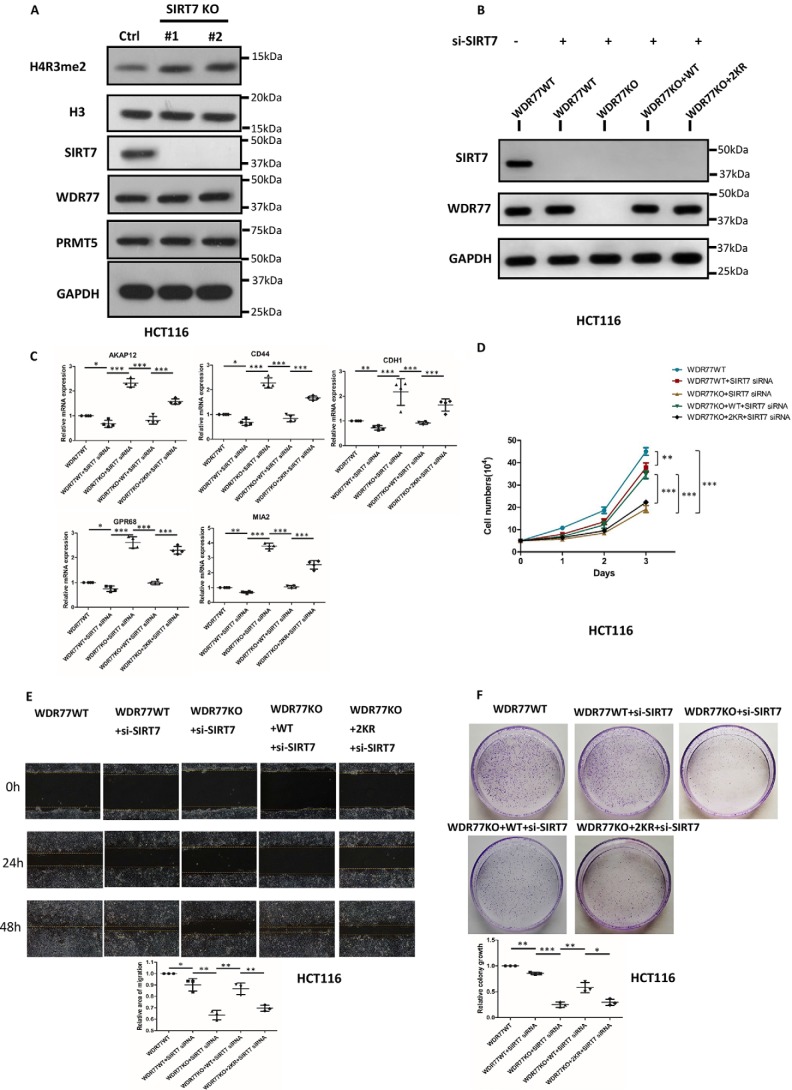
WDR77 deacetylation affects proliferation. A, whole cell lysates and histones extracted from WT, WDR77-KO, or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells were probed with the indicated antibodies. GAPDH and H3 are loading controls for soluble lysate and histone immunoblots, respectively. Gel code staining of extracted histones is also shown (bottom). B, RT-qPCR for the indicated genes from WT and WDR77-KO or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells (n = 4). C, WT, WDR77-KO, or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells were seeded into 6-well plates at the same density. Cell numbers were counted every day (n = 3). D, WT, WDR77-KO, or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells were analyzed using wound-healing assays. Relative gaps after wounding were measured at the indicated time point (n = 3). E, colony formation assays of WT, WDR77-KO, or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells are quantified as relative colony numbers (% of control) (n = 3). Data are shown as mean ± S.D. *, p < 0.05; ***, p < 0.001. p values are calculated by Student's t test.
Recently, PRMT5/WDR77 was reported to regulate cancer cell proliferation through histone H4R3me2 methylation (23). Thus, we measured the mRNA expression of a series of metastasis suppressor genes, which were reported to be regulated by H4R3me2. The results of RT-qPCR showed that these genes were indeed up-regulated in WDR77 knockout cells and that mRNA expression levels of these genes were very different in WDR77-WT and WDR77–2KR cells, consistent with the levels of H4R3me2. The mRNA expression levels of these genes were decreased in WDR77-WT cells but remained at higher levels in WDR77–2KR cells (Fig. 5B). These findings led us to investigate further the difference between WDR77-WT and WDR-2KR in metastasis. We observed that WDR77–2KR cells grew much slower compared with WDR77-WT cells. This observation was confirmed by a cell counting assay. WDR77-WT cells presented nearly the same growth rate as HCT116-WT cells, whereas the WDR77–2KR cells grew much slower and only a little faster than WDR77 KO cells (Fig. 5C). Moreover, we performed a wound-healing assay and demonstrated a significantly reduced rate of closure in the WDR77–2KR cells compared with that in the WDR77-WT cells (Fig. 5D). To examine the anchorage-independent growth behavior, we employed a colony formation assay and observed much less colony formation in WDR77–2KR cells compared with WDR77-WT cells (Fig. 5E). Thus, we identified a different behavior in cell proliferation between WDR77-WT and WDR77–2KR through a knockout-and-rescue approach. Although WDR77-WT and WDR77–2KR expression levels were almost the same (Fig. 5B), WDR77–2KR exerted a weaker effect than WDR77-WT in these assays.
To further investigate the role of SIRT7 in cell proliferation, we knocked down SIRT7 in HCT116 cells, WDR77 KO cells, WDR77-WT cells, and WDR77–2KR cells by using siRNAs (Table 1, Fig. 6B) and performed similar assays as presented in Fig. 5. We first measured the mRNA expression of those metastasis suppressor genes detected in Fig. 5B. The results of RT-qPCR showed that these genes were down-regulated in SIRT7 KD cells consistent with the up-regulated levels of H4R3me2 in SIRT7 KO cells (Fig. 6A), and the mRNA expression levels of these genes were decreased in WDR77-WT cells but remained at higher levels in WDR77–2KR cells (Fig. 6C). In the cell counting assay, the WDR77–2KR cells still grew much slower than WDR77-WT cells (Fig. 6D). Moreover, we performed a wound-healing assay and still observed a significantly reduced rate of closure in the WDR77–2KR cells compared with that in the WDR77-WT cells (Fig. 6E). We also observed much less colony formation in WDR77–2KR cells compared with WDR77-WT cells when performing colony formation assays (Fig. 6F). Taken together, these results demonstrated that WDR77 deacetylation influences cancer cell proliferation by altering WDR77/PRMT5 complex activity.
Table 1.
Sequences of siRNAs used in this study
| Targets | Sequence (5′-3′) |
|---|---|
| hSIRT7 | CUCACCGUAUUUCUACUACUA |
| hSIRT7 | GGGAGUACGUGCGGGUGUU |
| hSIRT7 | CCCUGAAGCUACAUGGGAA |
Figure 6.
SIRT7 affects proliferation through a WDR77-dependent method. A, whole cell lysates and histones extracted from WT or SIRT7-KO HCT116 cells were probed with the indicated antibodies. GAPDH and H3 are loading controls for soluble lysate and histone immunoblots, respectively. B, whole cell lysates from WT- or si-SIRT7-treated WT, WDR77-KO, or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells were probed with the indicated antibodies. C, RT-qPCR for the indicated genes from WT- or si-SIRT7-treated WT, WDR77-KO, or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells (n = 4). D, WT- or si-SIRT7-treated WT, WDR77-KO, or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells were seeded into 6-well plates at the same density. Cell numbers were counted every day (n = 3). E, WT- or si-SIRT7-treated WT, WDR77-KO, or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells were analyzed using wound-healing assays. Relative gaps after wounding were measured at the indicated time point (n = 3). F, colony formation assays of WT- or si-SIRT7-treated WT, WDR77-KO- or two types of rescued WDR77-KO (WT and 2KR) HCT116 cells are quantified as relative colony numbers (% of control) (n = 3). Data are shown as mean ± S.D. *, p < 0.05; ***, p < 0.001. p values are calculated by Student's t test.
Discussion
As the least studied Sirtuin family member, SIRT7 has attracted more attention recently. However, the targets of SIRT7 remain poorly understood. Identifying novel targets is therefore essential to better understand the functions of SIRT7. In this current study, we identified WDR77 as a novel target of SIRT7, and SIRT7 interacts with and deacetylates WDR77. We further identified the major acetylation sites of WDR77 at Lys-3 and Lys-243. WDR77-deficient cells complemented with WDR77–2KR, which mimics hypoacetylated WDR77, displayed a reduced ability to interact with PRMT5 compared with the WDR77-deficient cells complemented with WT WDR77. Significantly, the disruption of the interaction between WDR77 and PRMT5 reduced the PRMT5 enzymatic activity and hence influenced cancer cell proliferation. Our study increased the understanding of functions of SIRT7.
The role of SIRT7 in tumor progression remains the subject of debate. On the one hand, Barber et al. (6) showed that SIRT7 can selectively deacetylate H3K18ac and promote oncogenic transformation and tumor growth. SIRT7 levels are also reportedly elevated in thyroid and breast cancer tissues and may predict poor prognosis in colorectal and prostate cancers (24, 25). On the other hand, SIRT7 is negatively correlated with the progression of human head and neck squamous cell carcinoma (26). Moreover, previous work demonstrated that SIRT7 can antagonize transforming growth factor β signaling by destabilizing Smad4 to inhibit breast cancer metastasis. Transforming growth factor β signaling is frequently disturbed during tumor progression and metastasis. This complex signaling pathway converges with the epithelial-mesenchymal transition through the altered expression and regulation of key transcription factors. Oncogenic transformation, tumor growth, and metastasis are phenotypically distinct and mechanistically different processes, and SIRT7 seemingly plays a different role in metastasis. This dual role of SIRT7 in tumor development may arise from its various deacetylation targets in different cells, such as H3K18ac, PAF53, p53, NPM1, GABP-β1, and Smad4, with the overt phenotypes being attributed to the combined effects of various targets. Our study showed that SIRT7 may decrease proliferation by deacetylating WDR77 and thereby disrupting the WDR77/PRMT5 complex.
WDR77 is a core component of the WDR77/PRMT5 complex, which mediates interactions with binding partners and substrates. Many studies have shown that WDR77 and PRMT5 can regulate each other's expression. Down-regulation or up-regulation of WDR77 results in decreased or increased PRMT5 expression, respectively, which is consistent with our results (Fig. 5A). PRMT5 inhibition or loss disrupts cancer cell phenotypes, and its direct interaction with proteins commonly dysregulated or mutated in cancers indicates that PRMT5 acts as an oncogene. PRMT5-MEP50 is also essential for the transcriptional regulation that promotes cancer cell invasion in lung adenocarcinoma, lung squamous cell carcinoma, and breast carcinoma cancer cells (23). However, the regulation of WDR77 and PRMT5 activity is poorly understood. Our study demonstrates that WDR77/PRMT5 complex formation is indeed important for PRMT5-mediated H4R3 methylation. More importantly, by deacetylating WDR77, SIRT7 regulates WDR77/PRMT5 complex activity and participates in the regulation of cancer cell proliferation.
WDR77 acetylation has not been reported previously. In the current study, we found that WDR77 was primarily acetylated by CBP at lysines 3 and 243. More importantly, we further ascertained that WDR77 was deacetylated by SIRT7 both in vivo and in vitro. However, we cannot completely exclude the possibility that WDR77 is also deacetylated by other deacetylases because TSA treatment also slightly increased the acetylation levels of WDR77 (Fig. 2B). It is worth further investigation to find out whether the other histone deacetylases may deacetylate WDR77 and regulate its functions. Nevertheless, we found that SIRT7 acts as a major deacetylase for WDR77 and the deacetylation of WDR77 at Lys-3 and Lys-243 weakened the interaction between WDR77 and PRMT5 and further influenced WDR77/PRMT5 complex activity. Through this mechanism, SIRT7 regulates the activity of the WDR77/PRMT5 methyltransferase complex, which is important in cancer cell proliferation.
Experimental procedures
Cell culture
HEK293T and HCT116 (p53+/+) cells were obtained from ATCC between 2012 and 2016 and maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) FBS, 100 units/ml of penicillin, and 100 mg/ml of streptomycin (Sigma). All cell lines were tested for mycoplasma contamination and authenticated utilizing short tandem repeat profiling. The same batch of cells was thawed every 1 to 2 months.
Western blotting and antibodies
Proteins were analyzed by Western blotting according to standard methods. Visualization was performed using ECL (Thermo Fisher Scientific, number 32106). Antibodies specific to the following proteins were purchased commercially: mouse anti-SIRT7 (Santa Cruz, sc-365344, 1:1,000 dilution), rabbit anti-SIRT7 (Santa Cruz, sc-135055, 1:1,000 dilution), anti-WDR77 (Abcam, ab154190, 1:1,000 dilution), anti-PRMT5 (Abcam, ab151321, 1:1,000 dilution), anti-H3 (Abcam, ab1791, 1:1,000 dilution), anti-H4R3me2 (Abcam, ab5823, 1:1,000 dilution), anti-GAPDH (Cell Signaling Technology, number 5174S, 1:2,000 dilution), anti-FLAG (Sigma, F-3165, 1:10,000 dilution), anti-HA (Pierce, number 26183, 1:5,000 dilution), and anti-pan-acetyl-lysine (Cell Signaling Technology, number 9441L, 1:1,000 dilution).
Plasmids
SIRT7, WDR77, and PRMT5 cDNA were amplified and cloned into the pcDNA3.1 vector. SIRT7- and WDR77-mutant constructs were generated by using the KOD Plus Mutagenesis Kit (TOYOBO). WDR77 full-length (FL), WDR77-NT (NT), WDR77-M (M), and WDR77-CT (CT) cDNA were subcloned into the pGEX4T-3 vector. All expression constructs were verified by Sanger sequencing.
Co-immunoprecipitation
Total cells were lysed in BC100 buffer (20 mmol/liter of Tris-HCl (pH 7.9), 100 mmol/liter of NaCl, 0.2% Nonidet P-40, and 20% glycerol) containing 1 mmol/liter of dithiothreitol (DTT; Sigma), 1 mmol/liter of phenylmethylsulfonyl fluoride (PMSF; Sigma) and protease inhibitor mixture (Sigma). Whole cell lysates were incubated with DNase I (Invitrogen) for 45 min at 37 °C and then incubated with 1 μg of mouse anti-SIRT7 or rabbit anti-WDR77 or normal mouse/rabbit IgG (Santa Cruz) for 10–14 h at 4 °C. Protein A/G-agarose beads (Santa Cruz) were added and the reaction mixtures were further mixed for 2 h at 4 °C. Beads were washed by BC100 several times and eluted with 0.1 mol/liter of glycine (pH 2.5) and then neutralized with 1 mol/liter of Tris buffer. Eluted proteins were analyzed by Western blot analysis.
GST pulldown assay
For the GST pulldown, 1 μg of GST or GST-WDR77-FL (1–342 aa), GST-WDR77-NT (1–116 aa), GST-WDR77-M (117–250 aa), and GST-WDR77-CT (251–342 aa) protein were expressed and purified from Rosetta bacteria and incubated with FLAG-tagged SIRT7 purified from HEK293T cells for 8 h at 4 °C in BC100. Beads were washed and then bound proteins were eluted with GSH. Eluted proteins were analyzed by Western blotting.
Acetylation/deacetylation assay in cells
HEK293T cells were seeded in a 10-cm Petri dish at the density of 2 × 106 cells/dish the day before transfection. FLAG-tagged plasmids, as indicated, were transfected into HEK293T cells with polyethyleneimine method. At 24 h, for the acetylation assay, cells were incubated with 1 μmol/liter of TSA (Sigma) and 5 mmol/liter of nicotinamide (Sigma) for an additional 6 h before harvest. For the deacetylation assay, cells were not treated with any drugs before harvest. After collection of cells by centrifugation, whole cell lysates were prepared in FLAG lysis buffer (50 mmol/liter of Tris-HCl, pH 7.9, 137 mmol/liter of NaCl, 1% Triton X-100, 0.2% Sarkosyl, 1 mmol/liter of NaF, 1 mmol/liter of Na3VO4, and 10% glycerol) containing fresh protease inhibitors, 10 μmol/liter of TSA, and 5 mmol/liter of nicotinamide. Cell extracts were then incubated with anti-FLAG M2 beads (Sigma) at 4 °C overnight. After washing the beads five times with BC100 buffer (50 mmol/liter of Tris-HCl (pH 7.8), 100 mmol/liter of NaCl, 0.2% Triton X-100, and 10% glycerol), FLAG peptide was added, and the beads were incubated for an additional 2 h to elute the bound proteins. Immunoprecipitated proteins were subjected to SDS-PAGE and analyzed by Western blotting with different antibodies as indicated.
In vitro acetylation assay
GST-WDR77-FL, GST-WDR77-NT, GST-WDR77-M, and GST-WDR77-CT were expressed and purified from Rosetta bacteria. Reactions were performed in a 30-μl system containing 3 μl of 10× reaction buffer (200 mmol/liter of HEPES, pH 8.0, 10 mmol/liter of PMSF, 10 mmol/liter of DTT, 170 nmol/liter of acetyl-CoA), 3 μg of GST or GST fusion protein, 0.1 μg of HA-CBP, and double-distilled H2O as needed. The reaction mixture was then incubated for 2 h at 37 °C, followed by Western blotting. To confirm equal protein loading in the in vitro acetylation assays, a parallel SDS-PAGE was performed using the same amount and loading of GST or GST fusion proteins, followed by gel code blue staining (Thermo Scientific).
Immunoprecipitation and protein purification
Total cells were lysed in FLAG lysis buffer (50 mmol/liter of Tris-HCl, pH 7.9, 137 mmol/liter of NaCl, 1% Triton X-100, 0.2% Sarkosyl, 1 mmol/liter of NaF, 1 mmol/liter of Na3VO4, and 10% glycerol) containing protease inhibitor mixture, 1 mmol/liter of DTT, and 1 mmol/liter of PMSF. Immunoprecipitations were carried out by incubating with M2 affinity gel (Sigma) for over 4 h at 4 °C. After incubation, beads were washed four times with ice-cold BC100 buffer and eluted with FLAG peptide. GST-WDR77-FL, GST-WDR77-NT, GST-WDR77-M, and GST-WDR77-CT were expressed and purified from Rosetta bacteria and bound to GST-agarose columns (Novagen). After incubation, the beads were eluted with GSH and rapidly diluted with 4 volumes of BC100 buffer.
Identification of WDR77 acetylation sites
HEK293T cells were cotransfected with FLAG-WDR77 and HA-CBP plasmids and treated with or without 10 mmol/liter of nicotinamide and 1 μmol/liter of TSA for 6 h before being harvested. Cells were lysed in FLAG lysis buffer plus 10 mmol/liter of nicotinamide and 10 μmol/liter of TSA, and cell lysates were immunoprecipitated overnight with M2 beads. Immunoprecipitated WDR77 proteins were separated via 8% SDS-PAGE, and WDR77 bands were excised from the gel and analyzed by MS. Distinct modification sites and undetected sites were chosen for further analysis.
In vitro deacetylation assay
For the deacetylation assay, 2 μg of acetylated FLAG-WDR77 expressed and purified from HEK293T cells were incubated with or without 1 μg of HA-SIRT7 (as indicated) expressed and purified from HEK293T cells in deacetylation reaction buffer (10% glycerol, 4 mmol/liter of MgCl2, 100 mmol/liter of NaCl, 50 μmol/liter of NAD+ co-factor (Sigma) (when indicated)), at 37 °C for 2 h. The total volume of reaction mixtures was 30 μl. The reactions were stopped by adding SDS-PAGE sample buffer and resolved for Western blotting to detect the acetylation level of FLAG-WDR77.
CRISPR-Cas9 knockout and rescue cell lines
To generate HCT116 SIRT7, HCT116 WDR77 knockout (KO; down) cell lines, single guide RNA sequences were ligated into the LentiCRISPRv2 plasmid and then cotransfected with viral packaging plasmids (psPAX2 and pMD2G) into HEK293T cells. After transfection for 6 h, the medium was changed, and the viral supernatant was filtered through a 0.45-mm strainer after 42 h. Targeted cells were infected with the viral supernatant and selected with 1 mg/ml of puromycin for 2 weeks. Single guide RNA sequences targeting SIRT7 and WDR77 were designed using the CRISPR designer (http://crispr.mit.edu). 3 The guide sequences designed to target Exon 3 of human SIRT7 and Exon 1 of human WDR77 were as follows: SIRT7, 5′-CACCGCGTCCGGAACGCCAAATACT-3′; WDR77, 5′-AGCCTCCGTTTGGACTCCGG-3′
For WDR77-rescue cell lines, FLAG-tagged WDR77 wildtype (WT) and WDR77 2KR (K3R and K243R) mutant cDNA was subcloned into the pQCXIH retrovirus vector. The two plasmids were cotransfected with viral packaging plasmids (vsvg and gag) into HEK293T cells. Twenty-four hours after transfection, viral supernatants were filtered and infected into HCT116 WDR77 KO cells. The infected cells were selected with 150 mg/ml of hygromycin for 2 weeks.
RNA isolation and quantitative RT-PCR
Total RNA was extracted using TRIzol reagent (Sigma), and RNA was reverse transcribed into cDNA using the qPCR RT Kit (FSQ-101 TOYOBO). Quantitative PCR was performed on a 7500 Fast Real-time PCR System (Applied Biosystems) using qPCR SYBR Green Master Mix (Q131-03 Vazyme). mRNA quantity was calculated using the method ΔΔCt and normalized to actin. The real-time PCR primers are listed in Table 2.
Table 2.
RT-qPCR primers used in this study
| Gene name | Forward sequence | Reverse sequence |
|---|---|---|
| AKAP12 | 5′-TGGCAGGAAGACATTCTGTG-3′ | 5′-GCGGGTGGAATTTAAACAAA-3′ |
| CDH1 | 5′-CAAGTGCCTGCTTTTGATGA-3′ | 5′-GTTTTCTGTGCACACCTGGA-3′ |
| CD44 | 5′-AAGGAACCTGCAGAATGTGG-3′ | 5′-TCCAACGGTTGTTTCTTTCC-3′ |
| MIA2 | 5′-CAGAGCACATTCCCAAACCT-3′ | 5′-GCCCTGTATCCTCATCTCCA-3′ |
| GPR68 | 5′-CTGGGTCAGTGACATTGGTG-3′ | 5′-TGGGAAGCCAGTGTTTAAGG-3′ |
| ACTB | 5′-CGGGACCTGACTGACTACCTC-3′ | 5′-ATTGGAACGATACAGAGAAGATT-3′ |
Wound-healing assay
Different cells were seeded into 6-well cell culture plates and grown to confluence for 24 h. A scratch wound was introduced in the center of the cell monolayer using a sterile plastic pipette tip, and debris was removed by washing with PBS. Cell migration was observed and photographed by microscopy (Eclipse TS100, Nikon) at the indicated times.
Cell proliferation and colony formation
For all cell proliferation experiments, cells were initially seeded into 6-well plates at a density of 5.0 × 104 cells/well. Cells were trypsinized and counted every day (Countstar IC1000, Inno-Alliance Biotech). For colony formation assays, 1.0 × 104 cells were seeded in 6-cm dishes and kept in the same medium for 7 days. Cells were fixed with 4% paraformaldehyde (Sigma), stained with 0.2% crystal violet solution (Sigma) and imaged using a digital scanner. Relative growth was quantified by counting.
Statistical analysis
Statistical analyses were performed using two-tailed unpaired Student's t tests in SPSS version 20. All data are represented in the figures as the mean ± S.D. Differences were considered statistically significant at p < 0.05 (*, p < 0.05; **, p < 0.01; and ***, p < 0.001).
Author contributions
H. Q. and B. L. data curation; H. Q., X. S., M. Y., B. L., M. L., S. S., S. C., J. Z., W.-G. Z., and J. L. formal analysis; H. Q. and B. L. investigation; H. Q. and B. L. visualization; H. Q. and J. L. methodology; H. Q. and B. L. writing-original draft; J. Z. and J. L. project administration; W. -G. Z. resources; J. L. conceptualization; J. L. supervision; J. L. funding acquisition; J. L. validation; J. L. writing-review and editing.
Acknowledgments
We thank Dr. Ying Zhao for a gift of anti-PRMT5 antibody. We thank Thomas Luo for editing the manuscript. We also thank the members of the Luo lab for helpful comments on the manuscript. We thank the core facility at Peking University Health Science Center for MS analysis.
This work was supported by National Natural Science Foundation of China Grants 81270427, 81471405, 81671389, and 81621063 and National Research Program of China 973 Program Grant 2013CB530801 (to J. L.). The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PRMT5
- protein arginine methyltransferase 5
- WDR77
- WD repeat-containing protein 77
- TSA
- trichostatin A
- GST
- glutathione S-transferase
- qPCR
- quantitative PCR
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- FL
- full-length
- PMSF
- phenylmethylsulfonyl fluoride
- aa
- amino acid(s)
- NT
- amino-terminal
- CT
- carboxyl-termina
- CBP
- cAMP-response element-binding protein.
References
- 1. Hirschey M. D. (2011) Old enzymes, new tricks: sirtuins are NAD(+)-dependent deacylases. Cell Metab. 14, 718–719 10.1016/j.cmet.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Michan S., and Sinclair D. (2007) Sirtuins in mammals: insights into their biological function. Biochem. J. 404, 1–13 10.1042/BJ20070140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Osborne B., Bentley N. L., Montgomery M. K., and Turner N. (2016) The role of mitochondrial sirtuins in health and disease. Free Radic. Biol. Med. 100, 164–174 10.1016/j.freeradbiomed.2016.04.197 [DOI] [PubMed] [Google Scholar]
- 4. Taylor D. M., Maxwell M. M., Luthi-Carter R., and Kazantsev A. G. (2008) Biological and potential therapeutic roles of sirtuin deacetylases. Cell. Mol. Life Sci. 65, 4000–4018 10.1007/s00018-008-8357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nahálková J. (2015) Novel protein-protein interactions of TPPII, p53, and SIRT7. Mol. Cell. Biochem. 409, 13–22 10.1007/s11010-015-2507-y [DOI] [PubMed] [Google Scholar]
- 6. Barber M. F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R. I., Paredes S., Young N. L., Chen K., Struhl K., Garcia B. A., Gozani O., Li W., and Chua K. F. (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114–118 10.1038/nature11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen S., Blank M. F., Iyer A., Huang B., Wang L., Grummt I., and Voit R. (2016) SIRT7-dependent deacetylation of the U3–55k protein controls pre-rRNA processing. Nat. Commun. 7, 10734 10.1038/ncomms10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen S., Seiler J., Santiago-Reichelt M., Felbel K., Grummt I., and Voit R. (2013) Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell 52, 303–313 10.1016/j.molcel.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 9. Mohrin M., Shin J., Liu Y., Brown K., Luo H., Xi Y., Haynes C. M., and Chen D. (2015) Stem cell aging: a mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377 10.1126/science.aaa2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ryu D., Jo Y. S., Lo Sasso G., Stein S., Zhang H., Perino A., Lee J. U., Zeviani M., Romand R., Hottiger M. O., Schoonjans K., and Auwerx J. (2014) A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab. 20, 856–869 10.1016/j.cmet.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 11. Tang X., Shi L., Xie N., Liu Z., Qian M., Meng F., Xu Q., Zhou M., Cao X., Zhu W. G., and Liu B. (2017) SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat. Commun. 8, 318 10.1038/s41467-017-00396-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang C., Zhai Z., Tang M., Cheng Z., Li T., Wang H., and Zhu W. G. (2017) Quantitative proteome-based systematic identification of SIRT7 substrates. Proteomics 17, 10.1002/pmic.201600395 [DOI] [PubMed] [Google Scholar]
- 13. Ford E., Voit R., Liszt G., Magin C., Grummt I., and Guarente L. (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075–1080 10.1101/gad.1399706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kiran S., Anwar T., Kiran M., and Ramakrishna G. (2015) Sirtuin 7 in cell proliferation, stress and disease: rise of the seventh Sirtuin!. Cell. Signal. 27, 673–682 10.1016/j.cellsig.2014.11.026 [DOI] [PubMed] [Google Scholar]
- 15. De Nigris F., Cerutti J., Morelli C., Califano D., Chiariotti L., Viglietto G., Santelli G., and Fusco A. (2002) Isolation of a SIR-like gene, SIR-T8, that is overexpressed in thyroid carcinoma cell lines and tissues. Br. J. Cancer 87, 1479 10.1038/sj.bjc.6600636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ashraf N., Zino S., Macintyre A., Kingsmore D., Payne A. P., George W. D., and Shiels P. G. (2006) Altered sirtuin expression is associated with node-positive breast cancer. Br. J. Cancer 95, 1056–1061 10.1038/sj.bjc.6603384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hosohata K., Li P., Hosohata Y., Qin J., Roeder R. G., and Wang Z. (2003) Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription. Mol. Cell. Biol. 23, 7019–7029 10.1128/MCB.23.19.7019-7029.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bao X., Zhao S., Liu T., Liu Y., Liu Y., and Yang X. (2013) Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J. Histochem. Cytochem. 61, 206–217 10.1369/0022155413475452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ibrahim R., Matsubara D., Osman W., Morikawa T., Goto A., Morita S., Ishikawa S., Aburatani H., Takai D., Nakajima J., Fukayama M., Niki T., and Murakami Y. (2014) Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum. Pathol. 45, 1397–1405 10.1016/j.humpath.2014.02.013 [DOI] [PubMed] [Google Scholar]
- 20. Stopa N., Krebs J. E., and Shechter D. (2015) The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell. Mol. Life Sci. 72, 2041–2059 10.1007/s00018-015-1847-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Antonysamy S., Bonday Z., Campbell R. M., Doyle B., Druzina Z., Gheyi T., Han B., Jungheim L. N., Qian Y., Rauch C., Russell M., Sauder J. M., Wasserman S. R., Weichert K., Willard F. S., Zhang A., and Emtage S. (2012) Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. U.S.A. 109, 17960–17965 10.1073/pnas.1209814109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho M. C., Wilczek C., Bonanno J. B., Xing L., Seznec J., Matsui T., Carter L. G., Onikubo T., Kumar P. R., Chan M. K., Brenowitz M., Cheng R. H., Reimer U., Almo S. C., and Shechter D. (2013) Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PloS One 8, e57008 10.1371/journal.pone.0057008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen H., Lorton B., Gupta V., and Shechter D. (2017) A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 36, 373–386 10.1038/onc.2016.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu H., Ye W., Wu J., Meng X., Liu R. Y., Ying X., Zhou Y., Wang H., Pan C., and Huang W. (2014) Overexpression of sirt7 exhibits oncogenic property and serves as a prognostic factor in colorectal cancer. Clin. Cancer Res. 20, 3434–3445 10.1158/1078-0432.CCR-13-2952 [DOI] [PubMed] [Google Scholar]
- 25. Malik S., Villanova L., Tanaka S., Aonuma M., Roy N., Berber E., Pollack J. R., Michishita-Kioi E., and Chua K. F. (2015) SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci. Rep. 5, 9841 10.1038/srep09841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lai C. C., Lin P. M., Lin S. F., Hsu C. H., Lin H. C., Hu M. L., Hsu C. M., and Yang M. Y. (2013) Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol. 34, 1847–1854 10.1007/s13277-013-0726-y [DOI] [PubMed] [Google Scholar]