Abstract
Aim:
Members of the genus Citrobacter are important opportunistic pathogens responsible for high mortality rate. Therefore, in this study, we aimed to develop efficient and accurate Citrobacter typing schemes for clinical detection and epidemiological surveillance.
Materials & methods:
Using genomic and experimental analyses, we located the O-antigen biosynthesis gene clusters in Citrobacter genome for the first time, and used comparative genomic analyses to reveal the specific genes in different Citrobacter serotypes.
Results:
Based on the specific genes in O-antigen biosynthesis gene clusters of Citrobacter, we established experimental and in silico serotyping systems for this bacterium.
Conclusion:
Both serotyping tools are reliable, and our observations are biologically and clinically relevant for understanding and managing Citrobacter infection.
Keywords: : Citrobacter, molecular O-serotyping, O-antigen biosynthesis gene cluster
The genus Citrobacter, a group of facultative, anaerobic and Gram-negative bacilli, belongs to the family Enterobacteriaceae. Citrobacter strains are often isolated from water, soil, food and intestinal tracts of animals and humans [1], and can cause bacteremia, meningitis, sepsis, peritonitis, urinary tract infections, respiratory tract infections and postoperative infections, especially in infants, children under the age of 6 years and immunocompromised individuals [2–5]. In particular, invasive Citrobacter infections are associated with high mortality rate, with 33–48% of patients succumbing to Citrobacter bacteremia [6–8]. Owing to high isolation rate, and severe infection and high mortality in the clinic, Citrobacter is considered an important opportunistic pathogen [3].
The Citrobacter genus is divided into 11 different genomospecies [9]. C. freundii and C. koseri constitute the most important pathogenic species and have the highest isolation rate in the clinic [4,10,11]. Citrobacter is most closely related to Escherichia coli and Salmonella [12], albeit with higher level of diversity. For example, the branch lengths separating certain pairs of Citrobacter species are almost as long as those separating E. coli and Salmonella [13].
Several more recently developed molecular typing methods, such as pulsed-field gel electrophoresis, multilocus sequence typing and restriction fragment length polymorphism, have been used to detect and genotype the Citrobacter clinical isolates [8,14–16]. However, the serotyping scheme based on variations in O-antigens is still the ‘gold standard’ for Gram-negative pathogen detection and identification in clinical specimens and environmental samples [17].
West and Edwards first established an antigenic scheme comprising 32 O-serogroups for Citrobacter in 1954 [18,19], which was extended to 43 O-serogroups in 1966 by Sedlak and Slajsova [12,20]. Although this serotyping scheme has been widely accepted and applied for Citrobacter [19,21,22], it has not been updated since 1966. Furthermore, considering that O-antisera of Citrobacter are currently not commercially available, the majority of laboratories are unable to perform conventional antigenic typing. In addition, cross-reactions between Citrobacter and other genera of Enterobacteriaceae, such as Escherichia, Klebsiella, Hafnia and Salmonella, hinder accurate identification of Citrobacter [19]. Molecular methods are more specific and sensitive than traditional methods. Among these, microarrays allow large-scale detection of multiple targets because of their throughput power. In recent years, the Luminex-based suspension array has become an attractive approach for pathogenic detection of both protein and nucleic acids, and has been approved by the US FDA for clinical diagnosis in various applications [23,24].
The diversity of the O-antigen arises mainly from genetic variations in the O-antigen biosynthesis gene clusters (O-AGCs), which provides the basis for molecular O-serotyping [25]. Most genes in an O-AGC are involved in one of the following three functions: nucleotide sugar synthesis, sugar transfer and O unit processing [26]. In particular, the O-antigen processing genes wzy, wzx, wzm and wzt are highly serotype determinative, and have hence been widely used as target genes in molecular serotyping of many Gram-negative bacteria [27–29]. Notably, the consistency between O-AGC genetic variation-based molecular serotyping and the conventional serotyping scheme has been demonstrated in several genera [28,30–33].
However, studies on Citrobacter O-antigens have mainly focused on the structures [22,34–39] instead of diversities of O-AGCs. In the present study, we determined the location of O-AGC in Citrobacter genome and its function by deletion and complementation testing. We subsequently obtained 18 O-AGCs of Citrobacter by whole-genome sequencing. We then developed a molecular O-serotyping system using a microsphere-based suspension array (MSA) platform based on the specific genes of O-AGCs. In addition, we also established a valid, genome-based tool for in silico serotyping of Citrobacter. All 98 Citrobacter genome sequences available in GenBank (National Institutes of Health [NIH], MD, USA) were subjected to the genome-based serotyping tool, and 90 were classified into 33 novel O-AGC groups. We provided alternatives in this study to perform faster and easier O-serotyping of Citrobacter using both experimental and bioinformatic approaches. Finally, we demonstrated the potential of this method for epidemiological surveillance and clinical identification of Citrobacter.
Materials & methods
Bacterial strains & genomic DNA extraction
In total, 18 Citrobacter strains were obtained from the Polish Collection of Microorganisms (PCM) at the Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences (Wroclaw, Poland). The details of the bacterial strains used in this study are listed in Table 1. Citrobacter strains were grown in Luria broth on an orbital shaker at 180 rpm and 37°C overnight. The genomic DNAs were extracted using a bacteria extraction kit (CWBIO Co. Ltd, Beijing, China) and stored at -20°C.
Table 1. . Citrobacter strains sequenced in this study.
| Lab collection NO. | Species | Serovar | Structure reference | Gene cluster accession number |
|---|---|---|---|---|
| G3336 | C. youngae | O3 | 19 | MH325885 |
| G3337 | C. youngae | O4 | 19 | MH325886 |
| G3340 | C. braakii | O5 | 19 | MH325887 |
| G3341 | C. braakii | O6 | 22 | MH325888 |
| G3344 | C. braakii | O7 | 19 | MH325889 |
| G3518 | C. braakii | O8 | 19 | MH325890 |
| G3533 | C. gillenii | O9 | 19 | MH325891 |
| G3535 | C. gillenii | O12 | 19 | MH325892 |
| G3537 | C. werkmanii | O14 | 39 | MH325893 |
| G3541 | C. youngae | O16 | 19 | MH325894 |
| G3515 | C. freundii | O22 | 38 | MH325895 |
| G3516 | C. freundii | O23 | 19 | MH325896 |
| G3521 | C. werkmanii | O24 | 19 | MH325897 |
| G3522 | C. werkmanii | O26 | 19 | MH325898 |
| G3919 | C. braakii | O30 | 19 | MH325899 |
| G3543 | C. youngae | O32 | 19 | MH325900 |
| G3538 | C. werkmanii | O38 | 19 | MH325902 |
| G3915 | C. freundii | O41 | 19 | MH325903 |
Draft genome sequencing & analysis
Whole genomes of the 18 Citrobacter strains were sequenced using Solexa pair-end sequencing technology [40]. The Solexa Genome Analyzer IIx (Illumina, Little Chesterford, Essex) was used to a depth of 90- to 100-fold coverage. The Illumina data were de novo assembled using the Velvet Optimizer v2.2. Polymerase chain reaction (PCR) was used to close gaps within the major genes in O-antigen biosynthesis gene clusters, and the PCR products were sequenced using traditional Sanger sequencing. In total, 98 genome sequences of Citrobacter were downloaded from the GenBank database (listed in Supplementary Table 1). Detailed analyses of genes in each O-AGC were performed using BLAST search (NIH).
Construction of the wzm deletion mutant strain & complementary strain of C. werkmanii O26
wzm of the C. werkmanii O26 (G3522) strain was deleted as described previously [41], with some modifications. A common plasmid pKD46 (Novagen, Trento, Italy) was used instead of pKD46-Gm as G3522 is not resistant to ampicillin. The concentration of L-arabinose was increased to 4 mM to induce recombination. wzm was finally replaced by a kanamycin resistance cassette from plasmid pKD4. The O26 △wzm strain was termed H2659. A complementary strain with full ABC transporter function was also constructed using the plasmid pTRC99a harboring the original wzm from strain G3522. The recombinant plasmid was transferred into H2659 to complement for the loss of wzm. The complementary strain was termed H2660. Strains, plasmids and primers used for generating and identifying H2659 and H2660 are listed in Supplementary Table 2.
Extraction & silver staining of lipopolysaccharide
Lipopolysaccharide (LPS) was extracted using hot aqueous-phenol extraction as described previously [42]. In the present study, we pelleted the bacteria from 20 ml cultured Luria broth medium and quadrupled all the reagent dosages. The extracted LPSs of G3522, H2659 and H2660 were separated by 13% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE). Silver staining was performed using the Fast Silver Stain kit (no. P0017S, Beyotime, Shanghai, China).
Primer & probe design
Primers and probes were designed based on the sequences of wzx, wzy, wzm and wzt. As O4 does not contain any of these genes in its O-AGC, the gene encoding glycosyl transferase (GT) was selected as the target. For O7 and O26, another set of primers and probes were designed based on gtrC. Biotin was used to label to the 5′-end of reverse primers (Invitrogen Co. Ltd, Beijing, China). Probes were synthesized with an amino C-12 module at the 5′-end (AUGCT, Beijing, China). Primers and probes used in this study are listed in Table 2.
Table 2. . Primers and probes used in Citrobacter molecular typing.
| OAgc form | Target gene | Forward/reverse primers (5′-3′) | Product size (bp) | Probes |
|---|---|---|---|---|
| O3 | wzm | wl-72175:TTTTTCGCGATGTTGGG | 212 | OA-5600:TCCTATGGCTAGCGTGATTGAG |
| wl-72176:ACCAAGACTAAGTGGGTAGATTAA | ||||
| O7 | wzm | wl-72175:TTTTTCGCGATGTTGGG | 212 | OA-5600:TCCTATGGCTAGCGTGATTGAG |
| wl-72176:ACCAAGACTAAGTGGGTAGATTAA | ||||
| O7 | gtrC | wln-12418:TATCTTTTAGCGATGGCACTT | 236 | OAn-171:CCGCTGATGGGTATGGTTTC |
| wln-12419:GAGTATGGTGGGTAATAAGGGT | ||||
| O4 | GT | wl-72149:CCTGGTAGCCTCACGATAGA | 219 | OA-6027:GAGTTACCTGGTCATGTGGATAATG |
| wl-72150:AACTACGGTCTCCCGACAC | ||||
| O5 | wzm | wl-72155:ATCCTGGCATCTTTGGACT | 336 | OA-6031:GAAGGGTGTGTAACTTTCATATCGT |
| wl-72156:ACTATATTATGGCAAAGCATCATA | ||||
| O6 | wzm | wl-72157:CATGGCGGCGGTACA | 261 | OA-6034:GGTAGTGGCATACGCAGTCTTTT |
| wl-72158:CAGGATCGTTAGATGCGG | ||||
| O8 | wzm | wl-72187:TTCTGTCGGGCTGGGTAT | 258 | OA-5606:TTGATGCGCTACCGGAAAA |
| wl-72188:ATTCCACCGGATATTAGAACA | ||||
| O9 | wzm | wl-72193:TTGGCCTGTGCTGGATC | 273 | OA-6037:TGTGTTGATTGACTATAACCCTGTATATC |
| wl-72194:TAGGCCATTCGCCTTGTA | ||||
| O12 | wzx | wl-73677:CTGTATCTCTCCAAAGCCACTC | 252 | OA-6266:TACCGCAAGTAAGGTTAGTTTTTTATAG |
| wl-73678:AATTTCGGCGACTCTATATCAT | ||||
| O14 | GT | wl-72195:GGCTGCTGTAGCGAGGA | 249 | OA-5610:CGGCAACTGGACGGTTTATT |
| wl-72196:CAACCTATATCACAACCTCTAAGC | ||||
| O16 | wzy | wl-72165:AATCACGATGTGGAAGCTG | 308 | OA-5595:CTAATCACGTTACCCCTTATTGGT |
| wl-72166:CCAAATCTCCCGAAACTAAATA | ||||
| O22 | wzy | wln-743:GTTTCTCGCAAGGTGGAGTAG | 267 | OAn-35:CAATTACCCTTTCTAGTTAAGGTC |
| wln-744:CAAAAGCATAAGCAGAGTAAACAC | ||||
| O23 | wzy | wln-759:TTGTGGTTGGGATTTGTCAG | 471 | OAn38:ATTTATCAAGGTTTCAAAAGTCC |
| wln-760:GTAATCCTGCTAAGGTTGAAGATAG | ||||
| O24 | wzx | wl-72159: GGTCTGCTTGTAGGAGTATGG | 299 | OA-6043: TATGTTTAATCGTGGCAGTGGG |
| wl-72160: AATACTAGTCCACCACCAGCC | ||||
| O26 | wzm | wl-72179: GCAAGTAGATATCAAGGTTCAGTT | 328 | OA-5602: TTGGTCTGCAGATAGTACAGGAAGT |
| wl-72180: CCCAAACGACAAAGCTAATT | ||||
| O26 | gtrC | wln-12420: ATTAGGCGTGAGCAGTAGAGG | 444 | OAn-172: TGTAGATCCTCGCGGCCTAT |
| wln-12420: GGCTTCTCGAAATCATTTTGTA | ||||
| O30 | wzm | wl-72179: GCAAGTAGATATCAAGGTTCAGTT | 328 | OA-5602: TTGGTCTGCAGATAGTACAGGAAGT |
| wl-72180: CCCAAACGACAAAGCTAATT | ||||
| O32 | wzm | wl-72167: GTGTAGTATCGTCATCGTTGTTAG | 294 | OA-5596: CTATGGTATATCACCTACAGTAGAGTGGTT |
| wl-72168: AGGTTATAAAGCGTTCTAAATCC | ||||
| O38 | wzy | wl-72161: TTTGACCCGAGGGGC | 361 | OA-5593: CAGACATCACGTGGGTCTACG |
| wl-72162: TCCAGTTTCTAGTGCCATCC | ||||
| O41 | wzy | wl-72185: ATACTGGAGCAGCTAAGACAATT | 341 | OA-6040: TGCCGTTTTTACTAAGATTCAAGTT |
| wl-72186: ATTATTGAAGGGCCAACATG | ||||
Multiplex PCR
Multiplex PCR was performed in a 50 μl reaction mixture containing 100 ng DNA template, 1× Goldstar PCR buffer, 10 μM of each deoxynucleotide triphosphate (dNTP), 0.5 μM of each forward primer, 2 μM of each reverse primer and 2.5 units of Goldstar Taq polymerase (CWBIO Co. Ltd, Beijing, China). Initial denaturation of DNA was performed for 10 min at 95°C, followed by 35 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 30 s and a final extension at 72°C for 10 min. PCR products were stored at 4°C for use in the subsequent experiments.
Probe-microsphere coupling
Unique colored fluorescent microspheres can be detected in multiplex PCR reactions using the Luminex xMAP technology. The microspheres (Bio-Rad Laboratories, CA, USA) were conjugated to the 5¢-end amino C-12 modification according to the manufacturer's instructions (Luminex Corporation, TX, USA). Briefly, 80 μl microspheres was suspended in 10 μl 0.1 M 2-(N-morpholino) ethanesulfonic acid, pH 4.5 (MES); subsequently, 2 μl probes and 6 μl fresh 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (10 mg/ml) were added; After incubating for 1 h, the probe-microsphere conjugated products were washed in 1 ml 0.02% Tween-20 (Sigma-Aldrich, MO, USA) and 1 ml 0.1% SDS (Sigma-Aldrich). Finally, the coupled microspheres were resuspended in 25 μl Tris-EDTA buffer, pH 8.0 (Sigma-Aldrich) and stored at 4°C in the dark. Notably, the whole coupling process was performed in dark to avoid fluorescence quenching.
Hybridization & suspension array performance
A 2 μl sample of each 18 probe-microsphere conjugated product was mixed and diluted with 1.5× tetramethylammonium chloride (TMAC) solution (Sigma-Aldrich) that contained 4.5 M TMAC, 0.15% Sarkosyl, 75 mM Tris-HCl (pH 8.0) and 6 mM EDTA (pH 8.0). Further 33 μl bead mixture was added to 17 μl of each PCR product. The 50 μl hybridization reactions were denatured at 95°C for 5 min, hybridized at 60°C for 15 min, transferred to a multiscreen-HV filter plate (Millipore, MA, USA) and the supernatants removed after centrifuging at 1000 rpm for 1 min. The hybridized products were next washed twice with 70 μl 1× TMAC and resuspended in 80 μl of 4 μg/ml streptavidin labeled with R-phycoerythrin (SAPE; Invitrogen), which was diluted in 1.0× TMAC. Finally, the hybridized products were analyzed using a Bio-Plex 100 suspension array system (Bio-Rad Laboratories). The fluorescent signals were captured using a digital signal processor and Bio-Plex Manager 4.1 software. Background controls were set up using all ingredients except the template DNA. The background was subtracted from each sample signal, and the positive signals of specific probes were considered to possess at least triple the fluorescence intensity resulting from nonspecific binding.
Sensitivity detection
To determine the minimum detection level of purified DNA, tenfold serial dilutions of genomic Citrobacter DNA (100 to 10-6 ng) were prepared and used as templates for MSA detection. To determine the sensitivity of our MSA system for pure cultures, Citrobacter cultures were tenfold serially diluted to 108 to 100 colony forming units (CFU)/ml and used as templates for MSA detection. The multiplex PCR and suspension array detection procedures were identical to that described above.
Construction of in silico serotyping program
A Python script was constructed for prediction of Citrobacter serotypes with genome data (available in Supplementary Tool). The database was constructed based on the O-serotype specific genes wzx, wzy, wzm, wzt, gtrC and GT, for in silico O typing. All specific genes of Citrobacter strains assigned to different serotypes were stored in the database. Genome assemblies were subjected to a BLASTn search against the O-serotype specific gene database with an identity cutoff >98% and a minimum length of 95%. The script outputs the best-matching genes from the BLAST analysis, as well as percentage identity between the gene detected in the query genome and the O-serotype specific gene. In addition, the script outputs the predicted O serotypes, based on the best-matching genes.
Accession numbers
All 18 O-AGC sequences of Citrobacter are available from the GenBank database (accession numbers from MH325885 to MH325900, and from MH325902 to MH325903).
Results & discussion
Location & detailed analyses of O-AGC in Citrobacter O26 strain
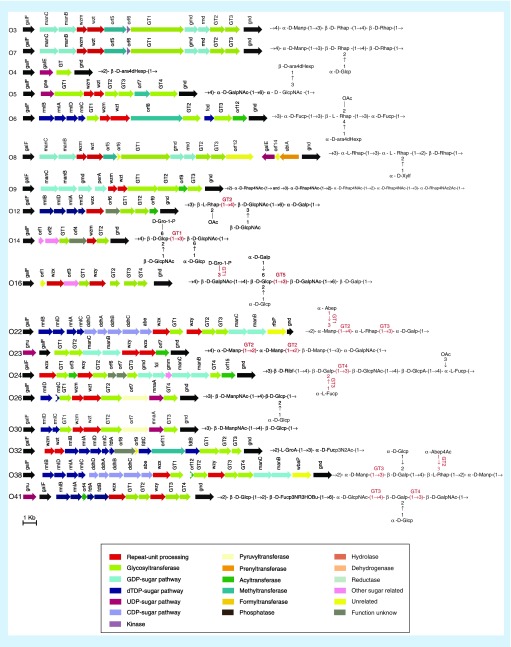
Whole-genome-sequencing of C. werkmanii O26 (strain G3522) predicted a putative O-AGC, consisting of nucleotide sugar biosynthesis genes, three sugar transfer genes (GTs) and the O unit processing genes wzm and wzt (Figure 1). The putative O-AGC is located between two housekeeping genes galF and gnd in the genome (Figure 1). We subsequently analyzed the putative O-AGC in detail to investigate whether it correlated to the O-antigen structure. The reported O-antigen structure showed that the main chain of the O26 antigen unit consists of a 2-acetamido-2-deoxy-D-mannose (D-ManNAc) residue and a D-glucose (D-Glc) residue, with a D-Glc residue on the side chain. orf8 in O26-AGC shares 77% identity with MnaA of Aeromonas hydrophila (accession number WP_101613945.1), which converts UDP-GlcNAc to UDP-ManNAc [43]. The biosynthesis pathway is shown in Supplementary Figure 1. We therefore predicted that orf8 is responsible for the biosynthesis of UDP-ManNAc in Citrobacter O26. In turn, the D-Glc residue is considered to comprise a common sugar that is involved in basic cell metabolism. As genes for D-Glc biosynthesis are not always located in the O-AGC but may be found elsewhere in the genome in other genera [44], we considered that the same condition may occur in Citrobacter.
Figure 1. . The O-antigen biosynthesis gene clusters of 18 Citrobacter strains.
The arrows represent transcription direction and location of putative genes in O-antigen gene clusters. Different colors represent different kinds of gene functions. The proposed functions of glycosyltransferases are shown.
Furthermore, early reports indicated that a gtr operon found in phages is responsible for the glucosylation of the basic O-antigen unit on the side chain [45]. gtrABs are highly conserved, whereas the third gene, gtrC, is always unique to different serotypes [46]. GtrA and GtrB add the glucose to a carrier lipid and flip it to the periplasm, whereas GtrC performs the modification by adding a glucosyl group to the O-antigen units [47]. A whole genome search of O26 revealed, two conserved genes, gtrA and gtrB, in the bacteriophage as expected. In addition, as the gene adjacent to gtrAB shares 41% similarity with gtrC in Edwardsiella tarda (accession number PVD63346.1), we annotated it as gtrC and proposed that it was responsible for the glucosylation of the O26 antigen unit on the side chain. Characteristics of each gene in the O-AGC are summarized in Supplementary Table 3. These analyses demonstrated that O26-AGC shows good correlation with the O26 antigen structure.
Function identification of the O-AGC in Citrobacter by deletion & complementation testing
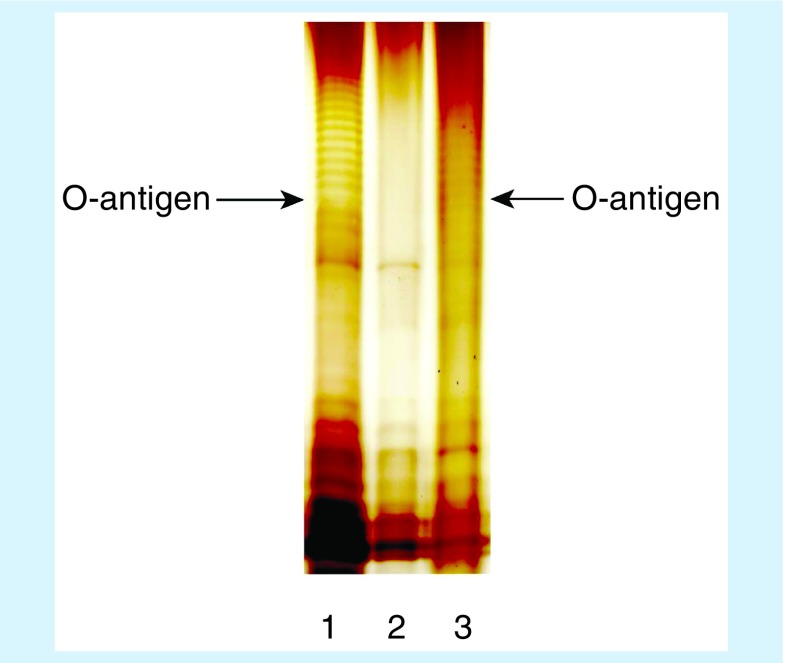
A deletion and complementation experiment was performed on C. werkmanii O26 to further confirm the function of the putative O-AGC. Specifically, we replaced wzm with a gene encoding chloramphenicol acetyltransferase, and the mutation was complemented by the plasmid pTrc99a containing the O26 wzm gene. The SDS-PAGE profiles of LPS showed that compared with the wild-type G3522, the △wzm strain (H2659) was unable to biosynthesize O-antigen, whereas the complemented strain H2660 showed restored function (Figure 2). These results confirmed that the putative O26-AGC is responsible for the biosynthesis of the O-antigen.
Figure 2. . Detecting lipopolysaccharide from the △wzm strain of C. werkmanii O26.
Lipopolysaccharide was extracted using the hot aqueous-phenol method. The extracts were electrophoresed on 13% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and stained by silver staining. Lane 1: G3522 (C. werkmanii O26 wild type strain); lane 2: H2659 (O26 with wzm deletion); lane 3: H2660 (O26 with plasmid pTRC99a harboring O26 wzm).
General features of putative O-AGCs in 18 Citrobacter strains
For establishing a molecular serotyping system using the MSA platform, we sequenced 17 other Citrobacter strains (Table 1), the O-antigen structures of which have been reported, to obtain the O-AGCs. Each of the 17 O-AGCs is located between galF and gnd (Figure 1). Including O26, the sizes of the 18 O-AGCs of Citrobacter range from 2415 to 20,559 bp, with the average %GC content being approximately 37%, which is significantly lower than that of the entire Citrobacter genome (50%), indicating that O-AGCs may have been derived from another genus and consistently show the typical features of O-AGCs in Enterobacteriaceae. Generally, the O-AGCs contain genes associated with nucleotide sugar synthesis, sugar transfer and O unit processing in each O-AGC of the 17 strains, with the exception of O4 (Supplementary Results). We reasoned that biosynthesis of the O4 antigen may differ from those of other Citrobacter strains. Seven serotypes, namely, O12, O16, O22, O23, O24, O38 and O41, contain wzx and wzy in their respective O-AGC, and process O-antigen in a Wzx/Wzy-dependent pathway. The other ten serotypes containing wzm and wzt process O-antigen in an ABC transporter-dependent pathway. Detailed analysis of each of the 17 O-AGCs of Citrobacter showed that they exhibited good correlation with their O-antigen structures (Supplementary Results).
Molecular serotyping system on the MSA platform
As the O-antigen processing genes wzy, wzx, wzm and wzt have been successfully utilized for molecular serotyping of many Gram-negative bacteria [27–29], these were selected as specific targets for Citrobacter serotyping in the present study. wzm in O3, O5, O6, O7, O8, O9, O14, O26, O30 and O32, wzx in O12 and wzy in O16, O22, O23, O24, O38 and O41 were selected. Furthermore, compared with genes involved in nucleotide sugar precursor synthesis, genes encoding glycosyl transferase are heterogeneous among serotypes owing to the wide range of possible monosaccharide linkages (although showing less serotype determination than the O-antigen processing genes) [29,48]. Since no O-antigen processing gene was present in O4-AGC, a GT (orf2) was selected for this serotype (Table 2). These selected genes were subjected to bioinformatic analysis, which revealed no sequence similarity after pairwise alignment of any two sequences, except for two pairs of strains (O3/O7 and O26/O30). As detailed analyses of the 18 O-AGCs indicated that O26 and O30 O-AGCs shared high identity (99%) (Supplementary Figure 2), a single set of O-AGC-specific-gene-based primers and probe was unable to distinguish these homologous strains. However, although O26 and O30 share the same main chain of the O-antigen unit, O26 carries a glucosylation on the side chain, encoded by the gtr operon. Therefore, to further distinguish O26 from O30, we designed another set of primers and probe based on the serotype-specific gene gtrC. Similarly, between O3 and O7, the O7-antigen is glucosylated on the side chain, whereas the main chain is identical to the linear O-antigen of O3. Hence, we also designed a set of primers and probe based on gtrC of O7.
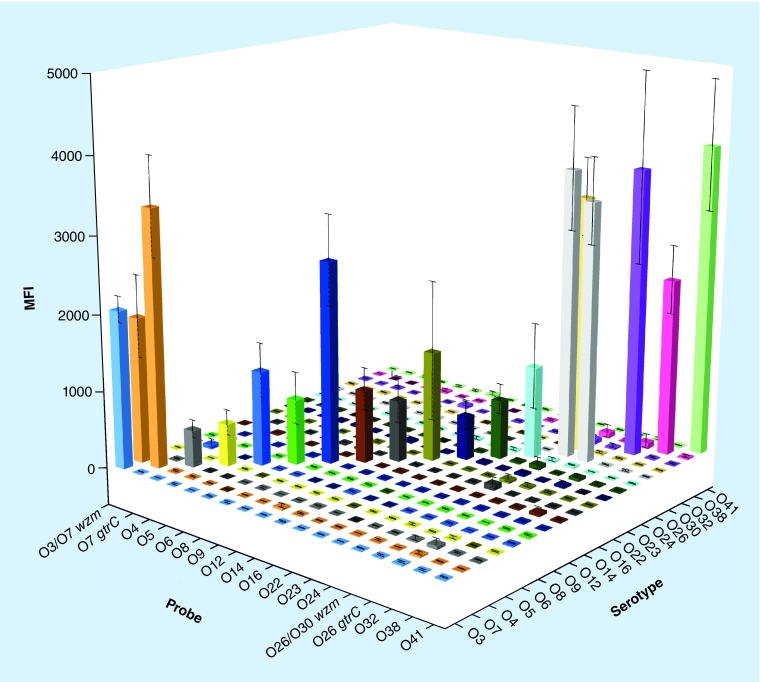
Strains representing all 18 serotypes, together with ten other Citrobacter strains of ten diverse serotypes, were used to test the specificity of these specific gene-based primers and probes on the MSA platform. The MSA results showed that each O-AGC-specific probe detected the homologous strains correctly and no signals corresponding to heterologous strains were observed, whereas both wzm-based and gtrC-based signals were detected in O7 and O26 (Figure 3). To test the reproducibility of the molecular serotyping system on the MSA platform, three parallel tests were performed independently and the error bars were shown in Figure 3.
Figure 3. . Fluorescence signals of the O-serotype system on the microsphere-based suspension array platform.
Each cube represents a fluorescence signal from mixed microspheres that had combined with a template PCR amplicon irrespective of whether it represented the correct target. The X-axis shows the probes, whereas the Y-axis shows the different O-serotype strains. The Z-axis shows the fluorescence signal values. The higher the cube, the more the combinations between microspheres and amplicons.
MFI: Median fluorescence intensity.
The MSA experiments revealed that each probe was specific to its homologous DNA, and the specific fluorescence signal was at least threefold more intense than the nonspecific signals (Figure 3). No cross-reaction was observed in the test. Several bacterial strains, namely, E. coli (n = 4), Shigella spp. (n = 4), Salmonella spp. (n = 3), Klebsiella pneumoniae (n = 3), Vibrio cholerae (n = 2) and Legionella spp. (n = 2), which were used to determine the specificity of primers and probes yielded negative results (data not shown). To assess the availability of our MSA technique, a double-blind test was conducted using 87 clinical isolates from Shanghai Municipal Center for Disease Control. Among them, two isolates were designated as O5, three as O8, seven as O22 and four as O41, with six isolates each representing individual serotype and 65 being nontypeable. All strains assigned certain serotypes were confirmed to be correct using ABI 3730 sequencing of each single PCR product. In addition, a sensitivity test of molecular serotyping system on the MSA platform was performed using genomic DNAs or pure cultures from O5, O8, O22 and O41, as they appeared at a relatively high proportion. Our test demonstrated that positive signals could be generated for templates containing 10-2 ng of genomic DNA or 103 CFU of pure culture.
O-serotyping in silico using genome data of Citrobacter
A program was next designed to develop an O-serotyping scheme in silico for Citrobacter using only the genome data. The database covered the 18 studied O-serotypes of Citrobacter, which were represented by the genes used in the serotyping system on the MSA platform as listed in Table 2. Most serotypes were represented only by a single wzy, wzx or wzm, whereas O7 and O26 were represented by both wzm and gtrC. To evaluate and extend the in silico serotyping program, 98 publicly available Citrobacter genomes in GenBank were subjected to the program. Among these, eight Citrobacter genomes were assigned to the 18 O serotypes contained in the database, with a threshold of 98% identity and a minimum length of 95%. We then extracted the O-AGCs from these eight genomes and conducted further analyses. The composition and arrangement of genes in each of the eight O-AGCs were identical to those in their assigned serotypes, which validated the credibility of the in silico serotyping method (Supplementary Figure 3).
We also extracted all the O-AGCs from the 90 unallocated Citrobacter genomes for further analysis. According to the composition and arrangement of genes, we classified the 90 O-AGCs into 33 novel O-AGC groups (Supplementary Figure 4), a portion of which may overlap with the reported 43 known serotypes of Citrobacter. This classification indicated the existence of hitherto-unrecognized genetic O variation in Citrobacter. Each of the O-AGC groups, which were temporarily termed O-AGC temp 1 to O-AGC temp 33, was represented by one of the wzy, wzx, wzm and wzt genes (Supplementary Table 4) and were added in the in silico serotyping program. Once their assignment to the exact O serotypes is established, the temporary name will be replaced with the widely recognized O-serotypes of Citrobacter. Furthermore, 25 Citrobacter genomes in GenBank were derived from a single strain and with identical O-AGCs [49]; these have been represented as RU2 in Supplementary Figure 4.
The O-serotyping system using the MSA platform based on Citrobacter O-AGCs was shown to correlate well with the conventional antigenic method. In addition, the program developed in this study offered another superior in silico serotyping method using genome data, which enabled the rapid and high-throughput serotyping of Citrobacter. Although we only targeted 18 serotypes of Citrobacter strains in this study, both the experimental and bioinformatic serotyping systems have the ability to be updated in real-time as novel specific primers, probes and genes can be easily added into our serotyping systems. In the future, we plan to sequence other Citrobacter strains belonging to different serotypes, the O-antigen structures of which have been previously reported, to determine whether their O-AGCs are also consistent with their O-antigen structures. Sequence alignments will be subsequently conducted to ascertain whether the serotype-specific genes of these novel Citrobacter O-AGCs are already present in the database of our in silico serotyping program, inclusion of any nonrepresented genes in our database, and addition of specific-gene-based primers and probes in the O-serotyping system of the MSA platform. Furthermore, for Citrobacter strains with glucosylation on the O-antigen side chain, gtrC can be considered as another serotype-specific gene and the relevant gene sequences, primers and probes will also be included in the serotyping tools to facilitate accurate identification. Overall, the two serotyping tools developed in this study represent potentially powerful approaches for detecting and identifying Citrobacter in environmental and clinical samples, as well as for epidemiological surveillance and tracing.
Conclusion
In this study, we first located the O-AGCs of Citrobacter by genomic and biochemical analyses and further characterized the O-AGCs of 18 serotypes, thereby providing the basis for the evolution of O-antigens of this pathogenic bacterium. Next, a highly sensitive and specific molecular serotyping assay based on the MSA platform was developed and an in silico serotyping method was presented. These tools show considerable potential for the clinical diagnosis and epidemiological surveillance of the genus Citrobacter.
Future perspective
The serotyping technique has been used as a standard tool for bacterial detection and epidemiological studies. With the emergence and development of whole genome sequencing technologies and automated data analysis pipelines, several genomics-based methods are being used for routine serotyping and surveillance of pathogens, showing potential advantages in public health microbiology. The O-AGCs of the remaining serotypes of Citrobacter have to be identified to promote its clinical application.
Summary points.
The widespread and fatal infections of Citrobacter highlight the necessity of establishing accurate and rapid molecular typing systems for effective surveillance and clinical detection of this bacterium. However, the lack of commercial Citrobacter O-antisera has limited the application of conventional serotyping. In the present study, we developed rapid and simple experimental and in silico serotyping systems for Citrobacter.
Here, we located the O-antigen biosynthesis gene cluster (O-AGC) in Citrobacter genome by genomic and molecular analyses, and the biological function was verified experimentally for the first time.
Subsequently, we sequenced the whole genome of 18 Citrobacter strains and obtained the O-AGCs. Based on the specific genes in these O-AGCs, we developed a sensitive molecular serotyping system using a microsphere-based suspension array platform.
Additionally, we established an in silico serotyping method for Citrobacter based solely on the whole genome sequencing data. All 98 available Citrobacter genomes in GenBank were used to test the reliability of this in silico serotyping method, and a previously unrecognized level of diversity was revealed.
Notably, the two tools developed in this study enabled us to perform Citrobacter O-serotyping using both experimental and bioinformatic approaches. The system can be conveniently updated in real-time, allowing addition of new O-AGCs identified in different laboratories, which will supplement the two serotyping tools.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: https://www.futuremedicine.com/doi/suppl/10.2217/fmb-2018-0187
Author contributions
C Qian wrote the manuscript. C Qian, Y Du and H Li designed and carried out the experiments, analyzed the 18 O-AGCs in detail and performed the in silico serotyping system for Citrobacter. P Wu, L Wang and Y Wei helped in analyzing Citrobacter O-AGCs from GenBank database. H Cao and Z Yin helped in downloading Citrobacter genomes from GenBank and provided guidance in bioinformatics. Y Zhang and Y Zhu helped in checking and modifying the manuscript. X Guo and B Liu provided guidance throughout the entire study. All authors have read and approved the manuscript.
Financial & competing interests disclosure
This work was supported by National Key Programs for Infectious Diseases of China (2017ZX10303405-001 and 2017ZX10104002-001-006), National Natural Science Foundation of China (NSFC) General Program Grant (81471904, 81772148 and 31470194) and a Tianjin Municipal Natural Science Foundation Grant (17JCYBJC24300). The funders had no role in study design, data collection and interpretation, or the decision to submit for publication. We declare no conflicts of interest. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Lipsky BA, Hook EW, 3rd, Smith AA, Plorde JJ. Citrobacter infections in humans: experience at the Seattle Veterans Administration Medical Center and a review of the literature. Rev. Infect. Dis. 1980;2(5):746–760. doi: 10.1093/clinids/2.5.746. [DOI] [PubMed] [Google Scholar]
- 2.Doran TI. The role of Citrobacter in clinical disease of children: review. Clin. Infect. Dis. 1999;28(2):384–394. doi: 10.1086/515106. [DOI] [PubMed] [Google Scholar]
- 3.Sedlak J. Present knowledge and aspects of Citrobacter . Curr. Top. Microbiol. Immunol. 1973;62:41–59. doi: 10.1007/978-3-642-65772-6_2. [DOI] [PubMed] [Google Scholar]
- 4.Samonis G, Karageorgopoulos DE, Kofteridis DP, et al. Citrobacter infections in a general hospital: characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2009;28(1):61–68. doi: 10.1007/s10096-008-0598-z. [DOI] [PubMed] [Google Scholar]
- 5.Chao CT, Lee SY, Yang WS, et al. Citrobacter peritoneal dialysis peritonitis: rare occurrence with poor outcomes. Int. J. Med. Sci. 2013;10(9):1092–1098. doi: 10.7150/ijms.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drelichman V, Band JD. Bacteremias due to Citrobacter diversus and Citrobacter freundii. Incidence, risk factors, and clinical outcome. Archv. Intern. Med. 1985;145(10):1808–1810. [PubMed] [Google Scholar]
- 7.Shih CC, Chen YC, Chang SC, Luh KT, Hsieh WC. Bacteremia due to Citrobacter species: significance of primary intraabdominal infection. Clin. Infect. Dis. 1996;23(3):543. doi: 10.1093/clinids/23.3.543. [DOI] [PubMed] [Google Scholar]
- 8.Pepperell C, Kus JV, Gardam MA, Humar A, Burrows LL. Low-virulence Citrobacter species encode resistance to multiple antimicrobials. Antimicrob. Agents Chemother. 2002;46(11):3555–3560. doi: 10.1128/AAC.46.11.3555-3560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner DJ, O'Hara CM, Grimont PA, et al. Biochemical identification of Citrobacter species defined by DNA hybridization and description of Citrobacter gillenii sp. nov. (formerly Citrobacter genomospecies 10) and Citrobacter murliniae sp. nov. (formerly Citrobacter genomospecies 11) J. Clin. Microbiol. 1999;37(8):2619–2624. doi: 10.1128/jcm.37.8.2619-2624.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih CC, Chen YC, Chang SC, Luh KT, Hsieh WC. Bacteremia due to Citrobacter species: significance of primary intraabdominal infection. Clin. Infect. Dis. 1996;23(3):543–549. doi: 10.1093/clinids/23.3.543. [DOI] [PubMed] [Google Scholar]
- 11.Kim BN, Woo JH, Ryu J, Kim YS. Resistance to extended-spectrum cephalosporins and mortality in patients with Citrobacter freundii bacteremia. Infection. 2003;31(4):202–207. doi: 10.1007/s15010-003-2176-8. [DOI] [PubMed] [Google Scholar]
- 12.Katzenellenbogen E, Staniszewska M, Kocharova NA, et al. Re-classification within the serogroups O3 and O8 of Citrobacter strains. BMC Microbiol. 2017;17(1):169. doi: 10.1186/s12866-017-1078-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Points out that heterogeneity of strains within particular serogroup is observed in Citrobacter.
- 13.Petty NK, Bulgin R, Crepin VF, et al. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli . J. Bacteriol. 2010;192(2):525–538. doi: 10.1128/JB.01144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Indicates that Citrobacter is a polyphiletic genus.
- 14.Liu LH, Wang NY, Wu AY, Lin CC, Lee CM, Liu CP. Citrobacter freundii bacteremia: risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 2017;S1684–1182(17):30059–2. doi: 10.1016/j.jmii.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Gaibani P, Ambretti S, Farruggia P, et al. Outbreak of Citrobacter freundii carrying VIM-1 in an Italian hospital, identified during the carbapenemases screening actions, June 2012. Int. J. Infect. Dis. 2013;17(9):e714–e717. doi: 10.1016/j.ijid.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Papasian CJ, Kinney J, Coffman S, Hollis RJ, Pfaller MA. Transmission of Citrobacter koseri from mother to infant documented by ribotyping and pulsed-field gel electrophoresis. Diagn. Microbiol. Infect. Dis. 1996;26(2):63–67. doi: 10.1016/s0732-8893(96)00177-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Ruan X, Wei D, et al. Development of a serogroup-specific multiplex PCR assay to detect a set of Escherichia coli serogroups based on the identification of their O-antigen gene clusters. Mol. Cell. Probes. 2010;24(5):286–290. doi: 10.1016/j.mcp.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 18.West MG, Edwards PR. The Bethesda–Ballerup group of paracolon bacteria. Public Health Monogr. 1954;69(10):1–35. [PubMed] [Google Scholar]
- 19.Knirel Y, Kocharova N, Bystrova O, Katzenellenbogen E, Gamian A. Structures and serology of the O-specific polysaccharides of bacteria of the genus Citrobacter . Arch. Immunol. Ther. Exp. (Warsz.). 2002;50(6):379–391. [PubMed] [Google Scholar]; •• An important review presents the structures of the O-antigen isolated from different Citrobacter species and serogroups.
- 20.Sedlak J, Slajsova M. Antigen structure and antigenic relationships of the species Citrobacter . Zentralbl. Bakteriol. Orig. 1966;200(3):369–374. [PubMed] [Google Scholar]
- 21.Katzenellenbogen E, Kocharova NA, Zatonsky GV, et al. Structure of the O-specific polysaccharide from the lipopolysaccharide of Citrobacter gillenii O11, strain PCM 1540. Carbohydr. Res. 2003;338(13):1389–1395. doi: 10.1016/s0008-6215(03)00166-6. [DOI] [PubMed] [Google Scholar]
- 22.Katzenellenbogen E, Kocharova NA, Zatonsky GV, et al. Structural and serological studies on a new 4-deoxy-d-arabino-hexose-containing O-specific polysaccharide from the lipopolysaccharide of Citrobacter braakii PCM 1531 (serogroup O6) Eur. J. Biochem. 2003;270(13):2732–2738. doi: 10.1046/j.1432-1033.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 23.Glushakova LG, Bradley A, Bradley KM, et al. High-throughput multiplexed xMAP Luminex array panel for detection of twenty two medically important mosquito-borne arboviruses based on innovations in synthetic biology. J. Virol. Methods. 2015;214:60–74. doi: 10.1016/j.jviromet.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silbereisen A, Tamborrini M, Wittwer M, Schurch N, Pluschke G. Development of a bead-based Luminex assay using lipopolysaccharide specific monoclonal antibodies to detect biological threats from Brucella species. BMC Microbiol. 2015;15:198. doi: 10.1186/s12866-015-0534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Knirel YA, Feng L, et al. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 2014;38(1):56–89. doi: 10.1111/1574-6976.12034. [DOI] [PubMed] [Google Scholar]; •• An important review that summarized the biosynthesis pathways of some rare sugar residues, which are also present in O-antigens of Citrobacter.
- 26.Feng L, Senchenkova SN, Yang J, et al. Synthesis of the heteropolysaccharide O antigen of Escherichia coli O52 requires an ABC transporter: structural and genetic evidence. J. Bacteriol. 2004;186(14):4510–4519. doi: 10.1128/JB.186.14.4510-4519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Reeves PR. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 1998;66(8):3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao B, Tian Z, Wang S, et al. Structural comparison of O-antigen gene clusters of Legionella pneumophila and its application of a serogroup-specific multiplex PCR assay. Antonie Van Leeuwenhoek. 2015;108(6):1405–1423. doi: 10.1007/s10482-015-0594-0. [DOI] [PubMed] [Google Scholar]
- 29.Ballmer K, Korczak BM, Kuhnert P, Slickers P, Ehricht R, Hachler H. Fast DNA serotyping of Escherichia coli by use of an oligonucleotide microarray. J. Clin. Microbiol. 2006;45(2):370–379. doi: 10.1128/JCM.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo X, Wang M, Wang L, et al. Establishment of a molecular serotyping scheme and a multiplexed Luminex-based array for Enterobacter aerogenes . Front. Microbiol. 2018;9:501. doi: 10.3389/fmicb.2018.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Wang M, Liu H, et al. Development of an O-antigen serotyping scheme for Cronobacter sakazakii . Appl. Environ. Microbiol. 2011;77(7):2209–2214. doi: 10.1128/AEM.02229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan Z, Niedziela T, Lugowski C, et al. Genetic diversity of O-antigens in Hafnia alvei and the development of a suspension array for serotype detection. PLoS ONE. 2016;11(5):e0155115. doi: 10.1371/journal.pone.0155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X, Torzewska A, Zhang X, et al. Genetic diversity of the O antigens of Proteus species and the development of a suspension array for molecular serotyping. PLoS ONE. 2017;12(8):e0183267. doi: 10.1371/journal.pone.0183267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ovchinnikova OG, Kocharova NA, Katzenellenbogen E, et al. Structures of two O-polysaccharides of the lipopolysaccharide of Citrobacter youngae PCM 1538 (serogroup O9) Carbohydr. Res. 2004;339(4):881–884. doi: 10.1016/j.carres.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 35.Kocharova NA, Katzenellenbogen E, Zatonsky GV, et al. Structure of the O-polysaccharide of Citrobacter youngae PCM 1503. Carbohydr. Res. 2010;345(17):2571–2573. doi: 10.1016/j.carres.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 36.Kocharova NA, Mieszala M, Zatonsky GV, et al. Structure of the O-polysaccharide of Citrobacter youngae O1 containing an alpha-D-ribofuranosyl group. Carbohydr. Res. 2004;339(2):321–325. doi: 10.1016/j.carres.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Mieszala M, Lipinski T, Kocharova NA, et al. The identity of the O-specific polysaccharide structure of Citrobacter strains from serogroups O2, O20 and O25 and immunochemical characterisation of C. youngae PCM 1507 (O2a,1b) and related strains. FEMS Immunol. Med. Microbiol. 2003;36(1–2):71–76. doi: 10.1016/S0928-8244(03)00081-6. [DOI] [PubMed] [Google Scholar]
- 38.Katzenellenbogen E, Kocharova NA, Toukach PV, et al. Structure of an abequose-containing O-polysaccharide from Citrobacter freundii O22 strain PCM 1555. Carbohydr. Res. 2009;344(13):1724–1728. doi: 10.1016/j.carres.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Katzenellenbogen E, Kocharova NA, Korzeniowska-Kowal A, et al. Structure of the glycerol phosphate-containing O-specific polysaccharide and serological studies on the lipopolysaccharides of Citrobacter werkmanii PCM 1548 and PCM 1549 (serogroup O14) FEMS Immunol. Med. Microbiol. 2008;54(2):255–262. doi: 10.1111/j.1574-695X.2008.00477.x. [DOI] [PubMed] [Google Scholar]
- 40.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maervoet VE, De Maeseneire SL, Avci FG, Beauprez J, Soetaert WK, De Mey M. 1,3-propanediol production with Citrobacter werkmanii DSM17579: effect of a dhaD knock-out. Microb. Cell. Fact. 2014;13:70. doi: 10.1186/1475-2859-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis M, Jr, Goldberg JB. Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J. Vis. Exp. 2012;63(63):e3916–e3916. doi: 10.3791/3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell RE, Mosimann SC, Tanner ME, Strynadka NC. The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases. Biochemistry. 2000;39(49):14993–5001. doi: 10.1021/bi001627x. [DOI] [PubMed] [Google Scholar]
- 44.Liu B, Knirel YA, Feng L, et al. Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 2008;32(4):627–653. doi: 10.1111/j.1574-6976.2008.00114.x. [DOI] [PubMed] [Google Scholar]; •• An important review that summarized the biosynthesis pathways of some rare sugar residues, which are also present in O-antigens of Citrobacter.
- 45.Kintz E, Davies MR, Hammarlof DL, Canals R, Hinton JC, Van Der Woude MW. A BTP1 prophage gene present in invasive non-typhoidal Salmonella determines composition and length of the O-antigen of the lipopolysaccharide. Mol. Microbiol. 2015;96(2):263–275. doi: 10.1111/mmi.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri . Trends. Microbiol. 2000;8(1):17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]; •• An important article indicated gtrC is a serotype-specific glucosyl transferase.
- 47.Korres H, Mavris M, Morona R, Manning PA, Verma NK. Topological analysis of GtrA and GtrB proteins encoded by the serotype-converting cassette of Shigella flexneri . Biochem. Biophys. Res. Commun. 2005;328(4):1252–1260. doi: 10.1016/j.bbrc.2005.01.087. [DOI] [PubMed] [Google Scholar]
- 48.Li Q, Reeves PR. Genetic variation of dTDP-L-rhamnose pathway genes in Salmonella enterica . Microbiology. 2000;146(Pt 9):2291–2307. doi: 10.1099/00221287-146-9-2291. [DOI] [PubMed] [Google Scholar]
- 49.Saxer G, Krepps MD, Merkley ED, et al. Mutations in global regulators lead to metabolic selection during adaptation to complex environments. PLoS Genet. 2014;10(12):e1004872. doi: 10.1371/journal.pgen.1004872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.