Abstract
Glycosaminoglycans (GAGs) and GAG-degrading enzymes have wide-ranging applications in the medical and biotechnological industries. The former are also an important nutrient source for select species of the human gut microbiota (HGM), a key player in host–microbial interactions. How GAGs are metabolized by the HGM is therefore of interest and has been extensively investigated in the model human gut microbe Bacteroides thetaiotaomicron. The presence of as-yet uncharacterized GAG-inducible genes in its genome and of related species, however, is testament to our incomplete understanding of this process. Nevertheless, it presents a potential opportunity for the discovery of additional GAG-degrading enzymes. Here, we investigated a gene of unknown function (BT_3328) from the chondroitin sulfate (CS) utilization locus of B. thetaiotaomicron. NMR and UV spectroscopic assays revealed that it encodes a novel polysaccharide lyase (PL), hereafter referred to as BtCDH, reflecting its source (B. thetaiotaomicron (Bt)) and its ability to degrade the GAGs CS, dermatan sulfate (DS), and hyaluronic acid (HA). When incubated with HA, BtCDH generated a series of unsaturated HA sugars, including Δ4,5UA-GlcNAc, Δ4,5UA-GlcNAc-GlcA-GlcNac, Δ4,5UA-[GlcNAc-GlcA]2-GlcNac, and Δ4,5UA-[GlcNAc-GlcA]3-GlcNac, as end products and hence was classed as endo-acting. A combination of genetic and biochemical assays revealed that BtCDH localizes to the cell surface of B. thetaiotaomicron where it enables extracellular GAG degradation. BtCDH homologs were also detected in several other HGM species, and we therefore propose that it represents the founding member of a new polysaccharide lyase family (PL29). The current discovery also contributes new insights into CS metabolism by the HGM.
Keywords: chondroitin sulfate, hyaluronan, hyaluronate lyase, carbohydrate metabolism, bacterial metabolism, polysaccharide, CAZymes, cell surface enzyme, glycosaminoglycan degradation, human gut microbiota, PL29
Introduction
Mammalian glycosaminoglycans (GAGs) 2 are complex carbohydrates consisting of repeating disaccharide units of uronic acids or galactose linked to a hexosamine sugar (1, 2). These include chondroitin sulfate (CS) made up of GlcA and GalNAc, dermatan sulfate (DS), or chondroitin sulfate B made up of IdoA and GalNAc, hyaluronic acid (HA) made up of GlcA and GlcNAc, heparin/heparan sulfate (HS) made up of a mixture of GlcA and IdoA linked to GlcNAc and keratan sulfate made up of galactose linked to GlcNAc. GAGs such as CS, DS, and HS are also characterized by highly diverse sulfation patterns (introduced during biosynthesis by specific sulfotransferase enzymes) that define various molecular subtypes of each glycan (3, 4). CS, for example, can be sulfated on GalNAc at different positions including C4, C6, or both to yield major CS subtypes namely CSA, CSC, and CSE, respectively (5, 6) or at both C2 of GlcA and C6 of GalNAc, yielding CSD (7). In contrast, HA is not sulfated and hence less structurally complex compared with the other GAG types.
GAGs are ubiquitously distributed in the human body and implicated in several important physiological processes such as cell signaling, inflammation, neuronal development, adhesion, and tumor progression (8–11). Dietary GAGs are also a nutrient source for the human gut microbiota (HGM), which is known to impact greatly on human health and disease status (12–17). Indeed, recent evidence suggests that CS is a priority nutrient for prominent gut microbes such as Bacteroides thetaiotaomicron and Bacteroides ovatus and that only a subset of HGM species are capable of metabolizing the GAG (13, 15, 17). Understanding how GAGs are metabolized could therefore inform strategies including nutrient-based approaches to manipulate the HGM and hence improve human health. In line with this, several studies (reviewed by Ndeh and Gilbert (18)) have examined the metabolism of dietary GAGs by the HGM. The results demonstrate that GAG degradation is largely orchestrated by a combination of three major enzyme types including polysaccharide lyases (PLs, EC 4.2.2.-), sulfatases (EC 3.1.6.-), and glycoside hydrolases (EC 3.2.1.-). In the model human gut microbe B. thetaiotaomicron, as well as other gut Bacteroidetes, the genes encoding these enzymes are typically located within polysaccharide utilization loci (PULs) (14, 19–21). These genetic loci are diverse and encode a variety of cell envelope–associated multiprotein systems involved in complex glycan metabolism. Although the mechanisms of various GAG PULs in these organisms have been extensively investigated, knowledge of their functioning is incomplete, evident by the presence of as-yet uncharacterized GAG-inducible genes within these PULs (21). In the CS PUL of B. thetaiotaomicron, three of such genes, BT_3328, BT_3329, and BT_3330 (all of unknown function), appear in a distinct operon (BT_3328–30) conserved in several HGM Bacteroidetes (14, 20, 21). The latter suggests that they might be an important adaptation for CS metabolism in these microbes. Here, we characterized the largest member of this operon (BT_3328), revealing that it encodes a novel polysaccharide lyase hereafter referred to as BtCDH, reflecting its source (B. thetaiotaomicron (Bt)) and ability to degrade the GAGs CS, DS, and HA. We also provide evidence suggesting that BtCDH is not only the founding member of a novel enzyme family but also a cell surface GAG-degrading enzyme, the first to be reported in B. thetaiotaomicron and related species. Taken together, our findings contribute new insights into our knowledge of CS metabolism by the HGM, a key player in host–microbial interactions.
Results
In silico data on native BtCDH lyase
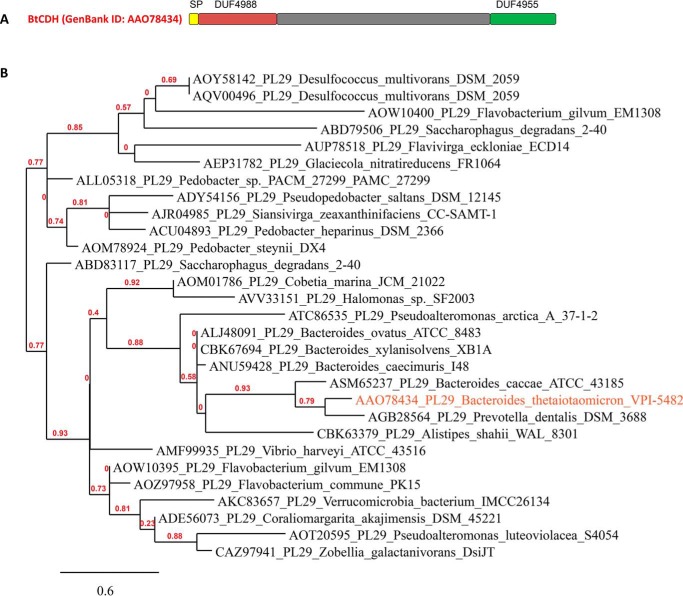
The gene encoding native BtCDH lyase (BT_3328 from B. thetaiotaomicron VPI-5482) is 2607 bp in length and has a GC content of 51%. BtCDH is a large protein of 868 amino acid residues, a molecular mass of 96 kDa, and an isoelectric pI of 5.33. A majority of the top hits (>50% identity) from a BLASTp search with native BtCDH are hypothetical/putative uncharacterized proteins mainly annotated as DUF4955 domain–containing. The DUF4955 domain whose function is unknown is present at the C terminus of the protein, whereas another domain DUF4988 is located directly after the putative lipoprotein signal peptide sequence at the N terminus of the protein (Fig. 1A). Top BtCDH lyase homologs were mostly detected in species from the Bacteroides, Alistipes, and Prevotella genera, all highly represented in the HGM. They were also prominent in species from marine environments including Saccharophagus degradans, Zobellia galactinovorans, Vibrio harveyi, and Coraliomargarita akajimensis. A phylogenetic tree of selected homologs of BtCDH is presented in Fig. 1B.
Figure 1.
Modular architecture of native BtCDH and phylogenetic analyses. A, BtCDH (GenBankTM accession number AAO78434) contains an N-terminal lipoprotein signal peptide sequence (SP) and a large central region sandwiched by two domains of unknown function (DUF4988 and DUF4955). B, phylogenetic tree showing the distribution of BtCDH homologs in selected species from various environments.
Activity of BtCDH
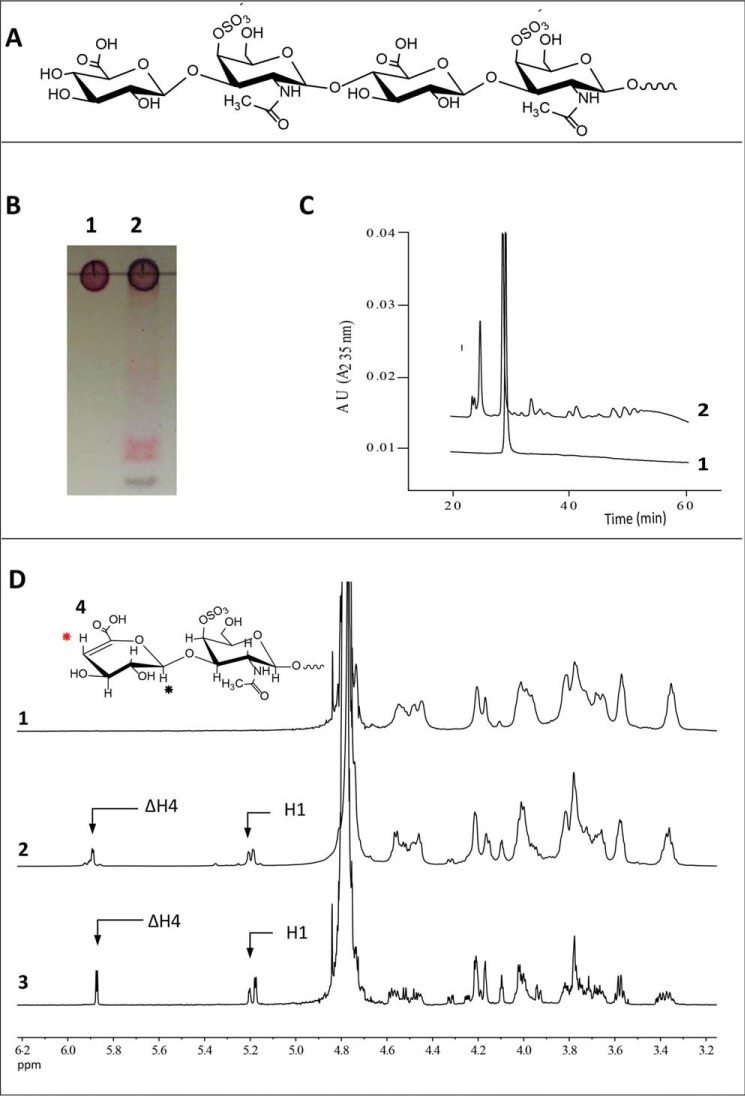
Purified recombinant BtCDH was initially assayed spectrophotometrically using CS from bovine trachea (which is mainly CS-4 sulfate or CSA (22)) (Fig. 2A) as substrate. BtCDH degraded CSA generating unsaturated products that were detected by TLC and high-performance anion-exchange chromatography (HPAEC) using a UV detector at 235 nm (A235) (Fig. 2, B and C, respectively). Limit products of the reaction were also resolved by size-exclusion chromatography (SEC) and analyzed by 1H NMR (Fig. 2D and Fig. S1). Distinct resonance signals were detected at 5.9 and 5.2 ppm in the spectra of the digested products as opposed to undigested CSA. The former is evidence of a H4 proton chemical shift consistent with glyosidic bond cleavage and the generation of C4,5 double bonds (Δ4,5) in the terminal nonreducing end GlcA or uronic acid (UA) residue (Δ4,5UA) of various products, whereas the latter peak at 5.2 ppm corresponds to resonance signals of the H1 anomeric proton of the same sugar (Fig. 2D) (23–26).
Figure 2.
Analyses of chondroitin 4-sulfate (CSA) degradation by BtCDH lyase. A, chemical structure of intact CSA. B, TLC of CSA before and after digestion by BtCDH lyase. For the reaction, 13.3 mg/ml of CSA from bovine trachea was treated with 0.3 μm of BtCDH lyase overnight. The reaction was stopped by boiling and 53.4 μg of digested CSA analyzed by TLC. Lane 1, CSA; lane 2, CSA treated with BtCDH lyase. C, HPAEC chromatogram of CSA digestion. Half the amount (26.7 μg) of sample in A was analyzed by HPAEC. D, 1H NMR results of CSA digestion by BtCDH lyase. Digested CSA products were purified by SEC and later analyzed by NMR. Panel 1, NMR spectrum of undigested CSA. Panel 2, spectra of species from all SEC fractions combined (fractions 1–43 in Fig. S1). Panel 3, spectra of species from selected homogenous fractions (pooled fractions 37–39 in in Fig. S1). Both spectra show resonance peaks for H1 and H4 (black and red) at 5.1 and 5.9 ppm, respectively, consistent with the formation of terminal unsaturated CSA oligosaccharides. Panel 4 (inset), a typical chemical rearrangement of components of the nonreducing end sugar in CSA after lyase digestion. H4 and H1 are marked with red and black circles, respectively.
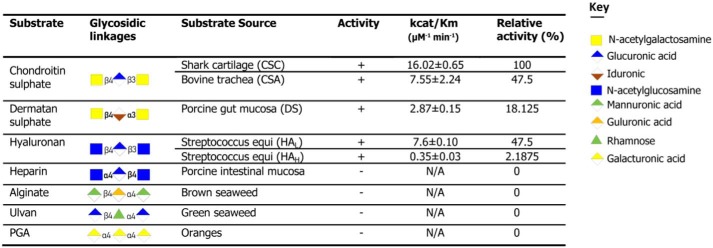
To explore the substrate specificity in BtCDH in more detail, the enzyme was also tested against a series of commercially available uronic acid-based substrates including CS from shark cartilage (mainly CS-6 sulfate or CSC), DS from porcine intestinal mucosa (mainly DS-4 sulfate), high and low molecular weight HA sodium salt from Streptococcus equi (10,000–30,000 (HAL) and 90,000–110,000 Da (HAH), respectively), heparin, alginate, ulvan, and polygalacturonic acid. BtCDH was active against CS DS and HA (in the order CSC > CSA/HAL > DS > HAH) but not heparin, alginate, ulvan, and polygalacturonic acid (Table 1). Previous studies analyzing CSA and CSC have reported major differences in the sulfated disaccharide composition of both substrates with the most prominent being the levels of the ΔUA-GalNAc4S, which is higher in CSA, and ΔUA-GalNAc6S, which is higher in CSC (12, 22, 27, 28). BtCDH activity thus appears to be affected by the type of sulfation in the GAG substrates.
Table 1.
Activity of BtCDH lyase against various uronate substrates
Degradation pattern and mode of BtCDH lyase activity
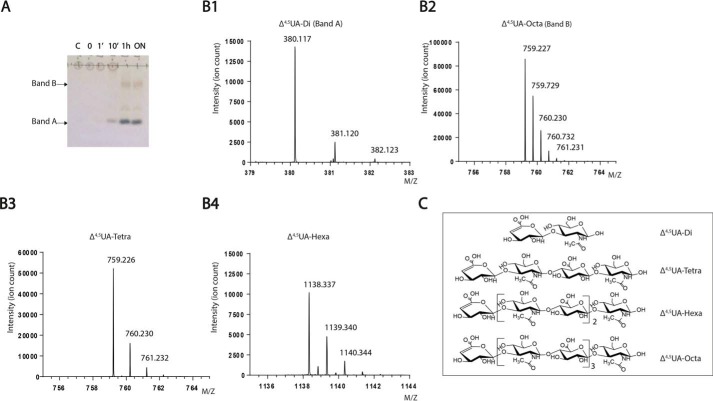
To determine whether BtCDH was endo- or exo-acting, we performed time-course experiments with HAL, a less chemically complex GAG compared with its sulfated counterparts. HAL was incubated with BtCDH over different times (0, 1, 5, and 60 min and ∼12 h (overnight), after which reactions were stopped by boiling and analyzed by TLC. BtCDH degraded HAL within 10 min, generating a fast migrating band (band A) (Fig. 3A). After 1 h of the reaction, the original HAL spot at the origin disappeared accompanied by the appearance another major band (band B), as well as low staining intermittent bands (Fig. 3A). The pattern of products remained unchanged even after overnight digestion, and hence the generated sugars represented the final limit products of HAL digestion. Bands A and B limit products were purified by SEC and analyzed by MS (HILIC LC-MS). Purified Band A yielded incremental masses of 380.117, 381.120, and 382.123 Da (differing by a mass of 1 Da) equivalent to the mass of singly charged ions of the unsaturated Δ4,5UA-GlcNAc disaccharide, whereas band B yielded incremental masses of 759.227, 759.729, and 760.230 Da (differing by a mass of 0.5 Da) etc. (Fig. 3B), equivalent to the mass of doubly charged fragments derived from the unsaturated Δ4,5UA-[GlcNAc-GlcA]3-GlcNAc octasaccharide parent ion. To detect underrepresented sugars, all SEC fractions were pooled in one sample and analyzed. The spectra revealed the presence of new products with incremental masses of 759.226, 760.230, and 761.232 (differing by a mass of 1 Da) and 1138.337, 1139.340, and 1140.344 (differing by a mass of 1 Da), corresponding to singly charged ions of the unsaturated Δ4,5UA-GlcNAc-GlcA-GlcNAc tetrasaccharide and Δ4,5UA-[GlcNAc-GlcA]2-GlcNAc hexasaccharide sugars, respectively (Fig. 3B). The generation of diverse molecular weight limit products thus suggests that BtCDH is an endo-acting enzyme. In addition, the accumulation of the high molecular weight HA octasaccharide suggests that BtCDH preferentially cleaves sugars with a degree of polymerization (DP) typically ≥10. The requirement for substrates with a DP ≥ 10 was also demonstrated using sulfated CS oligosaccharides as substrates with HPAEC data showing a major shift in the DP10 oligosaccharide profile (as opposed to a minor shift for DP8) after treatment with BtCDH (Fig. 4). The proportion of each HA product generated was also determined by UV with the results showing that over 70% of the generated products were Δ4,5UA-GlcNAc disaccharides (Fig. S2). Time-course experiments with sulfated GAGs CSA and CSC as substrates also yielded fairly similar patterns observed for HAL (Fig. S3). As with HAL, high amounts of the basic disaccharide building block (Δ4,5UA-GalNAc) were detected, suggesting high processivity in the enzyme.
Figure 3.
Evidence of BtCDH endo-lyase activity. A, TLC showing progressive degradation of hyaluronanic acid (HAL, 10–20 kDa) by BtCDH lyase over time. For the reaction, 32 mg/ml of HAL was digested with 2 μm of enzyme. The reaction was stopped at the various time points indicated, and 64 μg of each sample was analyzed by TLC. B, mass spectrometry data of purified HAL oligosaccharide species. Panel 1, spectrum for Δ4,5UA-disaccharide (Δ4,5UA-Di or band A). Panel 2, spectrum for Δ4,5UA-octasaccharide (Δ4,5UA-Octa or band B). Panel 3, spectrum for Δ4,5UA-tetrasaccharide (Δ4,5UA-Tetra). Panel 4, spectrum for Δ4,5UA-hexasaccharide (Δ4,5UA-Hexa). C, structural representation of various species detected in B.
Figure 4.
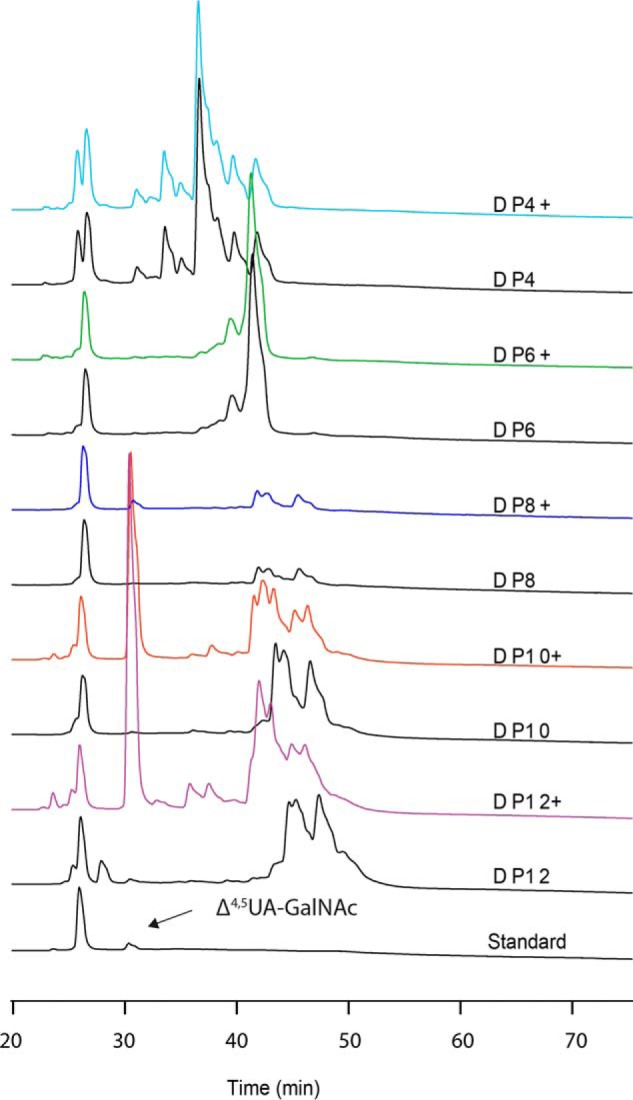
Activity of BtCDH against various chondroitin sulfate oligosaccharides. Substrates (1 mg/ml each) were treated with 0.5 μm of BtCDH (shown as DPx +, with x being the number of constituent monosaccharides) overnight or diluted in equivalent volume of buffer as control (DPx). HPAEC chromatograms show major shifts in the signals of sulfated DP10 and DP12 oligosaccharides following treatment, accompanied by the appearance of a strong signals for the Δ4,5UA-GalNAc disaccharide in each case.
Impact of temperature, pH, and metals on BtCDH
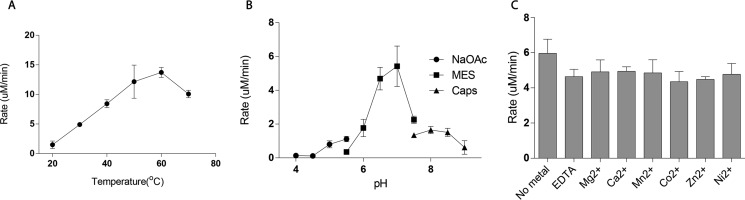
The effects of temperature, pH and metal ions on BtCDH activity were tested with CSA as substrate. BtCDH preferred higher reaction temperatures with the activity increasing linearly from 20 °C to a maximum rate of ∼15 μm/min at 60 °C (Fig. 5A). After this optimum temperature, the enzyme activity dropped sharply. To determine the optimum pH of BtCDH, enzyme activity was examined against CSA in different buffers over a pH range of 4–9. BtCDH activity rose gradually with increase pH between 4 and 6 and then sharply to an optimum pH of 7.0 in 10 mm MES buffer (Fig. 5B). BtCDH activity against CS was also tested in the presence of 5 mm EDTA and metal ions including Mg2+, Ca2+, Mn2+, Co2+, Zn2+, and Ni2+. Its activity was only slightly affected by metals with the most inhibition (27.42%) observed in the presence of Co2+ ions (Fig. 5C).
Figure 5.
Effect of temperature and pH and metals on BtCDH lyase activity. A, effect of temperature. Enzymatic activity was measured using 40 μm of CSA as substrate in 20 mm NaH2PO4 at pH 7.0 over different temperatures. B, effect of pH. BtCDH activity was measured at different pH levels using 20 mm of various buffers at 37 °C. C, effect of various metals. The reactions were carried out in 20 mm NaH2PO4 buffer containing 5 mm EDTA, Mg2+, Ca2+, Mn2+, and Co2+ and 1 mm Zn2+ and Ni2+ in separate experiments.
Growth experiments
A genetic mutant ΔBtCDH lacking the entire ORF for BtCDH (the BT_3328 gene) was created by counter selectable allelic exchange. Growth experiments with both B. thetaiotaomicron WT and ΔBtCDH strains were carried out with four different GAG substrates CSA, DS, CSC, HAL, and glucose as sole carbon source for at least 24 h. B. thetaiotaomicron WT and BtCDH cells grew at a very similar rates on various substrates, except for CSC where a minor defect in late phase growth was observed (Fig. S4).
Polyclonal antibodies and cellular localization assays
Polyclonal antibodies to BtCDH detect two forms of the protein
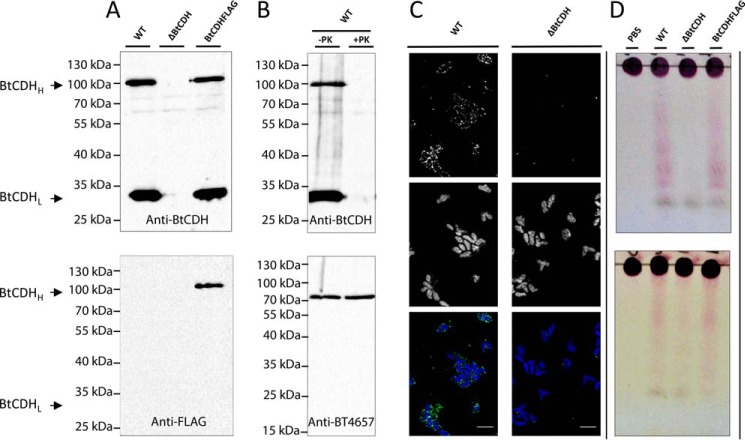
Rabbit anti-BtCDH polyclonal antibodies where generated using the recombinant BtCDH protein as immunogen. On Western blots containing cell-free extracts of B. thetaiotaomicron grown on CSA, two immunosensitive bands (BtCDHH and BtCDHL of high and low molecular weights, respectively) were detected (Fig. 6A). Despite its poor enrichment relative to BtCDHL, BtCDHH's molecular weight was close to 96 kDa, corresponding to the mass of native BtCDH, whereas the mass of BtCDHL was ∼30 kDa. Interestingly, when cell extracts of the ΔBtCDH strain lacking BtCDH were tested with the same antibodies, both bands were not detected, suggesting that BtCDHL is a subcomponent possibly a truncated form of BtCDH (Fig. 6A, top panel). To confirm this, BtCDH was FLAG-tagged (BtCDH FLAG) by genetically introducing a FLAG peptide sequence (DYKDDDK) at the C-terminal of the protein, thus allowing for its detection using rabbit anti-FLAG specific antibodies. Under these circumstances, only BtCDHH was detected by immunoblotting (Fig. 6A), suggesting that BtCDHL was an N-terminal truncated version of BtCDH.
Figure 6.
Localization of BtCDH and evidence of extracellular chondroitin lyase activity. A, Western blots showing the detection of full and truncated versions of BtCDH (BtCDHH and BtCDHL, respectively) in B. thetaiotaomicron WT, ΔBtCDH, and BtCDH FLAG cell extracts using polyclonal rabbit anti-BtCDH (top panel) and anti-FLAG tag antibodies (bottom panel). B, Western blotting analyses of proteinase K–treated and control cells using anti-BtCDH antibodies. The top results show loss of BtCDHH and BtCDHL after the treatment of whole cells with proteinase K. The bottom results show detection of an intact periplasmic control protein, the BT4657 heparinase of B. thetaiotaomicron (16). C, immunofluorescence detection of BtCDH in B. thetaiotaomicron WT and ΔBtCDH cells. The top panel shows green fluorescence signals of BtCDH. The middle panel shows blue signals from stained nuclei. The bottom panel is a merger of both signals. D, evidence of extracellular CS degradation in B. thetaiotaomicron by TLC. Top gel, TLC analyses of CS degradation when whole cells of B. thetaiotaomicron WT, ΔBtCDH, and BtCDH FLAG are incubated with CSA. Bottom gel, TLC analyses of CS degradation when whole cells of B. thetaiotaomicron WT, ΔBtCDH, and BtCDH FLAG are incubated with CSC.
Proteinase K, immunofluorescence, and whole-cell assays
As indicated earlier, BtCDH contains an N-terminal lipidation sequence (Fig. 1A) and hence is potentially localized extracellularly at the cell surface of B. thetaiotaomicron. To confirm this experimentally, we performed proteinase K (PK) and immunofluorescence assays. B. thetaiotaomicron WT cells were grown to exponential phase in 1% CSA, harvested, and treated with 0.5 mg/ml PK. Treated and untreated cells (controls) were lysed under reducing conditions, and cell extracts were analyzed by Western blotting using anti-BtCDH antibodies. The results showed degradation of BtCDHL and BtCDHH in PK-treated but not control cells (Fig. 6B). The experiment was also controlled by immunoblotting the same extracts with polyclonal antibodies targeting the constitutively expressed periplasmic heparin lyase BT4657 (16) whose bands remained intact in test and control extracts (Fig. 6B). The data thus suggest that BtCDH is localized to cell surface of B. thetaiotaomicron. The cell-surface localization of BtCDH was finally confirmed through immunofluorescence assays using anti-BtCDH antibodies (Fig. 6C). We also performed whole-cell aerobic assays on B. thetaiotaomicron WT and ΔBtCDH following growth on CSA. Under these conditions, transport across the outer membrane is limited, and only surface enzyme activity is detected. The cells were harvested at exponential phase and reincubated with CSA and CSC for 2 h, and the reaction was examined by TLC. The results showed extensive degradation of both CSA and CSC by the WT strain, visible as a smear of products (Fig. 6D). This was significantly reduced in the knockout strain especially for the CSA substrate in which just a single band of generated product was observed (Fig. 6D). Supernatants at exponential phase were also analyzed for secreted BtCDH by Western blotting with polyclonal rabbit anti-BtCDH antibodies, but only a faint band of the truncated form BtCDHL was detected (Fig. S5). Finally culture supernatants from B. thetaiotaomicron WT and ΔBtCDH at exponential phase were tested for secreted activity, but the TLC profiles of the treated and untreated CS substrates were very similar, except for a major band initially present in the supernatants (Fig. S5). We thus conclude that BtCDH activity is mainly present at the cell surface of B. thetaiotaomicron.
Discussion
In this study, we discovered and characterized the founding member of a novel family of GAG lyases from the prominent gut bacterium B. thetaiotaomicron. BtCDH is encoded by a gene from an operon (BT3328–30) of unknown function within the CS PUL of B. thetaiotaomicron (14), and hence until now, its function was unknown. We show that BtCDH exhibits polysaccharide lyase activity toward a variety of GAGs substrates including CS, DS, and HA. BtCDH alongside previously characterized CS ABC lyases from B. thetaiotaomicron WAL2926 (BactnABC) (22) and Proteus vulgaris (ChS ABC lyase I and ChS ABC lyase II) (29), named after their ability to degrade CSA, DS or CSB and CSC, are the only few lyases known to exhibit such broad GAG substrate specificity. BtCDH, however, displays very low catalytic efficiency (based on kcat/Km values) when compared with both enzymes. Bacteroides stercoris, Pedobacter saltans, Flavobacterium heparinum, and Vibrio sp. FC509 CS AC lyases degrade a variety of GAGs but are unable to degrade DS (2, 23, 30, 31), whereas B. stercoris ABC and F. heparinum ABC lyases, which are capable of degrading DS and CS, are unable to degrade HA (2). We also show, using HA as substrate, that BtCDH exhibits an endo-mode of activity, preferentially targeting oligosaccharides with DP ≥ 10. It may be possible that BtCDH contains over 10 subsites in its active site; however, further 3D structural studies of the enzyme in complex with the substrate (for which attempts were made in this study but were unsuccessful) will be required to determine this.
The enzyme also showed optimum activity at a very high temperature of ∼60 °C, contrary to most PLs with optimum temperatures between 30 and 40 °C (2, 22, 23, 29, 31). A notable exception is the ScPL8H lyase from Streptomyces coelicolor A3 (a nonpathogenic filamentous soil bacterium) with an optimum temperature of 57 °C (32). BtCDH activity was also only slightly affected by EDTA and the divalent metal ions tested in this study. Although it is possible that a metal ion inaccessible to EDTA chelation may be present and directly or indirectly involved in enzyme activity, the low sensitivity to diverse metals, broad CS substrate specificity, and thermophilic nature of BtCDH make it an attractive target from a biotechnological perspective. It is also worth noting that of the 28 PL families known to date, only 3 contain CS lyases (33), and hence BtCDH and its family members make an important addition to this group of enzymes in terms of the diversity of enzymatic options available for CS related investigations. The current family has now been designated polysaccharide lyase family 29 (PL29). Its presence in species from diverse environments including soil and marine bacteria also suggests that it may serve as an important adaptation for CS metabolism in these environments.
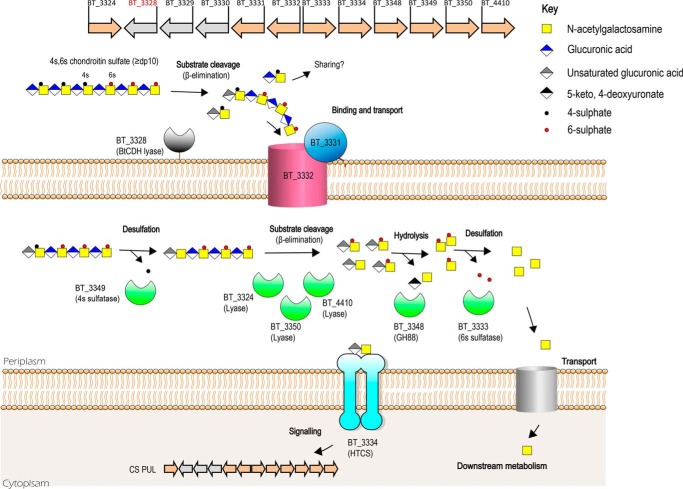
Another major finding in our study was the extracellular localization of BtCDH in B. thetaiotaomicron. The significance of this discovery lies in the fact that until now, models of CS metabolism did not include extracellular cell surface degradation of the substrate because no such enzymes had been reported in this organism before. In recent reports by Raghavan and Groisman (14) and Raghavan et al. (20), extracellular CS is bound and directly transported by a SusCD-like complex (BT3331–3332) into the periplasmic space without any initial extracellular cleavage. Hence to date, general consensus has held that enzymatic CS degradation begins in the periplasmic space of B. thetaiotaomicron. We not only present evidence of the extracellular localization of BtCDH but show through whole-cell assays that B. thetaiotaomicron is capable, with the aid of BtCDH of degrading CS extracellularly. Although our growth data did not show a significant defect in the ability of B. thetaiotaomicron to utilize most of the GAG substrates during growth, extracellular substrate degradation in a complex polymicrobial setting like the HGM could have significant implications for competition or cooperation between HGM species and hence microbial community structure. We also observed that the absence of BtCDH did not completely prevent extracellular degradation of CS in whole-cell assays, suggesting the presence of other unknown surface CS-degrading enzymes. This was more pronounced in whole-cell assays involving CSC where the mutant strain still showed significant CS degradation compared with the WT. Nevertheless, based on the evidence thus far for BtCDH, we propose a modification of the current model for CS metabolism in B. thetaiotaomicron to include the extracellular CS degrading activity encoded by BtCDH (Fig. 7). In this model, a complexly sulfated CS substrate such as CSC with a high degree of polymerization (DP ≥ 10) is initially cleaved on the cell surface generating oligosaccharides that are then bound and transported into the periplasmic space for further degradation by periplasmic enzymes as previously described (12, 14, 34) (also see Fig. 7). In future studies, we will also investigate the possibility that extracellular CS oligosaccharides generated by BtCDH, e.g. in the case of CSA, are the basis of cross-feeding and interbacterial interactions that may influence host health. To conclude, our current findings not only satisfy the growing demand for novel biocatalysts for modern biotechnological and medical research applications but also contribute new insights relevant to our understanding of CS or complex glycan metabolism by the HGM, a key player in host–microbial interactions.
Figure 7.
Proposed model of CS metabolism showing the context of BtCDH activity. BtCDH orchestrates extracellular degradation of sulfated CS (DP ≥ 10) to generate oligosaccharide products that are imported through an outer membrane protein complex (encoded by BT_3331 and BT_3332 genes) into the periplasmic space. They are further degraded by periplasmic sulfatases, lyases, and unsaturated glycoside hydrolase enzymes yielding end products 5-keto, 4-deoxyuronate, and GalNAc, which can be metabolized to provide energy for cell growth. Disaccharide intermediates generated during the process serve as signaling molecules activating expression of locus genes through binding to a hybrid two-component sensor protein (encoded by BT_3334) when CS is present.
Materials and methods
Plasmid construction and heterologous protein expression
The full-length BtCDH gene lacking the N-terminal lipoprotein signal sequence was amplified by PCR from B. thetaiotaomicron VPI-5482 genomic DNA using primers (BtCDHF, CGCGGGATCCgacgaacgggatgatctg; and BtCDHR, CGCGCTCGAGttatttaagtgcgttgagccatc) containing BamHI and XhoI restriction sites, respectively. The gene was cloned into pET-28a (+) vector (Novagen) which introduces a C-terminal His6 tag. For this, Escherichia coli TOP10 cells (Thermo Fisher Scientific) were used. The recombinant construct was sequenced (Eurofins Genomics) and later used to transform E. coli BL21 (DE3) expression cells (Thermo Fisher Scientific). The cells were cultured in Luria–Bertani medium containing 10 μg/ml kanamycin antibiotic to mid-log phase (A600 of 0.6). Protein expression was induced by adding 0.01 mm isopropyl β-d-1-thiogalactopyranoside to cells followed by growth overnight at 16 °C. The next day, the cells were harvested by centrifugation (4000 × g) and resuspended in Talon buffer (20 mm Tris/HCl, pH 8.0, plus 100 mm NaCl). The cells were then disrupted and centrifuged (16,000 × g) for 20 min at 4 °C. BtCDH was then purified from the resulting cell-free extract by immobilized metal affinity chromatography using fresh Talon resins (Clontech Laboratories Inc.). In the process, the CFE was loaded in a column containing the Talon resin and later washed with Talon buffer. Another wash was performed with Talon buffer containing 10 mm imidazole followed by recombinant protein elution with 100 mm imidazole. Purified proteins were analyzed by SDS-PAGE (12.5% gels) under reducing conditions. The analyses revealed a major protein band of ∼96 kDa, consistent with the predicted size of BtCDH. The samples were pooled, and buffer was exchanged into 20 mm NaH2PO4 buffer, pH 7.0, or other buffers of choice depending on the intended use.
Enzyme assays and biochemical properties of BtCDH
Activity assays against various GAGs were performed at 37 °C in 20 mm sodium phosphate buffer, pH 7.0. The final reaction volume was 600 μl containing 10 nm of enzyme in all cases except HA (containing 40 nm of enzyme) and varying concentrations of the substrate. The reactions were monitored using a Ultrospec 4000, UV-visible spectrophotometer at A235. To determine the optimum temperature of BtCDH activity, 40 μm of CSA was digested with 10 nm of BtCDH at different temperatures, and the activity was monitored. To determine the optimum pH, the same reaction was performed at different pH levels with 20 mm of each buffer. For metal dependence assays, various metals (5 mm for Mg2+, Ca2+, Mn2+, and Co2+ and 1 mm for Zn2+ and Ni2+), as well as EDTA (5 mm), were included in separate reactions, and their effects on enzyme activity were tested.
Thin-layer chromatography
Enzymatic reactions were stopped by heating at 98 °C for 3 min and later centrifuged at 16,000 × g for 1 min. Each reaction (2-μl drops) was spotted onto a Silicagel 60 TLC plate (Merck), and the components were resolved in a mobile phase containing butanol/acetic acid/water (2:1:1, v/v/v). The plates were later dried and developed with orcinol–sulfuric acid reagent (containing 0.1% orcinol and sulfuric acid, ethanol, water in the ratio 3.2:75:21.5 v/v/v). To visualize the sugar bands, the plates were gently warmed near a Bunsen flame.
SEC of polysaccharides
Samples (5 ml) were resolved using a Bio-Gel® P2 size-exclusion gels packed in Econo-Column® chromatography columns (Bio-Rad). The mobile phase was 50 mm acetic acid, and the pump flow rate was set at 0.2 ml min−1. For small scale (200 μl) purifications, elution was achieved by gravity. Fractions were later analyzed by TLC.
NMR
Digested products of CSA from bovine trachea were separated by SEC on a Bio-Gel® P2 size-exclusion system (Bio-Rad), and fractions were analyzed by TLC as described above. Purified fractions were collected and freeze-dried at −50 °C using a CHRIST Gefriertrocknung ALPHA 1–2 (Helmholtz-Zentrum) freeze-dryer. The samples were redissolved in 0.7 ml of deuterium oxide (99.9 atom % D) for NMR analyses. NMR spectra were obtained on a Bruker Avance III HD 700 MHz NMR spectrometer equipped with a TCI cryoprobe. Spectra were acquired at 298 K and were referenced to trimethylsilylpropanoic acid (TSP).
High-performance anion-exchange chromatography
HPAEC analyses were carried out using an automated Dionex ICS3000 system (Dionex) fitted with Spherisorb®-SAX columns and linked to an AD25 absorbance detector (Dionex). For HA samples, DionexTM CarboPacTM PA1 columns (Dionex) were used. The mobile phase consisted of 3 m NaCl applied as a 0–100% gradient over 110 min. The data were analyzed using the ChromeleonTM chromatography software (version 6.8) (Dionex).
BtCDH deletion mutant and FLAG tagging
ΔBtCDH and BtCDH FLAG strains were generated through counter-selectable allelic exchange using the pExchange vector as described by Koropatkin et al. (35).
Growth of B. thetaiotaomicron WT and mutant strains
B. thetaiotaomicron WT, ΔBtCDH, and BtCDH FLAG strains were cultured in 5 ml of TYG (tryptone–yeast extract–glucose) medium at 37 °C in an anaerobic cabinet (Whitley A35 work station; Don Whitley). The next day 10 μl of cells were added to 200 μl of minimal medium (38) containing 1% of various GAGs plus 1.2 mg/ml porcine hematin (Sigma–Aldrich). Cell growth was monitored at A600 using a Gen5 v2.0 microplate reader (Biotek).
Proteinase K experiments
The experiments were performed as described by Ndeh et al. (36) with the following modifications: 1) B. thetaiotaomicron WT cells were grown in 5 ml of minimal medium containing 1% CSA from bovine trachea to exponential phase (A600 of 0.8). Final cell pellets were dissolved in 300 μl of Laemmli buffer, and the samples were heated for 3 min. For each sample, 10 μl was analyzed by SDS-PAGE (7.5%). Immunoblotting was performed with rabbit anti-BtCDH antibodies at a 1:5000 dilution. For BtCDH FLAG cells, a 1:2000 dilution of rabbit anti-FLAG antibodies was used. In all cases, secondary antibodies were goat anti-rabbit IgG antibody, and (H+L) HRP conjugate (Invitrogen) at a 1:4000 dilution. The signals were developed using ClarityTM Western ECL substrate (Bio-Rad). To detect the native control protein (BT4657), the blots were washed overnight in PBS–Tween 20 (1× PBS containing 5% Tween 20) and reprobed with rabbit anti-BT4657 antibodies (1: 2000) as primary.
Immunofluorescence assays
B. thetaiotaomicron WT, ΔBtCDH, and BtCDH FLAG cells were grown to the exponential phase (A600 of 0.8) in 5 ml of minimal medium containing 1% CSA from bovine trachea. The cells were fixed by adding an equal volume of cell culture to a 1× PBS solution containing 3.7% formalin and rocking overnight at 4 °C. The cells were harvested by centrifugation at 4000 × g for 5 min and washed in 5 ml of PBS three times. The Cells (1 ml) were later incubated in blocking solution (2% goat serum, 0.02% NaN3 in 1× PBS) overnight at 4 °C while rocking. The next day cells were harvested by centrifugation at 16,000 × g for 1 min and incubated with primary rabbit anti-BtCDH antibodies (1:1000) for 2 h at room temperature. Another PBS wash was performed, and secondary goat anti-rabbit IgG (H+L) cross-adsorbed, Alexa Fluor 488 antibodies (Thermo Fisher Scientific, catalog no. R37116) were added according to the supplier's instructions. The cells were rocked for 30 min at room temperature and later washed two times in 1× PBS. The final pellet was resuspended in 100 μl of PBS alongside 20 μm Hoechst 33342 nuclear stain (Thermo Fisher Scientific, catalog no. 62249). The samples were mounted on agarose beds (1% w/v in PBS) in Gene Frames (Thermo Fisher Scientific, catalog no. AB0576) and visualized on a Zeiss Axio Observer Z1, with a LSM800 confocal scan head. The images were taken using a 63× (N.A. 1.4) oil immersion lens with 5× scan zoom applied. Excitation of the Alexa 488 was performed using the 488-nm laser line, and the reflected light was collected on a GaAsp detector between 510 and 700 nm through the variable beam splitter dichroic block. Excitation of the Hoechst was performed using the 405-nm laser line with emission detected between 400 and 510 nm. A z-section of 25 images was recorded with an interval of 0.1 μm using Zeiss Zen 2.3 software with identical settings for all images. Deconvolution was carried out using Huygens Professional (Scientific Volume Imaging) before a maximum intensity projection was carried out within ImageJ.
Whole-cell assays and supernatant analyses
B. thetaiotaomicron WT, ΔBtCDH, and BtCDH FLAG cells were cultured to exponential phase (A600 of 0.8) in 5 ml of minimal medium. The cells (1 ml) were harvested at 4000 × g for 5 min and washed once in 1× PBS by centrifuging again at 4000 × g for 5 min. The cells were then resuspended in 100 μl of PBS containing 4% of the GAG substrate. The samples were incubated overnight for 2 h followed by centrifugation 4000 × g for 5 min. The supernatants (2 μl) were then analyzed by TLC. To test for BtCDH secretion, 10 μl of the B. thetaiotaomicron WT supernatant was analyzed by Western blotting using polyclonal anti-BtCDH antibodies. For this, filtered and unfiltered aliquots including a 10× concentrated aliquot of the former were analyzed. Whole cells (10 μl) prepared by resuspending centrifuged cells in 1 ml of PBS were also analyzed alongside as controls. To confirm extracellular CS degradation during growth, cells were harvested at exponential phase (16,000 × g for 1 min), and supernatant (8 μl) was analyzed by TLC. Supernatants were also analyzed for secreted BtCDH activity. For this, supernatants were collected at exponential and centrifuged at 4000 × g for 5 min to remove cells. This was repeated once, and the supernatants were finally filtered through a 0.2-μm syringe cap filter (Pall Life Sciences). For each reaction, 20 μl of the supernatant was incubated overnight with 20 μl of 40 mg/ml CS substrate. The next day 4 μl of each sample was analyzed by TLC.
Mass spectrometry
After purification of HA oligosaccharides, 200-μl fractions were freeze-dried and dissolved in distilled water. The samples were diluted 1:10 (v/v) with buffer B (80% acetonitrile, 20% 50 mm ammonium formate in water, pH 4.7), and 2.0 μl was injected onto an LC-MS instrument configuration comprising a NanoAcquity HPLC system (Waters) and QTof mass spectrometer (Impact II, Bruker). Analysis was performed via gradient elution from a ZIC-HILIC (SeQuant®, 3.5 μm, 200 Å, 150 × 0.3 mm, Merck) capillary column heated to 35 °C and flowing at 5 μl/min. The elution gradient was as follows; 100% buffer B for 5 min, followed by a gradient to 0% B, 100% buffer A (50 mm ammonium formate in water, pH 4.7) over 40 min. 10 column volumes of buffer B equilibration was performed between injections. MS data were collected in positive ion mode, 50–2000 m/z, with capillary voltage and temperature settings of 2800 V and 200 °C, respectively, together with a drying gas flow of 6 liters/min and nebulizer pressure of 0.4 bar. The resulting MS data were analyzed using Compass DataAnalysis software (Bruker).
Phylogenetic analyses
Sequences were retrieved from the NCBI database (cut-off ≥ 35% identity) from a BlastP search with BtCDH (residues 308–708) as query. After trimming the N- and C-terminal domains of unknown function (DUFs), sequences were submitted to the http://www.phylogeny.fr/ server (37) in “one-click mode” to generate a phylogenetic tree.
Author contributions
D. N. conceptualization; D. N. and B. H. data curation; D. N., A. C., C. W., B. H., and J. G. formal analysis; D. N. supervision; D. N., J. M. M., D. B., C. W., and J. G. investigation; D. N. methodology; D. N. writing-original draft; D. N. project administration.
Supplementary Material
Acknowledgment
We thank Prof. Harry Gilbert for the guidance provided during the preparation of this manuscript.
This work was supported by the European Union's Seventh Framework Program Grant FP/2007/2013 and European Research Council Grant Agreement 322820. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- GAG
- glycosaminoglycan
- CS
- chondroitin sulfate
- PL
- polysaccharide lyase
- CS
- chondroitin sulfate
- DS
- dermatan sulfate
- HA
- hyaluronic acid
- HGM
- human gut microbiota
- PUL
- polysaccharide utilization locus
- HPAEC
- high-performance anion-exchange chromatography
- SEC
- size-exclusion chromatography
- UA
- uronic acid
- DP
- degree of polymerization
- PK
- proteinase K.
References
- 1. Pomin V. H., and Mulloy B. (2018) Glycosaminoglycans and proteoglycans. Pharmaceuticals (Basel) 11, E27 10.3390/ph11010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hong S. W., Kim B. T., Shin H. Y., Kim W. S., Lee K. S., Kim Y. S., and Kim D. H. (2002) Purification and characterization of novel chondroitin ABC and AC lyases from Bacteroides stercoris HJ-15, a human intestinal anaerobic bacterium. Eur. J. Biochem. 269, 2934–2940 10.1046/j.1432-1033.2002.02967.x [DOI] [PubMed] [Google Scholar]
- 3. Kusche-Gullberg M., and Kjellén L. (2003) Sulfotransferases in glycosaminoglycan biosynthesis. Curr. Opin. Struct. Biol. 13, 605–611 10.1016/j.sbi.2003.08.002 [DOI] [PubMed] [Google Scholar]
- 4. Gama C. I., Tully S. E., Sotogaku N., Clark P. M., Rawat M., Vaidehi N., Goddard W. A. 3rd, Nishi A., and Hsieh-Wilson L. C. (2006) Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat. Chem. Biol. 2, 467–473 10.1038/nchembio810 [DOI] [PubMed] [Google Scholar]
- 5. Vázquez J. A., Rodríguez-Amado I., Montemayor M. I., Fraguas J., González Mdel P., and Murado M. A. (2013) Chondroitin sulfate, hyaluronic acid and chitin/chitosan production using marine waste sources: characteristics, applications and eco-friendly processes: a review. Mar. Drugs 11, 747–774 10.3390/md11030747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., and Kitagawa H. (2003) Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 13, 612–620 10.1016/j.sbi.2003.09.011 [DOI] [PubMed] [Google Scholar]
- 7. Davidson S., Gilead L., Amira M., Ginsburg H., and Razin E. (1990) Synthesis of chondroitin sulfate D and heparin proteoglycans in murine lymph node-derived mast cells. The dependence on fibroblasts. J. Biol. Chem. 265, 12324–12330 [PubMed] [Google Scholar]
- 8. Kim S. H., Turnbull J., and Guimond S. (2011) Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 209, 139–151 10.1530/JOE-10-0377 [DOI] [PubMed] [Google Scholar]
- 9. Wight T. N., Kinsella M. G., and Qwarnström E. E. (1992) The role of proteoglycans in cell adhesion, migration and proliferation. Curr. Opin. Cell Biol. 4, 793–801 10.1016/0955-0674(92)90102-I [DOI] [PubMed] [Google Scholar]
- 10. Gill S., Wight T. N., and Frevert C. W. (2010) Proteoglycans: key regulators of pulmonary inflammation and the innate immune response to lung infection. Anat. Rec. (Hoboken) 293, 968–981 10.1002/ar.21094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iozzo R. V., and Sanderson R. D. (2011) Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J. Cell Mol. Med. 15, 1013–1031 10.1111/j.1582-4934.2010.01236.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ulmer J. E., Vilén E. M., Namburi R. B., Benjdia A., Beneteau J., Malleron A., Bonnaffé D., Driguez P. A., Descroix K., Lassalle G., Le Narvor C., Sandström C., Spillmann D., and Berteau O. (2014) Characterization of glycosaminoglycan (GAG) sulfatases from the human gut symbiont Bacteroides thetaiotaomicron reveals the first GAG-specific bacterial endosulfatase. J. Biol. Chem. 289, 24289–24303 10.1074/jbc.M114.573303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pudlo N. A., Urs K., Kumar S. S., German J. B., Mills D. A., and Martens E. C. (2015) Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. MBio 6, e01282–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raghavan V., and Groisman E. A. (2015) Species-specific dynamic responses of gut bacteria to a mammalian glycan. J. Bacteriol. 197, 1538–1548 10.1128/JB.00010-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tuncil Y. E., Xiao Y., Porter N. T., Reuhs B. L., Martens E. C., and Hamaker B. R. (2017) Reciprocal prioritization to dietary glycans by gut bacteria in a competitive environment promotes stable coexistence. MBio 8, e01068–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cartmell A., Lowe E. C., Baslé A., Firbank S. J., Ndeh D. A., Murray H., Terrapon N., Lombard V., Henrissat B., Turnbull J. E., Czjzek M., Gilbert H. J., and Bolam D. N. (2017) How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc. Natl. Acad. Sci. U.S.A. 114, 7037–7042 10.1073/pnas.1704367114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desai M. S., Seekatz A. M., Koropatkin N. M., Kamada N., Hickey C. A., Wolter M., Pudlo N. A., Kitamoto S., Terrapon N., Muller A., Young V. B., Henrissat B., Wilmes P., Stappenbeck T. S., Núñez G., et al. (2016) A Dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ndeh D., and Gilbert H. J. (2018) Biochemistry of complex glycan depolymerisation by the human gut microbiota. FEMS Microbiol. Rev. 42, 146–164 10.1093/femsre/fuy002 [DOI] [PubMed] [Google Scholar]
- 19. Martens E. C., Chiang H. C., and Gordon J. I. (2008) Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 10.1016/j.chom.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raghavan V., Lowe E. C., Townsend G. E. 2nd, Bolam D. N., and Groisman E. A. (2014) Tuning transcription of nutrient utilization genes to catabolic rate promotes growth in a gut bacterium. Mol. Microbiol. 93, 1010–1025 10.1111/mmi.12714 [DOI] [PubMed] [Google Scholar]
- 21. Terrapon N., Lombard V., Gilbert H. J., and Henrissat B. (2015) Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 31, 647–655 10.1093/bioinformatics/btu716 [DOI] [PubMed] [Google Scholar]
- 22. Shaya D., Hahn B.-S., Park N. Y., Sim J.-S., Kim Y. S., and Cygler M. (2008) Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926. Biochemistry 47, 6650–6661 10.1021/bi800353g [DOI] [PubMed] [Google Scholar]
- 23. Rani A., and Goyal A. (2016) A new member of family 8 polysaccharide lyase chondroitin AC lyase (Ps PL8A) from Pedobacter saltans displays endo- and exo-lytic catalysis. J. Mol. Catalysis B Enzymatic 134, 215–224 10.1016/j.molcatb.2016.11.001 [DOI] [Google Scholar]
- 24. Lemmnitzer K., Riemer T., Groessl M., Süss R., Knochenmuss R., and Schiller J. (2016) Comparison of ion mobility-mass spectrometry and pulsed-field gradient nuclear magnetic resonance spectroscopy for the differentiation of chondroitin sulfate isomers. Anal. Methods 8, 8483–8491 10.1039/C6AY02531E [DOI] [Google Scholar]
- 25. Huckerby T. N., Lauder R. M., Brown G. M., Nieduszynski I. A., Anderson K., Boocock J., Sandall P. L., and Weeks S. D. (2001) Characterization of oligosaccharides from the chondroitin sulfates. 1H-NMR and 13C-NMR studies of reduced disaccharides and tetrasaccharides. Eur. J. Biochem. 268, 1181–1189 10.1046/j.1432-1327.2001.01948.x [DOI] [PubMed] [Google Scholar]
- 26. Silva C., Novoa-Carballal R., Reis R. L., and Pashkuleva I. (2015) Following the enzymatic digestion of chondroitin sulfate by a simple GPC analysis. Anal. Chim. Acta 885, 207–213 10.1016/j.aca.2015.05.034 [DOI] [PubMed] [Google Scholar]
- 27. Muthusamy A., Achur R. N., Valiyaveettil M., Madhunapantula S. V., Kakizaki I., Bhavanandan V. P., and Gowda C. D. (2004) Structural characterization of the bovine tracheal chondroitin sulfate chains and binding of Plasmodium falciparum–infected erythrocytes. Glycobiology 14, 635–645 10.1093/glycob/cwh077 [DOI] [PubMed] [Google Scholar]
- 28. Malavaki C. J., Asimakopoulou A. P., Lamari F. N., Theocharis A. D., Tzanakakis G. N., and Karamanos N. K. (2008) Capillary electrophoresis for the quality control of chondroitin sulfates in raw materials and formulations. Anal. Biochem. 374, 213–220 10.1016/j.ab.2007.11.006 [DOI] [PubMed] [Google Scholar]
- 29. Hamai A., Hashimoto N., Mochizuki H., Kato F., Makiguchi Y., Horie K., and Suzuki S. (1997) Two distinct chondroitin sulfate ABC lyases. J. Biol. Chem. 272, 9123–9130 10.1074/jbc.272.14.9123 [DOI] [PubMed] [Google Scholar]
- 30. Shim K.-W., and Kim D.-H. (2008) Cloning and expression of chondroitinase AC from Bacteroides stercoris HJ-15. Protein Expr. Purif. 58, 222–228 10.1016/j.pep.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 31. Han W., Wang W., Zhao M., Sugahara K., and Li F. (2014) A novel eliminase from a marine bacterium that degrades hyaluronan and chondroitin sulfate. J. Biol. Chem. 289, 27886–27898 10.1074/jbc.M114.590752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elmabrouk Z. H., Vincent F., Zhang M., Smith N. L., Turkenburg J. P., Charnock S. J., Black G. W., and Taylor E. J. (2011) Crystal structures of a family 8 polysaccharide lyase reveal open and highly occluded substrate-binding cleft conformations. Proteins 79, 965–974 10.1002/prot.22938 [DOI] [PubMed] [Google Scholar]
- 33. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng Q., Yu M. C., Reeves A. R., and Salyers A. A. (1995) Identification and characterization of a Bacteroides gene, csuF, which encodes an outer membrane protein that is essential for growth on chondroitin sulfate. J. Bacteriol. 177, 3721–3727 10.1128/jb.177.13.3721-3727.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koropatkin N. M., Martens E. C., Gordon J. I., and Smith T. J. (2008) Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115 10.1016/j.str.2008.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ndeh D., Rogowski A., Cartmell A., Luis A. S., Baslé A., Gray J., Venditto I., Briggs J., Zhang X., Labourel A., Terrapon N., Buffetto F., Nepogodiev S., Xiao Y., Field R. A., et al. (2017) Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70 10.1038/nature21725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., and Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Larsbrink J., Rogers T. E., Hemsworth G. R., McKee L. S., Tauzin A. S., Spadiut O., Klinter S., Pudlo N. A., Urs K., Koropatkin N. M., Creagh A. L., Haynes C. A., Kelly A. G., Cederholm S. N., Davies G. J., et al. (2014) A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502 10.1038/nature12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.